Summary
Polyketides are important secondary metabolites, many of which exhibit potent pharmacological applications. Biosynthesis of polyketides is carried out by a single polyketide synthase (PKS) or multiple PKSs in successive elongations of enzyme‐bound intermediates related to fatty acid biosynthesis. The polyketide gene PKS306 from Pseudallescheria boydii NTOU2362 containing domains of ketosynthase (KS), acyltransferase (AT), dehydratase (DH), acyl carrier protein (ACP) and methyltransferase (MT) was cloned in an attempt to produce novel chemical compounds, and this PKS harbouring green fluorescent protein (GFP) was expressed in Saccharomyces cerevisiae. Although fluorescence of GFP and fusion protein analysed by anti‐GFP antibody were observed, no novel compound was detected. 6‐methylsalicylic acid synthase (6MSAS) was then used as a template and engineered with PKS306 by combinatorial fusion. The chimeric PKS containing domains of KS, AT, DH and ketoreductase (KR) from 6MSAS with ACP and MT from PKS306 demonstrated biosynthesis of a novel compound. The compound was identified with a deduced chemical formula of C7H10O3, and the chemical structure was named as 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione. The novel compound synthesized by the chimeric PKS in this study demonstrates the feasibility of combinatorial fusion of PKS genes to produce novel polyketides.
Introduction
Secondary metabolites that are obtained from microbes are widely applied in medicine, agriculture, environmental treatment and industrial processes (Cox and Simpson, 2009; Gerke and Braus, 2014). Metabolites of polyketides and non‐ribosomal peptides (NRP), such as lovastatin, erythromycin, rapamycin and cytochalasin E, have shown antibiotic, antifungal, antiviral, immune‐suppressant, anticholesterol and anticancer biological activities (Udagawa et al., 2000; Cane, 2010; Mulder et al., 2015; Waldner et al., 2016). Devising a feasible strategy for synthesis of secondary metabolites is important to contribute to further biological applications.
Polyketides are synthesized by the enzymes of polyketide synthases (PKSs) to form diverse chemical structures from the simplest monocyclic aromatic compounds, such as 6‐methylsalicylic acid (6‐MSA) (Beck et al., 1990), to complex compound such as 6‐deoxyerythronolide B (6‐dEB) (Katz, 2009). PKSs incorporate a wide variety of starter unit carboxylates via acyltransferase (AT) or ketosynthase (KS) domain onto acyl carrier protein (ACP) (Jenke‐Kodama et al., 2005; Tran et al., 2010). In the extension of the final β‐keto‐acyl chain, AT domain introduces the extender unit bound ACP, and KS domain containing growing polyketide catalyses decarboxylative Claisen condensation to afford a new intermediate. The elongation product is tailored by dehydratase (DH), ketoreductase (KR), enoyl reductase (ER) or thioesterase (TE) domains to yield the fully processed polyketides (Shen, 2003; Dutta et al., 2014; Weissman, 2014). Leading to increasing various patterns of the polyketides, the most common is programmed methylation by methyltransferase (MT) (Storm et al., 2017). Moreover, phosphopantetheinyl transferase (PPTase) uses a phosphopantetheine to modify a serine residue in the ACP, and the activated ACP may harbour acyl group intermediates in the biosynthesis (Wattana‐amorn et al., 2010; Wang et al., 2016). PKSs are characterized as modular or iterative PKSs based on the frequency of usage of their domains. Modular PKSs use their domains only once (Katz, 2009), whereas iterative PKSs may repeatedly use their domains to synthesize polyketides (Chen and Du, 2016).
Numerous PKS genes have been obtained from next‐generation sequencing (NGS) and data mining in many fungal species (Helfrich et al., 2014; Amoutzias et al., 2016). Most of the fungal PKS genes have a very long open reading frame (ORF) and are translated to form megaenzymes that have a high molecular weight (Tsunematsu et al., 2013). The first identified PKS gene, 6‐methylsalicylic acid synthase (6MSAS), produces 6‐methylsalicylic acid (6MSA) in Penicillium patulum and Aspergillus terreus (Dimroth et al., 1970; Beck et al., 1990). The gene with a 6200 bp ORF and harbouring KS‐AT‐DH‐KR‐ACP domains in the protein sequence was successfully and heterologously expressed with PPTase in Escherichia coli and Saccharomyces cerevisiae (Wattanachaisaereekul et al., 2007; Lee et al., 2009; Bond et al., 2016). Moreover, 6MSA, as an intermediate, may play a role in the polyketide biosynthesis of other structurally diverse compounds with comparable differences in biological activities (Holm et al., 2014; Huang et al., 2018; Kong et al., 2018). The enzyme is regarded as a multifunctional catalytic machine and operates sequentially in individual active sites to yield a polyketide product. Therefore, 6MSAS is a model gene that is suitable to develop cloning methodology and expression of PKS for the production of polyketide. Although 6MSAS is the first PKS discovered in the biosynthesis of secondary metabolites, its use in gene engineering to create the novel compound is not well explored.
Pseudallescheria boydii NTOU2362 was isolated from mangrove endophytic fungus and identified to produce several polyketides (Chang et al., 2013). The whole‐genome sequence of P. boydii NTOU2362 was obtained by NGS, and PKS genes were annotated by data mining. PKS306 and PKS64 from P. boydii NTOU2362 contain domains of KS‐AT‐DH‐ACP‐MT and KS‐AT‐DH‐ER‐KR‐ACP respectively. These PKSs were transformed into S. cerevisiae, but no novel compound was found. In an attempt to produce a novel compound in the transformed S. cerevisiae, chimeric 6MSAS was created by fusion of the domains of KS, AT, DH and KR from 6MSAS with ACP and MT from PKS306, and the engineered enzyme produced the novel compound, 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione, which is reported in this study.
Results
Cloning strategy, expression system and detection of 6MSAS
PKS and PPTase are required for the biosynthesis of polyketides, and a feasible strategy is therefore needed to establish a methodology to construct yeast expression system. The genes of PKS were generated by long‐range PCR to clone into a donor vector using an orientation‐directed TOPO ligation system, and then, the vectors used DNA recombination to transfer the PKS gene into a suite of Gateway cloning vectors (Fig. 1A) that have various promoters, auxotrophic selection markers, N‐ or C‐terminus GFP tags and replication origins (Fig. 1B). The constitutive promoter glyceraldehyde‐3‐phosphate dehydrogenase (GPD) and the inducible promoter GAL1 were used in this study. Auxotrophic selection markers ura3, leu2 and trp1 could combine multiple vectors to transform into S. cerevisiae to obtain multiple gene expression in yeast cells. Moreover, GFP may either maintain the efficiency of translation in the N‐terminus or detect full‐length expression in the C‐terminus. To set up the PKS expression system, 6MSAS from Aspergillus terreus and PPTase from A. nidulans were cloned and constructed.
Figure 1.
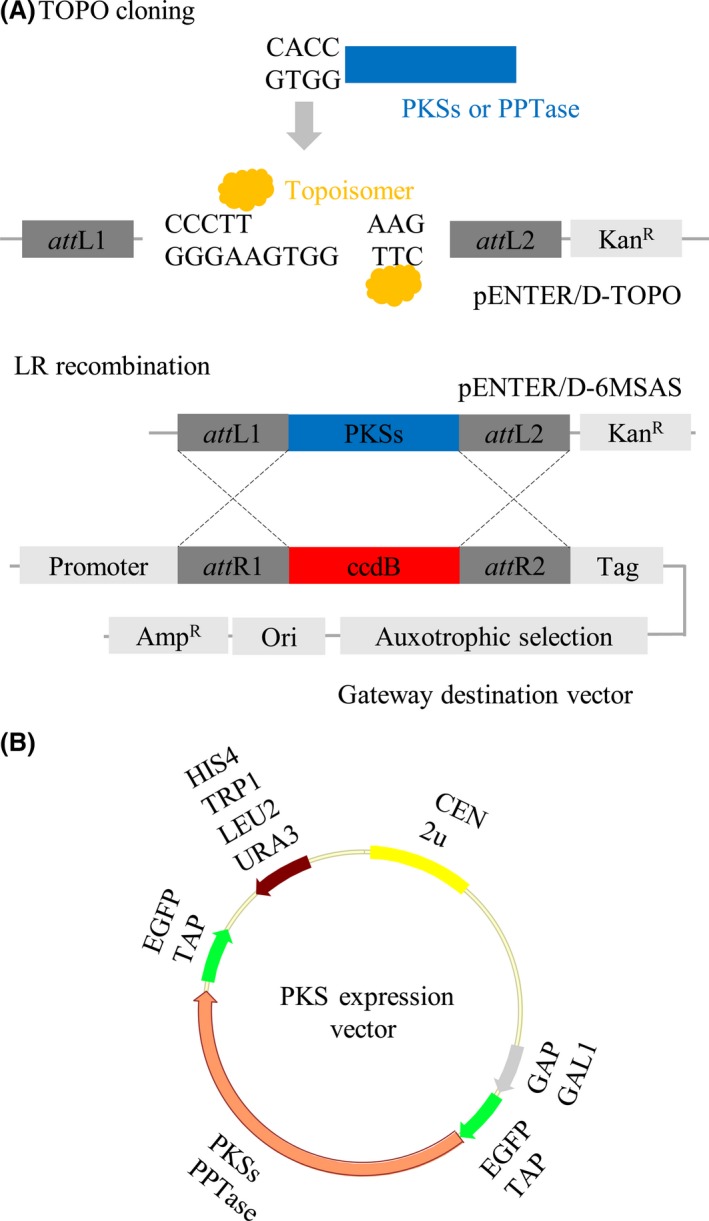
Strategy for the cloning and expression of PKS in S. cerevisiae.
A. Scheme for the construction of the PKS expression system. The genes of PKSs and PPTase were amplified by PCR with specific primers as indicated in Table S2. The expression vectors were generated by the Gateway system.
B. The yeast expression system contains the GAP or GAL1 promoter, ura3, his4, leu2 or trp1 auxotrophic selection markers, N‐ or C‐terminus GFP tags and replication origins as indicated.
Green fluorescent protein fused in frame with 6MSAS and the fusion protein 6MSAS‐GFP exhibited green fluorescence, as observed by fluorescence microscopy (Fig. 2A). Remarkably, spots were observed in the yeast cells instead of a dispersed pattern in the cytosol, implying that 6MSAS is localized to a specific organelle in S. cerevisiae. To examine the expression of 6MSAS, galactose was added to activate the GAL1 promoter, and the protein was detected by anti‐GFP antibody. The expression of 6MSAS with a molecular weight of 230 kDa was represented by the 194 kDa (6MSAS) and 28 kDa (GFP) band, as demonstrated in Fig. 2B after an induction of 96 h, showing that the vector is translated in frame and does not contain a frameshift or a stop codon before GFP. The S. cerevisiae BJ2168 harbouring either pGAL426‐6MSAS‐GFP (6MSAS‐GFP) only or pGAL426‐6MSAS‐GFP with or without pGAL425‐PPTase (PPTase) were cultured for 48 h and were induced expression by galactose for 96 h. The medium was collected and extracted with the same volume of ethyl acetate. The extract was concentrated by evaporation and was analysed by HPLC. In Fig. 2C, a distinct peak is shown in the yeast harbouring pGAL426‐6MSAS‐GFP and pGAL425‐PPTase, but the peak was not observed in the yeast harbouring pGAL426‐6MSAS‐GFP or pGAL425‐PPTase.
Figure 2.
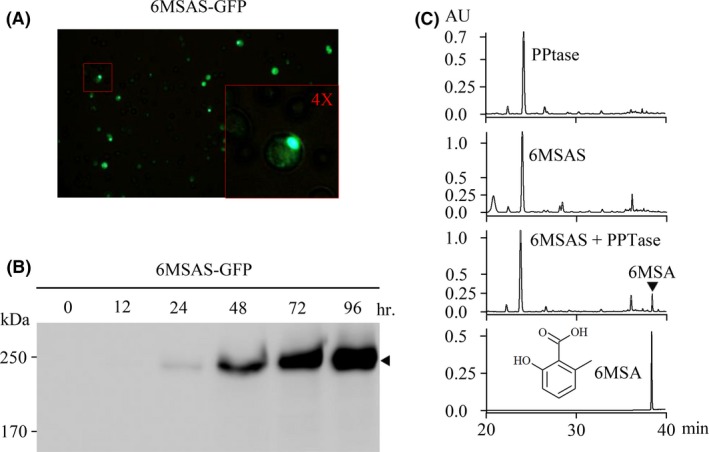
Detection of 6MSAS and 6MSA.
A. The yeast harbouring 6MSAS‐GFP was examined by fluorescence microscopy. The inset indicates the pattern of GFP observed forming a spot.
B. The expression of 6MSAS‐GFP was detected by Western blotting using an anti‐GFP antibody after the addition of galactose as indicated.
C. The product of 6MSAS was analysed by HPLC. The yeast harbouring vectors are indicated, and the last diagram shows the results of the chemical standard of 6MSA.
Heterologous expression of PKS from P. boydii NTOU2362
The genomic sequence of P. boydii NTOU2362 was obtained by NGS. PKS64 (MK134561) and PKS306 (MK134562) contain the functional domains, as shown in Fig. 3A. Compared with the functional domains of 6MSAS, PKS306 contained the MT domain in the carboxyl terminus, and PKS64 contained the ER domain. Because PKS64 harbours intron, cDNA was prepared, whereas the intronless PKS306 was obtained by PCR using genomic DNA as the template. By fluorescence microscopy, PKS64 and PKS306 showed a dispersed pattern of green fluorescence in the cytosol (Fig. 3B). Fusion of GFP with PKS306 and PKS64 in the amino (pGPD424‐GFP‐PKS306 and pGPD424‐GFP‐PKS64) or carboxyl terminus (pGAL426‐PKS306‐GFP and pGAL426‐PKS64‐GFP) shows a molecular weight of 250 and 280 kDa respectively. Despite the fluorescence signal, reduced protein expression was demonstrated by Western blotting using an anti‐GFP antibody (Fig. 3C). Moreover, no compound was identified by HPLC from the ethyl acetate extract (data not shown).
Figure 3.
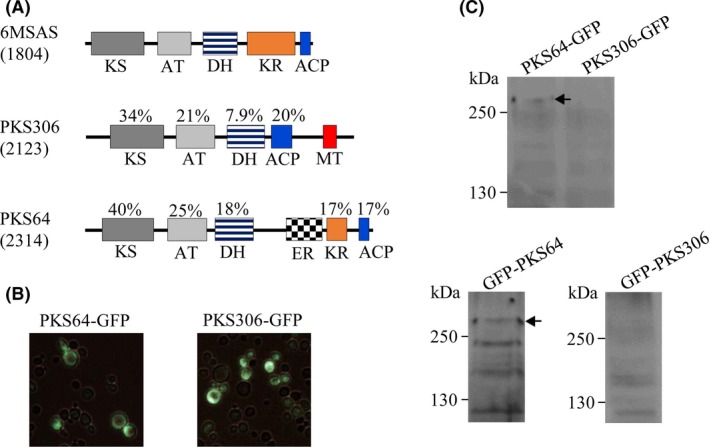
Analysis of the biosynthetic activity of PKS306 and PKS64.
A. The diagram shows the functional domains of 6MSAS, PKS306 and PKS64. The percentage value indicates the identity of the domain compared with 6MSAS.
B. The transformants as indicated were observed by fluorescence microscopy.
C. The expression of recombinant proteins was detected by Western blotting using an anti‐GFP antibody. GFP fused in the C‐ or N‐terminus of PKS is indicated. The arrow shows molecular weight of the PKS.
Chimeric PKS constructed by the fusion of 6MSAS with PKSs from P. boydii NTOU2362
To explore the diversity of 6MSAS, chimeric 6MSAS was constructed by replacing its ACP with the C‐termini of PKS64 and PKS306 to construct the expression vector of R6MSASΔ‐64‐ER‐KR‐ACP and R6MSASΔ‐306‐ACP‐MT, respectively, as shown in Fig. 4A. Remarkably, the pattern of green fluorescence of R6MSASΔ‐306‐ACP‐MT and R6MSASΔ‐64‐ER‐KR‐ACP was similar to that of 6MSAS analysed by fluorescence microscopy (Fig. 4B). By Western blotting, the fusion of GFP with R6MSASΔ‐64‐ER‐KR‐ACP and R6MSASΔ‐306‐ACP‐MT in the carboxyl terminus shows molecular weight of 250 and 280 kDa respectively (Fig. 4C). Regarding HPLC analysis, the extract from R6MSASΔ‐306‐ACP‐MT revealed a new peak (Fig. 4D), whereas R6MSASΔ‐64‐ER‐KR‐ACP was not observed a new peak. Fortunately, chimeric PKS showed the novel biosynthetic activity to differ from that of 6MSAS. To rule out variation during HPLC, the distinct peak was demonstrated by the mixture containing the extract from R6MSASΔ‐306‐ACP‐MT and R6MSAS (Fig. 4D). Using the GAL1 promoter, the peak was observed after galactose induction but not in the extract from the culture containing glucose (Fig. S1). Our findings showed that the novel PKS was constructed by the fusion of the domains of KS, AT, DH and KR from 6MSAS with those of ACP and MT from PKS306.
Figure 4.
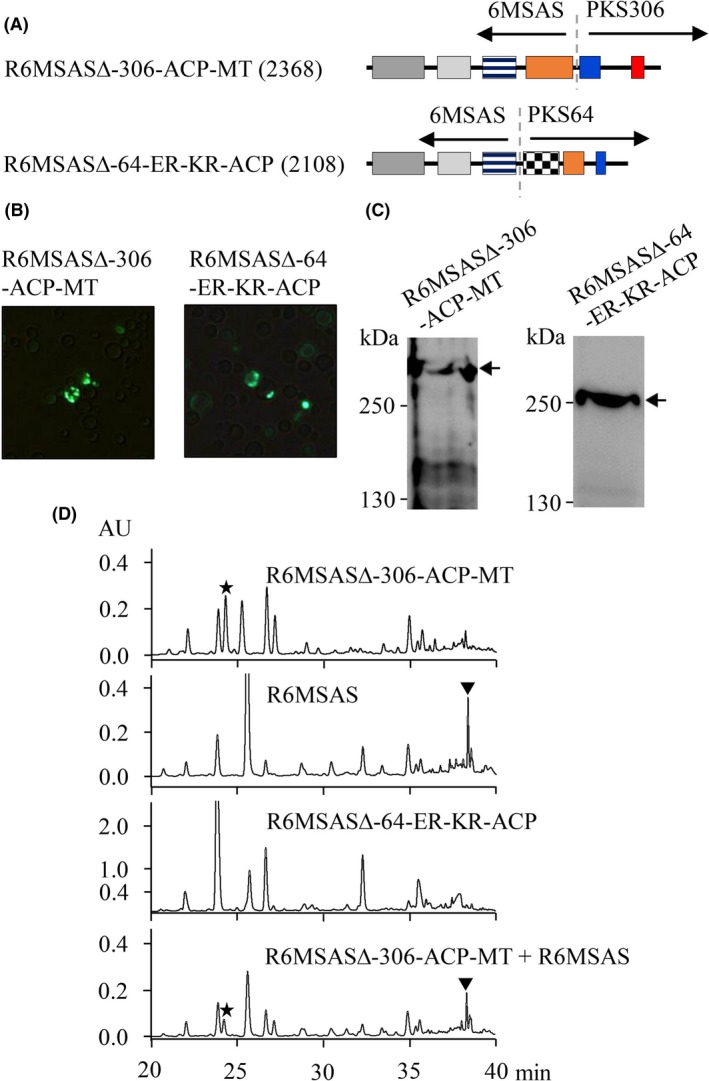
Domain of KS, AT, DH and KR from 6MSAS fused with ACP and MT from PKS306.
A. The diagram shows the functional domains of chimeric PKS generated by the fusion of 6MSAS with PKS306 or PKS64 as indicated.
B. The transformants as indicated were observed by fluorescence microscopy.
C. The expression of recombinant proteins was detected by Western blotting using an anti‐GFP antibody. “←”indicates the molecular weight of PKS.
D. The chemical product was analysed by HPLC.
The MT domain is involved in the biosynthesis of the compound
To analyse the role of the MT domain, R6MSASΔ‐306‐ACP was constructed by removal of the MT domain from R6MSASΔ‐306‐ACP‐MT (Fig. 5A). However, the pattern of green fluorescence of R6MSASΔ‐306‐ACP was similar to that of 6MSAS analysed by fluorescence microscopy (Fig. 5B). The product of this construction was not found in the ethyl acetate extract, showing that MT domain is required for the biosynthesis of the novel compound (Fig. 5D). To explore whether MT plays a role in the biosynthesis of 6MSA, R6MSAS‐306‐MT was constructed by the fusion of MT from PKS306 (Fig. 5A). Regarding the HPLC results, no difference was observed in the production of 6MSA (Fig. 5D). Our finding shows that MT is unable to change 6MSAS biosynthetic activity.
Figure 5.
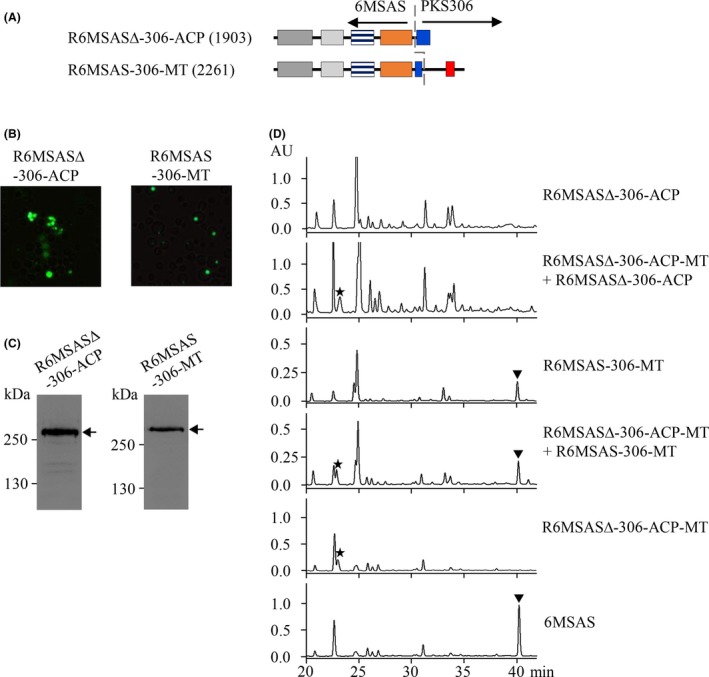
ACP‐MT contributes to the biosynthetic activity of R6MSASΔ‐306‐ACP‐MT.
A. The diagram shows the functional domains of chimeric PKS as indicated.
B. The transformants as indicated were observed by fluorescence microscopy.
C. The expression of recombinant proteins was detected by Western blotting using an anti‐GFP antibody. “←”indicates the molecular weight of PKS.
D. The chemical product was analysed by HPLC.
Chemical structure of the novel compound
To identify the molecular weight of the novel compound, a mass spectrum was generated [M]+ and [M‐H]+ at 142.0579 and 141.0546 respectively (Fig. 6A). Moreover, the FT‐IR spectrum revealed the presence of carbonyl (1746 cm−1) and hydroxyl (3372 cm−1) groups (Fig. 6B), but not benzyl and unsaturated acyl groups (2260–2000 cm−1), in the novel compound. Based on the elemental analysis result, carbon and hydrogen were confirmed to be the elements in this compound without nitrogen (data not shown). The results of NMR spectra for the product of R6MSASΔ‐306‐ACP‐MT including 1H, 13C (Fig. S2), 1H‐13C HSQC (Fig. S3), 1H‐13C HMBC (Fig. S4) and 1H‐1H COSY (Fig. S5) are summarized in Table 1. From the 1H‐13C HSQC spectrum, we conclude that two methyl groups (δ H 0.945, δ C 15.263, C‐7; δ H 0.992, δ C 16.135, C‐6), one methylene group (δ H 2.791, δ C 40.832, C‐4) and one methanetriyl group (δ H 2.046, δ C 35.288, C‐5), are involved in this novel compound. Additionally, three carbons were determined to have no proton attached (δ C 77.087, C‐2; δ C 173.326, C‐1; δ C 176.849, C‐3). The 1H‐13C HMBC (Fig. S3) and 1H‐1H COSY spectra (Fig. S5) showed two methyl groups correlated with C‐2, C‐5 and each other (C‐6 or C‐7); the methylene group correlated with C‐1, C‐2, C‐3 and C‐5; and methanetriyl group correlated with C‐2, C‐3, C‐6 and C‐7. The 1H‐1H COSY indicated two methyl groups interacted with the methanetriyl group that constituting the CH(CH3)2 group, and two protons showed interacting on the methylene group. According to the above data, the chemical structure of the product of R6MSASΔ‐306‐ACP‐MT included a cyclobutane‐like backbone, C‐1 and C‐3 were keto groups, and the CH(CH3)2 group was at the diagonal position of the methylene group. C‐2 contains a hydroxyl group and links with two carbonyl groups as well as the CH(CH3)2 group. The molecular formula is C7H10O3 termed 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione, and the chemical structure is shown in Fig. 7.
Figure 6.
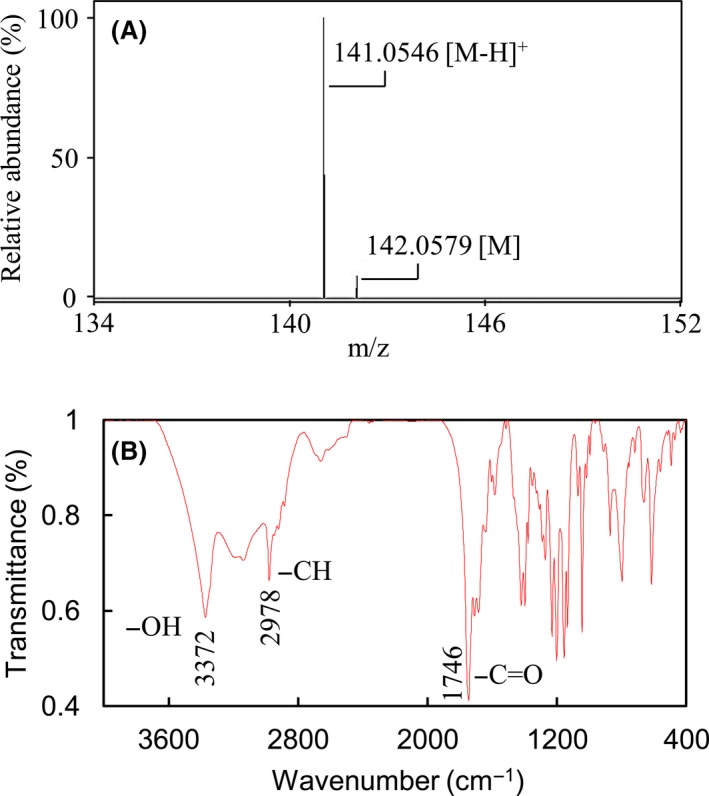
Characterization of the novel compound. Spectra of the mass.
A. and FT‐IR.
B. were performed to analyse the novel compound.
Table 1.
1H and 13C chemical shifts of 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione in methanol‐d 4
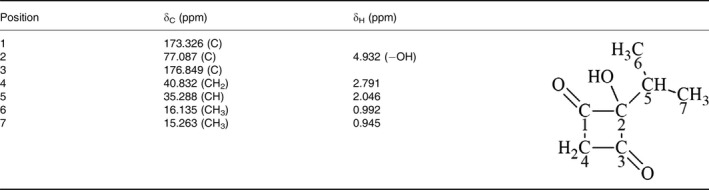
Figure 7.
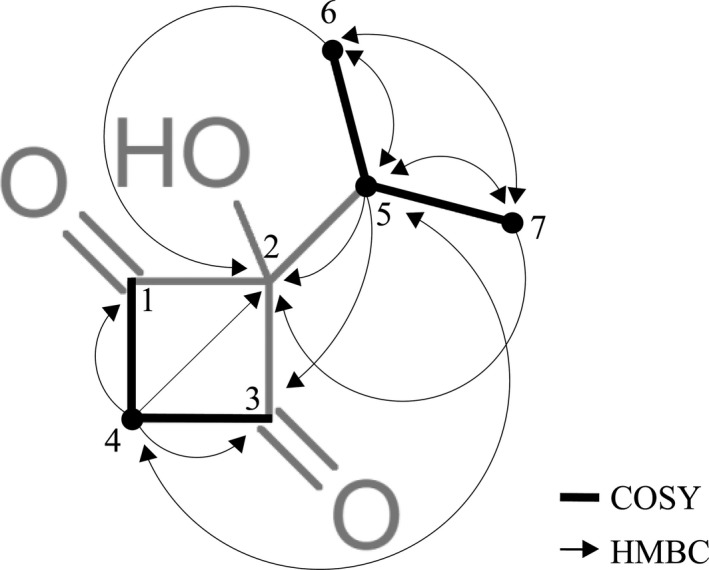
Selected key 1H‐1H COSY (bold bonds) and 1H‐13C HMBC (→) correlations of 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione by two‐dimensional NMR analysis.
Discussion
Although a reliable and feasible strategy for the heterologous expression of PKSs in S. cerevisiae is developed in this study, functional enzymatic activity and novel compounds were not demonstrated except by 6MSAS. The failure of recombinant PKS from P. boydii NTOU2362 to generate biosynthetic activity may be due to the following reasons. FAS, biosynthetic machinery for fungal melanin and mycotoxins biosynthesis localize in the endoplasmic reticulum, endosome and peroxisome, respectively, implying that the localization signal harbouring in PKSs may have important roles in the biosynthesis of polyketides (Tehlivets et al., 2007; Kistler and Broz, 2015; Upadhyay et al., 2016). However, localization signal of heterologous expression of recombinant PKS genes was not recognized in the machinery of protein transport in S. cerevisiae (Hesketh and Chater, 2003). Moreover, an oligomer is needed various PKSs to form a functional enzyme, whereas recombinant PKS expressed from one gene or inadequate PKS genes cannot form functional oligomers (Studt et al., 2015; Herbst et al., 2018).
Chimeric PKS comprising KS, AT, DH and KR from 6MSAS with ACP and MT from PKS306 can biosynthesize 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione. ACPs consist of 70‐100 amino acids typically and participate in the biosynthesis of polyketides to relay growing acyl chains through the assemble line of PKS (Chen et al., 2011; Crosby and Crump, 2012). Remarkably, ACP collaborates with the functional domains of PKS to add a specific chemical moiety such as a chain length of acyl groups (Moore and Hertweck, 2002; Crawford et al., 2006; Ray and Moore, 2016; Keatinge‐Clay, 2017). Compared with the protein sequence of ACP from 6MSAS, a low similarity of ACP from PKS306 is shown except at the phosphopantetheine attachment site, which is recognized by phosphopantetheinyl transferase (Fig. S6). ACP and MT from PKS306 showed cooperativity to biosynthesize the compound. Comparison of ACP and MT from PKS306 with Scedosporium apiospermum (XP_016643174.1), Oidiodendron maius Zn (KIM92947.1), Rutstroemia sp. NJR‐2017a BVV2 (PQE24163.1) and Madurella mycetomatis (KXX79217.1) reveals identities of 89%, 53%, 51% and 47% respectively. However, these PKSs are not characterized. The role of KS, AT, DH, KR, ACP and MT in the synthesis of the novel compound remains to be explored.
The MT domain is necessary for the biosynthesis of the compound because removal of the MT domain may deprive the biological activity of the chimeric PKS. Moreover, 6MSAS fused with the MT domain did not affect the biosynthesis of 6MSA and is not found in the novel compound, demonstrating that biosynthesis of the compound is not exclusively attributed to the cooperation of ACP and MT domain. Methyltransferase in PKS functional domains may perform iterative C‐methylation of fungal non‐reduced polyketides using S‐adenosylmethionine as the donor of methyl groups one or more times to a tetra‐ or pentaketide (Kroken et al., 2003; Ahuja et al., 2012). The novel compound containing an isopropyl group demonstrates activity of the MT domain that is essential for biosynthesis because removal of the domain abolishes the synthesis of the compound. Although 6MSAS fused with the MT domain, 6MSA was observed suggesting that the domain did not alter the biosynthetic pathway of the iterative PKS. AprA, a PKS involved in the biosynthesis of apratoxin A, contains two MT domains, MT1 and MT2 (Skiba et al., 2017). MT1 performs two methylations to produce Me2Mal‐ACP using Mal‐ACP as the substrate. MT2 is a bifunctional enzyme, carrying out both decarboxylation and methylation, and is active on multiple substrates to produce pivaloyl‐, isobutyl‐, propionyl‐ or acetoacetyl‐ACP. MT from PKS306, compared with MT2, shows 27% identity and 44% similarity, whereas MT1 shows < 15% and 29% identity and similarity (Fig. S7 and S8) respectively. Interestingly, ACP and MT from PKS306 provide unpredictable biological activity to rebuild the 6MSAS biosynthetic logic.
The first discovered PKS is 6MSAS, which has demonstrated remarkable biosynthesis to produce the secondary metabolite, 6MSA. Moreover, the PKS may provide an intermediate that is involved in the polyketide biosynthesis of other structurally diverse compounds with comparable differences in biological activities. The enzyme is regarded as a multifunctional catalytic machine and operates sequentially in individual active sites to yield a polyketide product. Our findings imply that biosynthesis of the novel compound involves the functional domain of KS, AT, DH and KR as well as ACP and MT. By replacing ACP in 6MSAS with ACP‐MT from PKS306, the chimeric PKS produces the novel compound 2‐hydroxy‐2‐(propan‐2‐yl) cyclobutane‐1,3‐dione leading to a remarkable possibility to general secondary metabolites using combinatorial functional domains from various PKSs.
Experimental procedures
Bacterial strains and culture conditions
The strains and plasmids used in this study are described in Table S1. E. coli, S. cerevisiae, A. nidulans and A. terreus cultures were grown at 37, 28 and 25°C respectively. E. coli Mach1TMT1® was used for cloning, and vectors containing ccdB used E. coli DB3.1 as the host. E. coli strains were cultured in Luria–Bertani medium. S. cerevisiae BJ2168, A. nidulans and A. terreus were grown in YPD (1% yeast extract, 2% peptone and 2% dextrose).
Preparation of genomic DNA and RNA
Aspergillus nidulans or P. boydii NTOU2362 was cultured in 3 ml YPD overnight and pelleted by centrifugation. Genomic DNA was prepared using the Puregene Yeast/Bac kit (Qiagen Hilden, Germany) according to the manufacturer's protocol. The amount of DNA was estimated by OD260 and gel electrophoresis by compared the loaded DNA markers. A. terreus or P. boydii NTOU2362 was cultured in 3 ml YPD for 48 h and harvested. To the pellet, 2 grams of acid‐washed glass beads in lysis buffer (50 mM Tris–HCl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% (v/v) Nonidet® P‐40, 1000 U ml−1 of RNase inhibitor and 1 mM DTT) was added and vortexed vigorously for 3 min. After the beads settled, 1 ml of the supernatant was transferred to a new 15 ml centrifuge tube, and then, 1 ml of lysis buffer was added to the sample to replace the aspirated supernatant. The sample was shaken, and the beads were allowed to settle again, and then, 1 ml of the supernatant was transferred the same 15 ml centrifuge tube. The lysate was centrifuged for 5 min at 3000 g. Next, 2 ml of 70% ethanol was added to the supernatant, mixed thoroughly and subjected to RNeasy midi column purification following the manufacturer's instructions (Qiagen, Hilden, Germany). The amount and quality of RNA were determined by OD260 and RNA electrophoresis respectively.
Reverse transcription and polymerase chain reaction
One microgram of the total RNA was used as the template for reverse transcription (RT) using random primers, and the mixture was incubated at 42°C for 30 min in a total volume of 20 μl. Following RT, 2 μl of the RT reaction mixture was utilized for PCR. Primer sequences are shown in Table S2. For directional cloning, 5′ end of the forward primer included the 4‐base pair sequence (CACC). KOD HOT Start DNA polymerase (TOYOBO) was used in this study due to the high fidelity and processivity of the enzyme. PCR was performed following GC‐rich and genomic DNA template protocol described by the manufacturer (TOYOBO). The polymerase was activated by heating at 95°C for 2 min followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 30 s and extension at 68°C for 1 kb min−1. To analyse the PCR products, a 10 μl sample was loaded and run on an agarose gel. The DNA was visualized after ethidium bromide staining using a UV illuminator.
Construction of donor vectors
The vector pENTR/D‐TOPO was used to construct donor vectors for the Gateway cloning technology. The PCR products were purified from agarose gel using the Gel Elution kit (Qiagen Hilden, Germany). To directionally clone the purified blunt‐end DNA harbouring CACC in the 5′‐end, the pENTR Directional TOPO Cloning kit (Invitrogen, Waltham, MA, USA) was used. The DNA was transformed into Mach1™T1® competent E. coli. The vectors were confirmed by DNA sequencing. The constructed plasmids are shown in Table S1.
Yeast expression vectors were constructed by LR recombination or Multisite Gateway technology
A suite of Gateway cloning vectors (pAGs) was used to establish PKS expression in S. cerevisiae (Alberti et al., 2007). The yeast expression vectors were constructed using the Gateway® LR Clonase™ II enzyme mix (Invitrogen) with the attL‐containing donor vector pENTR and attR‐containing destination vector. Briefly, the LR reaction was performed by mixing 150 ng of pENTR‐containing vectors with 150 ng of pAG‐containing vectors, 4 μl of TE buffer and 2 μl of LR Clonase enzyme to form a 10‐μl reaction mixture that was incubated at 25°C for 1 h. The reaction was stopped by adding 2 μl of Proteinase K solution at 37°C for 10 min. Chimeric PKSs were obtained to perform a BP recombination reaction between the attB‐flanked DNA fragment and the appropriate attP‐containing donor vector by 2‐fragment recombination in Multisite Gateway (Invitrogen). The generating donor might undergo LR recombination to construct the expression vector (Fig. S9). The expression vectors were obtained after DNA sequencing and are shown in Table S1. To perform a positive control of 6MSAS in a 2‐fragment Multisite Gateway reaction, pAG424GPD‐ccdB‐EGFP, pDONR‐6MSAS KS‐KR P1‐P5r and pDONR‐6MSAS ACP P5‐P2 were perform to construct the pGPD424‐RMSAS‐GFP and to generate R6MSAS (Fig. S10). The chimeric PKSs of R6MSASΔ‐64‐ER‐KR‐ACP, R6MSASΔ‐306‐ACP‐MT, R6MSASΔ‐306‐ACP and 6MSAS‐306MT were obtained as described in Table S1.
Saccharomyces cerevisiae transformation
Circular forms (1 μg) of expression vectors were transformed into S. cerevisiae using the lithium chloride method. The transformants were incubated without shaking at 30°C for 2 h, and then, 1 ml of medium was added before incubation at the same temperature for 2 h. The cells were centrifuged at 1500 g for 5 min and were spread onto an SDC (0.67% [w/v] yeast nitrogen base without amino acids, 0.192% [w/v] yeast synthetic dropout medium supplement without His, Leu, Try and uracil, and 2% [w/v] dextrose) agar plate. The colonies were collected, and PCR was performed to detect the plasmids harboured in these clones. Fluorescence of GFP fusion protein in transformants was examined by Olympus fluorescence microscopy (Olympus, Tokyo, Japan).
Western blot
The yeast cells were collected by centrifugation at 1500 g for 5 min at 4°C. The pellet was resuspended with 300 μl of lysis buffer (50 mM Tris–HCl pH 7.4, 1% Triton X‐100, 150 mM NaCl, 1% glycerol, 1 mM EDTA and 0.1 mM phenylmethylsulfonyl fluoride), and 1 gram of glass beads was added to break the cells under vigorous vortexing for 2 min. The supernatant was collected by centrifugation at 14 000 g for 10 min at 4°C. The protein concentration was determined using the Bio‐Rad protein kit. The lysate (20 μg) was resolved by SDS‐PAGE and electrotransferred onto nitrocellulose paper. The blot was hybridized with anti‐GFP antibody (MAB1083, Merck Millipore), followed by detection using ECL reagent (Merck Millipore). The signals were examined using the Fujifilm LAS‐4000 BioSpectrum system and were quantified using Fujifilm software (Akasaka, Tokyo, Japan).
Extraction of 6MSA
Saccharomyces cerevisiae transformants containing the expression vector of pGAL426‐6MASA‐GFP and pGAL425‐PPTase or pGAL426‐6MASA‐GFP only as the control were seeded into a 250 ml flask with 50 ml of SDC medium and were incubated at 25°C on a rotary shaker (200 rpm) for 48 h. The culture was transferred into 500 ml of SDC medium in a 2.5 litre flask and was incubated for 48 h, and then, the cells were collected by centrifugation and refreshed with 500 ml of SDC medium containing galactose (2%) to activate the GAL1 promoter for 96 h. The medium was collected by centrifugation at 500 g for 10 min. The supernatant was extracted with 500 ml of ethyl acetate (EA), and the sample was concentrated by an evaporator to 20 ml.
Detection of 6MSA by HPLC
The dried extract was redissolved with 200 μl of buffer containing 5% methanol and 0.05% TFA in water for HPLC analysis. The chromatographic separation was performed using a Waters e2695 HPLC system equipped with a reversed phase C18 column (Agilent ZORBAX Eclipse XDB‐C18, 9.4 mm I.D. × 250 mm, 5μm) and a Waters 2998 photodiode array detector. The column was equilibrated with 95% solvent A (0.05% TFA in water) and 5% solvent B (0.05% TFA in methanol) at a flow rate of 1 ml min−1. The gradient elution steps were as follows: 0–1 min, 5% solvent B; 1–30 min, a linear gradient to 50% solvent B; 30–40 min, to 99% solvent B, and the UV absorption was 254 nm.
Determination of the chemical compound
Nuclear magnetic resonance (NMR) spectra were recorded using a Bruker AV600 NMR spectrometer. All NMR spectra were resolved by iNMR software. The molecular weight was determined by Synapt G2 mass spectrometer (Waters). IR spectra were obtained using a Fourier transform infrared spectrometer (Bruker TENSOR II). Element analysis was performed using the Elementar Vario EL III system.
Conflict of interest
None declared.
Supporting information
Fig. S1. The product of the chemical compound was analyzed by HPLC after the induction using galactose or glucose as indicated.
Fig. S2. 1H NMR (600MHz) and 13C NMR (150MHz) spectrum of novel compound in Methanol‐d4.
Fig. S3. 1H‐13C HSQC (600MHz) spectrum of novel compound in Methanol‐d4.
Fig. S4. 1H‐13C HMBC (600MHz) spectrum of novel compound in Methanol‐d4.
Fig. S5. 1H‐1H COSY (600MHz) spectrum of novel compound in Methanol‐d4.
Fig. S6. Sequence alignment of the ACP from of 6MSAS and PKS306. The phosphopantetheine attachment site is labeled by “*”.
Fig. S7. Amino acid sequence alignment of the methyltransferase (MT) domain of PKS306 and MT1 domain of AprA (WP_075900460).
Fig. S8. Amino acid sequence alignment of the MT domain of PKS306 and MT2 domain of AprA.
Fig. S9. Strategy for the construction of the chimeric PKSs. Chimeric PKSs were generated by performing a BP recombination reaction between the attB‐flanked DNA fragment and appropriate attP‐containing donor vector by 2‐fragment recombination in Multisite Gateway system. The indicated peptide sequence from attB5 would replace the similar sequence in the target gene.
Fig. S10. Detection of R6MSAS after 2‐fragment recombinant of Multisite Gateway. (A) The diagram shows the functional domains of R6MSAS, and the indicated peptide sequence from attB5 would replace the sequence of 6MSAS to generate R6MSAS. (B) The yeast harboring R6MSAS‐GFP was examined by fluorescence microscopy. (C) The expression of R6MSAS‐GFP was detected by western blotting using an anti‐GFP antibody. (D) The product of 6MSAS or R6MSAS was analyzed by HPLC.
Table S1. Strains and plasmids used in this study.
Table S2. Primer sequences.
Acknowledgements
The work was supported by grants from the National Science Council (102‐2313‐B‐019‐013), Tri‐Service General Hospital and National Defense Medical Center (TSGHC104057) and Taipei City Hospital, Taiwan.
Microbial Biotechnology (2019) 12(5), 920–931
Funding Information
The work was supported by grants from the National Science Council (102‐2313‐B‐019‐013), Tri‐Service General Hospital and National Defense Medical Center (TSGHC104057) and Taipei City Hospital, Taiwan.
Contributor Information
Chii‐Cheng Hsieh, Email: chii.cheng@msa.hinet.net.
Shye‐Jye Tang, Email: tsj@mail.ntou.edu.tw.
References
- Ahuja, M. , Chiang, Y.‐M. , Chang, S.‐L. , Praseuth, M.B. , Entwistle, R. , Sanchez, J.F. , et al (2012) Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans . J Am Chem Soc 134: 8212–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti, S. , Gitler, A.D. , and Lindquist, S. (2007) A suite of Gateway® cloning vectors for high‐throughput genetic analysis in Saccharomyces cerevisiae . Yeast 24: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias, G.D. , Chaliotis, A. , and Mossialos, D. (2016) Discovery strategies of bioactive compounds synthesized by nonribosomal peptide synthetases and Type‐I polyketide synthases derived from marine microbiomes. Mar Drugs 14: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, J. , Ripka, S. , Siegner, A. , Schiltz, E. , and Schweizer, E. (1990) The multifunctional 6‐methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur J Biochem 192: 487–498. [DOI] [PubMed] [Google Scholar]
- Bond, C. , Tang, Y. , and Li, L. (2016) Saccharomyces cerevisiae as a tool for mining, studying and engineering fungal polyketide synthases. Fungal Genet Biol 89: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane, D.E. (2010) Programming of erythromycin biosynthesis by a modular polyketide synthase. J Biol Chem 285: 27517–27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.‐C. , Deng, T.‐S. , Pang, K.‐L. , Hsiao, C.‐J. , Chen, Y.‐Y. , Tang, S.‐J. , and Lee, T.‐H. (2013) Polyketides from the littoral plant associated fungus Pseudallescheria boydii . J Nat Prod 76: 1796–1800. [DOI] [PubMed] [Google Scholar]
- Chen, H. , and Du, L. (2016) Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl Microbiol Biotechnol 100: 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Kelly, E.E. , Masluk, R.P. , Nelson, C.L. , Cantu, D.C. , and Reilly, P.J. (2011) Structural classification and properties of ketoacyl synthases. Protein Sci 20: 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R.J. , and Simpson, T.J. (2009) Fungal type I polyketide synthases. Methods Enzymol 459: 49–78. [DOI] [PubMed] [Google Scholar]
- Crawford, J.M. , Dancy, B.C.R. , Hill, E.A. , Udwary, D.W. , and Townsend, C.A. (2006) Identification of a starter unit acyl‐carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci USA 103: 16728–16733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby, J. , and Crump, M.P. (2012) The structural role of the carrier protein – active controller or passive carrier. Nat Prod Rep 29: 1111–1137. [DOI] [PubMed] [Google Scholar]
- Dimroth, P. , Walter, H. , and Lynen, F. (1970) Biosynthesis of 6‐methylsalicylic acid. Eur J Biochem 13: 98–110. [DOI] [PubMed] [Google Scholar]
- Dutta, S. , Whicher, J.R. , Hansen, D.A. , Hale, W.A. , Chemler, J.A. , Congdon, G.R. , et al (2014) Structure of a modular polyketide synthase. Nature 510: 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke, J. , and Braus, G.H. (2014) Manipulation of fungal development as source of novel secondary metabolites for biotechnology. Appl Microbiol Biotechnol 98: 8443–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich, E.J.N. , Reiter, S. , and Piel, J. (2014) Recent advances in genome‐based polyketide discovery. Curr Opin Biotechnol 29: 107–115. [DOI] [PubMed] [Google Scholar]
- Herbst, D.A. , Townsend, C.A. , and Maier, T. (2018) The architectures of iterative type I PKS and FAS. Nat Prod Rep 35: 1046–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh, A. , and Chater, K.F. (2003) Evidence from proteomics that some of the enzymes of actinorhodin biosynthesis have more than one form and may occupy distinctive cellular locations. J Ind Microbiol Biotechnol 30: 523–529. [DOI] [PubMed] [Google Scholar]
- Holm, D.K. , Petersen, L.M. , Klitgaard, A. , Knudsen, P.B. , Jarczynska, Z.D. , Nielsen, K.F. , et al (2014) Molecular and chemical characterization of the biosynthesis of the 6‐MSA‐derived meroterpenoid yanuthone D in Aspergillus niger . Chem Biol 21: 519–529. [DOI] [PubMed] [Google Scholar]
- Huang, C. , Yang, C. , Zhang, W. , Zhang, L. , De, B.C. , Zhu, Y. , et al (2018) Molecular basis of dimer formation during the biosynthesis of benzofluorene‐containing atypical angucyclines. Nat Commun 9: 2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke‐Kodama, H. , Sandmann, A. , Muller, R. , and Dittmann, E. (2005) Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol 22: 2027–2039. [DOI] [PubMed] [Google Scholar]
- Katz, L. (2009) The DEBS paradigm for type I modular polyketide synthases and beyond. Methods Enzymol 459: 113–142. [DOI] [PubMed] [Google Scholar]
- Keatinge‐Clay, A.T. (2017) The uncommon enzymology of cis‐acyltransferase assembly lines. Chem Rev 117: 5334–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler, H.C. and Broz, K. (2015) Cellular compartmentalization of secondary metabolism. Front Microbiol 6: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, C. , Huang, H. , Xue, Y. , Liu, Y. , Peng, Q. , Liu, Q. , et al (2018) Heterologous pathway assembly reveals molecular steps of fungal terreic acid biosynthesis. Sci Rep 8: 2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken, S. , Glass, N.L. , Taylor, J.W. , Yoder, O.C. , and Turgeon, B.G. (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA 100: 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.K. , Da Silva, N.A. , and Kealey, J.T. (2009) Determination of the extent of phosphopantetheinylation of polyketide synthases expressed in Escherichia coli and Saccharomyces cerevisiae . Anal Biochem 394: 75–80. [DOI] [PubMed] [Google Scholar]
- Moore, B.S. , and Hertweck, C. (2002) Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep 19: 70–99. [DOI] [PubMed] [Google Scholar]
- Mulder, K.C. , Mulinari, F. , Franco, O.L. , Soares, M.S. , Magalhaes, B.S. , and Parachin, N.S. (2015) Lovastatin production: from molecular basis to industrial process optimization. Biotechnol Adv 33: 648–665. [DOI] [PubMed] [Google Scholar]
- Ray, L. , and Moore, B.S. (2016) Recent advances in the biosynthesis of unusual polyketide synthase substrates. Nat Prod Rep 33: 150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B. (2003) Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Skiba, M.A. , Sikkema, A.P. , Moss, N.A. , Tran, C.L. , Sturgis, R.M. , Gerwick, L. , et al (2017) A mononuclear iron‐dependent methyltransferase catalyses initial steps in assembly of the apratoxin a polyketide starter unit. ACS Chem Biol 12: 3039–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm, P.A. , Herbst, D.A. , Maier, T. , and Townsend, C.A. (2017) Functional and structural analysis of programmed c‐methylation in the biosynthesis of the fungal polyketide citrinin. Cell Chem Biol 24: 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt, L. , Janevska, S. , Niehaus, E.‐M. , Burkhardt, I. , Arndt, B. , Sieber, C.M.K. , et al (2015) Two separate key enzymes and two pathway‐specific transcription factors are involved in fusaric acid biosynthesis in Fusarium fujikuroi . Environ Microbiol 18: 936–956. [DOI] [PubMed] [Google Scholar]
- Tehlivets, O. , Scheuringer, K. , and Kohlwein, S.D. (2007) Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta 1771: 255–270. [DOI] [PubMed] [Google Scholar]
- Tran, L. , Broadhurst, R.W. , Tosin, M. , Cavalli, A. , and Weissman, K.J. (2010) Insights into protein‐protein and enzyme‐substrate interactions in modular polyketide synthases. Chem Biol 17: 705–716. [DOI] [PubMed] [Google Scholar]
- Tsunematsu, Y. , Ishiuchi, K. , Hotta, K. , and Watanabe, K. (2013) Yeast‐based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat Prod Rep 30: 1139–1149. [DOI] [PubMed] [Google Scholar]
- Udagawa, T. , Yuan, J. , Panigrahy, D. , Chang, Y.H. , Shah, J. , and D'Amato, R.J. (2000) Cytochalasin E, an epoxide containing Aspergillus‐derived fungal metabolite, inhibits angiogenesis and tumor growth. J Pharmacol Exp Ther 294: 421–427. [PubMed] [Google Scholar]
- Upadhyay, S. , Xu, X. , Lowry, D. , Jackson, J.C. , Roberson, R.W. , and Lin, X. (2016) Subcellular compartmentalization and trafficking of the biosynthetic machinery for fungal melanin. Cell Rep 14: 2511–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldner, M. , Fantus, D. , Solari, M. , and Thomson, A.W. (2016) New perspectives on mTOR inhibitors (rapamycin, rapalogs and TORKinibs) in transplantation. Br J Clin Pharmacol 82: 1158–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.Y. , Luo, H.D. , Zhang, X.S. , Lin, T. , Jiang, H. , and Li, Y.Q. (2016) The substrate promiscuity of a phosphopantetheinyl transferase SchPPT for coenzyme A derivatives and acyl carrier proteins. Arch Microbiol 198: 193–197. [DOI] [PubMed] [Google Scholar]
- Wattana‐amorn, P. , Williams, C. , Płoskoń, E. , Cox, R.J. , Simpson, T.J. , Crosby, J. , and Crump, M.P. (2010) Solution structure of an acyl carrier protein domain from a fungal type I polyketide synthase. Biochem 49: 2186–2193. [DOI] [PubMed] [Google Scholar]
- Wattanachaisaereekul, S. , Lantz, A.E. , Nielsen, M.L. , Andresson, O.S. , and Nielsen, J. (2007) Optimization of heterologous production of the polyketide 6‐MSA in Saccharomyces cerevisiae . Biotechnol Bioeng 97: 893–900. [DOI] [PubMed] [Google Scholar]
- Weissman, K.J. (2014) The structural biology of biosynthetic megaenzymes. Nat Chem Biol 11: 660–670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The product of the chemical compound was analyzed by HPLC after the induction using galactose or glucose as indicated.
Fig. S2. 1H NMR (600MHz) and 13C NMR (150MHz) spectrum of novel compound in Methanol‐d4.
Fig. S3. 1H‐13C HSQC (600MHz) spectrum of novel compound in Methanol‐d4.
Fig. S4. 1H‐13C HMBC (600MHz) spectrum of novel compound in Methanol‐d4.
Fig. S5. 1H‐1H COSY (600MHz) spectrum of novel compound in Methanol‐d4.
Fig. S6. Sequence alignment of the ACP from of 6MSAS and PKS306. The phosphopantetheine attachment site is labeled by “*”.
Fig. S7. Amino acid sequence alignment of the methyltransferase (MT) domain of PKS306 and MT1 domain of AprA (WP_075900460).
Fig. S8. Amino acid sequence alignment of the MT domain of PKS306 and MT2 domain of AprA.
Fig. S9. Strategy for the construction of the chimeric PKSs. Chimeric PKSs were generated by performing a BP recombination reaction between the attB‐flanked DNA fragment and appropriate attP‐containing donor vector by 2‐fragment recombination in Multisite Gateway system. The indicated peptide sequence from attB5 would replace the similar sequence in the target gene.
Fig. S10. Detection of R6MSAS after 2‐fragment recombinant of Multisite Gateway. (A) The diagram shows the functional domains of R6MSAS, and the indicated peptide sequence from attB5 would replace the sequence of 6MSAS to generate R6MSAS. (B) The yeast harboring R6MSAS‐GFP was examined by fluorescence microscopy. (C) The expression of R6MSAS‐GFP was detected by western blotting using an anti‐GFP antibody. (D) The product of 6MSAS or R6MSAS was analyzed by HPLC.
Table S1. Strains and plasmids used in this study.
Table S2. Primer sequences.


