Abstract
Squamous cell carcinoma of the head and neck is a lethal disease with suboptimal survival outcomes and standard therapies with significant comorbidities. Whole exome sequencing data recently revealed an abundance of genetic and expression alterations in a family of enzymes known as protein methyltransferases in a variety of cancer types, including squamous cell carcinoma of the head and neck. These enzymes are mostly known for their chromatin-modifying functions through methylation of various histone substrates, though evidence supports their function also through methylation of non-histone substrates. This review summarizes the current knowledge on the function of protein methyltransferases in squamous cell carcinoma of the head and neck and highlights their promising potential as the next generation of therapeutic targets in this disease.
Keywords: Protein methyltransferase, Protein methylation, Squamous cell carcinoma of the head and neck, NSD1, NSD2, NSD3, EHMT2, EZH2, PRMT1, PRMT5
Introduction
Over the past decade, multiple studies have uncovered the significance of protein methylation in cancer development and progression. Protein lysine (PKMTs) and arginine (PRMTs) methyltransferases comprise a group of 62 enzymes that mediate the reversible deposition of methyl groups on lysine and arginine residues, respectively, on either histone or non-histone substrates. Protein demethylases (PDMTs) comprise approximately 30 enzymes and they erase these marks. Studies conducted by the Cancer Genome Atlas (TCGA) consortium have revealed multiple genetic and expression level alterations in these enzymes in multiple cancer types, including squamous cell carcinoma of the head and neck (SCCHN) [1]. More specifically, 95% of patients with SCCHN in the TCGA database carry genetic or expression alterations in any of the protein methyltransferases (PMTs).
Interestingly, while PMTs are mostly known to regulate epigenetic and transcriptional events through histone methylation, preclinical studies have also revealed a number of non-histone protein substrates that are methylated either at lysine or arginine residues. Functionally, methylation of a non-histone substrate may compete with or enable neighbouring post-translational modifications, modify interactions with other proteins, regulate the stability of a protein, or its subcellular localization [2]. These effects may either enable or hinder oncogenesis and/or cancer progression in different cancer types. Based on these observations, PMTs and PDMTs have emerged as promising, novel anticancer targets, with multiple drug development programs focusing on the development of small molecule inhibitors, and a small number of inhibitors already being evaluated in clinical trials [3–6].
Squamous cell carcinoma of the head and neck (SCCHN) is the 6th most common malignancy with approximately 500,000 new cases worldwide every year [7]. It comprises a group of cancers deriving from the epithelium of the oral cavity, tonsils, pharynx (including the nasopharynx, oropharynx and hypopharynx), larynx, epiglottis, the paranasal sinuses and the nasal cavity. SCCHN is pathogenetically classified as human papilloma virus (HPV)-positive and HPV-negative disease. Standard surgery and/or chemoradiotherapy treatment options are associated with high morbidity and toxicity rates, deformities, chronic disability and high 5-year recurrence rates of approximately 50% in patients with HPV-negative SCCHN [8]. In the metastatic setting, survival rates are poor and, although immunotherapy with checkpoint inhibition was recently approved as a promising second line standard of care treatment option, only a small fraction of patients benefit from it [9, 10]. For this reason, the search for novel approaches to control and cure this disease is urgent.
In this review, we comprehensively summarize the results of preclinical studies with mechanistic insight in the role of PMTs in SCCHN with the goal to highlight the emerging potential of the protein methylome as a novel therapeutic avenue in SCCHN. As a clarification, although the top five PMTs with the highest frequency of genetic and expression alterations in SCCHN are MLL2 (KMT2D, 20%), SUV420H1 (KMT5B, 18%), SETD3 (18%), NSD1 (17%) and NSD3 (17%), we found literature with mechanistic insight pertaining to SCCHN only on NSD1 and NSD3, as well as NSD2, EHMT2, EZH2, PRMT1 and PRMT5.
Protein lysine methyltransferases in SCCHN
The NSD family of protein lysine methyltransferases
The NSD (nuclear SET-Suppressor of variegation 3–9, Enhancer of zeste and Trithorax-domain) family of PKMTs consisting of NSD1, NSD2 (MMSET/WHSC1, Wolf-Hirschhorn Syndrome Candidate 1) and NSD3 (WHSC1L1, Wolf-Hirschhorn Syndrome Candidate 1-Like 1) is a group of chromatin modifiers known to catalyze the deposition of mono- and di-methyl groups on lysine 36 of histone H3 (H3K36mel, H3K36me2), marks which induce active gene expression. They share 70–75% homology in amino-acid sequences and they contain a SET domain that possesses methyltransferase activity. Recently, the Cancer Genome Atlas (TCGA) revealed aberrancies of these enzymes in various cancer types, with some preclinical data supporting their role in oncogenesis [11–13] Each of the family members of the NSD PKMTs was recently reported to play an important role in SCCHN biology.
NSD1
NSD1 is among the top 10 most frequently mutated genes in SCCHN, with most mutations predicted to be inactivating. Per TCGA, 17% of patients with SCCHN have genetic and expression alterations in NSD1 (Table 1). A recent study [14] reported a subset of HPV-negative SCCHN cases with impaired methylation of H3K36 which was attributed to NSD1 inactivating mutations or recurrent histone 3 lysine 36 (H3K36M) mutations, indicating that loss of H3K36 methylation contributes to SCCHN oncogenesis. The authors initially performed unsupervised hierarchical clustering of DNA methylation data available on the TCGA database for 528 SCCHN samples, and found a DNA hypomethylation subgroup enriched in NSD1 damaging mutations (n = 44), large chromosomal deletions (n = 2), focal deletions (n = 2) or splicing defects (n = 1) of the NSD1 gene. 84% (51 out of 61) of the DNA hypomethylation samples had any of the above defects in the NSD1 gene. In silico analysis predicted that these NSD1 mutations are inactivating for the methyltransferase activity of this gene. Interrogation of other genes known to be involved in H3K36 methylation showed that 16% (10 out of 61) of the DNA hypomethylation samples had K36M mutations in histone H3 variant genes (H3.1, H3.2, H3.3), while other H3K36 methyltransferase genes, such as SETD2 and NSD2, were only rarely mutated. The DNA hypomethylation subgroup comprised 13% of the HPV-negative SCCHN samples and was also characterized by downregulation of the expression of genes involved in epidermal differentiation and keratinization processes, suggesting a turn towards a more undifferentiated state. NSD1-mutant tumors were noted to be significantly hypermutated and localized mostly in the larynx, whereas H3K36M-mutant tumors had similar number of mutations compared to the other DNA methylation clusters and were all localized in the oral cavity. No other clinicopathological features were significantly associated with this DNA hypomethylation cluster. The authors also detected the mutant H3K36M protein by immunohistochemical staining at a frequency of 2% in a tissue microarray of 158 oropharyngeal squamous cell carcinomas. These samples had concordant decrease in H3K36me2 and H3K36me3 levels. Patients with no NSD1 signal also had decreased H3K36me2 levels, but normal H3K36me3 levels, consistent with the di-methyltransferase but not tri-methyltransferase activity of NSD1. Accordingly, SCCHN cell lines with mutant NSD1 showed decreased levels of H3K36me2, eventhough the levels of NSD2 and NSD3 were normal, signifying a non-redundant role of NSD 1 in the production of H3K36me2 in SCCHN. Interestingly, ectopic expression of H3K36M in SCCHN cells lines with wild-type NSD1 also lead to decreased levels of H3K36me2, implying a dominant function and that H3K36M may operate as a “trap” for H3K36 methyltransferases in SCCHN [15]. These data support that reduced H3K36me2 levels contribute to the pathogenesis of this subtype of HPV-negative SCCHN.
Table 1.
Selected protein lysine and arginine methyltransferases (PMTs): frequency of genetic/expression alterations per TCGA and clinicopathologic significance in SCCHN.
| PMTs | Frequency of genetic and expression alterations per TCGA | Clinicopathologic significance in SCCHN |
|---|---|---|
| Protein lysine methyltransferases | ||
| NSD1 | 17% | • Wild-type NSD1 mRNA levels associated with poor OS and PFS in laryngeal carcinoma [16] |
| • Inactivated mutant NSD1 is associated with the immune cold phenotype [18] | ||
| NSD2 | 6% | High NSD2 protein levels associated with poor grade [19] |
| NSD3 | 17% | High NSD3 protein levels associated with poor grade and heavier smoking history [20,21] |
| EHMT2 | 9% | High EHMT2 protein levels associated with poor PFS and OS [22,23] |
| EZH2 | 5% | High EZH2 protein levels associated with poor OS and advanced tumor size, nodal and clinical stage in oral SCCHN [26,29,31] and nasopharyngeal carcinoma [30] |
| Protein arginine methyltransferases | ||
| PRMT1 | 6% | Not investigated |
| PRMT5 | 14% | High nuclear PRMT5 protein levels associated with advanced clinical, lymph node stage and poor OS in nasopharyngeal [37] and oropharyngeal carcinoma [38] |
In another report [16], inactivating NSD1 and NSD2 mutations were found to stratify patients with laryngeal cancer in two prognostically distinct subtypes, with patients with NSD1 or NSD2 mutations having a favorable prognosis. These results were obtained in the TCGA database and validated in a separate cohort of 63 laryngeal cancer patients. More specifically, the authors performed integrated clustering analysis that incorporated DNA mutation, copy number alteration, DNA methylation and gene expression data, and found that laryngeal cancers accumulated in two distinct clusters characterized by significant differences in overall (OS) and recurrence-free survival (RFS). The cluster with favorable OS and RFS had a significantly higher prevalence of inactivating NSD1 mutations. Also, NSD1 and/or NSD2 inactivating mutations in laryngeal cancer patients with more advanced stages (III, IV) rendered a more pronounced OS and PFS advantage. This survival impact was observed only in laryngeal cancers and not in other head and neck cancer anatomical sites. Also, there were no statistically significant differences between the two groups regarding nodal status or clinical stage. Interestingly, lower NSD1 mRNA levels were not associated with better survival outcomes, despite the fact that the majority of the NSD1 mutations in the cluster with the favorable prognosis in the TCGA database seemed to be truncating, rather than missense, Selected protein lysine and arginine methyltransferases (PMTs): frequency of genetic/expression alterations per TCGA and clinicopathologic significance in SCCHN. inactivating mutations. This discrepancy could be explained if the truncating mutations render a functional, truncated NSD1 variant protein which activates pathways that render a favorable prognosis. Alternatively, it is possible that the statistical power to detect differences based on mRNA expression levels was limited. The authors also report that the laryngeal cluster with wild-type NSD1 overexpressed genes involved in stem cell maintenance and hypoxia, which could explain the poorer survival in these patients. This finding seems to contrast with the findings by the Papillon-Cavanagh study, which reported decreased expression of genes involved in epidermal differentiation and keratinization, alluding to a more undifferentiated phenotype in NSD1-mutant SCCHN tumors. Another important point in this study is that missense or truncating NSD1 mutations are identified as a favorable prognostic factor in laryngeal cancers but not other head and neck cancers. This signifies that NSD1 may be functioning as an oncogene in laryngeal cancer, though functional studies are necessary to validate this possibility. Although NSD1 has been characterized to function as an oncogene in pediatric acute myeloid leukemias with recurrent NUP98-NSD1 translocations [11], it has also been reported as a putative tumor suppressor in neuroblastoma [17]. These data, as well as the fact that a recurrent hotspot mutation at Cl710 (C substituted for S or Y) was recently reported by TCGA allude to the possibility that the function of NSD1 is cell-context dependent. Preclinical studies to further dissect the function of NSD1 in SCCHN are warranted.
One more report [18] has described the NSD1-mutant SCCHN sub-type to be associated with widespread DNA hypomethylation and overexpression of stem-cell related or genes that are active during embryonic development. Furthermore, this study revealed a significant association of the NSD1-mutant SCCHN subtype with an immune cold phenotype in this disease. More specifically, the authors found that NSD1-mutant SCCHN demonstrated the lowest tumor associated leukocyte levels, including Ml macrophages, CD8+ and CD4+ memory T-cells. Accordingly, the expression of immune checkpoints, such as PD-L1 and PD-L2 were overall low compared to the other DNA methylation SCCHN subtypes. Additionally, the authors showed that NSD1 mRNA expression is positively correlated with T-cell infiltration and that knockdown of NSD1 in three SCCHN cell lines led to significant downregulation of the mRNA expression levels of a panel of multiple chemokines involved in immune cell recruitment, such as CXCL1 and CXCL3. This finding was corroborated by a mouse model of NOD-scid IL2Rgammanu11 (NSG) mice with flank subcutaneous SCCHN tumors treated with control versus NSD1 shRNAs, where after the infusion of human peripheral blood mononuclear cells, NSD1 knockdown tumors had decreased CD8+ T-cell infiltration compared to the control tumors. It is interesting to note that the Peri study showed that NSD1-mutant laryngeal SCCHN tumors are associated with better survival outcomes, even though NSD1 inactivation seems to induce an immune cold phenotype in SCCHN tumors. This could be explained if NSD1 promotes tumor infiltration not only by cytotoxic CD8+ T-cells, but also by other immune suppressive cell subtypes.
Overall, the above data support that NSD1 has a pathogenetic role in a subset of SCCHN patients. While its pattern of genetic alterations allude to its function as a tumor suppressor, the mechanism of action of NSD1 will need to be further dissected with in-depth functional studies.
NSD2
NSD2 or WHSC1/MMSET is a PKMT with an estimated 6% genetic and expression alteration rate in the SCCHN TCGA database (Table 1). While most of these alterations are deletions or missense mutations, a study published by our group [19] found that NSD2 is moderately or strongly overexpressed in 73% of SCCHN patients with locoregionally advanced disease and that this overexpression was significantly higher compared to normal squamous epithelium. The levels of NSD2 increased significantly with the transition from normal squamous to dysplastic epithelium and then to SCCHN, supporting its pathogenetic role in the initial stages of SCCHN oncogenesis. We also found a significant association of higher NSD2 expression with poor grade, but no associations were found with other clinicopathological characteristics. Knockdown of NSD2 led to significant decrease in the cell viability of four SCCHN cell lines and to decreased levels of H3K36 di- and tri-methylation, indicating its non-redundant role in establishing H3K36 methylation levels in SCCHN. Furthermore, NSD2 directly regulated the transcription of NIMA-related-kinase-7 (NEK7), a cell cycle regulator necessary for progression into cytokinesis and mitotic spindle formation, and, accordingly, its knockdown delayed the cell-cycle progression of SCCHN cells. Although this is, to our knowledge, the only study that supports a role for NSD2 in SCCHN oncogenesis, further investigation is warranted to dissect the whole-genome landscape regulated by NSD2 in SCCHN, to understand its interactions with other H3K36 methyl-transferases and to delineate the specific therapeutic setting in which NSD2 inhibition could be incorporated in the current treatment standards.
NSD3
NSD3 or WHSC1L1 is a PKMT encoded by a gene localized in the chromosomal locus 8p11.23, which shows recurrent amplification in the TCGA SCCHN cohort and a genetic/expression alteration rate of 17% (Table 1). Recently, our group reported that NSD3 is moderately or strongly overexpressed compared to normal squamous epithelium by immunohistochemical analysis in 58% of patients with locoregionally advanced SCCHN and its expression is significantly increased from normal squamous to dysplastic epithelium and then to SCCHN [20]. Interestingly, correlative analysis of the protein levels of NSD3 and NSD2 in the same tissue microarrays revealed only a mild correlation (rho = 0.37), potentially indicating non-redundant oncogenic roles in SCCHN. Significant associations were found between NSD3 protein levels with tumor grade and smoking status, in that higher NSD3 protein levels correlated positively with poor grade and heavier smoking history. No associations were found with survival outcomes or HPV-status, implying a significant role of NSD3 in the initial stages of SCCHN oncogenesis, similarly to NSD2. NSD3 knockdown caused a significant decrease in the cell viability of both HPV-positive and HPV-negative SCCHN cell lines, accompanied by a decrease in the global levels of H3K36 di-methylation. These findings support a non-redundant role of NSD3 as an oncogene and as a H3K36 methyltransferase in SCCHN. Mechanistically, this study showed that NSD3 is necessary for the transition of SCCHN cells from the G1 to the S phase through induction of H3K36 di-methylation in the gene body regions of two critical cell cycle regulators, cell division cycle 6 (CDC6) and cyclin-dependent kinase 2 (CDK2), and subsequent activation of their transcription.
In another study [21], our group reported the epidermal growth factor receptor (EGFR) as a novel non-histone substrate of NSD3. More specifically, we reported that NSD3 mono-methylated EGFR at lysine K721, which is located within the tyrosine kinase domain of EGFR, and that this methylation enhances the activating phosphorylation marks of serine 845, 1148 and 1173, and activating the downstream ERK cascade. This constitutive activation of EGFR and its downstream ERK cascade was independent of the presence of epidermal growth factor. Given that NSD3 is a nuclear protein, we hypothesized that the methylation of EGFR takes place in the nucleus of SCCHN cells in which EGFR translocates to the nucleus. K721 mono-methylation of nuclear EGFR potentiated its interaction with proliferating cell nuclear antigen (PCNA), stabilized PCNA and led to enhanced DNA replication in SCCHN cells. We hypothesized that a fraction of the K721 mono-methylated EGFR remains in the nucleus to exert its function through stabilization of PCNA, while another fraction translocates back to the cytoplasm and activates the membrane/cytoplasmic EGFR cascade. Furthermore, we showed that knockdown of NSD3 sensitizes SCCHN cells to Erlotinib. This indicates that methylation of lysine K721 of EGFR may allosterically enhance the affinity of the ATP-binding site of EGFR with ATP and thus decrease its affinity to Erlotinib, rendering resistance to this drug. Alternatively, NSD3-mediated methylation of nuclear EGFR may be responsible for resistance to EGFR inhibition through the potentiation of the functions of nuclear EGFR which is known confer resistance to EGFR inhibitors and cetuximab. As moderate or strong K721 EGFR mono-methylation was detected in 82% of patients with SCCHN, it is not unlikely that NSD3-mediated K721 EGFR mono-methylation is a major resistance mechanism to EGFR inhibition and that NSD3 inhibition could increase the therapeutic efficacy of EGFR inhibition in SCCHN.
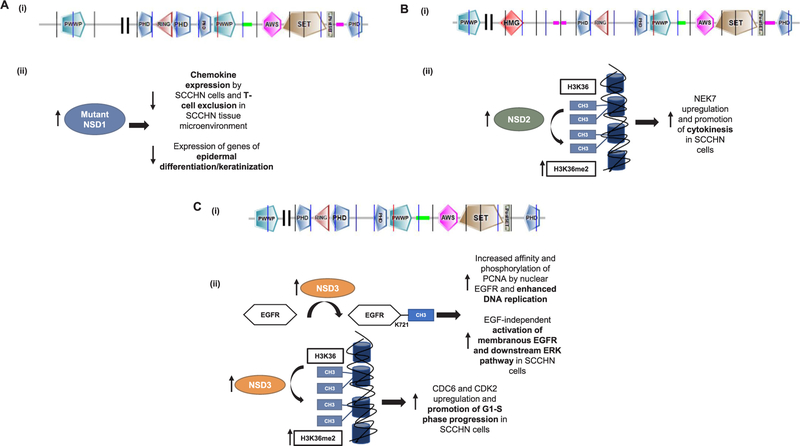
To summarize, the NSD family of protein lysine methyltransferases seems to have a distinct role in the pathogenesis and disease progression of SCCHN, and the above described studies already provide a biological rationale to target NSD2 and NSD3 in patients with SCCHN and aberrancies in these enzymes. Fig. 1 shows the protein structure and summarizes the mechanisms of function of the NSD family of PMTs in SCCHN. It is interesting to note that although all three NSDs are known to mono- and di-methylate H3K36, NSD2 and NSD3 seem to function as oncogenes, while NSD1 appears to behave as a tumor suppressor gene in SCCHN. This observation could potentially be explained if one considers that histone methylation marks are not the only enzymatic product of PMTs and that other methylation substrates or protein interactions may dictate their function. This could have critical implications in drug development, as non-histone substrates may need to be identified as more accurate pharmacodynamic biomarkers of activity of relevant inhibitors. Furthermore, the differential genomic landscape and the contribution of each of these NSDs to the H3K36 di-methylation levels in SCCHN should be delineated, and further studies should aim at defining the specific subsets of SCCHN patients that would benefit from interventions in NSD-driven oncogenic mechanisms. Finally, the role of other known H3K36 methyltransferases, such as ASH1L, SMYD3, SETD2, SETD3 and SETMAR, in SCCHN biology and their interplay with the NSDs have not been investigated and would constitute an important focus of research in order to further dissect the function of H3K36 methylation in SCCHN.
Fig. 1.
Protein structure and mechanisms of action of the NSD family of protein methyltransferases in SCCHN. A. (i) Protein structure of NSD1 (UniProt, 2696 aminoacids). PWWP: domain with conserved proline-tryptophan-tryptophan-proline motif, PHD: plant homeodomain zinc finger, RING: really interesting new gene finger domain, AWS: associated with SET domain, SET: Su(var)3–9, Enhancer-of-zeste, Trithorax domain, PostSET: cysteine-rich motif following the SET domain, (ii) Mechanisms of action of NSD 1. Truncating NSD1 mutations downregulate chemokine expression in SCCHN cells, inducing T-cell exclusion from the tissue microenvironment. NSD1 mutations are also associated with decreased expression of epidermal differentiation genes in SCCHN. B. (i) Molecular structure of the long isoform of NSD2 (UniProt, 1365 aminoacids). Domains described as per A(i) and HMG: High mobility group domain, (ii) Mechanism of action of NSD2 in SCCHN. NSD2 di-methylates H3K36 and induces transcriptional upregulation of NIMA-related kinase 7 (NEK7), leading to entry of SCCHN cells to cytokinesis. C. (i) Molecular structure of the long isoform of NSD3 (UniProt, 1437 aminoacids). Domains described as per A(i). (ii) Mechanisms of action of NSD3. NSD3 di-methylates H3K36 and induces transcriptional upregulation of cell-cycle related genes CDC6 and CDK2, leading to promotion of Gl-S phase progression in SCCHN cells. Additionally, NSD3 directly mono-methylates the epidermal growth factor receptor (EGFR) at lysine K721 within its tyrosine kinase domain. This induces EGF-independent activation of EGFR and its downstream ERK pathway, as well as increased affinity of nuclear EGFR for PCNA and subsequent enhancement of DNA replication in SCCHN cells.
EHMT2
EHMT2 (euchromatic histone lysine methyltransferase 2, G9a) is a protein lysine methyltransferase that induces mono- and di-methylation of H3K9 and silences the transcription of target genes. Per TCGA, 9% of patients with SCCHN have genetic or expression alterations in this enzyme (Table 1). Liu et al. [22] found that G9a interacts with Snail in a metastatic SCCHN cell line derived from lymph-node metastasis (HN12) and suppresses the expression of E-cadherin through H3K9 promoter di-methylation in these cells, but not the non-metastatic parental cell line (HN4). Additionally, the authors showed that TGF-β-induced epithelial-mesenchymal transition (EMT) of the HN4 cells was reversed by BIX01294, a G9a inhibitor, and decreased the levels of di-methylated H3K9 at the E-cadherin promoter, suggesting that G9a is essential for the EMT of the HN4 cells. Accordingly, knockdown of G9a in the metastatic HN12 cells led to a decrease in the expression of EMT markers N-cadherin and vimentin, restored the expression of E-cadherin and decreased the motility and invasiveness of HN12 cells. Furthermore, BIX01294 treatment of HN4 cells suppressed TGF-β-induced tumorsphere formation and CD44 protein expression, both markers of cancer sternness in SCCHN cells. Accordingly, knockdown of G9a in HN12 cells led to abolishment of these cancer sternness features, supporting that G9a promotes cancer sternness in SCCHN. These results support that G9a could be a rational drug target for cancer stem cells in SCCHN.
In another study by Li et al. [23], high protein levels of G9a were found to be associated with worse survival in patients with SCCHN. Knockdown of G9a by shRNAs or BIX01294 led to decreased cell proliferation and colony formation, and this was mediated through induction of autophagy. The authors also showed that G9a inhibition induced transcriptional upregulation of the ERK dephosphatase dual specificity phosphatase44 (DUSP4), leading to inactivation of the ERK pathway, which is one of the mechanisms of autophagy induction. Tumor growth suppression via autophagy by G9a inhibition was also demonstrated in a doxycycline-inducible xenograft mouse model. Similar results demonstrating both autophagy and induction of apoptosis with BIX01294 in oral squamous cell carcinoma cell lines were obtained by a different group [24].
A recently published study also showed that G9a promotes cisplatin resistance and is associated with poor disease-free survival in patients with locoregionally advanced SCCHN [25]. This observation was validated in two independent cohorts of SCCHN patients with locoregionally advanced disease who received neoadjuvant cisplatin-based chemotherapy. G9a knockdown or treatment with the G9a inhibitor UNC0638 led to sensitization of resistant SCCHN and sphere forming SCCHN cells to cisplatin. Mechanistically, the cisplatin resistance was attributed to G9a-mediated H3K9 mono-methylation and subsequent transcriptional upregulation of the glutamate-cysteine ligase catalytic subunit which promotes glutathione biosynthesis.
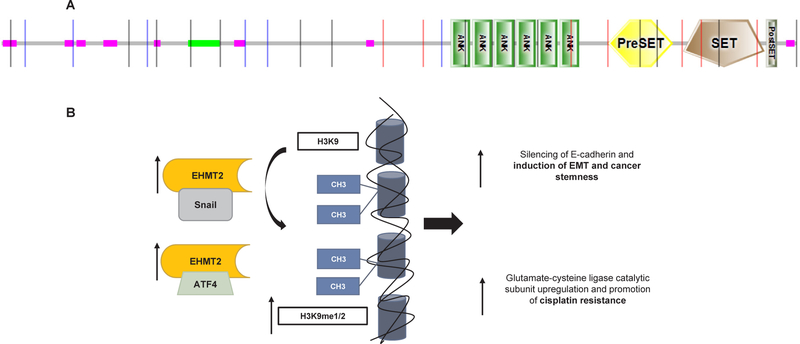
As efforts for the development of potent and selective G9a inhibitors are underway, the above studies may provide the rationale to introduce such inhibitors in clinical trials for patients with SCCHN. The protein structure and a summary of the reported mechanisms of action of EHMT2 in SCCHN are shown in Fig. 2.
Fig. 2.
Molecular structure and mechanisms of action of EHMT2 in SCCHN. A. Molecular structure of EHMT2 (UniProt, 1210 aminoacids). ANK: ankyrin repeats, PreSET: N-terminal to SET, cys-rich putative Zn2+-binding domain, SET: Su(var)3–9, Enhancer-of-zeste, Trithorax, PostSET: cysteine-rich motif following the SET domain. B. Mechanisms of action of EHMT2. EHMT2 associates with Snail and induces silencing of E-cadherin through H3K9 mono- and di-methylation, leading to induction of epithelial-mesenchymal transition (EMT) and cancer sternness features in SCCHN cells. Additionally, EHMT2 binds to ATF4 to induce mono-methylation of H3K9 and transcriptional upregulation of the glutamate-cysteine ligase catalytic subunit which promotes cisplatin resistance.
EZH2
EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) is a PKMT mostly known to tri-methylate H3K27 as the catalytic component of the polycomb repressive complex 2 (PRC2). EZH2 has been established as an oncogene in multiple cancer types, and an EZH2 inhibitor by Epizyme, Tazemetostat, is already being evaluated in a phase II trial of patients with relapsed/refractory non-Hodgkin lymphoma, synovial sarcoma and malignant mesothelioma. 5% of SCCHN patients in the TCGA database carry genetic or expression alterations in this PKMT (Table 1).
The first study to show an association of EZH2 protein expression with survival outcomes in patients with SCCHN was conducted by Kidani et al. [26]. Tissue sections from 102 patients with oral squamous cell carcinoma and available clinicopathological annotation were stained for EZH2. Analysis of the immunohistochemical scoring revealed that higher EZH2 protein levels were associated with poor survival and advanced tumor, nodal and clinical stage in patients with stage II-IV locoregionally advanced disease, supporting that EZH2 functions as an oncogene in SCCHN.
EZH2 has been implicated in the regulation of cell cycle in nasopharyngeal carcinoma (NPC). More specifically, Lu et al. [27] reported EZH2 as a direct downstream target of the tumor suppressive microRNA miR-26a in NPC, which is downregulated in NPC. miR-26a directly downregulates the translation of EZH2 in NPC, leading to decreased proliferation as well as G1 cell cycle arrest in NPC cells. The authors showed that reconstitution of EZH2 expression in NPC cells with suppressed miR-26a expression led to growth promotion, as well as overexpression of critical cell cycle regulators that regulate the G1-S transition, such as CDK4 and CDK6, implying that EZH2 enhances the expression of these cell cycle regulators. This study though did not examine the role of H3K27 tri-methylation in this regulatory network. Moreover, Huang et al. [28] also showed that overexpression of EZH2 in laryngeal cancer cells promotes entry of the cells into the S phase of the cell cycle, and promotes proliferation, tumorigenicity as well as resistance to cisplatin.
Furthermore, numerous studies have supported the importance of EZH2 in the invasiveness and epithelial-mesenchymal transition (EMT) of SCCHN, as well as its prognostic significance independently of established survival risk factors. Cao et al. [29] showed that knockdown of EZH2 led to a decrease in the proliferation rate and invasive potential of two SCCHN cell lines, and that higher EZH2 protein levels were significantly associated with the presence of perineural invasion, and worse overall survival in multivariate analysis of a cohort of 117 SCCHN patients with locoregionally advanced disease. The prognostic significance of EZH2 in SCCHN was also shown in another study [30] where high protein levels of EZH2 were associated with poor overall survival in a cohort of 209 nasopharyngeal cancer patients with locoregionally advanced disease independently of other known risk factors. Furthermore, this study supported that shRNA-mediated knockdown of EZH2 decreased the invasive potential of NPC cell lines and the metastatic burden of an in vivo tumor model of CNE2 nasopharyngeal cancer cells. Mechanistically, this was explained by the repression of E-cadherin, a cell adhesive molecule, by EZH2-mediated H3K27 tri-methylation, which enhanced the metastatic potential of CNE2 cells. The authors also found that the EZH2-mediated repression of E-cadherin required the synergistic action of histone deacetylases 1 and 2 (HDAC1/2) and of the transcription factor Snail, and that EZH2, HDAC1/2 and Snail form a co-repressive complex that silences E-cadherin in NPC cells. Similar results were reported by Wang et al. [31], who showed that overexpression of EZH2 was anti-correlated with E-cadherin expression and was associated with lymph node stage and poor overall survival in patients with oral tongue squamous cell carcinoma. Con-cordantly, they showed that ectopic expression of EZH2 reduced E-cadherin expression and enhanced the invasive potential in oral tongue SCCHN cell lines. This mechanism of EZH2-mediated silencing of E-cadherin and subsequent enhancement of the migration and invasive properties of SCCHN cells was further confirmed in another study [32] which also supported upregulation of N-cadherin and vimentin, and thus the acquisition of an EMT state by EZH2 in SCCHN cells. Additional studies have found that EZH2 promotes invasiveness and EMT through miRNA networks in SCCHN. For example, Li et al. [33] showed that EZH2 is downregulated by miR-630, which is in turns suppressed by HI9. HI9 suppresses miR-630, which then leads to upregulation of EZH2 expression, subsequent decrease in E-cadherin levels and thus induces invasive properties in SCCHN.
Gannon et al. [34] investigated whether H3K27 tri-methylation, which is known to maintain squamous cells in a de-differentiated state, may also play a vital role in the maintenance of de-differentiation features in SCCHN. Protein levels of EZH2 and H3K27 tri-methylation were evaluated in a tissue microarray of 59 patients with SCCHN (and 12 normal oral epithelial tissue sections). Interestingly, although higher levels of EZH2 would be expected to be associated with higher levels of H3K27 tri-methylation, no associations were found between these in this cohort of SCCHN patients. This finding could be explained either by the overexpression of H3K27 demethylases that reduce H3K27 methylation levels, or by the presence of other methyltransferases that tri-methylate H3K27, such as EZH1. The authors also found that EZH2 protein levels were significantly lower in normal oral mucosa compared to SCCHN tissue sections and that chromatin immunoprecipitation of tri-methylated H3K27 in normal oral mucosa versus SCCHN tissues revealed enrichment at the promoter of the differentiation gene involucrin in the SCCHN tissues. The authors also found that inhibition of EZH2-mediated H3K27 tri-methylation by EZH2-specific siRNAs or DZNep, an EZH2 inhibitor, increased the expression of squamous differentiation markers in SCCHN cell lines and induced cell cytotoxicity both in vitro and in in vivo xenograft mouse models. Although the global levels of H3K27 tri-methylation were not different between normal mucosa and SCCHN tissues, this study showed that it is likely that the distribution of the H3K27 tri-methylation mark is different, including the persistent presence of this mark on the promoters of differentiation genes in SCCHN tissues.
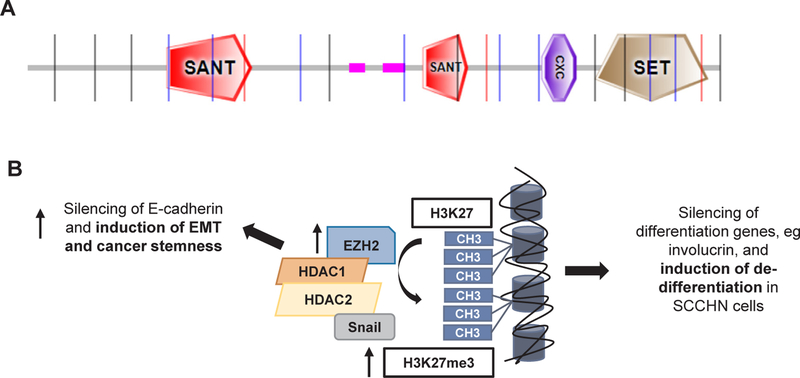
In summary, the above data suggest that EZH2 functions as an oncogene and independently predicts poor survival through promotion of cell cycle progression, EMT features, chemoresistance and de-differentiation in SCCHN, and could thus serve as a rational drug target, most likely as a cisplatin sensitizer or in the secondary prevention setting following curative-intent chemoradiotherapy. Fig. 3 shows the protein structure and summarizes the reported mechanisms of action of EZH2 in SCCHN.
Fig. 3.
Molecular structure and mechanisms of action of EZH2 in SCCHN. A. Molecular structure of EZH2 (UniProt, 751 aminoacids, isoform a). SANT: SANT SWI3, ADA2, N-CoR and TFIIIB” DNA-binding domain, CXC: Tesmin/TSOl-like CXC domain, SET: Su (var)3–9, Enhancer-of-zeste, Trithorax. B. Mechanisms of action of EZH2. EZH2 forms a repressive complex with histone deacetylase 1 (HDAC1), histone deacetylase 2 (HDAC2) and Snail and induces transcriptional silencing of E-cadherin through tri-methylation of H3K27 and EMT features in nasopharyngeal carcinoma cells. EZH2-in-duced tri-methylation of H3K27 also silences the expression of differentiation genes in SCCHN.
Protein arginine methyltransferases in SCCHN
PRMT1
PRMT1 (protein arginine methyltransferase 1) is a protein arginine methyltransferase that mono- and asymmetrically di-methylates various histone and non-histone substrates. Per TCGA, 6% of SCCHN patients have genetic or expression alterations in this gene (Table 1). Recently, PRMT1 was found to mono-methylate arginine R198 and R200 of the extracellular domain of the epidermal growth factor receptor (EGFR), enhancing EGF binding and subsequent EGFR dimerization and EGFR downstream activation [35]. Furthermore, PRMT1-mediated methylation of EGFR rendered colon cancer cells resistant to Cetuximab treatment and was associated with significantly higher recurrence rates and decreased overall survival in metastatic colon cancer patients treated with cetuximab.
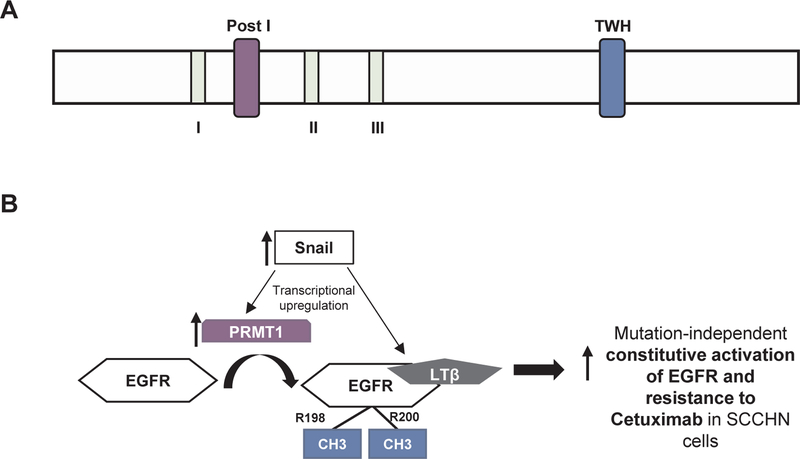
Given that Cetuximab is also a standard of care treatment option for SCCHN patients, Hsu et al. [36] investigated mechanisms of Cetuximab resistance in SCCHN, beyond mutations in EGFR, RAS, PIK3CA and ERBB2 amplification that are known to be culprits for Cetuximab resistance, and found that Cetuximab resistant SCCHN cell lines upregulated Snail and that this transcription factor directly bound to the promoter and increased the transcription of lymphotoxin-β which induced EMT transition. The authors also described that lymphotoxin-β directly and preferentially bound to R198/R200 methylated EGFR and that PRMT1, which induces this methylation, is directly upregulated by Snail. In summary, the authors suggest a mutation-independent model of Cetuximab resistance, whereby Snail induces the direct transcriptional upregulation of lymphotoxin-β and PRMT1, leading to the interaction of lymphotoxin-β and methylated EGFR, its subsequent constitutional activation and resistance to Cetuximab. The above data support PRMT1 inhibition as an approach to overcome Cetuximab resistance in SCCHN. Fig. 4 shows the protein structure and summarizes the reported mechanisms of action of PRMT1 in SCCHN.
Fig. 4.
Protein structure and mechanisms of action of PRMT1 in SCCHN. A. Protein structure of PRMT1 (UniProt, 361 aminoacids, isoform 1). Signature motif I, post I, post II, post III, conserved TWH loop: tandem winged-helix. B. Mechanisms of action of PRMT1. PRMT1 and lymphotoxin-β (LTβ) are upregulated by Snail in SCCHN cells with EMT features. PRMT1 then methylates EGFR at R198/R200, and LTβ preferentially interacts with R198/R200-methylated EGFR, inducing its activation and resistance to Cetuximab.
PRMT5.
PRMT5 (protein arginine methyltransferase 5) is a protein arginine methyltransferase that mono- and symmetrically di-methylates both histone and non-histone proteins, and has been implicated as an oncogene in a variety of cancer types, such as lung adenocarcinoma and colon cancer. 14% of SCCHN patients in the TCGA database have genetic or expression alterations in this gene (Table 1).
In NPC, Yang et al. [37] analyzed the protein expression levels in a tissue microarray of 112 patients with NPC at various stages and found that high nuclear PRMT5 protein levels were associated with more advanced clinical and lymph node stage, and poor overall survival. Furthermore, the authors report that PRMT5 enhances the radioresistance of NPC cells through the upregulation of fibroblast growth factor receptor 3 (FGFR3). Higher levels of nuclear PRMT5 have also been associated with poor survival in a cohort of 209 patients with oropharyngeal carcinoma [38].
PMT inhibitors and ongoing clinical trials
PMTs are increasingly being recognized as promising anticancer drug targets given their crucial role in the epigenetic regulation of cancer processes not only in SCCHN, as presented above, but also in multiple other cancer types. Thus, drug discovery programs to identify small-molecule inhibitors for these enzymes have already been initiated. The main inhibition strategies include blocking the binding either of S-adenosylmethionine, the methyl-donor in methyltransferase reactions, in its binding pocket, or of a specific enzymatic substrate to the respective binding site of a PMT [39]. A small number of inhibitors that disrupt protein-protein interactions of PMTs with partner proteins have also been reported, such as astemizole which disrupts the interaction of EZH2 with its binding partner EED (Embryonic Ectoderm Development protein), and a number of small-molecule compounds that inhibit protein-protein interactions of MLL proteins with their partners. The latter approach is important, especially in the context of PMTs that promote oncogenesis independent of their enzymatic activity, such as SMYD3 and the short isoform of NSD3 [40].
To date, the inhibitor Tazemetostat targeting the protein methyl-transferase EZH2 is the first-in-class to have reached phase II clinical trials. Tazemetostat (EPZ-6438) is an orally administered, S-adeno-sylmethionine (SAM) competitive EZH2 inhibitor with high selectivity of ≥20,000 fold towards EZH2 compared to other PMTs. The first-inhuman phase I/II trial investigating EPZ-6438 ( NCT01897571) is a multicenter single-agent study that was initiated in 2013 and is still enrolling patients. Eligible patients have advanced solid tumors or relapsed/refractory B-cell non-Hodgkin lymphoma (NHL) [41]. Tazemetostat demonstrated an acceptable safety profile, and objective responses were observed in 9 out of 15 evaluable patients with both wild-type and mutant EZH2 relapsed/refractory B-cell NHL, as well as in INI1 (integrase interactor 1)- or SMARCA4 (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A, Member 4)-negative tumors. INI1 or SMARCA4 loss has been shown to create oncogenic dependency on EZH2 in solid tumors, such as sarcomas and rhabdoid tumors [42]. In 2017, Tazemetostat received fast track designation by the Food and Drug Administration for the treatment of relapsed/refractory diffuse large B-cell lymphoma with EZH2 activating mutations and for relapsed/refractory follicular lymphoma regardless of EZH2 mutation status. Furthermore, Tazemetostat is being further investigated in phase II trials for pediatric or adult relapsed/refractory or INI-negative synovial sarcoma, in advanced solid tumors and NHL with EZH2 or SMARC4 mutations and in malignant mesothelioma.
Drug discovery programs are under way for multiple other PMTs, including DOT1L (disruptor of telomeric silencing 1-like protein), NSD2, EHMT2, PRMT1 and PRMT5 [40]. Such efforts are truly promising and portend the next generation of anticancer therapy targeting the protein methylome in cancer.
Conclusion
In this review, we have summarized the preclinical evidence and reported mechanisms of action of PMTs in SCCHN, and have highlighted relevant clinicopathological associations (Table 1). Despite the plethora of genetic and expression aberrations revealed in these enzymes by the TCGA, which underlines their importance in SCCHN, there is still a profound gap in our in-depth knowledge of the function of PMTs in this disease. To date, available studies have uncovered versatile functions of these enzymes in SCCHN biology, including cell cycle progression, epidermal differentiation, epithelial-mesenchymal transition, cancer sternness, DNA replication, constitutive activation of cellular signaling pathways (i.e. EGFR cascade) and more recently, immune cell infiltration of the tumor microenvironment. Of paramount importance is that methylation substrates are not only restricted to histone proteins, but also to non-histone proteins, and this pertains also to SCCHN, with EGFR being the first non-histone substrate reported in this cancer type. This realization will likely have a significant impact on the speed of progress towards accurate functional dissection of the protein methylome in SCCHN and thus towards the successful development and translation of novel therapeutics in this disease. Furthermore, given that methylation is a reversible mark, it is important to understand the balance between methylation “writers” and “erasers” and to characterize the differences generated in the phenotypes driven by the overactive state of a “writer” versus the hypoactive state of an “eraser” and vice versa. Also, it would be of interest to investigate differences in the function of each of these enzymes based on cell-context specificity derived from variations in the embryonal origin of each of the head and neck structures. Finally, as more selective, potent and better bioavailable PMT inhibitors are being developed, it is exciting to envision the possibility of combinatorial therapies which could enhance immunotherapy and chemoradiotherapy efficacy, and that resistance mechanisms may be more difficult to ensue compared to the more “targeted” kinase inhibitors, given the broader range of epigenetically and non-epigenetically mediated functions of these enzymes.
Acknowledgements
This work was supported by the Intramural Research Program of the NIH, Thoracic and Gastrointestinal Oncology Group, National Cancer Institute.
Footnotes
Conflict of interest
The authors declare no relevant conflicts of interest.
References
- [1].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer 2015;15(2):110–24. 10.1038/nrc3884. [DOI] [PubMed] [Google Scholar]
- [3].Wagner T, Jung M. New lysine methyltransferase drug targets in cancer. Nat Biotechnol 2012;30(7):622–3. 10.1038/nbt.2300. [DOI] [PubMed] [Google Scholar]
- [4].Kaniskan HÜ, Eram MS, Liu J, et al. Design and synthesis of selective, small molecule inhibitors of coactivator-associated arginine methyltransferase 1 (CARM1). Medchemcomm 2016;7(9):1793–6. 10.1039/C6MD00342 Epub 2016 Jul 13.: Paper not available online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Somervaille T, Salamero O, Montesinos P, et al. 4060 Safety, Phamacokinetics (PK), Pharmacodynamics (PD) and preliminary activity in acute leukemia of ory-1001, a first-in-class inhibitor of lysine-specific histone demethylase 1A (LSD1/KDM1A): initial results from a first-in-human phase 1 study. Oral and poster abstracts, session: 616. Acute myeloid leukemia: novel therapy, excluding transplantation: poster III; December 5, 2016, San Diego. [Google Scholar]
- [6].Morschhauser F, Salles G, McKay P et al. Initial report from a phase 2 multi-center study of tazemetostat (EPZ-6438), an inhibitor of enhancer of Zeste-Homolog 2 (EZH2), in patients with relapsed or refractory B-cell non-Hodgkin lymphoma (NHL). Abstract #88525. Presented at the 2016 ASH Meeting on Lymphoma Biology; June 20, 2016; Colorado Springs CO. [Google Scholar]
- [7].Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990–2015: a systematic analysis for the global burden of disease study. JAMA Oncol 2017;3(4):524–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ang KK, Sturgis EM. Human papillomavirus as a marker of the natural history and response to therapy of head and neck squamous cell carcinoma. Semin Radiat Oncol 2012;22(2): 128–42. [DOI] [PubMed] [Google Scholar]
- [9].Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase lb trial. Lancet Oncol 2016;17(7):956–65. [DOI] [PubMed] [Google Scholar]
- [10].Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375(19): 1856–67. Epub 2016 Oct 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 2011;118(13):3645–56. [DOI] [PubMed] [Google Scholar]
- [12].Toyokawa G, Cho HS, Masuda K, et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia 2011;13(10):887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kang D, Cho HS, Toyokawa G, et al. The histone methyltransferase Wolf-Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosom Cancer 2013;52(2): 126–39. [DOI] [PubMed] [Google Scholar]
- [14].Papillon-Cavanagh S, Lu C, Gay den T, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 2017;49(2):180–5. 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lu C, Jain SU, Hoelper D, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016;352(6287):844–9. 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peri S, Izumchenko E, Schubert AD, et al. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun 2017;8(1):1772. 10.1038/s41467-017-01877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berdasco M, Ropero S, Setien F, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSDlin human neuroblastoma and glioma. Proc Natl Acad Sei USA 2009;106(51):21830–5. 10.1073/pnas.0906831106. Epub 2009 Dec 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brennan K, Shin JH, Tay JK, et al. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep 2017;7(1):17064. 10.1038/s41598-017-17298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saloura V, Cho HS, Kiyotani K, et al. WHSCl promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res 2015;13(2):293–304. 10.1158/1541-7786.MCR-14-0292-T. Epub 2014 Oct 3. [DOI] [PubMed] [Google Scholar]
- [20].Saloura V, Vougiouklakis T, Zewde M, et al. WHSCILI drives cell cycle progression through transcriptional regulation of CDC6 and CDK2 in squamous cell carcinoma of the head and neck. Oncotarget 2016;7(27):42527–38. http://dx.doi.org/10. 18632/oncotarget.9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Saloura V, Vougiouklakis T, Zewde M, et al. WHSClLl-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci Rep 2017;19(7):40664. 10.1038/srep40664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu S, Ye D, Guo W, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget 2015;6(9):6887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li KC, Hua KT, Lin YS, et al. Inhibition of G9a induces DUSP4-dependent autophagic cell death in head and neck squamous cell carcinoma. Mol Cancer 2014;15(13):172. 10.1186/1476-4598-13-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ren A, Qiu Y, Cui H, et al. Inhibition of H3K9 methyltransferase G9a induces autophagy and apoptosis in oral squamous cell carcinoma. Biochem Biophys Res Commun 2015;459(1):10–7. http://dx.doi.Org/10.1016/j.bbrc.2015.01.068. Epub 2015 Jan 26. [DOI] [PubMed] [Google Scholar]
- [25].Liu CW, Hua KT, Li KC, et al. Histone methyltransferase G9a drives chemotherapy resistance by regulating the glutamate-cysteine ligase catalytic subunit in head and neck squamous cell carcinoma. Mol Cancer Ther 2017;16(7):1421–34. 10.1158/1535-7163.MCT-16-0567-T. Epub 2017 Mar 6. [DOI] [PubMed] [Google Scholar]
- [26].Kidani K, Osaki M, Tamura T, et al. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol 2009;45(1):39–46. http://dx.doi.Org/10.1016/j.oraloncology.2008.03.016. Epub 2008 Jul 10. [DOI] [PubMed] [Google Scholar]
- [27].Lu J, He ML, Wang L, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res 2011;71(1):225–33. 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- [28].Huang J, Zhou L, Chen H, et al. EZH2 is overexpressed in laryngeal squamous cell carcinoma and enhances the stem-like properties of AMC-HN-8 cells. Oncol Lett 2016;12(2):837–46. Epub 2016 Jun 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cao W, Feng Z, Cui Z, et al. Up-regulation of enhancer of zeste homolog 2 is associated positively with cyclin D1 overexpression and poor clinical outcome in head and neck squamous cell carcinoma. Cancer 2012;118(11):2858–71. 10.1002/cncr.26575. Epub 2011 Oct 11. [DOI] [PubMed] [Google Scholar]
- [30].Tong ZT, Cai MY, Wang XG, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene 2012;31(5):583–94. 10.1038/onc.2011.254. Epub 2011 Jun 20. [DOI] [PubMed] [Google Scholar]
- [31].Wang C, Liu X, Chen Z, et al. Polycomb group protein EZH2-mediated E-cadherin repression promotes metastasis of oral tongue squamous cell carcinoma. Mol Carcinog 2013;52(3):229–36. 10.1002/mc.21848. Epub 2011 Dec 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chang JW, Gwak SY, Shim GA, et al. EZH2 is associated with poor prognosis in head-and-neck squamous cell carcinoma via regulating the epithelial-to-mesenchymal transition and chemosensitivity. Oral Oncol 2016;52:66–74. 10.1016/j.oraloncology.2015.11.002. Epub 2015 Nov 18. [DOI] [PubMed] [Google Scholar]
- [33].Li X, Lin Y, Yang X, et al. Long noncoding RNA HI9 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun 2016;473(4):913–9. http://dx.doi.Org/10.1016/j.bbrc.2016.03.150. Epub 2016 Apr 1. [DOI] [PubMed] [Google Scholar]
- [34].Gannon OM, Merida de Long L, Endo-Munoz L, et al. Dysregulation of the repressive H3K27 trimethylation mark in head and neck squamous cell carcinoma contributes to dysregulated squamous differentiation. Clin Cancer Res 2013;19(2):428–41. 10.1158/1078-0432.CCR-12-2505. Epub 2012 Nov 27. [DOI] [PubMed] [Google Scholar]
- [35].Liao HW, Hsu JM, Xia W, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest 2015;125(12):4529–43. 10.1172/JCI82826. Epub 2015 Nov 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hsu DS, Hwang WL, Yuh CH, et al. Lymphotoxin-β interacts with methylated EGFR to mediate acquired resistance to Cetuximab in head and neck cancer. Clin Cancer Res 2017;23(15):4388–401. 10.1158/1078-0432.CCR-16-1955. Epub 2017 Feb 14. [DOI] [PubMed] [Google Scholar]
- [37].Yang D, Liang T, Gu Y, et al. Protein N-arginine methyltransferase 5 promotes the tumor progression and radioresistance of nasopharyngeal carcinoma. Oncol Rep 2016;35(3): 1703–10. 10.3892/or.2015.4513. Epub 2015 Dec 24. [DOI] [PubMed] [Google Scholar]
- [38].Kumar B, Yadav A, Brown NV, et al. Nuclear PRMT5, cyclin D1 and IL-6 are associated with poor outcome in oropharyngeal squamous cell carcinoma patients and is inversely associated with pi 6-status. Oncotarget 2017;8(9): 14847–59. 10.18632/oncotarget.14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schapira M Chemical inhibition of protein methyltransferases. Cell Chem Biol 2016;23(9): 1067–76. http://dx.doi.Org/10.1016/j.chembiol.2016.07.014. Epub 2016 Aug 25. [DOI] [PubMed] [Google Scholar]
- [40].Kaniskan HÜ, Martini ML, Jin J. Inhibitors of protein methyltransferases and demethylases. Chem Rev 2018;118(3):989–1068. 10.1021/acs.chemrev.6b00801. Epub 2017 Mar 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Morera L, Liibbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics 2016;8:57. 10.1186/sl3148-016-0223-. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, et al. SWI/SNF mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015;21 (12): 1491–6. 10.1038/nm.3968. Epub 2015 Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]