Abstract
Purpose:
FGFR1 amplification occurs in ~15% of ER+ human breast cancers.We investigated mechanisms by which FGFR1 amplification confers antiestrogen resistance to ER+ breast cancer.
Methods:
ER+ tumors from patients treated with letrozole before surgery were subjected to Ki67 immunohistochemistry, FGFR1 FISH, and RNA-sequencing. ER+/FGFR1 amplified breast cancer cells and patient-derived xenografts (PDXs) were treated with FGFR1 siRNA or the FGFR tyrosine kinase inhibitor lucitanib. Endpoints were cell/xenograft growth, FGFR1/ERα association by co-immunoprecipitation and proximity ligation, ER genomic activity by ChIP-sequencing, and gene expression by RT-PCR.
Results:
ER+/FGFR1 amplified tumors in patients treated with letrozole maintained cell proliferation (Ki67). Estrogen deprivation increased total and nuclear FGFR1 and FGF ligands expression in ER+/FGFR1-amplified primary tumors and breast cancer cells. In estrogen-free conditions, FGFR1 associated with ERα in tumor cell nuclei and regulated the transcription of ER-dependent genes. This association was inhibited by a kinase-dead FGFR1 mutant and by treatment with lucitanib. ChIP-seq analysis of estrogen-deprived ER+/FGFR1 amplified cells showed binding of FGFR1 and ERα to DNA. Treatment with fulvestrant and/or lucitanib reduced FGFR1 and ERα binding to DNA. RNA-seq data from FGFR1-amplified patients’ tumors treated with letrozole showed enrichment of estrogen response and E2F target genes. Finally, growth of ER+/FGFR1-amplified cells and PDXs was more potently inhibited by fulvestrant and lucitanib combined than each drug alone.
Conclusions:
These data suggest the ERα pathway remains active in estrogen-deprived ER+/FGFR1-amplified breast cancers. Therefore, these tumors are endocrine resistant and should be candidates for treatment with combinations of ER and FGFR antagonists.
Keywords: FGFR1, fulvestrant, drug resistance, breast cancer
INTRODUCTION
Amplification of the chromosomal region 8p11–12, the genomic location of fibroblast growth factor receptor 1 (FGFR1), has been reported in breast, ovarian, bladder, lung and oral squamous cancers, and in rhabdomyosarcoma (1–7). FGFR1 amplification occurs in ~10% of patients with ER+/HER2-negative breast cancer where it is associated with early relapse following adjuvant tamoxifen therapy and with poor survival (8). Blockade of FGFR1 signalling by pharmacological or genetic approaches in human breast cancer cells harboring FGFR1 amplification leads to decreased cell growth and survival, suggesting FGFR1 gene amplification is a surrogate of cancer cell dependence on aberrant FGFR activity (8).
FGFRs belong to the family of receptor tyrosine kinases (RTKs) that consist of an extracellular ligand-binding domain linked to an intracellular catalytic protein kinase core via a single-pass transmembrane domain (TMD) (9). Binding of FGF ligands induces receptor dimerization, activation of the kinase domain and phosphorylation of C-terminal tyrosines to which adaptor proteins dock, followed by activation of signal transduction pathways, including PI3K/AKT, RAS/RAF/MEK/ERK, phospholipase Cγ (PLCγ) and STATs (10). In addition, there is strong evidence that FGFRs traffic to the nucleus, where they may function in a different manner to classic transmembrane RTKs (11). For example, nuclear FGFR3 has been shown in the nucleus of malignant and non-malignant breast epithelial cells (12). A nuclear interaction of FGFR2, STAT5 and progesterone receptor (PR), associated with PR/STAT5-regulated gene expression and breast cancer progression was also reported (13). Other studies have reported nuclear localization and a nucleus-specific function of FGFR1 in non-mammary cells (14–17). Medulloblastoma cells transfected with FGFR1-eGFP and evaluated by immunofluorescence have shown FGFR1 is associated with cell membranes, cytosol and nuclear compartments (17). Substitution of the atypical TMD of FGFR1 (β-sheet containing polar amino acids) with the typical TMD of FGFR4 (α−helical, hydrophobic) prevents the nuclear localization of FGFR1 (18). Inability of both the full-length and cleaved forms of FGFR1 to localize to the nucleus results in reduced migration and invasiveness of cancer cells (15, 16). Finally, ChIP-seq studies revealed that FGFR1 binds nuclear transcription factors involved in neural and muscle development (19).
Amplification of the FGF3/4/19 ligand genes on chromosome 11q12–14 occurs in ~15% of human breast cancers (20, 21). Notably, one-third of FGFR1-amplified tumors also harbor amplification of CCND1, FGF3, FGF4 and FGF19 (22). This co-amplification has also been associated with resistance to estrogen deprivation in ER+ breast cancer and poor patient outcome (22), suggesting the possibility of ligand-receptor cooperativity.
Herein, we investigated mechanisms by which FGFR1 amplification confers resistance to antiestrogens in ER+ breast cancer. In a cohort of patients with ER+ breast cancer treated with the aromatase inhibitor letrozole, we observed that tumors with FGFR1 amplification maintained their proliferation despite drug-induced estrogen deprivation and exhibited nuclear localization of FGFR1. Estrogen deprivation also resulted in an increase of total and nuclear FGFR1 as well as FGF3/4/19 ligand expression in ER+/FGFR1-amplified breast cancer cells. FGFR1 coupled with ERα to drive estrogen-independent transcription of ERα-responsive genes. The association of FGFR1 with ERα was inhibited upon transfection with a kinase-dead FGFR1 mutant and by pharmacological inhibition of FGFR1. Finally, combined inhibition of FGFR1 and ERα with fulvestrant and lucitanib reduced the association of FGFR1 and ERα and growth of ER+/FGFR1-amplified PDXs. We propose a physical interaction between FGFR1 and ERα provides a mechanistic explanation for how FGFR1 amplification contributes to resistance to endocrine therapy in ER+ breast cancer.
MATERIALS AND METHODS
Clinical trial and tumor biopsies.
Tumor samples were obtained from patients with stage I-III operable ER+/HER2– breast cancer enrolled in a clinical trial of the aromatase inhibitor letrozole administered for 2 weeks prior to surgery () (23). Patients provided written informed consent according to a protocol approved by the Vanderbilt-Ingram Cancer Center Institutional Review Board. Intra-operative biopsies or surgical specimens, snap-frozen in liquid nitrogen and formalin-fixed paraffin-embedded (FFPE), were obtained from each patient’s tumor. Immunohistochemistry (IHC) was conducted in both the pretreatment biopsy and in the post-treatment surgical biopsy of both tumors for Ki67 (Dako #M7240), ER (Santa Cruz #sc542) and PR (Dako #M3569). IHC for ER and PR was conducted according to methods reported elsewhere (24). FFPE tumor sections were scanned at 100x magnification, and the area containing the highest number of positive cells was selected. Positive and negative tumor cells were manually counted at 400× the percentage of positive cells was calculated with at least 1,000 viable cells. Ki67 IHC was scored by two independent pathologists (MVE and JMG).
Cell Lines.
Cell lines were obtained from American Type Culture Collection (ATCC) between 2014–2016 and maintained in DMEM/10% fetal bovine serum (FBS; Gibco). Long-term estrogen-deprived (LTED) cells were generated upon long-term culture in phenol red-free IMEM/10% dextran-charcoal-treated FBS [DCC-FBS; Hyclone, contains <0.0367 pM 17β-estradiol (E2)] for 3–8 months until exponentially-growing, hormone-independent cells emerged as described previously (25). Cell lines were authenticated by ATCC prior to purchase by the STR method. Cell lines were not authenticated after purchase. Mycoplasma testing was conducted for each cell line before use. All experiments were performed less than 2 months after thawing early passage cells.
FGFR1 and CCND1 fluorescence in situ hybridization (FISH).
FGFR1 and CCND1 copy number was measured by FISH analysis in FFPE tumor sections (see Supplementary Methods).
Cell proliferation.
Cells were plated in 10% DCC-FBS ± FGF3 100 ng/mL or ± 2 μM lucitanib for 7 or 14 days before being trypsinized and counted using a Coulter Counter (Beckman Coulter), or fixed and stained with crystal violet followed by quantification by spectrophotometric detection at 490 nm using a plate reader (GloMax®-Multi Detection System, Promega; see Supplementary Methods).
Viral transduction.
FGFR1 wild-type and GFP-expressing lentiviral constructs were generated in the pLX302 Gateway vector (Open Biosystems); FGFR1/TK– (K514M) pLX302 was created by site-directed mutagenesis by Genewiz (New Jersey, USA). To generate stably-transduced lines, 4 μg of the FGFR1, FGFR1/TK– (K514M), and GFP-pLX302 constructs were co-transfected with 3 μg psPAX2 (plasmid encoding gag, pol, rev, and Tat genes), and 1 μg pMD2G envelope plasmid (Sigma Aldrich) into 293FT cells using Lipofectamine 2000 (Thermo Fisher). 293FT growth media was changed 24 h post-transfection; virus-containing supernatants were harvested 48 and 72 h post-transfection, passed through a 0.45-μm filter, diluted 1:4, and applied to target cells with 8 μg/mL polybrene (Sigma Aldrich). Virus-producing cells were selected in 1 μg/mL puromycin.
Proximity Ligation Assay (PLA).
PLA was performed in cultured cells and in FFPE primary tumor sections to detect FGFR1/ERα localization using the Duolink Detection Kit (#DUO92101, Sigma; see Supplementary Methods).
Gene Expression Analyses.
CAMA1 cells were plated in estrogen-free media and treated ± 100 ng/mL FGF3/19 (Sigma) for 6 h. Cells were harvested and RNA was purified using the RNeAsy kit (Qiagen, Valencia, CA). cDNA was generated using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Carlsbad, CA) followed by analysis of ERα pathway genes using the Estrogen Receptor PCR Array (Qiagen, PAHS-005Z). RNA sequencing data (see “RNA Sequencing and cDNA library construction” below) were aligned to human genome version 19 using the splice-aware aligner TopHat (v2.0.9), and isoform level expression was quantified using cufflinks. Expression levels were normalized across the data set using cuffnorm. We compared genes upregulated in FGFR1-amplified vs. FGFR1 non-amplified cancers [≥2.0 fold, false discovery rate (FDR)-adjusted p≤0.05]. These genes were entered into Gene Set Enrichment Analysis (GSEA) as a ranked list. Gene sets with an FDR of <0.01 were considered to be enriched in FGFR1-amplified vs. non-amplified tumors (see Supplementary Methods).
Chromatin immunoprecipitation (ChIP)/DNA sequencing.
ChIP was done using CAMA1 cells plated in estrogen free media ± 100 ng/mL FGF3 and treated with 2 μM lucitanib, 1 μM fulvestrant or the combination. Cells were grown to 80% confluency, washed 3× in ice-cold PBS, and then fixed for 10 min at room temperature using 7% formaldehyde followed by quenching with 2.5 M glycine. Cells were first lysed using Farnham lysis buffer and then with nuclei lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1% SDS). Chromatin was sonicated using a Covaris LE220 with the following conditions: 35 min at peak power 350, duty factor 15, 200 cycles/burst, and average power 52.5; 200 μL of the chromatin was saved for input. Sonicated chromatin was diluted using ChIP Dilution Buffer (50 mM Tris-HCl pH 8.0, 0.167 M NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate), RIPA-150, protease inhibitors, and sodium butyrate. ERα (sc-8002) and FGFR1 (ab10646) antibodies were linked to magnetic anti-mouse and anti-rabbit Dynabeads respectively, and then incubated with chromatin for >12 h at 4°C. Immunoprecipitates (IPs) were washed with the following buffers (RIPA-150, RIPA-500, RIPA-LiCl, and TE Buffer) for 5 min each. Chromatin-IPs were eluted from the beads, treated with RNAse A at 65°C with shaking for 4 h to reverse crosslinks, followed by Proteinase-K treatment at 55°C for 1 h. Next, DNA was purified using phenol-chloroform extraction followed by ethanol precipitation and subsequent quantification by Qubit. Standard Illumina ChIPseq library kits were used to build sequencing libraries. Libraries were sequenced at Vanderbilt Technologies for Advanced Genomics (VANTAGE) Core Resource as SR50 on a HiSeq3000. Each antibody pulldown and the corresponding matching input was sequenced. The resulting sequencing files were aligned to human genome version 19 by BWA (Burrows-Wheeler Aligner). For each replicate, peaks were called comparing to matched input, using MACS14 and default settings. The intersection of peak calls from each replicate was used to define the peak call set for each condition. Peaks were assigned to closest genes using annotatePeaks.pl in the HOMER analysis suite and heatmaps were generated using ngs.plot.
ChIP/quantitative PCR.
ChIP was performed in CAMA1 cells as described above. DNA was analyzed by real-time qPCR in triplicate with Sso Advanced SYBR Green Supermix (Bio-Rad) in a CFX qPCR machine (Bio-Rad). The fold-enrichment of ChIP samples was calculated using the 2ΔCt (threshold cycle) method. Ct values for ERα-ChIP and FGFR1-ChIP samples were normalized to input DNA Ct values, and then independently to respective negative control Ct values to account for antibody background. Primer sequences are listed in Table S1.
RNA Sequencing and cDNA library construction.
Core biopsies were flash frozen in liquid N2 and stored at −80°C until RNA extraction was performed as described elsewhere (26). Total RNA was quantified using the Quant-iT™ RiboGreen® RNA Assay Kit (Invitrogen) and normalized to 4 ng/μL; 200 ng of each sample were used for library preparation in an automated variant of the Illumina Tru Seq™ RNA Sample Preparation protocol (Revision A, 2010). This method uses oligo(dT) beads to select mRNA from the total RNA sample and is followed by heat fragmentation and cDNA synthesis from the RNA template. The resultant cDNA went through library preparation (end repair, base ‘A’ addition, adapter ligation, and enrichment) using Broad Institute-designed indexed adapters for multiplexing. After enrichment, libraries were quantitated with qPCR using the KAPA Library Quantification Kit for Illumina Sequencing Platforms and pooled equimolarly. The entire process was performed in a 96-well format with all pipetting done by either the Agilent Bravo or PerkinElmer JANUS Mini liquid handlers.
Non-stranded Illumina RNA-sequencing.
Pooled libraries were normalized to 2 nM and denatured using 0.2 N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocol using either the HiSeq 2000 v3 or HiSeq 2500. Each run was a 76-bp, paired-end run with an eight-base index barcode. Data were analyzed using the Broad Picard Pipeline, which includes de-multiplexing and data aggregation. TopHat spliced aligner software was used to map sequencing reads and to generate a BAM file for each tumor (27). RNAseq GCT files were generated from BAM files using RNA-SeQC (28).
Xenograft studies.
These studies were approved and performed in accordance with the Vanderbilt Institutional Animal Care and Use Committee. We used two ER+/HER2-/FGFR1-amplified patient-derived xenografts (PDXs). PDX T272 (XenTech) required estrogen supplementation in the drinking water (8.5 mg/L estrogen) to grow as tumors in female athymic nude mice (Envigo). The second PDX, TM00368 (Jackson Laboratory), was implanted in female ovariectomized SCID/beige mice (Jackson Laboratory) supplemented with a s.c. 21-day relase, 0.25-mg 17β-estradiol pellet (Innovative Research of America). Tumors were serially transplanted in athymic or SCID/beige mice under general anesthesia. When xenografts reached a volume ≥200 mm3, mice were randomized to treatment with vehicle, lucitanib (10 mg/kg/day p.o. for T272 or 7 mg/kg/day p.o. for TM00368), fulvestrant (5 mg/week s.c.) or both drugs (n= 10 per group for T272 and n= 8 per group for TM00368). Tumors diameters were measured using calipers twice a week, and volume in mm3 calculated with the formula: volume = width2 × length/2. When tumor volume exceeded 2 cm3 or at the end of treatment, mice were sacrificed and tumors harvested 1 h after the last dose of lucitanib. Portions of tumors were snap frozen or fixed in 10% neutral buffered formalin and embedded in paraffin for subsequent analyses. Five-μm paraffinized sections were used for IHC using Y653/54 phosphorylated FGFR1 (Abcam #111124) and ERα (Santa Cruz Biotech #8002). Sections were scored by an expert pathologist (MVE) blinded to treatment arm.
Statistics.
Results are representative of 3 independent experiments and are expressed as the mean ± SEM. A p value of less than 0.05, determined by Student’s t-test, was considered statistically significant.
RESULTS
FGFR1 amplification and overexpression is associated with endocrine resistance in ER+ breast cancer.
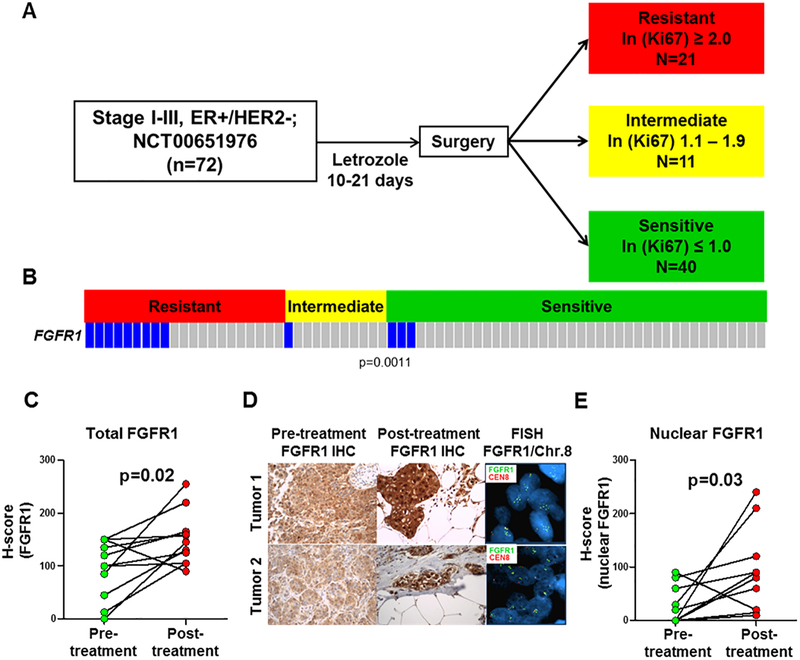
We studied 72 tumor biopsies from post-menopausal women with clinical stage I-III operable, ER+/HER2– breast cancer treated with the aromatase inhibitor letrozole for 2 weeks prior to surgery (). Earlier studies have demonstrated that a Ki67 score 2 weeks after antiestrogen therapy can be utilized to predict which tumors are endocrine sensitive or resistant, as measured by their odds of recurrence following adjuvant endocrine therapy (29). We applied these metrics to our tumor set and categorized 40 tumors as sensitive [natural log (ln) of post-letrozole Ki67 ≤1.0 or ≤2.4% tumor cells], 11 tumors as intermediate responders (ln=1.1–1.9 or 2.5–7.3% tumor cells), and 21 tumors as resistant (ln ≥2.0 or ≥7.4% tumor cells; Fig. 1A). FGFR1 copy number was determined in tumor sections by fluorescence in situ hybridization (FISH). We observed FGFR1 amplification in 9/21 (43%) resistant tumors compared to 3/40 (7%) sensitive tumors and 1/11 (10%) intermediate tumors (resistant vs. intermediate and sensitive tumors; p=0.0011; Fig. 1B). To correlate FGFR1 copy number with protein levels, we performed IHC. FGFR1 protein levels correlated with gene amplification by FISH. In FGFR1-amplified cancers, we observed a significant increase in total and nuclear FGFR1 in post-treatment compared to pre-treatment biopsies (p<0.05; Fig. 1C,E). A letrozole-induced increase in both total and nuclear FGFR1 was not observed in tumors without FGFR1 amplification (Suppl. Fig. 1A). There was no a statistical correlation between FGFR1 amplification and histological tumor grade in the letrozole resistant group (Suppl. Table 2).
Figure 1. FGFR1 amplification and overexpression associate with endocrine resistance in ER+ breast cancer.
A, Clinical trial schema: Patients with stage I-III, ER+/HER2– breast cancer were treated with letrozole for 10–21 days. Surgery was performed following treatment and tumor response was categorized by calculating the natural log (ln) of the post-letrozole Ki67 score as determined by IHC analysis. B, FGFR1 amplification, determined by FISH, was significantly associated with resistant vs. intermediate or sensitive tumors (p<0.05, Student’s t-test). C-E, FFPE sections from FGFR1-amplified tumors were stained for FGFR1; the percent of FGFR1-positive tumor cells and staining intensity were assessed in both the cytoplasmic and nuclear compartments by a blinded expert breast pathologist (M.V.E.) to generate an H-score (D). The percent of cytoplasmic and nuclear FGFR1+ tumor cells and their staining intensity were assessed by a blinded expert pathologist (M.V.E.) to generate an H-score. Total and nuclear FGFR1 H-scores are shown in C and E, respectively (Student’s t-test). Both total and nuclear FGFR1 staining was higher in post-treatment tumor sections.
Estrogen deprivation increases nuclear and cytosolic FGFR1 expression.
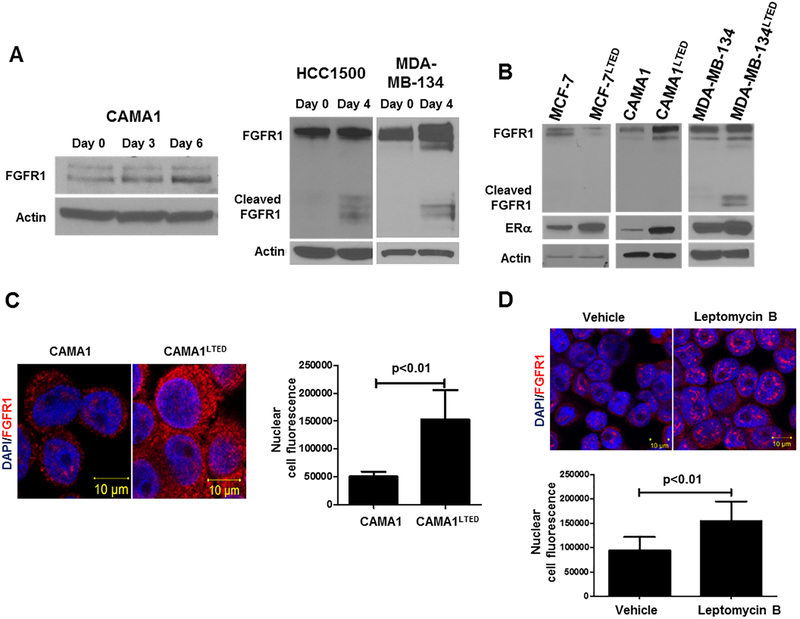
To examine whether this same modulation of FGFR1 levels occurred in more controlled experimental conditions, we tested five ER+/HER2– human breast cancer cell lines with and without FGFR1 gene amplification as determined by FISH: CAMA1, MDA-MB-134 and HCC1500 cells are FGFR1 amplified while MCF-7 and ZR75.1 cells are not (Suppl. Fig. 2A). FGFR1 amplification correlated with FGFR1 protein levels; MDA-MB-134 and HCC1500 cells express both full-length and cleaved FGFR1 while only full-length FGFR1 was detected in CAMA1 cells (Suppl. Fig. 2B). To mirror the acute estrogen deprivation induced by letrozole in primary tumors in the clinical trial (30), we cultured the FGFR1-amplified cell lines in estrogen-free medium for 4–6 days. Estrogen withdrawal resulted in an increase in FGFR1 expression in all FGFR1-amplified lines(Fig. 2A).
Figure 2. Estrogen deprivation increases nuclear and cytosolic FGFR1 expression.
A, Immunoblot analysis of lysates from CAMA1, HCC1500 and MDA-MB-134 cells exposed to short-term estrogen deprivation up to 6 days revealed an increase in FGFR1 expression over time. HCC1500 cells showed increased expression of the cleaved form of FGFR1. B, Immunoblot analysis of parental and LTED ER+ cell lines following 24 h of estrogen deprivation revealed an increase in FGFR1 and ERα in FGFR1-amplified CAMA1LTED and MDA-MB-134LTED cells but not in FGFR1 non-amplified MCF-7 cells. C, Proximity ligation assay (PLA) to detect FGFR1 expression. Analysis of red, amplified loci by confocal microscopy confirmed immunoblot and FISH results in that CAMA1LTED cells harbor more cytosolic and nuclear FGFR1 compared to CAMA1 parental cells. Each bar in the graph to the right of the PLA image represents the mean nuclear fluorescent signals ± SD of 3 wells. D, Immunofluorescence (IF) was performed in CAMA1 cells treated with vehicle or 30 ng/mL leptomycin B for 2 h. Nuclear localization of FGFR1 was detected by confocal microscopy. Each bar represents the mean nuclear fluorescent signals ± SD of 3 wells.
To determine whether long-term estradiol deprivation also affected FGFR1 expression, we generated three long-term estrogen-deprived (LTED) cell lines as previously described (25): CAMA1LTED and MDA-MB-134LTED (FGFR1-amplified) and MCF-7LTED (FGFR1 non-amplified). As we had observed with acute estrogen-deprivation, CAMA1LTED and MDA-MB-134LTED cells exhibited increased expression of full-length and cleaved FGFR1, respectively, whereas MCF-7LTED cells showed a reduction in FGFR1 expression compared to parental MCF-7 cells. The LTED lines also displayed an increase in ERα levels compared to their parental counterparts (Fig. 2B). Immunofluorescence (IF) by confocal microscopy highlighted the increase in total and nuclear FGFR1 in CAMA1LTED vs. parental cells (Fig. 2C). We next treated CAMA1 cells with nuclear export inhibitor leptomycin B (31); this resulted in an increase in nuclear FGFR1 as measured by IF (Fig. 2D). Knockdown of FGFR1 with siRNA confirmed the specificity of the FGFR1 antibody used for both immunoblot and IF analyses (Suppl. Fig. 2C,D).
FGF3/4/19 expression is upregulated upon estrogen deprivation.
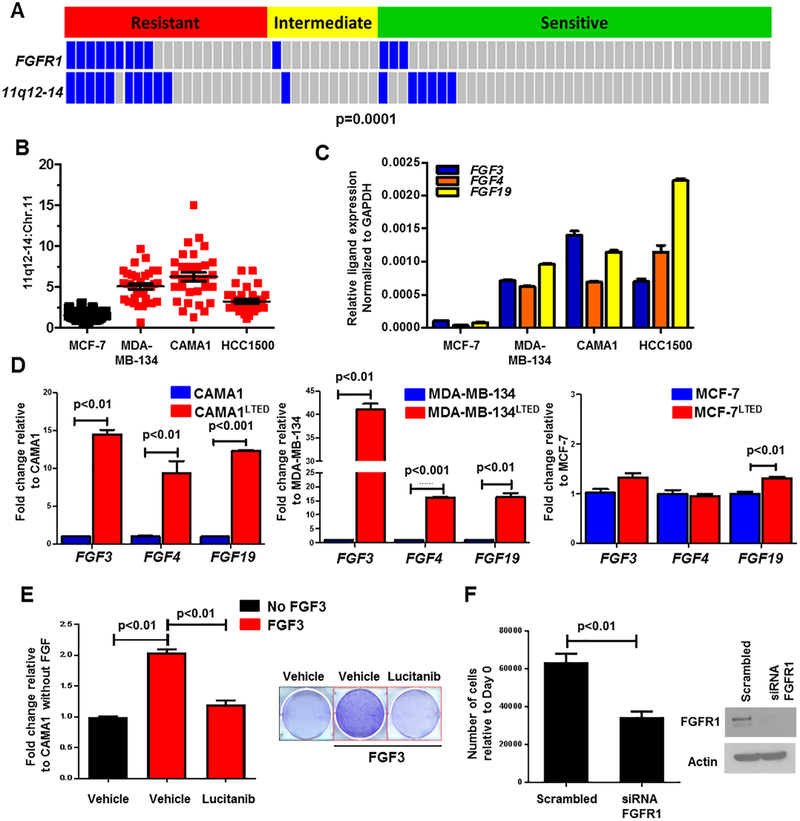
Approximately 30–40% of FGFR1-amplified breast cancers exhibit amplification of CCND1, FGF3, FGF4 and FGF19 in chromosome 11q12–14 (32). Co-amplification of these genes has been associated with reduced patient survival (22). By interrogating The Cancer Genome Atlas (TCGA), we found that among the 13% of breast cancers with FGFR1 amplification, 36% of these tumors also harbor 11q-12–14 amplification (Suppl. Fig. 3A,B) (33, 34). Outcomes analysis of Kaplan Meier-plotter [breast cancer] showed that patients with co-amplification of FGFR1 and CCND1/FGF3/FGF4/FGF19 treated with antiestrogen therapy exhibit a shorter time to relapse compared to patients without co-amplified tumors (hazard ratio=1.75; Suppl. Fig. 3C) (35). Thus, we next investigated co-amplification of FGFR1 and 11q12–14 in our cohort of patients treated with letrozole. In this study, 8 of 9 (90%) FGFR1-amplified tumors exhibited co-amplification of FGF3/4/19 and this co-amplification strongly correlated with resistance to estrogen deprivation with letrozole (p=0.0001; Fig. 3A). These data suggest that co-amplification of 11q12–14 and FGFR1 may play a causal role in endocrine resistance.
Figure 3. FGF3/4/19 expression is upregulated upon estrogen deprivation.
A, FISH analysis of primary tumor sections showed co-amplification of FGFR1 and 11q12–14 mainly in letrozole-resistant vs. intermediate and sensitive cancers (p=0.0001, Student’s t-test). B, Co-amplification of 11q12–14 was observed in ER+/FGFR1-amplified cell lines MDA-MB-134, CAMA1 and HCC1500; the Y axis shows the 11q12–14:Chr.11 ratio. C, Relative transcript expression of FGF3/4/19 in the indicated cell lines was determined by qPCR as described in Methods. D, Transcript levels of FGF3/4/19 were higher in FGFR1-amplified LTED cells (CAMA1 and MDA-MB-134) but not in FGFR1 non-amplified MCF-7LTED cells compared to their parental counterparts (Student’s t-test). E, CAMA1 cells were treated with 100 ng/mL FGF3 ± 2 μM lucitanib in estrogen-free medium. After 15 days, plates were washed and stained with crystal violet and their imaging intensity was quantified by spectrophotometric detection. Representative images and quantification of the integrated intensity values as % of vehicle-treated controls are shown (Student’s t-test). F, CAMA1 cells were plated in 100-mm dishes and transfected with FGFR1 or control siRNAs as described in Methods. Medium containing 100 ng/mL FGF3 was replenished every 3 days. Seven days later, monolayers were harvested and cell counts determined using a Coulter Counter. Each bar in the left panel represents the mean cell number ± SD of triplicate wells (Student’s t-test). FGFR1 knockdown was confirmed by immunoblot analysis of cell lysates from plates treated identically in parallel (right panel).
Notably, all FGFR1-amplified cell lines but not MCF-7 cells exhibited co-amplification of 11q12–14 (Fig. 3B, Suppl. Fig. 4A). All 11q12–14-amplified cell lines expressed markedly higher FGF3/4/19 mRNA levels by qRT-PCR compared to MCF-7 cells (Fig. 3C). Similar to the effect on FGFR1 protein levels, 24 h of estrogen deprivation increased FGF3/4/19 mRNA expression 1.5- to 2-fold in all FGFR1-amplified cells (Suppl. Fig. 4B). This increase in FGF3/4/19 was even more substantial in LTED FGFR1-amplified cells. In contrast, MCF7LTED cells exhibited little or no increase in FGF ligands mRNA compared to MCF-7 parental cells (Fig. 3D).
These results also suggested that FGFs can provide a growth advantage to ER+/FGFR1-amplified cells in estrogen-free conditions. To test this, we stimulated estrogen-starved CAMA1 cells with FGF3 in hormone-depleted media. Exogenous FGF3 enhanced estrogen-independent cell growth compared to unstimulated cells. Both treatment with the FGFR1 tyrosine kinase inhibitor (TKI), lucitanib (36) (Fig. 3E), and transfection with FGFR1 siRNA prevented this outgrowth (Fig. 3F).
Long-term estradiol deprivation increases the interaction of FGFR1 with ERα.
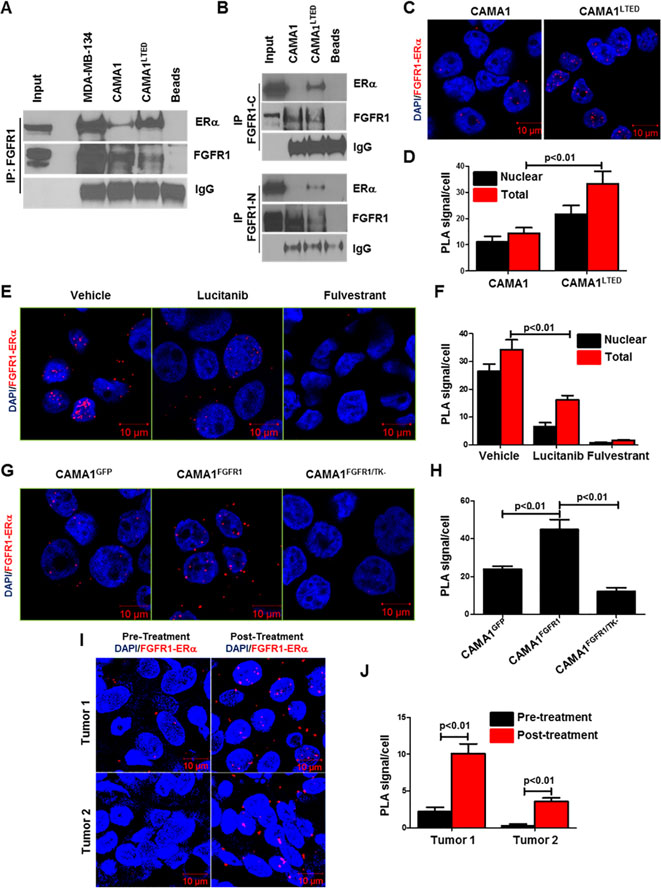
An association of FGFR1 with other nuclear proteins, such as ribosomal S6 kinase (RSK1) and CREB-binding protein (CBP), has been shown to be required for nuclear FGFR1 to induce gene expression in medulloblastoma and neuroblastoma cells (37). An interaction between FGFR1 and ERα has been reported to mediate lactotroph proliferation in the pituitary gland (38). Nuclear colocalization of progesterone receptor (PR), FGFR2 and STAT5 at DNA progesterone responsive elements with increased transcription of PR/STAT5 regulated genes were also reported in human breast cancer cells (13). Thus, we next investigated whether ER and FGFR1 interacted in ER+/FGFR1 amplified breast cancer cells. Antibody pulldown of FGFR1 from MDA-MB-134, CAMA1 and CAMA1LTED whole cell lysates co-precipitated ERα in all three cell lines (Fig. 4A). This association was stronger in MDA-134 and CAMA1LTED cells compared to parental CAMA1 cells. We next confirmed the FGFR1-ERα association in CAMA1 and CAMA1LTED nuclear extracts after precipitation with both C-terminal and N-terminal FGFR1 antibodies (Fig. 4B), suggesting with presence of full-length FGFR1 in cell nucleus. To quantitate this interaction, we performed proximity ligation assays (PLA). An interaction between FGFR1 and ERα was observed in the cytoplasm and nucleus of CAMA1 and CAMA1LTED cells by PLA, particularly in the latter (Fig. 4C,D), in line with the immunoprecipitation experiments. Treatment with lucitanib reduced FGFR1/ERα complexes (Fig. 4E,F), suggesting this interaction requires FGFR1 tyrosine kinase (TK) activity.
Figure 4. Long-term estradiol deprivation increases the interaction of FGFR1 with ERα.
A, FGFR1 was precipitated from MDA-MB-134, CAMA1 and CAMA1LTED cell lysates; immune complexes were separated by SDS-PAGE and subjected to immunoblot analysis with an ERα antibody. CAMA1LTED cells exhibited greater levels of FGFR1-ERα co-immunoprecipitation compared to CAMA1 cells. B, FGFR1 was precipitated from CAMA1 and CAMA1LTED nuclear extracts with C-terminal (Abcam) and N-Terminal (Cell Signaling) FGFR1 antibodies; immune complexes were separated by SDS-PAGE and analyzed by ERα immunoblot. C-D, PLA of CAMA1LTED cells showed greater nuclear co-localization of FGFR1 and ERα compared to parental CAMA1 cells. PLA foci/cell are quantified in D. E-F, CAMA1LTED cells were treated with 2 μM lucitanib or 1 μM fulvestrant for 6 h. Monolayers were subjected to PLA as described in Methods. Quantification of FGFR1-ERα complexes as PLA signals/cell is shown in (F). Each bar represents the mean ± SD of 3 wells. G-H, CAMA1 cells were stable transfected with expression vectors encoding GFP, FGFR1 and FGFR1/TK– (K514M TK mutant), as described in Methods, and then plated in chamber slides followed by PLA. Quantification of FGFR1-ERα complexes as PLA signals/cell is shown in (H). Each bar represents the mean ± SD of 3 wells. I-J, Paired pre- and post-letrozole primary tumor sections were subjected to PLA as described in Methods. Post-letrozole tumor cells exhibited more FGFR1-ERα complexes compared to pre-treatment tumor cells as quantitated in J. Each bar represents the mean PLA signals/cell ± SD of 20 cells counted in each of 4 high-power fields.
To explore further whether the TK function of FGFR1 is required for FGFR1-ERα complex formation, CAMA1 cells were transduced with constructs expressing GFP, wild-type FGFR1 or a TK dead K514M FGFR1 mutant (FGFR1/TK–). Overexpression of wild-type FGFR1 increased detectable FGFR1-ERα complexes while overexpression of FGFR1/TK– decreased them as measured by PLA (Fig. 4G,H). Steady-state levels of pFRS2 were upregulated in cells transduced with wild-type FGFR1 but not with the FGFR1/TK– mutant (Suppl. Fig. 5A). Importantly, the CAMA1FGFR1/TK– cells were not able to grow in the absence of estradiol (Suppl. Fig. 5B,C). These data suggest that FGFR1 TK activity is important for estrogen-independent growth and the association of FGFR with ERα. Finally, we observed an increase of FGFR1-ERα complexes in post-letrozole compared to paired pre-letrozole FFPE tumor sections from two breast cancer patients harboring tumor co-amplification of FGFR1 and 11q12–14 (Fig. 4I,J).
FGFs/FGFR pathway modulates ERα-DNA binding.
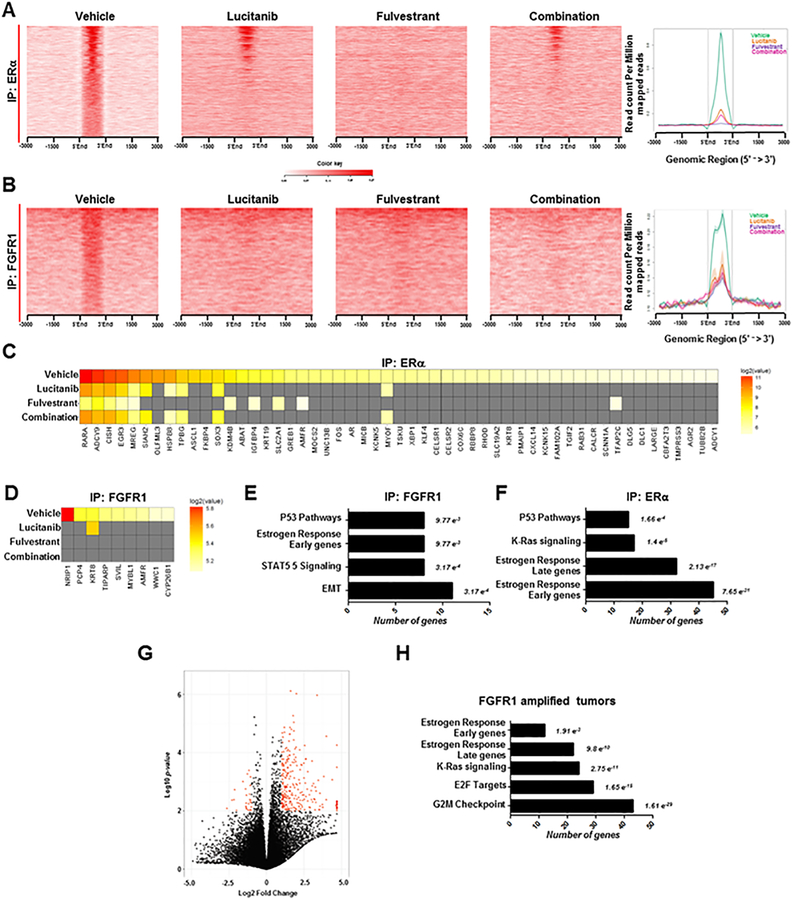
To evaluate estrogen-independent genomic functions of ERα in ER+/FGFR1 amplified cells, we performed chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) in estrogen-deprived CAMA1 cells ± FGF3. First, we confirmed by cells fractionation that parental CAMA1 cells exhibited nuclear soluble and chromatin-bound FGFR1 at steady state (Suppl. Fig. 6A). Treatment with FGF3 shifted both ERα and FGFR1 to new binding sites that were unoccupied in the absence of the FGF ligand. (Suppl. Fig. 6B,C).We identified 1120 and 553 regions (peaks) by ERα-ChIP and FGFR1-ChIP, respectively, that were significantly enriched upon FGF3 treatment. Treatment of CAMA1 cells with each fulvestrant or lucitanib alone or in combination reduced ERα or FGFR1 DNA binding to these new sites (Fig. 5A,B). These results were validated by ChIP-PCR (Suppl. Fig. 6D,E). As shown in Figure 5C,D, ERα and FGFR1 bound to different ERα-related genes, but treatment with lucitanib, fulvestrant or the combination reduced or abrogated this binding.
Figure 5. Identification of FGF-sensitive ERα and FGFR1 genomic binding sites.
A-B, CAMA1 cells were plated in estrogen-free medium and stimulated with 100 ng/mL FGF3 for 6 h in the presence of 1 μM fulvestrant, 2 μM lucitanib or the combination. Cells were harvested and subjected to ChIP-seq as described in Methods. Shown are heatmaps generated from ChIP-seq analysis of ERα (A) and FGFR1 (B) DNA binding. Treatment with fulvestrant, lucitanib or the combination reduced binding of ERα (A) or FGFR1 (B) binding to DNA. Heatmaps represent the mean of two different experiments. C-D, Heatmaps of ChIP-seq data showing the effects of fulvestrant, lucitanib or the combination on DNA/ERα-associated (C) and DNA/FGFR1-associated (D) genes, respectively, as shown in A-B. E-F, Gene set enrichment analysis (GSEA) of FGFR1- and ERα-associated genes. Numbers to the right of each bar represent the False Discovery Rate (FDR) q-value. G, Volcano plot analysis of differentially expressed genes in tumors from patients treated with letrozole in the clinical trial. Each data point represents the ratio of the average expression for a particular gene in FGFR1-amplified tumors (n=7) vs. FGFR1 non-amplified tumors (n=25). The red dots in the Volcano plot represent genes that are significantly up- or down-regulated >2-fold with p<0.01. H, GSEA of significantly enriched genes in FGFR1-amplified relative to FGFR1 non-amplified tumors showed that ERα-related pathways are still active in estrogen-deprived (by letrozole treatment) ER+/FGFR1-amplified primary tumors (G). Numbers to the right of each bar represent the FDR q-value.
To interrogate the functional output of estrogen-independent ERα activity, we classified the genes identified by FGFR1 and ERα ChIP-seq using gene set enrichment analysis (GSEA). The top enriched gene sets included epithelial mesenchymal transition, STAT5 signaling, estrogen response early genes and p53-pathways (all FDR <0.009) after FGFR1 ChIP-seq (Fig. 5E); and estrogen response early genes, estrogen response late genes, K-Ras signaling and p53-pathways (all FDR <0.0001) after ERα ChIP-seq (Fig. 5F). To apply these findings to primary ER+ breast cancers, we performed RNA sequencing analysis on 7 FGFR1-amplified and 25 FGFR1 non-amplified tumors treated with letrozole in the clinical trial. The Volcano-plot in Fig. 5G shows that, of >24,000 genes analyzed, 280 gene transcripts were increased >2-fold in FGFR1-amplified compared to FGFR1 non-amplified cancers (p<0.01; red dots in Fig. 5G). The top enriched genes by GSEA in FGFR1 amplified patients included G2M checkpoint genes, E2F target genes, estrogen response late genes and estrogen response early genes (all FDR <0.01; Fig. 5H and Suppl. Fig. 7). These results further suggest that the ERα pathway is still active in estrogen-deprived (upon letrozole treatment) ER+/FGFR1-amplified primary tumors.
To further elucidate the role of the FGF/FGFR1 axis on ERα signaling, we performed a qRT-PCR profiling assay including 84 ERα regulated genes. FGF3/19 stimulation of estrogen-deprived CAMA1 cells induced >2-fold expression of a subset of ERα target genes, including TFF1, CCND1, THSB1, CTGF, CCL2 and EGR3 (Suppl. Fig. 8A). Both FGF3 and FGF19 induced EGR3, CCND1 and THSB1 mRNA; this induction was inhibited by treatment with lucitanib, fulvestrant or the combination (Suppl. Fig. 8B–C), and also by transfection of a TK dead K514M FGFR1 mutant into CAMA1 cells (Suppl. Fig. 5D). In line with their higher levels of ERα, FGFR1 and FGF3/4/19 (Figs. 2C, 3D), CAMA1LTED cells expressed higher levels of ERα-regulate genes than CAMA1 parental cells (Suppl. Fig. 8D). Finally, to support our results with lucitanib were not due to off-target effects of the small molecule, we tested the FGFR inhibitor INCB054828 (39). Treatment with INCB054828 also blocked FGF3-induced pFRS2, CAMA1 cell growth and ERα target gene expression (Suppl. Fig. 9A–C).
We next examined the effect of the knockdown of FGFR1 on ERα transcriptional activity. Compared to scrambled control siRNA, knockdown of FGFR1, but not of its major signal transducer FRS2, reduced ERE-luciferase reporter activity (Suppl. Fig. 10A). FGFR1 and FRS2 downregulation were confirmed by immunoblot or RT-PCR, respectively (Suppl. Fig. 10B,C). The inability of siFRS2 to reduce ER reporter activity, suggested a MEK-independent and PI3K-independent role of the FGFR1 tyrosine kinase (TK) on ERα transcriptional function. Supporting this speculation, treatment of CAMA1 cells with lucitanib reduced mRNA levels of the ERα regulated genes CCND1 and THSB1 more potently than the MEK1/2 inhibitor trametinib (40) and the pan-PI3K inhibitor buparlisib (41) (Suppl. Fig. 10E). In parallel experiments, we confirmed by immunoblot analyses drug-mediated inhibition of their molecular targets: ERα for fulvestrant, pFRS2 and pERK for lucitanib, pERK for trametenib, and pAKT for buparlisib (Suppl. Fig. 10D).
Combined blockade of FGFR1 and ERα potently inhibits growth of ER+/FGFR1-amplified breast cancers.
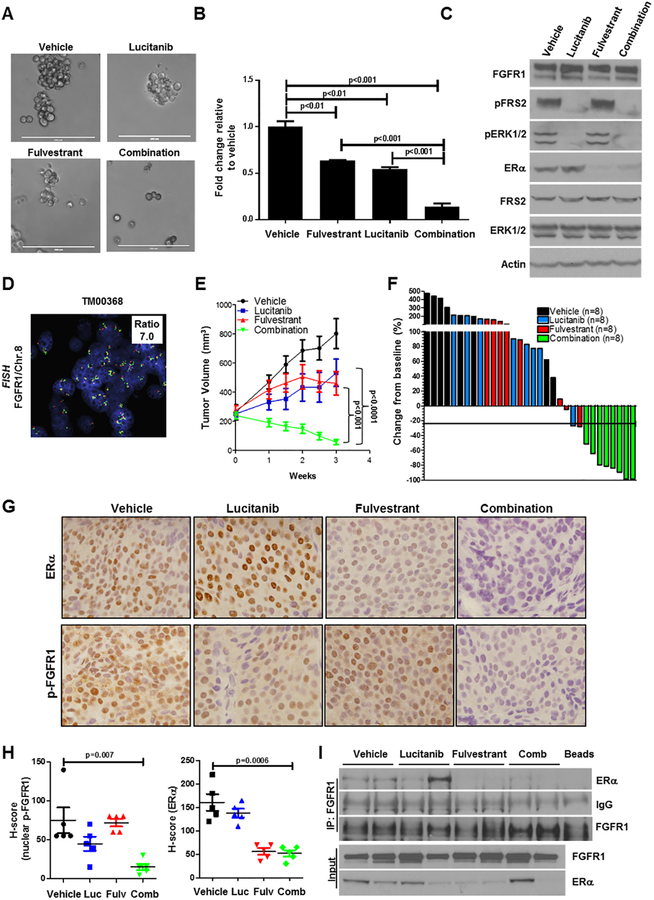
To follow the effect of fulvestrant and lucitanib on ER-dependent gene expression, we next examined whether FGFR1 and/or ERα inhibitors would have an effect on ER+/FGFR1 amplified tumor cell growth. Treatment with the combination of lucitanib and fulvestrant suppressed CAMA1 colony formation in 3D-matrigel significantly more potently than each drug alone (Fig. 6A,B). Western blot analysis of lysates from cells treated for 6 h showed that the only the combination simultaneously reduced levels of pFRS2, pERK1/2 and ERα (Fig. 6C). We next examined the effect of these drugs against two ER+/HER2–/FGFR1-amplified patient-derived xenografts (PDXs), T272 and TM00368 (Fig. 6D). Ovariectomized mice with established xenografts (≥250 mm3) were treated with vehicle, lucitanib, fulvestrant or both drugs. PDX T272 but not PDXTM00368 required brief estrogen supplementation to generate tumors. In mice bearing PDX T272, the dose of lucitanib was reduced from 10 to 7 mg/kg/day after 3 weeks of therapy due to toxicity in both lucitanib-containing arms. Mice with TM00368 PDXs were treated with 7 mg/kg/day lucitanib. Treatment with the combination of fulvestrant and lucitanib inhibited growth of both PDXs more potently than either drug alone (Fig. 6E and Suppl. Fig. 11A). All mice bearing TM00368 xenografts exhibited a ≥50% reduction in tumor size from baseline after 3 weeks of treatment with fulvestrant/lucitanib (Fig. 6F). Biomarkers of response were assessed by IHC in TM00368 tumors harvested at the completion of therapy. Treatment with the combination of lucitanib plus fulvestrant markedly reduced detectable levels of Y653/4 p-FGFR1 and total ERα (Fig. 6G,H). FGFR1 antibody pulldowns of tumor lysates from vehicle- and lucitanib-treated mice co-precipitated ERα. This was not observed in tumors treated with fulvestrant or the combination (Fig. 6I). No change in mouse weight was observed in any of the treatment arms (Suppl. Fig. 11B,C).
Figure 6. Combined blockade of FGFR1 and ERα potently inhibits growth of ER+/FGFR1-amplified breast cancers.
A-B, CAMA1 cells were cultured in 3D Matrigel as described in Methods and treated with vehicle, 2 μM lucitanib, 1 μM fulvestrant or the combination. After 15 days, images were captured from 3 different fields using a CK40 microscope. Quantitation of representative images is shown in (B). Each bar represents the fold change in colony number relative to vehicle ± SD of three replicate wells repeated twice (Student’s t-test). C, CAMA1 cells in were treated as in A & B for 6 h, after which lysates were prepared and subjected to immunoblot analyses with the indicated antibodies. E, ER+/HER2–/FGFR1-amplified TM00368 PDXs were established in ovariectomized SCID/beige mice implanted with a s.c. 21-day relase, 0.25-mg 17β-estradiol pellet. Once tumors reached ≥200 mm3, mice were randomized to treatment with vehicle, fulvestrant (5 mg/kg/week), lucitanib (7 mg/kg/day), or both drugs for 3 weeks. Each data point represents the mean tumor volume in mm3 ± SD (n=8 per arm; ANOVA test). F, Bar graph showing the % change in volume in individual TM00368 PDXs after three weeks of treatment relative to tumor volumes on day 0 (baseline). G-H, TM00368 tumors were harvested at the end of treatment. FFPE tumor sections were prepared and subjected to IHC with Y653/4 phosphorylated FGFR1 and ERα antibodies as described in Methods. The percent of phospho-FGFR1+ and ER+ tumor cells and their staining intensity was assessed by an expert breast pathologist (M.V.E.) blinded to treatment to generate an H-score. Nuclear phospho-FGFR1 and ERα H-scores are shown Student’s t-test). I, FGFR1 was precipitated from lysates of TM00368 tumors harvested at the end of treatment; immune complexes were separated by SDS-PAGE and subjected to immunoblot analysis with the indicated antibodies. Bottom two lanes show FGFR1 and ERα content in lysates before i.p.
DISCUSSION
We report herein a novel mechanism by which FGFR1 amplification confers resistance to antiestrogens in ER+ breast cancers. In a cohort of post-menopausal patients treated with the aromatase inhibitor letrozole, cancers with FGFR1 amplification retained tumor cell proliferation, suggesting aberrant FGFR1 signaling is associated with resistance to estrogen deprivation. Short and long-term estrogen deprivation increased total and nuclear FGFR1 and FGF ligand expression in ER+/FGFR1-amplified breast cancer cells and primary tumors. This was associated with an increase in nuclear FGFR1/ERα complexes and maintenance of estrogen-independent transcription of ER-responsive genes. The interaction between FGFR1 and ERα was blocked by a kinase-dead FGFR1 mutant or by FGFR TKIs. ChIP-seq analysis of FGF-stimulated FGFR1-amplified cells showed binding of FGFR1 and of ERα to DNA, which was inhibited by the FGFR TKI lucitanib and by the ER downregulator fulvestrant, respectively, suggesting a possible inter-dependence between FGFR1 and ERα at transcription start sites. Of note, RNA-seq data from ER+/FGFR1-amplified tumors from patients treated with letrozole suggested the ERα pathway is still active (Fig.5G,H), thus providing a plausible explanation for maintenance of proliferation in these estrogen-deprived cancers. Finally, dual pharmacological inhibition of FGFR1 and ERα potently inhibited growth of ER+/FGFR1-amplified breast cancer cells and PDX models, supporting the clinical development of this combination in patients with this subtype of breast cancer.
FGFR1 in association with nuclear proteins such as RSK1 and CBP has been shown to induce gene expression in other cancers (37). Since FGFR1 inhibition reduced the transcription of ERα related genes (Suppl. Fig. 8B–C), we speculated the previously reported transcriptional function of FGFR1 (16, 19, 37, 40) may play a role in resistance to estrogen deprivation. Of note, we precipitated both FGFR1 and ERα with C-terminal and N-terminal FGFR1 antibodies from FGFR1-amplified CAMA1 cell nuclei (Fig. 4B). These findings were supported by PLA and confocal microscopy studies (Fig. 4C). Inhibition of FGFR1 TK activity with lucitanib and expression of a TK dead K514M FGFR1 mutant into CAMA1 cells reduced ERα-dependent gene transcription (Suppl. Fig. 8B–C and Suppl. Fig. 5D) and inhibited the association of FGFR1 with ERα (Fig. 4E–H). Taken together, these data support a novel TK-dependent role of nuclear FGFR1 on ERα-dependent gene transcription in estrogen-independent ER+/FGFR1-amplified breast cancers.
To the best of our knowledge, this is the first report of a physical association of FGFR1 and ERα associated with antiestrogen resistance. It follows studies supporting both the nuclear localization and function of FGFR1. FGFR1 can enter the nucleus by retrograde transport from the endoplasmic reticulum lumen to the cytosol via Sec61p channels before endoplasmic vesicles deliver the receptor to the plasma membrane (17, 41). This process is possible because of the atypical TMD of FGFR1, which consists of non-polar amino acid chains interrupted by polar regions in a β-sheet structure, thus allowing mobilization of the receptor out of the membrane (17, 41). Cell surface biotinylation assays show that nuclear FGFR1 can also originate from the cell surface (42), suggesting FGFR1 is internalized and traffics to the nucleus via endosomal pathways. Indeed, FGFR1 and FGFR2 can translocate to the nucleus following ligand stimulation in pancreatic stellate cells; this process requires the interaction of FGFR1 with nuclear import proteins, like importin β (15, 43). Once in the nucleus, FGFR1 has been shown to regulate gene transcription (16, 19, 37, 40). Nuclear targeting of FGFR1 by substituting its signal peptide for a nuclear localization sequence (NLS) is sufficient to initiate DNA synthesis and transcription of c-Jun, an activator of cyclin D1. Removal of the kinase region of nuclear-targeted FGFR1 ablates this effect (44). These data suggest the TK function of FGFR1 is necessary for its transcriptional role, consistent with our data from ER+/FGFR1-amplified breast cancer cells shown herein.
In summary, we have identified a mechanism by which amplified FGFR1 can sustain estrogen-independent breast tumor growth. We propose this mechanism explains, in part, the limited effects of estrogen deprivation on ER+/FGFR1-amplified breast cancers in the clinical trial with letrozole. Based on these data, we propose combinations of ERα and FGFR antagonists should be tested in patients with ER+/FGFR1-amplified breast cancer.
Supplementary Material
Statement of Translational Relevance.
ER+/FGFR1-amplified breast cancer cells and tumors retain proliferation and ERα genomic activity despite estrogen deprivation therapy. Combined inhibition of ERα and FGFR1 induced regression of ER+/FGFR1-amplified breast cancer cells and patient-derived xenografts, supporting further development of combinations of ER and FGFR antagonists for the treatment of patients with this subtype of breast cancer.
ACKNOWLEDGMENTS
The authors thank Dr. Scott Hiebert and Dr. Charles Sanders for helpful discussions and review of this manuscript.
Financial Support: This study was funded by NIH Breast SPORE grant P50 CA098131, Vanderbilt-Ingram Cancer Center Support grant P30 CA68485, Susan G. Komen for the Cure Foundation grant SAC100013 (CLA), and a grant from the Breast Cancer Research Foundation (CLA). JMB is supported by NIH/NCI 4R00 CA181491 grant and Susan G. Komen Career Catalyst Research award CCR 299052. VMJ is supported by Conquer Cancer Foundation ASCO Young Investigator Award 8364 and Susan G. Komen Postdoctoral Fellowship Grant PDF 15329319. TPS is supported by NIH grant K08 CA148912.
Footnotes
Conflict of Interest: The authors have declared no conflicts of interest
REFERENCES
- 1.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–7. [PubMed] [Google Scholar]
- 2.Jacquemier J, Adelaide J, Parc P, Penault-Llorca F, Planche J, deLapeyriere O, et al. Expression of the FGFR1 gene in human breast-carcinoma cells. Int J Cancer. 1994;59(3):373–8. [DOI] [PubMed] [Google Scholar]
- 3.Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12(22):6652–62. [DOI] [PubMed] [Google Scholar]
- 4.Gorringe KL, Jacobs S, Thompson ER, Sridhar A, Qiu W, Choong DY, et al. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13(16):4731–9. [DOI] [PubMed] [Google Scholar]
- 5.Simon R, Richter J, Wagner U, Fijan A, Bruderer J, Schmid U, et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 2001;61(11):4514–9. [PubMed] [Google Scholar]
- 6.Missiaglia E, Selfe J, Hamdi M, Williamson D, Schaaf G, Fang C, et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosomes Cancer. 2009;48(6):455–67. [DOI] [PubMed] [Google Scholar]
- 7.Jiang T, Gao G, Fan G, Li M, Zhou C. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer. 2015;87(1):1–7. [DOI] [PubMed] [Google Scholar]
- 8.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer research. 2010;70(5):2085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6(3):743–50. [DOI] [PubMed] [Google Scholar]
- 10.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–29. [DOI] [PubMed] [Google Scholar]
- 11.Bryant DM, Stow JL. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic. 2005;6(10):947–54. [DOI] [PubMed] [Google Scholar]
- 12.Zammit C, Barnard R, Gomm J, Coope R, Shousha S, Coombes C, et al. Altered intracellular localization of fibroblast growth factor receptor 3 in human breast cancer. J Pathol. 2001;194(1):27–34. [DOI] [PubMed] [Google Scholar]
- 13.Cerliani JP, Guillardoy T, Giulianelli S, Vaque JP, Gutkind JS, Vanzulli SI, et al. Interaction between FGFR-2, STAT5, and progesterone receptors in breast cancer. Cancer Res. 2011;71(10):3720–31. [DOI] [PubMed] [Google Scholar]
- 14.Stachowiak MK, Birkaya B, Aletta JM, Narla ST, Benson CA, Decker B, et al. “Nuclear FGF receptor-1 and CREB binding protein: an integrative signaling module”. J Cell Physiol. 2015;230(5):989–1002. [DOI] [PubMed] [Google Scholar]
- 15.Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, et al. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med. 2014;6(4):467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197(6):801–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers JM, Martins GG, Ostrowski J, Stachowiak MK. Nuclear trafficking of FGFR1: a role for the transmembrane domain. J Cell Biochem. 2003;88(6):1273–91. [DOI] [PubMed] [Google Scholar]
- 18.Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, et al. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. Journal of cellular biochemistry. 2003;90(4):662–91. [DOI] [PubMed] [Google Scholar]
- 19.Terranova C, Narla ST, Lee YW, Bard J, Parikh A, Stachowiak EK, et al. Global Developmental Gene Programing Involves a Nuclear Form of Fibroblast Growth Factor Receptor-1 (FGFR1). PLoS One. 2015;10(4):e0123380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh M WNT and FGF gene clusters (review). Int J Oncol. 2002;21(6):1269–73. [PubMed] [Google Scholar]
- 21.Brady N, Chuntova P, Bade LK, Schwertfeger KL. The FGF/FGFR axis as a therapeutic target in breast cancer. Expert Rev Endocrinol Metab. 2013;8(4):391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuny M, Kramar A, Courjal F, Johannsdottir V, Iacopetta B, Fontaine H, et al. Relating genotype and phenotype in breast cancer: an analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000;60(4):1077–83. [PubMed] [Google Scholar]
- 23.Balko JM, Mayer IA, Sanders ME, Miller TW, Kuba MG, Meszoely IM, et al. Discordant cellular response to presurgical letrozole in bilateral synchronous ER+ breast cancers with a KRAS mutation or FGFR1 gene amplification. Molecular cancer therapeutics. 2012;11(10):2301–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11(2):155–68. [PubMed] [Google Scholar]
- 25.Miller TW, Balko JM, Fox EM, Ghazoui Z, Dunbier A, Anderson H, et al. ER alpha-Dependent E2F Transcription Can Mediate Resistance to Estrogen Deprivation in Human Breast Cancer. Cancer Discovery. 2011;1(4):338–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhola NE, Jansen VM, Bafna S, Giltnane JM, Balko JM, Estrada MV, et al. Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer. Cancer Res. 2015;75(2):405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, A’Hern R, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99(2):167–70. [DOI] [PubMed] [Google Scholar]
- 30.Dixon JM, Renshaw L, Young O, Murray J, Macaskill EJ, McHugh M, et al. Letrozole suppresses plasma estradiol and estrone sulphate more completely than anastrozole in postmenopausal women with breast cancer. J Clin Oncol. 2008;26(10):1671–6. [DOI] [PubMed] [Google Scholar]
- 31.Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4(2):139–47. [DOI] [PubMed] [Google Scholar]
- 32.Kwek SS, Roy R, Zhou H, Climent J, Martinez-Climent JA, Fridlyand J, et al. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28(17):1892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–31. [DOI] [PubMed] [Google Scholar]
- 36.Bello E, Colella G, Scarlato V, Oliva P, Berndt A, Valbusa G, et al. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011;71(4):1396–405. [DOI] [PubMed] [Google Scholar]
- 37.Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, et al. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell. 2009;20(9):2401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sosa LD, Gutierrez S, Petiti JP, Vaca AM, De Paul AL, Torres AI. Cooperative effect of E-2 and FGF2 on lactotroph proliferation triggered by signaling initiated at the plasma membrane. Am J Physiol-Endoc M. 2013;305(1):E41–E9. [DOI] [PubMed] [Google Scholar]
- 39.Liu PCC, Wu LX, Koblish H, Bowman K, Zhang Y, Klabe R, et al. Preclinical characterization of the selective FGFR inhibitor INCB054828. Cancer Research. 2015;75. [Google Scholar]
- 40.Somanathan S, Stachowiak EK, Siegel AJ, Stachowiak MK, Berezney R. Nuclear matrix bound fibroblast growth factor receptor is associated with splicing factor rich and transcriptionally active nuclear speckles. J Cell Biochem. 2003;90(4):856–69. [DOI] [PubMed] [Google Scholar]
- 41.Romisch K Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J Cell Sci. 1999;112 (Pt 23):4185–91. [DOI] [PubMed] [Google Scholar]
- 42.Bryant DM, Wylie FG, Stow JL. Regulation of endocytosis, nuclear translocation, and signaling of fibroblast growth factor receptor 1 by E-cadherin. Mol Biol Cell. 2005;16(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152(6):1307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiedlocha A, Falnes PO, Madshus IH, Sandvig K, Olsnes S. Dual mode of signal transduction by externally added acidic fibroblast growth factor. Cell. 1994;76(6):1039–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.