Abstract
Background
Cell contain diverse array of proteins with different molecular weight and isoelectric point (pI). The molecular weight and pI of protein play important role in determining the molecular biochemical function. Therefore, it was important to understand the detail regarding the molecular weight and pI of the plant proteins.
Results
A proteome-wide analysis of plant proteomes from 145 species revealed a pI range of 1.99 (epsin) to 13.96 (hypothetical protein). The spectrum of molecular mass of the plant proteins varied from 0.54 to 2236.8 kDa. A putative Type-I polyketide synthase (22244 amino acids) in Volvox carteri was found to be the largest protein in the plant kingdom. However, Type-I polyketide synthase was not found in higher plant species. Titin (806.46 kDa) and misin/midasin (730.02 kDa) were the largest proteins identified in higher plant species. The pI and molecular weight of the plant proteins showed a trimodal distribution. An acidic pI (56.44% of proteins) was found to be predominant over a basic pI (43.34% of proteins) and the abundance of acidic pI proteins was higher in unicellular algae species relative to multicellular higher plants. In contrast, the seaweed, Porphyra umbilicalis, possesses a higher proportion of basic pI proteins (70.09%). Plant proteomes were also found to contain selenocysteine (Sec), amino acid that was found only in lower eukaryotic aquatic plant lineage. Amino acid composition analysis showed Leu was high and Trp was low abundant amino acids in the plant proteome. Additionally, the plant proteomes also possess ambiguous amino acids Xaa (unknown), Asx (asparagine or aspartic acid), Glx (glutamine or glutamic acid), and Xle (leucine or isoleucine) as well.
Conclusion
The diverse molecular weight and isoelectric point range of plant proteome will be helpful to understand their biochemical and functional aspects. The presence of selenocysteine proteins in lower eukaryotic organism is of interest and their expression in higher plant system can help us to understand their functional role.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5983-8) contains supplementary material, which is available to authorized users.
Keywords: Proteome, Amino acids, Isoelectric point, Molecular weight, Selenocysteine, Pyrrolysine
Background
The isoelectric or isoionic point of a protein is the pH at which a protein carries no net electrical charge and hence is considered neutral [1–4]. The zwitterion form of a protein becomes dominant at neutral pH. The pI of polypeptides is largely dependent on the dissociation constant of the ionisable groups [5]. The major ionisable groups present in the amino acids are arginine, aspartate, cysteine, histidine, glutamate, lysine, and glutamate, where they play a major role in determining the pI of a protein [6–8]. Co-translational and post-translational modifications of a protein, however, can also play a significant role in determining the pI of a protein [9, 10]. The exposure of charged residues to the solvents, hydrogen bonds (diploe interactions) and dehydration also impact the pI of a protein [11, 12]. The inherent pI of protein, however, is primarily based on its native protein sequence. The pI of a protein is crucial to understanding its biochemical function and thus determining pI is an essential aspect of proteomic studies. During electrophoresis, the direction of movement of a protein in a gel or other matrix depends its’pI, hence numerous proteins can be separated based on their pI [13–16]. Given the impact of post-translational modifications and other biochemical alterations (phosphorylation, methylation, alkylation), however, the predicted pI of a protein will certainly be different than the predicted pI; the latter of which is based on the composition of amino acids in a protein [9, 17, 18]. Nonetheless, an estimated isoelectric point is highly important and a commonly identified parameter.
Several studies have been conducted to understand the pI of proteins/polypeptides [3, 19–21]. These studies have been mainly based on animal, bacteria, and virus models and databases containing the pI of experimentally verified proteins. None of these databases, however, contain more than ten thousand proteins sequences which is very few relative to the availability of proteomic data. Therefore, an analysis was conducted of the pI and molecular weight of proteins from 144 plant species which included 5.87 million protein sequences. This analysis provides an in-depth analysis of the pI and molecular mass of the proteins in the plant kingdom.
Results and discussion
Plant proteins range from 0.54 kDa to 2236.8 kDa
A proteome-based analysis of plant proteins of 144 plant species that included more than 5.86 million protein sequences was considered to study the molecular mass, pI, and amino acid composition of proteins that exist in plant proteomes (Additional file 2: Table S1). The analysis indicated that Hordeum vulgare possessed the highest number (248180) of protein sequences, while Helicosporidium sp. had the lowest number (6033). On average, plant proteomes possess 40988.66 protein sequences per species (Fig. 1). The analysis also revealed that the molecular mass of plant proteomes ranged from 0.54 kDa to 2236.8 kDa. The gross molecular weight of plant proteome was ranged from 9857470.16 kDa (Hordeum vulgare) to 178551.42 kDa (Helicosporidium sp.) (Additional file Table S1). Volvox carteri was found to possess the largest plant protein (XP_002951836.1) of 2236.8 kDa, containing 22244 amino acids (pI 5.94), while Citrus unshiu possessed the smallest protein of 0.54 kDa, containing only four amino acids (pI 5.98) (id: GAY42954.1). This is the first analysis to document the largest (2236.8 kDa) and smallest (0.54 kDa) protein in the plant kingdom. These smallest and largest proteins yet to be functionally annotated. The BLASTP analysis in the NCBI database did not identify suitable similarity with any other proteins. Some of the domains identified in the largest protein were matched with protein domains of Type-I polyketide synthase. The molecular mass of some other high molecular mass proteins were: 2056.44 kDa (id: XP_001698501.1, type-1 polyketide synthase, pI: 6.00, aa: 21004, Chlamydomonas reinhardtii); 1994.71 kDa (id: XP_001416378.1, polyketide synthase, pI: 7.38, aa: 18193, Ostreococcus lucimarinus); 1932.21 kDa (id: Cz02g22160.t1, unknown protein, pI: 5.7, aa: 18533, Chromochloris zofingiensis); 1814.1 kDa (id: XP_007509537.1, unknown protein, pI: 4.46, aa: 16310, Bathycoccus prasinos); 1649.26 kDa (id: XP_011401890.1, polyketide synthase, pI: 5.53, aa: 16440, Auxenochlorella protothecoides); 1632.35 kDa (id: XP_005650993.1, ketoacyl-synt-domain-containing protein, pI: 5.86, aa: 15797,Coccomyxa subellipsoidea); 1532.91 kDa (id: XP_002507643.1, polyketide synthase, pI: 7.07, aa: 14149, Micromonas commoda); 1370.23 kDa (id: GAX78753.1, hypothetical protein CEUSTIGMA, pI: 5.97, aa: 13200, Chlamydomonas eustigma); 1300.83 kDa (id: XP_022026115.1, unknown protein/filaggrin-like, pI: 11.75, aa: 12581, Helianthus annuus); 1269.42 kDa (id: XP_009350379.1, unknown protein, pI: 5.37, aa: 11880, Pyrus bretschneideri); 1237.34 kDa (id: XP_022840687.1, polyketide synthase, pI: 7.30, aa: 11265, Ostreococcus tauri); 1159.35 kDa (id: XP_005847912.1, polyketide synthase, pI: 5.91, aa: 11464, Chlorella variabilis); 1150.02 kDa (id: PKI66547.1, unknown protein, pI: 3.87, aa: 11234, Punica granatum); 1027.64 kDa (id: Sphfalx0133s0012.1, unknown protein, pI: 4.05, aa: 9126, Sphagnum fallax); 909.93 kDa (id: XP_002985373.1, unknown/titin-like protein, pI: 4.02, aa: 8462, Selaginella moellendorffii); 881.59 kDa (id: KXZ46216.1, hypothetical protein, pI: 5.80, aa: 8881, Gonium pectorale); 848.29 kDa (id: XP_003056330.1, pI: 6.12, aa: 7926, Micromonas pusilla); 813.31 kDa (id: GAQ82263.1, unknown protein, pI: 4.60, aa: 7617, Klebsormidium nitens), 806.46 kDa (id: XP_017639830.1, titin-like, pI: 4.21, aa: 7209, Gossypium arboreum); 806.12 kDa (id: OAE35580.1, pI: 4.83, hypothetical protein, aa: 7651, Marchantia polymorpha); and 802.74 kDa, (id: XP_012444755.1, titin-like, pI: 4.19, aa: 7181, Gossypium raimondii) (Additional file 2: Table S1).
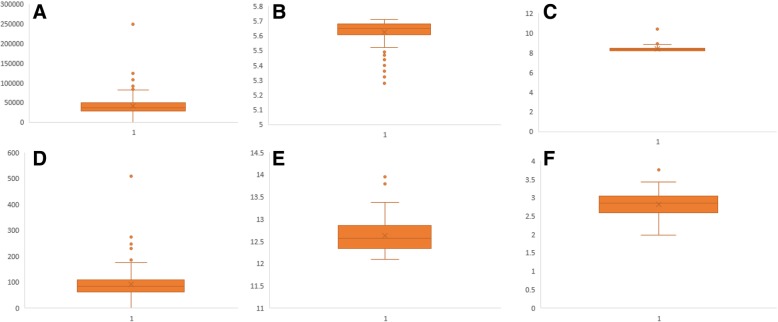
Fig. 1.
Box and Whisker plot of (a) average number of protein sequences encoded in plant proteome, b average acidic pI, c average basic pI of plant proteome, d average number of neutral pI proteins per plant proteome (e) average of highest pI protein, and (f) average of lowest pI proteins. Details can be found in Additional file 2: Table S1
On average, approximately 7.38% of the analysed proteins were found to contain ≥100 kDa proteins. Analysis of average molecular weight of proteins in a proteome revealed, the average molecular weight of individual protein of Chlamydomonas reinhardtii proteome was found to be 62.74 kDa, followed by 60.24 kDa (Volvox carteri), 58.64 kDa (Klebsmordium nitens), and 58.10 kDa (Bathycoccus prasinos). Similarly, Trifolium pratense (28.37 kDa) encode the lowest molecular weight protein in the plant kingdom. The algal species, V. carteri, was found to encode largest plant protein (putative polyketide synthase); while other unicellular algae, and multi-cellular lower eukaryotic plants, including bryophytes and pteridophytes, were also found to encode some of the larger proteins (e.g. ketoacyl synthase) in the plant kingdom. The higher eukaryotic plants, including gymnosperms and angiosperms, were not found to encode a high molecular mass polyketide synthase protein. Notwithstanding, they contain the high molecular mass proteins; titin (806.46 kDa), misin/midasin (730.02 kDa), futsch (622.14 kDa), filaggrin (644.4 kDa), auxin transport protein BIG (568.4 kDa), and von Willebrand factor (624.74 kDa) (Additional file 2: Table S1). Titin protein found in human striated muscle is a large protein that is higher than 1 μM in length [22, 23]. The largest titin protein found in plants, however, was only 806.46 kDa (Gossypium arboreum). The predicted formula of the 806.46 kDa titin protein was C33863H54610N9232O13061S200 and its estimated half-life was 10–30 h; whereas the predicted formula of the 2236.8 kDa protein of V. carteri was C97783H157401N28489O30265S637. Almost all of the higher eukaryotic plants were found to possess titin, misin/midasin, and auxin transport protein BIG proteins. Species of unicellular algae were not found to possess titin or misin/midasin proteins. This suggests that titin and misin/midasin proteins originated and evolved in more complex, multicellular organisms rather than unicellular organisms. Thus, the evolution of titin, misin/midasin proteins may also be associated with the evolution of terrestrial plants from aquatic plants. The Midasin protein.
The presence of the low molecular weight protein, other than the tripeptide glutathione (Cys, Gly, and Glu), was also determined. A 0.54 kDa molecular mass protein, containing only four amino acids (MIMF) and starting with methionine and ending with phenylalanine, was identified in Citrus unshiu (id: GAY42954.1) (Additional file 2: Table S1). Other low molecular mass plant proteins were 0.57 kDa (NP_001336532.1/ AT5G23115, Arabidopsis thaliana) and 0.63 kDa (AH003201-RA, Amaranthus hypochondriacus). Small proteins found in A. thaliana was MNPKS and that found in A. hypochondriacus was MLPYN, contained only five amino acids. The above mentioned low molecular mass proteins were missing in all of the studied species and their cellular and molecular functions have not been reported yet. One of the universal small molecular weight plant proteins, however, was identified as cytochrome b6/f complex subunit VIII (chloroplast) (MDIVSLAWAALMVVFTFSLSLVVWGRSGL) that contains only 29 amino acids. Cytochrome b6/f play important role in electron transfer system of photosystem II and administer photosynthesis [24–28]. It is commonly known that glutathione is the smallest functional polypeptide and that it plays diverse roles in cell signaling [29–31]. The tetra and penta peptides identified in the present analysis, however, were quite different from glutathione and none of them contained Cys, Gly, or Glu amino acids, as found in glutathione. Polypeptides with less than 100 amino acids are considered small proteins and studies indicate that considerable number of the small proteins are involved in cell signaling, DNA damage response, and cell growth [32–35]. During genome annotation, small protein-coding genes are overlooked constantly and hence they get buried amongst an enormous number of open reading frames [36]. Therefore, it is strenuous to find more numbers of small proteins in plants. The tetra and pentapeptides are involved in diverse signaling process [37, 38]. A bioactive tetrapeptide GEKG reported to boost extracellular matrix formation [39]. In bacteria, small peptides are act as pheromones [40]. The pentapeptide ERGMT regulate genetic competence in B. subtilis whereas ARNQT involved in sporulation [40].
A previously conducted comparative study revealed that plant proteins are comparatively smaller than animal proteins, as the former are encoded by fewer exons [41]. Longer proteins harbour more conserved domains and hence display a greater number of biological functions than short proteins. The average protein length of the studied plant species was 424.34 amino acids. A previous study reported the average length of eukaryotic proteins to be 472 amino acids and that the average length of plant proteins is approximately 81 amino acids shorter than animal proteins [41]. Our analysis indicates, however, that plant proteins are approximately 47.66 amino acid shorter than animal proteins. In addition, studies have also indicated that eukaryotic proteins are longer than bacterial proteins and that eukaryote genomes contain approximately 7 fold more proteins (48% larger) than bacterial genomes [42]. Although the average size of plant proteins was found to be 424.34 amino acids, the average protein size of lower, eukaryotic unicellular aquatic plant species; including Chlamydomonas eustigma, Volvox carteri, Klebsordium nitens, Bathycoccus prasinos, and Durio zibethinus, was found to be 576.56, 568.22, 538.73, 521.05, and 504.36 amino acids, respectively. This indicates that protein size unicellular plant species is larger than terrestrial multicellular complex plant species, suggesting that the evolution of plant proteins involved a loss of protein size and hence gene size. The reason for the alteration in protein length in the phylogenetic lineage of eukaryotic plants has yet to be elucidated. A multitude of evolutionary factors, including deletion (loss of exons) or fusion of multiple domains of proteins, most possibly played important roles in configuring the size of higher plant proteins. The insertion of transposon and splitting of genes expand the number of proteins but scale down the average size of the proteins [43–46]. Higher plants contain a very large number of transposable elements and therefore these elements are the most responsible factor to expect to have played a major role in increasing protein numbers and reducing the protein size in higher plants. The percentage of transposable elements in a genome is directly proportional to the genome size of the organism and varies from approximately 3% in small genomes to approximately 85% in large genomes [45]. Kirag et al. (2007) reported a significant correlation between protein length and the pI of a protein [19]. In our analysis, however, no correlation was found between protein length and the pI of a protein. For example, titin and misin are two of the larger proteins in the plants and they fall in the acidic pI range, but not in the alkaline pI range. Lack of sufficient proteomic data might be a possible factor for such results by Kirag et al., (2007).
Plant encode a higher number of proteins than animals and fungi
Our analysis identified an average of 40469.47 proteins per genome (Additional file 2: Table S1). Previously the number of proteins in plant species was reported as 36795 per genome [41]. On average, animals and fungi encode 25189 and 9113 proteins per genome, respectively [41]. An average of 40469.47 proteins per plant genome is 62.24% higher than in animals and 444.08% higher than in fungi. Although, plant species encode a higher number of proteins, their size is smaller than the average size of animal proteins. Notably, green algae contain a smaller number of proteins than higher plants but their average protein size is 1.27 times larger. The average protein size (low to high) in the species of green algae ranged from 273.08 (Helicosporidium sp.) to 576.56 (Chlamydomonas eustigma) amino acids, dicots ranged from 253.34 (Trifolium pratense) to 498.49 (Vitis vinifera), and monocots ranged from 111.54 (Hordeum vulgare) to 473.35 (Brachypodium distachyon) amino acids. The average protein size of monocot proteins (431.07 amino acids), however, is slightly larger than dicots (424.3 amino acids). In addition to transposons, previous studies have reported that endosymbiosis may have also played an important role in the reduction of protein size in plant genomes [41, 47, 48]. This would have been due to the post endosymbiosis acquisition of thousands of genes from the chloroplast, since cyanobacterial proteins are smaller than eukaryotic proteins and cyanobacteria are the ancestors of plastids [41, 49]. In this hypothesis, the intermediate size of plant proteins would be the result of the migration of proteins from cyanobacteria (chloroplast) to the plant nucleus, thereby reducing the overall average size of the protein by a dilution effect [50, 51].
The pI of plant proteins ranges from 1.99 to 13.96
Results indicated that the pI of analysed plant proteins ranged from 1.99 (id: PHT45033.1, Capsicum baccatum) to 13.96 (id: PKI59361.1, Punica granatum). The protein with the lowest pI (1.99) was epsin and the protein with the highest pI (13.96) was a hypothetical protein. We are the first to report on the plant proteins with the lowest and highest pI. The C. baccatum protein with pI 1.99 contains 271 amino acids, whereas the P. granatum protein with pI 13.96 contains 986 amino acids. The epsin protein (pI 1.99) is composed of 16 amino acid repeats (GWIDGWIDGWIDGW), while the hypothetical protein (pI 13.06) is composed of 64 QKLKSGLT and 31 TRRGLTAV repeats. From among the 20 essential amino acids, the epsin protein only contained five amino acids, namely Asp (68), Gly (68), Ile (65), Met (3), and Trp (67). The amino acids were arranged in a repeating manner within the full-length epsin protein. We are the first to report a full-length protein conists of such a minimum number of essential amino acids. Similarly, the hypothetical protein with the highest pI (13.96) was composed of only nine amino acids, namely Ala (62), Gly (132), Lys (127), Leu (197), Met (M), Pro (4), Gln (64), Arg (132), and Ser (66). Intriguingly, cysteine, which is one of the most important amino acids as it is responsible for the formation of disulphide bonds, was not found in either the smallest or largest protein. Disulphide bonds provide the conformational stability of protein and are commonly found in extracellular proteins and only rarely in intracellular proteins [52]. The absence of Cys amino acids in the above-mentioned proteins suggests, they are localized to the intracellular compartments within the cell.
The plant proteome is primarily composed of acidic pI proteins rather than basic pI proteins (Additional file 2: Table S1). Approximately, 56.44% of the analysed proteins had a pI within the acidic pI range with an average of pI 5.62 (Fig. 1, Additional file 2: Table S1). The average (%) of acidic pI proteins was comparatively higher in the lower eukaryotic plants, algae, and bryophytes, than in the higher land plants. A total of 64.18% of proteins in Chlamydomonas eustigma were found in the acidic pI region, followed by Ostreococcus lucimarinus (64.17%), Micromonas commoda (63.30%), Helicosporium sp. (62.97%), Gonium pectoral (62.76%), Chromochloris zofingiensis (62.41%), Coccomyxa subellipsoidea (62.12%), and Sphagnum fallax (61.83%). The algal species, Porphyra umbilicalis, had the lowest percentage (29.80%) of acidic pI proteins. The dicot plant, Punica granatum, and the algal species, Botrycoccus braunii, had a significantly lower percentage of acidic pI proteins (45.72 and 47.18%, respectively) relative to other plants. Principal component analysis (PCA) of acidic pI protein content revealed that the acidic proteins of bryophytes and monocots cluster closely to each other compared to algae and eudicot plants (Fig. 2). Similarly, in the case of basic pI proteins, a great variation was observed for algae, eudicot and monocot plants (Fig. 3). The basic pI proteins of bryophytes, however, were found to be consistent. A previous study reported that protein pI values are correlated with the sub-cellular localization of the proteins, and that the pI of cytosolic proteins fall below 7 [21]. Among cytosolic proteins are those involved in 26S proteasome degradation, oxidative pentose phosphate pathway, actin/tubulin, mevalonate pathway, sugar and nucleotide biosynthesis, glycolysis, RNA processing, and several other cellular process. Our analysis indicated that the pI of all cytosolic proteins does not fall in the acidic pI range. Ribosomal proteins, pre-mRNA splicing factors, transcription factors, auxin induced protein, extensin, senescence associated protein, cyclin dependent protein kinase and other cytoplasmic proteins had a pI greater than 7.
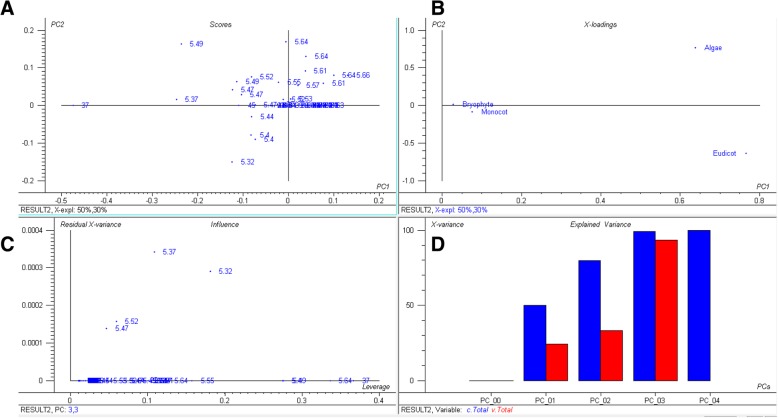
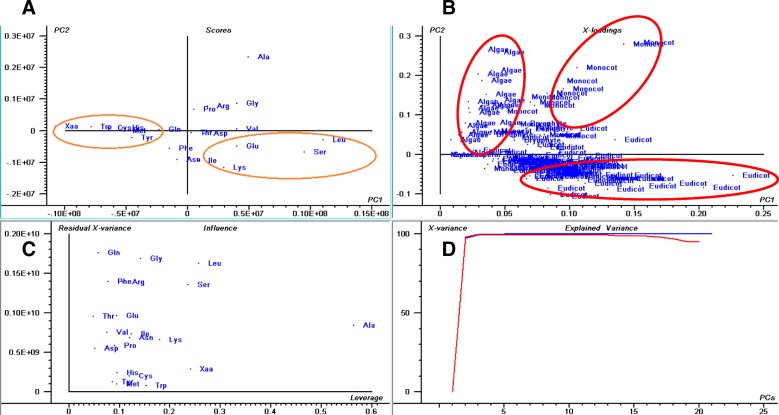
Fig. 2.
Principal component analysis (PCA) of acidic pI proteins. The PCA plot illustrates the relationship between the acidic pI of bryophytes and monocot plants which exhibit a linear correlation relative to algae and eudicots. In the figure (a) scores: show the similarities in sample grouping, b loading: represents the relative position of a variables and how it relates the samples to the different variables (c). Influence plot: represents the Q- or F-residuals vs. Leverage or Hotelling T2 statistics that show the residual statistics on the ordinate axis of sample distance to model, and (d) variance: represents the variation in the data described by the different components. Total residual variance is computed as sum of square of residuals for all the variables, divided by the number of degrees of freedom. The green colour indicates the calibration and the red indicates the validation
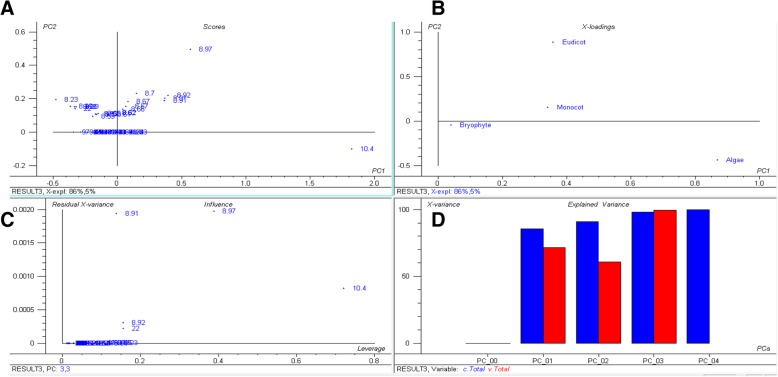
Fig. 3.
Principal component analysis (PCA) of basic pI proteins. The PCA plot illustrates that the basic pI of algae, bryophytes, eudicots, and monocot plants cluster distinctly from each other and that there is no lineage-specific correlation with basic pI proteins. In the figure (a) scores: show the similarities in sample grouping, (b) loading: represents the relative position of a variables and how it relates the samples to the different variables (c) Influence plot: represents the Q- or F-residuals vs. Leverage or Hotelling T2 statistics that show the residual statistics on the ordinate axis of sample distance to model, and (d) variance: represents the variation in the data described by the different components. Total residual variance is computed as sum of square of residuals for all the variables, divided by the number of degrees of freedom. The green colour indicates the calibration and the red indicates the validation
In contrast to acidic pI proteins, plants possess a comparatively low number of basic pI proteins. On average, 43.34% of the analysed plant proteins possessed a pI in the basic range with an average pI of 8.37 (Fig. 1, Additional file 2: Table S1). The highest percentage of basic pI proteins was found in Porphyra umbilicalis, where 70.09% of the proteins had a basic pI (Additional file 2: Table S1). Punica granatum also had a high percentage (54.11%) of basic pI proteins (Additional file 2: Table S1). The lowest percentage of basic pI proteins was found in the algal species, Chlamydomonas eustigma (35.56%), followed by Ostreococcus lucimarinus (35.65%), Micromonas commoda (36.52%), Helicosporidium sp. (36.89%), and Gonium pectorale (37.04%). It is difficult to establish the reason that algal species contain more acidic pI and less basic pI proteins. Porphyra umbilicalis is a cold-water seaweed within the family, Bangiophyceae, and it is the most domesticated marine algae. The 87.7 Mbp haploid genome of P. umbilicalis has a 65.8% GC content and an evolutionary study reported that the genome of Porphyra umbilicalis had undergone a reduction in size [53]. Since this species is found in the intertidal region of the ocean, it has developed the ability to cope with mid-to-high levels of tidal stress. Porphyra is also tolerant to UV-A and UV-B radiation [53–55]. The high GC content in Porphyra umbilicalis is directly proportional to the high percentage of basic proteins. The GC content of algal species is higher relative to other plant species and algal species possess a lower percentage of basic pI proteins. This suggests that, in algae, percentage GC content is inversely proportional to percentage of proteins with a basic pI. However, this is not true in the case of higher plants.
The pI of plant proteomes exhibits a trimodal distribution
The pI of the analysed plant proteins ranged from 1.99 to 13.96 and exhibited a trimodal distribution (Fig. 4). Schwartz et al., previously reported a trimodal distribution of the pI of eukaryotic proteins [21], however, they did not provide information on the number of sequences/species considered in their study. Proteins are typically soluble near their isoelectric point and the cytoplasm possesses a pH that is close to neutral. This might be the possible reason for the trimodal distribution of pI. Although the pI values of proteins estimated in silico or experimentally might be different in vivo, they are typically in close agreement and represents a virtual 2-DE gel [56]. However, the acidic peak was more prominent than the alkaline (Fig. 4). Kiraga et al., (2006) reported a bimodal distribution of the pI of proteins from all organisms, citing acidic and basic pI as the basis of the modality [19], where modality is defined as the set of data values that appears most often. They reported that taxonomy, ecological niche, proteome size, and sub-cellular localization are correlated with acidic and basic proteins. Notwithstanding, no correlation was found in the present study between either acidic or basic pI of proteins with regard to taxonomy, ecological niche, or proteome size. For example, Hordeum vulgare and Brassica napus possess the largest proteomes among the studied plant species, possessing 248180 and 123465 proteins, respectively. In H. vulgare, 53.28% of the proteins fall in the acidic and 46.50% fall in the basic pI ranges; while in B. napus, 55.28% of the proteins have an acidic pI and 44.48% have a basic pI. Other species with smaller proteomes, however, possess a higher percentage of acidic or basic proteins (Additional file 2:Table S1). Therefore, no correlation exists between the percentage of either acidic or basic proteins and proteome size, taxonomy, or the ecological niche of an organism. Knight et al. also addressed a negative correlation between the pI of a protein and phylogeny of the organism [57]. The presence of a trimodal distribution of the pI of the plant proteome can be considered as a virtual 2D-gel of a plant’s proteins where the pI of the protein is plotted against the molecular weight of the protein. On average, 0.21% of the analysed proteins were found to have a neutral pI (pI 7), while only 0.09% of the proteins in O. lucimarinus fall in neutral pI. The modal distribution of protein pI shows its physiochemical properties rather than sequence evolution [58].
Fig. 4.
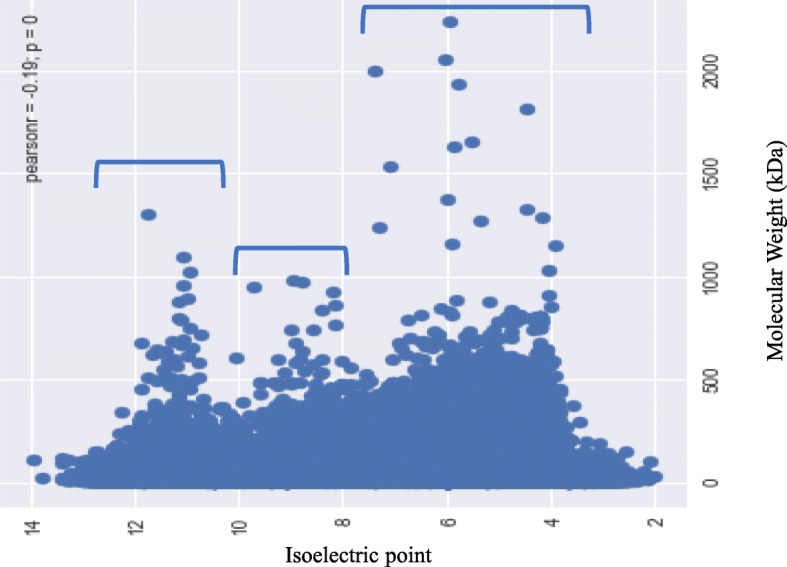
Trimodal distribution of isoelectric points (pI) and the molecular mass (kDa) of plant proteins. The pI of plant proteins ranged from 1.99 (epsin) to 13.96 (hypothetical protein), while the molecular mass ranged from 0.54 (unknown) to 2236.8 (type I polyketide synthase) kDa. The X-axis represents the pI and the Y- axis represents the molecular mass of the proteins
Leu is a high- and Trp is a low-abundant amino acid in the plant proteome
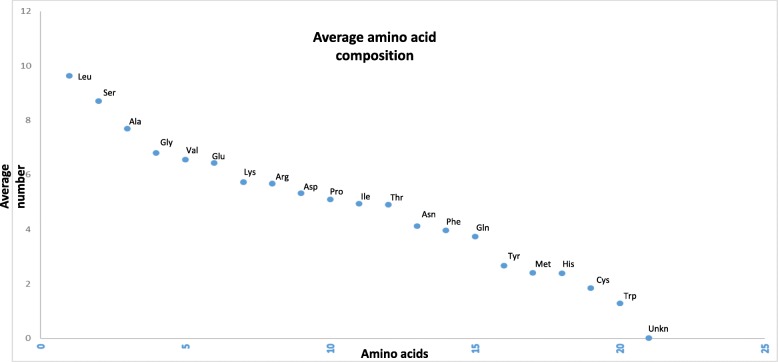
The plant-kingdom-wide proteome analysis of amino acid composition revealed that the Leu was the most (9.62%) while Trp was the least (1.28%) abundant amino acid (Fig. 5, Additional file 1). Leu is a nonpolar amino acid, whereas Trp contains an aromatic ring. The distribution of amino acids indicates that the synthesis of nonpolar amino acids is more favoured in the plant proteomes than the polar amino acids or those containing an aromatic ring. The average abundance of other nonpolar amino acids Ala, Gly, Ile, Met, and Val was 6.68, 6.80, 4.94, 2.40, and 6.55%, respectively (Table 2, Additional file 1). Trp and Tyr amino acid contain an aromatic ring and the abundance of these two proteins is relatively low in the plant proteome compared to other amino acids. Results of the conducted analysis indicated that the abundance of Ala (17.58%), Gly (11.76%), Pro (9.2%), and Arg (9.81%) were the highest; whereas, Tyr (1.33%), Gln (2.04%), Asn (1.53%), Met (1.45%), Lys (7.07%), Lys (2.08%), Ile (1.77%), Phe (2.01%), and Glu (3.52%) were the lowest in Porphyra umbilicalis. In a few algae and seaweeds Ala, Asp, Glu, Gly, Pro, Gln, Arg, Thr, and Val were found in high percentage while Asp, Glu, Phe, His, Ile, Lys, Leu, Met, Asn, Gln, and Ser were found in low percentage (Additional file 3: Table S2). These findings indicate that the composition of amino acids in unicellular algae, seaweeds, and gymnosperms are more dynamic and variable than in angiosperms and other terrestrial land plants. Principal component analysis revealed that the low abundant amino acids, Trp, Tyr, His, Met, Cys, and Xaa (unknown), cluster in one group while the high abundant amino acids, Leu, Glu, Ile, Lys, and Ser, cluster in another group (Fig. 6). None of the terrestrial land plants were located in the high- and low-abundant amino acid clusters. This put forward the fact that the amino acid composition and proteome of the land plants are more conserved and stable relative to the algae and seaweeds. In addition, the PCA analysis revealed that the pI of algae, eudicots, and monocots are lineage specific. The pI of algae, monocots, and eudicots were strongly correlated and clustered together (Fig. 6). The question arises, however, as to why the plant proteome contains the highest percentage of Leu and of the lowest percentage of Trp amino acids. Do the energy requirements of the different biosynthetic pathways play a pivotal role in deciding the abundance of amino acids in a proteome? To address this question, an study was conducted to figure out the role of amino acid biosynthetic pathways in determining the abundance of specific amino acids in the proteome. The amino acid composition are used to deduced the properties of proteins that are selected in evolution for their chemical properties [59]. Most importantly, longer protein helps to form complex secondary and tertiary structure. In addition, amino acid composition influence the core and surface protein structure.
Fig. 5.
Average amino acid composition of proteins in the plant kingdom. Leu is the most abundant while Trp is the least abundant. The amino acid, Sec, was only found in a few species of algae and was absent from all other species. Ambiguous amino acids were found in a few species as well
Fig. 6.
Principal component analysis (PCA) of amino acid abundance in plant proteomes. The PCA plot shows that Tyr, Trp, Cys, His, Met, and Xaa (unknown) amino acids are low-abundance and cluster together. The abundance of Leu, Ser, Ile, Lys, and Gln was higher and grouped together. The plot shows that the abundance of amino acids is lineage specific. Algae, eudicots, and monocot plants exhibit a lineage specific correlation. In the figure (a) scores: show the similarities in sample grouping, (b) loading: represents the relative position of a variables and how it relates the samples to the different variables (c) Influence plot: represents the Q- or F-residuals vs. Leverage or Hotelling T2 statistics that show the residual statistics on the ordinate axis of sample distance to model, and (d) variance: represents the variation in the data described by the different components. Total residual variance is computed as sum of square of residuals for all the variables, divided by the number of degrees of freedom. The green colour indicates the calibration and the red indicates the validation
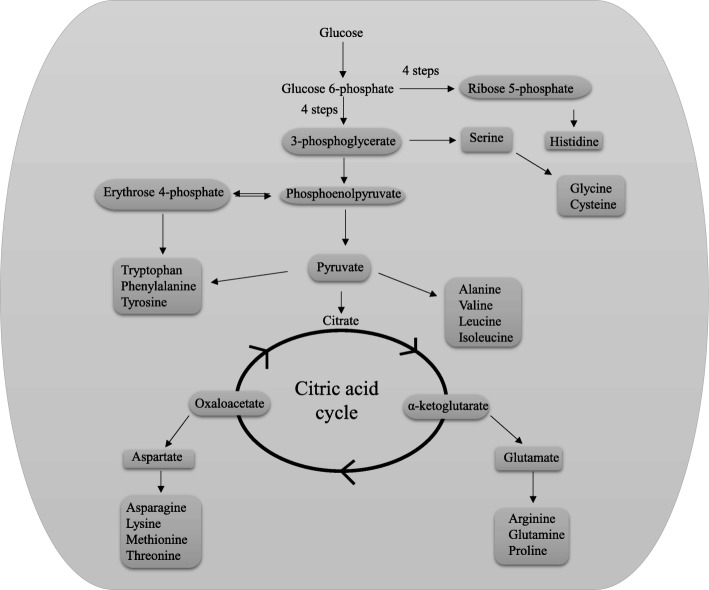
Various amino acids are produced in different biosynthetic pathways [60–64] (Fig. 7). In few cases, some of the amino acids act as the substrate for the biosynthesis of other amino acids; whereas in other cases, allosteric inhibition of the biosynthesis of amino acids occurs [65–67]. In all of these amino acid biosynthetic pathways, along with substrate, ATP or NADH/NADPH are used as a source of energy. Overall, the biosynthesis of 20 essential amino acid families are grouped by metabolic precursors [68] (Table 1); namely α-ketoglutarate (Arg, Gln, Glu, Pro), pyruvate (Ala, Ile, Leu, Val), 3-phosphoglycerate (Cys, Gly, Ser), phosphoenolpyruvate and erythrose 4-phosphate (Phe, Trp, Tyr), oxaloacetate (Asn, Asp, Lys, Met, Thr), and ribose 5-phosphate (His) (Table 1) [68]. Ile, Alan, Leu, and Val are synthesized from pyruvate; Glu, Arg, Gln, and Pro are synthesized from α-ketoglutarate whereas Gly and Ser are synthesized from 3-phosphoglycerate [68]. The abundance of these amino acids in plant proteomes are considerably higher relative to the other amino acids (Fig. 7, Table 3). 3-phosphoglycerate and pyruvate are act as transient product of glycolysis and the amino acids produced from these intermediates maintain a high abundance in the plant proteome. The intermediate, 3-phosphoglycerate, is formed in an early step of glycolysis [68]. Gly and Ser are produced from 3-phosphoglycerate and quite bountiful in the plant proteome (Fig. 7, Table 3). The amino acid Cys, which is also synthesized from 3-phosphoglycerate [68], however, is present in low abundance (1.85%) in the plant proteome. The low richness of Cys possibly due to the allosteric inhibition. Phe (3.97), Trp (1.28%), and Try (2.67%) contain an aromatic ring and are synthesized via phosphoenolpyruvate and erythrose 4-phospahte. The aromatic amino acids including Trp, Tyr, and Phe are in low abundance in the plant proteome (Table 1). Since Phe also plays a role in the biosynthesis of Tyr, the abundance of Phe is relatively higher than Trp and Tyr. Glucose 6-phosphate gives rise to ribose 5-phosphate in a complex reaction of four steps [68] and His gets subsequently synthesized from ribose 5-phosphate. It is possible that the complexity of the biosynthetic pathways of amino acids containing ring compounds might be the reason for their low abundance in the plant proteome.
Fig. 7.
Schematic illustration of the biosynthetic pathway of amino acids. The abundance of aromatic ring containing amino acids is lower relative to other amino acids. The average abundance of the aromatic ring containing amino acid, Trp, is the lowest amongst others that are biosynthesized via phosphoenolpyruvate and erythrose 4-phosphate. Similarly, the abundance of Cys is relatively low compared to other amino acids. Ser is biosynthesized from 3-phosphoglycerate and Ser is subsequently used to produce Gly and Cys amino acids. The abundance of Cys is lower relative to Gly, suggesting the existence of allosteric feed-back inhibition of the biosynthesis of Cys by Ser
Table 1.
Average abundance of different amino acids in plant proteome. Leu was high abundant whereas Trp was the low abundant amino acid in the plant kingdom. The average amino acid composition includes 5.8 million protein sequences from 145 plant species
| Biosynthetic pathways/Substrate | Amino acids | Average abundance (%) in Proteome |
|---|---|---|
| α-Ketoglutarate | Arginine | 6.68 |
| Glutamate | 6.43 | |
| Glutamine | 3.74 | |
| Proline | 5.10 | |
| Pyruvate | Alanine | 7.68 |
| Isoleucine | 4.94 | |
| Leucine | 9.62 | |
| Valine | 6.55 | |
| 3-Phosphoglycerate | Glycine | 6.80 |
| Cysteine | 1.85 | |
| Serine | 8.71 | |
| Oxaloacetate | Asparagine | 4.13 |
| Aspartate | 5.32 | |
| Lysine | 5.73 | |
| Methionine | 2.40 | |
| Threonine | 4.90 | |
| Phosphoenolpyruvate & Erythrose 4-phosphate | Phenylalanine | 3.97 |
| Tryptophan | 1.28 | |
| Tyrosine | 2.67 | |
| Ribose 5-phosphate | Histidine | 2.4 |
Plants possess selenocysteine (sec) and other novel amino acids
A few of the plant proteomes that were analysed had proteins containing the amino acid, selenocysteine (Sec) encoded by TGA (opal) codon [69]. C. reinhardtii, M. pusilla, and V. carteri contained 9, 16, and 11 Sec amino acids in their proteome, respectively. Selenium containing proteins are frequently found in the proteome of bacterial and animal kingdom [69–71]. However, it has been reported to be present in the plant species as well. Novoselov et al., (2002) reported the presence of a selenoprotein in C. reinhardtii [72]. In our analysis, nine selenoproteins (selenoprotein H, selenoprotein K1, selenoprotein M2, selenoprotein T, selenoprotein U, selenoprotein W1, selenoprotein W2, NADPH-dependent thioredoxin reductase 1, and glutathione peroxidase) were identified in C. reinhardtii. In addition, M. pusilla was found to possess 14 Sec-containing proteins [DSBA oxidoreductase (2 no.), glutathione peroxidase (4 no.), selenoprotein T, selenoprotein W, selenoprotein, selenoprotein M, selenoprotein H, selenoprotein O, methyltransferase, and peroxiredoxin). In addition, V. carteri was found to possess 10 Sec containing proteins (selenoprotein T, selenoprotein K1, selenoprotein H, selenoprotein W1, selenoprotein M2, selenoprotein U, glutathione peroxidase, membrane selenoprotein, NADPH-dependent thioredoxin reductase, and peptide methionine-S-sulfoxide reductase). According to our observation, this is the first report regarding the presence of selenoproteins in M. pusilla and V. carteri and the first to report the presence of H, K1, T, U, M, M2, O, W1, and W2 selenoprotein families in C. reinhardtii, M. pusilla, and V. carteri. The I, N, P, R, S, and V selenoprotein family members are commonly found in the animal kingdom [73] but are absent in C. reinhardtii, M. pusilla, and V. carteri. This is also the first report of the Sec-containing proteins, DSBA oxidoreductase, methyltransferase, peroxiredoxin, peptide methionine-S-sulfoxide reductase, and membrane selenoprotein in plants lineage (algae). Notably, the selenoproteins DSBA oxidoreductase, methyltransferase, peroxiredoxin, peptide methionine-S-sulfoxide reductase, and membrane selenoprotein have not been reported in animal species. Outside of algal species, no other plant species; including bryophytes, pteridophytes gymnosperms and angiosperms, were found to possess a selenoprotein. Selenoprotein is superimposed in the translation machinery and more specifically selenoprotein mRNA contain selenocysteine insertion sequence element (SECIS) downstream to the Sec-encoding UGA codon [74, 75]. The SECIS-sequence recognise and binds the selenocysteine-specific elongation factor (selB) and forms complex with tRNASec (selC). The tRNASec acylated with serine by seryl-tRNA synthastase and subsequently transformed to Sec-tRNASec by Sec synthase (selA). Later, selA utilizes selenophosphate as the selenium donor that synthesized by selenophosphate synthetase (selD) [71, 75]. The selenium is found in the active site of selenoproteins, more specifically involved in redox reactions [71]. In human, selenoprotein H regulates cell cycle progression and proliferation of human colorectal cancer cells [76]. Selenoprotein K enhances protein palmitoylation [77, 78], selenoprotein T attenuates the development of left ventricular dysfunction after myocardial infraction in rats [79].
Some plant proteomes were also found to possess a few unknown or unspecified amino acids, commonly designated as Xaa (X). Among the analysed plant species, Aegilops tauschii, Amaranthus hypochondriacus, and Amborella trichocarpa encoded 149377, 55412, and 25843 X amino acids, respectively. Solanum lycopersicum was found to contain only one X amino acid, while at least 18 species (Solanum pennellii, Solanum tuberosum, Sorghum bicolor, Sphagnum fallax, Spinacia oleracea, Spirodela polyrhiza, Tarenaya hassleriana, Theobroma cacao, Trifolium pratense, Trifolium subterraneum, Triticum aestivum, Triticum urartu, Vigna angularis, Vigna radiata, Vigna anguiculata, Vitis vinifera, Volvox carteri, and Zostera marina) were found to lack any Xaa amino acids in their proteome. Xaa amino acids are commonly considered as non-protein amino acids as they are yet to be reported as protein coding amino acids. Among the studied plant species, ten were found to contain amino acid B (Asx) that codes for the ambiguous amino acid Asn or Asp that is translated as Asp. Species that were found to possess an Asx amino acid included Arachis duranensis (1), Brachypodium stacei (40), Dichanthelium oligosanthes (20), Dunaliella salina (31), Glycine max (1), Malus domestica (4080), Momordica charantia (98), Nelumbo nucifera (64), Prunus persica (1), and Trifolium pratense (76). At least six species were found to possess a J (Xle) amino acid. Xle amino acid can encode either Leu or Ile but during translation produces Leu. Species that were found to possess Xle amino acids included Arabidopsis thaliana (10), Dichanthelium oligosanthes (11), Malus domestica (2175), Momordica charantia (39), Nelumbo nucifera (29), and Trifolium pratense (39). At least seven species were found to possess a Z (Glx) amino acid that codes for either Glu or Gln, which is subsequently translated as Glu. Species that were found to encode a Glx amino acid included Brachypodium stacei (20), Dichanthelium oligosanthes (16), Dunaliella salina (7), Malus domestica (1552), Momordica charantia (28), Nelumbo nucifera (14), and Trifolium pratense (25). Among the studied species, Malus domestica was found to contain highest number of ambiguous amino acids (Asx, Xle, and Glx). Bodley and Davie (1966) reported the incorporation of ambiguous amino acids in a peptide chain [80]. The presence of ethanol or streptomycin or a high magnesium ion concentration induces ambiguous coding in the peptide chain [80]. They reported that poly-U (uridylic acid) in the presence of a high concentration of magnesium ions or ethanol or streptomycin induces the incorporation of Leu/Ile amino acids in a peptide chain [80]. This explains how the specificity of the protein translation process can be altered by the presence of environmental factors. A high concentration of magnesium ions, organic solvents, antibiotics, pH, and low temperature have the ability to modify the coding specificity of a peptide chain [80]. Under some conditions poly-U triggers the incorporation of Leu and Ile or Phe [80]. Malus domestica is rich in magnesium ions (1%) and this might explain the presence of such a high number of ambiguous amino acids in its proteome. The presence of D-Xaa-D-Xaa-tRNA in the P-site lead to ribosomal stalling of elongation factor G (EF-G) [81].
Conclusion
A proteome-wide analysis of the plant kingdom identified proteins with a great range of molecular mass and isoelectric points. Isoelectric points ranged from 1.99 to 13.96, covering almost the entire pH range. It is quite intriguing to think about the functions of protein at pI 1.99 or 13.96. Proteins with an acidic pI predominate over the proteins with an alkaline pI, and the presence of proteins with a pI that is near neutral is very negligible. The percentage of proteins with acidic or basic pI is not related to the host cell, alkalinity or acidity of the environment. The trimodal distribution of pI of plant proteome is due to the differential composition of amino acids in different species and this might be associated with the environmental and ecological pressure. Rate of mutation and nucleotide substitution can also be the other responsible factors associated with composition of the amino acids and hence the pI. Additionally, the GC content of a genome or size distribution of the overall proteome are not directly proportional to the distribution (percent basic vs. percent acidic) of the pI in the proteome. The presence of Sec-containing proteins in some algal species needs to be further investigated to determine their functional role. Similarly, the presence of ambiguous amino acids in plant species should be further evaluated individually. The presence of a pyrrolysine amino acid in the plant kingdom was not observed in the present study.
Methods
Protein sequences of the entire proteome of the analysed plant species were downloaded from the National Center for Biotechnology Information (NCBI) and Phytozome, DOE Joint Genome Institute (https://phytozome.jgi.doe.gov/pz/portal.html). All of the studied sequences were annotated nuclear-encoded proteins. The isoelectric point of each protein of each of the analysed plant species was calculated individually using the Python-based command line “Protein isoelectric point calculator” (IPC Python) in a Linux platform [2]. The source code used was as written by Kozlowski (2016).
Once the molecular mass and isoelectric point of the proteins in each species was determined, they were separated into acidic and basic pI categories. Subsequently, the average pI and percentage of proteins in each category was calculated using a Microsoft excel worksheet. A graph comparison of isoelectric point versus molecular mass was prepared using a python-based platform. Pearson-correlation (r = 0.19, p = 0) was used for the association analysis of molecular mass and isoelectric point. The X-axis data statistics were as follows: mean, 4.717365e+ 01; std., 3.662983e+ 01; min, 8.909000e-02; 25%, 2.279452e+ 01; 50%, 3.874486e+ 01; 75%, 5.999628e+ 01, and max, 2.236803e+ 03. The Y-axis data statistics were: mean, 6.840657e+ 00; std., 1.594912e+ 00; min, 1.990000e+ 00; 25%, 5.537000e+ 00; 50%, 6.605000e+ 00; 75%, 8.053000e+ 00, and max, 1.396300e+ 01.
Principal component analysis
Principal component analysis of the plant proteome parameters was carried out using a portable Unscrambler software version 9.7 using the excel file format. For acidic and basic pI, the plant proteome data were grouped according to the plant lineage algae, bryophyte, monocot, and eudicot plants. The average of acidic and basic pI was used to construct the PCA plot. Similarly, amino acid abundance was also analysed in relation to algae, bryophyte, eudicot, and monocot lineage.
Additional files
Supplementary file containing average amino acid composition of plant proteomes. The file is present in excel sheet and individual sheet represents details of different amino acid. (XLSX 252 kb)
Table S1. Details of plant proteome. Table shows acidic pI of proteins predominates the basic pI. However, in sea weed Porphyra umbilicalis, basic pI predominates over the acidic pI. Putative polyketide synthase type I found in lower eukaryote Volvox carteri was found to be the largest protein in the plant lineage. However, titin was found to be the largest protein in the higher eukaryotic land plants. Asterisks represents no specific data available for the said item. (DOCX 61 kb)
Table S2. Abundance of various amino acids in different species. The second column represents the average abundance whereas the third and fourth column represent variation (high and low) in amino acid composition in different species from the average. (DOCX 16 kb)
Acknowledgements
The authors would like to extend their sincere thanks to University of Nizwa, Oman for the support towards the research. The authors would also like to extend their sincere appreciation to the deanship of scientific research at King Saud University for its funding to the research group number (RGP-271).
Availability of data materials
All the studied data were taken from publicly available databases and data associated with the manuscript is provided in supplementary file.
Competing of interest
The authors declare that they have no competing interests.
Abbreviations
- 60S RP L41
60S ribosomal protein subunit L41
- AAPT
Aminoalcoholphosphotransferase
- ATF7IP
Activating transcription factor 7-interacting protein 1
- CDPK
Cyclin-dependent serine/threonine-protein kinase
- CSF
Cleavage stimulation factor subunit 2 tau variant-like
- CWP
Cell wall protein
- DDG
Dolichyl-diphosphooligosaccharideprotein glycosyltransferase subunit 4A
- DDGT
Dolichyl-diphosphooligosaccharideprotein glycosyltransferase subunit 4A
- DDHGT 4A
Dolichyl-diphosphooligosaccharideprotein glycosyltransferase subunit 4A
- ER TF
ethylene responsive transcription factor
- FS CAYBR BP
Fibrous sheath CABYR-binding protein-like
- GRCW protein
Glycine-rich cell wall structural protein
- IMP
inosine-5′-Monophosphate cyclohydrolase
- LEA
Late embryogenesis associated protein
- NFD
Nuclear fusion defective
- PERK2
Proline-rich receptor-like protein kinase PERK2
- PKDP
Polycystic kidney disease protein 1-like 3
- PKS
Polyketide synthase
- PPCD VHS3-like
Phosphopantothenoylcysteine decarboxylase subunit VHS3-like isoform X2
- PPCD
Phosphopantothenoylcysteine decarboxylase
- PS-I RC N
Photosystem I reaction center subunit N, chloroplastic, partial
- RBP
RNA binding protein
- RNA Pol. II Med 17
Mediator of RNA polymerase II subunit 17
- RPB1
DNA-directed RNA polymerase II subunit RPB1-like isoform X3
- SARMP
Serine/arginine repetitive matrix protein 2-like
- SCP
Spore coat protein
- SR45
Serine/arginine-rich splicing factor SR45-like
- SVC
Satellite virus coat protein
- ZRF
Zinc ring finger-type
Authors’ contributions
TKM: conceived the idea, collected the protein sequences, analysed and interpreted the data, drafted the manuscript, ALK: revised the manuscript, AH: drafted and revised the manuscript, EFA: drafted and revised the manuscript, AAH: revised the manuscript. All authors read and approved the final manuscript.
Funding
Not available.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree and have consent for publication.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tapan Kumar Mohanta, Email: nostoc.tapan@gmail.com, Email: tapan.mohanta@unizwa.edu.om.
Abdullatif Khan, Email: abdullatif@unizwa.edu.om.
Abeer Hashem, Email: habeer@ksu.edu.sa.
Elsayed Fathi Abd_Allah, Email: eabdallah@ksu.edu.sa.
Ahmed Al-Harrasi, Email: aharrasi@unizwa.edu.om.
References
- 1.Kirkwood J, Hargreaves D, O’Keefe S, Wilson J. Using isoelectric point to determine the pH for initial protein crystallization trials. Bioinformatics. 2015;31:1444–1451. doi: 10.1093/bioinformatics/btv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozlowski LP. IPC – Isoelectric Point Calculator. Biol Direct. 2016;11:55. doi: 10.1186/s13062-016-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozlowski LP. Proteome-pI: proteome isoelectric point database. Nucleic acids res. 2016/10/26. Oxford University Press. 2017;45:D1112–D1116. doi: 10.1093/nar/gkw978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stekhoven F, Gorissen M, Flik G. The isoelectric point, a key to understanding a variety of biochemical problems: a minireview. Fish Physiol Biochem. 2008;34:1–8. doi: 10.1007/s10695-007-9145-6. [DOI] [PubMed] [Google Scholar]
- 5.Kenneth W, Kenneth G, Raymond E. General Chemistry. 4th ed. Saunders College Publishing; 1992.
- 6.Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2008/12/02. Wiley subscription services, Inc. A Wiley Company. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masunov A, Lazaridis T. Potentials of mean force between Ionizable amino acid side chains in water. J Am Chem Soc American Chemical Society. 2003;125:1722–1730. doi: 10.1021/ja025521w. [DOI] [PubMed] [Google Scholar]
- 9.Zhu K, Zhao J, Lubman DM, Miller FR, Barder TJ. Protein pI shifts due to posttranslational modifications in the separation and characterization of proteins. Anal Chem American Chemical Society. 2005;77:2745–2755. doi: 10.1021/ac048494w. [DOI] [PubMed] [Google Scholar]
- 10.Locke D, Koreen IV, Harris AL. Isoelectric points and post-translational modifications of connexin26 and connexin32. FASEB J Federation of American Societies for Experimental Biology. 2006;20:1221–1223. doi: 10.1096/fj.05-5309fje. [DOI] [PubMed] [Google Scholar]
- 11.Youden WJ, Denny FE. Factors influencing the pH equilibrium known as the isoelectric point of plant tissue. Am J Bot Botanical Society of America. 1926;13:743–753. [Google Scholar]
- 12.Shaw KL, Grimsley GR, Yakovlev GI, Makarov AA, Pace CN. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci Cold Spring Harbor Laboratory Press. 2001;10:1206–1215. doi: 10.1110/ps.440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saraswathy N, Ramalingam P. Introduction to proteomics. In: Saraswathy N, Ramalingam PBT-C and T in G and P, editors. Woodhead Publ Ser Biomed. Woodhead Publishing; 2011. p. 147–58.
- 14.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci. 2000;97:9390 LP–9399395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabilloud T, Lelong C. Two-dimensional gel electrophoresis in proteomics: a tutorial. J Proteome. 2011;74:1829–1841. doi: 10.1016/j.jprot.2011.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Kumar M, Singh R, Meena A, Patidar BS, Prasad R, Chhabra SK, et al. An Improved 2-Dimensional Gel Electrophoresis Method for Resolving Human Erythrocyte Membrane Proteins. Proteomics Insights. SAGE Publications Ltd STM; 2017;8:1178641817700880. [DOI] [PMC free article] [PubMed]
- 17.Anderson JC, Peck SC. A simple and rapid technique for detecting protein phosphorylation using one-dimensional isoelectric focusing gels and immunoblot analysis. Plant J. 2008;55:881–885. doi: 10.1111/j.1365-313X.2008.03550.x. [DOI] [PubMed] [Google Scholar]
- 18.Jones LR, Simmerman HK, Wilson WW, Gurd FR, Wegener AD. Purification and characterization of phospholamban from canine cardiac sarcoplasmic reticulum. J Biol Chem. 1985;260:7721–7730. [PubMed] [Google Scholar]
- 19.Kiraga J, Mackiewicz P, Mackiewicz D, Kowalczuk M, Biecek P, Polak N, et al. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. BMC Genomics. BioMed Central. 2007;8:163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carugo O. Isoelectric points of multi-domain proteins. Bioinformation. 2007;2:101–104. doi: 10.6026/97320630002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz R, Ting CS, King J. Whole proteome pl values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.GR-1587R. [DOI] [PubMed] [Google Scholar]
- 22.Labeit S, Barlow DP, Gautel M, Gibson T, Holt J, Hsieh C-L, et al. A regular pattern of two types of 100-residue motif in the sequence of titin. Nature. Nat Publ Group. 1990;345:273. doi: 10.1038/345273a0. [DOI] [PubMed] [Google Scholar]
- 23.Kurzban GP, Wang K. Giant polypeptides of skeletal muscle titin: sedimentation equilibrium in guanidine hydrochloride. Biochem Biophys Res Commun. 1988;150:1155–1161. doi: 10.1016/0006-291X(88)90750-4. [DOI] [PubMed] [Google Scholar]
- 24.Kurisu G. Structure of the Cytochrome b6f Complex of Oxygenic Photosynthesis: Tuning the Cavity. Science. 2003;302(5647):1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 25.Leister D. Schneider ABT-. From genes to photosynthesis in Arabidopsis thaliana. Int rev Cytol. Academic Press. 2003;228:31–83. doi: 10.1016/s0074-7696(03)28002-5. [DOI] [PubMed] [Google Scholar]
- 26.Barbagallo RP, Finazzi G, Forti G. Effects of inhibitors on the activity of the cytochrome b6f complex: evidence for the existence of two binding pockets in the Lumenal site. Biochem Am Chem Soc. 1999;38:12814–12821. doi: 10.1021/bi990424+. [DOI] [PubMed] [Google Scholar]
- 27.Cramer WA, Soriano GM, Ponomarev M, Huang D, Zhang H, Martinez SE, et al. Some new structural aspects and old controversies concerning the cytochrome B6f complex of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol Annual Reviews. 1996;47:477–508. doi: 10.1146/annurev.arplant.47.1.477. [DOI] [PubMed] [Google Scholar]
- 28.Kallas T. The cytochrome b6f complex. In: Bryant DA, editor. Mol Biol Cyanobacteria. Dordrecht: Springer Netherlands; 1994. pp. 259–317. [Google Scholar]
- 29.Filomeni G, Rotilio G, Ciriolo MR. Cell signalling and the glutathione redox system. Biochem Pharmacol. 2002;64:1057–1064. doi: 10.1016/S0006-2952(02)01176-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Hongqiao, Forman Henry Jay. Glutathione synthesis and its role in redox signaling. Seminars in Cell & Developmental Biology. 2012;23(7):722–728. doi: 10.1016/j.semcdb.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aquilano K, Baldelli S, Ciriolo MR. Glutathione: new roles in redox signaling for an old antioxidant. Front Pharmacol. 2014;5:196. doi: 10.3389/fphar.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su M, Ling Y, Yu J, Wu J, Xiao J. Small proteins: untapped area of potential biological importance. Front Genet Frontiers Media S.A. 2013;4:286. doi: 10.3389/fgene.2013.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol Elsevier. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Schalk Catherine, Cognat Valérie, Graindorge Stéfanie, Vincent Timothée, Voinnet Olivier, Molinier Jean. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proceedings of the National Academy of Sciences. 2017;114(14):E2965–E2974. doi: 10.1073/pnas.1618834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue Yuan, Shen Lian, Cui Ying, Zhang Huabing, Chen Qi, Cui Anfang, Fang Fude, Chang Yongsheng. Tff3, as a Novel Peptide, Regulates Hepatic Glucose Metabolism. PLoS ONE. 2013;8(9):e75240. doi: 10.1371/journal.pone.0075240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basrai MA, Hieter P, Boeke JD. Small open Reading frames : beautiful needles in the haystack small open Reading frames : beautiful needles in the haystack. Genome Res. 1997;7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 37.Ludovic W, Jérôme B, Jian-Miao L, Delphine M, ET G, José V, et al. Tetrapeptide AcSDKP induces Postischemic neovascularization through monocyte chemoattractant Protein-1 signaling. Arterioscler Thromb Vasc biol. Am Heart Assoc. 2006;26:773–779. doi: 10.1161/01.ATV.0000203510.96492.14. [DOI] [PubMed] [Google Scholar]
- 38.Goldstein JL, Brown MS, Stradley SJ, Reiss Y, Gierasch LM. Nonfarnesylated tetrapeptide inhibitors of protein farnesyltransferase. J Biol Chem [Internet]. 1991;266:15575–15578. [PubMed]
- 39.Farwick M, Grether-Beck S, Marini A, Maczkiewitz U, Lange J, Köhler T, et al. Bioactive tetrapeptide GEKG boosts extracellular matrix formation: in vitro and in vivo molecular and clinical proof. Exp Dermatol. John Wiley & Sons, Ltd (10.1111); 2011;20:602–604. [DOI] [PubMed]
- 40.Lazazzera BA, Grossman AD. The ins and outs of peptide signaling. Trends Microbiol. 1998;6:288–294. doi: 10.1016/S0966-842X(98)01313-4. [DOI] [PubMed] [Google Scholar]
- 41.Ramírez-Sánchez Obed, Pérez-Rodríguez Paulino, Delaye Luis, Tiessen Axel. Plant Proteins Are Smaller Because They Are Encoded by Fewer Exons than Animal Proteins. Genomics, Proteomics & Bioinformatics. 2016;14(6):357–370. doi: 10.1016/j.gpb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiessen A, Pérez-Rodríguez P, Delaye-Arredondo LJ. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res Notes. BioMed Central. 2012;5:85. doi: 10.1186/1756-0500-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purugganan M, Wessler S. The splicing of transposable elements and its role in intron evolution. Genetica. 1992;86:295–303. doi: 10.1007/BF00133728. [DOI] [PubMed] [Google Scholar]
- 44.Huff Jason T., Zilberman Daniel, Roy Scott W. Mechanism for DNA transposons to generate introns on genomic scales. Nature. 2016;538(7626):533–536. doi: 10.1038/nature20110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S-I, Kim N-S. Transposable elements and genome size variations in plants. Genomics inform. 2014/09/30. Korea Genome Organ. 2014;12:87–97. doi: 10.5808/GI.2014.12.3.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plasterk RHA. Transposable elements. Brenner S, Miller JHBT-E of G, editors. Encycl. Genet. New York: Academic Press; 2001.
- 47.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002/09/06. National Academy of Sciences; 2002;99:12246–12251. [DOI] [PMC free article] [PubMed]
- 48.Sloan DB, Moran NA. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 2012;29:3781–3792. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohanta Tapan Kumar, Pudake Ramesh N., Bae Hanhong. Genome-wide identification of major protein families of cyanobacteria and genomic insight into the circadian rhythm. European Journal of Phycology. 2017;52(2):149–165. doi: 10.1080/09670262.2016.1251619. [DOI] [Google Scholar]
- 50.Reyes-Prieto A, Weber APM, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet Annual Reviews. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- 51.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik K V. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. Macmillan Magazines Ltd.; 1998;393:162. [DOI] [PubMed]
- 52.Mallick P, Boutz DR, Eisenberg D, TO Y. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci U S A. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brawley SH, Blouin NA, Ficko-Blean E, Wheeler GL, Lohr M, Goodson HV, et al. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta) Proc Natl Acad Sci. 2017;114:E6361 LP–E63E6370. doi: 10.1073/pnas.1703088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dring MJ, Wagner A, Boeskov J, Lüning K. Sensitivity of intertidal and subtidal red algae to UVA and UVB radiation, as monitored by chlorophyll fluorescence measurements: influence of collection depth and season, and length of irradiation. Eur J Phycol Taylor & Francis. 1996;31:293–302. doi: 10.1080/09670269600651511. [DOI] [Google Scholar]
- 55.Gröniger A, Hallier C, Häder DP. Influence of UV radiation and visible light on Porphyra umbilicalis: Photoinhibition and MAA concentration. J Appl Phycol. 1999;11:437. doi: 10.1023/A:1008179322198. [DOI] [Google Scholar]
- 56.Sillero A, Ribeiro JM. Isoelectric points of proteins: theoretical determination. Anal Biochem. 1989;179:319–325. doi: 10.1016/0003-2697(89)90136-X. [DOI] [PubMed] [Google Scholar]
- 57.Knight CG, Kassen R, Hebestreit H, Rainey PB. Global analysis of predicted proteomes: functional adaptation of physical properties. Proc Natl Acad Sci U S A. 2004;101:8390–8395. doi: 10.1073/pnas.0307270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiller GF, Caraux G, Sylvester N. The modal distribution of protein isoelectric points reflects amino acid properties rather than sequence evolution. Proteomics. John Wiley & Sons. Ltd. 2004;4:943–949. doi: 10.1002/pmic.200200648. [DOI] [PubMed] [Google Scholar]
- 59.Brüne D, Andrade-Navarro MA, Mier P. Proteome-wide comparison between the amino acid composition of domains and linkers. BMC Res Notes. 2018;11:117. doi: 10.1186/s13104-018-3221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson F, Pittard J. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol Rev. 1968;32:465–492. [PMC free article] [PubMed] [Google Scholar]
- 61.Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant. 2010;3:956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- 62.Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 63.Galili G, Höfgen R. Metabolic engineering of amino acids and storage proteins in plants. Metab Eng. 2002;4:3–11. doi: 10.1006/mben.2001.0203. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida S. Biosynthesis and conversion of aromatic amino acids in plants. Annu Rev Plant Physiol Annual Reviews. 1969;20:41–62. doi: 10.1146/annurev.pp.20.060169.000353. [DOI] [Google Scholar]
- 65.Light SH, Anderson WF. The diversity of allosteric controls at the gateway to aromatic amino acid biosynthesis. Protein Sci. Wiley subscription services, Inc., A Wiley Company; 2013;22:395–404. [DOI] [PMC free article] [PubMed]
- 66.Berg J, Tymoczko J, Stryer L. Amino acid biosynthesis is regulated by feedback inhibition. Biochemistry. 5th ed. New York: WH Freeman; 2002. [Google Scholar]
- 67.Nelson D, Cox M. Amino acid biosynthesis is under allosteric regulation. In: Lehninger AL, editor. Lehlingers Princ Biochem. 4th ed. 2005. p. 833–80.
- 68.Nelson D, Cox M. Biosynthesis of amino acids, nucleotides, and related molecules. Lehlinger Princ Biochem. 4th ed. New York: W.H. Freeman and Company; 2005. p. 842.
- 69.Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, et al. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 70.Mousa R, Notis Dardashti R, Metanis N. Selenium and Selenocysteine in Protein Chemistry. Angew Chemie Int Ed. 2017;56:15818–15827. doi: 10.1002/anie.201706876. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, et al. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reeves MA, Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell Mol Life Sci. 2009;66:2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–208. doi: 10.1016/S0968-0004(96)80016-8. [DOI] [PubMed] [Google Scholar]
- 75.Zinoni F, Heider J, Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci U S A. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bertz M, Kühn K, Koeberle SC, Müller MF, Hoelzer D, Thies K, et al. Selenoprotein H controls cell cycle progression and proliferation of human colorectal cancer cells. Free Radic Biol Med. 2018;127:98–107. doi: 10.1016/j.freeradbiomed.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 77.Fredericks Gregory, Hoffmann FuKun, Hondal Robert, Rozovsky Sharon, Urschitz Johann, Hoffmann Peter. Selenoprotein K Increases Efficiency of DHHC6 Catalyzed Protein Palmitoylation by Stabilizing the Acyl-DHHC6 Intermediate. Antioxidants. 2017;7(1):4. doi: 10.3390/antiox7010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fredericks GJ, Hoffmann PR. Selenoprotein K and protein Palmitoylation. Antioxid Redox Signal. 2015;23:854–862. doi: 10.1089/ars.2015.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boukhalfa I, Dumesnil A, Nicol L, Henry J-P, Thuillez C, Ouvrard-Pascaud A, et al. Selenoprotein T attenuates the development of left ventricular dysfunction after myocardial infarction in rats. Arch Cardiovasc Dis Suppl. 2015;7:141. [Google Scholar]
- 80.Bodley JW, Davie EW. A study of the mechanism of ambiguous amino acid coding by poly U: the nature of the products. J Mol Biol. 1966;18:344–355. doi: 10.1016/S0022-2836(66)80252-8. [DOI] [PubMed] [Google Scholar]
- 81.Katoh T, Tajima K, Suga H. Consecutive elongation of D-amino acids in translation. Cell Chem Biol. 2017;24:46–54. doi: 10.1016/j.chembiol.2016.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file containing average amino acid composition of plant proteomes. The file is present in excel sheet and individual sheet represents details of different amino acid. (XLSX 252 kb)
Table S1. Details of plant proteome. Table shows acidic pI of proteins predominates the basic pI. However, in sea weed Porphyra umbilicalis, basic pI predominates over the acidic pI. Putative polyketide synthase type I found in lower eukaryote Volvox carteri was found to be the largest protein in the plant lineage. However, titin was found to be the largest protein in the higher eukaryotic land plants. Asterisks represents no specific data available for the said item. (DOCX 61 kb)
Table S2. Abundance of various amino acids in different species. The second column represents the average abundance whereas the third and fourth column represent variation (high and low) in amino acid composition in different species from the average. (DOCX 16 kb)