Abstract
Our purpose is to evaluate the microstructural and metabolism property in the white matter that later become white matter hyperintensity (WMH), and of WMH that later disappeared. Forty subjects with two-year follow-up were included. Each subject had 3DT1, T2FLAIR, DTI and FDG-PET scans. White matter was classified into: constant WMH, growing WMH, shrinking WMH and normal appearing white matter (NAWM). The average DTI (FA and MD) and FDG-PET (standardized FDG-PET rSUV) of each of the above-mentioned region were extracted and compared. At baseline, the growing WMH had lower FA and FDG-PET rSUV than NAWM, but had higher FA than the constant WMH. Longitudinally, in NAWM, there was a more rapid decline in metabolism compared to WMH areas, while in the growing WMH, a progression in diffusion was found. Finally, we discovered that the shrinking WMH had a similar microstructural and metabolism property and progression to the constant WMH. Our results suggest there are dynamic changes in microstructural and metabolism in WMH. The metabolic change was mainly found in NAWM, while the microstructural change was mainly found in WMH region. Besides, the reduced volume in WMH, to a larger extent, is irrelevant to the microstructural or metabolism recovery.
Keywords: White matter hyperintensities, DTI, FDG-PET, longitudinal analysis, lesion classification
Introduction
White matter hyperintensities (WMH) are a marker of cerebral small vessel disease (CSVD). It is highly prevalent among the elderly. WMH progression is associated with cognitive decline and increased risk of stroke or even mortality.1 But until now, the mechanisms underlying normal appearing white matter (NAWM) developing into white matter hyperintensity are still not fully understood. In previous studies, the progression of WMH is usually presented as volumetric changes between two time points.2 Histopathologically, the progression of WMH is associated with disruption of white matter microstructural integrity, demyelination, axon loss, arteriolosclerosis and capillary loss. Besides, reduced glucose uptake in white matter has also been attributed to neuroglial dysfunction.3 But less is known about the precise microstructural and metabolic changes in the NAWM that later progressed into WMH (referred as “the growing WMH”). By longitudinally examining the diffusion and metabolic property in the growing WMH and comparing it with that of NAWM and the WMH area that is stable during the entire follow-up (constant WMH), we may have a better understanding of the underlying mechanisms of WMH growth.
Furthermore, several recent studies have demonstrated that the progression of WMH may be more dynamic than we previously realized, showing that there could be a regression in WMH volume, which referred to WMH areas that later became NAWM (referred as “shrinking WMH”).4,5 Some longitudinal studies reported negative WMH volume growth, or the range of their WMH progression included negative value. But these studies made no comment on such discoveries and made no further attempt to discover the nature of these shrinking WMH.6,7 Since volumetric analysis alone cannot solve these problems, analyzing the microstructural changes and metabolism changes in these shrinking WMH may provide more insight into its nature.
In view of these gaps, our study sought to compare the WMH areas between initial scan and two-year follow-up scan, identified growing WMH, shrinking WMH, constant WMH and NAWM. We further analyzed the differences in microstructure and metabolism between these areas in different time points. Our first goal is to examine the microstructural and metabolism changes in growing WMH and provide insight into the mechanisms and the pattern of WMH development from MAWM to growing WMH then to constant WMH. Second, we aimed to examine the microstructural and metabolism alteration in the shrinking WMH and help determine its nature.
Methods
Alzheimer's disease neuroimaging initiative
Dataset used in this study was obtained from the Alzheimer's disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, five-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians in developing new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
Subjects
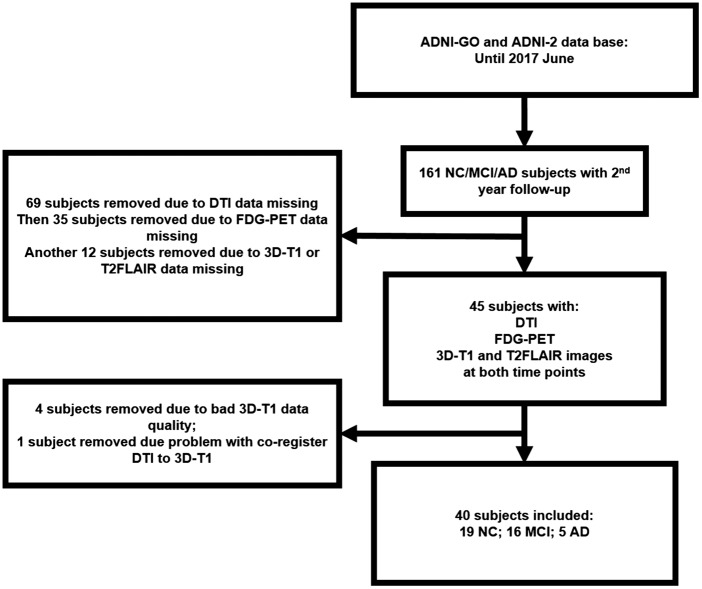
This study was approved by the Institutional Review Boards of all of the participating institutions and informed written consent was obtained from all participants at each site. Currently, the ADNI study includes 59 acquisition sites which can be viewed in detail at this web site: adni.loni.usc.edu/about/centers-cores/study-sites/. The PI of each site is responsible of communications with IRB. The detailed requirement of each site to begin the screening can be found at http://adni.loni.usc.edu/wp-content/uploads/2008/07/adni2-procedures-manual.pdf. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki Declaration and its later amendments. Imaging data and neuropsychological assessment data for each subject were gathered at two time points (baseline and two years follow-up). Using the ADNI GO and ADNI 2 databases, 40 subjects including 5 AD, 16 MCI and 19 cognitive intact elderly were identified in June 2017. According to ADNI manual, diagnosis of the AD was made if the subject had an mini-mental state examination (MMSE) score between 24 and 26 (inclusive), a clinical dementia rating (CDR) of 0.5 or 1.0, and meets NINCDS/ADRDA criteria for the AD. Individuals who were classified as amnestic MCI had an MMSE score between 24 and 30 (inclusive), a memory complaint, objective memory loss measured by Wechsler Memory Scale Logical Memory II, a CDR of 0.5, while essentially preserved activities of daily living and non-demented. On the other hand, all normal control individuals met the following criteria: an MMSE score between 24 and 30 (inclusive), a CDR of 0, non-depressed, non-MCI, and non-demented. The age range of normal subjects will be roughly matched to that of MCI and AD subjects with minimal enrollment under the age of 70. For up-to-date information, see www.adni-info.org. The subject inclusion and exclusion steps are shown in Figure 1.
Figure 1.
Showing the step we used to filter for our subjects. Only subjects with complete T1, T2FLAIR, DTI and FDG-PET scans at both baseline and two-year follow-up were selected.
MRI acquisition
All MR image data were downloaded from the ADNI site in the raw DICOM format. PET image data were preprocessed on the ADNI site (detailed below). All participants were scanned using a 3.0-Tesla MRI scanner. The 3D MPRAGE T1-weighted sequence was acquired using the following parameters: repetition time (TR) =2300 ms; echo time (TE) = 2.98 ms; inversion time (TI) =900 ms; 170 sagittal slices; within plane FOV = 256 ×240 mm2; voxel size = 1.1 × 1.1 × 1.2 mm3; flip angle = 9 °; bandwidth = 240 Hz/pix. The T2 FLAIR scans were obtained using an echo-planar imaging sequence with the following parameters: TR = 9000 ms, TE = 90 ms, and TI = 2500 ms, number of slices = 42, slice thickness = 5 mm. As for the DTI: 256 × 256 matrix; voxel size: 2.7 × 2.7 × 2.7 mm3; TR = 9000 ms; scan time = 9 min. There are 46 separate images acquired for each DTI scan, specifically, 5 T2-weighted images with no diffusion sensitization (b0 images) and 41 diffusion-weighted images (b = 1000 s/mm2). FDG-PET acquisition details have been described elsewhere.8 Briefly, FDG-PET images were acquired by either a 30-min six frame scan acquired 30 to 60 min post-injection or a static 30 min single-frame scan acquired 30 to 60 minute post-injection. Dynamic scans were co-registered to the first frame and averaged to create a single average image. Static or single-frame averaged images were then aligned along the AC-PC line to a standard 160 × 160 × 96 voxel image grid. For PiB-PET, the following imaging parameters were used: Image matrix = 128 × 128, number of slices = 63, pixel resolution = 2 × 2 × 2 mm3; slice thickness = 2.4 mm; radiopharmaceutical = 11C-PiB, reconstruction method =iterative. The methods regarding acquisition of PET was described in detail in Landau et al.8
MRI post-processing
All the post-processing was conducted by our researchers (YJ, XL). First, T2FLAIR, DTI, and FDG-PET image was each co-registered to the 3DT1 image of the same subject. Then, within each subject, the longitudinal image was co-registered to their corresponding baseline imagine to make sure that the images at baseline and follow-up scans were in a common space. The co-registration process was done using an affine co-registration in SPM.
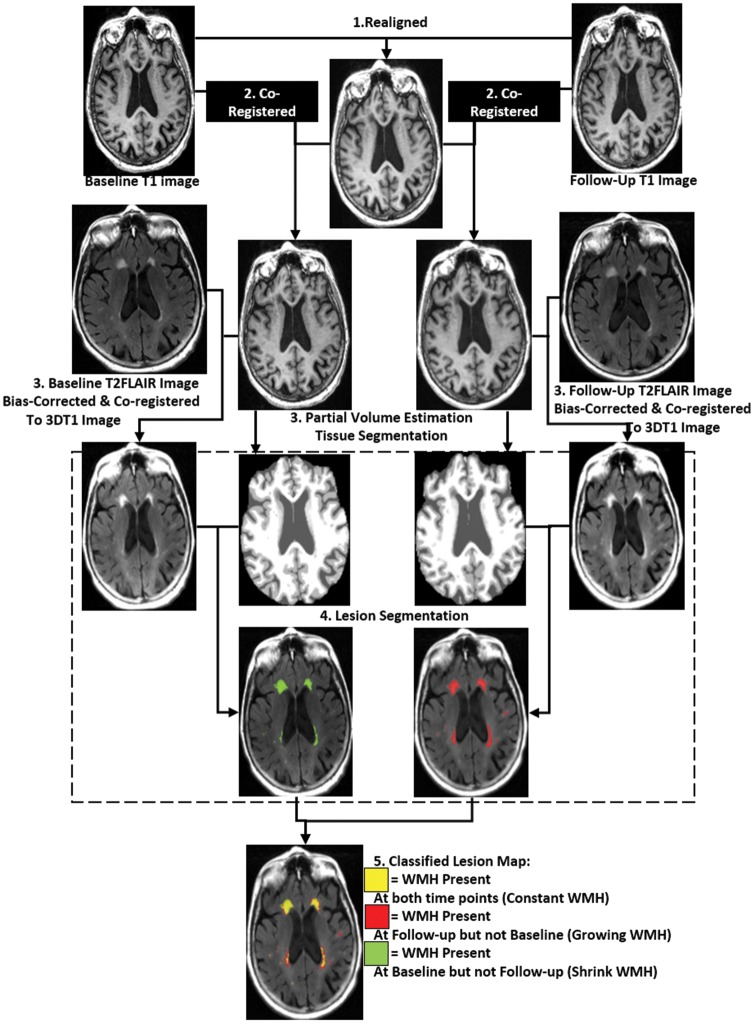
3DT1 and T2FLAIR images were used for tissue segmentation and WMH lesion segmentation. 3DT1 images were used first to segment the brain tissue into three classes: Gray matter, white matter and cerebrospinal fluid. Then T2FLAIR image was used to segment the WMH lesion in white matter tissue. All the previous procedures were completed automatically using the Lesion Segmentation Tool in SPM 12 (Statistical Parametric Mapping). The automatically created WMH segmentation was then manually corrected for: Scalp and tissue at the base of the brain falsely classified as WMH – WMH at the white matter of cerebellum and brain stem. The radiologist (XF Y) was blind to the patient's clinical data or the time point information of the image while manually correcting the WMH segmentation. Then the baseline WMH segmentation and follow-up WMH segmentation were combined; the WMH area existed only in the baseline but not in the follow-up scan was classified as the shrinking WMH. The shrinking WMH around the ventricular wall was removed from the shrinking WMH class because it may be caused by enlarged ventricle. The WMH existed only in the follow-up but not in the baseline scan were classified as the Growing WMH. The WMH existed at both time points were classified as Constant WMH. Furthermore, white matter area that contained no WMH at both time points was considered NAWM (Figure 2.)
Figure 2.
Flowchart demonstrating our longitudinal process for each subject. The 3DT1 images at different time points were first realigned and a with-in subject mean T1 map was created. Then images at different time points were co-registered to the mean T1 map. WMH segmentation was based on T2FLAIR image and partial volume estimated tissue segmentation map.
DTI images were first removed of the scalp and corrected for eddy current, then FA and MD map was calculated based on DTI image using FSL (bet, eddy current, and DTI fit command respectively). Then we applied the growing, constant and shrinking WMH and NAWM ROIs on FA and MD map to extract the average FA and MD value in each region.
FDG-PET preprocessing details have been described elsewhere.9 Each frame of a given baseline image series was co-registered to the first acquired frame. The image series was aggregated into a dynamic image set. The image set was then averaged, reoriented to a standard 160 × 160 × 96 voxel spatial matrix of resliced 1.5 mm3 voxels, intensity normalized, and smoothed with an 8-mm full width at half maximum kernel. We then normalized FDG-PET pixel intensity to the cerebellum, due to its preserved glucose metabolism in AD, to derive the standardized uptake value ratio. This step removes inter-individual variability in tracer metabolism. The growing, constant and shrinking WMH and NAWM ROIs were also applied to the FDG-PET image and standardized-rSUV in each region.
As for the amyloid PET data, composite baseline A-beta deposition value readings were retrieved from the latest available dataset (‘UCBERKELEYAV45_10_17_16’), at ADNI site.
Statistical analysis
ANOVA test and post hoc analysis were first used to determine the differences in FA, MD and FDG-PET rSUV value between different regions (Constant, growing and shrinking WMH and NAWM). A pair-wise t-test between the baseline and follow-up parameter in each region was done to determine if there was significant progression of parameters in each region. Furthermore, repetitive measurements ANOVA were performed to compare if there are group differences between the progressions of each parameter in each region. Linear regression analyses were used to determine the variables that can influence the volume of the expanding WMH. The volume of expanding WMH was the dependent variable of the linear regression model, while the following variables were selected as independent variables: Age, Sex, FA, MD and FDG-PET rSUV in NAWM and expanding WMH. A stepwise method was used to determine in which the independent variable will enter the linear regression model. Furthermore, in order to determine the possible effect of the interaction between clinical diagnose in the subjects (Cognitive normal NC, MCI or AD) on the diffusion and perfusion parameters, we performed another ANOVA test to test the differences in FA, MD and FDG-PET rSUV value between NC, MCI and AD subjects in each region. Also, a Pearson correlation test was conducted to determine the correlation between amyloid deposition and diffusion/metabolism parameters in different WMH regions. Pearson correlations were used to determine the possible correlation between shrinking and growing WMH with brain volume alteration.
Results
The average age of the subjects was 73 ( ± 6.5) year-old and included 35% of female subjects. The average follow-up year was 2.08 ( ± 0.09) year. Between AD, MCI and NC subject group, there were no significant differences in FA, MD and FDG-PET rSUV value in any region. But AD subjects had more WMH volume than MCI subjects, while NC subjects had the least WMH volume. The differences in APOE genotype also had no significant impact on the parameters and WMH volumes. These results are further elaborated in Table 1.
Table 1.
Demographic data.
| Total | Stratified by AD diagnose |
Stratified by APOE genotype |
|||||
|---|---|---|---|---|---|---|---|
| NC | MCI | AD | ɛ23 | ɛ33 | ɛ34 | ||
| N | 40 | 19 | 16 | 5 | 3 | 20 | 17 |
| Sex (female%) | 35.00% | 36.80% | 31.30% | 40.00% | 33.33% | 30.00% | 41.20% |
| Age ± SD (year) | 73.77 ( ± 6.50) | 72.42 ( ± 4.90) | 73.14 ( ± 6.2) | 80.88 ( ± 9.74) | 75.47 ( ± 7.43) | 74.45 ( ± 6.83) | 72.66 ( ± 6.19) |
| Follow-up period ± SD (year) | 2.08( ± 0.10) | 2.1 ( ± 0.11) | 2.07 ( ± 0.08) | 2.04 ( ± 0.05) | 2.13 ( ± 0.15) | 2.1 ( ± 0.11) | 2.05 ( ± 0.06) |
| WMH volume ± SD (mL) | |||||||
| Baseline WMH | 15.57 ( ± 20.75) | 11.11 ( ± 22.81) | 14.37 ( ± 15.63) | 36.17 ( ± 16.24) | 10.02 ( ± 13.53) | 9.11 ( ± 8.75) | 23.84 ( ± 28.14) |
| Follow-up WMH | 20.70 ( ± 26.09) | 15.23 ( ± 28.06) | 16.98 ( ± 17.01) | 53.40 ( ± 21.74) | 11.65 ( ± 14.15) | 13.31 ( ± 13.17) | 30.99 ( ± 35.09) |
| Composite florbetapir rSUV ± SD | |||||||
| Baseline rSUV | 1.15 ( ± 0.21) | 1.03 ( ± 0.08) | 1.24 ( ± 0.22) | 1.34 ( ± 0.27) | 1.02 ( ± 0.07) | 1.06( ± 0.14) | 1.29 ( ± 0.23) |
| Follow-up rSUV | 1.16 ( ± 0.21) | 1.04 ( ± 0.08) | 1.24 ( ± 0.23) | 1.34 ( ± 0.28) | 1.02 ( ± 0.04) | 1.06 ( ± 0.16) | 1.30 ( ± 0.22) |
WMH: white matter hyperintensity; MCI: mild cognitive impairment; AD: Alzheimer's disease.
At baseline, the ANOVA test and the following post hoc analysis of FA, MD and standardized FDG-PET rSUV between different regions (constant, growing, shrinking WMH and NAWM) revealed that there were significant differences between NAWM and all the WMH regions (constant, growing WMH shrinking WMH). Between the WMH regions, the growing WMH had significantly lower MD and higher FA compared to constant WMH, while shrinking WMH had no significant differences in any parameters compared to constant WMH (Tables 2 and 3.)
Table 2.
The average parameter in each regions.
| Constant |
Growing |
Shrink |
NAWN |
P
|
||
|---|---|---|---|---|---|---|
| Number | 37 | 37 | 35 | 40 | ||
| FA (SD) | Baseline | 0.28 (0.05) | 0.32 (0.04) | 0.28 (0.06) | 0.34 (0.03) | <0.01 |
| Follow-up | 0.26 (0.07) | 0.30 (0.08) | 0.26 (0.08) | 0.32 (0.09) | 0.02 | |
| MD (SD) | Baseline | 1.17 (0.16)×10−3 | 1.03 (0.16)×10−3 | 1.25 (0.24)×10−3 | 0.84 (0.04)×10−3 | <0.01 |
| Follow-up | 1.1 9(0.34)×10−3 | 1.06 (0.30)×10−3 | 1.26 (0.39)×10−3 | 0.83 (0.21)×10−3 | <0.01 | |
| FDG-PET rSUV(SD) | Baseline | 0.52 (0.13) | 0.55 (0.13) | 0.56 (0.16) | 0.83 (0.15) | <0.01 |
| Follow-up | 0.52 (0.14) | 0.55 (0.14) | 0.56 (0.17) | 0.82 (0.20) | <0.01 | |
FA: fractional anisotropy; MD: mean diffusivity; FDG-PET: [18F]fluorodeoxyglucose-positron emission tomography; rSUV (SD): standardized uptake value ratio (standard deviation).
Table 3.
Post hoc analysis.
| P value | Constant vs. growing | Constant vs. shrinking | Constant vs. NAWM | Growing vs. shrinking | Growing vs. NAWM | Shrinking VS. NAWM | |
|---|---|---|---|---|---|---|---|
| FA | Baseline | <0.001† | 0.591 | <0.001† | <0.001† | 0.02† | <0.001† |
| Follow-up | 0.043† | 0.684 | 0.003† | 0.013† | 0.33 | 0.001† | |
| MD | Baseline | <0.001† | 0.031† | <0.001† | <0.001† | <0.001† | <0.001† |
| Follow-up | 0.075 | 0.338 | <0.001† | 0.007† | 0.001† | <0.001† | |
| FDG-PET rSUV | Baseline | 0.265 | 0.166 | <0.001† | 0.785 | <0.001† | <0.001† |
| Follow-up | 0.345 | 0.273 | <0.001† | 0.878 | <0.001† | <0.001† | |
†p < 0.05. NAWM: normal appearing white matter; FA: fractional anisotropy; MD: mean diffusivity; FDG-PET: [18F]fluorodeoxyglucose-positron emission tomography; rSUV (SD): standardized uptake value ratio (standard deviation).
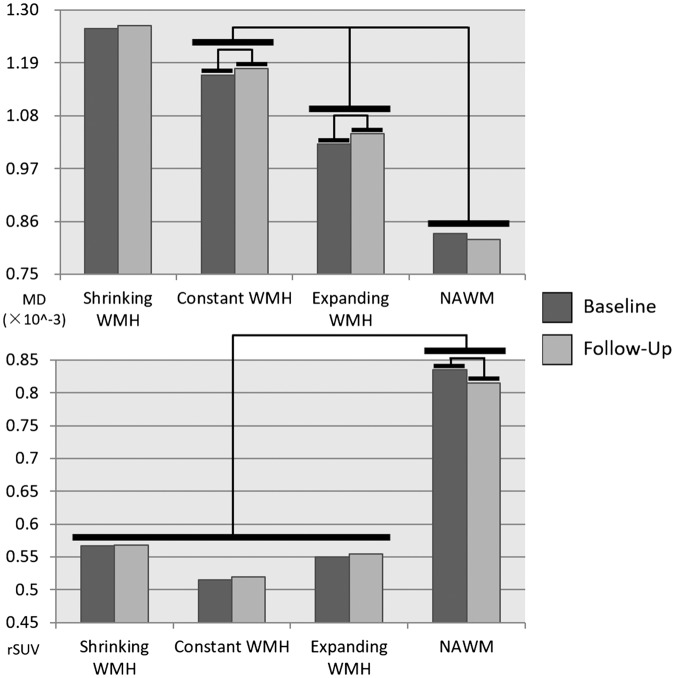
The Pair-wise t-test revealed that, longitudinally, MD had significant progression in growing (P = 0.001), shrinking (P = 0.026) and constant WMH (P = 0.003), while had no significant progression in NAWM (P = 0.659); FDG-PET rSUV had significant negative progression in NAWM (P = 0.005), while had no significant progression in any WMH regions (all P values > 0.05). Further repetitive measurement ANOVA test revealed that there are differences in the progression of MD and rSUV between different regions. (F = 32.075, P < 0.001, Figure 3) Further post hoc analysis revealed that, regarding MD, the constant WMH had the most rapid progression while the growing WMH had relatively slower progression; of FDG-PET rSUV, NAWM had relatively faster progression than any WMH regions (Table 4.)
Figure 3.
Bars showing the differences between different regions and different time points. There were significant differences in MD between constant WMH and Growing WMH and NAWM. While shrinking WMH and constant WMH have similar MD property. And there were significant differences in FDG-PET rSUV between NAWM and WMH regions. There were significant progressions in MD in Constant and Growing WMH, and a significant reduction in FDG-PET rSUV in NAWM region.
Table 4.
Post hoc analysis of longitudinal data.
| Pair(i–j) | FA | MD | rSUV | |
|---|---|---|---|---|
| NAWM-constant | Δmean | 0.057 | −0.00035 | 0.31 |
| Sig. | <0.001† | <0.001† | <0.001† | |
| NAWM-growing | Δmean | 0.021 | −0.00021 | 0.27 |
| Sig. | 0.457 | <0.001† | <0.001† | |
| NAWM-shrink | Δmean | 0.064 | −0.00042 | 0.26 |
| Sig. | <0.001† | <0.001† | <0.001† | |
| Constant-growing | Δmean | −0.037 | 0.00013 | −0.04 |
| Sig. | 0.002† | 0.026† | 1 | |
| Constant-shrink | Δmean | 0.006 | −0.00007 | −0.04 |
| Sig. | 0.582 | 0.665 | 1 |
Note: P value was corrected by Bonferroni method.
NAWM: normal appearing white matter; FA: fractional anisotropy; MD: mean diffusivity; rSUV: standardized uptake value ratio.
Linear regression analysis revealed that among the subjects with expanding WMH, the FA value in the NAWM (standardized β = −2.634, P = 0.002) and age (standardized β = 3.329, P<0.001) had entered the linear regression model. The regression model (R = 0.791, P = 0.002) is: Y(Expanding WMH volume) =0.566X(Age) – 94.437X(Baseline FA in NAWM)
Correlation results revealed that the volume of shrinking WMH (R = −0.03, P = 0.854) and expanding WMH (R = −0.104, P = 0.522) is not significantly correlated with the brain parenchyma volume change between baseline and follow-up scan. And overall, the composite florbetapir rSUV detected by PiB-PET was not significantly correlated with any of the diffusion or metabolism parameter in any region (all P values > 0.05).
Discussion
In the present longitudinal study, we observed the metabolic and diffusion parameters in the NAWM, growing WMH, constant WMH and shrinking WMH region. Specifically, at baseline, the growing WMH had significantly lower FA and FDG-PET rSUV and higher MD than NAWM, and had significantly higher FA and lower MD than the constant WMH. Longitudinally, we found that NAWM had no significant progression in diffusion parameters between the two time points, and the growing WMH had a relatively slower progression than the constant WMH. Regarding metabolism, NAWM had more rapid metabolic progression than any WMH regions (growing, shrinking and constant), while in WMH regions there was no significant progression. Linear regression model also revealed that the baseline microstructural disruption in the NAWM was correlated with the future volume of the growing WMH. Finally, we discovered that the shrinking WMH had a similar microstructural and metabolism property and progression pattern to the constant WMH both before and after the shrinking WMH appears to be normal white matter.
At baseline, we discovered that from NAWM to growing WMH then to constant WMH, each component had more severe diffusion and metabolic damage than the last one. Moreover, the result of the subsequent linear model suggested the disruption of microstructure in NAWM at baseline was associated with the future volume of the growing WMH. In line with previous DTI studies, our results demonstrated that lower FA and higher MD at baseline are good predictors for WMH formation over time.10,11 According to the previous study, the alteration in diffusion suggested compromised white matter integrity and altered water mobility in the interstitial space, which was associated with histopathological findings of diffuse demyelination and axon loss.12 Taken together, these results suggested at the baseline where the growing WMH appeared as normal, the metabolic and microstructural damages were already present. A previous study has also discovered that NAWM in the patients with CSVD has less perfusion compared to controls.13 In addition, our study confirmed that the microstructural and metabolic changes in the NAWM that later became WMH. This suggested a constantly progressing pattern of WMH development. Despite appearing on the T2FLAIR as a homogeneous patch of hyperintensities with a clear border with surrounding white matter, WMH may actually represent a continuous spectrum of white matter damage. Occurring in NAWM, small vessel damage, demyelination and axon loss may slowly turning NAWM into newly formed WMH. Within WMH area, the newly formed WMH has significantly less severe pathological changes than the “older” constant WMH.
Previous studies have discovered that the NAWM may have changes in perfusion and diffusion parameters associated with WMH severity.14,15 Also, a previous study has reported that the NAWM that is closer to WMH have less blood perfusion than average NAWM.16 In line with these previous findings, our longitudinal results showed differences in the rate at which the metabolism and microstructural changes can occur. NAWM have a markedly faster rate of decline in metabolism, but no significant progression in diffusion. On the other hand, regarding the growing WMH and the constant WMH, the declining rate of metabolism is not significant, whereas the microstructures were found to deteriorate more rapidly. These results suggest that the main changes occurring in NAWM are decreasing metabolism, while its microstructure remains relatively intact. As for the WMH regions where the metabolism is already at a lower level, the microstructural changes are more significant.
Interestingly, our findings suggested the shrinking WMH may actually have no significant difference than constant WMH, either metabolism-wise or microstructural-wise, before or after the shrinking WMH appears like normal white matter. A previous study discovered that the edema and cerebrospinal fluid leakage (due to blood-brain barrier disruption) might cause a reversible shift in the water content which may appear hyperintensities on T2FLAIR image.4 Accounting these temporary shifts in water content as WMH may potentially cause WMH regression to appear. But our results suggested however temporary these shifts may be, the microstructural and metabolism alteration in the shrinking WMH area are still on par with that in the WMH area. As such, these white matter areas that appeared normal are actually still damaged. We also noticed that the changes in brain volume may impact the progression and regression of WMH. Thus, we took extra care in the classification of shrinking WMH by removing the shrinking WMH around the periventricular region.
Limitation
There are several potential limitations of our study. First, the T2FLAIR data used in the current study had a slice thickness of 5 mm, which may affect lesion segmentation. Although great effort was taken to account for the partial volume effect, future study with higher resolution image is warranted. Second, the involving of MCI and AD subjects in our study may potentially be a confounding factor. However, our results suggested there were no significant differences of the diffusion and metabolism in all NAWM and WMH regions between three groups. We further tested the correlation relationship between amyloid deposition detected by PiB-PET and diffusion/metabolism values. There were no significant correlations found. Therefore, the lack of significant correlation suggested the potential AD pathology had relatively limited impact on our results. Due to the limited number of AD patients, the relationship between CSVD and AD pathology cannot be determined. Future study should consider recruiting cognitively intact subjects or with large enough AD/MCI samples to determine the effect of AD pathology on CSVD.
Conclusion
There is a dynamic change in microstructural and metabolism in WMH. Specifically, the metabolic change was mainly found in NAWM, while the microstructural change was mainly found in WMH region. Besides, the reduced volume in WMH, to a larger extent, is irrelevant to the microstructural or metabolism recovery.
Acknowledgements
The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuroimaging at the University of Southern California.
Authors' note
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the National Key Research and Development Program of China (NO.2016YFC1306600), and supported by the Fundamental Research Funds for the Central University, NO.2017XZZX001-01), Zhejiang Provincial Natural Science Foundation of China (Grant No. LZ14H180001, Grant No. LY16H090010). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; Bio Clinica, Inc.; Biogen; Bristol-Myers Squibb Company; Cere Spir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; Euro Immun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; MesoScale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org).
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' contributions
Yeerfan Jiaerken: writing the manuscript, data analysis, statistical analysis. Xiao Luo: writing the manuscript, data interpretation. Xinfeng Yu: data interpretation, revising the manuscript of intellectual content. Peiyu Huang: data analysis, statistical analysis. Xiaojun Xu: data interpretation, revising the manuscript of intellectual content. Minming Zhang: supervising of the study, approval of the submission.
References
- 1.Wardlaw JM, Smith EE, Biessels GJ, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 2013; 12: 822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt R, Berghold A, Jokinen H, et al. White matter lesion progression in LADIS: frequency, clinical effects, and sample size calculations. Stroke 2012; 43: 2643–2647. [DOI] [PubMed] [Google Scholar]
- 3.Weber B, Fouad K, Burger C, et al. White matter glucose metabolism during intracortical electrostimulation: a quantitative [(18)F]Fluorodeoxyglucose autoradiography study in the rat. Neuroimage 2002; 16: 993–998. [DOI] [PubMed] [Google Scholar]
- 4.van Leijsen E, de Leeuw FE, Tuladhar AM. Disease progression and regression in sporadic small vessel disease-insights from neuroimaging. Clin Sci 2017; 131: 1191–1206. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez J, McNeely AA, Berezuk C, et al. Dynamic progression of white matter hyperintensities in Alzheimer's disease and normal aging: results from the Sunnybrook dementia study. Front Aging Neurosci 2016; 8: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo X, Jiaerken Y, Yu X, et al. Associations between APOE genotype and cerebral small-vessel disease: a longitudinal study. Oncotarget 2017; 8: 44477–44489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert C, Benjamin P, Zeestraten E, et al. Longitudinal patterns of leukoaraiosis and brain atrophy in symptomatic small vessel disease. Brain 2016; 139: 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landau SM, Breault C, Joshi AD, et al. Amyloid-β imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nucl Med 2013; 54: 70–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jagust WJ, Bandy D, Chen K, et al. The Alzheimer's disease neuroimaging initiative positron emission tomography core. Alzheimers Dement 2010; 6: 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maillard P, Carmichael O, Harvey D, et al. FLAIR and diffusion MRI signals are independent predictors of white matter hyperintensities. Am J Neuroradiol 2013; 34: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot M, Verhaaren BF, de Boer R, et al. Changes in normal-appearing white matter precede development of white matter lesions. Stroke 2013; 44: 1037–1042. [DOI] [PubMed] [Google Scholar]
- 12.Erten-Lyons D, Woltjer R, Kaye J, et al. Neuropathologic basis of white matter hyperintensity accumulation with advanced age. Neurology 2013; 81: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marstrand JR, Garde E, Rostrup E, et al. Cerebral perfusion and cerebrovascular reactivity are reduced in white matter hyperintensities. Stroke 2002; 33: 972–976. [DOI] [PubMed] [Google Scholar]
- 14.van der Veen PH, Muller M, Vincken KL. Longitudinal relationship between cerebral small-vessel disease and cerebral blood flow: the second manifestations of arterial disease-magnetic resonance study. Stroke 2015; 46: 1233–1238. [DOI] [PubMed] [Google Scholar]
- 15.Croall ID, Lohner V, Moynihan B, et al. Using DTI to assess white matter microstructure in cerebral small vessel disease (SVD) in multicentre studies. Clin Sci 2017; 131: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Promjunyakul N, Lahna D, Kaye JA, et al. Characterizing the white matter hyperintensity penumbra with cerebral blood flow measures. Neuroimage Clin 2015; 8: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]