This validation study assesses the clinical validity of molecular relapse detection with circulating tumor DNA analysis in early-stage breast cancer.
Key Points
Question
What is the clinical validity of molecular relapse detection with circulating tumor DNA analysis in early-stage breast cancer?
Findings
This independent, prospective, multicenter, validation study of 101 women early-stage breast cancer assessed circulating tumor DNA mutation tracking and found that detection of circulating tumor DNA during follow-up had a median lead time of 10.7 months compared with clinical relapse, anticipating relapse in all major breast cancer subtypes. Brain-only metastasis was detected less frequently by circulating tumor DNA analysis, potentially requiring alternative surveillance.
Meaning
The findings suggest that molecular relapse detection has high levels of clinical validity, and clinical trials of treatment initiated at molecular relapse without waiting for incurable metastatic disease to develop are needed.
Abstract
Importance
Current treatment cures most cases of early-stage, primary breast cancer. However, better techniques are required to identify which patients are at risk of relapse.
Objective
To assess the clinical validity of molecular relapse detection with circulating tumor DNA (ctDNA) analysis in early-stage breast cancer.
Design, Setting, and Participants
This prospective, multicenter, sample collection, validation study conducted at 5 United Kingdom medical centers from November 24, 2011, to October 18, 2016, assessed patients with early-stage breast cancer irrespective of hormone receptor and ERBB2 (formerly HER2 or HER2/neu) status who were receiving neoadjuvant chemotherapy followed by surgery or surgery before adjuvant chemotherapy. The study recruited 170 women, with mutations identified in 101 patients forming the main cohort. Secondary analyses were conducted on a combined cohort of 144 patients, including 43 patients previously analyzed in a proof of principle study.
Interventions
Primary tumor was sequenced to identify somatic mutations, and personalized tumor-specific digital polymerase chain reaction assays were used to monitor these mutations in serial plasma samples taken every 3 months for the first year of follow-up and subsequently every 6 months.
Main Outcomes and Measures
The primary end point was relapse-free survival analyzed with Cox proportional hazards regression models.
Results
In the main cohort of 101 female patients (mean [SD] age, 54 [11] years) with a median follow-up of 35.5 months (interquartile range, 27.9-43.0 months), detection of ctDNA during follow-up was associated with relapse (hazard ratio, 25.2; 95% CI, 6.7-95.6; P < .001). Detection of ctDNA at diagnosis, before any treatment, was also associated with relapse-free survival (hazard ratio, 5.8; 95% CI, 1.2-27.1; P = .01). In the combined cohort, ctDNA detection had a median lead time of 10.7 months (95% CI, 8.1-19.1 months) compared with clinical relapse and was associated with relapse in all breast cancer subtypes. Distant extracranial metastatic relapse was detected by ctDNA in 22 of 23 patients (96%). Brain-only metastasis was less commonly detected by ctDNA (1 of 6 patients [17%]), suggesting relapse sites less readily detectable by ctDNA analysis.
Conclusions and Relevance
The findings suggest that detection of ctDNA during follow-up is associated with a high risk of future relapse of early-stage breast cancer. Prospective studies are needed to assess the potential of molecular relapse detection to guide adjuvant therapy.
Introduction
Breast cancer is the most frequently diagnosed cancer worldwide, with approximately 95% of women presenting with early-stage breast cancer without macroscopic metastatic disease. Better tools to establish individuals at risk of relapse are needed. Detecting which patients have molecular residual disease (MRD) that has not been eradicated by treatment would allow clinical trials of adjuvant therapies focused on those at highest risk. Several small proof of principle studies have also found that detection of circulating tumor DNA (ctDNA) may present a strategy to identify MRD in patients with breast,1,2 colon,3,4 and lung cancer.5,6 This study assessed the potential of MRD detection in a prospective, multicenter series of patients with primary breast cancer.
Methods
Patients and Sample Collection
A total of 170 patients were recruited from November 24, 2011, to October 18, 2016, from 5 United Kingdom hospitals into 2 prospective ctDNA sample collection studies: the ChemoNEAR study and the Plasma DNA study. Both studies were approved by the East of England Health Research Authority ethics committees (Essex and London as well as Bromley). Written informed consent was obtained from all participants. All data were deidentified. All patients had primary breast cancer without evidence of distant metastatic disease, with staging scans conducted according to local guidelines. Patients scheduled to receive standard treatment with neoadjuvant chemotherapy followed by surgery (n = 140) consented for sample collection before chemotherapy, and patients scheduled to receive adjuvant chemotherapy (n = 30) consented after surgery and before chemotherapy. Plasma samples were collected every 3 months for the first year of follow-up and subsequently every 6 months for 5 years (eMethods and eFigure 1 in the Supplement).
Sample Analysis
Tumor DNA was extracted from the diagnostic biopsy samples and sequenced to identify somatic mutations to track in plasma with a breast cancer driver gene panel (eMethods in the Supplement). Personalized digital polymerase chain reaction (dPCR) assays were designed to track individual somatic mutations in plasma samples. Plasma DNA was extracted and analyzed on a Bio-Rad QX-200 system (eFigure 2 in the Supplement). The dPCR analysis criteria were prespecified.1
Statistical Analysis
The primary study objective was to assess whether patients with ctDNA detected in follow-up blood samples had worse relapse-free survival than patients without ctDNA detected using standard and time-dependent Cox proportional hazards regression models (eMethods in the Supplement). Secondary end points included lead time between ctDNA detection and relapse using Kaplan-Meier methods and association between detection of ctDNA in the diagnosis sample before neoadjuvant chemotherapy using a Cox proportional hazards regression model. Stata, version 13.1 (StataCorp) and GraphPad Prism, version 7 (GraphPad Inc) were used for statistical analyses. All P values were 2-sided and considered to be significant at P < .05.
Results
Patient Cohort
Primary tumor samples from the 170 patients were sequenced to identify somatic mutations, identifying mutations in 101 patients (mean [SD] age, 54 [11] years), which formed the primary analysis cohort (eFigure 3 and eTable 1 in the Supplement). In total, 165 mutations were identified: 78 patients (77.2%) with 1 mutation and 23 patients (22.8%) with multiple mutations, with median allele frequency of 26% (eFigure 4A in the Supplement). Validated personalized dPCR assays were developed for 150 mutations (91.5%) from 101 patients (eFigure 5 in the Supplement).
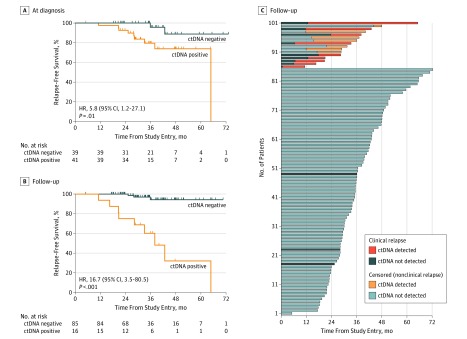
Plasma DNA was extracted from 695 samples (median per patient, 7 samples; interquartile range [IQR], 5-8 samples) and analyzed for the presence of ctDNA. Buffy coat DNA was analyzed to control for clonal hematopoiesis of indeterminate potential (CHIP) (eMethods in the Supplement), with CHIP detected in 3 of the 101 patients (3.0%) (eFigure 6 in the Supplement). In blood samples obtained at diagnosis before any treatment, ctDNA was detected in 41 of 80 patients (51.2%), at a median allele frequency of 0.36% (eFigure 4B in the Supplement). Detection of ctDNA at diagnosis was associated with relapse-free survival (hazard ratio [HR], 5.8; 95% CI, 1.2-27.1) (Figure 1A).
Figure 1. Relapse-Free Survival Among Patients With Circulating Tumor DNA (ctDNA)–Detected Molecular Residual Disease.
A, Relapse-free survival by ctDNA detection at diagnosis before any treatment in patients who subsequently received neoadjuvant chemotherapy. B, Relapse-free survival in 101 patients with ctDNA-detected molecular residual disease during follow-up after neoadjuvant chemotherapy and surgery and in patients without ctDNA-detected molecular residual disease. The population consisted of 35 estrogen receptor–positive and ERBB2-negative cancers, 41 ERBB2-positive cancers, and 25 triple negative breast cancers (eTable 1 in the Supplement). C, Relapse-free survival among individual patients with or without ctDNA detection during the study. Censored patients did not have a clinical relapse at the time of the data collection. HR indicates hazard ratio.
Mutation Tracking to Identify MRD and Anticipate Relapse
At a median follow-up of 35.5 months (IQR, 27.9-43.0 months), MRD was detected in 16 patients at a median allele frequency of 0.16% (eFigure 4C in the Supplement). Median relapse-free survival among patients with ctDNA-detected MRD was 38.0 months (95% CI, 20.8 months to undetermined), with the median not reached in patients without ctDNA-detected MRD (standard HR, 16.7; 95% CI, 3.5-80.5; P < .001) (Figure 1B). Most patients with ctDNA-detected MRD tested negative at the first time point during follow-up and tested positive in a follow-up sample (Figure 1B and C). To account for this, a Cox proportional hazards regression time-dependent model was fitted (time-dependent HR, 25.2; 95% CI, 6.7-95.6; P < .001) (eFigure 7 in the Supplement). Detection of MRD remained highly prognostic in a multivariable model (time-dependent HR, 35.7; 95% CI, 6.0-212.0; P < .001) (eTable 2 in the Supplement) adjusted for clinicopathologic factors (subtype, tumor size, nodal status, and tumor grade), pathologic complete response, and ctDNA detection at diagnosis.
Mutation Tracking in Breast Cancer Subtypes
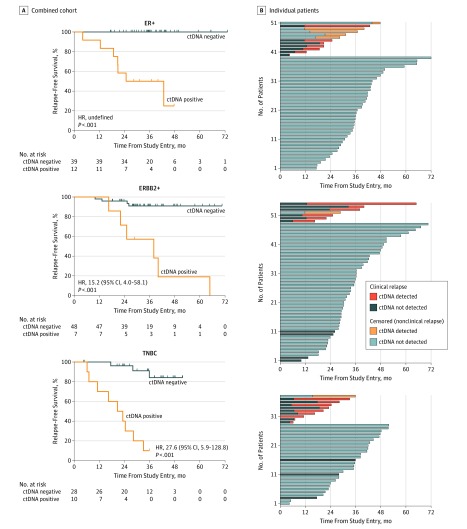
To investigate individual breast cancer subtypes, we conducted a combined analysis of the current study with a prior proof of principle study (eFigure 8 in the Supplement).1 The combined cohort of 144 patients had 210 trackable mutations (eTable 3 and eFigure 9 in the Supplement) and a median follow-up of 36.3 months (IQR, 24.7-41.9 months). Molecular residual disease was detected in 29 patients, which was highly prognostic in a standard (HR, 17.4; 95% CI, 6.3-47.8; P < .001) (eFigure 10 in the Supplement) and time-dependent model (HR, 32.8; 95% CI, 13.5-79.2; P < .001) (eFigure 10 in the Supplement), with a median lead time between ctDNA detection and relapse of 10.7 months (95% CI, 8.1-19.1 months) (eFigure 10 in the Supplement). Detection of ctDNA in follow-up samples was highly prognostic in all major breast cancer subtypes (Figure 2).
Figure 2. Relapse-Free Survival by Tumor Subtype in Patients With Circulating Tumor DNA (ctDNA)–Detected Molecular Residual Disease.
A, Relapse-free survival in the major subtypes of breast cancer in the combined cohort. For the 51 patients with estrogen receptor–positive (ER+) and ERBB2-negative breast cancer, the hazard ratio (HR) was not definable because no patients experienced relapse in the ctDNA-negative group, with a median lead time of 13.3 months (95% CI, 2.1 months to undefined; P < .001). For the 55 patients with ERBB2-positive (ERBB2+) breast cancer, the HR was 15.2 (95% CI, 4.0–58.1, P < .001), with a median lead time of 14.5 months (95% CI, 7.5, months to undefined). For the 38 patients with triple negative breast cancer (TNBC), the HR was 27.6 (95% CI, 5.9-128.8; P < .001), with a median lead time of 10.6 months (95% CI, 0.6-19.1 months). B, Relapse-free survival among patients, in the major subtypes of breast cancer, from study entry and during follow-up. Censored patients did not have a clinical relapse at the time of the data collection.
We investigated the characteristics associated with ctDNA detection at diagnosis in samples obtained before treatment. Patients with triple negative breast cancer (TNBC) had the highest level of ctDNA (median, 4.96 copies/mL; IQR, 0-17.0 copies/mL), those who tested positive for ERBB2 (formerly HER2 or HER2/neu) had intermediate levels (median, 0.81 copies/mL; IQR, 0-5.4 copies/mL), and those who tested positive for estrogen receptor and negative for ERBB2 had the lowest levels (median, 0 copies/mL; IQR, 0-4.4 copies/mL) (P = .004) (eFigure 11 and eTable 4 in the Supplement). Detection at diagnosis was also associated with larger tumor size and higher grade (eTable 4 in the Supplement).
Metastatic Sites Not Detected by Mutation Tracking
Of the 29 patients who relapsed, 23 (88.4%) relapsed with prior ctDNA detection, whereas 6 (21.6%) relapsed without ctDNA detection before or at the time of relapse. All 6 patients had a single site of relapse: 3 brain-only relapses without extracranial relapse, 1 ovarian solitary metastasis, and 2 solitary locoregional relapse (P = .02, Table). Brain-only relapses were unlikely to be detected (Table), similar to the low rates of ctDNA detection in primary brain tumors.7,8,9
Table. Clinicopathologic Factors Associated With Lack of ctDNA Detection Before Disease Relapsea.
| Factor | Recurrence Without ctDNA Detection (n = 6) | ctDNA Detected Recurrence (n = 23) | P Valueb |
|---|---|---|---|
| Age, median (range), y | 55 (43-65) | 51 (45-59) | .72c |
| Sites of recurrence | |||
| Single | 6 (100) | 8 (35) | .02 |
| Multiple | 0 | 14 (61) | |
| Brain only | 3 (50) | 0 | .006 |
| Extracranial | 3 (50) | 23 (100) | |
| Brain only or locoregional | 5 (83) | 1 (4) | <.001 |
| Distant extracranial | 1 (17) | 22 (96) | |
| Pathologic findings | |||
| IDC | 5 (83) | 19 (83) | >.99 |
| Non-IDC | 1 (17) | 4 (17) | |
| Histologic grade | |||
| 2 | 0 | 7 (30) | .28 |
| 3 | 5 (83) | 15 (65) | |
| Subtype | |||
| Estrogen receptor positive and ERBB2 negative | 0 | 7 (30) | .25d |
| ERBB2 positive | 3 (50) | 6 (26) | |
| Triple negative | 3 (50) | 10 (43) | |
| Clinical size at presentation (cT) | |||
| cT2 | 4 (67) | 12 (52) | .66 |
| cT3/4 | 2 (33) | 11 (48) | |
| Nodal status at presentation | |||
| Positive | 3 (50) | 6 (26) | .34 |
| Negative | 3 (50) | 17 (74) |
Abbreviations: ctDNA, circulating tumor DNA; IDC, invasive ductal carcinoma.
Data are presented as number (percentage) of patients unless otherwise indicated.
P values are derived from the Fisher exact text unless otherwise indicated.
Mann-Whitney test.
χ2 Test.
Discussion
We present the results of an independent, prospective, multicenter, validation study of mutation tracking. Detection of ctDNA in follow-up samples was associated with future relapse overall and for all major breast cancer subtypes, with ctDNA detected before relapse in 22 of 23 patients (95.7%) with extracranial distant metastatic relapse. The TNBCs had the highest ctDNA level at diagnosis, likely representing high proliferative rates and cell turnover. Detection of ctDNA at diagnosis, before any treatment, was also associated with risk of relapse, suggesting the potential for incorporation of this feature into future prognostic models if validated in future studies.
CHIP is common with increasing age,10,11 potentially causing false-positive results in ctDNA analysis.12,13,14 We prospectively assessed controls detecting CHIP, all TP53 mutations, in 3 patients who would otherwise have had generated false-positive ctDNA results. These patients remained relapse free after 18.4, 42.3, and 50.7 months of follow-up.
Limitations
Our results demonstrate clinical validity for ctDNA mutation tracking with dPCR but do not demonstrate clinical utility. Without evidence that mutation tracking can improve patient outcome, our results should not be recommended yet for routine clinical practice. For example, protein tumor marker assessment with lead times of a few months did not improve overall survival when assessed in a large study.15
Conclusions
Prospective clinical trials are now required to assess whether detection of ctDNA can improve outcomes in patients, and a phase 2 interventional trial in TNBC has been initiated.16 This trial may develop a new treatment paradigm for treating breast cancer, in which treatment is initiated at molecular relapse without waiting for symptomatic incurable metastatic disease to develop.
eMethods. Supplementary methods
eReferences.
eFigure 1. Personalized digital PCR assays for mutation tracking of circulating tumor DNA in plasma of patients with early breast cancer
eFigure 2. Reproducibility of digital PCR between replicates
eFigure 3. CONSORT diagram for the study
eFigure 4. Identified mutations by massive parallel sequencing.
eFigure 5. Personalized mutation specific digital PCR assays accurately quantify DNA
eFigure 6. Patients with CHIP identified in plasma cfDNA
eFigure 7. Time dependent relapse free survival in patients with ctDNA detected in follow up
eFigure 8. Relapse free survival and overall survival of extension of proof-of-principle study
eFigure 9. CONSORT diagram for the combined analysis
eFigure 10. Time dependent relapse free survival in patients with ctDNA detected in follow up for the combined analysis (A) and (B) relapse free survival for individual patients in the combined cohort from study entry and during follow-up
eFigure 11. Level of ctDNA in diagnosis plasma samples, prior to any treatment, according to subtype in 122 patients that received neoadjuvant chemotherapy
eTable 1. Clinical and pathological characteristics of the study cohort
eTable 2. Covariates used in the multi-variable Cox regression analysis
eTable 3. Clinical and pathological characteristics of combined analysis
eTable 4. Clinical and pathological factors associated with ctDNA level at diagnosis prior to treatment
References
- 1.Garcia-Murillas I, Schiavon G, Weigelt B, et al. . Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021 [DOI] [PubMed] [Google Scholar]
- 2.Olsson E, Winter C, George A, et al. . Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7(8):1034-1047. doi: 10.15252/emmm.201404913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tie J, Wang Y, Tomasetti C, et al. . Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinert T, Schøler LV, Thomsen R, et al. . Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625-634. doi: 10.1136/gutjnl-2014-308859 [DOI] [PubMed] [Google Scholar]
- 5.Abbosh C, Birkbak NJ, Wilson GA, et al. ; TRACERx consortium; PEACE consortium . Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446-451. doi: 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. . Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7(12):1394-1403. doi: 10.1158/2159-8290.CD-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Mattos-Arruda L, Mayor R, Ng CKY, et al. . Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merker JD, Oxnard GR, Compton C, et al. . Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36(16):1631-1641. doi: 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 10.Xie M, Lu C, Wang J, et al. . Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. doi: 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, Kähler AK, Handsaker RE, et al. . Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. doi: 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, Ulrich BC, Supplee J, et al. . False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24(18):4437-4443. doi: 10.1158/1078-0432.CCR-18-0143 [DOI] [PubMed] [Google Scholar]
- 13.Ivey A, Hills RK, Simpson MA, et al. ; UK National Cancer Research Institute AML Working Group . Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374(5):422-433. doi: 10.1056/NEJMoa1507471 [DOI] [PubMed] [Google Scholar]
- 14.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. . Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378(13):1189-1199. doi: 10.1056/NEJMoa1716863 [DOI] [PubMed] [Google Scholar]
- 15.Henry NL, Hayes DF, Ramsey SD, Hortobagyi GN, Barlow WE, Gralow JR. Promoting quality and evidence-based care in early-stage breast cancer follow-up. J Natl Cancer Inst. 2014;106(4):dju034. doi: 10.1093/jnci/dju034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ClinicalTrials.gov A trial using ctDNA blood tests to detect cancer cells after standard treatment to trigger additional treatment in early stage triple negative breast cancer patients (c-TRAK-TN). NCT03145961. https://clinicaltrials.gov/ct2/show/NCT03145961. Accessed June 21, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Supplementary methods
eReferences.
eFigure 1. Personalized digital PCR assays for mutation tracking of circulating tumor DNA in plasma of patients with early breast cancer
eFigure 2. Reproducibility of digital PCR between replicates
eFigure 3. CONSORT diagram for the study
eFigure 4. Identified mutations by massive parallel sequencing.
eFigure 5. Personalized mutation specific digital PCR assays accurately quantify DNA
eFigure 6. Patients with CHIP identified in plasma cfDNA
eFigure 7. Time dependent relapse free survival in patients with ctDNA detected in follow up
eFigure 8. Relapse free survival and overall survival of extension of proof-of-principle study
eFigure 9. CONSORT diagram for the combined analysis
eFigure 10. Time dependent relapse free survival in patients with ctDNA detected in follow up for the combined analysis (A) and (B) relapse free survival for individual patients in the combined cohort from study entry and during follow-up
eFigure 11. Level of ctDNA in diagnosis plasma samples, prior to any treatment, according to subtype in 122 patients that received neoadjuvant chemotherapy
eTable 1. Clinical and pathological characteristics of the study cohort
eTable 2. Covariates used in the multi-variable Cox regression analysis
eTable 3. Clinical and pathological characteristics of combined analysis
eTable 4. Clinical and pathological factors associated with ctDNA level at diagnosis prior to treatment