Abstract
A novel, common, and potent cardiovascular risk factor has recently emerged: clonal hematopoiesis of indeterminate potential (CHIP). CHIP arises from somatic mutations in hematopoietic stem cells that yield clonal progeny of mutant leukocytes in blood. Individuals with CHIP have a doubled risk of coronary heart disease and ischemic stroke, and worsened heart failure outcomes independent of traditional cardiovascular risk factors. The recognition of CHIP as a non-traditional risk factor challenges specialists in hematology/oncology and cardiovascular medicine alike. Should we screen for CHIP? If so, in whom? How should we assess cardiovascular risk in people with CHIP? How do we manage the excess cardiovascular risk in the absence of an evidence base to guide us? This document explains CHIP, explores the clinical quandaries, strives to provide reasonable recommendations for the multidisciplinary management of cardiovascular risk in individuals with CHIP, and highlights current knowledge gaps.
Keywords: leukocyte, cardio-oncology, cancer, leukemia, cardiovascular risk
Condensed Abstract:
A common, potent, and independent cardiovascular risk factor has recently emerged: clonal hematopoiesis of indeterminate potential (CHIP). Somatic mutations in hematopoietic stem cells cause CHIP, yielding clones of mutant leukocytes in blood. CHIP doubles the risk of atherosclerotic events and worsens heart failure outcomes. Should we screen for CHIP? If so, in whom? How should we assess cardiovascular risk in people with CHIP? How do we manage the excess cardiovascular risk? This document explains CHIP, explores the clinical quandaries, strives to provide reasonable recommendations for the multidisciplinary management of cardiovascular risk in individuals with CHIP, and highlights current knowledge gaps.
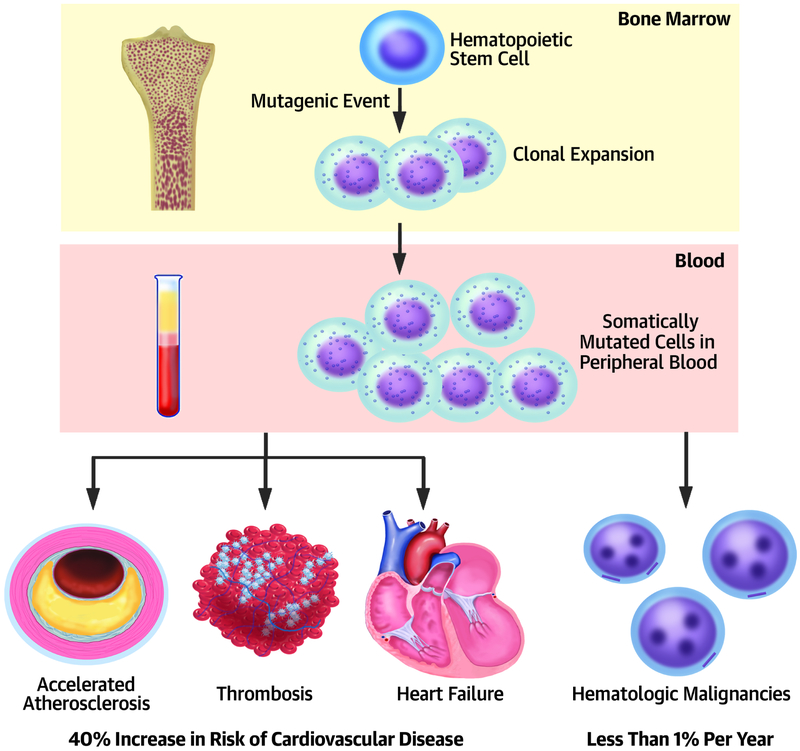
A powerful, previously unrecognized, and independent cardiovascular risk factor lies at the interface of aging, heart disease, and cancer: clonal hematopoiesis of indeterminate potential (CHIP, see Tables 1-3 for definitions).(1-3) With age, we can acquire somatic mutations. When bone marrow hematopoietic stem cells sustain some such genetic alterations in specific genes (Central Illustration, top), these cells can give rise to clones of mutated leukocytes that populate peripheral blood (Central Illustration, middle). This situation differs from cancer, but can be viewed as one step down the path to leukemia (Central Illustration, lower right). Most individuals who harbor these circulating clones of mutated white blood cells will never develop leukemia, hence the term “indeterminate potential.” The transition to acute leukemia usually requires the acquisition of two or three successive mutations in leukemia driver genes in the same leukocyte clone, a relatively rare occurrence that arises only 0.5% -1% per year in CHIP carriers. Yet, CHIP confers a 40% increase in cardiovascular risk, independent of traditional risk factors (Central Illustration, lower left). As up to 20% of septuagenarians have CHIP, this condition comprises a newly recognized, common, and potent cardiovascular risk factor that links with aging and predisposition to hematologic malignancy.
Table 1:
Definitions of some CHIP-related terms
| Name | Abbreviation | Definition |
|---|---|---|
| Variant allelefraction | VAF | The percentage of sequence reads of variant DNA at a locus divided by the overall coverage at that locus.In cancer genetics studies, these sequence variants are tumor-specific somatic mutations not found in germline DNA. |
| Clonal hematopoiesis | CH | Somatic (acquired) mutations in the bone marrow or peripheral blood that can lead to clonal expansion |
| Idiopathic Cytopenias of Undetermined Significance | ICUS | Patients with one or more unexplained cytopenias, who do not meet the diagnostic criteria for myelodysplastic syndrome or other hematologic disorders and do not have known clonal hematopoiesis. |
| Clonal Hematopoiesis of Indeterminate Potential | CHIP | See Table 3 |
| Idiopathic Dysplasia of Undetermined Potential | IDUS | Patients with an unexplained morphological blood cell dysplasia, without cytopenia and do not have clonal hematopoiesis determined either from unrevealing testing or no testing was performed. |
| Clonal Cytopenia of Undetermined Significance | CCUS | Patients with clonal hematopoiesis with somatic mutations associated with hematologic neoplasia, at ≤ 2% variant allele frequency, that also have one or more unexplained cytopenias but do not meet the diagnostic criteria for MDS or another hematologic disorder. |
Table 3:
Current Diagnostic Criteria for Clonal Hematopoiesis of Indeterminate Potential (CHIP)
| – Absence of definitive morphological evidence of a hematological neoplasm |
| – Does not meet diagnostic criteria for paroxysmal nocturnal hemoglobinuria (PNH), Monoclonal gammopathy of unknown significance (MGUS) or Monoclonal B-cell lymphocytosis (MBL) |
| – Presence of a somatic mutation associated with hematological neoplasia at a variant allele fraction of at least 2% |
| – Odds of progression to overt neoplasia are approximately 0.5-1% per year, similar to MGUS Adapted from Steensma et al. (2) |
Central Illustration: Clonal hematopoiesis: a potent newly recognized risk factor for atherothrombosis and adverse heart failure outcomes.
A mutation in a hematopoietic stem cell in the bone marrow confers a proliferative advantage that yields a clone of mutant leukocytes (top panel) that appear in peripheral blood (middle panel). The presence of these clones in blood associates with a heightened risk of atherothrombotic events and with worsened outcomes in patients predisposed to ischemic cardiomyopathy (lower panel, left). Individuals with CHIP transition to acute leukemia only at an annual rate of 0.5-1% (lower panel, left) Thus, for an individual, CHIP may entail a greater risk of cardiovascular events than for cancer.
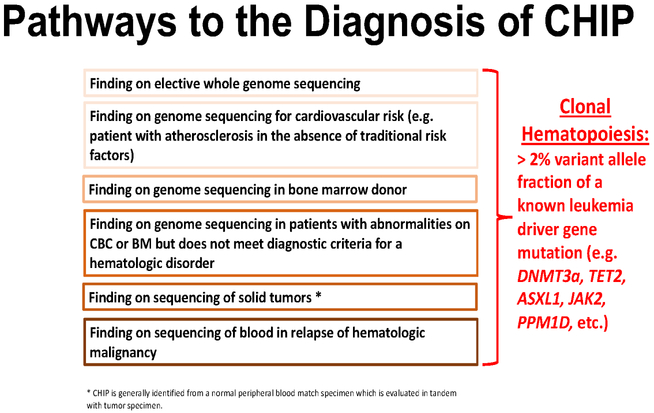
CHIP can be detected through DNA sequencing of peripheral blood, saliva, and tumor samples (through blood contamination) (4-7). Cancer centers increasingly perform DNA sequencing of tumor samples and blood either as a matched normal sample or for the purpose of cancer predisposition germline testing. When blood serves as a control for solid tumor sequencing, 25% of patients have mutations present in the blood and not the tumor, and ~5% of patients have mutations in putative leukemia drivers that define CHIP.(8) The genes most commonly mutated in CHIP lie within the panel of genes mutated in hematologic malignancies (Table 2). Thus, cancer sequencing studies or unbiased genome or exome sequencing studies can incidentally identify individuals with CHIP.(5) As DNA sequencing becomes routine in cancer care, growing numbers of survivors of solid and liquid malignancies will be found to have CHIP. DNA sequencing is increasing in individuals without cancer as well. From genetic predisposition testing to direct-to-consumer genetics products, millions of Americans have undergone genomic profiling.(9) Thus, the detection of CHIP can arise through several portals, ranging from an incidental finding in apparently well individuals to patients with known malignancy (Figure 1). Hence, the management of incidental findings such as CHIP will present an increasing challenge to clinicians of various specialties.
Table 2:
Genes commonly mutated in CHIP
| Gene | Name | Description |
|---|---|---|
| TET2 | Ten-eleven-translocation-2 | A methylcytosine dioxygenase that catalyzes the conversion of 5-methylcytosine to 5-hydroxymethylcytosine. An epigenetic regulator that can activate or repress transcription. |
| DNMT3A | DNA methyltransferase 3A | A de novo DNA methyltransferase. |
| ASXL1 | Additional sex combs-like 1 | Polycomb chromatin-binding protein that is involve in the transcriptional regulation of Hox genes. |
| PPMD1 | Protein phosphatase, magnesium/manganese-dependant 1D | Protein phosphatase involved in dephosphorylating and inactivating proteins in the DNA damage response pathway. |
| SF3B1 | Splicing factor 3B, subunit 1 | A component of the U2 small nuclear riboprotein that binds to the 3’ branch site in pre-mRNA splicing and processing. |
| SRSF2 | Serine/Arginine rich splicing factor 2 | Required for 5’ and 3’ spliceosome assembly, splice-site selection, U1 and U2 snRNP interactions with pre-mRNA, and alternative splicing. |
| TP53 | Transformation-related protein 53 | Tumor suppressor transcription factor that responds to cellular stress and DNA damage. |
| JAK2 | Janus kinase 2 | Receptor tyrosine kinase involved in hematopoietic cytokine signalling and myelopoiesis. |
Figure 1: Pathways to the Diagnosis of CHIP.
The detection of CHIP can arise through several portals: ranging from an incidental finding in apparently well individuals, to patients with known malignancy. This diversity of presentations highlights the need for multidisciplinary approaches to the counseling and management of individuals found to bear CHIP mutations by physicians of different specialties.
Individuals found to have CHIP require expert management of their long-term cardiovascular risk. In addition, as the condition becomes more widely known to physicians and the public, apparently well individuals who seek comprehensive risk assessment, or those with premature atherosclerosis (age less than 60) without apparent risk factors to account for their disease burden may undergo DNA sequencing to identify CHIP. While we do not recommend routine testing for CHIP at this time, these later categories of “worried well” or of secondary prevention patients will present for evaluation by cardiovascular specialists.
Specialists in cardiovascular medicine and in hematology/oncology will need to incorporate the very new research findings that link acquired DNA mutations in blood cells with cardiovascular events into their practices. As a community, we need to counsel and care for individuals with this risk factor, despite the current lack of a firm evidence base. This statement, developed by an expert panel of physicians, aims to provide a summary of our current understanding of CHIP, and proposes a working framework on how to approach screening, diagnosis, and the management of patients with this finding. We also highlight the critical need for further investigations to develop evidence-based screening, surveillance, and management strategies.
Clonal Hematopoiesis of Indeterminate Potential (CHIP): A Newly Recognized and Potent Risk Factor for Cardiovascular Disease
Somatic mutations accumulate during the human lifespan in a wide variety of healthy tissue including normal esophageal tissue(10), the skin(11), and blood.(1,12) In hematopoietic stem cells, certain mutations, all of which are also found in hematologic malignancies, can drive a clonal expansion (Central Illustration, top). Since these mutations do not block hematopoietic differentiation, the mutant progeny of these hematopoietic cells circulate in the peripheral blood (Central Illustration, middle). As expected, the probability of having such a mutant clone in the blood increases with advancing age: by age 70, 10-20% of individuals harbor a leukocyte clone in peripheral blood with a variant allele fraction of at least 2%.(3,12) The consistent increase in the prevalence of CHIP with age may reflect the cumulative duration of exposure to age-ependent mutational processes, environmental mutagens such as radiation (ambient, occupational, diagnostic or therapeutic(8)), tobacco smoke, or air pollutants. Exposure to mutagenic drugs provide a selective pressure for particular CHIP clones. (8,13,14) In addition, impaired DNA repair and altered telomere dynamics may contribute to accumulation of CHIP-associated mutations with age.(15) Recent work has identified these specific mutations as occurring in more than 20 genes commonly implicated in the pathogenesis of myelodysplastic syndrome and acute myeloid leukemia, with the majority of cases of CHIP caused by mutations in only a handful of genes, including DNMT3A, TET2, ASXL1, PPM1D, JAK2, TP53, SF3B1, and SRSF2 (Table 2).
These mutations confer a relatively modest risk of 0.5-1% per year of developing a hematological neoplasm, and most individuals who carry such mutations in hematopoietic cells will never develop hematologic malignancies and will remain asymptomatic with normal blood counts. As noted above, hematologic malignancies generally require the successive acquisition of several subsequent mutations in the same clone.(2) The condition characterized by a mutation associated with a hematological neoplasm in the absence of a hematological neoplasm was defined in 2015 as “clonal hematopoiesis of indeterminate potential” (CHIP), indicating the variable consequences for an individual, ranging from no apparent manifestation to a precursor state for hematologic neoplasms (Table 3). This situation resembles the more familiar case of monoclonal gammopathy of unknown significance (MGUS), wherein an incidentally noted paraprotein spike can presage the development of multiple myeloma. Most individuals with MGUS, however, like those with CHIP, will never progress to a frank malignancy.
The current criteria for the diagnosis of CHIP include a normal peripheral blood count, and a population of mutant cells of at least 2% of the peripheral blood leukocytes (a variant allele fraction or VAF, of > 2 %) (Table 3). This definition excludes several types of clonal hematopoiesis that currently have unclear significance. First, a much larger proportion of individuals have very small clones with a VAF <2%; these have a less well-established clinical impact and also lie beneath the analytical sensitivity (level of detection) of most clinically available, next-generation sequencing assays. Second, some individuals have evidence of an expanded hematopoietic clone without a known leukemia-associated driver mutation; this state is more difficult to detect with targeted sequencing panels, and its impact remains unclear. Finally, the definition of CHIP excludes individuals with overt hematologic abnormalities or malignancies, although all such patients have an expanded clonal population of cells and may also be at increased risk for a cardiovascular event, in part related to chronic anemia and transfusional hemosiderosis.
The identification of asymptomatic carriers of these mutations will doubtless increase as the frequency of people grows who undergo sequencing of their genome, and who have the subsequent incidental detection of blood-restricted mutations. Such individuals have already begun to present to practitioners in search of advice about the implications of the results for their heath and counsel regarding steps that they can take to manage the cardiovascular and oncologic risk.
Different mutations appear to confer a variable risk of transition to acute leukemia. Recent studies show that healthy individuals who carry somatic mutations in DNMT3A, TET2, JAK2, and spliceosome genes such as U2AF1 and SRSF2, and IDH1/2 and TP53, have increased risk of developing acute myeloid leukemia (AML)(16-18). Moreover, the risk of developing AML differed based on the particular CHIP gene mutated in the individual’s clone. For example, TP53 and the spliceosome gene U2AF1 associate with high risk of subsequent myelodysplastic syndromes (MDS) or acute myeloid leukemia (AML), while DNMT3A and TET2 mutations confer a lower risk(16). Despite the less than 1%/year chance of developing leukemia, individuals with CHIP have a 40 % increase in all-cause mortality.(1,12) This increased risk of death by far outstrips that attributable to hematologic malignancy. A series of large populations analyzed by whole exome sequencing revealed that bearers of CHIP have a high prevalence of cardiovascular events and deaths due to myocardial infarction and stroke. Moreover, recent data show that survivors of myocardial infarction with CHIP have increased mortality and worsened heart failure outcomes.(19,20) Thus, the major adult cardiovascular diseases account for the bulk of the mortality associated with CHIP (Central Illustration, bottom left).
These findings present urgent clinical challenges to practitioners. Should we routinely screen broad populations for the presence of CHIP, as we do with individuals who have traditional cardiovascular risk factors such as hypertension and hyperlipidemia? Should we evaluate for the presence of CHIP only in selected populations – those over 65 years of age, or patients with cardiovascular disease without apparent traditional risk factors? After identification of CHIP, what cardiovascular risk factor testing and monitoring should CHIP bearers have? Should all individuals with CHIP undergo further cardiac testing to detect atherosclerosis or myocardial ischemia? What other cardiac or vascular imaging strategies should we consider for monitoring individuals with CHIP? Should cancer patients and survivors with CHIP be managed differently from screened populations? As genome sequencing becomes more prevalent, we must prepare to confront these issues as clinicians who will encounter CHIP bearers with increasing frequency, despite the paucity of presently available data to guide clinical management.
Important questions that require elucidation include whether different CHIP-causing mutations vary in the type, presentation, and severity of cardiovascular complications, similar to the varied risk of leukemia depending on the particular gene mutated in the expanded clone. Should each of the several common CHIP mutations receive the same management strategies to address cardiovascular risk? Does the presence of other risk factors for cardiovascular disease— such as diabetes, dyslipidemia, or tobacco use—modify the effect of CHIP on future risk? Finally, the frequency of follow-up examinations presents additional and ongoing clinical challenges.
Potential mechanisms by which CHIP mutations augment cardiovascular risk
To inform this discussion, we consider some of the mechanisms postulated to link CHIP to cardiovascular events. Age strongly associates with both CHIP and atherosclerotic cardiovascular disease (ASCVD). One might therefore question whether CHIP mutations contribute causally to cardiovascular conditions, merely accompany aging, or reflect a common risk factor (e.g., polymorphisms in DNA repair leading to a greater likelihood of acquisition of mutations, leading to expansion of hematopoietic clones, which in turn can cause accelerated vascular endothelial injury). While there are certainly germline predispositions to CHIP,(14) experimental evidence in mice and cultured cells instead suggest a direct causal relationship between CHIP and cardiovascular events (12).
Mice engineered to bear common mutations in CHIP, e.g. Tet2, on a background of atherosclerosis susceptibility (low-density lipoprotein [LDL] receptor deficiency) show accelerated lesion formation.(12,21) RNA sequencing of cells with loss-of-function mutations in Tet2 show augmented expression of pro-inflammatory mediators implicated in the pathogenesis of atherosclerosis including the cytokines interleukin-1-beta (IL-1nβ) and interleukin-6 (IL-6) when appropriately stimulated with a classical cardiovascular risk factor, LDL (12).
Mice that bear blood cells with loss-of-function mutations of Tet2 have increased plasma concentration of a cluster of chemokines and cytokines. Thus, at both the RNA and protein level, Tet2 appears to potentiate atherogenesis by stimulating inflammation (12,22).
The activation of IL-1β involves the cytoplasmic supramolecular assembly known as the NLRP3 inflammasome. Inhibitors of the inflammasome can limit CHIP-associated accelerated atherogenesis and ischemia-induced heart failure in mice.(21,23) Approved strategies exist for human use of antibodies that inhibit active IL-1β, a key product of the inflammasome or IL-6, another pluripotent pro-inflammatory cytokine itself induced by IL-1.(24) Convincing human genetic data also support the causality of IL-6 signaling in atherosclerotic risk.(25,26) Overall, these findings not only indicate causality of one CHIP mutation with cardiovascular disease, but also have immediate translational implications.
The driver genes that cause CHIP include Janus kinase 2 (JAK2), although mutations in this gene causes CHIP much less frequently than DNMT3A and TET2. The most common CHIP JAK2 mutation is the V617F variant associated with polycythemia vera, other myeloproliferative neoplasms, and idiopathic thromobcytosis. The granulocytes bearing this mutation show heightened sensitivity to the formation of neutrophil extracellular traps (NETs).(27) These structures, comprised of extruded nuclear DNA decorated with proteins implicated in inflammation and coagulation, participate in thrombosis. Granulocytes bearing Jak2V617F exhibit activation of the β1 and β2 integrins that mediate binding to endothelial-leukocyte adhesion molecules, another link with vascular inflammation.(28) Furthermore, introduction of Jak2V617F leukocytes into the bone marrow of atherosclerosis-prone mice enhances the formation of the plaque’s lipid-rich necrotic core due to a defect in clearance of dead leukocytes (a process termed efferocytosis).(29,30) Jak2V617F macrophages also engulf red cells more voraciously compared to wild-type phagocytes. These observations indicate that CHIP due to mutant JAK2 promotes cardiovascular events, at least to some degree, through mechanisms distinct from TET2 mutations that associate with enhanced expression of pro-inflammatory mediators. The observations with Jak2 also have immediate translatability, as a JAK1/2 inhibitor, ruxolitinib, has received approval for treatment of primary myelofibrosis and polycythemia vera. Other JAK inhibitors are in late stages of development.
Should we screen for CHIP?
Despite a lack of consensus guidelines for CHIP screening, diagnosis, and management, a growing need exists for clinical recommendations and building a strategy for evidence in multi-disciplinary settings for patients with CHIP mutations. DNA sequencing has now become routine in the diagnostic workup of patients with established and suspected hematological malignancies, patients with solid tumors, and inherited disorders, and for genetic predisposition testing.(8) While age has the strongest association with CHIP, other factors increase the frequency of CHIP, including smoking, and even germline polymorphisms, e.g. in the telomere protein TERT and in JAK2 and TET2, among many others (25).
Survivors of non-myeloid malignancies also have an increased prevalence of CHIP, particularly those who received cytotoxic chemotherapy and radiation. Individuals with cancer more frequently have CHIP due to mutations in genes involved in DNA damage response, such as TP53 and PPM1D, than do people with CHIP in the absence of malignancy.(6) These patients have a heightened risk of therapy-related MDS or AML, especially in the case of TP53 mutations.(6) Pre-treatment identification of CHIP also associates with increased relapse, poorer outcomes post autologous or allogeneic marrow transplantation, and overall increased mortality (see my review article for all these citations). Cardiovascular specialists who participate in the evaluation and management of cancer survivors with CHIP should integrate overall prognosis and quality of life in their shared decision-making. Cancer patients with limited life expectancy may not require cardiovascular testing and intense risk reduction interventions.
As we lack evidence to guide us in the management of cardiovascular risk and other complications associated with CHIP, we do not presently recommend screening of unselected individuals for CHIP mutations. Yet, obtaining sequencing for CHIP mutations from a cardiovascular perspective might be appropriate for selected individuals with premature or unexpected coronary artery disease in the absence of traditional risk factors. Genotyping for CHIP should involve shared decision-making between an informed individual and the practitioner. Appropriate counseling should be available in the event of identification of a CHIP mutation. Individuals who choose to undergo genotyping for CHIP require full disclosure of the lack of current evidence regarding management of this situation. Targeted next-generation sequencing panels inclusive of CHIP-associated mutations currently cost hundreds of US dollars. At present insurers will often not cover the cost of such tests. The price of testing will certainly fall in the coming years, augmenting the access to detection of CHIP. The clinical effectiveness and cost/benefit aspects of testing in individuals of various risk categories requires further exploration.
Management of cardiovascular risk in individuals with CHIP
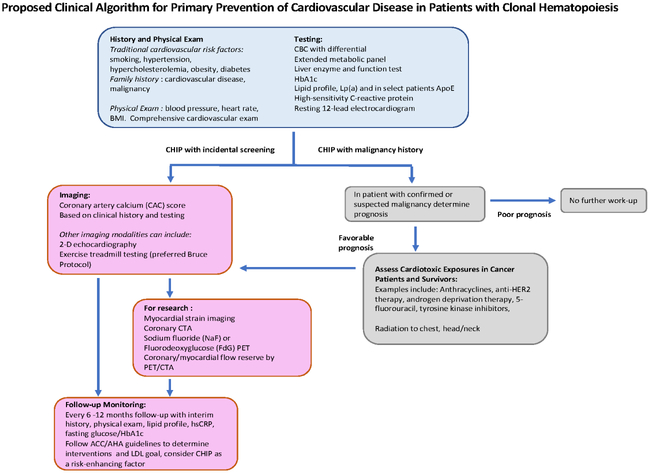
Current guidelines regarding the management of cardiovascular risk factors and the several risk calculators promulgated by various professional organizations do not take into account the enhanced risk of CHIP. However, growing scientific data and cohort studies and advances in the field suggest that CHIP may eventually integrate into the landscape of cardiovascular risk stratification guidelines. Currently, we suggest adopting shared decision-making with CHIP carriers, including the stringent treatment of all modifiable risk factors. We have developed a standard protocol for all individuals with CHIP (Figure 2). We perform a thorough assessment of traditional cardiovascular risk factors, among them tobacco use, family history of premature (age less than 60) atherosclerotic cardiovascular disease, systolic and diastolic blood pressure measurements, physical examination (including body mass index), a lipid panel including LDL, triglycerides, high-density lipoprotein (HDL), very low-density lipoprotein (VLDL), high-sensitivity C-reactive protein (hsCRP), and hemoglobin A1c (HbA1c) and fasting glucose. Further workup includes discussion with the patient of the uncertainties we face in the management of cardiovascular risk due to CHIP, takes into account individual preferences, and offers the most informed advice regarding aggressive control of the aforementioned actionable risk factors, including hypertension, high LDL, diabetes or impaired glucose tolerance, obesity, or tobacco use. Our recommendations to individuals with CHIP emphasize lifestyle measures including tobacco use cessation, weight control, encouragement of regular physical activity, and a heart-healthy diet. An exercise prescription consistent with AHA guidelines consists of at least 150 mins/wk of moderate-intensity exercise or at least 75 mins of vigorous exercise or a combination thereof.
Figure 2: Proposed Clinical Algorithm for Primary Prevention of Cardiovascular Disease in Patients with Clonal Hematopoiesis.
We propose the following pathway for management of individuals with CHIP. We lack an evidence base for such recommendations, as recognition of the relationship between CHIP and cardiovascular disease has only recently become apparent. Yet, clinicians must be able to offer assistance and counseling to CHIP carriers while building a body of evidence.
Lifestyle recommendations in most cases should accompany the discussion of and shared decision-making regarding pharmacologic treatment, including consideration of statins or the cholesterol-absorption inhibitor ezetimibe. When using guidelines to help manage blood cholesterol we suggest consideration of CHIP as an additional “risk-enhancing factor” favoring the initiation of a statin.(31) Two classes of glucose-lowering agents have shown the ability to improve cardiovascular outcomes (certain glucagon-like peptide (GLP) agonists and sodium-glucose co-transporter-2 blockers [SGLT2].) Thus, practitioners may wish to consider these agents that have demonstrated cardiovascular risk reduction for individuals with CHIP and diabetes. The use of aspirin in primary prevention has come under considerable scrutiny in light of recent clinical trials. A possible association of CHIP with intracerebral hemorrhage further discourages aspirin use in those with CHIP who have not had an ischemic event.
Should we image individuals with CHIP?
Current guidelines generally do not endorse screening of unselected individuals with modalities that assess atherosclerotic burden such as coronary artery calcium scoring (CAC) or computed tomographic angiography (CTA). Indeed, the detection of subclinical atherosclerosis in asymptomatic individuals can initiate a chain of events that typically involve further testing, which is often invasive, with obvious consequences including complications and anxiety. The 2018 US Cholesterol Guidelines suggest CAC score as an option to aid decision-making in cases of uncertainty about statin treatment.(31) Although we have no information regarding the regulation of atheroma calcification in individuals with CHIP mutations, implementing the use of CAC score in cases of doubt or discussion regarding statin therapy seems reasonable. The evidence base supporting the use of CTA has grown, and evidence that its judicious use can improve outcomes has begun to emerge (32). Nonetheless, current data do not warrant routine imaging to assess cardiovascular risk in people with CHIP in usual practice.
Yet, in CHIP, we have virtually no understanding of the prevalence of atherosclerosis in various arterial beds compared with individuals matched for age and traditional cardiovascular risk factors. Nor do we have data on the tempo of increase of atherosclerosis burden. To address these knowledge gaps research centers could employ a tiered imaging strategy for CHIP carriers and collate the data systematically to learn more of the natural history of cardiovascular disease from an investigative perspective. Such research protocols might include CAC to assess the presence and overall burden of calcified coronary plaque (Figure 2). Indeed, CHIP associates with increased CAC (12).
Further cardiovascular testing merit consideration in designing research protocols for learning more about the cardiovascular complications associated with CHIP. Such studies include baseline echocardiogram with strain imaging to provide an estimate of ventricular function using a widely available modality, a variable of particular relevance given the emerging relationship between CHIP and heart failure prognosis. A baseline electrocardiographically-monitored exercise stress test can detect silent ischemia, and offer an objective assessment of exercise capacity as well as effort-induced blood pressure response. In research centers, additional imaging with coronary CTA could permit assessment of total plaque volume, both calcified and non-calcified, the severity of any stenoses, and other non-coronary measures such as peri-coronary fat volume and peri-coronary fat attenuation, which may offer a quantitative measure of coronary arterial inflammation. For individuals with known atherosclerosis, myocardial perfusion imaging using PET or CTA can assess the presence and severity of ischemia, as well as calculate myocardial blood flow reserve. Impaired myocardial blood flow reserve can reveal diffuse atherosclerosis and microvascular dysfunction.
Other molecular-imaging techniques such as fluorodeoxyglucose (FdG) uptake or sodium fluoride uptake remain unvalidated with respect to assessing risk or guiding therapy even in patients without CHIP. Nevertheless, their exploratory use may help expand our understanding of CHIP, and thus merit consideration a research environment.
The urgent need for more research
As described above, identification of individuals with CHIP will outstrip our evidence base for managing these individuals. Research centers can meet this challenge by following patients with CHIP, building cohorts, and devising strategies to provide an evidence base for the management of cardiovascular risk in individuals with CHIP (Table 4). Such data will be required before duly constituted organizations will be able to formulate formal guidelines for the managing cardiovascular risk in CHIP carriers.
Table 4:
Selected Research Questions Regarding CHIP-Associated Cardiovascular Complications
| • Does cardiovascular risk and pathogenic mechanisms in CHIP vary with the gene mutated or the specific mutation? |
| • Can therapeutic interventions (e.g. lifestyle modification, medications) alter cardiovascular risk in CHIP, and do so in a mutation dependent manner? |
| • Should the VAF of the mutant gene in CHIP be used in clinical decision making? |
| • To what degree does CHIP interact with other risk factors for cardiovascular disease, including genetic predisposition. |
| • What populations can benefit for screening for CHIP? |
| • At what intervals should individuals with CHIP have follow-up from a clinical effectiveness perspective? |
| • What pathways prove most clinically effective for CHIP carriers with a known/prior malignancy? Should these pathways differ from patients without a cancer diagnosis? |
We are establishing a multi-center registry of individuals with CHIP that will follow a common collection of genetic and clinical information and imaging studies (Table 4). We advocate the acquisition of specimens for storage for future genetic and biomarker analyses, with appropriate informed consent and ability to re-contact individuals. We plan long-term follow-up of individuals in the registry. As we document the natural history of CHIP mutations and acquire non-randomized data regarding the influence of different variants on evolution of cardiovascular biomarkers and events, the registry could provide a foundation for recruiting individuals who might participate in randomized evaluations of different therapies. The presence of particular CHIP mutations might predicate the interventions to be studied.
From the perspective of laboratory investigation, the recognition of the relationship of CHIP to cardiovascular disease opens up a new vista of mechanistic studies. The role of the functions of distinct leukocyte subclasses in atherosclerosis and the ischemically-injured myocardium has burgeoned recently.(33-36) CHIP brings a new dimension to this rapidly growing field, as it identifies the associated somatic mutations beyond heterogeneity defined by cell surface markers as new area to mine mechanistically in the context of cardiovascular disease.
Broadening the Perspective of Cardio-Oncology
The nascent field of cardio-oncology has evolved from the growing recognition of cardiovascular disease in cancer patients and cancer survivors. Both traditional and novel cancer therapies associate with diverse cardiovascular complications during therapy.(37) Cardiovascular disease also represents a major health consideration in the growing number of cancer survivors, numbering nearly 17,000,000 in the United States in 2019. This coalescence calls for cooperation among cardiovascular specialists and oncologists and hematologists to assess and mitigate these risks. Identifying those cancer survivors at elevated risk of cardiovascular disease presents a major challenge. CHIP offers an opportunity to implement personalized medicine in this population based on genotype. CHIP may also add to the growing appreciation of common risk factors that predispose to both cardiovascular diseases and cancer.(38) Reciprocally, conventional cardiovascular risk factors—classically smoking but also obesity, dyslipidemia, and diabetes—may enhance the risk of cancer. In contrast to inherited Mendelian germline mutations, these acquired somatic mutations present a new challenge, but also an opportunity for ongoing collaborations within the community. The recognition of CHIP strengthens the link between oncology and cardiovascular disease, and enlarges the purview of cardio-oncology to embrace prospective management of cardiovascular risk by close collaboration between hematologists, oncologists, and cardiovascular specialists.(38)
Bullet Points.
Aging humans commonly develop leukocyte clones in blood due to somatic mutations in stem cells.
Clonal hematopoiesis constitutes an independent cardiovascula risk factor.
People with clonal hematopoiesis will increasingly present to cardiovascular specialists for management.
We review clonal hematopoiesis and present an approach for dealing with this condition in practice.
Research is needed to obtain evidence to guide management of this newly recognized entity.
Acknowledgments
Funding: PL is funded by the National Heart, Lung, and Blood Institute (R01HL080472); the American Heart Association (18CSA34080399); and the RRM Charitable Fund. RS is supported by the NIH/NCI Cancer Center Support Grant (P30 CA008748). AEL is supported by the John S. LaDue Memorial Fellowship in Cardiology. LWJ is supported by research grants from the National Cancer Institute, the KavliTrust, AKTIV Against Cancer, and the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748). JM is funded by the NIH (R56HL141466), Pfizer, and Bristol Myers Squibb. AZ is supported by the German Research Foundation (DFG) and the German Center for Cardiovascular Research (DZHK). SJ is supported by the Burroughs Wellcome Foundation and The Edward P. Evans Foundation. CS is supported by the German Research Foundation (DFG) and the German Center for Cardiovascular Research (DZHK). RB is supported by Amgen Inc, and Astellas Inc. KLB received funding from GRAIL. DS reports funding from the Edward P Evans Foundation and the James and Lois Champy Fund. RLL was supported in part by MSKCC Support Grant/Core Grant (P30 CA008748). BLE reports funding from the Leducq Foundation and the Howard Hughes Medical Institute. DG reports no funding.
Abbreviations:
- CHIP
clonal hematopoiesis of indeterminate potential
- AML
acute myeloid leukemia
- MGUS
monoclonal gammopathy of unknown significance
- ASCVD
atherosclerotic cardiovascular disease
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- VLDL
very low-density lipoprotein
- hsCRP
high-sensitivity C-reactive protein
- HbA1c
hemoglobin A1c
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: PL is an unpaid consultant to, or involved in clinical trials for Amgen, AstraZeneca, Esperion Therapeutics, Ionis Pharmaceuticals, Kowa Pharmaceuticals, Novartis, Pfizer, Sanofi-Regeneron, and XBiotech, Inc. PL is a member of scientific advisory board for Amgen, Corvidia Therapeutics, DalCor Pharmaceuticals, IFM Therapeutics, Kowa Pharmaceuticals, Olatec Therapeutics, Medimmune, Novartis, and XBiotech, Inc. Dr. Libby’s laboratory has received research funding in the last 2 years from Novartis. LWJ reports stock ownership in Pacylex, Inc. JM is on the advisory board for Pfizer, Novartis, Bristol Myers Squibb, Takeda, Myokardia, and Deciphera. AZ is an advisory Board Member for Sanofi, Pfizer, Boehringer Ingelheim and Amgen. SJ reports a relationship with GRAIL and patent applications related to the subject. BLE has received consulting fees from GRAIL and research funding from Celgene and Deerfield. DS reports Independent Data Safety Monitoring Committee service for Onconova, Janssen, Takeda, and Pharmessentia; Institutional research funding for trials on which he is local or overall Principal Investigator: H3 Biosciences, Aprea, Celgene Consulting: Sensei, Otsuka, Stemline. RLL is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics and Isoplexis, which each include an equity interest. He receives research support from and consulted for Celgene and Roche, he has received research support from Prelude Therapeutics, and he has consulted for Incyte, Novartis, Astellas, Morphosys and Janssen. He has received honoraria from Lilly and Amgen for invited lectures and from Gilead for grant reviews. DG, KLB, RS, AEL, CS, and RB report no disclosures.
References
- 1.Jaiswal S, Fontanillas P, Flannick J et al. Age-related clonal hematopoiesis associated with adverse outcomes. New England Journal of Medicine 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steensma DP, Bejar R, Jaiswal S et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busque L, Buscarlet M, Mollica L, Levine RL. Concise Review: Age-Related Clonal Hematopoiesis: Stem Cells Tempting the Devil. Stem Cells 2018;36:1287–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleppe M, Comen E, Wen HY et al. Somatic mutations in leukocytes infiltrating primary breast cancers. NPJ Breast Cancer 2015;1:15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ptashkin RN, Mandelker DL, Coombs CC et al. Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors. JAMA Oncol 2018;4:1589–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolton KL, Gillis NK, Coombs CC et al. Managing Clonal Hematopoiesis in Patients With Solid Tumors. J Clin Oncol 2018:JCO1800331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinds DA, Barnholt KE, Mesa RA et al. Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood 2016;128:1121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombs CC, Zehir A, Devlin SM et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017;21:374–382 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krier JB, Kalia SS, Green RC. Genomic sequencing in clinical practice: applications, challenges, and opportunities. Dialogues Clin Neurosci 2016;18:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martincorena I, Fowler JC, Wabik A et al. Somatic mutant clones colonize the human esophagus with age. Science 2018;362:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martincorena I, Roshan A, Gerstung M et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaiswal S, Natarajan P, Silver AJ et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. New England Journal of Medicine 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TN, Miller CA, Jotte MRM et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat Commun 2018;9:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh P-R, Genovese G, Handsaker RE et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018;559:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aviv A, Levy D. Hemothelium, Clonal Hematopoiesis of Indeterminate Potential, and Atherosclerosis. Circulation 2019;139:7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abelson S, Collord G, Ng SWK et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai P, Mencia-Trinchant N, Savenkov O et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 2018;24:1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sellar RS, Jaiswal S, Ebert BL. Predicting progression to AML. Nat Med 2018;24:904–906. [DOI] [PubMed] [Google Scholar]
- 19.Dorsheimer L, Assmus B, Rasper T et al. Association of Mutations Contributing to Clonal Hematopoiesis With Prognosis in Chronic Ischemic Heart Failure. JAMA Cardiol 2019;4:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Libby P, Jaiswal S, Lin AE, Ebert BL. CHIPping Away at the Pathogenesis of Heart Failure. JAMA Cardiol 2018. [DOI] [PubMed] [Google Scholar]
- 21.Fuster JJ, MacLauchlan S, Zuriaga MA et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Libby P, Ebert B. CHIP (Clonal Hematopoiesis of Indeterminate Potential): Potent and Newly Recognized Contributor to Cardiovascular Risk. Circulation 2018;138:666–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-Mediated Gene Editing to Assess the Roles of Tet2 and Dnmt3a in Clonal Hematopoiesis and Cardiovascular Disease. Circ Res 2018;123:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Libby P Interleukin-1 Beta as a Target for Atherosclerosis Therapy: Biological Basis of CANTOS and Beyond. J Am Coll Cardiol 2017;70:2278–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swerdlow DI, Holmes MV, Kuchenbaecker KB et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 2012;379:1214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarwar N, Butterworth AS, Freitag DF et al. IL6R Genetics Consortium Emerging Risk Factors Collaboration. Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet 2012;379:1205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolach O, Sellar RS, Martinod K et al. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelmann B, Gupta N, Schnoeder TM et al. JAK2-V617F promotes venous thrombosis through β1/β2 integrin activation. J Clin Invest 2018;128:4359–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Liu W, Fidler T et al. Macrophage Inflammation, Erythrophagocytosis, and Accelerated Atherosclerosis in Jak2V617F Mice. Circulation Research 2018;123:e35–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libby P, Molinaro R, Sellar Rob S, Ebert Benjamin L. Jak-ing Up the Plaque’s Lipid Core… and Even More. Circulation Research 2018;123: 1180–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundy SM, Stone NJ, Bailey AL et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2018. [Google Scholar]
- 32.Investigators S-H. Coronary CT Angiography and 5-Year Risk of Myocardial Infarction. New England Journal of Medicine 2018. [DOI] [PubMed] [Google Scholar]
- 33.Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science 2013;339:161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libby P, Nahrendorf M, Swirski FK. Leukocytes Link Local and Systemic Inflammation in Ischemic Cardiovascular Disease. Journal of the American College of Cardiology 2016;67:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swirski FK, Nahrendorf M, Libby P. Mechanisms of Myeloid Cell Modulation of Atherosclerosis. Microbiol Spectr 2016;4. [DOI] [PubMed] [Google Scholar]
- 36.Lavine KJ, Pinto AR, Epelman S et al. The Macrophage in Cardiac Homeostasis and Disease: JACC Macrophage in CVD Series (Part 4). J Am Coll Cardiol 2018;72:2213–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campia U, Moslehi JJ, Amiri-Kordestani L et al. Cardio-Oncology: Vascular and Metabolic Perspectives: A Scientific Statement From the American Heart Association. Circulation 2019: CIR0000000000000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libby P, Kobold S. Inflammation: a common contributor to cancer, aging, and cardiovascular diseases-expanding the concept of cardio-oncology. Cardiovasc Res 2019;115:824–829. [DOI] [PMC free article] [PubMed] [Google Scholar]