Significance
Posttranslational protein modification by ubiquitin (Ub) regulates aspects of biology, including protein turnover and the cell cycle. Proteins and enzymes that promote Ub conjugation are therapeutic targets because they are sometimes dysregulated in cancer, neurodegenerative diseases, and other disorders. Ub conjugation is initiated by a Ub-activating enzyme that adopts different conformations to catalyze Ub activation, Ub-activating enzyme thioester bond formation, and thioester bond transfer to Ub-conjugating enzymes. Here, we illuminate 2 uncharacterized states for Ub-activating enzyme, one bound to pyrophosphate prior to thioester bond formation and one captured during thioester bond formation. These structures reveal key differences and similarities among activating enzymes for Ub and SUMO with respect to conformational changes that accompany thioester formation.
Keywords: ubiquitin, adenylation, thioester, X-ray, E1
Abstract
The ubiquitin (Ub) and Ub-like (Ubl) protein-conjugation cascade is initiated by E1 enzymes that catalyze Ub/Ubl activation through C-terminal adenylation, thioester bond formation with an E1 catalytic cysteine, and thioester bond transfer to Ub/Ubl E2 conjugating enzymes. Each of these reactions is accompanied by conformational changes of the E1 domain that contains the catalytic cysteine (Cys domain). Open conformations of the Cys domain are associated with adenylation and thioester transfer to E2s, while a closed conformation is associated with pyrophosphate release and thioester bond formation. Several structures are available for Ub E1s, but none has been reported in the open state before pyrophosphate release or in the closed state. Here, we describe the structures of Schizosaccharomyces pombe Ub E1 in these two states, captured using semisynthetic Ub probes. In the first, with a Ub-adenylate mimetic (Ub-AMSN) bound, the E1 is in an open conformation before release of pyrophosphate. In the second, with a Ub-vinylsulfonamide (Ub-AVSN) bound covalently to the catalytic cysteine, the E1 is in a closed conformation required for thioester bond formation. These structures provide further insight into Ub E1 adenylation and thioester bond formation. Conformational changes that accompany Cys-domain rotation are conserved for SUMO and Ub E1s, but changes in Ub E1 involve additional surfaces as mutational and biochemical analysis of residues within these surfaces alter Ub E1 activities.
Ubiquitin (Ub) and Ub-like (Ubl) modifiers constitute a family of small proteins that regulate signaling, localization, and turnover of proteins through posttranslational modification (PTM) of substrates via conjugation of their C termini to substrates (1, 2). Conjugation most often occurs on lysine side chains to form an isopeptide bond between the Ub/Ubl C-terminal glycine and the ε-nitrogen of the substrate lysine (3, 4). Each Ub/Ubl family member requires a cascade of enzyme activities to promote conjugation to particular substrates (5–10). Ub/Ubl signaling can be reversed or regulated by deconjugation via proteases that remove Ub/Ubls from substrates (11).
Canonical Ub/Ubl conjugation cascades entail adenosine 5′-triphosphate (ATP)-dependent Ub/Ubl adenylation by an E1 activating enzyme (AE), formation of a high-energy thioester bond between a Ub/Ubl and AE, thioester transfer to an E2 conjugating enzyme, and formation of an amide bond after an amine substrate attacks the E2∼Ub/Ubl thioester. This last step can be catalyzed by E3 protein ligases either noncovalently or by formation of an E3∼Ub/Ubl thioester bond before conjugation (12–14). Adenylate-forming enzymes that use ATP to activate carboxylic acid substrates for subsequent conversion to thioesters and other metabolic intermediates are widely distributed outside the Ub/Ubl pathway, for example, in prokaryotic nonribosomal peptide synthetases, acyl-coenzyme A (CoA) synthetases, and firefly luciferase (15–18). Early structural characterization of acyl-CoA synthetases revealed that they use domain alternation to remodel active sites and switch between adenylation to thioesterification activities (19).
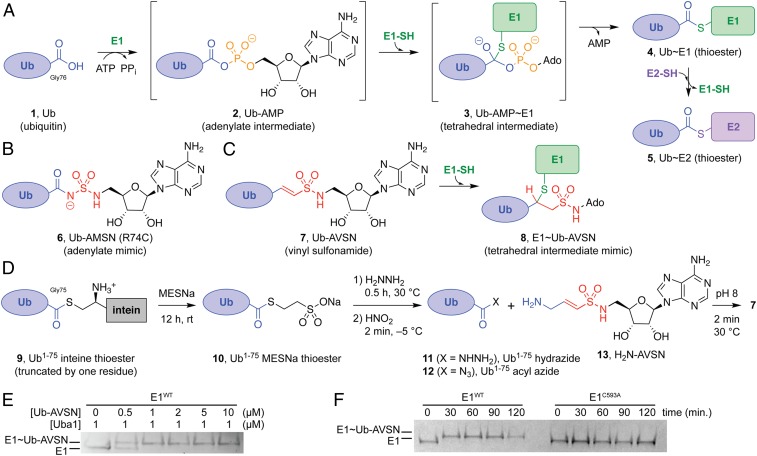
Uba1 is the Ub AE (UAE) for Ub, although Ub can also be activated by the Uba6 E1 in vertebrates (5). Similarly to AEs for the Ubl proteins SUMO, NEDD8, FAT10, and ISG15 (5), UAE binds ATP, Mg2+, and Ub to catalyze adenylation of the Ub C-terminal glycine (1; Fig. 1), forming a Ub-adenylate [Ub-adenosine 5′-monophosphate (Ub-AMP); 2] and pyrophosphate (PPi) (20, 21). After PPi release, Ub is transferred to the E1 catalytic cysteine by nucleophilic attack on the Ub-AMP via a tetrahedral intermediate (3), forming a thioester bond (E1∼Ub; 4) with loss of AMP. After AMP release from the active site, the adenylation active site can bind a second equivalent of Ub, ATP, and Mg2+ to create a doubly loaded E1 complex, with one Ub covalently bound to the second catalytic cysteine half-domain (SCCH) (Uba1∼Ub) and a second Ub bound noncovalently in the adenylation active site. This E1 ternary complex is best able to transfer the thioester from the E1 catalytic cysteine (E1∼Ub) to an E2 catalytic cysteine (E2∼Ub, 5) (Fig. 1A; see also SI Appendix, Fig. S1) (22).
Fig. 1.
Biochemistry and probes of Ub E1 AEs. (A) Ub E1s catalyze adenylation of the Ub C-terminal glycine-76 (1 → 2), thioesterification with an E1 catalytic cysteine (2 → 3 → 4), and transthioesterification to an E2 catalytic cysteine (4 → 5). (B) Ub-AMSN (6) mimics the adenylate intermediate (2); the R74C mutation results from the native chemical ligation protocol used in its synthesis (29, 37). (C) Ub-AVSN (7) reacts with the E1 catalytic cysteine to form a mimic (8) of the tetrahedral intermediate (3). (D) Ub-AVSN (7) is semisynthesized from a truncated Ub1–75 intein fusion protein (9) expressed in E. coli and synthetic H2N-AVSN (13) (see SI Appendix for full details). (E) Titration of Ub-AVSN in Uba1 E1 cross-linking reaction: 1 μM Uba1 was incubated with the indicated concentrations of Ub-AVSN for 2 h at room temperature. (F) Uba1∼Ub-AVSN thioether formation was complete after 30 min at room temperature, using a 2:1 ratio of Ub-AVSN:Uba1, but did not occur with a Uba1(C593A) mutant that lacks the catalytic cysteine. Ado, adenosine.
The first representative structures for E1 AEs for NEDD8 (NAE), SUMO (SAE), and UAE revealed commonalities with respect to domain architectures and conformations (23–25). This includes pseudosymmetric active (AAD) and inactive (IAD) adenylation domains that bind ATP, Mg2+, and Ub/Ubl (23); a Ub-fold domain (UFD) that participates in E2 selection (24, 26, 27); and a Cys domain that contains the catalytic cysteine required for thioester bond formation (23–25). Although UAE shares many features with other canonical AEs, its Cys domain is larger and has been divided into first and second catalytic cysteine half-domains (FCCH and SCCH), with the SCCH domain containing the active site Cys residue.
Canonical E1s adopt at least 2 conformations, an open conformation that promotes adenylation (1 → 2) and thioester bond transfer to E2 (4 → 5) and a closed conformation that transits the E1 cysteine more than 30 Å to promote thioester bond formation (2 → 3 → 4) (SI Appendix, Fig. S1) (28, 29). Many structures of canonical E1s have been captured in open conformations as apo enzymes or bound to Ub/Ubl, ATP, Mg2+, and/or E2s (23–27, 29–36), but only one has been captured for a canonical E1 (SUMO E1) in the closed conformation (29). This was accomplished by using a semisynthetic probe (SUMO-AVSN) having a vinylsulfonamide electrophile in place of the glycyl-phosphate linkage in the native SUMO-AMP intermediate (37). Reaction of the SUMO E1 with SUMO-AVSN generated a stable thioether bond between the E1 cysteine and the vinylsulfonamide to form a tetrahedral intermediate mimic (29). The resulting crystal structure revealed that thioester bond formation involved a 130° rotation of the SCCH domain and remodeling of key structural elements that were required for adenylation in step 1 of the E1 catalytic cycle. Aside from the SAE–SUMO-AVSN structure, canonical E1 enzymes have only been captured in open conformations, with the exception of 2 structures in which the Cys domain of UAE or SAE was observed in nonproductive conformations with small-molecule inhibitors (32, 38). Considerable variation within domain architectures also raises the question as to whether other AEs adopt similar conformations to that observed for SAE during thioester bond formation.
Several therapeutic candidates target proteins in the Ub–proteasome system (39–41), including NAE (42), UAE (43), and SAE (44). Further development of these agents might benefit from a complete understanding of the catalytic cycle for each type of E1. Here, we address 2 gaps in our understanding of the Ub E1 catalytic cycle by determining the structure of Uba1 bound to a Ub-adenylate mimetic (Ub-AMSN, 6; Fig. 1B) before release of pyrophosphate and by developing methods to generate a Ub-vinylsulfonamide electrophile (Ub-AVSN, 7; Fig. 1C) with suitable yields and specific activity to capture and structurally characterize a mimetic of Uba1 during thioester bond formation. Relative to a previously reported UAE/Ub/ATP/Mg2+ structure (27), the UAE/Ub-AMSN/pyrophosphate structure reveals changes in the active site that are consistent with in-line attack by the C-terminal Ub carboxylate at the ATP α-phosphate. The UAE∼Ub-AVSN structure reveals conformational changes that are analogous to SAE∼SUMO-AVSN (29), but the larger SCCH and FCCH domains in UAE involve additional contacts between E1 domains in both open and closed conformations that contribute to the UAE catalytic cycle. Comparison of the SUMO and Ub E1s suggests that other canonical E1s, such as those for NEDD8, ISG15, and FAT10 (5), adopt similar closed conformations during thioester bond formation.
Results
Generation of Ub Adenylate Mimetics.
Our prior studies reported generation of Ub/Ubl-adenylate mimetics through intein-mediated ligation of truncated Ub/Ubl proteins having a C-terminal thioester with synthetic peptidyl-sulfamoyladenosine derivatives having an N-terminal cysteine (29, 37). These studies resulted in 2 adenylate mimetics that could be used to characterize E1s after adenylation and during thioester bond formation. While this approach yielded sufficient quantities of Ub-AMSN (6) for structural studies herein, the yield and activity of Ub-AVSN (7) was insufficient for structural studies. To address this problem, we developed an alternative method for the semisynthesis of Ub/Ubl-AVSN probes that does not rely on a cysteine-mediated ligation, thus avoiding potential intra- or intermolecular attack of the cysteine thiol on the vinylsulfonamide electrophile. Wilkinson and coworkers have previously reported the synthesis of a C-terminally modified Ub analog by enzymatic conversion of Ub to a one-residue truncated Ub1–75 ethyl ester, conversion to the corresponding acyl azide, and aminolysis with the desired C-terminal fragment (45, 46). Merging this chemical approach with our intein thioester method, we expressed an Ub1–75 intein fusion protein in Escherichia coli to generate the thioester intermediate 9 (Fig. 1D) (47). Treatment with mercaptoethanesulfonic acid, sodium salt (MESNa) afforded the Ub1–75 MESNa thioester 10, which was then converted to the corresponding hydrazide 11 and acyl azide 12 (45, 46). Synthesis of H2N-AVSN (13) was achieved via straightforward modification of our established synthetic route to peptidyl-AVSN fragments (37) (SI Appendix, Fig. S2 and SI Appendix, Synthesis of H2N-AVSN). Finally, coupling of Ub1–75 acyl azide (12) with H2N-AVSN (13) proceeded rapidly and efficiently as assessed by liquid chromatography–mass spectrometry (LC-MS) analysis.
This aminolysis strategy provides 3 advantages over our previous cysteine-mediated ligation approach: (i) the Ub-AVSN conjugate is formed within 2 min, compared with 12 to 18 h using the cysteine-mediated ligation strategy; (ii) the resulting Ub-AVSN conjugate retains the native Arg74 residue, avoiding potential artifacts due to substitution of this position with the cysteine used in the cysteine-mediated ligation strategy (37); and (iii) the H2N-AVSN probe can be prepared in fewer synthetic steps than the corresponding cysteine-functionalized AVSN fragment. The Ub-AVSN probe appears to have high specific activity, as cross-linking was complete at a 1:1 ratio of E1:Ub-AVSN (Fig. 1E), and high specificity, in that no cross-linking was observed to a UAE C593A mutant that lacks the catalytic cysteine (Fig. 1F).
Structures of Uba1/Ub-AMSN/PPi/Mg2+ and Uba1∼Ub-AVSN.
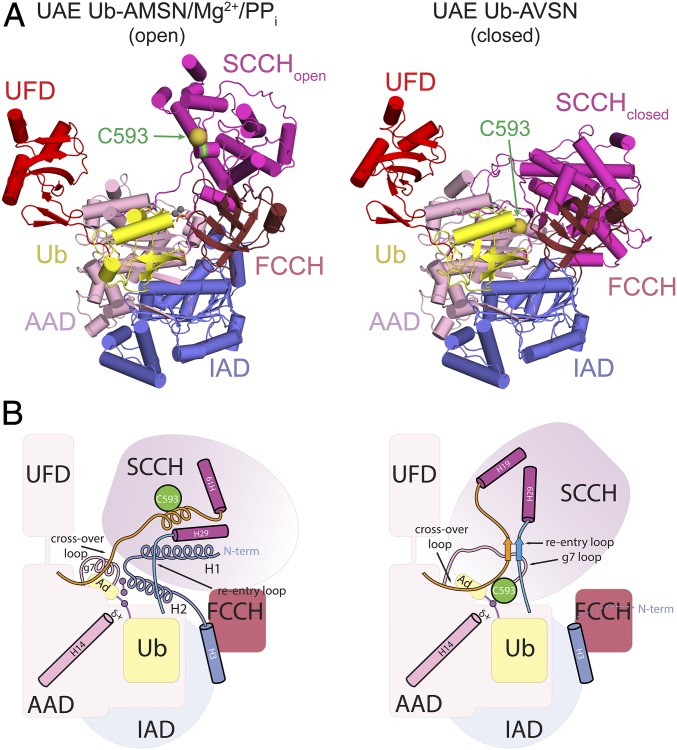
To obtain a structure of Uba1 bound to the nonreactive Ub-AMSN (6) adenylate mimetic and pyrophosphate, Ub-AMSN was combined with Schizosaccharomyces pombe Uba1, purified, and incubated with pyrophosphate (PPi) and magnesium before and during crystallization. A crystal of Uba1/Ub-AMSN/PPi/Mg2+ diffracted x-rays to 2.6-Å resolution, and the structure was determined by molecular replacement (SI Appendix, Table S1) (48). The asymmetric unit includes 2 Uba1/Ub-AMSN complexes. Superposition of the 2 complexes reveals differences in FCCH and UFD conformations that results in a 1.02-Å rmsd over 1,047 C⍺ atoms, differences that are reduced when FCCH and UFD domains are excluded (0.638-Å rmsd over 846 C⍺ atoms) or when isolated FCCH (0.343-Å rmsd over 88 C⍺ atoms) or UFD (0.557-Å rmsd over 113 C⍺ atoms) domains are compared (SI Appendix, Table S3). Figures depicting this structure use the complex that exhibits better electron densities and lower B-factors (chains A and B). With the exception of reactants in the adenylation active site, the overall architecture for the Uba1/Ub-AMSN/PPi/Mg2+ complex is similar to that observed for S. pombe Uba1 bound to Ub/ATP/Mg2+ (0.41-Å rmsd over 1,062 C⍺ atoms) (27) (Fig. 2A).
Fig. 2.
Structures of UAE/Ub complexes in open and closed states. (A) Cartoon representations of crystal structures of UAE Ub-AMSN/Mg2+/PPi in the open E1 conformation and UAE∼Ub-AVSN in the closed E1 conformation. IAD domains are in blue; AAD, FCCH, and SCCH domains are in shades of pink; Ub/SUMO is in yellow; UFD domain is in red; and active site cysteine residues are in green. The cross-over loop comprising residues 583 to 591 and reentry loop comprising residues 849 to 853 are included and colored the same as the SCCH domain. Representations were made with PyMOL (The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC.). (B) Schematic illustrations depicting states of UAE shown in A.
The Ub-AVSN probe (7) was incubated with Uba1 to generate a thioether-linked Uba1∼Ub-AVSN adduct (8), which was purified and crystallized. A crystal of Uba1∼Ub-AVSN diffracted x-rays to 3.15-Å resolution, and the structure was determined by molecular replacement (SI Appendix, Table S1) (49). While packing differs from that in the Uba1/Ub-AMSN/PPi/Mg2+ complex, the asymmetric unit also includes 2 Uba1∼Ub-AVSN complexes that are similar (0.75-Å rmsd over 1,029 C⍺ atoms); figures depicting this structure herein use the complex that exhibits better electron densities and lower B-factors (chains A and B). Similar to Uba1/Ub-AMSN/PPi/Mg2+, the 2 complexes are more similar with the FCCH and UFD domains removed (SI Appendix, Table S3). The Uba1∼Ub-AVSN structure reveals rotation of the SCCH domain, active site remodeling, and conformational changes that juxtapose the E1 active site cysteine and Ub-AVSN adduct (Fig. 2).
Pyrophosphate Conformation Consistent with In-Line Attack.
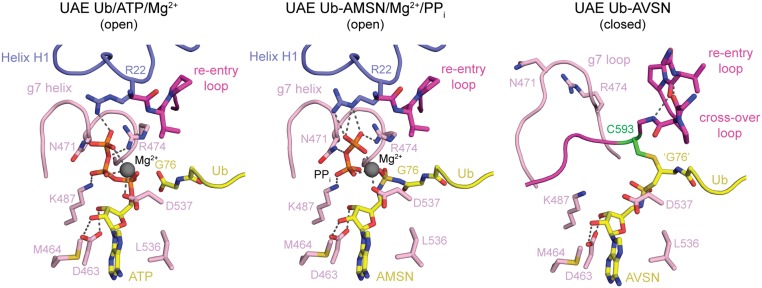
Comparing active sites in the present adenylate mimic-bound structure Uba1/Ub-AMSN/PPi/Mg2+ and the previously reported substrate-bound structure Uba1/Ub/ATP/Mg2+ (27) reveals similar conformations for the purine and ribose moieties of ATP and AMSN and the side chains that coordinate them (Asp463, Met464, Leu536) (Fig. 3). The pyrophosphate molecule is coordinated by Mg2+ and side chains of residues that coordinated to the β and ɣ phosphates of ATP in the substrate structure (Arg22, Asn471, Arg474, Lys487). The instructive aspects of this complex lie in the differences between the relative orientation of pyrophosphate compared with the β and ɣ phosphates of ATP and the residues that coordinate them, and differences in the relative orientation of the Ub C-terminal carboxylate compared with the acyl sulfamide linkage in Ub-AMSN.
Fig. 3.
Active sites of UAE/Ub complexes in open and closed states. Shown are close-ups of the active sites of UAE Ub/ATP/Mg2+ (PDB ID code 4II3), UAE Ub-AMSN/Mg2+/PPi, and UAE∼Ub-AVSN, with regions shown in ribbon cartoon with side chains and relevant backbone atoms shown in stick representation. Colors are as in Fig. 2.
In the ATP-bound structure, the 3 nonbridging ɣ-phosphate oxygen atoms are within hydrogen-bonding distance to the side-chain η-nitrogen of Arg22, the side-chain δ-nitrogen of Asn471, and the magnesium ion, while the β-ɣ bridging oxygen is coordinated by the side-chain η-nitrogen of Arg474 (Fig. 3, Left). The 2 nonbridging oxygen atoms of the β-phosphate are coordinated by the magnesium ion or the side-chain ζ-nitrogen of Lys487. In the Ub-AMSN/PPi structure, the ε-nitrogen and η-nitrogen atoms of Arg22 now contact 2 of the nonbridging oxygens of the PPi at the position equivalent to the ɣ-phosphate in ATP with the third oxygen coordinated by the magnesium ion (Fig. 3, Middle). Instead of contacting the β-ɣ bridging oxygen, the η-nitrogen of Arg474 now contacts a nonbridging oxygen of PPi at the ɣ-phosphate equivalent in ATP, while the PPi bridging oxygen is now coordinated by the δ-nitrogen of Asn471. While the orientations are different, the 2 nonbridging oxygen atoms of PPi at the β-phosphate equivalent in ATP remain coordinated by the magnesium ion and the side-chain ζ-nitrogen of Lys487.
The differences described between ATP and PPi are mainly due to the respective rotations of β-ɣ phosphate equivalents, which move the β-ɣ bridging oxygen toward the side chain of Asn471 by 1.2 Å. The magnesium ion moves less than 1 Å and remains coordinated by the δ-oxygen of Asp537 as well as one nonbridging oxygen from each of the α, β, and ɣ phosphates of ATP (which become one nonbridging oxygen from each phosphate of PPi, and a sulfone oxygen from Ub-AMSN). Similar to the magnesium ion, the C-terminal carbonyl carbon of Gly76 in the ATP-bound structure and its equivalent in Ub-AMSN nearly overlap, as do the α-phosphorus of ATP and sulfamide sulfur in Ub-AMSN. Perhaps more instructive are the changes in distances between the Gly76 carbonyl carbon, ATP α-phosphorus, and β-phosphorus atoms and their equivalents in the Ub-AMSN/PPi product complex. In the substrate complex, the distance between the Gly76 carbonyl carbon and the ATP α-phosphorus is 3.7 Å, while the corresponding distance in the Ub-AMSN adenylate mimic is 2.5 Å. Similarly, the distance between the ATP α-phosphorus and β-phosphorus is 3.0 Å in ATP, while the distance between analogous atoms in the Ub-AMSN/PPi complex is 4.0 Å. Taken together, these structural observations are fully consistent with the proposed mechanism of an in-line attack on the ATP α-phosphate by the Gly76 carboxylate (24, 25, 50–52).
Conformational Changes Required for Thioester Bond Formation.
Analysis of Uba1 structures in the open (ATP- or Ub-AMSN–bound) or closed (Ub-AVSN–bound) conformations show that the SCCH domain of Uba1 rotates 124° from open to closed (Fig. 2A). Rotation of the Uba1 SCCH domain is accompanied by a 0.8-Å translation relative to its center of mass [DynDom (53)]. In comparison, the SCCH domain of SAE rotates 132° with a 2.3-Å domain translation (29). Although the values for rotation and translation differ slightly, the overall trajectory of the conformational change appears similar based on alignments of the open and closed states of these E1s. With that said, the SCCH domain of Uba1 is larger than the SCCH domain of SAE due to several insertions that are conserved within the Ub E1 family (27). These include Uba1 residues 618 to 669 that incorporate helices H20 to H23, an extended loop between helices H24 and H25 (residues 690 to 706), and residues 737 to 810 that incorporate helices H26 and H27. Helices H20, H21, H22, and H27 form a 4-helix bundle. As a result, the closed conformation of Uba1 buries a surface area of 3,830 Å2 between the SCCH domain and the rest of the enzyme, while the smaller SCCH of SAE buries a surface area of 3,340 Å2. For comparison, the other Uba1 chain in crystallographic asymmetric unit, denoted as chain C, buries a surface area of 3,756 Å2.
Additional conformational changes that accompany rotation of the SCCH domain to its closed conformation enable the catalytic cysteine (Cys593) to come into proximity of the acyl-adenylate (Fig. 3). Similar to SAE (29), the N-terminal helices of Uba1 are displaced from the adenylation active site in the closed conformation and are presumed disordered as electron density is not observed before Gln37. These helices include a conserved arginine (Arg22), which binds the γ-phosphate of ATP (23–25, 27). An SAE variant in which the analogous arginine was mutated to alanine (SAE R21A) was unable to form the SUMO1-adenylate or SAE∼SUMO1 thioester, but its SUMO1-AVSN cross-linking activity was unaffected compared with wild type (WT) (29). In the same study, an S. pombe Uba1 variant lacking the first 27 amino acids was unable to form a UAE∼Ub thioester, but its Ub-AVSN cross-linking activity was unaffected. This suggests that the arginine residue in the N-terminal helix is necessary for ATP-binding and adenylation activity but unimportant for productive closure of the SCCH domain. The ATP-binding pocket is further dismantled through remodeling of the g7 helix (Fig. 3). In the open conformation, this element provides Asn471 and Arg474 side-chain contacts to ATP. Consistent with its contacts to ATP, a Uba1 N471A mutant is unable to form a UAE∼Ub thioester under our assay conditions (SI Appendix, Uba1∼Ub Thioesterification Gels). In the closed conformation, the g7 helix melts into a loop, rotating the conserved phosphate-coordinating residues away from the nucleotide-binding pocket. Melting of the N-terminal helices and g7 helix appears necessary to accommodate the SCCH domain in the closed conformation as elements within the cross-over loop and active-site cysteine occupy positions in the closed conformation that were previously occupied by the N-terminal helices and g7 helix in the open conformation.
The SCCH domain of Uba1 is tethered to the AAD adenylation domain via a cross-over loop comprising residues 583 to 591 and a reentry loop comprising residues 849 to 853. These elements change conformations when the SCCH domain transits from open to closed conformations (Fig. 4A). While the path of the Uba1 cross-over loop changes by 115°, the reentry loop changes by 59° (Fig. 4B). For comparison, the SAE cross-over and reentry loops change by 125 and 68°, respectively (Fig. 4C). Similar to that observed in SAE, the closed Uba1 structure reveals a short parallel 2-stranded β-sheet between the cross-over and reentry loops, a feature that may contribute to thioester bond formation by locking the cross-over loop in a position that places the catalytic cysteine (Cys593) proximal to the C-terminal adenylate (Figs. 2B and 3).
Fig. 4.
Rearrangement of E1 cross-over and reentry loops connecting SCCH and AAD domains. (A) Cartoon representations of UAE in the open and closed conformations, with Ub in ribbon and ATP or AVSN in stick, showing locations of cross-over and reentry loops in blue ribbon. Helices H14 and H27 are shown in orange for each structure to illustrate movement of the SCCH domain. (B) Ribbon representations of the cross-over and reentry loops of Uba1 in the open (Left) and closed (Right) conformations. Ub is displayed in yellow. Active site cysteine residues are colored green, with the nucleotide labeled and shown in stick representation. Orientation is as in A. (C) Cross-over and reentry loops of Uba1 (blue) and SAE (red) superimposed in the open (Left) and closed (Right) conformations. Active site cysteine residues colored green. Uba1 open conformation: PDB ID code 4II3; Uba1 closed conformation: this work; SAE open conformation: PDB ID code 1Y8R; SAE closed conformation: PDB ID code 3KYD. Color scheme for domains is as described for Fig. 2.
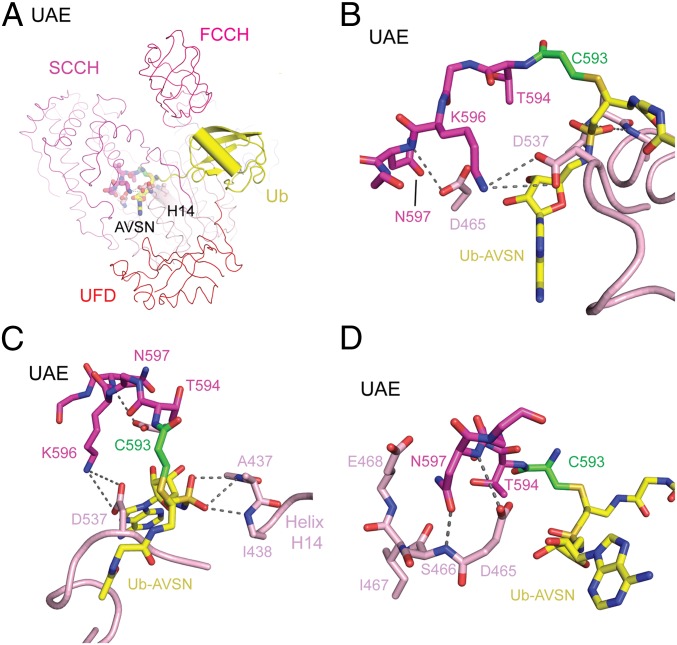
The closed conformation involves additional interdomain contacts between the AAD and SCCH domains near the active site (Fig. 5). While the AAD Asp465 side-chain carboxylate is within hydrogen-bonding distance of the backbone amide of Asn597 from SCCH, the AAD Asp537 side chain is within hydrogen-bonding distance of the ζ-nitrogen of Lys596 of the SCCH (Fig. 5B). Similar contacts were observed in the SAE∼SUMO-AVSN structure (SI Appendix, Fig. S4) (29). Asp465 and Asp537 are invariant among E1s, while Lys596 is conserved as lysine or arginine.
Fig. 5.
Adenosine-proximal interdomain interactions in a closed UAE conformation. (A) Ribbon and cartoon representation of structures of UAE in the closed conformation, with residues shown in B–D depicted in sticks. (B) Close-up of the active site depicting interactions between side chains of conserved residues in UAE. Segments are shown in ribbon, with side chains and relevant backbone atoms shown in stick representation. Potential hydrogen bonds are indicated by dashed gray lines, with relevant amino acid positions labeled. (C) A different orientation from B showing contacts between the N terminus of helix H14 and the sulfone of AVSN. (D) Contacts between the side chain of Uba1 Asn597 and the backbone amide of Ser466. Color scheme for domains is as described for Fig. 2.
We previously proposed that contacts between the Ubl C-terminal carboxylate and N-terminal backbone amides of the helix that resides under the active site might provide electrostatic stabilization of transition states during adenylation and thioester bond formation (29). Consistent with this, the backbone amides of Uba1 Ala437 and Ile438 at the N terminus of helix H14, comprising the positive end of the helix dipole, contact the sulfone oxygens of the AVSN moiety (Fig. 5C). A similar configuration is observed in SAE between the backbone amides of Gly27 and Ile28 in helix H2 and the corresponding AVSN sulfone oxygens (SI Appendix, Fig. S4C).
Conserved Residues Unique to Ubiquitin E1 Are Important for Domain Alternation.
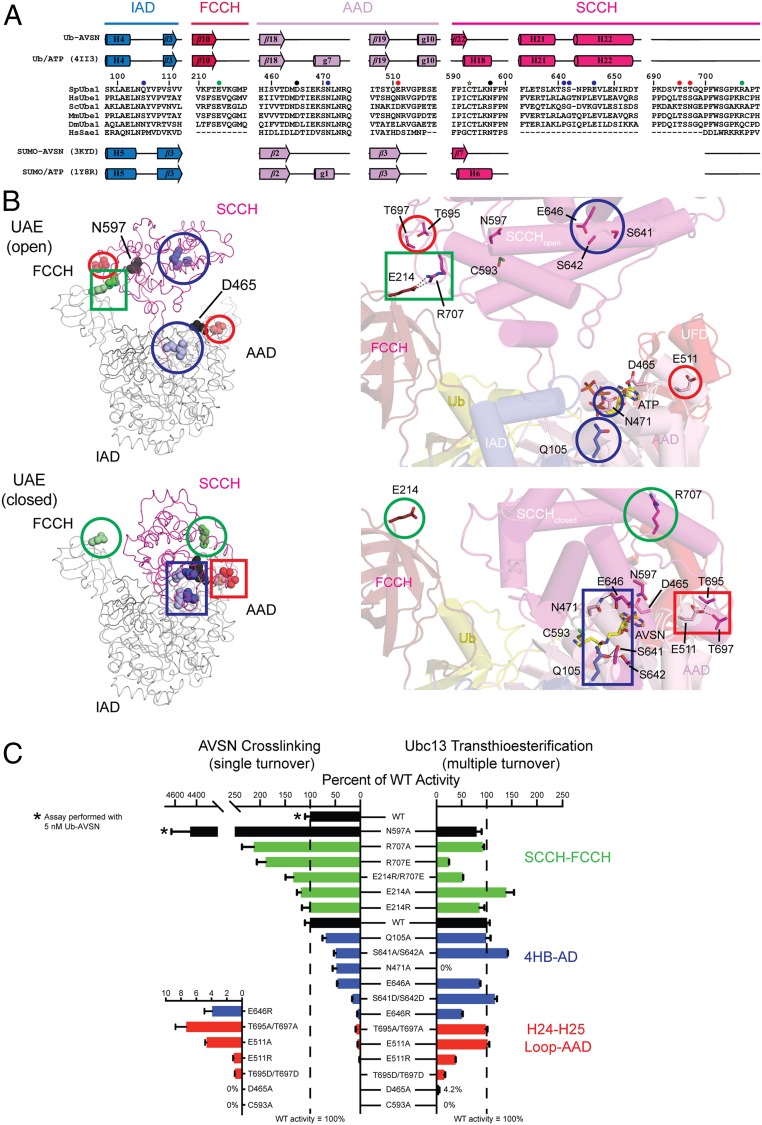
The SCCH domain of Uba1 is larger and buries additional surfaces within the SCCH-IAD/AAD interface compared with the closed SAE structure, including putative interdomain contacts that appear conserved and unique to Ub E1 (Figs. 2B and 6 A and B). These include a 4-helix bundle between Uba1 residues 619 to 653 and 786 to 792 that is not present in the SCCH domains of NAE or SAE but is retained in E1s for FAT10 and ISG15, an extended loop between Uba1 residues 690 to 706 that is present in the E1s for FAT10 and ISG15 but not the E1s for SUMO and NEDD8, and an FCCH domain between residues 173 to 260 that is present in E1s for FAT10 and ISG15 but absent or structurally dissimilar in SAE and NAE, respectively (23, 24, 33). Given the limited resolution of the crystallographic data, we interpreted these contacts with caution and assayed them biochemically.
Fig. 6.
Mutational analysis of interdomain contacts unique to the closed conformation of Uba1. (A) Structure-based alignment of Ub E1 amino acid sequences from (top to bottom) S. pombe, H. sapiens, Saccharomyces cerevisiae, M. musculus, Drosophila melanogaster, and SAE from H. sapiens. Residues at the interface between SCCH and FCCH domains are indicated by green circles; residues at the interface between the 4-helix bundle (4HB) of the SCCH domain and the adenylation domain (IAD/AAD) are denoted by blue circles; residues at the interface between the H24-H25 loop and the AAD domain are indicated by red circles. Asp465 and Asn597 are marked by black circles. Active site cysteine residue is marked with a star. Secondary structure elements are indicated for Uba1 (top) and SAE (bottom) in the open (ATP) and closed (AVSN) conformations. (B) Ribbon representation of crystal structures of Uba1/Ub in the open (Top) and closed (Bottom) conformations are shown on the Left, with residues highlighted in A in colored spheres. Circles surround residues not making interdomain contacts; rectangles enclose interdomain contacts. On the Right, a close-up view showing residue contacts with structures depicted in cartoon, with relevant side chains in stick. Potential hydrogen-bonding interactions are indicated by gray dashed lines. Residues, boxes, and circles are color coded as described for A. Close-up views at Right are in the same orientations as global views at Left. Domain colors are as in Fig. 2. (C) Bar graph depicting measured rates of cross-linking between Ub-AVSN and Uba1 harboring substitutions in interdomain interfaces (left) or measured rates for Uba1-Ubc13 transthioesterification using Ub with native C terminus for the same substitutions (right). Values represent the average of 3 independent experiments and are displayed as percentages of the average WT value; error bars are 1 SD between experiments and are displayed as percentages of the average WT value. Colors are as described for A.
The importance of unique interdomain contacts observed in Uba1 were tested using 2 assays (SI Appendix, Table S2). The first employs the Ub-AVSN probe to bypass adenylation and to measure formation of a productive closed conformation by virtue of cross-linking the catalytic cysteine to the probe. The second tests the entire E1 catalytic cycle from adenylation to thioester bond transfer to E2. While the Ub-AVSN thioether bond assay serves as a surrogate for domain closure, the absence or presence of a thioether bond can be interpreted in several ways. For instance, if a mutation increases cross-linking, it could mean that the closure rate increased or that it increases the time spent in the closed conformation or decreases the time spent in the open conformation. A decrease in cross-linking to Ub-AVSN can also be interpreted multiple ways. If a mutation decreases cross-linking, it could mean that the closure rate decreased, that mutation leads to nonproductive closed conformations as observed in other systems (32, 38), or that it spends increased time in the open conformation or decreased time in the closed conformation.
In contrast to the single turnover Ub-AVSN cross-linking assay where mutations might increase or decrease activity, the E2 thioester bond transfer assay requires that E1 undergo multiple rounds of productive adenylation, thioester bond formation, and thioester bond transfer. If mutations decrease Ub-AVSN cross-linking by altering its ability to form the closed conformation, these mutations would also be predicted to decrease E2 thioester bond formation. Importantly, mutations that increase Ub-AVSN cross-linking might also decrease E2 thioester bond formation if mutations cause the enzyme to spend more time in the closed conformation or less time in the open conformation. This would only be true if opening and closing of the SCCH domain becomes rate-limiting, a relevant point as transthioesterification between E1 and E2 was found to be rate-limiting in several studies (20, 54–56).
As controls for the aforementioned assays, we selected 2 residues that were previously shown to be important for E1 catalytic activity, the catalytic cysteine (Cys593) and Asp465 whose side chain is predicted to stabilize a productive closed conformation. As expected based on prior data (29), the C593A mutation results in no detectable activity in Ub-AVSN cross-linking or E2 thioester bond formation, while the D465A mutation results in no detectable activity in cross-linking and a 25-fold decrease in E2 thioester bond formation (Fig. 6C; see also SI Appendix).
In the closed conformation, the Asp465 side-chain carboxylate is within hydrogen-bonding distance of the Asn597 backbone amide whose side-chain carbonyl is within hydrogen-bonding distance of the Ser466 backbone amide (29) (Fig. 5D). In SAE, Asn177 (equivalent to Uba1 Asn597) points away from the backbone (SI Appendix, Fig. S4D); however, the Asp50 (equivalent to Uba1 Asp465) carboxylate contributes 2 potential hydrogen bonds to the backbone amides of Asn177 and Thr178. While proximal in the closed conformation, Uba1 Asn597 and Ser466 are separated by 39 Å in the open conformation. As noted earlier and published previously, Uba1 Asp465 and SAE Asp50 are important for thioester bond formation (29). We anticipated that the Asn597 mutation might disrupt the closed conformation in a manner analogous to that observed for Asp465. In contrast, N597A substitution led to a high rate of cross-linking while maintaining nearly WT activity in the E2 thioester bond assay (Fig. 6C; see also SI Appendix, Uba1∼Ub-AVSN Assay Gels).
In the 4-helix bundle, the Ser641 and Ser642 side-chain hydroxyls are proximal to the IAD Gln105 side-chain carbonyl in the closed conformation but separated by 46 Å in the open conformation (Fig. 6B). In addition, side chains of 4-helix bundle residue Glu646 and AAD residue Asn471 are within hydrogen-bonding distance in the closed conformation, but in the open conformation the Asn471 side chain contacts the ɣ-phosphate of ATP (Fig. 3) and is more than 31 Å away from Glu646 (Fig. 6B). While not broadly conserved, substitutions at these amino acid positions would be predicted to disrupt the closed conformation, and for Asn471, adenylation. Consistent with this, alanine substitutions at Ser641, Ser642, Gln105, and Asn471 resulted in a 2-fold decrease in cross-linking to Ub-AVSN, while S641D/S642D or E464R substitutions decreased activity by 7- or 25-fold, respectively (Fig. 6C; see also SI Appendix, Uba1∼Ub-AVSN Assay Gels). Most substitutions had a minimal effect on E2 thioester bond formation with the exception of N471A and E464R (Fig. 6C; see also SI Appendix, Ubc13∼Ub Transthioesterification Assay Gels). For N471A, no activity was detected for E1 thioester bond formation (SI Appendix, Uba1∼Ub Thioesterification Assay Gels) or E2 thioester bond formation, consistent with our hypothesis that it plays a role in adenylation. For E464R, a 2-fold decrease was observed for E2 thioester bond formation. Collectively, these data are consistent with transthioesterification as the rate-limiting step in the E2∼Ub charging cycle for these mutants.
Thr695 and Thr697 reside in an extended loop in the SCCH domain between helices H24 and H25. In the closed conformation, their side-chain hydroxyl groups are within hydrogen-bonding distance of the side-chain carboxylate of Glu511 within the AAD (Fig. 6B). In the open conformation, the Thr695 and Thr697 are 57 Å away from Glu511. Thr695 is conserved as threonine or a polar residue among several UAE orthologs, while Glu511 is sometimes substituted to Asp or Asn (Fig. 6A). Mutations at these positions would be predicted to disrupt productive SCCH domain closure. Consistent with this, amino acid substitutions to alanine, aspartate (Thr695/Thr697), or arginine (Glu511) reduced the rate of cross-linking to Ub-AVSN from 13- to 100-fold (Fig. 6C; see also SI Appendix, Uba1∼Ub-AVSN Assay Gels). In contrast, alanine substitutions had no effect in E1-E2 transthioesterification (Fig. 6C; see also SI Appendix, Ubc13∼Ub Transthioesterification Assay Gels). However, E511R and T695D/T697D substitutions decreased E2 charging by 3- and 7-fold, respectively.
Contacts between the SCCH and FCCH domains include a salt bridge between the side-chain carboxylate of Glu214 and side-chain guanidinium of Arg707 (Fig. 6B). Both residues are conserved in Uba1 proteins from human, Saccharomyces cerevisiae, Mus musculus, and the human FAT10 AE. It is worth noting that this interaction appears intact in crystals structures of UAE from S. cerevisiae (23), S. pombe (27), and Homo sapiens (31). This interaction is broken in the closed conformation of Uba1 with Arg707 and Glu214 separated by 41 Å (Fig. 6B). In this case, mutations at these positions would be predicted to disrupt the open configuration. Consistent with this, substitution at these positions resulted in Ub-AVSN cross-linking rates up to 2-fold faster than WT, with R707A, R707E, and E214R/R707E mutants showing the greatest increases (Fig. 6C; see also SI Appendix, Uba1∼Ub-AVSN Assay Gels). In the transthioesterification assay, most Uba1 variants exhibited activity comparable to WT, while Uba1R707E was 4-fold slower (Fig. 6C; see also SI Appendix, Ubc13∼Ub Transthioesterification Assay Gels).
Of the mutations characterized, most are distal from known E1–E2 interfaces for S. pombe Uba1 bound to Ubc4 (27) and Ubc15 (30). The exceptions are Ser642, which is within hydrogen-bonding distance of Ubc15 Lys73, and Thr695, which is within hydrogen-bonding distance of Asp116 of Ubc4 and Asp133 of Ubc15. Individual substitutions at Ser642 or Thr695 had no effect on E1–E2 transthioesterification, but substituting Thr695 and Thr697 to aspartate diminished the rate of transthioesterification. While T695D/T697D decreased cross-linking to Ub-AVSN, it remains possible that T695D/T697D might also disrupt interactions with Ubc13.
Discussion
The conformational changes and active-site remodeling associated with adenylation and thioester bond formation appear conserved between the SUMO and Ub E1 enzymes. These changes include a ∼130° rotation of the SCCH domain, remodeling of the g1 helix in SAE or g7 helix in UAE, remodeling of the ⍺-helix containing the catalytic cysteine, displacement of the N-terminal helices of the IAD domain that coordinate ATP or pyrophosphate, and rearrangement of the cross-over and reentry loops (Fig. 2B). As SUMO and Ub E1s represent divergent members of the canonical Ub/Ubl AEs, our data suggest that similar domain alternations will underlie cycles of adenylation and thioester bond formation for other canonical E1s.
Many aspects of the E1 catalytic cycle appear conserved, but our study also suggests that Uba1 contains unique elements that contribute to thioester bond formation that are either absent or poorly conserved among other E1s. These include contacts in the closed or open conformation that, when mutated, decreased or increased Ub-AVSN cross-linking, respectively. Mutations that increased or decreased cross-linking to Ub-AVSN either had little effect or decreased E2 thioester bond transfer. These results are consistent with the E2 thioester bond transfer being the rate-limiting step in the overall catalytic cycle and, because the E1 catalytic cycle requires E1 to open and close to transfer Ub to E2, mutations that destabilize either open or closed states would be expected to result in inhibition of E2 thioester bond formation. It is perhaps more difficult to reconcile the dramatic increase in cross-linking observed for the N597A mutant, as it exhibited normal E2 thioester transfer activity. While the mechanistic basis for this increase remains unclear, perhaps this mutation increases the rate of open-to-closed motion while decreasing rates for closed-to-open motion. In this scenario, the catalytic rate would remain constant. It is also worth noting that, despite the usefulness of the Ub-AVSN probe for biochemical and structural characterization of the closed state of E1, cross-linking to AVSN is on the order of minutes, while rates of thioester bond formation are on the order of fractions of a second (54–56). This may be due to the absence of the C-terminal glycine carbonyl carbon in Ub-AVSN (Fig. 1C).
Our results support a general mechanism for thioester bond formation among canonical E1s. However, the molecular details of this process have not been resolved for noncanonical E1s, such as those for Ubls involved in autophagy, as well as URM1 and UFM1 enzymes that lack Cys or SCCH domains (5). Instead, these enzymes harbor their catalytic cysteine residues within short, unstructured elements proximal to the active site for adenylation. As such, their mechanism of E1∼Ubl thioester bond formation remains unclear (57–64). Semisynthetic Ubl-AVSN probes analogous to those reported here may be useful in further characterizing these noncanonical E1s.
Materials and Methods
Reagents.
Reagents were obtained from Aldrich Chemical or Acros Organics and used without further purification. Optima or HPLC grade solvents were obtained from Fisher Scientific, degassed with Ar, and purified on a solvent drying system as described (65) unless otherwise indicated.
Reactions.
All small-molecule reactions were performed in flame-dried glassware under positive Ar pressure with magnetic stirring unless otherwise noted. Liquid reagents and solutions were transferred through rubber septa via syringes flushed with Ar before use. Cold baths were generated as follows: 0 °C, wet ice/water.
Chromatography.
Thin-layer chromatography was performed on 0.25-mm E. Merck silica gel 60 F254 plates and visualized under UV light (254 nm) or by staining with potassium permanganate (KMnO4), cerium ammonium molybdenate, phosphomolybdic acid, iodine (I2), or p-anisaldehyde. Silica flash chromatography was performed on E. Merck 230- to 400-mesh silica gel 60 or with RediSep silica gel normal phase columns. Lyophilization of larger aqueous samples was performed using a Labconco Freezone 2.5 instrument.
Analytical Instrumentation.
Infrared (IR) spectra were recorded on a Bruker Optics Tensor 27 Fourier-transform IR spectrometer using an attenuated total reflection attachment with peaks reported in reciprocal centimeters. NMR spectra were recorded on a Bruker UltraShield Plus 500 MHz Avance III NMR or UltraShield Plus 600 MHz Avance III NMR with DCH CryoProbe at 24 °C in CDCl3 unless otherwise indicated. Chemical shifts are expressed in parts per million (ppm) relative to TMS (1H, 0 ppm) or solvent signals: CDCl3 (13C, 77.0 ppm), C6D6 (1H, 7.16 ppm; 13C, 128.0 ppm) or acetone-d6 (13C, 206.2 ppm); coupling constants are expressed in Hz. NMR spectra were processed with Mnova software (www.mestrelab.com/software/mnova-nmr). Mass spectra were obtained at the MSKCC Analytical Core Facility on a Waters Acuity SQD LC MS or PE SCIEX API 100 by electrospray ionization (ESI) or atmospheric pressure chemical ionization. High-resolution mass spectra were obtained on a Waters Acuity Premiere XE TOF LC-MS by ESI.
Nomenclature.
Atom numbers shown in chemical structures in the SI Appendix may not correspond to International Union of Pure and Applied Chemistry nomenclature, which was used solely to name each compound. Compounds not cited in the paper are numbered in the SI Appendix from S1.
Cloning.
Cloning of S. pombe Uba1, Ub, the Ub1–73-inteinCBD construct used to generate Ub-AMSN, and Ubc13 were described in previous publications (27, 29, 37). All mutations were introduced using PCR-based mutagenesis. To generate Ub1–75-inteinCBD for preparing Ub-AVSN, residues 1 to 75 of the S. pombe Ubi5 gene were inserted into vector pTXB1 using the NdeI and SapI restriction sites. To generate Cys-Ub1–75-inteinCBD for preparing Alexa488-Ub-AVSN, a primer was used to add Met-Cys-Gly immediately N-terminal to Ub’s Met1 residue, and the resulting MCG-Ub1–75 gene was inserted into pTXB1 using NdeI and SapI. To generate Cys-Ub for preparing Alexa488-Ub for transthioesterification assays, the full-length S. pombe Ubi5 gene was inserted into a Smt3-pET28 vector using NdeI and XhoI, generating a His6-Smt3-Ser-His-Ub gene, where Smt3 is the SUMO ortholog from S. cerevisiae. PCR-based mutagenesis was then used to convert the serine to a cysteine, generating His6-Smt3-Cys-His-Ub in a pET28 backbone.
Protein Expression and Purification.
S. pombe Uba1, Ub, Ubc13, and Ub-inteinCBD were expressed in E. coli and purified as described previously, and Ub∼MESNa was prepared from Ub-inteinCBD as described previously (27, 29, 37). His6-Smt3-Cys-His-Ub was expressed in E. coli using the same protocol. After cell lysis and clearing, the protein was purified by nickel immobilized metal affinity chromatography before being treated with Ulp1403–621 (which was also His-tagged) to remove the N-terminal His-Smt3 (66). The sample was subjected to size exclusion chromatography on a HiLoad 26/600 Superdex 75 prep grade column (GE) equilibrated in 20 mM tris(hydroxymethyl)aminomethane (Tris), pH 8.0, 350 mM NaCl, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP). The Ub peak was passed back over nickel–nitrilotriacetic acid agarose to remove any remaining His-tagged proteins. Cys-Ub-AVSN and Cys-Ub were labeled with Alexa488 maleimide (Life Technologies) as recommended by the manufacturer. A Superdex 75 Increase 10/300 GL gel filtration column (GE) was used to separate fluorescently tagged Ub from excess Alexa488.
Preparation of Ub-AMSN and Ub-AVSN.
Ub-AMSN (R74C mutant) was prepared as described previously (29, 37). H2N-AVSN (13) was prepared as described in the SI Appendix. To produce Ub-AVSN, S. pombe Ub1–75∼intein-CBD (where CBD is chitin-binding domain) was expressed in E. coli strain BL21 (DE3) codon plus (Stratagene). After lysis and pelleting by centrifugation at 40,000 × g, 4 °C, 30 min, the soluble fraction of the lysate was applied to chitin resin (New England BioLabs) equilibrated in 20 mM bis(tris[hydroxymethyl]aminomethane) (BIS-Tris), pH 6.5, 350 mM NaCl. After washing the resin with the same buffer, MESNa (sodium 2-mercaptoethanesulfonate) was added to the resin to a final concentration of 200 mM. The resin was incubated overnight at room temperature. Ub∼MESNa was eluted with 20 mM BIS-Tris, pH 6.5, 350 mM NaCl, 20 mM MESNa. Ub∼MESNa was treated with 500 mM hydrazine in a 30 °C water bath for 30 min and then purified by size exclusion chromatography on a HiLoad 26/600 Superdex 75 prep grade column (GE) equilibrated in 20 mM BIS-Tris, pH 6.5, 350 mM NaCl. Ub-NHNH2 was concentrated, flash-frozen, and stored at −80 °C. To convert Ub-NHNH2 to Ub-N3, a solution of 3 mM Ub-NHNH2, 0.5 M NaNO2, 100 mM citrate, pH 3, was incubated for 2 min in an ice/brine bath. The solution was immediately added to an equal volume of 1.5 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 8.0, and a 10-fold molar excess of H2N-AVSN (13) (trifluoroacetic acid salt), incubated for 2 min at 30 °C, and then moved to ice. Ub-AVSN was purified on a Superdex 75 column equilibrated in 20 mM Tris, pH 8, 50 mM NaCl, and then flash-frozen and stored at −80 °C. Ub∼MESNa, Ub-NHNH2, and Ub-AVSN were distinguished via ultraperformance LC-MS (UPLC-MS) using a Waters Acquity SQD UPLC-MS-PDA UPLC-MS instrument, using a C8 column with a water/acetonitrile gradient. Cys-Ub-AVSN was prepared in the same way, but 0.5 mM TCEP was included in all buffers with the exception of the Cys-Ub-NHNH2 → Cys-Ub-N3 → Cys-Ub-AVSN aminolysis reaction.
E1 Cross-Linking Assays and Cross-Linking for Crystallization.
To prepare Uba1∼Ub-AVSN for crystallization trials, 10 μM S. pombe Uba1 (lacking the first 12 amino acids) (27) was incubated with 20 μM Ub-AVSN in 20 mM Tris, pH 8, 50 mM NaCl, 0.5 mM TCEP for 1 h in a 25 °C water bath. Uba1∼Ub-AVSN was separated from excess Ub-AVSN by size-exclusion chromatography on a HiLoad 26/600 Superdex 200 prep grade column (GE) equilibrated in the same buffer at 4 °C. Uba1∼Ub-AVSN was concentrated to 20 mg/mL (167 μM). Aliquots were flash-frozen in liquid nitrogen and stored at −80 °C. For Ub-AVSN cross-linking assays, 200 nM Alexa-Ub-AVSN (20 mM Hepes, pH 7.5, 50 mM NaCl, 0.1% Tween-20) was added to an equal volume of 20 nM Uba1 in the same buffer to start the reaction, for final assay concentrations of 100 nM Alexa-Ub-AVSN and 10 nM Uba1. For the N597A variant and the WT assay with which it was compared, 5 nM Alexa-Ub-AVSN and 10 nM Uba1 were used, because the reaction with Uba1N597A was too fast to measure using 100 nM Alexa-Ub-AVSN. Assays were conducted at room temperature. Reactions were quenched in equal volumes of 4× lithium dodecyl sulfate NuPAGE loading dye (Life Technologies), resolved by nonreducing 4 to 12% BIS-Tris sodium dodecyl sulfate–polyacrylamide gel electrophoresis with 2-(N-morpholino)ethanesulfonic running buffer (Life Technologies), and imaged on Typhoon FLA 9500 with a 473-nm laser and a longpass blue filter. All gels were imaged with a serial dilution of a known quantity of Uba1-Alexa-Ub-AVSN so that each gel contained a standard curve to convert band intensity to nanomoles of conjugate with ImageJ (NIH). This was done by manually defining lanes in ImageJ and then plotting intensity vs. gel migration using ImageJ’s “Plot Lanes” function. Gel bands appeared as peaks on these plots. The background was manually defined for each peak, and then the area under the curve for each peak was quantified using ImageJ’s Wand tool. The area under the curve for each gel band was normalized to nanomoles using the Uba1∼Alexa-Ub-AVSN standard curve. Dividing by volume gave product concentration in nM. For each of 3 triplicates per mutant nM product (y axis) was plotted against time (x axis), and the slope was calculated with the y-intercept set to 0 (0 nM Uba1∼Ub-AVSN at time t = 0). Average rate and SD were calculated from 3 separate slopes for each of 3 triplicate reactions performed for each Uba1 variant. Values and error bars in Fig. 6C represent average rate ± 1 SD, normalized to percentage of WT activity by dividing values by the average WT rate and multiplying by 100.
Uba1-Ub E2 Ub Thioester Transfer Assays.
Reactions included 15 nM Uba1, 10 nM Ubc13, 50 nM Alexa-Ub, 10 mM MgCl2, 20 mM Hepes, pH 7.5, 50 mM NaCl, and 0.1% Tween-20. Full-length S. pombe proteins were used for all assays; 2 mM ATP was used to initiate the reaction, which was run at room temperature. Reactions were quenched and quantified as described for Alexa-Ub-AVSN assays. Values and error bars in Fig. 6D represent average rate ± 1 SD, normalized to percentage of WT activity by dividing values by the average WT rate and multiplying by 100.
Crystallization and Data Collection.
To obtain the Uba1/Ub-AMSN complex, 22 μM Uba1 was incubated with 123 μM Ub-AMSN in 20 mM Tris, pH 8.0, 75 mM NaCl, 1 mM TCEP on ice for 1 h. The Uba1/Ub-AMSN complex was separated from excess Ub-AMSN using a HiLoad 26/600 Superdex 200 prep grade column (GE) equilibrated in the same buffer. The purified Uba1/Ub-AMSN complex was concentrated to 10 mg/mL and subjected to sparse matrix crystallization screening using the sitting drop vapor diffusion method with a mosquito robot (Molecular Dimensions). Crystal hits appeared in various conditions containing polyethylene glycol (PEG). After refinement, the best crystals were grown in 20% PEG 3350, 150 mM MgSO4, 5 mM MgCl2, and 1 mM pyrophosphate, pH 8.0. Crystals were cryoprotected in mother liquor plus 20% ethylene glycol and flash cooled by plunging into liquid nitrogen before data collection at the Advanced Photon Source (APS) (Argonne, IL), Northeastern Collaborative Access Team (NE-CAT) beamline 24-ID-E.
The cross-linked, purified Uba1∼Ub-AVSN was concentrated to 20 mg/mL (167 μM). Aliquots were flash-frozen in liquid nitrogen and stored at −80 °C. Before crystallization, an aliquot was rapidly thawed in a room temperature water bath and spun for 10 min. at 4 °C in a tabletop centrifuge (Eppendorf) at 18,213 × g and then incubated on ice. Uba1∼Ub-AVSN was diluted to 15 mg/mL (125 μM) in 20 mM Tris, pH 8, 50 mM NaCl, 0.5 mM TCEP and then subjected to sparse matrix crystallization screening using the hanging-drop vapor diffusion method with a mosquito robot (100 nL of well solution added to 100 nL of protein). The initial crystallization hit appeared within 2 d in 30% (+/−)-2-methyl-2,4-pentanediol, 5% PEG 4000, 100 mM Hepes, pH 7.5, 20 °C (microcrystals were also observed at 4 °C) and was refined to 27.3% MPD, 6.6% PEG 8000, 100 mM Bis-Tris propane, pH 7.5. The single crystal that was used to collect the Uba1∼Ub-AVSN dataset was generated by combining 1 μL of well solution to 1 μL of protein (15 mg/mL protein, 20 mM Tris, pH 8.0, 50 mM NaCl, 0.5 mM TCEP) on an 18-mm siliconized glass circle cover slide (Hampton Research) without mixing. The coverslip was placed over 500 μL of well solution in a 24-well hanging-drop crystallization tray with preapplied sealant (Hampton Research). The sealed tray was stored at 18 °C. The crystal was flash-cooled in liquid nitrogen without further cryoprotection 8 d after the tray was set. Data were collected at APS, NE-CAT beamline 24-ID-C.
Structure Determination and Refinement.
Datasets for Uba1/Ub-AMSN and Uba1∼Ub-AVSN crystal structures were indexed, integrated, and scaled using HKL2000 (67). For Uba1/Ub-AMSN, PHASER (68) was used for molecular replacement using the S. cerevisiae Uba1/Ub structure (PDB ID code 3CMM) as a search model. Coordinates were refined via iterative rounds of refinement and rebuilding using Crystallography & NMR System (69), Collaborative Computational Project Number 4 (70), Python-based Hierarchical Environment for Integrated Crystallography (PHENIX) (71), and Crystallographic Object-Oriented Toolkit (COOT) (72).
For Uba1∼Ub-AVSN, a molecular replacement solution was found using the IAD and AAD domains from S. pombe Uba1 (PDB ID code 4II3). The remaining domains were fitted into the electron density using COOT and, in the case of SCCH, rotated to the closed position using the SAE-SUMO-AVSN structure (PDB ID code 3KYD) as a guide. Coordinates were refined via iterative rounds of refinement and rebuilding using PHENIX and COOT.
Supplementary Material
Acknowledgments
We thank A. M. Levinson for assistance with developing and optimizing the Ub-AVSN semisynthetic strategy and for assistance with UPLC-MS analysis of semisynthetic species. This research was supported in part by NIH National Institute of General Medical Sciences (NIGMS) Grants R01 GM065872 (to C.D.L.), R35 GM118080 (to C.D.L.), and R01 GM100477 (to D.S.T.) and NIH National Cancer Institute–Cancer Center Support Grant P30 CA008748. M.C.L. was supported by NSF Graduate Research Fellowship 2015190598 and NIH Chemistry–Biology Interface Training Grant T32 GM115327. The work presented here was also based in part on research conducted at the NE-CAT beamlines (NIH NIGMS Grant P41 GM103403 and NIH Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205). Beamline research used resources of the APS, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not represent the official views of the NIH. C.D.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been in the Protein Data Bank, www.rcsb.org [PDB ID codes 6O82 (Ub-AMSN) and 6O83 (Ub-AVSN)].
See Commentary on page 15319.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905488116/-/DCSupplemental.
References
- 1.Taherbhoy A. M., Schulman B. A., Kaiser S. E., Ubiquitin-like modifiers. Essays Biochem. 52, 51–63 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Streich F. C. Jr, Lima C. D., Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Herr R. A., Hansen T. H., Ubiquitination of substrates by esterification. Traffic 13, 19–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A., Ciechanover A., The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Schulman B. A., Harper J. W., Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Wijk S. J. L., Timmers H. T. M., The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 24, 981–993 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Gareau J. R., Lima C. D., The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K., Suzuki T., Chiba T., The ligation systems for ubiquitin and ubiquitin-like proteins. Mol. Cells 8, 503–512 (1998). [PubMed] [Google Scholar]
- 9.Buetow L., Huang D. T., Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cappadocia L., Lima C. D., Ubiquitin-like protein conjugation: Structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mevissen T. E. T., Komander D., Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Freemont P. S., Hanson I. M., Trowsdale J., A novel cysteine-rich sequence motif. Cell 64, 483–484 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M., A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563–2567 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E., UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundlov J. A., Shi C., Wilson D. J., Aldrich C. C., Gulick A. M., Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem. Biol. 19, 188–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell C. A., Shi C., Aldrich C. C., Gulick A. M., Structure of PA1221, a nonribosomal peptide synthetase containing adenylation and peptidyl carrier protein domains. Biochemistry 51, 3252–3263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulick A. M., Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lux M. C., Standke L. C., Tan D. S., Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors. J. Antibiot. (Tokyo) 72, 325–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reger A. S., Wu R., Dunaway-Mariano D., Gulick A. M., Structural characterization of a 140° domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase. Biochemistry 47, 8016–8025 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas A. L., Rose I. A., The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 (1982). [PubMed] [Google Scholar]
- 21.Haas A. L., Warms J. V. B., Hershko A., Rose I. A., Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 257, 2543–2548 (1982). [PubMed] [Google Scholar]
- 22.Pickart C. M., Kasperek E. M., Beal R., Kim A., Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1). J. Biol. Chem. 269, 7115–7123 (1994). [PubMed] [Google Scholar]
- 23.Lee I., Schindelin H., Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Lois L. M., Lima C. D., Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walden H., et al. , The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Huang D. T., et al. , Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445, 394–398 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen S. K., Lima C. D., Structure of a ubiquitin E1-E2 complex: Insights to E1-E2 thioester transfer. Mol. Cell 49, 884–896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulman B. A., Haas A. L., Structural biology: Transformative encounters. Nature 463, 889–890 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Olsen S. K., Capili A. D., Lu X., Tan D. S., Lima C. D., Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv Z., et al. , S. pombe Uba1-Ubc15 structure reveals a novel regulatory mechanism of ubiquitin E2 activity. Mol. Cell 65, 699–714.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv Z., Williams K. M., Yuan L., Atkison J. H., Olsen S. K., Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity. J. Biol. Chem. 293, 18337–18352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv Z., et al. , Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme. Nat. Commun. 9, 5145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walden H., Podgorski M. S., Schulman B. A., Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422, 330–334 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Huang D. T., et al. , A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 11, 927–935 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schäfer A., Kuhn M., Schindelin H., Structure of the ubiquitin-activating enzyme loaded with two ubiquitin molecules. Acta Crystallogr. D Biol. Crystallogr. 70, 1311–1320 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Misra M., et al. , Dissecting the specificity of adenosyl sulfamate inhibitors targeting the ubiquitin-activating enzyme. Structure 25, 1120–1129.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Lu X., et al. , Designed semisynthetic protein inhibitors of Ub/Ubl E1 activating enzymes. J. Am. Chem. Soc. 132, 1748–1749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv Z., et al. , Domain alternation and active site remodeling are conserved structural features of ubiquitin E1. J. Biol. Chem. 292, 12089–12099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bedford L., Lowe J., Dick L. R., Mayer R. J., Brownell J. E., Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X., Dixit V. M., Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nalepa G., Rolfe M., Harper J. W., Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5, 596–613 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Soucy T. A., et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Hyer M. L., et al. , A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 24, 186–193 (2018). [DOI] [PubMed] [Google Scholar]
- 44.He X., et al. , Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat. Chem. Biol. 13, 1164–1171 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson K. D., et al. , A specific inhibitor of the ubiquitin activating enzyme: Synthesis and characterization of adenosyl-phospho-ubiquitinol, a nonhydrolyzable ubiquitin adenylate analogue. Biochemistry 29, 7373–7380 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Borodovsky A., et al. , A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 5187–5196 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muir T. W., Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 72, 249–289 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Olsen S. K., Lima C. D., S. pombe ubiquitin E1 complex with a ubiquitin-AMP mimic. Protein Data Bank. https://www.rcsb.org/structure/6O82. Deposited 24 January 2019.
- 49.Hann Z. S., Lima C. D., S. pombe ubiquitin E1∼ubiquitin-AMP tetrahedral intermediate mimic. Protein Data Bank. https://www.rcsb.org/structure/6O83. Deposited 24 January 2019.
- 50.Tokgöz Z., Bohnsack R. N., Haas A. L., Pleiotropic effects of ATP.Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme. J. Biol. Chem. 281, 14729–14737 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Lake M. W., Wuebbens M. M., Rajagopalan K. V., Schindelin H., Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414, 325–329 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Lehmann C., Begley T. P., Ealick S. E., Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry 45, 11–19 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayward S., Berendsen H. J. C., Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins 30, 144–154 (1998). [PubMed] [Google Scholar]
- 54.Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., Haas A. L., Protein interactions within the N-end rule ubiquitin ligation pathway. J. Biol. Chem. 278, 9448–9457 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Huang D. T., Zhuang M., Ayrault O., Schulman B. A., Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat. Struct. Mol. Biol. 15, 280–287 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Tokgöz Z., et al. , E1-E2 interactions in ubiquitin and Nedd8 ligation pathways. J. Biol. Chem. 287, 311–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bacik J.-P., Walker J. R., Ali M., Schimmer A. D., Dhe-Paganon S., Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: Mechanistic insights into a minimalistic E1 enzyme. J. Biol. Chem. 285, 20273–20280 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noda N. N., et al. , Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Taherbhoy A. M., et al. , Atg8 transfer from Atg7 to Atg3: A distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44, 451–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong S. B., et al. , Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat. Struct. Mol. Biol. 18, 1323–1330 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Kaiser S. E., et al. , Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat. Struct. Mol. Biol. 19, 1242–1249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaguchi M., et al. , Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat. Struct. Mol. Biol. 19, 1250–1256 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Oweis W., et al. , Trans-binding mechanism of ubiquitin-like protein activation revealed by a UBA5-UFM1 complex. Cell Rep. 16, 3113–3120 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi M., et al. , Atg7 activates an autophagy-essential ubiquitin-like protein Atg8 through multi-step recognition. J. Mol. Biol. 430, 249–257 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Pangborn A. B., Giardello M. A., Grubbs R. H., Rosen R. K., Timmers F. J., Safe and convenient procedure for solvent purification. Organometallics 15, 1518–1520 (1996). [Google Scholar]
- 66.Mossessova E., Lima C. D., Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast, Mol. Cell. 5, 865–876 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 68.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brunger A. T., Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Collaborative Computational Project, Number 4 , The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 71.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.