Significance
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths. Genome sequencing studies have provided comprehensive CRC genomic datasets; however, functional validation for most candidate CRC driver genes has not been performed. Here, we describe a platform for functionally validating CRC driver genes that utilizes CRISPR-Cas9 in mouse intestinal tumor organoids and human CRC-derived organoids. These studies showed Acvr1b, Acvr2a, and Arid2 could function as tumor suppressors in CRC. We also show that co-occurrent mutations in receptors for activin and transforming growth factor-β synergistically promote tumorigenesis, and shed light on the role of activin receptors. This experimental system can also be applied to organoids derived from other organs, which could further contribute to identification of novel cancer driver genes.
Keywords: colorectal cancer, organoid, activin, driver gene, CRISPR-Cas9
Abstract
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths worldwide. Several genome sequencing studies have provided comprehensive CRC genomic datasets. Likewise, in our previous study, we performed genome-wide Sleeping Beauty transposon-based mutagenesis screening in mice and provided comprehensive datasets of candidate CRC driver genes. However, functional validation for most candidate CRC driver genes, which were commonly identified from both human and mice, has not been performed. Here, we describe a platform for functionally validating CRC driver genes that utilizes CRISPR-Cas9 in mouse intestinal tumor organoids and human CRC-derived organoids in xenograft mouse models. We used genetically defined benign tumor-derived organoids carrying 2 frequent gene mutations (Apc and Kras mutations), which act in the early stage of CRC development, so that we could clearly evaluate the tumorigenic ability of the mutation in a single gene. These studies showed that Acvr1b, Acvr2a, and Arid2 could function as tumor suppressor genes (TSGs) in CRC and uncovered a role for Trp53 in tumor metastasis. We also showed that co-occurrent mutations in receptors for activin and transforming growth factor-β (TGF-β) synergistically promote tumorigenesis, and shed light on the role of activin receptors in CRC. This experimental system can also be applied to mouse intestinal organoids carrying other sensitizing mutations as well as organoids derived from other organs, which could further contribute to identification of novel cancer driver genes and new drug targets.
Colorectal cancer (CRC) develops through the stepwise selection of genetic changes. The initiating or gatekeeping mutation is primarily 2-hit mutations in APC, which occur in >80% of the cases (1, 2). Subsequent activating mutations in KRAS occur in the early to intermediate adenoma stage, while loss-of-function mutations in SMAD4 (transforming growth factor-β [TGF-β] signaling) and TP53 occur in the intermediate to late adenoma and late adenoma to adenocarcinoma stages, respectively (3, 4). In addition to the commonly mutated APC, KRAS, SMAD4, and TP53 genes (5–7), large-scale CRC genome sequencing studies have identified numerous infrequently mutated genes in CRC, revealing extensive intertumor and intratumor genetic heterogeneity in cancer tissues (8). The existence of large numbers of infrequently mutated genes has made it a challenge to sort out driver genes from passenger genes using genomic data alone. In addition, as the number of CRC genomes sequenced has increased, the list of significantly mutated genes has also increased, including genes that seem highly suspicious on the basis of their biological functions, such as genes that encode olfactory receptors (9).
To understand the complexity of CRC, it will be important to identify all driver genes involved in CRC, including infrequently mutated ones, since infrequently mutated genes can provide seeds for metastasis or confer drug resistance (8). To comprehensively identify CRC driver genes, we previously performed Sleeping Beauty (SB) transposon-based mutagenesis screens in wild-type mice and in mice carrying sensitizing mutations in Apc, Kras, Smad4, or Trp53. Collectively, these screens identified 1,333 candidate driver genes for CRC (10, 11). By comparing these genes with the datasets of genes mutated in human CRC, we were then able to identify 128 genes that were mutated in both humans and mice, and represented potential potent candidate driver genes (10). To validate the function of these genes in CRC, a common approach would normally be to generate knockout mice for candidate tumor suppressor genes (TSGs) or transgenic mice for candidate oncogenes and to examine cancer development or progression in these mice in vivo. However, these strategies would be time-consuming and expensive; therefore, novel approaches are required to validate such a large number of candidate driver genes.
In the study described here, we show that CRISPR-Cas9 screening in mouse intestinal organoids can provide an efficient platform for CRC driver gene validation. Previous studies employing a genome-wide, loss-of-function CRISPR-Cas9 screen in non–small-cell lung cancer cell lines showed that CRISPR-Cas9 could identify genes involved in tumor growth and metastasis (12). Additionally, a CRISPR-Cas9 screen using a customized pool of guide RNAs (gRNAs) in a melanoma cell line identified Ptpn2 as a novel gene involved in PD-1 resistance (13). Therefore, we reasoned that by using a customized pool of gRNAs composed of a relatively small number of focused candidate CRC TSGs, we might be able to validate some of these genes in CRC. Instead of using cancer cell lines, we decided to perform the CRISPR-Cas9 screens in intestinal tumor-derived organoids, since organoids have already been shown to provide a powerful approach for modeling malignant CRC progression through the step-wise introduction of mutations in genes important for CRC progression in a xenograft model (14–17). Moreover, it has been shown that tumor-derived organoids more faithfully recapitulate human CRC with stromal reactions when transplanted in mice compared with cancer cell lines (18).
We performed the CRISPR-Cas9 screens and validation in organoids derived from intestinal tumors that developed in mice carrying a heterozygous loss-of-function mutation in Apc and an activating mutation in Kras (ApcΔ716:Kras+/G12D; AK organoids) (19). Loss of the wild-type Apc gene occurs in the initiation step of tumor development in Apc+/Δ716:Kras+/G12D mice; therefore, only the mutant allele for Apc (ApcΔ716) is retained in AK organoids, while Kras retains both a wild-type allele and a mutant allele. In human CRC, APC and KRAS are mutated in about 80% and 40% of cases, respectively; therefore, AK organoids mimic the initial basal tumorigenic characteristics of the major population of human CRC. AK organoids are also less transplantable in subcutis (20), thus allowing the tumorigenic potential of candidate cancer driver genes to be validated by introduction of gene mutations in AK organoids and examination of their tumorigenic potential in transplanted mice. This experimental system can also be applied to mouse intestinal organoids carrying other initiating mutations as well as organoids derived from other organs, which will further contribute to identification of novel cancer driver genes for new drug targets to cure cancer.
Results
A Platform for Functional Validation of CRC TSGs.
Twenty-nine candidate CRC TSGs were selected for functional validation in intestinal tumor organoids based on the following criteria: (i) Genes are commonly mutated in mouse intestinal tumors by SB transposon mutagenesis and in human CRC (10) (Fig. 1A), (ii) the transposon insertion patterns in mouse intestinal tumors predicted “loss of function,” (iii) the genes were frequently mutated in SB mutagenesis screens (P < 0.01), and (iv) several transcriptional termination mutations have been identified in human CRC. These genes include well-established CRC TSGs such as Pten, Smad4, and Trp53 as well as genes with unknown functions (Dataset S1, part A). We then established Cas9-expressing AK organoid clones by transduction with a plasmid carrying complementary DNAs (cDNAs) encoding Cas9 and GFP (Fig. 1 A–C). Stable expression of Cas9 is important to achieve efficient genome editing (21); therefore, we selected 2 organoid clones (C2 and C3) that showed high expression of Cas9 (Fig. 1D). We further performed functional analysis to confirm Cas9 nuclease activity in these clones by transducing them with Trp53 gRNA and culturing them in the presence of nutlin3, which induces apoptosis in p53 wild-type cells. The number of nutlin3-resistant organoids was significantly higher in C2 and C3 organoids transduced with Trp53 gRNA compared with control organoids (SI Appendix, Fig. S1), confirming that these organoid clones have highly functional Cas9.
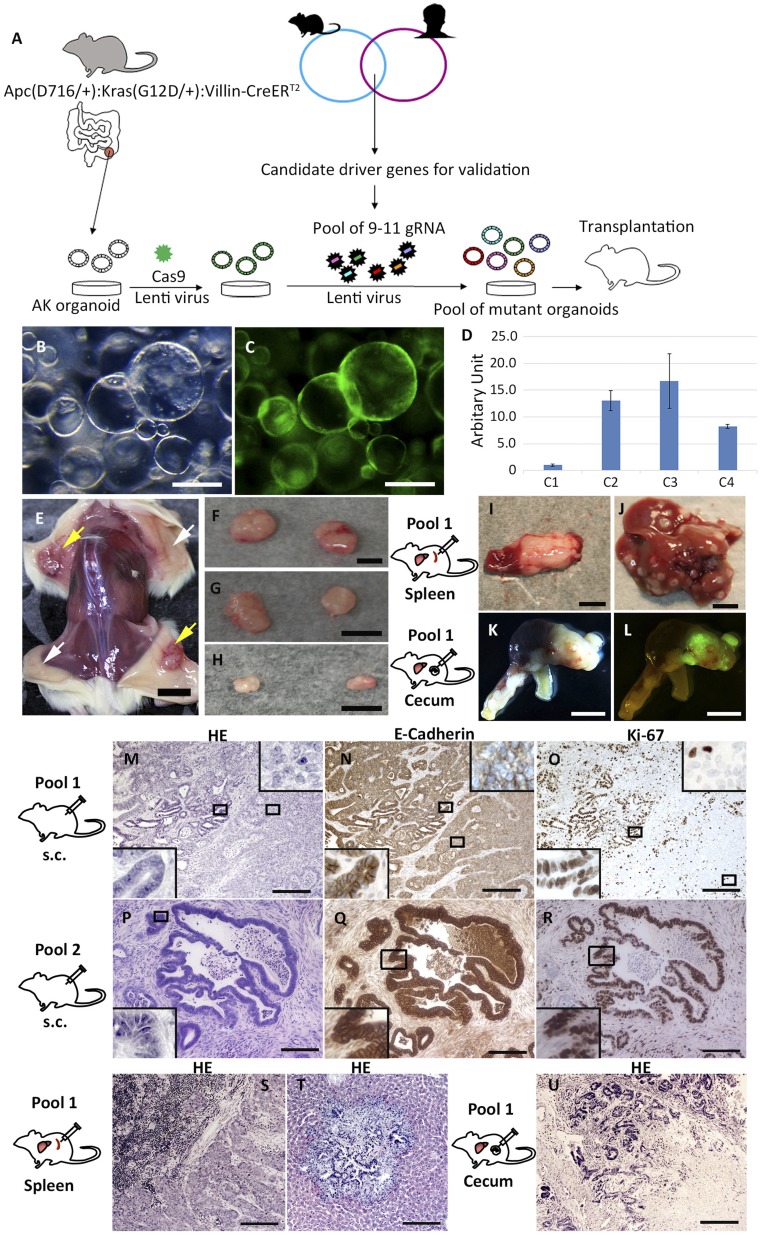
Fig. 1.
Candidate colorectal driver gene validation using mouse intestinal tumor organoids and CRISPR-Cas9. (A) Schematic view of the experimental system. Mouse intestinal tumor organoids were established from ApcΔ716:Kras+/G12D:Villin-CreERT2 mice (19). A Cas9 lentivirus containing a GFP expression cassette was introduced into the intestinal organoids. Pools of gRNAs targeting 9 to 10 unique candidate TSGs were then introduced into the GFP(+) organoids and subsequently transplanted into mice and monitored for tumor formation. (B) Macroscopic view of Cas9-expressing AK organoids. (C) GFP-positive organoids indicate that the plasmid encoding Cas9 was introduced into the organoids. (D) qPCR analysis for Cas9 messenger RNA showed that C2 and C3 organoids showed more than 10-fold higher expression compared with C1 organoids. (E and F) Macroscopic views of s.c. tumors derived from organoids with pool 1. Yellow arrows indicate s.c. tumors derived from organoids with pool 1. White arrows indicate where AK-Cas9-gRNA (nontarget) organoids were transplanted. (G) s.c. tumors derived from organoids with pool 2. (H) s.c. tumors derived from organoids with pool 3. A spleen tumor (I) and liver tumors (J), both derived from organoids with pool 1, are shown. (K) Bright-field photograph of a primary cecum tumor derived from organoids with pool 1. (L) Tumor cells derived from organoids were GFP-positive. Hematoxylin/eosin (HE) staining of s.c. tumors derived from organoids with pool 1 (M) and pool 2 (P) is shown. Immunohistochemistry (IHC) for E-cadherin on a s.c. tumor derived from organoids with pool 1 (N) and pool 2 (Q) is shown. IHC for Ki-67 on a s.c. tumor derived from organoids with pool 1 (O) and pool 2 (R) is shown. (M–R, Insets) Photographs with higher magnification. HE staining of a splenic tumor (S) and a metastatic liver tumor (T) derived from organoids with pool 1 is shown. (U) HE staining of a primary cecum tumor derived from organoids with pool 1 in an orthotopic model. (Scale bars: B and C, 250 μm; E, 1 cm; F–L, 5 mm; M–U, 200 μm.)
Next, we divided the 29 genes into 3 groups and then generated 2 different pools of gRNAs targeting the 9 to 10 genes in each group (pool 1-1, pool 1-2, pool 2-1, pool 2-2, pool 3-1, and pool 3-2; Dataset S1, part A). The gRNA pools were then transduced into AK-Cas9 organoids using lentivirus (Fig. 1A). Pool 1 gRNAs included well-known CRC TSGs such as Trp53, Smad4, and Pten, and it was used to confirm the reliability of the screening system. The gRNA vector also contained a puromycin-2A-BFP expression cassette (21); thus, puromycin selection for 2 wk was used to enrich organoids carrying both Cas9 and gRNAs, which are GFP(+) and BFP(+) (SI Appendix, Fig. S2).
Enhanced Tumorigenicity of Organoids by Introduction of Cas9 and gRNA Pools.
Mice transplanted with AK organoids transduced with pool 1, 2, or 3 gRNAs developed subcutaneous (s.c.) tumors (Fig. 1 E–H), while mice transplanted with parental AK organoids did not, showing that pool 1, 2, and 3 gRNA organoids have tumorigenic potential. To determine whether they also have metastatic potential, we transplanted pool 1 organoids to the mouse spleen. Not only did mice transplanted with pool 1 organoids develop splenic tumors (Fig. 1 I and S and SI Appendix, Fig. S3A) but they also developed liver metastases (Fig. 1 J and T and SI Appendix, Fig. S3B). In contrast, mice transplanted with pool 2 or pool 3 organoids did not develop spleen tumors or liver metastases (Dataset S1, part B). We also orthotopically transplanted pool 1 organoids to the cecum, which induced both large primary cecal tumors (Fig. 1 K, L, and U) and liver metastases (SI Appendix, Fig. S3C and Dataset S1, part B). In contrast, mice transplanted with control AK organoids developed primary cecal tumors but no liver metastases (SI Appendix, Fig. S4). Pool 1 organoids thus also have metastatic potential in the orthotopic model. These data suggest that loss of function of 1 or more genes targeted by the gRNAs in pool 1 is responsible for metastasis in both splenic and orthotopic transplantation models.
Histologically, s.c. tumors derived from pool 1 (Fig. 1M) and pool 2 (Fig. 1P) showed branched glandular structures, consisting of E-cadherin–positive cells (Fig. 1 N and Q), and the tumor cells proliferated intensively, as shown by Ki67-positive nuclei (Fig. 1 O and R). We also found tumor cells with weak expression of E-cadherin in pool 1 organoid-derived tumors, suggesting that these tumors are polyclonal in nature (Fig. 1 N, Insets). These results show that introduction of pooled gRNAs targeting candidate CRC TSGs enhances the tumorigenicity of AK organoids.
Identification of Candidate TSGs in s.c. Tumors.
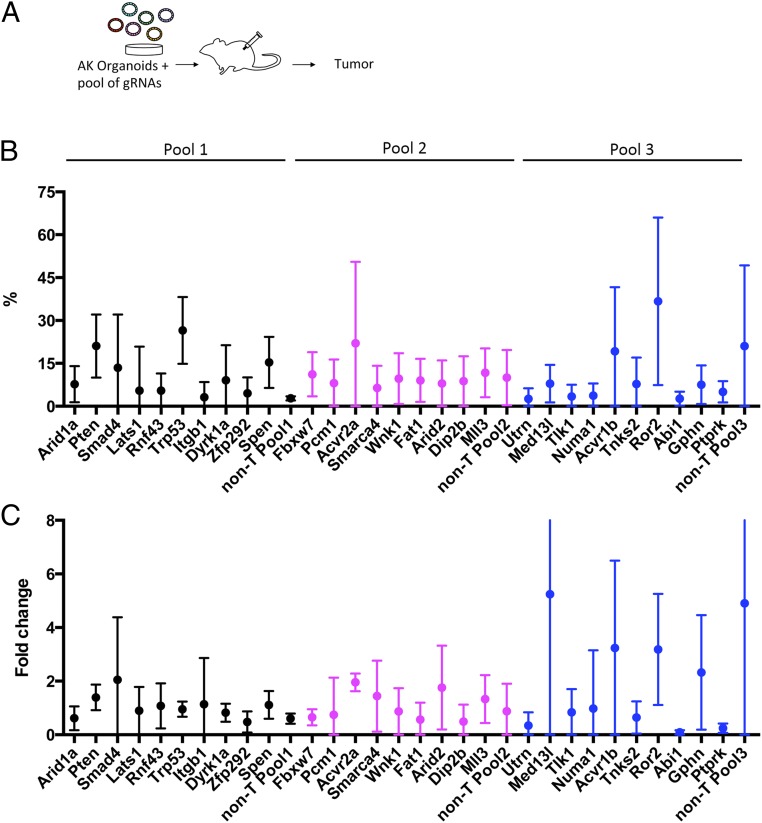
We first examined the fraction of each gRNA in preinjected organoids by deep sequencing to determine the distribution of each gRNA in the preinjected organoids (Dataset S1, parts C–E). We found that the fraction of each gRNA in preinjected organoids was heterogeneous. This is probably due to the low efficiency of lentivirus transduction to organoids; therefore, we generated 3 infectious replicates for transplantation to achieve sufficient coverage of all gRNAs. Next, to identify the genes responsible for s.c. tumor development, we looked for overrepresented gRNAs in s.c. tumors by deep sequencing (Fig. 2A). To validate the experimental system, we first examined tumors generated with pool 1 organoids, which contained established CRC TSGs. We analyzed the fraction of each gRNA in tumor tissues derived from organoids with 3 independent infectious replicates (n = 14, n = 6, and n = 8; Dataset S1, part F) and found that the dynamics of gRNA enrichment differed among independent experiments and among genes. We therefore calculated the average ratio (Fig. 2B) and fold increase (Fig. 2C) of each gRNA in tumor tissues compared with those in preinjected organoids, and identified the top TSG candidates using the following thresholds for gRNA enrichment: (i) the fraction of gRNA in tumors was 15% in ≥15% of tumors, (ii) the fold increase of gRNA in tumors compared with preinjected organoids was ≥2 in ≥15% of tumors, and (iii) these 2 criteria were met for 2 independent gRNAs for the same gene. In pool 1 tumors, Trp53, Smad4, and Pten as well as Spen, a gene with unknown functions in CRC, met these criteria (Fig. 2 B and C and Dataset S1, parts F and I). In pool 2 tumors derived from organoids for 3 independent infectious replicates (n = 5, n = 5, and n = 10), Fbxw7, Acvr2a, Arid2, and Mll3 met these criteria (Fig. 2 B and C and Dataset S1, parts G and I), while in pool 3 tumors derived from organoids for 3 independent infectious replicates (n = 6, n = 4, and n = 2), Acvr1b and Ror2 met these criteria (Fig. 2 B and C and Dataset S1, parts H and I). In total, 10 genes met these criteria from the screen in organoids.
Fig. 2.
Analysis of gRNA frequencies in tumors by next-generation sequencing. (A) Scheme describing the experimental flow. (B) Average fraction of each gRNA in tumor tissues. (C) Average fold change of each gRNA fraction in tumors compared with preinjected organoids.
Identification of TSGs Promoting Liver Metastasis.
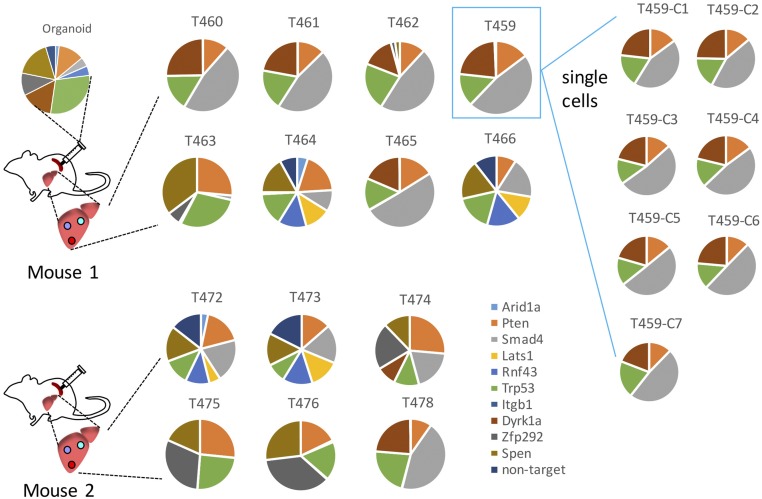
Only organoids transduced with pool 1 gRNAs induced liver metastases when transplanted to the spleen. To determine which gRNAs in pool 1 were responsible for liver metastasis, we analyzed the patterns of each gRNA fraction in metastatic liver tumors of the same and different mice. We found that the gRNA patterns in different metastatic tumors were complex, making it difficult to determine which gRNA(s) were responsible for liver metastasis. We also found that some of the metastatic tumors from the same mouse contained the same mixture of gRNAs targeting Trp53, Pten, Smad4, and Dyrk1a (Fig. 3 and Dataset S1, part J). These results suggested either of 2 possibilities: (i) These metastatic tumors were all clonally related and derived from the same primary tumor cell, or (ii) these tumors were polyclonal and composed of different tumor cells, each containing a different gRNA. To discriminate between these possibilities, we established organoids from one of these metastatic tumors (T459) and then dissociated the organoids into single cells. We then produced 10 subclones from these single cells. When we analyzed the gRNAs carried in these single cell-derived organoids, we found that they all carried the same 4 gRNAs in approximately the same ratios, showing that they are all clonally related and derived from a single tumor cell carrying 4 different gRNAs (Fig. 3). We also analyzed the gRNA patterns in metastatic liver tumors produced in the orthotopic transplantation model (SI Appendix, Fig. S5). These metastatic tumors all contained gRNAs for Pten, Trp53, Zfp292, and Spen, again suggesting that these metastatic tumors are clonally related and derived from a single tumor cell.
Fig. 3.
Analysis of gRNA frequencies. The pie charts show the fraction of gRNA in organoids, liver tumors, and organoids derived from single cells of tumor T459.
To further identify the gene(s) responsible for metastasis, we generated AK organoids carrying single gRNAs for each of the 11 genes in pool 1 and transplanted a mixture of the 11 different organoids at the same ratio to the spleen. Analysis of liver foci revealed that all tumors were composed of organoids carrying Trp53 gRNA, showing that Trp53 loss alone can confer metastatic ability to AK organoids among pool 1 genes (Dataset S1, part L) in a spleen transplantation model. Taken together, these results show that our organoid model can be used to identify genes involved in liver metastasis.
Acvr2a, Acvr1b, and Arid2 Are CRC TSGs.
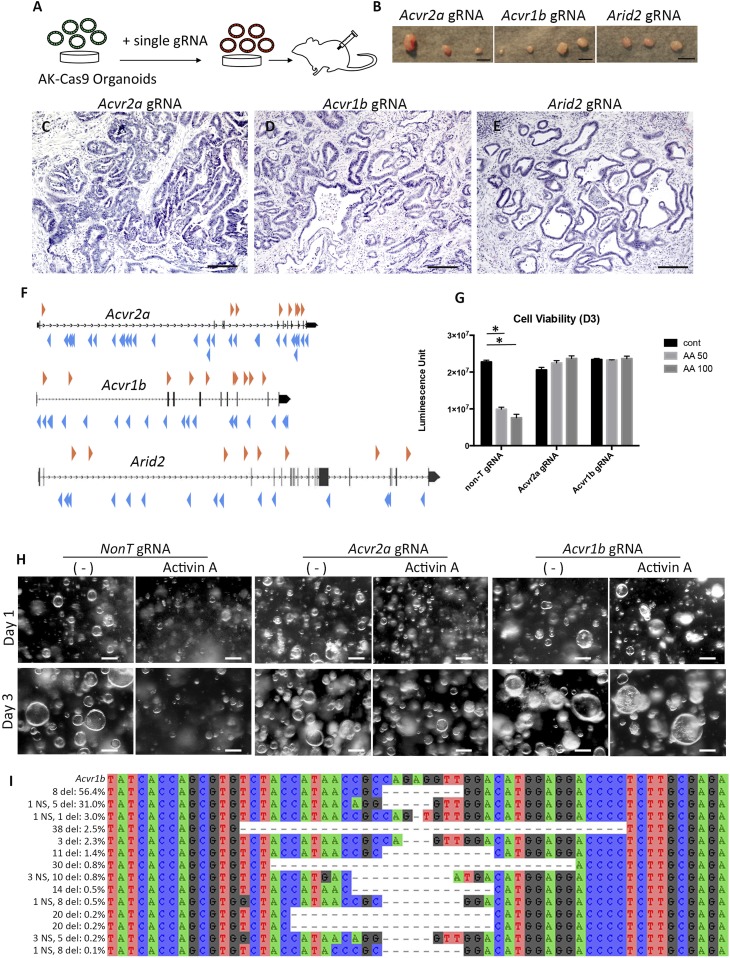
Among the 10 genes identified as candidate TSGs in s.c. tumors, we focused on Acvr2a, Acvr1b, Ror2, Spen, and Arid2, since the functions of these genes in CRC are not yet validated, and asked whether disruption of these genes individually could promote tumor development by a 1-by-1 approach. We transduced a single gRNA for each gene into AK organoids and then transplanted them s.c. into mice (Fig. 4A). The incidence of tumor formation for Trp53 and Pten gRNA organoids was 100%, while the tumor incidence for Acvr2a, Acvr1b, and Arid2 gRNA organoids was significantly increased compared with control organoids (Fig. 4B and Table 1). This is in contrast to the tumor incidence for Spen or Ror2 gRNA organoids, which was the same as for control organoids. These results show that the tumor-inducing ability of TSG dysfunction can be validated in this model. However, we failed to validate the tumor-suppressive function of Ror2 and Spen. In human CRC, termination mutations for ROR2 and SPEN have been observed, although less frequently than those for ACVR2A, ACVR1B, and ARID2. Therefore, it is possible that Ror2 and Spen have a weak tumor suppressive ability that cannot be validated in our platform.
Fig. 4.
Knockout of Acvr2a, Acvr1b, or Arid2 in AK organoids promotes s.c. tumor development. (A) Method used to test the ability of single Acvr2a, Acvr1b, or Arid2 knockouts in AK organoids to induce s.c. tumors following transplantation to NSG mice. (B) Macroscopic appearance of s.c. tumors derived from AK organoids carrying Acvr2a gRNA (Left), Acvr1b gRNA (Center), and Arid2 gRNA (Right). Hematoxylin/eosin (HE) staining of s.c. tumors derived from AK organoids containing gRNA for Acvr2a (C), Acvr1b (D), or Arid2 (E) is shown. (F) SB transposon insertion patterns were obtained from a previous study (10) for Acvr2a, Acvr1b, and Arid2. Orange arrowheads represent transposon insertions with a forward orientation relative to the transcript. Blue arrowheads represent transposon insertions with a reverse orientation relative to the transcript. (G) Bar graph indicates cell viability in the absence of activin A or in the presence of activin A (50 ng/mL or 100 ng/mL). AA, activin A; cont, control. *P < 0.05 by t test. (H) Photographs show colonic AK organoids with gRNA for non-T, Acvr2a, or Acvr1b in the presence or absence of activin A (50 ng/mL) at day 1 (Upper) and day 3 (Lower). (I) Deep sequencing analysis of the CRISPR on-target site in the Acvr1b gene. The majority of sequence reads showed deletions of insertions that resulted in frameshift mutations. del, deletion; NS, nucleotide substitution. (Scale bars: B, 5 mm; C–E, 200 μm; H, 100 μm.)
Table 1.
Efficiency for s.c. tumor development of AK organoids carrying a gRNA
| Gene symbol | Mutations in human CRC, % | P value in SB screening | gRNA-1 efficiency for tumor development | gRNA-2 efficiency for tumor development |
| Acvr2a | 12 | 2.24 × 106 | 83.3% (5/6)* | 42.9% (6/14)* |
| Acvr1b | 8 | 0 | 100% (10/10)* | 70% (7/10)* |
| Arid2 | 7 | 3.18 × 107 | 66.7% (8/12)*,† | 100% (4/4)*,† |
| Spen | 6 | 4.33 × 105 | 0% (0/6)‡ | 25% (1/4)‡ |
| Ror2 | 2.4 | 2.92 × 105 | 10% (1/10)‡ | 0% (0/4)‡ |
| Trp53 | 53 | 6.68 × 106 | 100% (8/8)* | — |
| Pten | 6 | 0 | 100% (4/4)* | — |
| Nontarget (2/14) + AK-Cas9-C2 (0/6) | — | — | 10.0% (2/20)§ | — |
AK-Cas9-C2 or AK-Cas9-C2 + gRNA (non-T) did not develop detectable tumors within 60 d.
P < 0.05, compared with nontarget by χ2 test.
Data from 1 mo after transplantation.
P > 0.05, compared with nontarget by χ2 test.
Data include both AK-Cas9-C2 and AK-Cas9-C2 + gRNA (nontarget).
Histologically, s.c. tumors were composed of glandular structures with branching similar to that of intestinal tumors observed in Apc mutant mice (19) (Fig. 4 C–E). We further confirmed that deletions or insertions were introduced by CRISPR-Cas9 by deep sequencing of gRNA target loci in Acvr2a, Acvr1b, and Arid2 in preinjected organoids (Fig. 4F and Dataset S1, part M).
In our previous SB mutagenesis screens in the intestine (10), transposons were frequently inserted in Acvr2a, Acvr1b, and Arid2. The transposon insertion patterns showed that each gene was likely disrupted by the transposon insertions; insertions with both forward and reverse orientation relative to the transcript were identified within each gene, predicting that Acvr2a, Acvr1b, and Arid2 are TSGs (Fig. 4G).
We used small intestinal tumor organoids in our experiments because they were easy to culture due to a reduced requirement for growth factors compared with colonic tumor organoids. Since ACVR2A, ACVR1B, and ARID2 are primarily mutated in colonic epithelium in human CRC, we tested whether knockout of Acvr2a, Acvr1b, and Arid2 in mouse colonic AK organoids could also induce tumor development in transplanted mice (SI Appendix, Fig. S6). These experiments showed that disruption of Acvr2a, Acvr1b, and Arid2 in mouse colonic AK organoids could also induce tumor development in transplanted mice (Dataset S1, part N), which is consistent with our results using small intestinal tumor-derived organoids.
ACVR2A and ACVR1B encode activin receptors type IIA and IB, respectively, which belong to the TGF-β superfamily-signaling pathway (SI Appendix, Fig. S7). Recent studies have shown that activin A signaling induces growth suppression of human colon cancer cells (22). We therefore treated mouse colonic AK organoids with activin A to determine whether activin A also suppressed growth suppression of mouse colonic AK organoids. Consistent with the previous report, activin A treatment of control mouse colonic AK organoids also resulted in growth suppression, whereas mouse colonic AK organoids containing Acvr1b or Acvr2a gRNA showed no growth suppression upon activin A treatment (Fig. 4 H and I). It has been reported that activin A levels are increased in the serum of CRC patients (23), probably to promote the differentiation of T cells and macrophages (24). Accordingly, these results suggest that colonic cancer cells might evade these growth-suppressive effects by acquiring mutations in activin receptors.
ACVR1B, ACVR2A, and ARID2 Mutations in Human CRC.
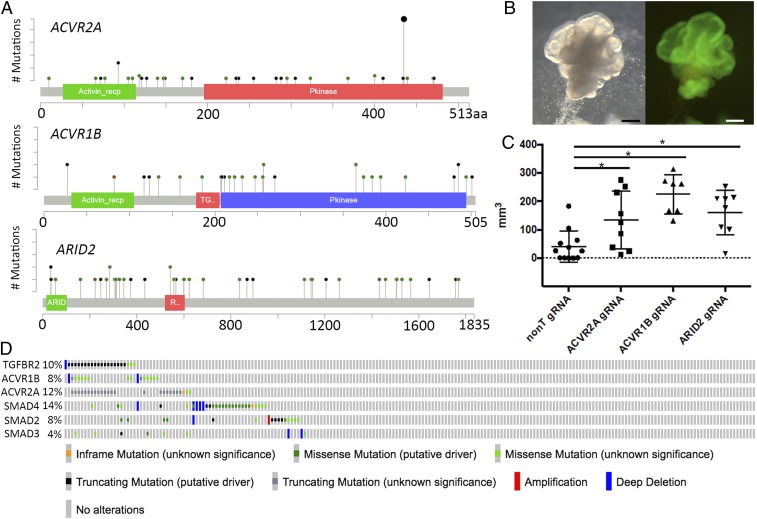
The cBioPortal describes the patterns of somatic mutations in individual genes obtained from cancer genome sequencing studies (25, 26). We therefore examined 2 large-scale CRC genome sequencing datasets (5, 6) for mutations in ACVR1B, ACVR2A, and ARID2 (Fig. 5A). We found numerous transcriptional termination mutations scattered throughout each gene in human CRC; therefore, it is highly likely that loss of function of these genes also contributes to human CRC development, consistent with our mouse results. In small bowel adenocarcinomas, similar mutations in ARID2, ACVR2A, and ACVR1B have also been reported in 7%, 9%, and 5% of tumors, respectively (27), further supporting these results.
Fig. 5.
Analysis of ACVR2A, ACVR1B, and ARID2 mutations in human CRC. (A) Spectrum of mutations in ACVR2A, ACVR1B, and ARID2 in human CRC. The mutational results are based on the data generated by the The Cancer Genome Atlas Research Network study (5) (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Black circles denote termination mutations, while green circles denote nonsynonymous mutations. (B) Transduction of Cas9 to human CRC-derived organoids. The plasmid contains both a Cas9 cDNA and a GFP cDNA; therefore, Cas9-expressing organoids can be visualized by GFP. (Left) Bright field. (Right) GFP. (C) Transduction of ARID2 gRNA or ACVR1B gRNA or ACVR2A into Cas9-expressing human CRC organoids promoted tumor growth compared with parental organoids. *P < 0.05 by t test. (D) Oncoprint showing the frequencies of mutations for genes belonging to the TGF-β superfamily in CRC. (Scale bars: 200 μm.)
To test whether the tumor-suppressive roles of ARID2, ACVR2A, and ACVR1B are also observed in the context of human CRC, we disrupted these genes by CRISPR-Cas9 in human CRC-derived organoids. We established organoids from human CRC and then transduced them with a Cas9-GFP plasmid (Fig. 5B and SI Appendix, Fig. S6) in addition to gRNAs for ARID2, ACVR2A, or ACVR1B. Notably, the mean size of tumors generated from the s.c. transplantation of human organoids carrying ARID2 (n = 8), ACVR2A (n = 10), or ACVR1B (n = 7) gRNA was significantly larger than that of tumors generated from control organoids, confirming that ARID2, ACVR2A, and ACVR1B are TSGs for human CRC (Fig. 5C). AIRD2 (AT-rich interacting domain 2) belongs to the SNF/SWI family and encodes a PBAF complex subunit, while ARID1A encodes a BAF complex subunit (28, 29). Recent studies have shown that Arid2 functions as a TSG in the liver through the disruption of the DNA repair process (30). Our results show that ARID2 also functions as a TSG in the colon.
Other TGF-β superfamily pathway-related genes such as TGFBR2 (TGF-β receptor type II) and SMAD4 are also known TSGs in CRC (31–33). Notably, in 2 CRC datasets (5, 6), we found that ACVR2A or ACVR1B mutations co-occurred significantly with TGFBR2 mutations (Fig. 5D and Dataset S1, parts O and P), but not with SMAD4 mutations. These data suggest a functional synergy between activin receptors and TGF-β receptors in CRC, and raise the possibility that once a tumor cell acquires a mutation in a single TGF-β superfamily receptor, mutations in other TGF-β superfamily receptors might be preferentially selected to promote tumorigenesis.
Cooperation of Acvr2a or Acvr1b Mutations with Tgfbr2 Mutations in CRC.
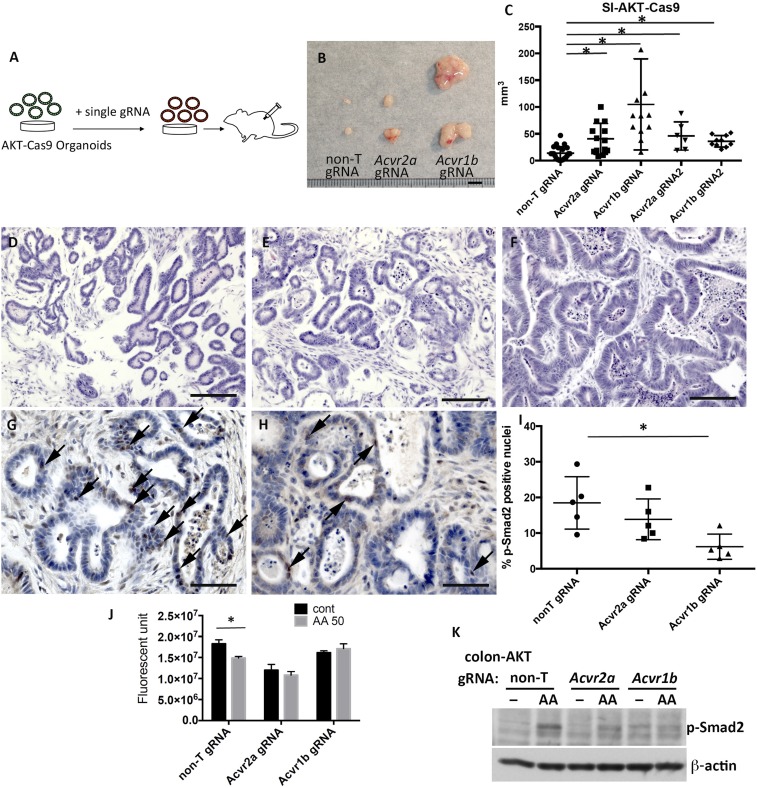
To examine the functional significance of co-occurring mutations in activin and TGF-β receptors, we disrupted Acvr2a or Acvr1b in AK organoids, which also carried homozygous deletions in the TGFBR2 (AKT organoids) (19) (Fig. 6A). The size of the s.c. tumors generated in transplanted mice was significantly increased when Acvr1b or Acvr2a was disrupted in AKT organoids in 2 independent experiments with different gRNAs (Fig. 6 B and C). Similarly, disruption of Acvr1b in colonic tumor-derived AKT organoids also promoted tumor development compared with control colonic AKT organoids (SI Appendix, Figs. S6 and S8). Histologically, disruption of Acvr2a or Acvr1b did not cause increased malignancy and the tumors were the typical glandular-type tumors seen with small intestinal AK organoids (Fig. 6 D–F).
Fig. 6.
Acvr2a or Acvr1b mutations synergistically cooperate with Tgfbr2 mutations in AKT organoids in tumor development. (A) Scheme describing the experimental design to knock out Acvr1b and Acvr2a in AKT organoids. (B) Macroscopic view of the s.c. tumors derived from AKT-Cas9 organoids carrying non-T gRNA, Acvr2a gRNA, or Acvr1b gRNA. (C) Graph shows the tumor size distribution for AKT-Cas9 organoids carrying non-T gRNA, Acvr2a gRNA, or Acvr1b gRNA. SI, small intestine. *P < 0.05 by χ2 test. Hematoxylin/eosin staining of s.c. tumors derived from AKT-Cas9 organoids carrying non-T gRNA (D), Acvr2a gRNA (E), or Acvr1b gRNA (F) is shown. Immunohistochemistry of p-Smad2 on the s.c. tumors derived from AKT-Cas9 non-T gRNA organoids (G) or AKT-Cas9 Acvr1b gRNA organoids (H) is shown. Black arrows indicate p-Smad2–positive nuclei in tumor epithelial cells. (I) Percentage of p-Smad2–positive nuclei in tumor epithelial cells. Five views were taken from 3 independent tumors, and >1,000 cells in total were counted. *P < 0.05. (J) Bar graph indicates cell viability in the absence of activin A or in the presence of activin A (AA; 50 ng/mL). cont, control. *P < 0.05 by t test. (K) Twelve hours after activin A (50 ng/mL) treatment, the level of p-Smad2 was analyzed by Western blotting. *P < 0.05 by t test. (Scale bars: B, 5 mm; D–F, 100 μm; G and H, 50 μm.)
Next, we tested whether the functional synergy between Tgfbr2 and Acvr1b in tumor development was mediated via Smad2/3 phosphorylation. Since canonical signaling of activin A and TGF-β commonly activates Smad4-dependent transcription via phosphorylation of Smad2 or Smad3 (24), we performed immunohistochemical analysis for phosphorylated Smad2 (p-Smad2). The number of tumor cells with p-Smad2–positive nuclei was significantly decreased in tumors produced by AKT Acvr1b gRNA organoids compared with tumors produced by control organoids (Fig. 6 G–I), further confirming that Acvr1b signals through Smad2. Activin A also showed a growth-suppressive function in control colonic AKT organoids, but not in colonic AKT organoids containing Acvr2a or Acvr1b gRNA (Fig. 6J). Control colonic AKT organoids also showed increased levels of p-Smad2 in response to activin A, whereas AKT organoids containing Acvr2a or Acvr1b gRNA showed a weak or no increase in p-Smad2 levels (Fig. 6K). These results, taken together, show that disruption of Tgfbr2 and Acvr1b synergistically enhances tumor development via the canonical TGF-β signaling pathway.
Discussion
Here, we describe a platform for functionally validating CRC driver genes that utilizes CRISPR-Cas9 in intestinal tumor organoids. Most CRISPR-Cas9–based screens performed to date have used cancer cell lines (12, 13, 34); however, we show that intestinal tumor organoids can also provide a platform for CRC driver gene validation. Cancer cell lines carry numerous genetic alterations, which often confer strong tumorigenic ability. In addition, cell lines carry different combinations of gene mutations; therefore, the effect of single gene knockouts is sometimes not consistent between cell lines. The organoid culture system makes it possible to culture normal intestinal epithelial cells as well as benign tumor-derived epithelial cells that cannot be cultured in ordinary culture systems. Taking advantage of this, we used genetically defined benign tumor-derived organoids carrying a minimum combination of gene mutations so that we could clearly evaluate the tumorigenic ability of mutations in a single gene. In addition, organoids can more precisely recapitulate human CRC when transplanted s.c., with a similar stromal cell reaction. In this study, we used “ground-state” AK organoids; however, organoids carrying other combinations of mutations could also be used to identify driver genes and pathways involved in cancer progression, metastasis, and drug resistance. The disadvantage of using intestinal tumor organoids, however, is that organoids are hard to engineer due to the low efficiency of lentivirus transduction. This is the main reason why we used a minipool, which was composed of 9 to 10 gRNAs; made 3 infectious replicates to analyze the dynamics of the fraction of all gRNAs in tumor development; and confirmed the tumorigenic ability, again by a single gene knockout approach, to overcome this disadvantage.
The tumorigenic potential of AK organoids transduced with 1 of 3 gRNA pools targeting 29 candidate TSG genes, including well-known CRC TSGs such as Trp53, Smad4, and Pten, was demonstrated by s.c. injection. Subsequent transplantation experiments showed that pool 1 organoids containing Trp53, Smad4, and Pten could also induce splenic tumors and liver metastases when transplanted to the spleen. Similarly, orthotopic transplantation of pool 1 organoids to the cecum could also induce large cecal tumors and metastases. These results indicated that loss of function of 1 or more genes targeted by a gRNA in pool 1 is responsible for the liver metastasis observed in the spleen transplantation and orthotopic models.
To identify the gene(s) responsible for liver metastases, we generated AK organoids carrying single gRNAs for each of the 11 genes contained in pool 1 and transplanted them to the spleen. Analysis of the liver metastases showed that all metastases were derived from organoids carrying Trp53 gRNA, indicating that Trp53 loss alone could confer metastatic ability to AK organoids. These results are consistent with human data showing the TP53 mutations occur in 40 to 50% of sporadic CRC, primarily at the transition from the late adenoma stage to the adenocarcinoma stage. Loss of TP53 has been shown to de-repress the Wnt pathway and epithelial–mesenchimal transition through microRNA-34 (35, 36) and to correlate with the site, biological behavior, and outcome of CRC (37). TP53 mutations that lose transcriptional activation ability are also more common in advanced CRC and associated with poor survival (38).
The role of activin receptors in cancer development has not been fully elucidated compared with the other receptors in the TGF-β superfamily; however, mutations in ACVR2A and ACVR1B have been reported in several cases of CRC (39–41). In a mouse pancreatic cancer model, ablation of Acvr1b in pancreatic ductal cells was shown to promote cancer development (42), indicating that activin receptor type IB can function as a TSG in the pancreas. Activin signaling has also recently been shown to suppress growth of human colon cancer cells (22), similar to our studies demonstrating that activin can suppress growth of mouse colonic AK organoids. Paradoxically, activin serum levels increase in CRC patients (23), perhaps to promote tumor malignancy by stimulating the differentiation of T cells and macrophages. Our studies suggest that colon cancer cells might evade these growth-suppressive effects by acquiring mutations in activin receptors.
Other TGF-β superfamily-related genes such as TGFBR2 and SMAD4 are also CRC TSGs (31, 32). By examining 2 CRC datasets (5, 6), we found that TGFBR2 mutations co-occur with ACVR2A or ACVR1B mutations in human CRC, suggesting a functional synergy between activin and TGF-β receptors in CRC. We subsequently confirmed this synergy by showing that AK organoids, which also carried homozygous deletions in the TGFBR2 (AKT organoids), generated larger tumors in transplanted mice when Acvr2a or Acvr1b was also disrupted. Interestingly, recent studies have shown that serum levels of TGF-β and activin are significantly correlated with each other in CRC patients and lead to decreased disease-free survival (43). The main source of TGF-β and activin is the stroma (43), raising the possibility that selective pressure from the tumor microenvironment could be the driving force behind the selection of mutations in these genes.
Materials and Methods
Generation of Lentiviral Vectors and Particles.
The Cas9 cDNA was excised by AgeI/XhoI digestion from pX330 (Addgene), blunt-ended using a Blunting Kit (Takara), and inserted into SwaI-digested pCDH1-MSCV-MCS-EF1-GFP (CD711B; System Biosciences) to generate pCDH1-MSCV-Cas9-EF1-GFP. To generate gRNA lentiviral vectors, we followed the protocol previously described (21). Briefly, the “TOP” oligo and the “BOTTOM” oligo (Dataset S1, part A) were annealed and ligated into the BbsI-digested pKLV vector (Addgene). For generation of lentivirus, HEK293FT cells were plated on polylysine-coated 6-cm dishes in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS) and penicillin/streptomycin (Invitrogen). The pCDH1-MSCV-Cas9-EF1-GFP was cotransfected with pCMV-dR8-2DVP and pCMV-VSV-G (both from Addgene) to generate Cas9 lentiviral particles. The pKLV vector with gRNA was cotransfected with packaging vectors (pLP1, pLP2, and pVSV-G) using polyethylenimine. To generate pooled gRNA virus, equal amounts of gRNA plasmids were pooled and transfected to 293T cells. On the next day, the culture medium was replaced with 5 mL of DMEM containing high glucose supplemented with B27 and N2. Forty-eight hours later, viral supernatants were collected and concentrated to 50 μL using a Lenti-X concentrator (TAKARA).
Transduction of Lentivirus to Mouse Tumor Organoids.
Organoids were dissociated into single cells using TrypLE (Invitrogen). A total of 1 × 105 cells were mixed with polybrene (Millipore) and virus particles in a 48-well plate, centrifuged at 600 × g for 1 h at 37 °C, and incubated for 6 h at 37 °C in a CO2 incubator. Organoids were then embedded in 30 μL of Matrigel (BD Sciences) and 300 μL of medium overlaid. The organoid culture medium contained Y-27635 and CHIR99021. To establish Cas9-expressing clones, Cas9 lentivirus was transduced into organoids, and Cas9-expressing clones were confirmed by qPCR (SI Appendix, Fig. S6).
At 24 to 48 h after transduction of gRNAs, puromycin (0.8 μL/mL) was added to select pKLV-positive cells. We made 3 independent infectious replicates with the different combinations of Cas9 clones and gRNAs for each pool. The gRNA-1 was used in experiments 1 and 2, while gRNA-2, which targets another region of the gene, was used in experiment 3. AK-Cas9 clone 2 was used in experiments 1 and 3, and AK-Cas9 clone 3 was used in experiment 2. To monitor the dynamics of AK organoids, nontarget gRNA was newly added in experiment 3. We sequenced several organoid samples, calculated the average ratios of each gRNA, and used this information for comparison (Dataset S1, parts C–E).
Mouse and Human Organoid Cultures.
Establishment and maintenance of mouse organoids were performed as previously described (44). For establishment of human CRC-derived organoids, a small portion of CRC was minced and put into a collagenase solution in which 0.02 g of collagenase type I (Gibco) was dissolved in 1 mL of 0.1% bovine serum albumin/phosphate-buffered saline (PBS). After 30 min of incubation at room temperature, the sample was washed with PBS, filtered with a 100-μm filter, and embedded in 30 μL of Matrigel, and 300 μL of medium was then overlaid. The medium for human organoids contained 50% of L-WRN–conditioned medium (45), 20% FBS (Biosera), penicillin/streptomycin (1:100), GlutaMax (1:100), Y-27632 (final 10 nM), and A83-01 (final 5 nM) in advanced DMEM-F12. For maintenance, organoids were dissociated from Matrigel using cell recovery solution, washed with PBS, and passaged once a week at a 1:3 to 1:4 ratio. All experiments involving human samples were approved by the Human Genome/Gene Analysis Research Ethics Committee of Kanazawa University [2016-086(433)]. Written informed consent was obtained from the patients for establishment of patient-derived organoids. Pictures of organoids were taken using a stereoscopic microscope (LEICA M205 FCA; Leica) with a LEICA DFC 7000T camera (Leica) or an inverted microscope system (IX73) with a DP73 camera system (Olympus).
Fluorescence-Activated Cell Sorting.
Organoids were dissociated into single cells using TrypLE (Invitrogen) and dissolved in PBS for sorting.
qPCR.
RNA was purified from organoids using the FastGene RNA Basic Kit (Nippon Genetics Co., Ltd). cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara) following the manufacturer’s protocol. The primer set for Gapdh was used as an internal control (MA050371; Takara). To quantitate the amount of Cas9 messenger RNA, hCas9-RT-L1:TCCTGCAGACAGTGAAGGTG and hCas9-RT-R3: TTCTGGGTGGTCTGGTTCTC were used. qPCR was performed using SYBR Premix Ex Taq (Takara) on Stratagene Mx3000P (Agilent Technology). The level of Cas9 RNA was compared using the delta cycle threshold method.
Transplantation.
Organoids were dissociated using TrypLE at 37 °C for 8 min. Cells were then washed using PBS and counted. A total of 1 × 105 cells dissolved in 100 μL of organoid culture medium containing 50% Matrigel were transplanted s.c. into recipient NOD.Cg-Prkdc<scid>Il2rg<tmlWjl>/SzJ (NSG) mice (Charles River Laboratories) using 27-gauge syringes. We used NSG mice, which lack T, B, and natural killer cells, since the transplantation efficiency is much better in NSG mice compared with C57BL/6 mice (19). For spleen transplantation, 1 × 105 cells dissolved in 10 μL of organoid culture medium containing 100% Matrigel were injected into the spleen of NSG mice using 30-gauge syringes. For cecum transplantation, 4 to 6 × 105 cells dissolved in 30 μL of organoid culture medium containing 100% Matrigel were injected into the subserosa of the cecum. All animal experimental protocols were approved by the Committee on Animal Experimentation of Kanazawa University.
Cell Titer Assays.
A total of 1 × 104 cells were plated into each well of a 96-well plate. Two or 3 d after the addition of activin A, cell viability was assayed using the CellTiter-Glo 3D Cell Viability Assay (Promega). Luminescence was measured by a Centros XS3 LB960 instrument using MicroWin2000 software.
Illumina Sequencing.
The primer sets HT-gLibrary-Miseq_150bp-PE-F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGcttgaaagtatttcgatttcttgg and HT-gLibrary-Miseq_150bp-PE-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGactcggtgccactttttcaa were used to amplify the gRNA locus. The PCR condition was 95 °C (20 s), 55 °C (20 s), and 72 °C (30 s) for 26 cycles. Q5 taq polymerase (New England Biolabs) was used. Secondary PCR was performed using Nextra XT index primers (Illumina). The secondary PCR condition was 95 °C (20 s), 55 °C (20 s), and 72 °C (30 s) for 12 cycles. PCR products were purified using a PCR purification kit (Hokkaido System Science) and sequenced on a Next-Seq 500 (Illumina) system (75-bp single-end reads). For CRISPR on-target site sequencing, primary PCR was performed using region-specific primers and secondary PCR was performed using index primers. The PCR condition was the same as above. PCR products were pooled and purified using a QIAprep kit (Hokkaido System Science). Paired-end sequencing (150-bp reads) was performed on an Illumina NextSeq 500 or HiSeq sequencer. Sequence reads were trimmed and quality-filtered using Trim Galore! (version 0.4.4), Trimmomatic (ver. 0.36), and Cutadapt (ver. 1.16), and then mapped to the mouse genome (genome assembly: GRCm38.p6) using the Burrows–Wheeler Aligner (BWA-SW; version 0.7.17-r1188) with default parameters. Reads mapping to target gene regions were extracted using SAMtools (version 1.8). Merged sequences of paired-end reads (contigs) were generated using FLASH (version 1.2.11) with default parameters. Frequency of occurrence of each contig sequence was calculated using the table function of R (version 3.4.1). Unique contig sequences were aligned with the corresponding regions of the mouse genome using MAFFT (version 7.407) with the following parameters: –adjustdirection –anysymbol –preservecase –genafpair –maxiterate 1000.
Histology.
Tumor tissues were fixed in 4% paraformaldehyde and processed to make paraffin blocks, following a standard protocol. Four-micrometer-thick sections were made and stained with hematoxylin/eosin. For immunohistochemistry, sections were blocked using horse serum and incubated with antibody for Ki67 (Life Technologies), E-Cadherin (R&D Systems), or phosphor-Smad2 (Millipore) overnight at 4 °C. The sections were then incubated with horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) for 30 min at room temperature. To visualize the signal, 3,3′-diaminobenzidine was used. Sections were stained with hematoxylin and then embedded with Eukit. The pictures were taken using a BZ-9000 microscope (Keyence).
Western Blotting.
Organoids were dissociated from Matrigel using cell recovery solution (Corning), washed with PBS, lysed with TNE solution (10 mM Tris⋅HCl [pH 7.6], 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% Nonidet P-40, protease inhibitor [Roche], and phosphatase inhibitor [Wako]). The solution was mixed with 5× sodium dodecyl sulfate sample buffer and boiled for 5 min at 95 °C. Western blotting was performed following the standard protocol. Phospho-Smad2/3 (Cell Signaling) and β-actin (Sigma) were used.
Supplementary Material
Acknowledgments
We thank the The Cancer Genome Atlas Research Network for sharing the CRC somatic mutational data. We also thank A. Tsuda and M. Watanabe for technical assistance. This research was supported by a Grand-in-Aid for Scientific Research (Grant 17H03586), the Japan Agency for Medical Research and Development (Grant 19cm0106533h0002), the Leading Initiative for Excellent Young Researchers, the Naito Foundation, the Takeda Foundation, the MSD Life Science Foundation, and the Hokkoku Cancer Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904714116/-/DCSupplemental.
References
- 1.Kinzler K. W., Vogelstein B., Lessons from hereditary colorectal cancer. Cell 87, 159–170 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Pinto D., Clevers H., Wnt, stem cells and cancer in the intestine. Biol. Cell 97, 185–196 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Bos J. L., ras oncogenes in human cancer: A review. Cancer Res. 49, 4682–4689 (1989). [PubMed] [Google Scholar]
- 4.Baker S. J., et al. , p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 50, 7717–7722 (1990). [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannakis M., et al. , Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 15, 857–865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seshagiri S., et al. , Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B., et al. , Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence M. S., et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda H., et al. , Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat. Genet. 47, 142–150 (2015). [DOI] [PubMed] [Google Scholar]
- 11.March H. N., et al. , Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat. Genet. 43, 1202–1209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S., et al. , Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manguso R. T., et al. , In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matano M., et al. , Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Drost J., et al. , Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015). [DOI] [PubMed] [Google Scholar]
- 16.O’Rourke K. P., et al. , Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 35, 577–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roper J., et al. , In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S. H., et al. , Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai E., et al. , Combined mutation of Apc, Kras, and Tgfbr2 effectively drives metastasis of intestinal cancer. Cancer Res. 78, 1334–1346 (2018). [DOI] [PubMed] [Google Scholar]
- 20.de Sousa e Melo F., et al. , A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Koike-Yusa H., Li Y., Tan E. P., Velasco-Herrera Mdel. C., Yusa K., Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Bauer J., Sporn J. C., Cabral J., Gomez J., Jung B., Effects of activin and TGFβ on p21 in colon cancer. PLoS One 7, e39381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S., et al. , Activin A as a novel biomarker for colorectal adenocarcinoma in humans. Eur. Rev. Med. Pharmacol. Sci. 19, 4371–4378 (2015). [PubMed] [Google Scholar]
- 24.Chen W., Ten Dijke P., Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 16, 723–740 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hänninen U. A., et al. , Exome-wide somatic mutation characterization of small bowel adenocarcinoma. PLoS Genet. 14, e1007200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson B. G., Roberts C. W., SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Hodges C., Kirkland J. G., Crabtree G. R., The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oba A., et al. , ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. J. Hepatol. 66, 942–951 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Oshima H., et al. , Suppressing TGFβ signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res. 75, 766–776 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Takaku K., et al. , Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92, 645–656 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Muñoz N. M., et al. , Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 66, 9837–9844 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Tzelepis K., et al. , A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim N. H., et al. , p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle 12, 1578–1587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim N. H., et al. , p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 4, ra71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russo A., et al. ; TP53-CRC Collaborative Study Group , The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: Influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 23, 7518–7528 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Iacopetta B., et al. ; TP53-CRC Collaborative Group , Functional categories of TP53 mutation in colorectal cancer: Results of an International Collaborative Study. Ann. Oncol. 17, 842–847 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Jung B., et al. , Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology 126, 654–659 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Jung B., Staudacher J. J., Beauchamp D., Transforming growth factor β superfamily signaling in development of colorectal cancer. Gastroenterology 152, 36–52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roerink S. F., et al. , Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Qiu W., et al. , Loss of activin receptor type 1B accelerates development of intraductal papillary mucinous neoplasms in mice with activated KRAS. Gastroenterology 150, 218–228.e2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staudacher J. J., et al. , Activin signaling is an essential component of the TGF-β induced pro-metastatic phenotype in colorectal cancer. Sci. Rep. 7, 5569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato T., et al. , Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Miyoshi H., Ajima R., Luo C. T., Yamaguchi T. P., Stappenbeck T. S., Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.