Significance
The remains of 2 Neanderthals were found in Gibraltar: the first at Forbes’ Quarry in 1848 and the second at Devil’s Tower in 1926. Since their discovery, present-day human DNA contamination has accumulated in the specimens. By developing a DNA library preparation method that reduces modern contamination before sequencing, we were able to isolate enough endogenous DNA from the specimens to determine their sex and to infer that the Forbes’ Quarry Neanderthal is more similar to 60,000- to 120,000-y-old Neanderthal specimens in Europe and western Asia than to younger Neanderthals. The laboratory protocols presented here improve access to ancient DNA from specimens that are highly contaminated with present-day human DNA.
Keywords: Gibraltar Neanderthal, Forbes’ Quarry, paleogenetics, ancient DNA, library preparation
Abstract
The Forbes’ Quarry and Devil’s Tower partial crania from Gibraltar are among the first Neanderthal remains ever found. Here, we show that small amounts of ancient DNA are preserved in the petrous bones of the 2 individuals despite unfavorable climatic conditions. However, the endogenous Neanderthal DNA is present among an overwhelming excess of recent human DNA. Using improved DNA library construction methods that enrich for DNA fragments carrying deaminated cytosine residues, we were able to sequence 70 and 0.4 megabase pairs (Mbp) nuclear DNA of the Forbes’ Quarry and Devil’s Tower specimens, respectively, as well as large parts of the mitochondrial genome of the Forbes’ Quarry individual. We confirm that the Forbes’ Quarry individual was a female and the Devil’s Tower individual a male. We also show that the Forbes’ Quarry individual is genetically more similar to the ∼120,000-y-old Neanderthals from Scladina Cave in Belgium (Scladina I-4A) and Hohlenstein-Stadel Cave in Germany, as well as to a ∼60,000- to 70,000-y-old Neanderthal from Russia (Mezmaiskaya 1), than to a ∼49,000-y-old Neanderthal from El Sidrón (El Sidrón 1253) in northern Spain and other younger Neanderthals from Europe and western Asia. This suggests that the Forbes’ Quarry fossil predates the latter Neanderthals. The preservation of archaic human DNA in the warm coastal climate of Gibraltar, close to the shores of Africa, raises hopes for the future recovery of archaic human DNA from regions in which climatic conditions are less than optimal for DNA preservation.
The retrieval of complete (1, 2) and partial (3, 4) nuclear genome sequences from Neanderthals has begun to provide insights into their population history. Apart from the ∼430-thousand-year (ka)-old Sima de los Huesos individuals, from whom only very little DNA has been sequenced (5), the genetically most divergent Neanderthal lineage known to date is represented by the ∼130-ka-old Altai Neanderthal (Denisova 5) from Denisova Cave in the Altai Mountains in Russia, which has yielded a high-quality genome sequence (1) and is one of the eastern-most Neanderthal specimens found. All other Neanderthal individuals from which substantial parts of the nuclear genome have been sequenced (1, 3, 4, 6) are more closely related to a >44-ka-old individual from Vindija Cave, Croatia (Vindija 33.19), the other individual from whom a high-quality genome sequence has been published (2). These include the Hohlenstein-Stadel specimen from Southern Germany (4, 7, 8), as well as Scladina I-4A from Belgium (4, 9), both of which are similar in age or slightly younger than the Altai Neanderthal (1, 4, 8); a ∼60- to 70-ka-old specimen from Mezmaiskaya Cave, Russia (Mezmaiskaya 1) (1, 10); and a ∼80-ka-old specimen (Chagyrskaya 8 from Chagyrskaya Cave, Russia) (6, 11), which was also recently sequenced to high coverage (6). The genomes of 4 late Neanderthals, which were dated to between 47 and 39 ka [Goyet Q56-1 (12) and Spy 94a (13), both from Belgium; Les Cottés Z4-1514 (14) from France; and Mezmaiskaya 2 from Russia (15)] (3) are even more similar to Vindija 33.19 than those of the aforementioned individuals (Hohlenstein-Stadel, Chagyrskaya 8, Mezmaiskaya 1, Scladina I-4A) (1, 3, 4, 16), despite spanning a geographic range from western Europe to western Asia (for an overview of all specimens, see SI Appendix, Table S1). Thus, there is currently no evidence for the existence of substantial genetic substructure in the Neanderthal population after ∼90 ka ago (4), the time at which the “Altai-like” Neanderthals in the Altai had presumably been replaced by more “Vindija 33.19-like” Neanderthals (17).
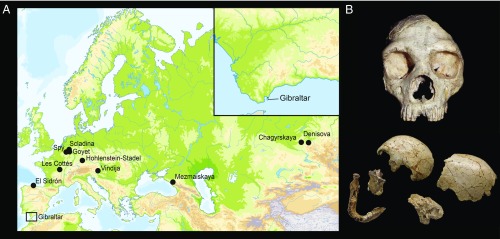
The Neanderthal fossils of Gibraltar are among the most prominent finds in the history of paleoanthropology. In 1848, a partial cranium (“Gibraltar 1”) was found at Forbes’ Quarry. Unsurprisingly, in the historical context of its discovery, it was not immediately recognized as belonging to a distinct type of hominin (18) (Fig. 1). A few years later, the discovery of the Feldhofer Neanderthal-type specimen sparked interest in and investigations of the Forbes’ Quarry cranium, which is assumed to have belonged to a female Neanderthal (19, 20). A form of benign bone overgrowth on the interior of the cranium (endocranial hyperostosis) (19) suggests that the individual died at a relatively advanced age. In 1926, an excavation of a nearby Mousterian rock shelter yielded the parts of a partial second Neanderthal cranium and an associated mandible (“Gibraltar 2”), which belong to a child (18) with an estimated age of 3 to 5 y at time of death (21, 22). While the Devil’s Tower child was excavated more systematically, the exact archaeological context of the Forbes’ Quarry specimen was not recorded. In the absence of direct chronological data for both fossils, no consensus has been reached on the age of the specimens, which have been suggested to date to anywhere between marine isotope stage 3 and 5 (∼30 to ∼130 ka ago) (23). It has been proposed that Neanderthals inhabited Gibraltar until as late as 24,000 uncalibrated radiocarbon years ago (24, 25), based on dates of charcoal from layers containing Mousterian artifacts. However, the accuracy of these dates has been questioned (26–29).
Fig. 1.
(A) Geographic locations of Gibraltar and other sites that are discussed in this study. (B) The Forbes’ Quarry cranium (Top) and the cranium and mandible of an infant found at Devil’s Tower (Bottom) on Gibraltar. Reprinted with permission from ref. 18.
The Iberian Peninsula, and Gibraltar in particular, are located on one of the extremes of the Neanderthal distribution and are thought to have served as a refuge for Neanderthals during glaciations (25, 30). In addition, it has been suggested that Neanderthals may have persisted in Gibraltar thousands of years after they were replaced by modern humans in other parts of Eurasia (24, 25). Genetic analysis of the Forbes’ Quarry and Devil’s Tower specimens would help to determine their relatedness to other Neanderthals in western and central Eurasia. However, the warm Mediterranean climate in Gibraltar is unfavorable for DNA preservation (31). Fortunately, advances in sample preparation techniques (32–35) are continuously improving the ability to retrieve highly degraded DNA. This, and the fact that both Gibraltar specimens preserve petrous bones, in which ancient DNA is particularly likely to be preserved (36, 37), warrant an attempt to investigate DNA preservation in the Gibraltar Neanderthals.
Results
The State of DNA Preservation.
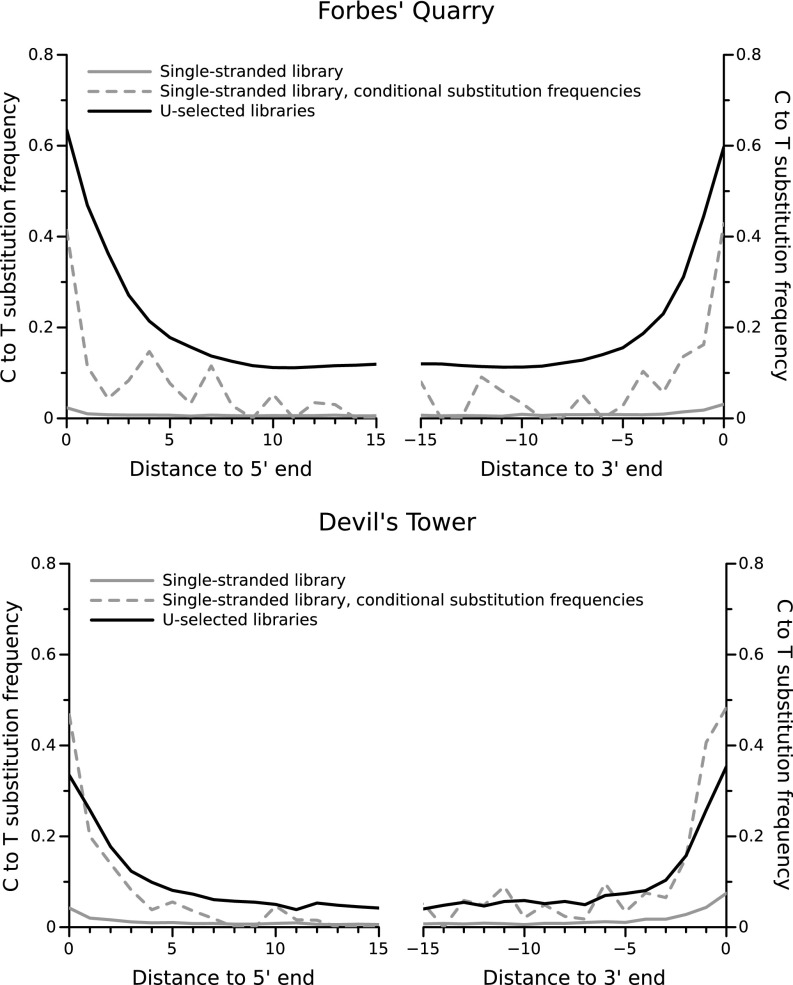
To assess the state of DNA preservation, we removed 36 and 20 mg of powder from the petrous bones from Forbes’ Quarry and Devil’s Tower, respectively. DNA was extracted from approximately half the powder of the Forbes’ Quarry specimen and all available powder of the Devil’s Tower specimen, and an aliquot of each extract was converted into single-stranded libraries for sequencing (34). Approximately 2 million DNA sequences were generated from each library, of which 2.9% and 11.6% could be mapped to the human reference genome when restricting to sequences 35 bp or longer (SI Appendix, Table S2). As these sequences could be derived from genuine Neanderthal DNA, recent human contamination, or both, we investigated whether they showed evidence of typical ancient DNA base damage resulting from the deamination of cytosine (C). This chemical reaction produces uracils (U), which accumulate predominantly at the end of ancient DNA molecules and are read by DNA polymerases as thymines (T) (38). Less than 5% of the sequences that start or end at a position at which the reference genome carries a C showed a T, suggesting that the vast majority, if not all, of these sequences derive from contemporary human contamination (Fig. 2).
Fig. 2.
Frequencies of C-to-T substitutions at the ends of sequence alignments for the Forbes’ Quarry and Devil’s Tower individuals. Only sequences of length ≥35 bp were used in this analysis. Conditional substitution frequencies were computed by filtering for sequences that carry a C-to-T substitution on the opposing end.
To test whether small amounts of authentic ancient DNA could be present in these sequences, we computed the frequency of C-to-T substitutions at the 5′-end for sequences that show a 3′ C-to-T substitution, and vice versa (conditional substitution frequencies) (39). We found that among such sequences, C-to-T substitution frequencies were higher than 40% (Fig. 2). We thus concluded that a population of highly deaminated ancient DNA molecules is present in both specimens, but that these molecules are present in a large excess of less deaminated recent human DNA.
An Improved Method for Uracil-Selective Library Preparation.
Useful sequence data can, in principle, be recovered from hominin fossils even in the presence of recent human DNA contamination if analyses are restricted to molecules showing evidence of deamination (39, 40). Such molecules can be selected in silico by filtering for sequences with terminal C-to-T substitutions, or in vitro using a library preparation method that specifically targets uracil-containing molecules (41). Here we favored the second approach, not only because it avoids the cost of sequencing noninformative molecules but also because it may be more suitable for very heavily contaminated samples, in which C-to-T substitutions derived from sequence error or evolutionary differences between the reference genome and the contaminant may confound the identification of deaminated molecules in a computational approach.
The U selection method described by Gansauge and Meyer (the Gansauge method) (41) achieves enrichment for uracil-containing molecules, but the yield of library molecules is lower than with single-stranded library preparation. In an effort to maximize the retrieval of genetic information from the limited Gibraltar material, we developed 2 uracil-enrichment library preparation methods, subsequently referred to as the A-tailed method and the simple method. Similar to the previous method, both methods rely on converting single-stranded DNA molecules into double-stranded library molecules that carry an adapter on each end and their immobilization on streptavidin-coated beads. Uracils in the template strand are then excised by the combined action of uracil-DNA-glycosylase and endonuclease IV, and the resulting nicks are repaired by a strand-displacing polymerase, which releases DNA strands produced from uracil-containing DNA strands from the beads. Although uracils are removed in this step, the C-to-T change remains encoded in the reverse library strand (SI Appendix, Fig. S1). The methods differ mainly in the strategies used for adapter ligation: in the Gansauge and A-tailed methods (SI Appendix, Figs. S1 and S2), a single-stranded adapter is ligated to the 3′-end of the molecules, followed by synthesis of a complementary strand and either blunt-end ligation (the Gansauge method) or sticky-end ligation (the A-tailed method) of a double-stranded second adapter. In the simple method (SI Appendix, Fig. S3), which involves fewer reaction steps, 2 single-stranded adapters are attached simultaneously to both ends of the molecules before the complementary strand is synthesized.
To test the efficiency of the methods in retrieving deaminated DNA molecules, we compared their performance with each other and with the regular single-stranded library preparation, using DNA extracts prepared from 3 cave bear specimens. As estimated by quantification of the library molecules and shallow shotgun sequencing, the highest number of deaminated molecules is obtained from single-stranded libraries, in the absence of uracil selection (SI Appendix, Fig. S4). Yields were almost as high with the uracil-selective A-tailed method (88% on average compared with single-stranded library preparation), substantially lower with the simple method (43%), and lowest with the original Gansauge method (23%). The selectivity of the method, as measured by the proportion of sequences that carried a C-to-T substitution indicative of deamination, was highest with the A-tailed method (98.2% on average), and was around 85% with the other 2 methods (SI Appendix, Fig. S5). We conclude that the A-tailed method performs best among the U selection methods, but that the simple method may nonetheless represent a viable alternative for projects in which the quantity of sample material is less limited, because it involves fewer steps and is therefore less time-consuming.
Nuclear DNA Sequencing and Sex Determination.
To generate as much DNA sequence data as possible from the Gibraltar specimens, we extracted DNA from the remaining bone powder of Forbes’ Quarry using a recently described extraction method that is particularly efficient at retrieving DNA molecules from highly degraded material when coupled with single-stranded library preparation methods (35). We then converted several aliquots of the new and existing extracts into DNA libraries, using U selection, mainly the A-tailed method (SI Appendix, Table S3). As expected, the uracil-selected libraries show more evidence of cytosine deamination than the regular single-stranded libraries (Fig. 2). However, for the Devil’s Tower specimen, the frequency of C-to-T substitutions remains lower (33% and 35% on 5′ and 3′-ends, respectively) than expected from the conditional substitution frequencies (47% and 48%, respectively). This observation indicates that some contaminant human DNA was recovered in the libraries, presumably due to deamination that had accumulated in the contaminant DNA since the specimen was found and a small leakage of nondeaminated DNA into the uracil-enriched libraries.
After deep sequencing of the libraries (SI Appendix, Table S3), we first determined the minimal sequence length at which genuine hominin sequences can be reliably differentiated from spurious alignments of microbial sequences (5, 42). At a cutoff of ≥30 bp, <1% of the alignments are spurious (SI Appendix, Fig. S6). To minimize mapping bias toward modern human alleles, we aligned all sequences both to the human reference genome and to an archaic version of the human genome, which carries alternative alleles identified in the high-coverage Neanderthal [Altai Neanderthal (1), Vindija 33.19 (2), and Chagyrskaya 8 (16)] and the Denisovan (Denisova 3) (32) genomes, and merged both sets of alignments (4). After restricting to sequences that are unique and carry a C-to-T substitution in the first or last 3 positions, where the deamination-induced increase in C-to-T substitutions is most pronounced, we obtained a total of 70 Mbp of DNA sequence for the Forbes’ Quarry individual (∼2% of the genome) and 0.4 Mbp for the Devil’s Tower child (0.1% of the genome), respectively (SI Appendix, Table S3).
Based on the sharing of the human state at sites where the high-coverage archaic genomes are homozygous ancestral and ≥90% of present-day human genomes show the derived state (5), we estimate human contamination among the nuclear sequences to ∼3.1% (95% confidence interval [CI], 2.09–4.37%) in the Forbes’ Quarry data and to 0% in the Devil’s Tower data, albeit with a wide confidence interval for the latter (95% CI, 0–26.5%) due to the sparse data that were obtained (Table 1). According to the coverage of the X chromosome and the autosomes, the Forbes’ Quarry individual was female and the Devil’s Tower child was male (SI Appendix, Fig. S7), which is consistent with the inferences from morphology of the Forbes’ Quarry specimen (20).
Table 1.
Contamination estimated by 2 methods, using all sequences or only those carrying a C-to-T substitution in the first or last 3 alignment positions (i.e., showing evidence of deamination)
| Contamination measurement | Forbes’ Quarry | Forbes’ Quarry deaminated | Devil’s Tower | Devil’s Tower deaminated |
| Nuclear | 8.9% (7.8–10.3%) | 3.1% (2.1–4.4%) | 81.4% (72.5–88.4%) | 0% (0–26.5%) |
| Mitochondrial | 46% (43.1–49%) | 15.4% (12.1–19.2%) | N/A | N/A |
95% binomial confidence intervals are provided in brackets. Mitochondrial contamination estimates are not available (‘N/A’) for the Devil’s Tower specimen due to a lack of data.
Lineage Assignment Based on Mitochondrial and Nuclear DNA.
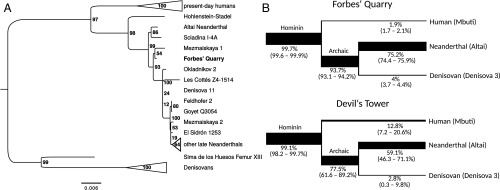
To determine the position of the Forbes’ Quarry Neanderthal in the hominin mitochondrial tree, all U-selected libraries were enriched for mitochondrial DNA (mtDNA) by hybridization capture (43). Combining unique sequences from deaminated DNA fragments obtained by capture and shotgun sequencing resulted in 11-fold coverage of the Forbes’ Quarry mtDNA genome. We did not attempt to analyze the mtDNA of the Devil’s Tower individual due to extremely poor DNA preservation. The mitochondrial contamination estimate for the Forbes’ Quarry specimen, as determined by the sharing of present-day human-specific mutations (positions at which 99% of a set of 311 present-day humans differ from 23 Neanderthals) was 15.4% (95% CI, 12.1–19.2%), which is higher than for the nuclear genome (Table 1). Using the sites that were covered by at least 5 fragments and with a minimum support of 80%, we called a consensus for ∼85% of the mtDNA genome of the Forbes’ Quarry individual (SI Appendix, SI Text, section 4). In a tree relating the mtDNAs of the Forbes’ Quarry individual, 23 other Neanderthals, 25 modern humans, 4 Denisovans, and the Sima de los Huesos hominin (SI Appendix, Table S4), the Forbes’ Quarry individual falls within the Neanderthal variation with a bootstrap support of 100%, but its exact position within the Neanderthal clade cannot be confidently resolved (Fig. 3A).
Fig. 3.
(A) Maximum-likelihood tree of the mitochondrial DNA sequences from Forbes’ Quarry, 25 modern humans (collapsed), 23 Neanderthals, 4 Denisovans, and the Sima de los Huesos individual (see SI Appendix, Table S4 for a complete list of mitochondrial genomes and accession numbers). The chimpanzee mitochondrial genome was used as an outgroup (not shown). Support from 100 bootstrap replicates is shown next to the nodes. (B) Derived allele sharing with human (Mbuti), Neanderthal (Altai), and the Denisovan (Denisova 3) for sequences of Forbes’ Quarry and Devil’s Tower showing signs of deamination. 95% binomial confidence intervals are shown in parentheses.
To analyze the nuclear genomes, we used the high-coverage nuclear genome sequences of the Altai Neanderthal, a Denisovan (Denisova 3), a present-day African (Mbuti), and 4 great apes (chimpanzee, bonobo, gorilla, and orangutan) to identify sites that are derived in at least 1 of the hominin genomes (5). Using DNA fragments overlapping these sites, we then determined the support of the derived state for each branch in the hominin tree (Fig. 3B). DNA fragments recovered from the Forbes’ Quarry specimen share more alleles with the Altai Neanderthal (75.2%; 95% CI, 74.4–75.9%) than with the Denisovan and modern human genomes. There is also substantial allele sharing with the Altai Neanderthal in the Devil’s Tower individual (59.1%; 95% CI, 46.3–71.1%), providing genetic evidence for its assignment to the Neanderthal lineage. However, the sharing of archaic alleles (i.e., alleles derived in both the Altai Neanderthal and Denisova 3) is lower for the Devil’s Tower individual than for the Forbes’ Quarry individual (77.5% [95% CI, 61.6–89.2%] vs. 93.7% [95% CI, 93.1–94.2%]). Together with an elevated support of the human lineage, this observation suggests a larger residual level of human contamination among the sequences from this specimen. Due to this and the small amount of data recovered, we concluded that we have reached the limits of what can be achieved for this specimen, even with uracil-selective library preparation, and did not analyze the Devil’s Tower sequences further.
Relationship to Other Neanderthals.
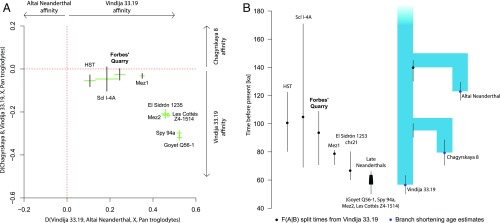
To explore the relatedness of the Forbes’ Quarry individual to other Neanderthals, we used D statistics (44) to determine the extent to which the high-coverage Neanderthal genomes (the Altai Neanderthal, Chagyrskaya 8, and Vindija 33.19) share derived alleles with the Forbes’ Quarry individual and other Neanderthals from which genome-wide data have been generated (SI Appendix, Table S1; i.e., Hohlenstein-Stadel [Germany], Scladina I-4A [Belgium], Goyet Q56-1 [Belgium], Spy 94a [Belgium], Les Cottés Z4-1514 [France], and Mezmaiskaya 1 and Mezmaiskaya 2 [both from Russia]) (3, 4). We also included chromosome 21 and exome capture data of El Sidrón 1253 (45, 46), a Neanderthal from northern Spain (47). The Forbes’ Quarry individual as well as all of the other Neanderthals tested share significantly (Z ≥ 3) more derived alleles with both Vindija 33.19 and Chagyrskaya 8 (Fig. 4A and SI Appendix, Table S5) than with the Altai Neanderthal. However, unlike the ∼49-ka-old El Sidrón 1253 individual and more recent Neanderthals from western Europe and Asia, the Forbes’ Quarry individual does not share significantly more alleles with Vindija 33.19 than with Chagyrskaya 8 (Z = 0.94; Fig. 4A and SI Appendix, Table S5), suggesting she was equally closely related to those 2 Neanderthals.
Fig. 4.
(A) D-statistics of low-coverage Neanderthals compared with high-quality genomes of Vindija 33.19, Chagyrskaya 8, and the Altai Neanderthal. Bars indicate 1 SD, as obtained by weighted-block jackknife. Green bars indicate that D is significantly (Z ≥ 3) different from zero. Only transversions were considered to suppress the influence of deamination-induced substitutions. (B) Population split times inferred by F(A|B) statistics. Blue circles indicate ages of the Neanderthal individuals estimated from branch shortening, black circles denote population split times from Vindija 33.19, and black lines correspond to 95% confidence intervals. Scl I-4A, Scladina I-4A; Mez1, Mezmaiskaya 1; Mez2, Mezmaiskaya 2; HST, Hohlenstein-Stadel.
We next estimated the population split times of the Forbes’ Quarry Neanderthal from modern humans, Denisovans, and other Neanderthal populations, using F(A|B) statistics (Fig. 4B and SI Appendix, Table S6) (44). The estimated split times between the Forbes’ Quarry individual and modern humans (522.3 ka; 95% CI, 476.3–574.2 ka) and between the Forbes’ Quarry individual and Denisova 3 (345 ka; 95% CI, 303.7–402.8 ka) are consistent with those previously estimated using other Neanderthal genomes (2). The population split time from Vindija 33.19 is estimated at 94 ka ago (95% CI, 70.8–108.9 ka) and overlaps with the 95% CI of the Forbes’ Quarry split from Chagyrskaya 8 (101.6 ka; 95% CI, 87.2–121.9 ka).
Both the analyses of allele sharing and the estimated population split times indicate that the relationships of the Forbes’ Quarry Neanderthal to the 3 Neanderthals sequenced at high-coverage are more similar to those of older Neanderthals Scladina I-4A, Hohlenstein-Stadel and Mezmaiskaya 1 than to the ∼49-ka-old El Sidrón 1253 individual from northern Spain or other younger Neanderthals in western Europe. These observations suggest that the Forbes’ Quarry individual is older than El Sidrón 1253. Alternatively, substantial population structure may have existed among Neanderthals on the Iberian Peninsula, resulting in the Forbes’ Quarry population being distinct from El Sidrón 1253 and other late Neanderthals.
Discussion
Until today, genetic analyses of archaic human DNA have been confined to material from Europe and the northern parts of continental Asia. No cases of ancient DNA preservation older than ∼15 ka have been recorded in Africa (48), the Middle East (49), or southeast Asia (50). By combining optimal sampling strategies with highly sensitive methods for sample preparation, including an improved method for uracil selection, we show that it is possible to retrieve archaic human DNA from specimens that were preserved in a warm coastal Mediterranean climate. The proximity of Gibraltar to Africa opens the prospect of retrieving DNA sequences of similar age from the latter continent in the future, which would represent a substantial advance in the study of recent human evolution.
Reliable dates are often not available for Middle Paleolithic sites that harbor the remains of archaic humans. Furthermore, in the case of Gibraltar, attempts to date the material are hampered by the fact that the finds were made at a time and under circumstances that did not favor preservation or documentation of the archeological context. However, as more genetic data become available from Neanderthals across their geographic and temporal range and our understanding of Neanderthal population history improves, it will become possible to put specimens into a temporal context based on their genetic affinity to other Neanderthals. Given our current state of knowledge and assuming that no major population structure existed in Neanderthals after 90 ka ago, we tentatively suggest that the Forbes’ Quarry specimen predates the ∼49-ka-old El Sidrón 1253 individual and possibly even the 60- to 70-ka-old Mezmaiskaya 1.
Materials and Methods
Sampling, DNA Extraction and Library Preparation.
Sampling of the Forbes’ Quarry and Devil’s Tower specimens was performed under clean room conditions in a dedicated ancient DNA facility at the Natural History Museum, London, United Kingdom, and further work was conducted at the Max-Planck Institute for Evolutionary Anthropology in Leipzig. DNA was extracted using silica-based methods (33, 35). Aliquots of the extracts were converted into single-stranded DNA libraries (34), using a liquid handling system (Bravo NGS Workstation, Agilent) (51) or subjected to uracil-selective library preparation using protocols developed in this study. Libraries were amplified with primers carrying unique combinations of 7-bp indices (52) and sequenced on an Illumina HiSeq 2500, using a 2x 76-bp configuration with 2 index reads (SI Appendix, Tables S2 and S3, and SI Text, sections 1 and 2, for further details on DNA extraction, library preparation, and sequencing). DNA extracts and libraries were also created from 3 cave bear bones to evaluate the performance of uracil-selective library preparation (SI Appendix, SI Text, section 3).
MtDNA Capture and Phylogenetic Reconstruction.
An aliquot of each amplified uracil-selected library was enriched for mitochondrial DNA, using baits designed based on the revised Cambridge Reference Sequence and the Altai Neanderthal mitochondrial genome (GenBank acc. no. KC879692) (1) as described (43), except that hybridization and wash temperatures were adjusted to 55 °C and the blocking oligonucleotide 11 (BO11) was replaced with BO12 for A-tailed libraries (see SI Appendix, Table S7 for oligonucleotide sequences). Captured libraries were pooled and sequenced on an Illumina MiSeq (see SI Appendix, Table S8 for summary statistics). Sequences were processed, contamination was estimated, and an mtDNA consensus sequence reconstructed for the Forbes’ Quarry specimen as described in SI Appendix, SI Text, section 4. A maximum-likelihood tree was constructed with phyML (53), using the Forbes’ Quarry consensus sequence, 25 present-day human mtDNAs, 23 Neanderthal mtDNAs, 4 Denisovan mtDNAs, the Sima de los Huesos mtDNA, and the chimpanzee mtDNA (see SI Appendix, SI Text, section 4 and Table S4, for further details).
Nuclear DNA Analyses.
Following raw sequence data processing and mapping (SI Appendix, SI Text, section 4), derived allele sharing [D-statistics (44)] of the Forbes’ Quarry Neanderthal and a set of other Neanderthal genomes with high-quality Neanderthal genomes (Altai, Chagyrskaya 8, Vindija 33.19) were calculated using AdmixTools (54). To reduce the effect of present-day human contamination, only sequences with deamination-induced C-to-T changes in the terminal positions and only transversions were considered. Furthermore, triallelic observations were disregarded. Split times from Vindija 33.19 were estimated using F(A|B) statistics (44). Specifically, we used the 3 high-coverage Neanderthal genomes to compute the fraction of derived alleles that the Forbes’ Quarry individual (population A) shares with the respective high-quality genome (population B) at heterozygous sites. The inferred demographic histories of the high-quality genomes were used in simulations to estimate the split times that would generate values of F(A|B) compatible with the observed values. To estimate the split time in years before present (ka), the age of the genome of population B, estimated via branch shortening, was added (2). It is important to note that the confidence intervals for the split times do not reflect uncertainties about the parameters used in the model, such as the Neanderthal mutation rate and effective population size of Neanderthals through time. All nuclear and mitochondrial data produced in this study are deposited on the European Nucleotide Archive under accession number PRJEB31410.
Supplementary Material
Acknowledgments
We thank Robert Kruszynski and Laura Buck for help with the sampling in London, Antje Weihmann and Barbara Schellbach for help with DNA sequencing, Anatoli P. Derevianko, Michael V. Shunkov, Gernot Rabeder, Pavao Rudan, Ivan Gušić, and Željko Kućan for providing cave bear bones for technical experiments. This study was funded by the Max Planck Society and the European Research Council (Grant 694707 to S.Pä.). Chris Stringer’s research is supported by the Calleva Foundation and the Human Origins Research Fund. I.B. and S.B. were supported by a Wellcome Trust Investigator Award (Project 100713/Z/12/Z).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (accession no. PRJEB31410).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903984116/-/DCSupplemental.
References
- 1.Prüfer K., et al. , The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prüfer K., et al. , A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajdinjak M., et al. , Reconstructing the genetic history of late Neanderthals. Nature 555, 652–656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peyrégne S., et al. , Nuclear DNA from two early Neandertals reveals 80,000 years of genetic continuity in Europe. Sci. Adv. 5, eaaw5873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer M., et al. , Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Mafessoni F., et al. , “A high-coverage Neandertal Genome from Chagyrskaya Cave” in The Origins of the Upper Paleolithic in Eurasia and the Evolution of the Genus Homo - Proceedings of the International Symposium, Klimenkova T. A., Ed. (IAET SB RAS Publishing, Denisova Cave, Altai, Russia, 2018), pp. 51–55. [Google Scholar]
- 7.Kunter M., Wahl J., Das Femurfragment eines Neandertalers aus der Stadelhöhle des Hohlenstein im Lonetal (Landesamt für Denkmalpflege im Regierungspräsidium Stuttgart, 1992), vol. 17. [Google Scholar]
- 8.Posth C., et al. , Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat. Commun. 8, 16046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toussaint M., Bonjean D., The Scladina I-4A Juvenile Neandertal, Andenne, Belgium: Palaeoanthropology and Context (Université de Liège, 2014) ERAUL Editions. [Google Scholar]
- 10.Skinner A. R., et al. , ESR dating at Mezmaiskaya Cave, Russia. Appl. Radiat. Isot. 62, 219–224 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Derevianko A. P., et al. , Chagyrskaya cave:A middle paleolithic site in the Altai. Archaeol. Ethnol. Anthropol. Eurasia 41, 2–27 (2013). [Google Scholar]
- 12.Rougier H., et al. , Neandertal cannibalism and Neandertal bones used as tools in Northern Europe. Sci. Rep. 6, 29005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semal P., et al. , New data on the late Neandertals: Direct dating of the Belgian Spy fossils. Am. J. Phys. Anthropol. 138, 421–428 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Soressi M., et al. , Préhistoire entre Vienne et Charente hommes et sociétés du Paléolithique, Buisson-Catil J., Primault J., Eds. (Association des publications chauvinoises, Chauvigny, France, 2010), pp. 221–234. [Google Scholar]
- 15.Pinhasi R., Higham T. F., Golovanova L. V., Doronichev V. B., Revised age of late Neanderthal occupation and the end of the Middle Paleolithic in the northern Caucasus. Proc. Natl. Acad. Sci. U.S.A. 108, 8611–8616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mafessoni F., High coverage Chagyrskaya Neandertal. 2019. https://www.eva.mpg.de/genetics/genome-projects/chagyrskaya-neandertal/home.html. Accessed 29 January 2019.
- 17.Slon V., et al. , The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561, 113–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buck L. T., Stringer C. B., A rich locality in South Kensington: The fossil hominin collection of the Natural History Museum, London. Geol. J. 50, 321–337 (2015). [Google Scholar]
- 19.Antón S. C., Endocranial hyperostosis in Sangiran 2, Gibraltar 1, and Shanidar 5. Am. J. Phys. Anthropol. 102, 111–122 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Rose E. P. F., Stringer C. B., Gibraltar woman and Neanderthal man. Geol. Today 13, 179–184 (1997). [Google Scholar]
- 21.Skinner M., Age at death of Gibraltar 2. J. Hum. Evol. 32, 469–470 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Stringer C. B., Dean M. C., Age at death of Gibraltar 2—A reply. J. Hum. Evol. 32, 471–472 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Daura J., et al. , Stratigraphic context and direct dating of the Neandertal mandible from Cova del Gegant (Sitges, Barcelona). J. Hum. Evol. 59, 109–122 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Finlayson C., et al. , Late survival of Neanderthals at the southernmost extreme of Europe. Nature 443, 850–853 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Finlayson C., et al. , Gorham’s Cave, Gibraltar—The persistence of a Neanderthal population. Quat. Int. 181, 64–71 (2008). [Google Scholar]
- 26.Zilhão J., Pettitt P., On the new dates for Gorham’s Cave and the late survival of Iberian Neanderthals. Before Farming 2006, 1–9 (2006). [Google Scholar]
- 27.Wood R. E., et al. , Radiocarbon dating casts doubt on the late chronology of the middle to upper Palaeolithic transition in southern Iberia. Proc. Natl. Acad. Sci. U.S.A. 110, 2781–2786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higham T., et al. , The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 512, 306–309 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Douka K., No hard borders for humans. Nat. Ecol. Evol. 3, 157–158 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Carrión J. S., et al. , Last Neanderthals in the warmest refugium of Europe: Palynological data from Vanguard cave. Rev. Palaeobot. Palynol. 259, 63–80 (2018). [Google Scholar]
- 31.Kistler L., Ware R., Smith O., Collins M., Allaby R. G., A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Res. 45, 6310–6320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer M., et al. , A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dabney J., et al. , Complete mitochondrial genome sequence of a middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gansauge M. T., et al. , Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. 45, e79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glocke I., Meyer M., Extending the spectrum of DNA sequences retrieved from ancient bones and teeth. Genome Res. 27, 1230–1237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamba C., et al. , Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinhasi R., et al. , Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10, e0129102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briggs A. W., et al. , Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer M., et al. , A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Skoglund P., et al. , Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. U.S.A. 111, 2229–2234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gansauge M. T., Meyer M., Selective enrichment of damaged DNA molecules for ancient genome sequencing. Genome Res. 24, 1543–1549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Filippo C., Meyer M., Prüfer K., Quantifying and reducing spurious alignments for the analysis of ultra-short ancient DNA sequences. BMC Biol. 16, 121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Q., et al. , DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U.S.A. 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green R. E., et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castellano S., et al. , Patterns of coding variation in the complete exomes of three Neandertals. Proc. Natl. Acad. Sci. U.S.A. 111, 6666–6671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhlwilm M., et al. , Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530, 429–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood R., et al. , A new date for the Neanderthals from El Sidrón Cave (Asturias, Northern Spain). Archaeometry 55, 148–158 2013. [Google Scholar]
- 48.van de Loosdrecht M., et al. , Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Lazaridis I., et al. , Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McColl H., et al. , The prehistoric peopling of Southeast Asia. Science 361, 88–92 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Slon V., et al. , Neandertal and Denisovan DNA from Pleistocene sediments. Science 356, 605–608 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Kircher M., Sawyer S., Meyer M., Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindon S., Gascuel O., A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Patterson N., et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.