Significance
The temporal control of DNA methylation by the CcrM DNA methyltransferase mediates cell cycle progression in the asymmetrically dividing bacterium Caulobacter crescentus. Limiting the presence of CcrM to a specific time in the cell cycle is achieved through temporally controlled transcription and Lon-mediated proteolysis. We discovered that a chromosomal DNA-based platform stimulates CcrM proteolysis by Lon. Upon cell division, CcrM is completely degraded by Lon in progeny swarmer cells. In progeny stalked cells, which initiate DNA replication immediately upon division, CcrM proteolysis is incomplete and the remaining enzyme is sequestered away from DNA at the cell pole. Thus, the cell ensures that once DNA replication starts, chromosomal DNA cannot serve as a substrate for remethylation until chromosome replication is complete.
Keywords: Lon protease, epigenetics, DNA methyltransferase, protein sequestration
Abstract
The cell cycle-regulated methylation state of Caulobacter DNA mediates the temporal control of transcriptional activation of several key regulatory proteins. Temporally controlled synthesis of the CcrM DNA methyltransferase and Lon-mediated proteolysis restrict CcrM to a specific time in the cell cycle, thereby allowing the maintenance of the hemimethylated state of the chromosome during the progression of DNA replication. We determined that a chromosomal DNA-based platform stimulates CcrM degradation by Lon and that the CcrM C terminus both binds to its DNA substrate and is recognized by the Lon protease. Upon asymmetric cell division, swarmer and stalked progeny cells employ distinct mechanisms to control active CcrM. In progeny swarmer cells, CcrM is completely degraded by Lon before its differentiation into a replication-competent stalked cell later in the cell cycle. In progeny stalked cells, however, accumulated CcrM that has not been degraded before the immediate initiation of DNA replication is sequestered to the cell pole. Single-molecule imaging demonstrated physical anticorrelation between sequestered CcrM and chromosomal DNA, thus preventing DNA remethylation. The distinct control of available CcrM in progeny swarmer and stalked cells serves to protect the hemimethylated state of DNA during chromosome replication, enabling robustness of cell cycle progression.
Epigenetic regulation of gene expression by DNA methylation is a conserved mechanism in all domains of life (1–3). In bacteria, DNA methylation was originally discovered as a component of restriction-modification (R-M) systems consisting of an endonuclease and an associated DNA methyltransferase, which differentiate the genome DNA from invading phage DNA (4). However, several solitary DNA methyltransferases without apparent cognate restriction enzymes were later identified in many bacterial species (5, 6). These orphan N6-adenine DNA methyltransferases were found to regulate the initiation of chromosome replication, DNA mismatch repair, gene expression, and cell cycle progression (6–11). The 2 best-studied examples are the Escherichia coli Dam enzyme (methylating the adenine of GATC) and the Caulobacter crescentus CcrM enzyme (methylating the adenine of GANTC).
The α-proteobacterium C. crescentus (hereafter referred to as Caulobacter) is a model organism that has been of value in elucidating the mechanisms that maintain an asymmetric cell division. Caulobacter produces 2 morphologically distinct progeny at each cell division: a motile swarmer (SW) cell and a sessile stalked (ST) cell (Fig. 1). The progeny swarmer cell cannot initiate chromosome replication until it differentiates into a stalked cell, whereas the progeny stalked cell immediately initiates chromosome replication. DNA replication only commences on a fully methylated chromosome (adenine of GANTC sites is methylated on both strands) and the movement of the replication fork generates 2 hemimethylated chromosomes (adenine of GANTC is methylated on only 1 of the 2 strands) (12) which are maintained in the hemimethylated state until the completion of chromosome replication. A burst of CcrM synthesis then allows the hemimethylated chromosomes to be converted back into 2 fully methylated chromosomes enabling a new round of DNA replication initiation in the progeny stalked cell and in the progeny swarmer cell 30 min later, after it differentiates into a stalked cell (13, 14).
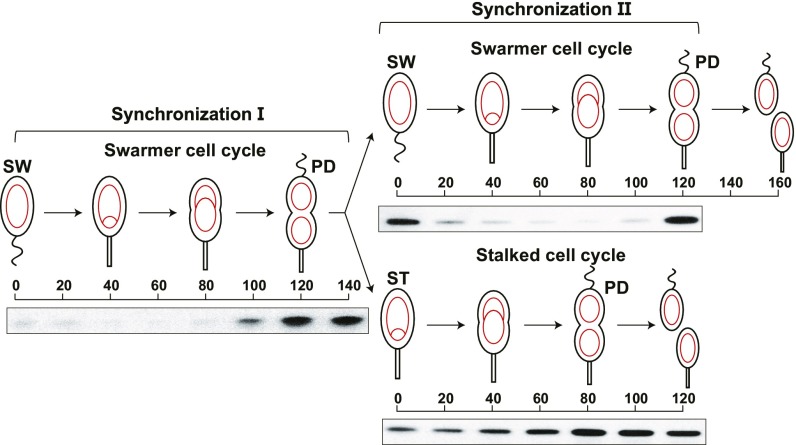
Fig. 1.
Presence of CcrM during the swarmer cell cycle and the stalked cell cycle. Each cell division is asymmetric yielding a SW cell and a ST cell. Circles and theta structures within the cells (red) indicate the single circular chromosome. The swarmer cell chromosome can only initiate replication once it has differentiated into a stalked cell, whereas the chromosome in the stalked cell that arises from a cell division can immediately initiate replication. Shown are immunoblots of protein samples from synchronized wild-type cultures using anti-CcrM antibody. Isolated swarmer cells were incubated in M2G minimal media and allowed to progress through the cell cycle. Upon cell division, the culture was subjected to a second synchronization. The swarmer and stalked cell fractions collected from the second synchronization were released in M2G for swarmer and stalked cell cycle analyses, respectively.
The methylation state of GANTC motifs within a subset of promoters regulates the transcription of genes that drive the cell cycle (11). For example, DnaA serves both as an initiator of chromosome replication and a transcription factor that controls the transcription of multiple cell cycle-regulated genes (15). Efficient transcription of dnaA (located close to the origin of replication) requires the GANTC site within its promotor to be in the fully methylated state. Upon replication initiation, the passage of the replication fork converts the dnaA promoter from the fully methylated state to the hemimethylated state, thus turning down the transcription of dnaA (16). The CtrA response regulator, serves as both an inhibitor of the initiation of DNA replication and as a transcription factor for a large number of cell cycle-regulated genes. In the case of the ctrA promoter, it is activated when in the hemimethylated state (17). As replication proceeds, the ctrA gene, which is positioned further from the replication origin, transitions from the fully methylated state to the hemimethylated state. Thus, the methylation state of the chromosome, which is temporally modulated by the passage of the replication fork that generates hemimethylated DNA, controls the sequential expression of the DnaA and CtrA master transcription factors. Together, this regulatory hierarchy activates or represses >199 cell cycle-regulated genes (13).
The Lon protease, which is present throughout the cell cycle, is known to degrade CcrM (18). Only when being actively synthesized upon completion of DNA replication does CcrM synthesis win the war against Lon degradation, resulting in CcrM accumulation and conversion of the hemimethylated chromosomes back to full methylation. In a lon-deficient strain, CcrM remains detectable throughout the cell cycle, leading to disabled cells that accumulate multiple chromosomes (18, 19). This phenotype can be partially ascribed to the misregulation of both DnaA and CtrA accumulation (16).
Because the initiation of DNA replication in the progeny swarmer cell is blocked until it differentiates into a stalked cell, the swarmer cell has sufficient time to degrade CcrM before its differentiation and coincident initiation of DNA replication (Fig. 1, swarmer cell cycle). However, the progeny stalked cell initiates DNA replication immediately upon cell division, with inadequate time for complete degradation of CcrM (Fig. 1, stalked cell cycle), yet the replicating DNA during the stalked cell cycle must be maintained in the hemimethylated state. Here, we have asked the critical question of how the replicating DNA in the progeny stalked cell escapes DNA remethylation by the remaining CcrM methyltransferase.
In this study, we report that the degradation of CcrM by Lon is stimulated by a DNA-based platform. We observed a 10-fold higher affinity of both CcrM and Lon to DNA than the affinity of CcrM to Lon. Further, we show that the C terminus of CcrM binds to its DNA substrate and also serves as the recognition site for the Lon protease. In the progeny swarmer cell, CcrM is completely degraded by Lon before its differentiation into a stalked cell after a 30-min delay. The progeny stalked cell sequesters remaining CcrM away from DNA at the cell pole, thus protecting the newly replicated chromosomes from remethylation. Our findings provide insight into robust regulation of DNA methylation during an asymmetric cell cycle that generates distinct progeny cells.
Results
CcrM Protein Turnover Undergoes Distinct Dynamics in Swarmer and Stalked Progeny Cells.
Using immunoblots with anti-CcrM antibody, we followed the accumulation of CcrM as the progeny SW and ST cells proceed through the cell cycle (Fig. 1). In synchronization I, we observed CcrM accumulation was confined to the predivisional cell. Upon cell division, we separated swarmer and stalked cell progeny shown as synchronization II. Microscopic observation showed that the swarmer cell population was pure but that ∼10% of the stalked cell progeny population was made up of late predivisional cells. This was confirmed by imaging progeny stalked cells harboring GFP-tagged TipN as a marker of the division plane. Each of the progeny cell populations was allowed to proceed through the cell cycle. In the progeny swarmer cell, CcrM was completely cleared within 20 min as it differentiated into a new stalked cell. However, CcrM was not completely cleared from the progeny stalked cell population (Fig. 1). Based on quantitative immunoblots, 42% of CcrM in the progeny stalked cell remained undigested, compared with CcrM levels observed in the predivisional cell before cell division.
Although it is known that Lon protein abundance does not change during the Caulobacter cell cycle (18), we asked if the activity of Lon on specific substrates is cell cycle regulated. To address this question, we assayed Lon activity as a function of cell cycle progression using a constitutively expressed substrate that is degraded directly by Lon in the absence of accessory factors. We found that the degradation rate of the constitutively expressed cytoplasmic Lon substrate, eYFP-sul20C, was not significantly different throughout the swarmer cell cycle (SI Appendix, Fig. S1), suggesting that cytoplasmic Lon activity is cell cycle independent.
DNA Binding Facilities CcrM-Lon Recognition.
It is known that the Lon protease is capable of binding to DNA (20). We confirmed that Caulobacter Lon binds chromosomal DNA in vivo (SI Appendix, Fig. S2). Given that CcrM and Lon are both DNA binding proteins, we asked whether a DNA platform mediates CcrM degradation by Lon. To address this question, we reconstituted the interactions of CcrM, its DNA substrate, and the Lon protease in vitro using 3 different DNA probes (Fig. 2A). Probe 1 contains the natural sequence upstream of the ccrM gene with WT GATTC motif that is recognized by CcrM. Probe 2 was generated by mutating probe 1’s CcrM’s recognition site from GATTC to AATAC. Probe 3 was constructed using a DNA fragment upstream of the pilA gene lacking any GANTC motif. Gel shift assays demonstrated that both purified CcrM and Lon proteins can individually bind probes 1, 2, and 3, without observable selectivity (SI Appendix, Fig. S3A). Because unmethylated DNA is absent in vivo, we sought to investigate the DNA binding capabilities of CcrM and Lon using hemimethylated and fully methylated DNA probes. We obtained fully methylated DNA by incubating PCR-generated probe 1 with purified CcrM protein. The hemimethylated DNA probe was generated by hybridization of fully methylated and unmethylated DNA probes. The methylation states of these DNA probes were confirmed by overlapping restriction digestions (SI Appendix, Fig. S3 B and C). We found that the binding capabilities of DNA to CcrM, as well as to the Lon protease, are methylation state independent (SI Appendix, Fig. S3D). Our results demonstrate that both CcrM and Lon are capable of binding to the same DNA probe, independent of methylation state.
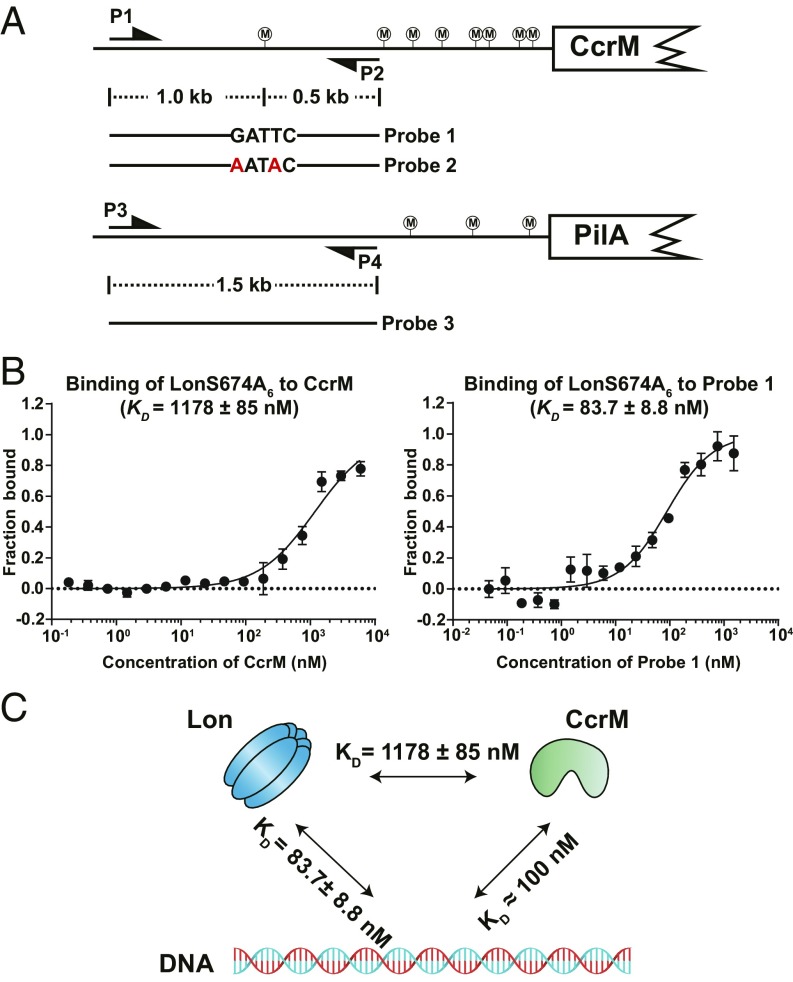
Fig. 2.
DNA binding facilities CcrM-Lon recognition. (A) Schematic view of DNA probe designs according to genome locus. Probe 2 is the same as probe 1 except with the mutation of GATTC to AATAC. Probe 3 contains the upstream sequence of pliA. P1-P2 and P3-P4 are primer pairs to amplify probe 1 (or probe 2) and probe 3, respectively. CcrM methylation sites are shown circled “M.” (B) The direct binding of purified LonS674A to CcrM or probe 1 was measured in vitro by microscale thermophoresis. LonS674A was fluorescently labeled with Atto-488 dye. The concentration of LonS674A6 was held constant at 20 nM while CcrM or probe 1 was titrated in 2-fold serial dilutions against it. The purified proteins were incubated at room temperature for 10 min before the binding assay. The data report the fraction of LonS674A6 that is bound at each concentration of CcrM or probe 1. See Materials and Methods for description of curve fits. (C) A schematic view showing affinities between CcrM, Lon, and DNA. CcrM and Lon have higher affinities to DNA than that of direct interaction.
To measure the affinities of Lon binding to DNA and Lon binding to CcrM, we used microscale thermophoresis (MST) assays (21). We first created a Lon mutant, LonS674A, that lacks proteolytic activity but retains DNA binding activity (functionally characterized later in SI Appendix, Fig. S7C). As shown in Fig. 2B, we measured the change in the thermophoresis of LonS674A over a 2-fold serial dilution of either CcrM or probe 1. Direct binding was observed between LonS674A and CcrM (KD = 1178 ± 85 nM) and between LonS674A and probe 1 (KD = 83.7 ± 8.8 nM). Lon exhibited an ∼14-fold higher affinity to DNA than to CcrM. Recent studies of DNA recognition by CcrM reported an equilibrium dissociation constant of 108 ± 20 nM for double-stranded DNA (22). A quantitative Western blot was performed to determine the in vivo concentration of CcrM and Lon at 120 min into the swarmer cell cycle, using purified CcrM and Lon to calibrate a standard curve. We determined that the intracellular concentration of CcrM ranged from 950 to 1,280 nM over 3 measurements, averaging 1,090 ± 135 nM for predivisional cells (SI Appendix, Fig. S4A). The intracellular concentration of the Lon monomer ranged from 2,040 to 2,820 nM over 3 measurements, averaging 2,480 ± 325 nM for predivisional cells (SI Appendix, Fig. S4B). The highest intracellular concentration of CcrM approached the KD value of the CcrM-Lon direct interaction. We also performed coimmunoprecipitation (Co-IP) of the reconstituted reactions to demonstrate that CcrM-Lon-DNA can form nucleoprotein complexes under physiological concentrations (SI Appendix, Fig. S4C). Taken together, these findings argue that both Lon and CcrM associate with DNA at physiological concentrations rather than directly interacting with each other (Fig. 2C).
C-Terminal Domain of CcrM Is Required for Its DNA Binding Activity and Lon Recognition.
ATP-dependent proteases usually rely on terminal sequences for substrate recognition (23, 24). To identify the CcrM degradation tag, we first fused an M2 epitope to the N or C terminus of CcrM that was expressed from the native CcrM chromosomal site. Western blots were carried out using antibody against CcrM on cell extracts of samples at multiple times during the cell cycle (SI Appendix, Fig. S5A). We observed that the C-terminal M2 tag interfered with Lon recognition of CcrM. Using in vivo degradation assays of CcrM C-terminal truncations lacking its C-terminal 65 amino acids we confirmed that the CcrM C terminus is recognized by Lon (SI Appendix, Fig. S5B). We further determined that the C-terminal 24 amino acids were sufficient for recognition by Lon (SI Appendix, Fig. S5C). We propose that when CcrM encounters Lon on the DNA, the CcrM C-terminal 24 amino acids become accessible to the protease.
Deletion of the CcrM C-terminal 65 amino acids abolished both CcrM DNA methyltransferase activity and DNA binding activity (SI Appendix, Figs. S5D, S5E, and S7A). Within the 65-amino acid C terminus of CcrM we observed 4 highly conserved motifs (SI Appendix, Fig. S6A). We generated single amino acid substitutions within each of these conserved motifs and found that a S315A mutation disrupted the DNA binding activity and caused severe cell cycle defects, including loss of viability and filamentous morphology, as was observed for the W332A mutation in conserved motif C, but not in conserved motif D within the 24 amino acids required for Lon recognition (SI Appendix, Fig. S6 B–D). We performed circular dichroism spectroscopy to confirm that the mutated proteins CcrMΔC65 and CcrM S315A are folded correctly (SI Appendix, Fig. S7B). Combined, our results demonstrate that the 24 amino acids at the CcrM C terminus are required for proteolysis by the Lon protease and conserved motifs B and C (shown in SI Appendix, Fig. S6A) are required for CcrM DNA binding activity and methyltransferase activity.
Chromosomal DNA Serves as a Platform That Stimulates CcrM Proteolysis by Lon.
Because both CcrM and Lon bind DNA (SI Appendix, Fig. S4C), we asked if DNA stimulates CcrM proteolysis by Lon. Accordingly, in vitro degradation assays were performed in the presence of the DNA probes described in Fig. 2A. The addition of probe 1, containing the GATTC methylation recognition site, dramatically boosted CcrM degradation (Fig. 3A and SI Appendix, Fig. S8A). Strikingly, the addition of probe 2 (same as probe 1 but with a scrambled DNA methylation site) or probe 3 (with a nonspecific DNA sequence) similarly boosted CcrM degradation (Fig. 3A and SI Appendix, Fig. S8A). To test whether stimulated proteolysis requires both CcrM and Lon to bind DNA, we used purified DNA binding-deficient mutants of both CcrM and Lon, CcrMS315A and LonQM, in the in vitro assays. The abolished DNA binding activity of LonQM was recently characterized (25), and verified in Caulobacter (SI Appendix, Fig. S7C). The LonQM mutant lacking DNA binding activity exhibited intact proteolytic activity on β-casein, a non-DNA binding substrate (SI Appendix, Fig. S8B). However, degradation of CcrMS315A by Lon or degradation of wild-type CcrM by LonQM was not stimulated by the addition of DNA (SI Appendix, Fig. S8C).
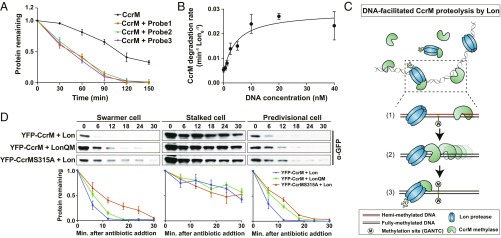
Fig. 3.
DNA serves as a platform in stimulating CcrM proteolysis by Lon. (A) In vitro degradation assays showing the stimulatory effect of DNA on CcrM degradation by Lon. CcrM (1 µM) was incubated with Lon6 (0.25 µM) in the absence or presence of DNA probes (10 nM). The intensity of the CcrM band from 3 independent experiments was quantified. Protein gels are shown in SI Appendix, Fig. S8A. The means ± SDs (n = 3) are plotted. (B) DNA stimulates CcrM degradation by Lon. Degradation rates of CcrM (1 µM) by Lon (0.25 µM) are shown with increasing concentration of DNA probe 1. See Materials and Methods for description of curve fits. (C) A proposed model of DNA-facilitated CcrM degradation by Lon. The Top shows the presence of CcrM, Lon, and DNA fragments in a mixed reaction. A zoomed-in schematic view shows the 3 steps of CcrM degradation by Lon on DNA: first, binding of CcrM and Lon to DNA fragment due to individual high affinity; second, enhanced-intermolecular collision frequency driven by CcrM processivity; third, substrate unfolding and proteolysis. (D) In vivo degradation assays showing the proteolysis of eYFP-CcrM by LonQM and the proteolysis of eYFP-CcrMS315A by Lon in isolated populations of swarmer, stalked, and predivisional (PD) cells in which CcrM and its variant are constitutively expressed. Cells expressing eYFP-CcrM or eYFP- CcrMS315A controlled by Pxyl were grown in M2G and induced with 0.3% xylose, synchronized, and harvested at 0 (swarmer cells), 45 (stalked cells), and 120 (predivisional cells) mps. Samples were treated with chloramphenicol (200 µg/mL) to shut off protein synthesis. Relative protein levels were monitored by immunoblots using anti-CcrM antibody (Top). Band intensities were quantified (Bottom) and error bars represent SDs (n = 4).
Titration of DNA showed that increasing concentrations of DNA stimulated the rate of CcrM proteolysis by Lon but reached a maximum rate of degradation at a concentration of 10 nM DNA (Fig. 3B). It has been reported that DNA stimulates Lon ATPase activity (20, 26, 27). We found that either DNA or CcrM could stimulate Lon ATPase activity, but that addition of both did not further stimulate the ATPase activity (SI Appendix, Fig. S8D). Taken together, we conclude that chromosomal DNA serves as a platform stimulating CcrM proteolysis by enhanced Lon-CcrM recognition (Fig. 3C), and this stimulation does not depend on the presence of a methylation site or a specific DNA sequence. DNA-facilitated proteolysis might be restricted to DNA binding substrates since we did not observe the stimulated degradation of β-casein, a non-DNA binding Lon substrate, in the presence and absence of DNA (SI Appendix, Fig. S8E).
To verify that DNA-facilitated CcrM proteolysis by Lon occurs in vivo, we constitutively expressed eYFP-CcrM or eYFP-CcrM S315A (with disrupted DNA binding activity) in merodiploid strains containing the WT CcrM gene. In these strains, the substrate degradation rates could be quantified in a synchronized cell population independent of the time of substrate synthesis (Fig. 3D). These in vivo assays were carried out on cultures in which eYFP-CcrM or eYFP-CcrM S315A were coexpressed with Lon or LonQM. We observed slower degradation of eYFP-CcrM by the Lon mutant, LonQM, that does not bind DNA. Similarly, the CcrM mutant, eYFP-CcrMS315A, that does not bind DNA, also exhibits a slower degradation rate in swarmer or predivisional cells (Fig. 3D). In a stalked cell population that originated from a swarmer-to-stalked cell transition, both eYFP-CcrM and eYFP-CcrMS315A were significantly more stable than the degradation observed in either swarmer or predivisional cells. As we show below, we determined that a separate mechanism is used by stalked cells to protect CcrM from degradation by Lon.
Dynamic Polar Sequestration of CcrM in Progeny Stalked Cells.
Considering the immediate initiation of DNA replication in progeny stalked cells, we hypothesized that the nondegraded CcrM was either inactivated or sequestered away from chromosomal DNA. Accordingly, we imaged a strain expressing a sole chromosomal copy of eyfp-ccrM under the control of its native promotor by fluorescence microscopy. Among 444 imaged cells in a mixed population, 40.99% of cells (n = 182) showed no detectable florescent signal, as expected for the percentage of cells expected to be cleared of CcrM by Lon. Of the remaining cells, 29.50% (n = 131) exhibited a unipolar focus (Fig. 4A). We also observed a diffuse signal in 18.92% of cells (n = 84), suggesting that polar localization of CcrM that is not degraded is dynamic (Fig. 4A). eYFP-CcrM formed a focus at the pole opposite to the SpmX stalked pole marker (28, 29), demonstrating that eYFP-CcrM accumulated specifically at the new cell pole (Fig. 4B). We used time-lapse microscopy to track cells (n > 100) that had an eYFP-CcrM florescent focus at the new pole. An eYFP-CcrM focus was consistently detected at the new pole of the progeny stalked cell and faded away during the transition to a predivisional cell (Fig. 4C). Upon cell division, the eYFP-CcrM focus appeared again at the incipient new pole of the progeny stalked cell, while no detectable signal was observed in the progeny swarmer cell (Fig. 4C).
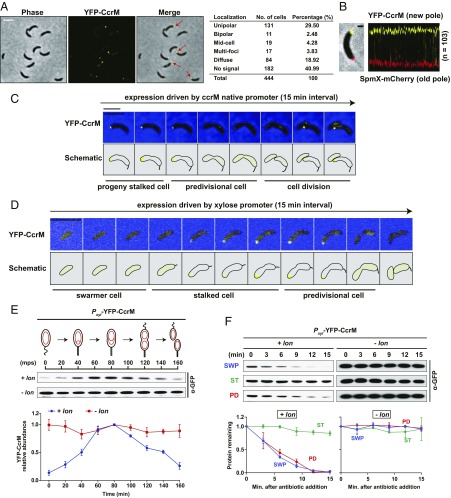
Fig. 4.
Dynamic sequestration of CcrM in Caulobacter. (A) Cells expressing the sole chromosomal copy of eyfp-ccrM under the control of the CcrM native promotor were grown in M2G media to exponential phase and imaged by phase contrast and epifluorescence microscopy. Arrows indicate the polar localization of eYFP-CcrM. The table shows the subcellular distribution of CcrM in 444 cells. (B) Cells coexpressing eYFP-CcrM and SpmX-mCherry, an old pole marker, under the control of their native promotors were grown in M2G to exponential phase and imaged by phase contrast and epifluorescence microscopy. A representative cell overlaid (Left) with phase, eYFP, and mCherry channels is shown. (Scale bar, 1 μm.) A fluorescent profile (Right) is shown by an alignment of 103 cells with their fluorescent channels of pole marker SpmX-mCherry, localized at the old cell pole, and eYFP-CcrM, localized at the new cell pole. (C) Time-lapse microscopy of cells producing chromosome-encoded eYFP-CcrM under the control of its native promotor. Images, starting with progeny stalked cells, were taken every 15 min as the cells progressed through the cell cycle culminating in cell division. eYFP-CcrM is localized to the new pole of the progeny stalked cell. (Scale bar, 5 μm.) (D) Time-lapse microcopy of cells constitutively producing eYFP-CcrM under the control of the xylose-inducible Pxyl promoter. Upon transition of the swarmer cell to a stalked cell, constitutively produced eYFP-CcrM was sequestered to the new pole of the stalked cell. (Scale bar, 5 μm.) (E) Immunoblots of protein samples from synchronized cells constitutively expressing eYFP-CcrM under the control of Pxyl. Merodiploid strains expressing eYFP-CcrM in the background of wild-type (+lon) or ∆lon (−lon) were grown in M2G with 0.3% xylose, synchronized, and released into M2G with 0.3% xylose for cell cycle progression. Samples were taken every 20 min and relative protein levels were monitored by immunoblot using anti-GFP antibody (Top). Band intensities were quantified (Bottom) and error bars represent SDs (n = 4). eYFP-CcrM is stable during the stalked cell stage coincident with that of its polar sequestration. (F) In vivo degradation assays showing eYFP-CcrM stabilities in swarmer, stalked, and predivisional cells. eYFP-CcrM stabilities in lon mutant are shown for comparison. Merodiploid strains expressing eYFP-CcrM controlled by Pxyl were grown in M2G with 0.3% xylose, synchronized, and harvested at 0 (swarmer cell), 60 (stalked cell), and 120 (predivisional cell) mps. Samples were treated with chloramphenicol (200 µg/mL) to shut off protein synthesis. Protein levels were monitored by immunoblot using anti-GFP antibody (Top). Band intensities were quantified (Bottom) and error bars represent SDs (n = 4). eYFP-CcrM is significantly stable in stalked cells.
To image CcrM’s polar presence in relation to known cell cycle milestone events, we carried out time-lapse microscopy of cells coexpressing eYFP-CcrM and either ParB-mCherry or TipN-GFP. ParB is a DNA-partitioning protein that binds to the centromeric parS locus near the chromosomal origin of replication. Localization of ParB reflects the movement of the ParB-bound centromere from the old pole to the new pole immediately upon the initiation of DNA replication (30). We observed the coappearance of the eYFP-CcrM focus and the ParB-parS complex at the new pole of the progeny stalked cell, suggesting that the sequestration of CcrM and initiation of chromosome replication begins at approximately the same time (SI Appendix, Fig. S9A). TipN-GFP, a marker for the new cell pole that orients the polarity axis (31, 32), colocalized with eYFP-CcrM at the pole of stalked cells (SI Appendix, Fig. S9B). During the transition from the progeny stalked cell to the predivisional cell, eYFP-CcrM started releasing from the pole before the relocating of TipN-GFP to midcell, demonstrating that CcrM polar sequestration ends before the formation of the division plane (SI Appendix, Fig. S9B). We observed neither an interaction between CcrM and TipN in bacterial 2-hybrid assays nor CcrM mislocalization in a tipN mutant (SI Appendix, Fig. S11A), suggesting that the release of CcrM from the cell pole might be TipN independent.
We posit that the appearance of CcrM at the pole of the progeny stalked cell sequesters remaining CcrM away from DNA. This would serve to prevent remethylation of the replicating chromosome, while paradoxically decreasing CcrM proteolysis by Lon. The difference between the stalked cell that arises from differentiation from the swarmer cell and the progeny stalked cell is that the latter inherits abundant CcrM from the dividing cell. Only the progeny stalked cell that results from cell division sequesters CcrM to the cell pole. To ascertain the in vivo stability of CcrM when sequestered at the pole, we were able to generate a “proxy” from the stalked cell arising from swarmer cell differentiation that sequestered CcrM to the new cell pole, by expressing ccrM from a constitutive promoter (Fig. 4D). Time-lapse microscopy of cells constitutively expressing eYFP-CcrM showed that only the stalked cell sequesters CcrM to the cell pole, and only to the new cell pole, as observed in the progeny stalked cell (Fig. 4D). Cell cycle regulation of CcrM degradation enabled by sequestration was assayed by immunoblots using merodiploid strains expressing eYFP-CcrM under the control of Pxyl in a wild-type strain or a lon deletion mutant. Xylose-inducible expression provided constitutive levels of eyfp-ccrM transcription throughout the swarmer cell cycle rather than only in the late predivisional cells, so that changes in protein abundance reflect changes in protein degradation. In the synchronized merodiploid cells containing Lon, eYFP-CcrM levels were low in swarmer cells at 0 min postsynchrony (mps), increased to the highest amount at ∼80 mps, and decreased again during later stages of the cell cycle (Fig. 4E). In contrast, the eYFP-CcrM levels were constant during cell cycle progression in the lon deletion mutant. We verified that mRNA levels of eYFP-CcrM are constant in both strains, thus the changes of protein amount assayed by immunoblots reflect the protein turnover (SI Appendix, Fig. S9C). To confirm differential CcrM turnover when CcrM was constitutively present during the cell cycle, in vivo stability assays were carried out in specific cell types. We observed robust degradation of eYFP-CcrM protein in progeny swarmer cells and predivisional cells with measured half-lives of ∼6 min in the presence of Lon (Fig. 4F). In stalked cells, however, eYFP-CcrM was quite stable (Fig. 4F). Our results demonstrate that CcrM sequestered to the new cell pole is protected from degradation by Lon.
Cell-Pole Sequestration of CcrM Prevents Physical Contact with Chromosomal DNA.
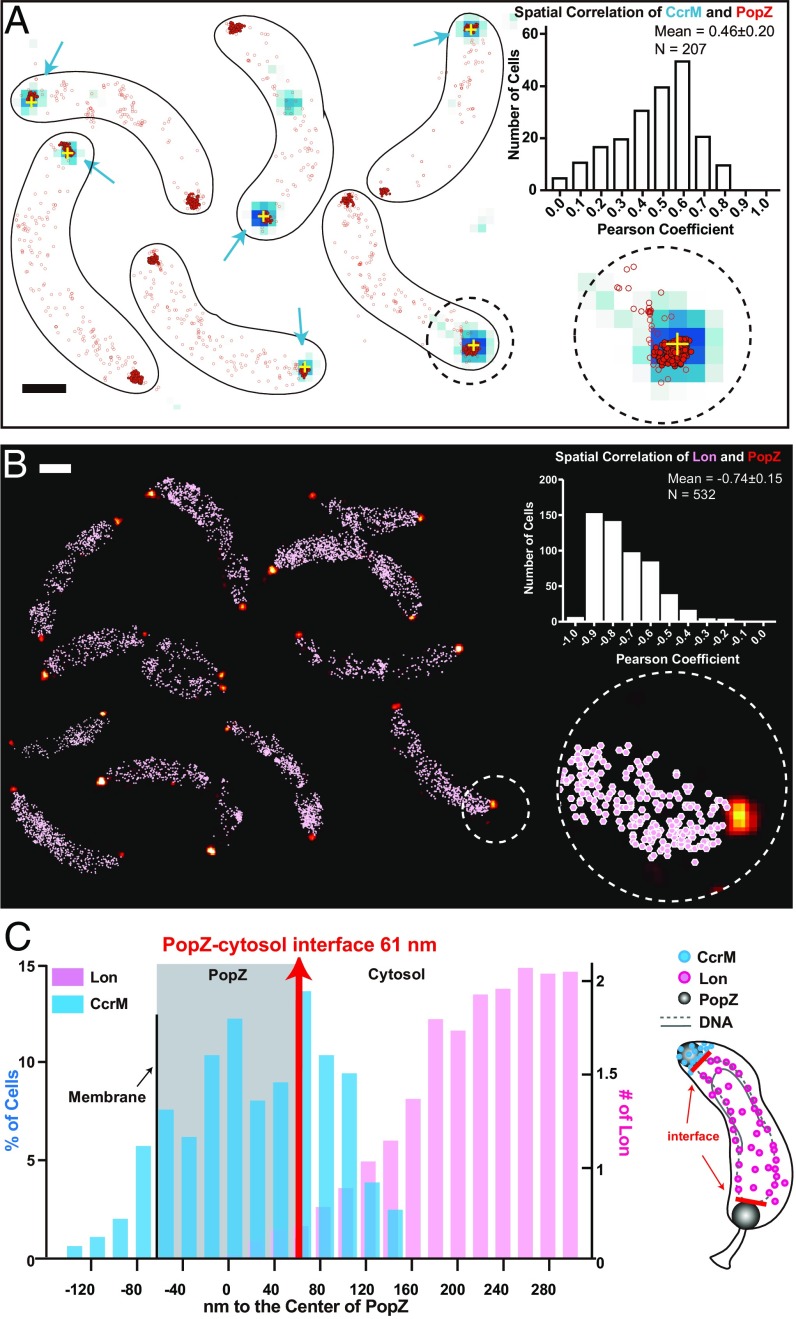
The fact that chromosome replication initiates immediately in the progeny stalked cell (33), and our observation that CcrM was still detected in the progeny stalked cells (Figs. 1 and 4), suggest that the transient sequestration of CcrM to the new cell pole might prevent remaining CcrM from interacting with DNA and thus not be available to methylate the replicating chromosome. The polar organizing protein PopZ assembles into a microdomain at the cell poles, which excludes DNA and ribosomes (34). To determine if CcrM is associated with the polar PopZ microdomain, we used single-molecule localization microscopy and diffraction-limited imaging to visualize strains coexpressing PAmCherry-PopZ and eYFP-CcrM. Single PAmCherry-PopZ locations (high-density polar localizations are represented by filled red scatter points; the rest are shown in empty red circles) and the diffraction-limited eYFP-CcrM clusters (blue pixels, with yellow cross at centroid) are shown in Fig. 5A. We quantified the Pearson’s correlation coefficient between the distributions of eYFP-CcrM and PAmCherry-PopZ within each cell and found an average of r = 0.46 ± 0.2 (error is SD determined from 207 cells), indicating that they are positively correlated. We calculated the distance between a CcrM cluster and the center of the PopZ domain along the cellular axis (Fig. 5 C, blue) and found a wide distribution with an average distance of 20 ± 62 nm (error is SD determined from 207 cells). CcrM and PopZ overlapped in space and CcrM could be found even toward the membranous periphery of the PopZ cluster at the cell poles (negative values).
Fig. 5.
Cell-pole sequestration separates CcrM from chromosomal DNA and the Lon protease. (A) Cells are shown with CcrM-eYFP diffraction-limited data overlaid with its centroid location (blue signal with yellow cross) and single-molecule localizations of PAmCherry-PopZ (red scatterplot). PAmCherry-PopZ is concentrated at the poles; localizations with more than 30 neighbors within a 100-nm radius are shown in filled red scatters with black outlines. The rest of the PAmCherry-PopZ that are typically nonpolar are shown in empty red circles. (Inset) The spatial correlation of CcrM-eYFP and PAmCherry-PopZ across 207 cells exhibited an average Pearson’s correlation coefficient of 0.46. (Scale bar, 600 nm.) This is a montage of different fields of view. (B) PAmCherry-PopZ localizations in 25-nm bins (yellow-red color scale) along with all Lon-eYFP localizations (pink scatterplot). The spatial correlation of PAmCherry-PopZ and Lon-eYFP across 270 cells (532 values) exhibited an average Pearson’s correlation coefficient of −0.74. (Scale bar, 600 nm.) This is a montage of a different field of views. (C) Distributions of the distance between the centroid of the CcrM-eYFP cluster to the center of PAmCherry-PopZ (blue) and the averaged distribution of distances between Lon-eYFP to the center of PAmCherry-PopZ (pink). The average distance of ParB-eYFP to PopZ was marked to show the PopZ-cytosol interface (red). The area that PopZ covers is marked by the gray shading, assuming symmetry along the cellular axis. A cartoon (Right) shows the physical separation between CcrM (blue) and Lon (pink). The cytosolic boundary of the PopZ domain is marked by red lines.
It was previously shown by two-color single-molecule imaging that ParB anchors the chromosome tightly to the cytosolic periphery of the PopZ microdomain (35). We visualized the spatial separation between the diffraction-limited ParB-eYFP position and single PAmCherry-PopZ molecules and found that ParB displayed an average distance of 61 ± 39 nm to the center of PopZ (error is SD determined from 183 cells) (Fig. 5C). As the location of ParB marked the interface of the cytosol and the PopZ DNA-free zone, 79% of CcrM was sequestered into the DNA-void volume at the new pole.
We performed 2-color single-molecule imaging of Lon-eYFP and PAmCherry-PopZ to determine if Lon has access to CcrM during polar sequestration of CcrM. We observed a distinct spatial anticorrelation between the 2 molecules with the average Pearson’s correlation coefficient to be −0.74 ± 0.15 (error is SD determined from 270 cells). Moreover, Lon-eYFP rarely diffused into the PopZ polar domain (Fig. 5B). The average distribution of distances from Lon molecules to the center of PopZ across all cells is shown in Fig. 5C. Combined with the ParB spatial measurements, we estimate that on average less than 1 Lon molecule was present within the PopZ domain (integral from the membrane to the PopZ-cytosol interface at 61 nm). As CcrM was measured to be about 20 nm away from the center of PopZ, we concluded that cell-pole sequestration of CcrM prevented its interaction with the chromosome and with DNA-bound Lon.
Polar Sequestration Is an Alternative Mechanism to Prevent Remethylation of the Replicating Chromosome from Excessive Intracellular CcrM.
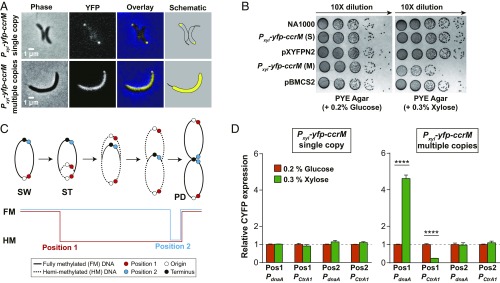
Excess intracellular CcrM could cause premature methylation of the replicating chromosome, resulting in abnormal cell cycle-regulated gene expression. We proposed polar sequestration as an alternative mechanism to prevent the function of excessive CcrM in the cell. When cells harboring a single chromosomal copy of a Pxyl-eyfp-ccrM were induced by 0.3% xylose to constitutively express CcrM throughout the cell cycle, we observed CcrM polar sequestration and a normal cell cycle and normal cell morphology (Fig. 6 A and B). To determine if constitutive overproduction of CcrM might saturate polar sequestration and result in premature methylation of the replicating chromosome, we examined CcrM localization in cells harboring Pxyl-eyfp-ccrM on a multicopy plasmid. Upon xylose induction, the constitutive expression of multiple copies of eyfp-ccrM disrupted the cell-pole sequestration and yielded filamentous cells (Fig. 6A and SI Appendix, Fig. S10A). The strain expressing multicopy eyfp-ccrM also exhibited defects in cell growth (Fig. 6B). When using a 10-fold dilution series of xylose to induce a controllable amount of eYFP-CcrM, we found that in the presence of 0.003% xylose, ∼20% of cells containing multicopy eyfp-ccrM had a unipolar focus and that cells exhibited morphology and growth similar to cells containing a single-copy eyfp-ccrM or under glucose repression (SI Appendix, Fig. S10 B and C). As the xylose concentration increased, we observed an increasing percentage of cells that had a diffuse eYFP-CcrM signal and a decreasing percentage of cells exhibiting unipolar eYFP-CcrM (SI Appendix, Fig. S10B). The cell length increased with the increasing concentrations of xylose (SI Appendix, Fig. S10C). Thus, increasing loss of polar sequestration of eYFP-CcrM is accompanied by filamentous cell formation.
Fig. 6.
Polar sequestration enhances cell cycle robustness by maintaining the hemimethylated (HM) state of newly synthesized chromosome. (A) Cells expressing single copy or multicopy of eyfp-ccrM under the control of Pxyl were grown in M2G + 0.3% xylose to exponential phase and imaged by phase contrast and epifluorescence microscopy. Expression of a higher amount of eYFP-CcrM impairs its polar localization. (B) Spot dilutions of strains in A. Wild-type C. crescentus NA1000 and strains harboring a single copy of Pxyl-eyfp-ccrM (S), multicopies of Pxyl-eyfp-ccrM (M), single copy of empty vector (pXYFPN2), and multicopies of empty vector (pBMCS2) were diluted to an OD600 of 0.03, serially diluted, and spotted onto the PYE agar plate supplied with either 0.2% glucose or 0.3% xylose and incubated at 30 °C for 2 d before photography. Induced expression of multicopy eyfp-ccrM resulted in impaired growth. (C) Schematic of the Caulobacter chromosome showing position 1 and position 2 where the CYFP reporter cassettes are inserted. Chromosome methylation states are shown as a function of DNA replication and cell cycle progression. Fully methylated (FM, solid line) state position 1 is progressively converted to a HM (dotted line) state following the SW-to-ST cell transition and the initiation of DNA replication. Fully methylated position 2 is converted to a hemimethylated state at the end of DNA replication in the predivisional cell (PD). (D) Relative CYFP expressions in synchronized stalked cells treated with 0.2% glucose (eYFP-CcrM repression) or 0.3% xylose (eYFP-CcrM induction). The CYFP reporter cassettes driven by either PdnaA or PctrA1 were inserted at 2 different positions on the chromosome of strains harboring single copy of Pxyl-eyfp-ccrM or multicopies of Pxyl-eyfp-ccrM. The qPCR assays were performed using primers targeting the transcribed promotor region of PdnaA or PctrA1 and CYFP linker, so that the assayed transcriptional levels represent the expression of CYFP gene, not eYFP-CcrM. Results suggest that eYFP-CcrM polar sequestration prevents remethylation of daughter chromosome in the strain constitutively expressing eYFP-CcrM. ****P < 0.0001, by unpaired Student’s t test.
One consequence of attenuated CcrM sequestration could be the mistimed expression of dnaA and ctrA induced by the premature methylation of newly replicated DNA in the progeny stalked cell. To test this hypothesis, we created 2 reporter cassettes bearing a partial eYFP gene (CYFP) expressed from either a dnaA promotor (PdnaA) or a ctrAP1 promoter (PctrA1). The PdnaA requires the fully methylated state for activation (16), while the PctrA1 requires the hemimethylated state for activation (17). We inserted the reporter cassettes at 2 positions on the chromosome: position 1 near the origin of replication and position 2 near the replication terminus. Following the initiation of replication in the stalked cell, position 1 was converted to the hemimethylated state, whereas position 2 remained in the fully methylated state until later in the cell cycle (Fig. 6C). As a control, we assayed cells harboring a single chromosomal copy of eyfp-ccrM transcribed from the inducible xylose promoter. The mRNA levels of CYFP exhibited no dramatic change in all tested positions for both promoters, suggesting that sequestration of eYFP-CcrM at the pole maintains the cell cycle-dependent methylation state of newly synthesized chromosomal DNA. For cells harboring the multicopy eyfp-ccrM induced with xylose, the mRNA level of CYFP driven by PdnaA at position 1 increased ∼5-fold (Fig. 6D). Presumably, this overexpression of CYFP mRNA was due to the fact that eYFP-CcrM wasn’t fully sequestered to the pole (Fig. 6A) and thus the dnaA promoter remained in the fully methylated, highly expressed state. In contrast, the mRNA level of CYFP driven by PctrA1 decreased ∼3-fold at position 1 (Fig. 6D). The expression of CYFP driven by either promoter when placed at position 2 were similar to that of glucose-treated cells (Fig. 6D). These results support the argument that cell-pole sequestration of remaining CcrM in progeny stalked cells prevents remethylation of newly synthesized chromosomal DNA, deactivates dnaA expression in a cell cycle-dependent manner, and thereby enhances cell cycle robustness.
Discussion
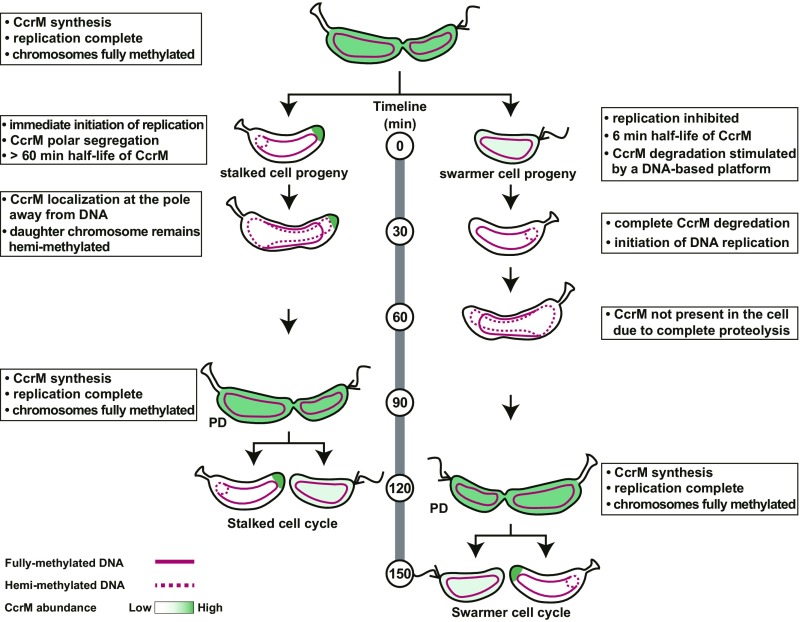
In this study, we present evidence that CcrM activity is controlled by coordinated DNA-facilitated protein degradation and by CcrM sequestration to the cell pole that occurs specifically in the progeny stalked cell (Fig. 7). DNA binding of both CcrM and Lon stimulates robust CcrM degradation. The affinity between CcrM or Lon to DNA is ∼14-fold higher than that of the CcrM-Lon interaction, contributing to the high efficiency of DNA methylation of ∼4,500 GANTC sites by only ∼600 CcrM molecules during a short time window of the cell cycle (SI Appendix, Fig. S4A) (9). In the progeny stalked cell, however, immediate chromosomal replication does not provide adequate time for proteolysis of remaining CcrM inherited from the predivisional cells. Instead of relying solely on protein degradation, CcrM is sequestered to the new cell pole, concurrent with the immediate initiation of chromosome replication (Fig. 7). This sequestration prevents CcrM-mediated DNA remethylation during replication by eliminating physical contact with the chromosomal DNA. The sequestered CcrM is released from the pole later in the cell cycle at the time of new CcrM synthesis in the predivisional cell. We propose that the different patterns of CcrM degradation and sequestration in progeny cells following an asymmetric division provide a fine-tuning mechanism, ensuring that the immediate onset of chromosome replication in the progeny stalked cell can proceed in the absence of remethylation and consequently, that DNA replication occurs only once per cell cycle.
Fig. 7.
Model of the distinct control of active CcrM in progeny cells following an asymmetric cell division. CcrM transcription and translation are confined to the predivisional cell, methylating GANTC sites on both hemimethylated daughter chromosomes (13, 14). Cell division generates 2 types of progeny cells: a motile swarmer cell and a sessile stalked cell. In swarmer cells, CcrM is completely proteolyzed by Lon, which is stimulated on a DNA-based platform, so by the time it differentiates into a stalked cell and DNA replication is initiated, CcrM is cleared from the cell. In progeny stalked cells, however, immediate chromosomal replication does not provide adequate time for proteolysis of remaining CcrM inherited from the predivisional cells. Instead of clearance by protein degradation, CcrM is sequestered to the new cell pole away from the chromosome, concurrent with the immediate initiation of chromosome replication. Sequestered CcrM has minimum contact with chromosome DNA and Lon, therefore preventing remethylation of newly synthesized chromosomal DNA. The fully methylated chromosome is indicated by a solid red line, and the hemimethylated chromosome is indicated by a dashed red line.
A DNA Platform Facilitates Proteolysis of CcrM by Lon.
Compared with the ClpXP protease that utilizes diverse adaptors for substrate delivery, Lon protease appears to process its substrate by directly recognizing clusters of exposed hydrophobic residues within a given polypeptide with little sequence specificity (36). The first Lon substrate-specific adaptor, SmiA (swarming motility inhibitor A), was recently identified in Bacillus subtilis (37). In Caulobacter, Lon has been shown to degrade several DNA binding proteins, including DnaA and SciP (38, 39). Given that adaptor-mediated proteolytic specificity for Lon protease is quite varied, Lon may employ multiple distinct mechanisms to regulate substrate specificity and degradation. A recent study revealed that several positively charged residues within the ATPase domain are critical for DNA binding activity of Lon (25). Although several lines of evidence suggest that Lon binding to DNA can stimulate its ATPase activity (20) and substrate degradation (26), the roles of these functions in regulating substrate specificity and degradation remained to be elucidated. We showed here that the stimulation of CcrM degradation requires the binding of substrate and protease to DNA (Fig. 3). Notably, DNA-mediated activation of Lon degradation of CcrM cannot be ascribed to stimulated ATPase activity upon binding to DNA. The presence of substrate alone can induce the ATPase activity to a level similar to that induced by the copresence of substrate and DNA (SI Appendix, Fig. S8D) and the presence of DNA does not stimulate degradation of non-DNA binding substrates (SI Appendix, Fig. S8E). Our results demonstrate that DNA serves as a platform for Lon-mediated CcrM proteolysis by facilitating substrate recognition rather than allosterically regulating Lon proteolytic activity. DNA moonlights as an adaptor aiding CcrM delivery to the Lon protease, which also prevents degradation of CcrM before its binding to DNA for chromosomal methylation. In prokaryotes, Lon is known to degrade multiple transcriptional regulators controlling the cell cycle, biofilm formation, motility and stress tolerance, and virulence (18, 40–42).
The known Lon substrates in Caulobacter, CcrM, SciP, and DnaA (18, 38, 39), all contribute to cell cycle regulation by their DNA binding activities. A recent study suggests that the presence of DNA accelerates the degradation of DnaA by Lon (43). It is therefore conceivable that DNA-facilitated proteolysis may be a universal regulatory mechanism for specific recognition and degradation of DNA binding substrates. However, it should be noted that cells bearing a DNA binding mutant of Lon, LonQM, do not exhibit cell morphology defects similar to a lon deletion mutant (43). Zeinert et al. (43) also showed that CcrM and DnaA degradation still can be observed in cells expressing LonQM, suggesting that both CcrM and DnaA degradation does not require DNA-bound Lon. Here, we quantified CcrM turnover and showed that the CcrM degradation rate is significantly slower in a lon deletion strain complemented with LonQM than the wild-type strain. However, this difference in degradation rate may not be sufficient to induce cell cycle defects caused by undegradable CcrM because polar sequestration adds an additional regulatory layer to CcrM clearance upon cell division. Other accessory factors or additional mechanisms could be induced in cells expressing LonQM to facilitate the degradation of DNA binding substrates.
Transient CcrM DNA Methyltransferase Sequestration to the Cell Pole.
Bacterial cells employ multiple mechanisms to drive protein localization to the cell poles (44, 45). Caulobacter has been shown to recruit proteins to the cell poles through interaction with proteins or protein complexes that are already positioned at the pole. For example, the polar PopZ protein forms a microdomain that anchors the chromosome origin via its interaction with the chromosome partition complex ParB-parS (34, 46). In addition, the stalked, or old pole-localized protein, SpmX, serves as a bridge to direct the interaction between the DivJ histidine kinase and PopZ protein in the polar microdomain (29). Although the mechanism that localizes PopZ to the pole is not known, the PopZ microdomain captures multiple proteins, thereby integrating several cellular processes within this membraneless organelle (47–49). Some CcrM polar foci were observed in ΔpopZ strains, arguing that CcrM recruitment to the pole might be PopZ independent (SI Appendix, Fig. S11). PopZ deletion strains exhibit significant morphological defects and misregulate the CtrA transcription factor, which is known to control ccrM transcription as part of its role as the master regulator of the cell cycle. Thus, our observation of decreased, but not loss of CcrM polar localization in popZ deletion strains could be the consequence of downstream effects associated with cell cycle defects in this strain. Assays of CcrM polar localization in strains lacking new pole-located proteins, including ΔmopJ, ΔpodJ, and a truncated divL (divLΔ28), demonstrated that these proteins did not mediate CcrM sequestration (SI Appendix, Fig. S11). Bacterial 2-hybrid assays showed that CcrM does not interact with the PleC, TipN, or TipF polar proteins (SI Appendix, Fig. S11A). Although it is unlikely that these proteins play a direct role in polar sequestration of CcrM, the striking specificity for localization to the new pole in stalked cells argues that the CcrM protein may be recognized by an as yet unidentified factor in polar organization. Notably, if CcrM is constitutively overexpressed throughout the cell cycle, and cannot be completely degraded by Lon, the stalked cell that arises from differentiation of the swarmer cell, but not the swarmer cell or the predivisional cell, is forced to sequester CcrM to the new cell pole (Fig. 4D). This cell type specificity of CcrM localization suggests that something specific about the new pole of the stalked cell dictates CcrM subcellular organization, and that CcrM undergoes cell type-specific spatiotemporal regulation. This result guides the way for future analysis of the polar functional topology of this cell type.
Another example of dynamically separating an enzyme from its substrate when it is not needed, was reported for the Escherichia coli MurG protein (50). MurG catalyzes the last step in peptidoglycan subunit synthesis. When its cellular concentration exceeds the need for new peptidoglycan synthesis, MurG is sequestered to the cell pole. The polar pool of MurG was shown to differ from inclusion bodies and to be released in active form when environmental conditions foster growth. Since mechanisms sequestering both CcrM and MurG to the new pole have remained enigmatic, the presence of alternative mechanisms other than protein interaction may give rise to polar localization of enzymatic proteins in bacterial cells.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used in this study are listed in SI Appendix, Table S1. E. coli strains were routinely grown in LB medium at 37 °C with appropriate antibiotics (100 µg mL−1 ampicillin, 50 µg mL−1 kanamycin). Caulobacter strains were grown in PYE (rich medium) or M2G (minimal medium) at 37 °C, supplemented with 0.3% xylose when necessary. Antibiotics were supplemented as needed for solid or liquid media, respectively, with the following concentration: kanamycin (25 µg mL−1 or 5 µg mL−1), spectinomycin (50 µg mL−1 or 25 µg mL−1), oxytetracycline (2 µg mL−1 or 1 µg mL−1), gentamycin (10 µg mL−1 or 5 µg mL−1), and chloramphenicol (1 µg mL−1 or 1 µg ml−1).
Microscopy.
C. crescentus strains were grown to exponential phase (OD600 < 0.3) and spotted on agarose pads (1.5%) containing M2G before imaging. Phase-contrast and fluorescence microscopy images were obtained using a Leica DMi8 microscope with an HC PL APO 100×/1.40 oil PH3 objective, Hamamatsu electron-multiplying charge-coupled device (EMCCD) C9100 camera, and Leica Application Suite X software. For all image panels, the brightness and contrast of the images were balanced with ImageJ (NIH) to represent foci or diffuse fluorescent signal. For computational image analyses, MicrobeJ (51) was used to determine cell outlines and lengths from phase images. Oufti (52) was used to determine normalized fluorescence intensities from each single cell. The data were plotted and statistically analyzed using Prism 7 (GraphPad).
Electrophoretic Mobility Shift Assay (EMSA).
DNA‐binding capacity of CcrM was evaluated by incubation of purified CcrM with 20 nM of indicated DNA probe in the presence of 200 µM sinefungin (Sigma) in EMSA buffer (50 mM Hepes pH 7.0, 200 mM NaCl, 1 mM EDTA, 1 mM DTT) for 30 min at room temperature and subjected to electrophoresis in 4 to 15% Mini-PROTEAN TGX precast protein gels (Bio-Rad) at constant 80 V for 3 h at 4 °C in 1× Tris glycine native gel buffer (25 mM Tris base, 192 mM glycine). Lon DNA binding capacity was assayed similarly to CcrM, except that 10 mM MgCl2 was added instead of 200 µM sinefungin. The protein-DNA complexes were stained with ethidium bromide and imaged with a Bio-Rad ChemiDoc XRS+ system.
In Vitro Ni-NTA Pull-Down Assay.
Purified LonS674A6 (0.2 µM) was incubated with 20 nM probe 1 and 200 µl buffer-equilibrated Ni-NTA beads at room temperature for 30 min in protein pull-down (PPD) buffer (protein storage buffer containing 10 mM MgCl2). One unit of DNase I was added when necessary to cleavage probe 1. The beads were washed once with 1 mL PPD buffer and resuspended in another 200 µl PPD buffer containing a low amount of CcrM (0.4 µM) or high amount of CcrM (4 µM). A 20-µl aliquot of reaction (input) was taken, suspended in SDS loading buffer, boiled for 10 min, followed by incubation at 65 °C for 5 min, and subjected to analyses by SDS/PAGE and 1% agarose gel. The content of remaining reaction was incubated at room temperature for 1 h, washed with PPD buffer extensively, and eluted with 100 µl PPD buffer containing 325 mM imidazole. The eluted protein samples were analyzed by SDS/PAGE for detection of the presences of LonS674A6-CcrM-DNA nucleoprotein complex via silver staining.
Supplementary Material
Acknowledgments
We thank all members of the Shapiro lab for helpful discussions throughout the project; Drs. Thomas Mann, Saumya Saurabh, and Darshankumar Pathak for their critical reading of the manuscript; and Dr. Jinfan Wang for help on modeling protein degradation data. Funding: This work was supported by the National Institute of General Medical Sciences, National Institutes of Health (R35-GM118071 to L.S.) and (R35-GM118067 to W.E.M.). L.S. is a Chan Zuckerberg Biohub Investigator. J.W. is a Mona M. Burgess Fellow through Stanford Bio-X.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906119116/-/DCSupplemental.
References
- 1.Casadesús J., Low D., Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He X.-J., Chen T., Zhu J.-K., Regulation and function of DNA methylation in plants and animals. Cell Res. 21, 442–465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith Z. D., Meissner A., DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Bickle T. A., Krüger D. H., Biology of DNA restriction. Microbiol. Rev. 57, 434–450 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Romero M. A., Cota I., Casadesús J., DNA methylation in bacteria: From the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Collier J., Epigenetic regulation of the bacterial cell cycle. Curr. Opin. Microbiol. 12, 722–729 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Val M.-E., Skovgaard O., Ducos-Galand M., Bland M. J., Mazel D., Genome engineering in Vibrio cholerae: A feasible approach to address biological issues. PLoS Genet. 8, e1002472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L., DNA mismatch repair: Functions and mechanisms. Chem. Rev. 106, 302–323 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez D., Kozdon J. B., McAdams H. H., Shapiro L., Collier J., The functions of DNA methylation by CcrM in Caulobacter crescentus: A global approach. Nucleic Acids Res. 42, 3720–3735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reisenauer A., Kahng L. S., McCollum S., Shapiro L., Bacterial DNA methylation: A cell cycle regulator? J. Bacteriol. 181, 5135–5139 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier J., Cell cycle control in Alphaproteobacteria. Curr. Opin. Microbiol. 30, 107–113 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Kozdon J. B., et al. , Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc. Natl. Acad. Sci. U.S.A. 110, E4658–E4667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou B., et al. , The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 11, e1004831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrader J. M., et al. , Dynamic translation regulation in Caulobacter cell cycle control. Proc. Natl. Acad. Sci. U.S.A. 113, E6859–E6867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hottes A. K., Shapiro L., McAdams H. H., DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58, 1340–1353 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Collier J., McAdams H. H., Shapiro L., A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 17111–17116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reisenauer A., Shapiro L., DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21, 4969–4977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright R., Stephens C., Zweiger G., Shapiro L., Alley M. R., Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 10, 1532–1542 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Zweiger G., Marczynski G., Shapiro L., A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235, 472–485 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Charette M. F., Henderson G. W., Doane L. L., Markovitz A., DNA-stimulated ATPase activity on the lon (CapR) protein. J. Bacteriol. 158, 195–201 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S., Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Woodcock C. B., Yakubov A. B., Reich N. O., Caulobacter crescentus cell cycle-regulated DNA methyltransferase uses a novel mechanism for substrate recognition. Biochemistry 56, 3913–3922 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Joshi K. K., Chien P., Regulated proteolysis in bacteria: Caulobacter. Annu. Rev. Genet. 50, 423–445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer R. T., Baker T. A., AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Karlowicz A., et al. , Defining the crucial domain and amino acid residues in bacterial Lon protease for DNA binding and processing of DNA-interacting substrates. J. Biol. Chem. 292, 7507–7518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung C. H., Goldberg A. L., DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 79, 795–799 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zehnbauer B. A., Foley E. C., Henderson G. W., Markovitz A., Identification and purification of the Lon+ (capR+) gene product, a DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 78, 2043–2047 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang C., Brown P. J. B., Ducret A., Brun Y. V., Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506, 489–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez A. M., et al. , A localized complex of two protein oligomers controls the orientation of cell polarity. MBio 8, e02238-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ptacin J. L., et al. , A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12, 791–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam H., Schofield W. B., Jacobs-Wagner C., A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124, 1011–1023 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Huitema E., Pritchard S., Matteson D., Radhakrishnan S. K., Viollier P. H., Bacterial birth scar proteins mark future flagellum assembly site. Cell 124, 1025–1037 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Collier J., Regulation of chromosomal replication in Caulobacter crescentus. Plasmid 67, 76–87 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Bowman G. R., et al. , A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134, 945–955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ptacin J. L., et al. , Bacterial scaffold directs pole-specific centromere segregation. Proc. Natl. Acad. Sci. U.S.A. 111, E2046–E2055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gur E., Sauer R. T., Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 22, 2267–2277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee S., et al. , Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc. Natl. Acad. Sci. U.S.A. 112, 250–255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonas K., Liu J., Chien P., Laub M. T., Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154, 623–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gora K. G., et al. , Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 87, 1277–1289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsui H., et al. , Oral immunization with ATP-dependent protease-deficient mutants protects mice against subsequent oral challenge with virulent Salmonella enterica serovar typhimurium. Infect. Immun. 71, 30–39 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breidenstein E. B. M., Bains M., Hancock R. E. W., Involvement of the lon protease in the SOS response triggered by ciprofloxacin in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 56, 2879–2887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers A., et al. , The LonA protease regulates biofilm formation, motility, virulence, and the type VI secretion system in Vibrio cholerae. J. Bacteriol. 198, 973–985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeinert R. D., et al. , A legacy role for DNA binding of Lon protects against genotoxic stress. bioRxiv:10.1101/317677 (9 May 2018).
- 44.Laloux G., Jacobs-Wagner C., How do bacteria localize proteins to the cell pole? J. Cell Sci. 127, 11–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudner D. Z., Losick R., Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2, a000307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebersbach G., Briegel A., Jensen G. J., Jacobs-Wagner C., A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134, 956–968 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergé M., Viollier P. H., End-in-sight: Cell polarization by the polygamic organizer PopZ. Trends Microbiol. 26, 363–375 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Holmes J. A., et al. , Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc. Natl. Acad. Sci. U.S.A. 113, 12490–12495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lasker K., et al. , Phospho-signal flow from a pole-localized microdomain spatially patterns transcription factor activity. bioRxiv:10.1101/220293 (21 February 2018).
- 50.Michaelis A. M., Gitai Z., Dynamic polar sequestration of excess MurG may regulate enzymatic function. J. Bacteriol. 192, 4597–4605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ducret A., Quardokus E. M., Brun Y. V., MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paintdakhi A., et al. , Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 99, 767–777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.