Significance
Glioblastoma, the most common and lethal of primary brain tumors, can diffusely infiltrate brain. Because the components of the cytoskeleton that drive tumor invasion are nonredundant, they have been attractive targets for blocking glioblastoma dispersion. However, we find that while targeting one of these—the molecular motor myosin IIA—effectively blocks glioblastoma invasion, it enhances tumor proliferation and lethality. In this study, we have found that myosin IIA functions as a tumor suppressor by reducing signaling of two oncogenes—ERK1/2 and NF-κB—in a manner regulated by the mechanics of the tumor and surrounding brain. Our results also argue that effective treatment of glioblastoma will require inhibiting targets that drive both invasion and proliferation.
Keywords: myosin, glioblastoma, signaling, invasion
Abstract
The ability of glioblastoma to disperse through the brain contributes to its lethality, and blocking this behavior has been an appealing therapeutic approach. Although a number of proinvasive signaling pathways are active in glioblastoma, many are redundant, so targeting one can be overcome by activating another. However, these pathways converge on nonredundant components of the cytoskeleton, and we have shown that inhibiting one of these—the myosin II family of cytoskeletal motors—blocks glioblastoma invasion even with simultaneous activation of multiple upstream promigratory pathways. Myosin IIA and IIB are the most prevalent isoforms of myosin II in glioblastoma, and we now show that codeleting these myosins markedly impairs tumorigenesis and significantly prolongs survival in a rodent model of this disease. However, while targeting just myosin IIA also impairs tumor invasion, it surprisingly increases tumor proliferation in a manner that depends on environmental mechanics. On soft surfaces myosin IIA deletion enhances ERK1/2 activity, while on stiff surfaces it enhances the activity of NFκB, not only in glioblastoma but in triple-negative breast carcinoma and normal keratinocytes as well. We conclude myosin IIA suppresses tumorigenesis in at least two ways that are modulated by the mechanics of the tumor and its stroma. Our results also suggest that inhibiting tumor invasion can enhance tumor proliferation and that effective therapy requires targeting cellular components that drive both proliferation and invasion simultaneously.
Glioblastoma (GBM), the most common malignant primary brain tumor, has remained lethal despite years of investigation into novel therapeutics (1). Although GBM rarely spreads outside the central nervous system (CNS), its ability to diffusely disperse within the CNS contributes to its highly malignant behavior (2, 3). Brain invasion requires cell motility, which is stimulated by the many growth factor receptors that are amplified or dysregulated in GBM (4). Although these receptors and their second messengers can be targeted with highly specific drugs, their efficacy in GBM has been limited by signal cascade redundancy, which allows cells to circumvent blockade of one pathway by activating another (5–7). A more promising approach would be to target downstream components where these redundant signaling pathways converge. These targets include members of the nonmuscle myosin II (NMII) family of molecular motors (8). We had previously shown that while the antiinvasive effects of EGFR blockade on GBM could be readily overcome with PDGFRα stimulation, and vice versa, pharmacologic inhibition of NMII effectively paralyzes GBM invasion even when both receptors are stimulated simultaneously (9). Nevertheless, our in vitro and ex vivo assays do not recapitulate the complexities of tumor behavior in a living host. This motivated us to determine whether targeting NMII function in vivo has similar effects. The NMII family consists of three members—IIA, IIB, and IIC—each with distinct functions (10–12). While their importance in cell motility makes NMII family members appealing targets to block tumor invasion, nearly all tissues express at least one member of this family. Hence, a pan-NMII targeting strategy would likely be toxic. An approach that only targets one of the three NMII isoform might still be effective and associated with fewer side effects. However, two reports have shown that targeting NMIIA induces squamous cell carcinoma in mice (13, 14). The mechanism underlying this effect remains unclear. While one report (13) found that NMIIA stabilizes the tumor suppressor p53, a second did not observe this (14).
In this study, we have examined the effects of targeting NMIIA and NMIIB together in GBM in vivo and in vitro. We find that genetic deletion of both NMIIA and NMIIB reduces tumorigenesis and impairs tumor proliferation. While a therapeutic approach that targets NMIIA might be less toxic than a pan-NMII approach, and while targeting NMIIA does markedly impair GBM invasion, we also find paradoxically that it enhances GBM growth and lethality in an in vivo rodent model of the disease. These effects are not associated with alterations in p53 levels or function. Instead, they occur hand in hand with enhancement in the activity of two signaling molecules, ERK1/2 and NF-κB, in a manner that is modulated by the stiffness of the tumor microenvironment. Our results indicate that the tumor-suppressive effects of NMIIA are a reflection of this molecular motor’s roles in shaping cell mechanics. In conjunction with previous work (15), they also suggest that in GBM efforts to target tumor invasion can enhance tumor proliferation, and vice versa, and that effective therapy of GBM requires targeting nonredundant cellular components that drive both of these malignant phenotypes simultaneously.
Results
A Retrovirally Driven Murine Model of GBM Provides a Way to Observe the Effect of NMII Deletion on Tumor Biology In Vivo.
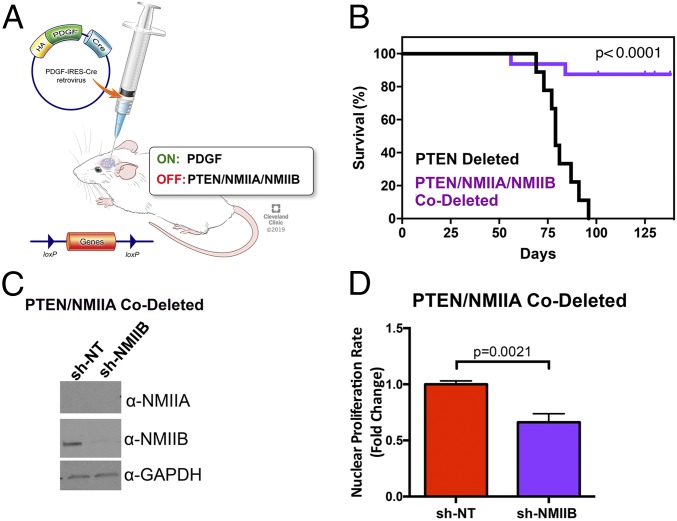
We have used an immunocompetent murine model of GBM that we have extensively described (16). It involves injection a bicistronic retrovirus encoding for PDGF and the cre recombinase into the white matter of mice with a floxed allele for either PTEN or TP53. The injected virus integrates into the genome of proliferative, PDGFRα-expressing glial progenitors (17), and tonic stimulation with PDGF, combined with cre-mediated deletion of the tumor suppressor, leads to development of tumors that closely resemble the proneural subtype of human GBM (16). For the studies described below, we have focused on mouse strains containing floxed alleles for PTEN (Fig. 1A). We measured the stoichiometry of the three NMII isoforms in this GBM model by using liquid chromatography and tandem mass spectrometry (LC/MS/MS). NMIIA is the predominant isoform, accounting for 56% of the total NMII, while NMIIB accounts for ∼35% (SI Appendix, Fig. S1A). NMIIC represents a small fraction (5 to 7%) of the total, and we elected not to examine this isoform further.
Fig. 1.
Deletion of NMIIA and NMIIB in GBM increases survival and reduces proliferation. (A) Mouse model of GBM. Reprinted with permission, Cleveland Clinic Center for Medical Art & Photography © 2019. (B) Kaplan–Meier curves comparing the survival of retroviral-induced GBMs in mice with floxed alleles for PTEN (n = 9) and for NMIIA, NMIIB, and PTEN (n = 16). The significance of differences in survival were assessed by both log-rank (Mantel–Cox) test and Gehan–Breslow–Wilcoxon tests and both give a P value of <0.0001. (C) Western blot demonstrating depletion of NMIIB heavy chain isoform using shRNA lentivirus from glioma cell lines genetically deleted for NMIIA with PDGF-IRES-cre retrovirus. (D) Nuclear doubling rate as measured by nuclear count every 3 h for 5 d, using Histone 2B-RFP–expressing glioma cells (n = 3). Statistical significance was assessed with a two-tailed t test.
Deleting NMIIA and NMIIB Together in a Murine GBM Model Impairs Tumor Proliferation and Reduces Tumorigenesis.
Our prior studies (16, 17) demonstrated that pharmacologic inhibition of NMII abolishes tumor invasion in vitro and in an ex vivo brain slice assay. However, these studies did not examine if prolonged targeting of NMII in vivo enhances survival. We therefore generated a mouse strain with floxed alleles for PTEN, NMIIA, and NMIIB. Injecting the PDGF-IRES-cre retrovirus in the white matter of these mice leads to deletion of PTEN, NMIIA, and NMIIB in the infected glial progenitor cells. We followed these mice for survival and results are depicted in Fig. 1B. They demonstrate that out of 16 NMIIA/NMIIB/PTEN triple-floxed mice, only 2 (12.5%) succumbed to tumor. Necropsy of the remaining mice shows no evidence of PDGF+ tumors while 100% of mice with only a PTEN-floxed allele develop tumors when injected with the same retrovirus (SI Appendix, Fig. S2). To determine what effect codeletion of NMIIA and NMIIB has on mitotic function, we injected retrovirus intracerebrally into mice with floxed alleles for PTEN and NMIIA to generate GBMs deleted for NMIIA and PTEN (SI Appendix, Fig. S2). Using LC/MS/MS, we found that NMIIA is barely detectable in these tumors (SI Appendix, Fig. S1A). Both these measurements (SI Appendix, Fig. S1A) as well as Western blots (SI Appendix, Fig. S1B) reveal that NMIIA deletion does not alter the content of either NMIIB or NMIIC to a statistically significant degree (P = 0.08 and 0.13, respectively). The PTEN/NMIIA-codeleted tumor cells are mononuclear, express one pair of γ tubulin-positive centrosomes (SI Appendix, Fig. S3), and undergo normal cytokinesis, as monitored by expression of an H2B-RFP nuclear reporter (Movie S1). We then suppressed NMIIB in these cells with short hairpin RNA (shRNA) (Fig. 1C). The resulting cells are multinucleated, contain multiple centrosomes (SI Appendix, Fig. S3), and are defective in cytokinesis (Movie S2). We measured the rate of nuclear proliferation by transfecting these cells with H2B-RFP. Deletion/suppression of NMIIA and NMIIB, respectively, reduces this rate by ∼30% (Fig. 1D). Since cells deleted for both NMIIA and NMIIB are multinucleated, the reduction in cellular proliferation rate is considerably greater than 30%.
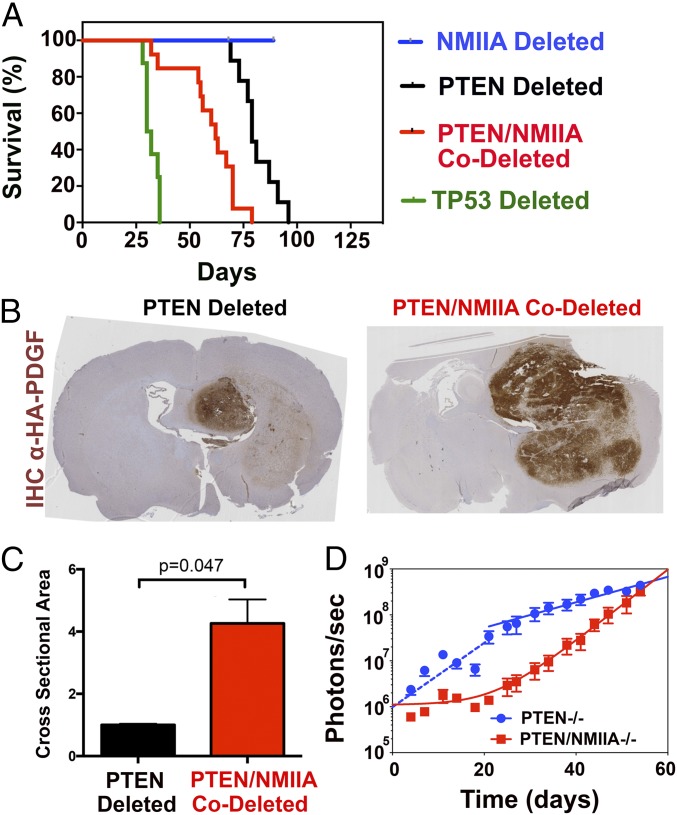
Deleting NMIIA in Murine GBM Impairs Tumor Dispersion but Enhances Tumor Lethality.
Since targeting both NMIIA and NMIIB would likely be toxic, we wondered if targeting only NMIIA might still prevent GBM dispersion, since it is a minor component of the NMII in brain (18). We accomplished this by injecting retrovirus into the white matter of mice with floxed alleles for NMIIA and PTEN. Since the PDGF encoded by our retrovirus is fused to the HA epitope, we were able to identify PDGF-expressing tumor cells within the host brain with αHA immunohistochemistry. Although both NMIIA-intact and -deleted tumor cells are found in white matter, those deleted for NMIIA were impaired in migrating through cerebral cortex (Fig. 2 A and B), which contains the smallest intercellular spaces in the brain (19–21). This implies, consistent with our prior study (22), that NMII is needed for cells to insinuate themselves through tight intercellular spaces. We tested this by examining migration through a 3-μm Transwell (Fig. 2 C and D) and found that tumor cells deleted (red) or shRNA-depleted (gray) for NMIIA were impaired in in vitro invasion. However, we paradoxically found that NMIIA deletion significantly enhances tumor lethality (Fig. 3A). Two previous studies also noted a relationship between NMIIA deletion and tumor progression (13, 14). One of these (13) concluded that NMIIA is a tumor suppressor that stabilizes p53. It reported that NMIIA suppression in normal keratinocytes prevents the physiologic increase in p53 protein levels seen in response to doxorubicin. To determine if this explains our results, we injected PDGF-IRES-cre–encoding retrovirus into the white matter of mice with floxed alleles for only NMIIA and compared survival to those with TP53 floxed alleles (Fig. 3A). As we previously reported (16), TP53-deleted murine GBM tumors led to mortality within 30 to 40 d after viral injection (Fig. 3A, green). By contrast, viral-injected NMIIA-floxed mice do not develop any signs of morbidity over 3 mo (Fig. 3A, blue). We also examined the effect of doxorubicin treatment on p53 protein expression in PTEN-deleted GBM cells that are intact or deleted for NMIIA (SI Appendix, Fig. S4 A and B). As the figure shows, p53 levels do not change appreciably after doxorubicin treatment in NMIIA-intact cells (n = 6). By contrast, in NMIIA-deleted cells, p53 levels rise significantly after 2, 3, and 6 h of doxorubicin treatment (n = 6; P < 0.01) and return to baseline at 12 h. We also tested the functionality of apoptosis by treating PTEN-deleted and PTEN/NMIIA-codeleted cells with doxorubicin and measuring expression of cleaved caspase 3. As SI Appendix, Fig. S4C shows, although cleaved caspase 3 can be detected in both cell lines, this effect is appreciably more robust in NMIIA-deleted cells. We had previously shown that tumor formation in our model of PTEN-deleted GBM requires subsequent mutations of p53 or deletion of p53-regulated transcriptional targets and concluded that loss of p53 activity is essential for tumor progression (23). The lack of increase in p53 and the meager rise in cleaved caspase 3 following doxorubicin treatment of our PTEN-deleted GBM cells (SI Appendix, Fig. S4) support this. By contrast, the rise in p53 levels and robust increase in cleaved caspase 3 following doxorubicin treatment of PTEN/NMIIA-codeleted cells leads us to conclude that p53-mediated apoptosis is much more functional in PTEN/NMIIA-codeleted tumors and deletion of NMIIA provides other protumorigenic stimuli that obviate the need for loss of p53.
Fig. 2.
NMIIA is necessary glioma migration and dispersion. (A) Immunohistochemistry of end-stage tumor sections stained for the HA epitope on infected tumor cells. (B) Higher magnification of A. T indicates tumor and CX indicates cerebral cortex. While tumors deleted for both PTEN and NMIIA are able to disperse through white matter, their invasion into the cortex is impaired. (C) Migration through 3-μm Transwell pore membrane toward 10% FBS (Scale bar, 200 μm). (D) Mean number of DAPI-stained nuclei per high power field (n = 8–10) migrating through 3 μm Transwells for PTEN-deleted and PTEN, NMIIA-codeleted tumors, along with corresponding results for murine PTEN-deleted GBMs where NMIIA expression was suppressed with shRNA, compared with PTEN-deleted tumor cells transfected with nontargeting (NT) shRNA. Statistical significance was assessed with a two-tailed t test.
Fig. 3.
Loss of NMIIA decreases survival in GBM and creates larger tumors. (A) Kaplan–Meier curves of mice bearing retrovirally induced GBMs deleted for PTEN (black, n = 9), NMIIA and PTEN (red, n = 13), TP53 (green, n = 6), and NMIIA (blue, n = 8). Median survivals are 79, 62, and 31 d after injection and not determined, respectively. Log-rank P values are PTEN−/− vs. PTEN−/−NMIIA−/− P = 0.0003, NMIIA−/− vs. PTEN−/−NMIIA−/− P < 0.0001, p53−/− vs. PTEN−/−NMIIA−/− P < 0.0001, and p53−/− vs. NMIIA−/− P < 0.0001. (B) Anti-HA immunohistochemistry of brains from mice with floxed alleles for PTEN or PTEN and NMIIA that were injected 35 d prior with retrovirus encoding a PDGF-HA fusion protein and the cre recombinase. (C) Area of HA-positive tumor measured by number of HA-positive pixels for PTEN-deleted (black) compared with PTEN/MNNIIA codeleted (red) tumors. (D) Fifty thousand luciferase-expressing PTEN-deleted and PTEN/NMIIA-codeleted tumor cells were injected into the white matter of NSG mice. Luminescence was monitored over the subsequent 54 d, and photon flux is plotted on a logarithmic scale versus days after injection. For both tumor types, growth follows a biphasic relationship. For PTEN-deleted cells (blue), this consists of an initial fast phase (blue dashed line) which transitions to a slower phase at ∼3 wk after injection. For PTEN/NMIIA-codeleted cells (red), growth fits a single exponential process that follows an initial lag phase, also lasting about 3 wk. The single exponential fits to the terminal growth phase describe tumor doubling times of 10.9 d (PTEN-deleted) and 3.9 d (PTEN/NMIIA-codeleted).
NMIIA deletion in our GBM model might instead be enhancing tumor proliferation. To test this, we implanted 50,000 luciferase-expressing PTEN-deleted and PTEN/NMIIA-codeleted tumor cells into the white matter of NSG mice. Luminescence was monitored over the subsequent 54 d, and the results are depicted in Fig. 3D. For PTEN-deleted tumors (blue), a plot of luminescence versus time fit two single exponential growth curves, with a faster phase (dashed blue line) for the first 3 wk converting to a slower phase (solid blue line). By contrast, PTEN/NMIIA-deleted tumors showed the opposite effect (red solid line)—with luminescence fitting a single exponential process that follows a lag of approximately 3 wk. Due to the lag, PTEN/NMIIA-codeleted tumors are initially smaller than the corresponding PTEN-deleted tumors, but they catch up and then exceed PTEN-deleted tumors by 60 d. For PTEN-deleted tumors, the fit of the terminal phase describes a doubling time of 10.9 d, while that for the PTEN/NMIIA-codeleted tumors is 3.9 d. Our data thus suggest that tumor growth evolves over time, and in opposite directions for NMIIA-intact versus -deleted tumors. One explanation is that tumor cells alter the brain over time in a way that makes growth more favorable for tumors that are NMIIA-deleted and less so for tumors that are not. One such alteration may be in the mechanics of the brain microenvironment, which as we will show in the next section has a significant influence on tumor growth.
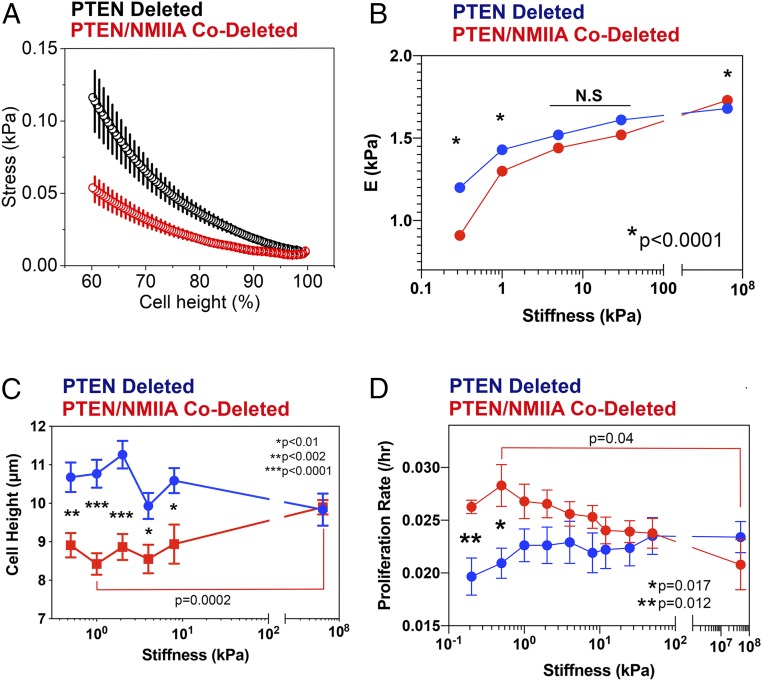
Deleting NMIIA in Murine GBM Alters Tumor Cell Morphology and Mechanics and Shifts the Stiffness Optimum for Proliferation.
One of the roles that NMIIA plays is in driving retrograde actin flow (24, 25), which counters actin-induced outgrowth of cellular processes. We examined the consequences of NMIIA suppression on cell morphology by using a microfabricated 3D invasion assay that has been previously described (26). This device (SI Appendix, Fig. S5A) maintains a stable chemotactic gradient as cells crawl through defined micrometer-sized constrictions formed by polydimethylsiloxane posts. We monitored migration of GFP-expressing, PTEN-deleted tumor cells that were treated with nontargeting shRNA or shRNA directed against NMIIA (SI Appendix, Fig. S5B). Results are depicted in Movies S3 and S4. Suppression of NMIIA markedly elongates cellular processes that entangle cells as they migrate toward the chemoattractant. To quantify this effect, we transfected PTEN-deleted and PTEN/NMIIA-codeleted GBM cells with H2B-RFP and we found that NMIIA deletion prolongs nuclear transit time through the 2-μm constriction by more than fivefold (SI Appendix, Fig. S5C).
NMIIA also can cross-link actin filaments, and it might be expected to shape cell mechanics. Indeed, pharmacologic inhibition of NMII results in a decrease in the apparent Young’s modulus of fibroblasts (27). We therefore subjected PTEN-deleted and PTEN/NMIIA-codeleted GBM cells grown on collagen-coated glass coverslips to atomic force microscopy (AFM). In our first AFM study, our goal was to compress the entire tumor cell body (diameter ∼10 µm) to generate the global stress curves depicted in Fig. 4A. To accomplish this, we used a 25-µm probe so that the contact area with the cell would be approximately flat. We applied a force of 50 nN to ensure that we would compress the cell by ∼50%. The resulting AFM data demonstrate that NMIIA-deleted GBM cells are less capable of resisting the probe-induced global deformation, suggesting that NMIIA deletion reduces actin cross-linking and the internal stress that leads to stiffening of the actin-based cytoskeleton. In the second study, we measured the stiffness of the cell cortex by using a smaller probe (1 µm), lower force (2 nN), and less compression over 25 positions per cell to calculate an apparent Young’s modulus as a function of substrate stiffness (Fig. 4B and SI Appendix, Fig. S6). As this figure shows, the apparent Young’s modulus for both NMIIA-intact and deleted cells increases—implying an increase in cortical stiffness—with increasing substrate stiffness. This feature has also been reported for nontransformed cells (28, 29). However, on a soft surface (0.3 kPa) the apparent Young’s modulus for NMIIA-deleted cells is significantly lower than for NMIIA-intact cells. This difference diminishes on stiffer surfaces (5 kPa) and reverses on glass (∼50 GPa).
Fig. 4.
NMIIA deletion alters GBM cell mechanics in a manner influenced by the stiffness of the tumor environment. (A) Global cell deformation was performed using a JPK Nanowizard 4 atomic force microscope equipped with cantilevers of a nominal stiffness of 2.4 N/m with a 25-µm-diameter sphere attached (Novascan). Measurements of cell deformability were made on PTEN-deleted (black) and PTEN/NMIIA-codeleted cells (red) grown on glass coverslips coated with collagen I. Force versus distance curves were converted into stress versus percentage of unstressed cell height curves with the assumption that normal stress can be calculated as the ratio of the applied force to the area of deformation. The percent cell height was calculated as the percentage of the total cell height that underwent indentation at a given force. (B) The local Young’s modulus was calculated from local cell cortex deformation measurements on PTEN-deleted (blue) and PTEN/NMIIA-codeleted (red) murine GBM cells, cultured on a range of substrate stiffness. Data are plotted as mean ± SEM. This reveals that NMIIA deletion reduces Young’s modulus compared with NMIIA-intact cells on soft surfaces and increases it on hard surfaces. Statistical significance was determined with a two-tailed t test. (C) Cell height was measured on NMIIA-intact (blue) and -deleted (red) tumor cells cultured on fibronectin-coated hydrogels (Softwell; Matrigen) covering a range of stiffness from 0.2 kPa to plastic. NMIIA-deleted cells are shorter than NMIIA-intact ones, and this difference decreases with increasing stiffness. Cell height for NMIIA-deleted cells on plastic is also significantly increased compared with the same cells grown on a 1-kPa surface. Statistical significance was determined with a two-tailed t test. (D) Plot of proliferation rate constant versus substrate stiffness for PTEN-deleted (blue) and PTEN/NMIIA-codeleted (red) tumor cells grown on fibronectin-coated Softwells (Matrigen). Growth kinetics were fit to a single exponential growth equation to yield rate constants, and each point represents the mean ± SEM of five replicates. Differences in rate constants between PTEN-deleted and PTEN/NMIIA-codeleted were significant on substrates of 0.2 and 0.5 kPa (***P = 0.012; **P = 0.017) using a two-tailed t test. Furthermore, the difference in rate constant for PTEN/NMIIA-codeleted cells grown on 0.5 kPa and on plastic is also significant (*P = 0.04).
We measured the the effect of NMIIA deletion on surface area, volume, and cell height (SI Appendix, Fig. S7 A–C) by culturing GFP-expressing PTEN-deleted and PTEN/NMIIA-codeleted cells on Matrigel-coated polyacrylamide gels of defined stiffness. Over a range of 0.7 to 50 kPa, NMIIA-deleted cells consistently have a larger surface area and volume than NMIIA-intact cells. PTEN/NMIIA-codeleted cells also appear to be decorated with longer and more numerous cellular processes over a range of substrate stiffness (SI Appendix, Fig. S8). NMIIA deletion also significantly flattens GBM cells on substrates of low (0.7 kPa) and intermediate (4.6 kPa) stiffness relative to NMIIA-intact cells, but this difference disappears on a stiff substrate (50 kPa). To ensure that this result is not affected by the complex matrix composition of Matrigel, we repeated this experiment using a 96-well plate that contains hydrogels of defined stiffness (Softwell; Matrigen), which we coated with fibronectin. As shown in Fig. 4C, we again observe that NMIIA deletion flattens GBM cells on soft, but not hard, surfaces.
Mih et al. (30) reported that pharmacologic inhibition of NMII or siRNA suppression of NMIIA enhances proliferation of fibroblasts on soft (1 kPa) but not on rigid (glass) substrates. To test this, we measured proliferation of PTEN-deleted and PTEN/NMIIA-codeleted GBM cells on the same fibronectin-coated Softwell plates that we used in Fig. 4C. We found that NMIIA-intact tumor cells (Fig. 4D, blue) show a broad optimum in proliferation between 1 and 20 kPa. By contrast, NMIIA deletion shifts this optimum to softer surfaces (Fig. 4D, red). At 0.2 and 0.5 kPa, NMIIA deletion enhances proliferation by ∼50% (P = 0.012 and 0.017, respectively), while at a higher range of stiffness differences become smaller and not statistically significant. Likewise, proliferation of NMIIA-deleted cells on 0.5-kPa substrates is significantly faster than that for the same cells on plastic (P = 0.04), while corresponding differences for NMIIA-intact cells are not (P = 0.15).
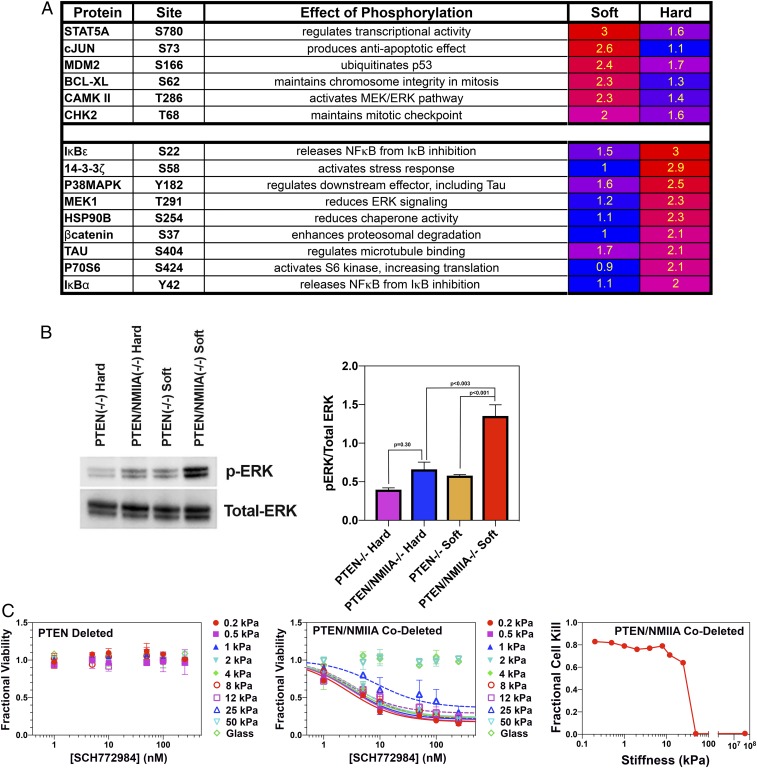
Deleting or Suppressing NMIIA in Murine GBM Alters the Activity of Signaling Effectors in a Mechanically Sensitive Manner.
We wondered if the effect of NMIIA deletion on proliferation (Fig. 4D) reflects changes in intracellular signaling. To test this, we generated lysates of PTEN-deleted and PTEN/NMIIA-codeleted cells grown on plastic and on a 0.5-kPa, fibronectin-coated Softwell substrate and subjected them to a phospho-antibody array (Cancer System Signaling; Full Moon Biosystems). We corrected for differences in protein loading by dividing each signal by the median overall signal. We then divided these signals by the corresponding total signal for the phosphorylated + nonphosphorylated species. Finally, we divided this normalized phosphorylation for NMIIA-deleted cells by that for NMIIA-intact cells on both soft (0.5 kPa) and plastic surfaces to generate ratios that reflect how NMIIA deletion alters phosphorylation as a function of stiffness. Fig. 5A, Upper lists the ratios for signaling components whose phosphorylation in NMIIA-deleted cells is elevated (≥2.0) only on a soft surface relative to NMIIA-intact cells, and Fig. 5A, Lower lists the corresponding ratios that are elevated only on a hard, plastic surface. Each of the proteins whose phosphorylation is up-regulated in NMIIA-deleted GBM cells on a soft surface is connected to the Ras-Raf-MEK-ERK pathway (31–36). Western blots of these lysates demonstrate that when grown on a soft (0.5 kPa) substrate, NMIIA-deleted cells enhance ERK1/2 phosphorylation approximately fourfold compared with NMIIA-intact cells. This enhancement disappears on a stiff substrate (Fig. 5B). By contrast, AKT (SI Appendix, Fig. S9) does not show any modulation with either NMIIA deletion or with alteration of substrate stiffness.
Fig. 5.
NMIIA deletion alters phosphorylation of signaling molecules in a manner that is modulated by substrate stiffness. (A) Ratios of normalized phosphorylation signal for PTEN/NMIIA-codeleted to PTEN-deleted tumors. (A, Upper) Phosphoproteins with ratios ≥2.0 on fibronectin-coated soft (0.5 kPa) but not hard (plastic) surfaces. (A, Lower) Phosphoproteins with ratios ≥2.0 on hard but not soft surfaces. (B, Left) Lysates from PTEN/NMIIA-codeleted (PTEN/NMIIA−/−) and PTEN-deleted (PTEN−/−) cells cultured on 0.5-kPa Softwells (Soft) and plastic (Hard) were blotted with antibodies to ERK1/2 and to pT202/pY204 ERK1/2. (B, Right) Plot of the phospho-ERK/total ERK ratio for PTEN-deleted and PTEN/NMIIA-codeleted cells cultured on 0.5-kPa and plastic substrates (n = 4). Significance was measured using a two-tailed t test. (C, Left) NMIIA-intact murine GBM cells were plated on 96-well Softwell plates, with substrate stiffness ranging from 0.2 kPa to glass, and treated with a range of concentrations of the ERK1/2 inhibitor SCH772984. Fractional cell viability relative to vehicle (DMSO)-treated cells is plotted as a function of drug concentration and over the range of substrate stiffness. This shows that SCH772984 has no effect over this stiffness range. (C, Middle) The corresponding experiment using NMIIA-deleted cells. Data are fit to hyperbolic dose–response relationships for all but the 50-kPa and glass surfaces, and the fitting defines maximum extrapolated degrees of cell kill. (C, Right) Plot of maximum cell kill versus substrate stiffness from the data in C, Middle.
If the enhanced growth we see with NMIIA deletion is ERK1/2-dependent, then an ERK1/2 inhibitor should be selectively toxic to NMIIA-deleted cells on soft surfaces. We treated NMIIA-deleted and -intact cells grown on Softwell plates with the ERK1/2 inhibitor SCH772984. Results are depicted in Fig. 5C. NMIIA-intact cells show no sensitivity to SCH772984 over the entire range of stiffness (Fig. 5 C, Left), implying little dependence on ERK for viability. By contrast, this drug is toxic to NMIIA-deleted cells (Fig. 5 C, Middle), and more so on soft surfaces. To quantify this effect, we fit the dose–response relationships to SCH772984 for NMIIA-deleted cells to a set of dose–response curves, one for each stiffness, and plotted the extrapolated degree of cell kill against substrate stiffness (Fig. 5 C, Right). As the figure shows, SCH772984 sensitivity drops sharply above 10 kPa, suggesting that NMIIA-deleted cells depend on ERK1/2 for viability over the stiffness range that has been measured in brain (37–40).
We also performed Western blots for ERK1/2 and phosphoERK1/2 in NMIIA-intact and -deleted tumors (SI Appendix, Fig. S10A). These demonstrate that NMIIA deletion is associated with an approximately twofold increase in ERK1/2 phosphorylation in our murine GBM, consistent with our in vitro results in Fig. 5. ERK1/2 is regulated by other signaling pathways, and one that is particularly relevant to cell stiffness sensing is YAP (41). Dephosphorylation of YAP at serine 127 translocates it to the nucleus, where it induces transcription of a number of genes. One of these encodes for RHAMM, which is necessary for ERK1/2 phosphorylation (42, 43). Thus, the enhanced ERK1/2 phosphorylation that we see in NMIIA-deleted tumors may be a consequence of reduced YAP phosphorylation. To test this, we performed Western blots for total YAP and pS127YAP in NMIIA-intact and -deleted tumors. These show that NMIIA deletion is associated with a 57% reduction in YAP phosphorylation (P = 0.019; SI Appendix, Fig. S10B), consistent with our proposal that a soft microenvironment, such as characterizes brain white matter, induces NMIIA-deleted GBM cells to enhance YAP and ERK1/2 signaling.
NMIIA-deleted GBM cells on a hard surface show a twofold or greater increase in phosphorylation of a number of regulators (Fig. 5 A, Lower), including MEK1, whose phosphorylation at T291 reduces ERK1/2 signaling (44), and two IκB isoforms, IκBα and IκBε. Phosphorylation of IκB dissociates it from NF-κB, releasing the latter to induce transcription of a wide array of genes (45–47). To confirm that NMIIA deletion enhances NF-κB activity in cells on a hard surface, we transfected GBM cells with a luciferase gene under control of the NF-κB response element, along with a mammalian expression vector for β-galactosidase (pCMV-βgal). Since we were unable to efficiently transfect our NMIIA-deleted GBM cells with this NF-κB reporter system, we utilized PTEN-deleted GBM cells infected with nontargeting or NMIIA targeting shRNA-expressing lentiviruses to knock down NMIIA >95% (SI Appendix, Fig. S5B). We also utilized this approach to determine if NMIIA suppression similarly alters NF-κB activity in a human triple-negative breast carcinoma (MDA-MB-231) and in a nontransformed primary human keratinocyte line cultured on a plastic substrate (SI Appendix, Fig. S5 D and E). As Fig. 6 A–C demonstrate, NMIIA suppression enhances NF-κB activity three- to eightfold in our PTEN-deleted murine GBM cells, five- to sevenfold in MDA-MB-231 cells, and five- to sixfold in human keratinocytes. We also examined whether expression of an shRNA-resistant NMIIA heavy chain could restore baseline NF-κB activity. MDA-MB-231 cells that expressed an shRNA targeting human NMIIA were stably transfected with a shRNA-resistant human NMIIA-GFP fusion protein. As Fig. 6D demonstrates, this restores NF-κB activity to near-control levels. We were not able to perform a similar experiment our PTEN-deleted GBM cells, as they could not tolerate stable transfection of an NMIIA-GFP construct. NF-κB plays an important role in the biology of tumor-initiating cells (48), in part by regulating the expression of stem cell transcription factors, including Sox2 (49–51), which is expressed in our murine GBM cell lines (52), and Nanog, which is expressed in MDA-MB-231 cells (53). We therefore examined the effect of shRNA-mediated NMIIA suppression on the activity of these transcription factors, utilizing a luciferase reporter under the control of either the Sox 2 or the Nanog promoter. NMIIA depletion increased Sox2 activity six- to eightfold in GBM cells (Fig. 6E) and Nanog activity 1.5- to twofold in MDA-MB-231 cells (Fig. 6F). We also examined the effect of targeting NMIIA on tumorsphere formation—a surrogate marker for tumor stem cell content—by applying a limiting dilution assay to both our rodent GBM as well as to human MDA-MB-231 cells (49). As illustrated in SI Appendix, Fig. S11, deletion or shRNA depletion of NMIIA increases tumorsphere size and tumorsphere frequency, implying that targeting NMIIA enhances proliferative and tumor initiating capacity, respectively.
Fig. 6.
NMIIA deletion enhances the activity of NF-κB, Sox2, and Nanog in cells grown on a stiff substrate. (A) Two PTEN-deleted GBM cell lines were infected with nontargeting (NT) or NMIIA-targeting shRNA-encoding lentiviruses. They were transiently transfected with a luciferase gene under control of an NF-κB response element and a mammalian expression vector for β-galactosidase (pCMV-βgal). NF-κB–dependent luciferase activity is plotted as mean ± SEM (n = 12). (B) NF-κB luciferase reporter assays were performed MDA-MB-231 cells expressing nontargeting or either of two NMIIA-targeting shRNA as above. Data are plotted as mean ± SEM (n = 9). (C) NF-κB luciferase reporter assays were performed on nontransformed keratinocytes that were treated with either NT shRNA or two different NMIIA-targeting shRNAs. (n = 8). (D) The MDA-MB-231 cells depleted for NMIIA were stably transfected with an shRNA-resistant GFP-NMIIA heavy-chain fusion protein and sorted for GFP expression. While NF-κB luciferase activity was significantly increased in cells depleted of NMIIA, expression of the GFP-NMIIA heavy chain returned NF-κB activity to baseline. Data are plotted as mean ± SEM (n = 17). (E) GBM cell lines treated with NT or NMIIA-targeting shRNA were transiently transfected with the Sox2 response element luciferase reporter and normalized luciferase activity was measured (n = 6). (F) MDA-MB-231 cells were transiently transfected with luciferase reporter construct under the control of Nanog transcriptional response element along with a control vector (pCMV-Renilla). Promoter activity was measured by luciferase activity. Values were normalized to Renilla activity to correct for transfection efficiency (n = 9). Data were analyzed with two-tailed t test. Significant at ****P < 0.0001, *** P = 0.0001 to 0.001, and **P = 0.001 to 0.01. Data are presented as the mean ± SEM.
Discussion
Myosin II Represents a “Point of Convergence” for Signaling Pathways That Are Dysregulated in Cancer.
We previously showed in an ex vivo murine model of GBM invasion that the promigratory effects produced by simultaneously activating EGFR and PDGFR could be overcome with pharmacologic blockade of NMII (9). However, tumors can adapt to long-term inhibition of therapeutic targets, and this motivated us to examine the effect of deleting both NMIIA and NMIIB in our murine GBM model. We found that this markedly reduces tumorigenesis and enhances survival (Fig. 1). Tumor cells that are deleted/suppressed for both NMIIA and NMIIB are polyploid (SI Appendix, Fig. S3), consistent with the role NMII plays in driving cytokinesis (54). Polyploidy and high degrees of aneuploidy enhance tumor cell fragility (55–57), and this effect, combined with the inhibition of tumor cell motility, explains the pronounced survival advantage that combined deletion of NMIIA and IIB provides (Fig. 1B). While dual inhibition of both NMIIA and IIB would likely be toxic, NMIIA is the predominant NMII isoform in our GBM model (SI Appendix, Fig. S1A) and is a minor component of the NMII expressed in normal brain (18, 22). This motivated us to examine the consequences of targeting just NMIIA.
Targeting NMIIA Impairs GBM Invasion but Enhances Tumor Lethality.
We found that while deleting NMIIA effectively blocks GBM invasion (Fig. 2), it also enhances GBM lethality and proliferation (Fig. 3). Several possibilities may explain this effect. First, NMIIA deletion could produce a defect in p53-mediated apoptosis. However, we find that in the presence of doxorubicin, NMIIA deletion neither prevents up-regulation of p53 nor blocks cleavage of caspase 3 (SI Appendix, Fig. S4). A second explanation is that since NMIIA plays a role in cytokinesis, its deletion might lead to aneuploidy. While severe degrees of aneuploidy place a metabolic burden that would slow tumor proliferation, lesser degrees can be associated with enhanced proliferation and malignant behavior (55–57). We therefore examined G-banded metaphase spreads from 20 PTEN-deleted and 20 PTEN/NMIIA-codeleted tumor cells (Creative Bioarray). While normal diploid mouse cells have 40 chromosomes, we found that 30% of PTEN-deleted cells have 41 to 53 chromosomes and another 50% have >67 chromosomes, with 10% containing >100. A corresponding analysis of PTEN/NMIIA-codeleted cells showed that 20% have 41 to 44 chromosomes and 70% have 72 to 76 chromosomes. Thus, both NMIIA-intact and -deleted tumor cells are characterized by significant but similar degrees of aneuploidy. Finally, by reducing tumor invasion we would expect that NMIIA deletion would make GBMs more compact and nodular (Fig. 2 A and B). A recent report has demonstrated that nodular brain tumors generate more local brain compression, resulting in reduced vascular perfusion and and increased disability (58). We therefore conclude that the effects of NMIIA deletion reflect enhanced proliferation in a tumor that is also more likely to damage normal neurologic structures because of its impairment in tumor dispersion.
The Roles That NMIIA Plays in Tumor Biology Are Modulated by the Mechanics of the Tumor Microenvironment.
Our data imply that NMIIA acts as a tumor suppressor by antagonizing ERK1/2 and NF-κB in a manner regulated by environmental mechanics. We conclude that ERK1/2 is the major driver of the increase in proliferation that we see for NMIIA-deleted cells on soft surfaces, since this increase can be nearly completely blocked with an ERK1/2 inhibitor (Fig. 5 C, Middle and Right). That SCH772984 is appreciably toxic for NMIIA-deleted cells over a fairly broad mechanical range (Fig. 5 C, Right) also implies that ERK activation in these cells is likely to also present over a similarly broad range of stiffness. Measures of the Young’s modulus of brain and of GBM both range from 0.1 to 10 kPa (37–40, 59). Nevertheless, these macroscopic measures do not necessarily reflect the microscopic mechanical features that individual cells experience within a tumor—features that could be dominated by cell processes that are filled with relatively incompressible microtubules. Thus, it seems likely that individual GBM cells could experience a broad range of stiffness in situ.
NMIIA deletion has significant effects on cell morphology, including height, volume, and surface area (SI Appendix, Figs. S7 and S8), and prior studies have shown that cellular geometry correlates with proliferation (30, 60, 61). Three possible mechanisms could explain this. First, ERK is well known to be regulated by integrin activity (62), which in turn depends on engagement with the extracellular matrix (ECM). Consequently, cells that have a larger surface area and volume might be expected to have enhanced integrin–ECM attachment with subsequent increased downstream signaling to ERK. However, while surface area and volume of NMIIA-deleted cells remain larger than for NMIIA-intact cells from soft to stiff surfaces (0.7 to 50 kPa), proliferation is only enhanced on the softest end of this range (0.2 to 1.0 kPa). Second, both proliferation rate and cell height vary with substrate stiffness (Fig. 4 B and C), and a mechanism that could explain this correlation has been proposed (63). It argues that phosphorylated second messengers have to “run a gauntlet” of cytoplasmic phosphatases to reach their targets, and cell flattening enhances the probability that these signaling molecules could phosphorylate their targets before colliding with a phosphatase. This model is consistent with our observation that cell flattening is most pronounced in NMIIA-deleted cells on low stiffness substrates, where both proliferation and ERK1/2 phosphorylation are increased (Figs. 4 B and C and 5B). However, it would also predict that there should be a corresponding increase in phosphorylation of other second messengers, such as AKT, which we do not see (Fig. 5C). Finally, the Young’s modulus of our GBM cells also varies with substrate stiffness (Fig. 4A) and is lowest on the softest surfaces. We could therefore explain our results if we argue that the activity of ERK1/2 is fine-tuned to cortical stiffness, with greater ERK1/2 phosphorylation in cells that are relatively soft. Metastatic tumor cells typically are softer than nonmetastatic tumor or normal cells (64, 65), and our results suggest that this softening selects for ERK1/2-driven tumor proliferation and invasion. This explanation is also consistent with a recent report, which found that while stretching of the tissue in the Drosophila pupal notum enhances ERK activity, compaction of this tissue inhibits it (66).
As substrate stiffness increases, the Young’s modulus of NMIIA-deleted tumor cells correspondingly increases and on glass it exceeds that of NMIIA-intact cells. A plausible explanation for this effect is that while on soft substrates most cells, including gliomas, have few actin bundles, these structures increase as substrate stiffness increases. In the more labile and thinner meshwork of actin bundles seen on soft substrates, NMIIA would play a dominant role, and so its loss would have a larger impact on stiffness. Since NMIIB has a dominant role in ventral stress fibers, the activity of NMIIA would be less important in determining the stiffness that is now dominated by contractile actin fibers (67).
On a stiff surface, NMIIA deletion does not activate ERK1/2. This may reflect the increased phosphorylation of MEK1 at threonine 291 (Fig. 5A), which regulates ERK1/2 activity through a negative feedback loop (44). However, we do observe increased phosphorylation of two members of the IκB family, which explains the enhanced NF-κB transcriptional activity (Fig. 6 A–D). In endothelial cells, the NMII regulatory light chain tonically suppresses NF-κB activity through its antagonism of TRAF2 binding to the TNF receptor superfamily (68). In cells such as GBM, where NMIIA is the major isoform, this would predict enhanced NF-κB activity with NMIIA deletion over the entire range of substrate stiffness (Fig. 6). However, in endothelial cells, ERK can block the activation of NF-κB (69). Were this mechanism active in in our murine GBM cells as well, then the increased ERK activity that we see on soft surfaces might explain why we do not see an increase in NF-κB activity under these conditions. NF-κB has been shown to play a central role in the maintenance of tumor stem cells (48, 70–73), and this is consistent with our finding that on a hard surface NMIIA suppression enhances the activity of Sox2 in GBM and Nanog in breast carcinoma (Fig. 6 E and F). That NMIIA deletion leads to activation of stem cell transcription factors is consistent with reports that NMII inhibition enhances survival of pluripotent stem cells in vitro (74, 75). SI Appendix, Fig. S12 summarizes these relationships between NMIIA expression, substrate stiffness, and ERK1/2 and NF-κB signaling.
In both neurons and GBM, cell migration varies with substrate stiffness in a manner that is characterized by a stiffness optimum. We have previously shown that two factors determine the position of this optimum—the internal, retrograde force generated by myosin motors on actin filaments and the compliance of cell–ECM attachments (76). Our current data suggest that there is a similar optimality in the dependence of growth on substrate stiffness—one that is also modulated by myosin II (Fig. 4D). The relevance of microenvironmental mechanics to tumor growth is also consistent with evidence that enhanced ECM stiffness can make GBM more aggressive (77).
Tumor invasion, like other forms of cell motility, requires tight spatial and temporal control of NMII activity, which is regulated by rho kinase (ROCK)-mediated phosphorylation of the NMII regulatory light chain. While we have shown that suppression of NMII activity can block tumor invasion, so too can overexpression of NMII or of a constitutively active rho kinase (77). Furthermore, mice bearing orthotopic human GBMs that express a constitutively active form of RhoA survive longer than those with control GBMs. While transfection with NMII or rho kinase may not be therapeutically practical, a small-molecule activator of NMIIB and IIC, 4 hydroxyacetophenone, also blocks invasion of pancreatic adenocarcinoma (78), suggesting that enhancement of tumor contractility may have therapeutic potential.
Our results imply that in GBM tumor proliferation and tumor invasion are linked, and that effective therapy requires targeting both phenotypes. Our finding that there is a dichotomy between ERK1/2 and NF-κB signaling in GBM that is connected to environmental mechanics is reminiscent of an earlier study (79) which demonstrated that there is a dichotomy in the behavior of GBM tumor cells between two phenotypes—one characterized by high proliferation and the other by high dispersion. This dichotomy, referred to as “go or grow,” is associated with corresponding differences in the expression of key transcriptional factors (15). The proliferation-dominant GBM cells, found in the core of the tumor, up-regulate several transcription factors that are downstream of ERK1/2, including MYC, CREB, and CRE-ATF, while the invasion-dominant GBM cells, found at the tumor/brain interface, up-regulate NF-κB. Furthermore, prior studies with antiangiogenics have found that this antiproliferative therapy enhances tumor invasion (80, 81). Combined with our results, this suggests that targeting GBM proliferation may enhance GBM invasion, and vice versa. If this premise is correct, we would predict that for a GBM therapy to be effective it must inhibit both invasion and proliferation simultaneously. Our finding that codeletion of NMIIA and IIB prevents tumorigenesis supports this premise. While a pan-NMII targeting approach is likely to be too toxic to be clinically practical, we note that a number of other molecular motors, including several mitotic kinesins (82), drive both tumor proliferation and invasion and may serve as effective targets for blocking the two defining phenotypes of GBM.
Materials and Methods
Retrovirus production, intracranial injection methods, and establishment of primary cell lines from our PTEN-deleted and PTEN/NMIIA-codeleted tumors were performed as described previously (16, 17). The manufacture and use of migration manifolds was performed as described in ref. 26. Transwell assays were performed as described in ref. 22. A complete discussion of all remaining methods, including generation of murine tumors, reporter assays, tumor invasion assays, and histologic analysis is included in SI Appendix.
All mouse procedures were performed with adherence to protocols approved by the Institute Animal Care and Use Committees at the Lerner Research Institute of the Cleveland Clinic and the Mayo Clinic.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants CA172986, CA210910, and NS073610 (to S.S.R.), GM050009 (to T.E.), NS073610, and NS052738 (to P.C.); CA210184 and HL082792 (to J.L.); GM111942 and CA193417 (to P.A.J.); and CA210190 and CA172986 (to D.J.O.); awards from the Department of Defense Breast Cancer Research Program (Breakthrough Award BC150580 to J.L.); and the National Science Foundation CAREER Award CBET-1254846 (to J.L.). This work was performed in part at the Cornell NanoScale Facility, an NNCI member supported by NSF Grant NNCI-1542081. We thank Drs. Robert Adelstein and Mary A. Conti (NHLBI, NIH) for their gift of NMIIAlox/lox and NMIIAlox/lox/NMIIBlox/lox mice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 15322.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902847116/-/DCSupplemental.
References
- 1.Puduvalli V. K., Hoang N., Chemotherapy of high-grade astrocytomas in adults. Prog. Neurol. Surg. 31, 116–144 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Giese A., Westphal M., Glioma invasion in the central nervous system. Neurosurgery 39, 235–250, discussion 250–252 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Burger P. C., Kleihues P., Cytologic composition of the untreated glioblastoma with implications for evaluation of needle biopsies. Cancer 63, 2014–2023 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Gschwind A., Fischer O. M., Ullrich A., The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat. Rev. Cancer 4, 361–370 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Ahluwalia M. S., et al. , Phase II trial of ritonavir/lopinavir in patients with progressive or recurrent high-grade gliomas. J. Neurooncol. 102, 317–321 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Raizer J. J., et al. ; Brain Tumor Trials Collaborative , A phase II study of bevacizumab and erlotinib after radiation and temozolomide in MGMT unmethylated GBM patients. J. Neurooncol. 126, 185–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot J. F., Gilbert M. R., New molecular targets in malignant gliomas. Curr. Opin. Neurol. 20, 712–718 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R., Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivkovic S., et al. , Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol. Biol. Cell 23, 533–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovács M., Wang F., Hu A., Zhang Y., Sellers J. R., Functional divergence of human cytoplasmic myosin II: Kinetic characterization of the non-muscle IIA isoform. J. Biol. Chem. 278, 38132–38140 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Wang F., et al. , Kinetic mechanism of non-muscle myosin IIB: Functional adaptations for tension generation and maintenance. J. Biol. Chem. 278, 27439–27448 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Billington N., Wang A., Mao J., Adelstein R. S., Sellers J. R., Characterization of three full-length human nonmuscle myosin II paralogs. J. Biol. Chem. 288, 33398–33410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schramek D., et al. , Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343, 309–313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conti M. A., et al. , Conditional deletion of nonmuscle myosin II-A in mouse tongue epithelium results in squamous cell carcinoma. Sci. Rep. 5, 14068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhruv H. D., et al. , Reciprocal activation of transcription factors underlies the dichotomy between proliferation and invasion of glioma cells. PLoS One 8, e72134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei L., et al. , Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One 6, e20041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assanah M. C., et al. , PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia 57, 1835–1847 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Ma X., Adelstein R. S., The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture 4, 88–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorne R. G., Nicholson C., In vivo diffusion analysis with quantum dots and dextrans predicts the width of brain extracellular space. Proc. Natl. Acad. Sci. U.S.A. 103, 5567–5572 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuddapah V. A., Robel S., Watkins S., Sontheimer H., A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci. 15, 455–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson C., Hrabětová S., Brain extracellular space: The final Frontier of Neuroscience. Biophys. J. 113, 2133–2142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beadle C., et al. , The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell 19, 3357–3368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonabend A. M., et al. , The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res. 74, 1440–1451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bangasser B. L., Rosenfeld S. S., Odde D. J., Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys. J. 105, 581–592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan C. E., Odde D. J., Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Davidson P. M., Sliz J., Isermann P., Denais C., Lammerding J., Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. 7, 1534–1546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schäfer A., Radmacher M., Influence of myosin II activity on stiffness of fibroblast cells. Acta Biomater. 1, 273–280 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Liu H., Sun Y., Simmons C. A., Determination of local and global elastic moduli of valve interstitial cells cultured on soft substrates. J. Biomech. 46, 1967–1971 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Georgouli M., et al. , Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 176, 757–774.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mih J. D., Marinkovic A., Liu F., Sharif A. S., Tschumperlin D. J., Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J. Cell Sci. 125, 5974–5983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pircher T. J., Petersen H., Gustafsson J. A., Haldosén L. A., Extracellular signal-regulated kinase (ERK) interacts with signal transducer and activator of transcription (STAT) 5a. Mol. Endocrinol. 13, 555–565 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Malmlo M., et al. , MEK-ERK-mediated phosphorylation of Mdm 2 at serine 166 in hepatocytes. J. Biol. Chem. 282, 2288–2296 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Deng Z., Sui G., Rosa P. M., Zhao W., Radiation-induced c-Jun activation depends on MEK1-ERK1/2 signaling pathway in microglial cells. PLoS One 7, e36739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baruah P. S., Beauchemin M., Hébert J., Bertrand R., Dynamic Bcl-xL (S49) and (S62) phosphorylation/dephosphorylation during mitosis prevents chromosome instability and aneuploidy in normal human diploid fibroblasts. PLoS One 11, e0159091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cipolletta E., et al. , Targeting the CAMKII/ERK interaction in the heart prevents cardiac hypertrophy. PLoS One 10, e0130477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei F., Xie Y., Tao L., Tang D., Both ERK1 and ERK2 kinases promote G2/M arrest in etoposide-treated MCF7 cells by facilitating ATM activation. Cell. Signal. 22, 1783–1789 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Pepin K. M., et al. , MR elastography analysis of glioma stiffness and IDH1-mutation status. AJNR Am. J. Neuroradiol. 39, 31–36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Streitberger K.-J., et al. , High-resolution mechanical imaging of glioblastoma by multifrequency magnetic resonance elastography. PLoS One 9, e110588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng S., Clarke E. C., Bilston L. E., Rheological properties of the tissues of the central nervous system: A review. Med. Eng. Phys. 30, 1318–1337 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Chatelin S., Constantinesco A., Willinger R., Fifty years of brain tissue mechanical testing: From in vitro to in vivo investigations. Biorheology 47, 255–276 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Mason D. E., et al. , YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J. Cell Biol. 218, 1369–1389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z., et al. , Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc. Natl. Acad. Sci. U.S.A. 111, E89–E98 (2014), Correction in: Proc. Natl. Acad. Sci. U.S.A.113, E7641–E7642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S., et al. , The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J. Biol. Chem. 273, 11342–11348 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Eblen S. T., et al. , Mitogen-activated protein kinase feedback phosphorylation regulates MEK1 complex formation and activation during cellular adhesion. Mol. Cell. Biol. 24, 2308–2317 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solt L. A., May M. J., The IkappaB kinase complex: Master regulator of NF-kappaB signaling. Immunol. Res. 42, 3–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayden M. S., Ghosh S., NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 26, 203–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karin M., Lin A., NF-kappaB at the crossroads of life and death. Nat. Immunol. 3, 221–227 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Rinkenbaugh A. L., Baldwin A. S., The NF-κB pathway and cancer stem cells. Cells 5, E16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lathia J. D., Mack S. C., Mulkearns-Hubert E. E., Valentim C. L. L., Rich J. N., Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gangemi R. M. R., et al. , SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 27, 40–48 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Suvà M. L., et al. , Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gill B. J., et al. , MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 111, 12550–12555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han J., et al. , RNA interference-mediated silencing of NANOG reduces cell proliferation and induces G0/G1 cell cycle arrest in breast cancer cells. Cancer Lett. 321, 80–88 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Robinson D. N., Spudich J. A., Mechanics and regulation of cytokinesis. Curr. Opin. Cell Biol. 16, 182–188 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Y. C., Amon A., Gene copy-number alterations: A cost-benefit analysis. Cell 152, 394–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegel J. J., Amon A., New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 28, 189–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon D. J., Resio B., Pellman D., Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Seano G., et al. , Solid stress in brain tumours causes neuronal loss and neurological dysfunction and can be reversed by lithium. Nat. Biomed. Eng. 3, 230–245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miroshnikova Y. A., et al. , Tissue mechanics promote IDH1-dependent HIF1α-tenascin C feedback to regulate glioblastoma aggression. Nat. Cell Biol. 18, 1336–1345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Folkman J., Moscona A., Role of cell shape in growth control. Nature 273, 345–349 (1978). [DOI] [PubMed] [Google Scholar]
- 61.Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E., Geometric control of cell life and death. Science 276, 1425–1428 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Schwartz M. A., Assoian R. K., Integrins and cell proliferation: Regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 114, 2553–2560 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Meyers J., Craig J., Odde D. J., Potential for control of signaling pathways via cell size and shape. Curr. Biol. 16, 1685–1693 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Swaminathan V., et al. , Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 71, 5075–5080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu W., et al. , Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS One 7, e46609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno E., Valon L., Levillayer F., Levayer R., Competition for space induces cell elimination through compaction-driven ERK downregulation. Curr. Biol. 29, 23–34.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuragano M., Uyeda T. Q. P., Kamijo K., Murakami Y., Takahashi M., Different contributions of nonmuscle myosin IIA and IIB to the organization of stress fiber subtypes in fibroblasts. Mol. Biol. Cell 29, 911–922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chandrasekharan U. M., et al. , Release of nonmuscle myosin II from the cytosolic domain of tumor necrosis factor receptor 2 is required for target gene expression. Sci. Signal. 6, ra60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maeng Y.-S., et al. , ERK is an anti-inflammatory signal that suppresses expression of NF-kappaB-dependent inflammatory genes by inhibiting IKK activity in endothelial cells. Cell. Signal. 18, 994–1005 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Guzman M. L., et al. , Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 98, 2301–2307 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Rajasekhar V. K., Studer L., Gerald W., Socci N. D., Scher H. I., Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-κB signalling. Nat. Commun. 2, 162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myant K. B., et al. , ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 12, 761–773 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao L., et al. , Osteopontin promotes a cancer stem cell-like phenotype in hepatocellular carcinoma cells via an integrin-NF-κB-HIF-1α pathway. Oncotarget 6, 6627–6640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker A., et al. , Non-muscle myosin II regulates survival threshold of pluripotent stem cells. Nat. Commun. 1, 71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe K., et al. , A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Bangasser B. L., et al. , Shifting the optimal stiffness for cell migration. Nat. Commun. 8, 15313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong S. Y., et al. , Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. Cancer Res. 75, 1113–1122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Surcel A., et al. , Pharmacological activation of myosin II paralogs to correct cell mechanics defects. Proc. Natl. Acad. Sci. U.S.A. 112, 1428–1433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie Q., Mittal S., Berens M. E., Targeting adaptive glioblastoma: An overview of proliferation and invasion. Neurooncol. 16, 1575–1584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu K. V., et al. , VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell 22, 21–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Norden A. D., et al. , Bevacizumab for recurrent malignant gliomas: Efficacy, toxicity, and patterns of recurrence. Neurology 70, 779–787 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Venere M., et al. , The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci. Transl. Med. 7, 304ra143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.