Significance
The formation of ridge-like trabeculae from the myocardium is central to heart formation, as it allows for the increase in muscle mass before the formation of coronary circulation. We found that cardiomyocytes display spatiotemporal orientation during trabecular formation. Cardiomyocytes in the Numb and Numblike double knockout display abnormal cellular orientation. The knockout hearts display reduced membrane localized N-cadherin due to attenuated N-cadherin recycling to the membrane. When chick N-cadherin is overexpressed in the knockouts via a mosaic model, the defects of cellular orientation and trabeculation were at least partially rescued, suggesting that Numb and Numblike regulate cellular orientation and trabeculation via N-cadherin. Cellular orientation might modulate the directionality of cellular migration, mitotic spindle orientation, and cellular organization during heart morphogenesis.
Keywords: Numb family proteins, trabecular morphogenesis, endocytosis, cellular orientation, single-cell lineage tracing
Abstract
The roles of cellular orientation during trabecular and ventricular wall morphogenesis are unknown, and so are the underlying mechanisms that regulate cellular orientation. Myocardial-specific Numb and Numblike double-knockout (MDKO) hearts display a variety of defects, including in cellular orientation, patterns of mitotic spindle orientation, trabeculation, and ventricular compaction. Furthermore, Numb- and Numblike-null cardiomyocytes exhibit cellular behaviors distinct from those of control cells during trabecular morphogenesis based on single-cell lineage tracing. We investigated how Numb regulates cellular orientation and behaviors and determined that N-cadherin levels and membrane localization are reduced in MDKO hearts. To determine how Numb regulates N-cadherin membrane localization, we generated an mCherry:Numb knockin line and found that Numb localized to diverse endocytic organelles but mainly to the recycling endosome. Consistent with this localization, cardiomyocytes in MDKO did not display defects in N-cadherin internalization but rather in postendocytic recycling to the plasma membrane. Furthermore, N-cadherin overexpression via a mosaic model partially rescued the defects in cellular orientation and trabeculation of MDKO hearts. Our study unravels a phenomenon that cardiomyocytes display spatiotemporal cellular orientation during ventricular wall morphogenesis, and its disruption leads to abnormal trabecular and ventricular wall morphogenesis. Furthermore, we established a mechanism by which Numb modulates cellular orientation and consequently trabecular and ventricular wall morphogenesis by regulating N-cadherin recycling to the plasma membrane.
The heart is the first functional organ formed in mammalian embryonic development (1). During cardiogenesis, cardiac progenitor cells from the cardiac crescent migrate toward the ventral midline to form a linear heart tube with a smooth inner surface (2, 3). When the linear heart tube undergoes looping, myocardium along the outer curvature of the tube will extend inward to form ridge-like structures (2, 3), which are the newly initiated trabeculae (4, 5). As the early embryonic heart does not have a coronary circulatory system to perfuse itself, the ridge-like trabeculae increase surface area to facilitate nutrient and oxygen exchange. The trabeculae will coalesce and compact with the compact zone once the coronary system is established (6, 7). A lack of trabecular formation results in lowered availability of oxygen and nutrients in the myocardial tissue and leads to embryonic demise, while excess trabeculation leads to left ventricular noncompaction (LVNC) cardiomyopathy and subsequent heart failure in both mice and humans (8–10). Despite the fundamental nature of this morphogenetic process and its significant clinical implications, the molecular and cellular mechanisms of trabecular morphogenesis and compaction are still not fully understood, especially in mammals (11–19).
Numb, the first discovered cell-fate determinant in the process of asymmetric cell division for multiple progenitor cells (20–26), is essential for a variety of biological processes (20, 21, 23, 27–34). In mice, there are 2 homologs: Numb and Numblike, collectively known as the Numb family proteins, abbreviated as NFPs (35, 36). Recently, Numb has been revealed to regulate cardiac progenitor cell differentiation and cardiac morphogenesis in different species (32, 37–40). In mice, Nkx2.5Cre/+ (41) mediated myocardial-specific NFPs double-knockout (MDKO) hearts display a variety of defects including trabecular formation and LVNC (31, 32, 42). The key molecular players and the related signaling pathways altered that are responsible for each defect in MDKO hearts have not been identified.
Tissue morphogenesis depends on the spatiotemporal arrangement of cells during development. The best-known mechanism that contributes to the final shape of a tissue is cellular intercalation in response to morphogens (43). A less-well-known but essential mechanism is oriented cell division (OCD) (43). OCD is typically determined by evaluating the alignment of the mitotic spindles relative to organ and anatomical axes (44, 45). OCD is a potential asymmetric cell division that contributes to cellular diversity, differentiation, and organogenesis during development (16, 46, 47). A third mechanism is cellular elongation-mediated cellular orientation, which has not been previously studied during organ morphogenesis. We discovered the defect of abnormal cellular orientation in the MDKO heart, which might be responsible for the morphogenetic defects observed in the trabeculae and ventricular wall.
Mechanistically, we found that MDKO hearts display reduced levels and abnormal subcellular localization of N-cadherin. Cadherins are a large family of cell–cell adhesion molecules that mediate calcium-dependent and homophilic adhesion, thereby promoting association of cells expressing the same cadherin family members (48). In the heart, N-cadherin is the major component of adherens junctions. N-cadherin is required for the formation and maintenance of intercalated discs in mature cardiomyocytes (49) and its deletion leads to conduction slowing and arrhythmogenesis (50). In the early embryonic heart, N-cadherin is required for adherens junction-mediated cell–cell association (51), which is required for multiple biological processes, including the establishment and maintenance of tissue architecture and coordination of cell behavior within the tissue. Work in zebrafish shows that dynamic N-cadherin subcellular localization correlates with trabecular initiation (13), N-cadherin is required for cardiomyocyte transmural migration in chicken hearts (52), and N-cadherin is required for transmural migration and mitotic spindle orientations during trabecular morphogenesis in mouse hearts (16). In this study, using the mCherry:Numb knockin (mCherry:NumbKI) line, we found that Numb mainly colocalizes with the recycling endosomes, and loss of NFPs disrupts N-cadherin recycling to the plasma membrane. The N-cadherin mislocalization contributes to the defects in cellular orientation and trabeculation in the MDKO, which can be partially rescued by N-cadherin overexpression.
Results
NFPs Are Required for Cellular Orientation and Organization during Trabecular and Ventricular Wall Morphogenesis.
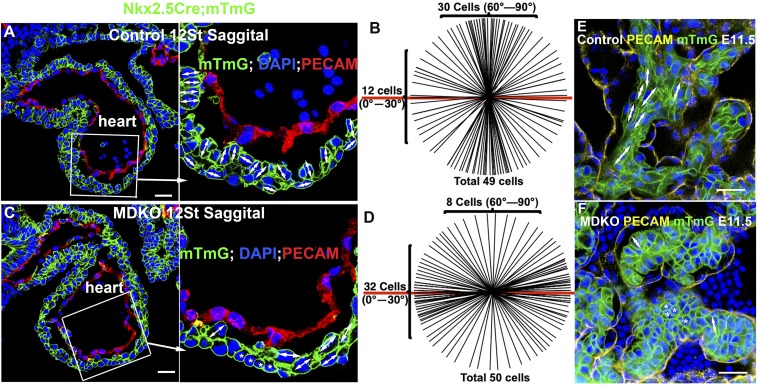
Tissue morphogenesis depends on spatiotemporal cellular arrangement. The cellular arrangement within an organ can be determined by cellular orientation, the magnitude of which can be evaluated by the ratio of length to width of a cell (47). To examine whether cardiomyocytes display cellular orientation, embryos of different ages with the genotype of Nkx2.5Cre/+ (41); ROSA26TMDtdTomato-EGFP (mTmG) (53) or cTnT-Cre (54); mTmG were stained for PECAM, a marker for endocardial and endothelial cells, and then cleared and 3D-imaged (55, 56). The cell membrane of the Cre-expressing cell is outlined by the membrane localized green fluorescent protein (GFP) and is used to determine the cellular orientation. We did not observe an obvious pattern of oriented cellular orientation at the 3 or 6 somite pair stages (st), as a majority of the cells in the myocardium are round before trabecular initiation (SI Appendix, Fig. S1 A and B). However, at 12 st, cardiomyocytes in the outer curvature of the left ventricle display orientation with the ratio of length to width of a cell being greater than 1.2. We found that 61% and 24% of cells in the inner layer at the outer curvature oriented perpendicularly and parallel, respectively, to the heart wall surface in the control hearts (n = 49 from 3 hearts) (Fig. 1 A and B). By contrast, 16% and 64% of cells in the inner layer at the outer curvature oriented perpendicularly and parallel, respectively, in the MDKO heart (Fig. 1 C and D). The patterns of cellular orientation between control and MDKO are significantly different based on the χ2 test (P < 0.01). The cells in the outer layer mainly oriented parallel to the heart wall in the control heart, but this orientation was lost in the MDKO hearts (Fig. 1 A and C). The cells of the inner layer orient perpendicularly to the heart wall in the control but randomly or parallel in the MDKO hearts at embryonic day (E)9.25 (SI Appendix, Fig. S1 C and D). After trabecular initiation, for example at E11.5, most cardiomyocytes in the trabecular zone of control hearts are elongated with a spindle shape and align parallel to the trabeculae (Fig. 1E and Movie S1), while in MDKO hearts most cardiomyocytes in trabeculae are round and do not align parallel along the trabeculae (Fig. 1F and Movie S2). The shapes of trabecular cardiomyocytes were further examined with reconstructed Z-stack images via the Imaris 9.1.2 software (Bitplane; Oxford Instruments). The surface module of the software was used to create 3D-reconstructed cells and then to determine the cell shape (Movies S3 and S4). The shape of control cells is columnar and the length, width, and height of the reconstructed trabecular cells were measured via the Imaris software and are 13.9, 6.9, and 6.9 µm, respectively (n = 13 cells), MDKO; mTmG cells have an average length, width, and height of 9.1, 7.8, and 7.2 μm, respectively (n = 13 cells), indicating a rounded morphology in MDKO (SI Appendix, Fig. S1E). The volumes of the control and knockout cardiomyocytes are significantly different (SI Appendix, Fig. S1F).
Fig. 1.
NFPs are required for cellular orientation and organization during cardiac morphogenesis. Most of cardiomyocytes in the inner layer of the compact zone from a control heart orient perpendicularly to the heart wall (A and B), while most of the cells in the MDKO display no or parallel orientation to the heart wall (C and D). The double-headed arrows indicate the orientation of a cell and the asterisk indicates no orientation of a cell in A and C. B shows the cellular orientation of 49 cells of control hearts and D shows 50 cells in MDKO hearts. Each line represents an orientation of a cell in the inner layer in A or C and the red line represents the heart surface reference line in B and D. (E and F) The cells in control trabeculae display a long spindle shape and align parallel to trabecula (E), while cells in MDKO trabeculae do not show orientation and are round, indicated by an asterisk (F). (Scale bars in A, C, E, and F: 20 μm.)
To establish whether NFPs regulate cardiomyocyte orientation autonomously, we performed an ex vivo Numb overexpression experiment to determine whether Numb rescues the cellular orientation defect in MDKO. MDKO embryos at E9.25 were cultured ex vivo, with the myocardium exposed to medium containing adenovirus for overexpression of histidine (His)-tagged Numb (Ad-Numb:His) or Ad-GFP control. Examination 24 h after infection revealed that 20% of the Ad-Numb:His infected cells localized to inner layer of the left ventricle and oriented perpendicularly to the heart wall (a total of 113 cells from 3 hearts), while only 3% Ad-GFP expressing cells localized to the inner layer of the myocardium and oriented parallel to the heart wall (a total of 152 cells from 3 hearts) (SI Appendix, Fig. S1 G and H). Furthermore, our previous study showed that Tie2Cre-mediated NFPs deletion did not cause trabeculation defects or other obvious defects (31). These results indicate that NFPs regulate cardiomyocyte orientation and trabecular initiation in a cell-autonomous manner.
NFPs Regulate OCD and Directional Migration.
Our previous study showed that MDKO hearts display thicker trabeculae with fewer numbers of trabeculae per unit length, indicating defective trabecular initiation (31). The thicker trabeculae are possibly caused by higher proliferation rates of cardiomyocytes in the compact and trabecular zones (31, 42, 57). Previous studies demonstrated that both OCD and directional migration contribute to trabecular initiation in the mouse (SI Appendix, Fig. S2A), while only apical constriction-mediated directional migration contributes to trabecular initiation in zebrafish (12, 15, 16, 18, 58). We asked whether NFPs regulate trabecular initiation through these mechanisms. We measured the spindle orientations of mitotic cardiomyocytes at the outer layer of left ventricular myocardium of ∼E9.25 control and MDKO hearts and the cells at anaphase or early telophase with both centrosomes in the same focal plane were quantified (16, 45). We found that cardiomyocytes of the MDKO displayed different division patterns with fewer perpendicular but more parallel divisions compared with the control, indicating that NFPs regulate mitotic spindle orientation (Fig. 2 A–C).
Fig. 2.
NFPs regulate oriented cell division (OCD) and directional migration. Cardiomyocytes of single-layer myocardium undergo perpendicular OCD in A and parallel division in B. The mitotic spindles are identified by acetylated α-tubulin and endocardial cells are identified by endomucin. (C) The angle between mitotic spindles and the myocardial plain was measured and compared between control and MDKO at about E9.25. The patterns of mitotic spindle orientation between control and MDKO are different based on χ2 analysis. (D) Via single-cell lineage tracing, the GFP-labeled iDKO clone, indicated by the white arrow, does not express Numb based on ISH. (E) The clonal patterns between control and iDKO are significantly different, and the iDKO clones contain smaller percentages of trabecular and transmural clones and higher percentage of surface clones. (F and G) The MDKO hearts display a smaller number of trabeculae and the trabeculae in MDKO are shorter than control trabeculae. White arrows indicate trabeculae. (Scale bars in A, B, and D: 20 μm and in F and G: 50 μm.)
Sparse cell labeling via autonomous intragenic recombination or virally labeling gave hints of coherent cell growth (59, 60) and ventricular wall morphogenesis (59, 61, 62). However, interpretations of these data are limited by the uncontrolled timing of cell labeling and by 2D analysis. To obtain a more comprehensive understanding of the mechanisms of trabecular morphogenesis, 3D imaging of the labeled clone is required to determine the transmural growth during ventricular wall morphogenesis. We applied sparse cell/multicolor lineage tracing, whole-heart clearing, and 3D imaging to determine the division patterns and cellular behaviors during trabecular morphogenesis (16, 56). Specifically, we crossed ROSA26-CreERT2 (iCre); Nbfl/+; NlD/D, in which iCre nuclear localization can be induced by tamoxifen (63), with the reporter female mouse ROSA26-Confetti (Conf); Nbfl/fl; NlD/D. Conf reporter mice can stochastically generate nuclear green, cytoplasmic yellow, cytoplasmic red, or membrane-bound blue cells upon Cre-mediated recombination (16, 64). Tamoxifen at a concentration of 20 μg/g body weight was delivered to pregnant females via gavage when embryos were at E7.75, a stage when the myocardium is a monolayer and the trabeculae are not yet initiated (SI Appendix, Fig. S2B); 72 or 120 h after induction, single labeled cells have undergone several rounds of cell division to exhibit specific geometric patterns (SI Appendix, Fig. S2C) (16). Based on the geometric distribution and anatomical annotation of each clone, the clones were categorized into 4 different patterns as previously reported (16): 1) surface clones, in which most of the cells localize to outmost layer of myocardium and likely are derived from parallel division of the labeled cell (SI Appendix, Fig. S2D and Movie S5); 2) compact clones, in which multiple cells are in the compact zone and some are in the trabecular zone (SI Appendix, Fig. S2E and Movie S6); 3) trabecular clones, in which all of the cells of the clone localize to the trabecular zone and likely are derived from directional migration (SI Appendix, Fig. S2F and Movie S7); and 4) transmural clones, in which the cells localize to both compact and trabecular zones with only 1 or 2 cells remaining in the outer compact zone (SI Appendix, Fig. S2G and Movie S8). The extent of Numb deletion in ROSA26Cre/+; Nbfl/fl; NlD/D (iDKO) was determined by in situ hybridization (ISH) with probes complementary to the sequence of the deleted exon 4 but not other exons, as the truncated mRNA of Numb in MDKO hearts was transcribed at a similar level to full-length Numb mRNA in control hearts based on qPCR and ISH (SI Appendix, Fig. S2 H–K). About 85% of the iDKO clones (n = 41) showed absence of Numb (Fig. 2D). We compared and found that the clonal patterns between control (iCre;Nbfl/+; NlD/D; Conf) and iDKO were significantly different based on a χ2 test (Fig. 2E). iDKO clones display a higher percentage of surface clones and lower percentages of transmural and trabecular clones, suggesting that NFPs play roles in OCD and directional migration. The abnormal cellular orientation (Fig. 1) and cellular behaviors (Fig. 2 A–E) in MDKO hearts might contribute to the trabecular initiation defect in the MDKO heart (Fig. 2 F and G).
NFPs Regulate Transmural Growth and Cardiomyocyte Proliferation Autonomously.
To determine if NFPs regulate ventricular wall transmural growth, clonal growth was monitored in embryos that were induced with low-dose tamoxifen at E7.75 and examined at E12.5 instead of E10.5 as described above. This concentration induced ∼30 separated clones in the hearts of both control and iDKO (Fig. 3 A and B). Because the heart is a 1-cell-thick tube when cells are labeled around E8.0 (16), the first labeled cell of each clone would be at the surface of the myocardium. Therefore, the number of its progeny and their geometric location can be used to infer the clone’s proliferation, migration, and dispersion to different transmural depths (16). The distance from the innermost cell of the clone to the myocardial surface, where the first labeled cell was localized, defines the transmural growth or invasion depth. We examined the clonal patterns and transmural growth via whole-heart clearing and 3D imaging (16). In contrast from the clonal patterns examined at E10.5 (Fig. 2E and SI Appendix, Fig. S2C), only 3 clonal patterns were observed: compact clones, likely derived from the surface clone at E10.5 (Fig. 3C and Movies S9 and S10); trabecular clones (Fig. 3D and Movie S11); and transmural clones, in which the cells localize to both compact and trabecular zones and are likely derived from the compact and transmural clones at E10.5 (Fig. 3E and Movie S12). The different clonal patterns examined at E10.5 and E12.5 are possibly explained by a higher proliferation rate of cardiomyocytes in the compact zone (31) and by transmural growth of the labeled clones. The percentage of trabecular clones in the iDKO is reduced by half compared with the control clones (Fig. 3F). The invasion depth of transmural clones and trabecular clones examined at E12.5 is triple the distance of those examined at E10.5 (Fig. 3G and SI Appendix, Fig. S3A) (16). We then compared the transmural growth between control and iDKO clones and found that the iDKO clones displayed a shorter invasion depth in both trabecular and transmural clones (Fig. 3G), suggesting that NFPs are involved in transmural growth during trabecular morphogenesis. We also quantified cell numbers in the different clonal types at E12.5. The trabecular clones had the smallest while the transmural clones had the largest numbers of cells among the 3 clonal types (Fig. 3H). The number of cells in compact and transmural clones increased 3.3 and 2.1 times, respectively, compared with the cell numbers examined at E10.5 (16). The number of cells in the trabecular clone did not increase significantly. To examine this further, we compared the proliferation of control and iDKO clones by quantifying the number of cells in the clones. We found that the iDKO clones contain more cells than the control, especially the trabecular clones (Fig. 3H), suggesting that NFPs regulate cardiomyocyte proliferation. These results are consistent with previous reports that compact cardiomyocytes proliferate at a higher rate than trabecular cardiomyocytes and NFPs regulate cardiomyocyte proliferation via Notch (31, 32, 42) and ErbB2 (57).
Fig. 3.
NFPs regulate transmural growth and cardiomyocyte proliferation autonomously. (A) Control hearts and (B) iDKO hearts show sporadic clones labeled with different colors. Cardiomyocytes in hearts were induced to be labeled at E7.75 with dosage of tamoxifen at 20 μg/g body weight and the hearts at E12.5 were imaged via Leica M205 FA, which can detect fluorescent protein-labeled cells at single-cell resolution. (C–E) Individual clones in the cleared hearts were 3D-imaged to identify the geometric pattern of each of the clones. C, D, and E show compact, trabecular and transmural clones, respectively. (F) The clonal patterns between control and iDKO. (G) The transmural growth of the clones and that the iDKO clones grow significantly shorter distances compared with the control clones. (H) The number of cells in each clone; the iDKO clones contain significantly more cells than the control clones across all 3 types of clones. (Scale bars in A and B: 200 μm and in C–E: 50 μm.)
NFPs Regulate Trabecular Morphogenesis through N-Cadherin.
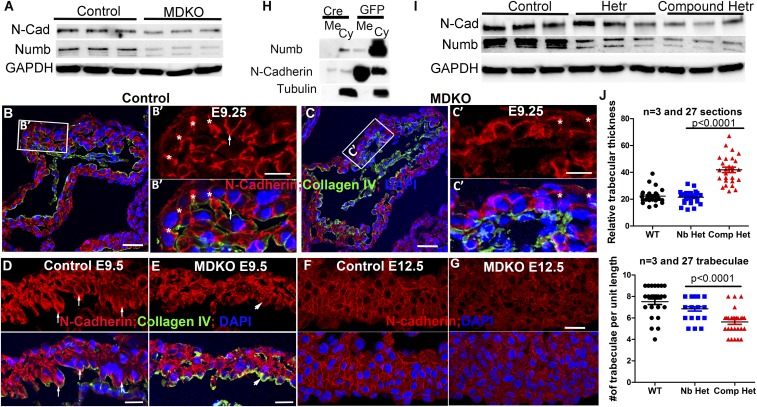
Previous work has shown that N-cadherin regulates directional migration during trabecular initiation (16, 18, 53). We further found that N-cadherin is required for cardiomyocyte orientation, as the cardiomyocytes in the inner layer in control hearts display perpendicular orientation, while the corresponding cardiomyocytes in Nkx2.5Cre/+; Cdh2fl/fl hearts display random or no orientation and are disorganized (SI Appendix, Fig. S4 A and B). We hypothesize that NFPs regulate N-cadherin to modulate cellular orientation and organization. We first examined the protein levels of N-cadherin and found that it was reduced in MDKO hearts by 42 ± 5% (n = 3) compared with control hearts (Fig. 4A). Protein expression of N-cadherin was reduced in cultured cardiomyocytes as well, and we determined that the central proline-rich (PRR) domain and NPF/DPF motifs are required to maintain N-cadherin protein levels in the cultured cardiomyocytes (SI Appendix, Fig. S4C). Previous work has shown that dynamic localization of N-cadherin correlates with cellular behaviors during trabecular initiation in zebrafish (13). Therefore, we examined the subcellular localization of N-cadherin and found that N-cadherin localizes to the lateral membrane of the cardiomyocyte in the outer layer of the myocardium (Fig. 4 B and B′) and its membrane localization is reduced in the MDKO (Fig. 4 C and C′). The ratio of membrane to cytoplasmic N-cadherin based on fluorescence intensity is 12.2 ± 1.1 (n = 30 spots) in control (Fig. 4B) and 5.2 ± 1.6 (n = 30 spots) in MDKO (Fig. 4C). In cardiomyocytes at the inner layer of the myocardium that display perpendicular orientation and abut the cardiac jelly (as indicated by collagen IV staining), N-cadherin is localized to both lateral and basal membranes and the ratio of membrane to cytoplasmic N-cadherin is 9.7 ± 1.1 (n = 30 spots) (Fig. 4D). Basal localization in the oriented cells is reduced in the MDKO (Fig. 4E) with a ratio of membrane to cytoplasmic N-cadherin of 3.4 ± 0.8 (n = 30 spots) (Fig. 4E). The reduced membrane localization of N-cadherin was observed at later stages in both trabecular and compact cardiomyocytes with a ratio of 4.5 ± 0.9 (n = 30 spots) in control and 1.7 ± 1.1 (n = 30 spots) in MDKO (Fig. 4 F and G). To further determine that less N-cadherin localizes to the membrane in MDKO hearts, cardiomyocytes from the Nbfl/fl;NlD/D heart at postnatal days 0 to 2 (p0-2) were cultured and then treated with Ade-Cre to delete Numb or treated with Ade-GFP as a control. The membrane and cytoplasmic N-cadherin were fractionated via EZ-Link NHS-Biotin. We found that the ratio of membrane to cytoplasmic N-cadherin is 8.5 ± 2.51 (n = 3) in control cardiomyocytes, while in Numb and Numblike DKO it is reduced to 2.6 ± 1.25 (n = 3) (Fig. 4H), indicating that NFPs play important roles in localization of N-cadherin to the membrane.
Fig. 4.
NFPs regulate trabecular morphogenesis through N-cadherin. (A) MDKO hearts display lower levels of Numb and N-cadherin than control hearts at E13.5. (B) N-cadherin localizes to the lateral domain of the cardiomyocytes in the myocardium at E9.25, indicted by the asterisk, and to the lateral and apical domains of some cardiomyocytes that face toward the heart lumen, indicated by the arrow. (C) N-cadherin localization to the lateral domain is weaker in the MDKO, as indicated by the asterisk. (D and E) N-cadherin localizes to the apical domain strongly in the control heart at E9.5 but weakly in the MDKO heart. The weaker membrane localization in the MDKO is further demonstrated in E12.5 hearts. (F and G) The membrane fractionation shows that the membrane N-cadherin in cultured NFP null cardiomyocytes is significantly reduced compared with the controls (H). (I) Levels of Numb and N-cadherin proteins are reduced in the compound heterozygotes. (J) Compound heterozygous hearts (Nkx2.5Cre/+;Nbfl/fl; Nlfl/+;Cdh2fl/+ or Nkx2.5Cre/+; Nbfl/+; Nlfl/fl; Cdh2fl/+) display thicker and less-dense trabecula compared with the single heterozygotes, which are not significantly different from control. (Scale bars in B, C, F, and G: 20 μm and in D and E: 10 μm.)
To determine the epistatic relationship between N-cadherin and NFPs during trabeculation, we examined trabecular morphogenesis and N-cadherin protein levels in hearts of compound heterozygotes (Nkx2.5Cre/+; Numb+/fl; Nlfl/fl; Cdh2+/fl or Nkx2.5Cre/+; Numbfl/fl; Nlfl/+; Cdh2+/fl). The N-cadherin protein level of the compound heterozygotes is reduced compared with that of single heterozygotes (Fig. 4I). The compound heterozygotes can survive to adulthood, but 3 out of 5 exhibited growth retardation (SI Appendix, Fig. S4D). The compound heterozygotes display a smaller number of trabeculae per unit length and thicker trabeculae compared with the controls (Fig. 4J and SI Appendix, Fig. S4E), indicating a trabeculation defect. These results suggest that N-cadherin protein levels correlate with the trabecular morphogenetic defect and that NFPs regulate trabecular morphogenesis through N-cadherin.
Numb Is Localized to Endocytic Organelles and Might be Required for Endosome Biogenesis and Transition.
To study the mechanism of how NFPs regulate N-cadherin protein levels and subcellular localization in cardiomyocytes, we generated the mCherry:NumbKI, in which the coding sequences of Flag and mCherry were inserted after the start codon of Numb (SI Appendix, Fig. S5A). The precise insertion of mCherry was confirmed by Southern blot (SI Appendix, Fig. S5 A and B) and genomic DNA sequencing, and the fusion protein of mCherry:NumbKI was confirmed by Western blot with antibodies against Flag or mCherry (Fig. 5A). The mCherry:NumbKI homozygotes can survive to adulthood without obvious defects, while the Numb global KO died at about E12.5 (65), suggesting that the fusion protein mCherry:Numb functions similarly to Numb. With this mCherry:NumbKI line, we examined the localization of Numb and found that there was no obvious asymmetric distribution of Numb in the cardiomyocytes oriented perpendicular to the heart wall (SI Appendix, Fig. S5C), suggesting that Numb is not a component of the polarity complex in the polarized myocardium (12, 58). Numb is enriched in epicardial cells (SI Appendix, Fig. S5D), in which NFPs play essential roles in epithelial–mesenchymal transition (45). Numb is asymmetrically localized to the leading edge of migrating mouse embryonic fibroblasts (SI Appendix, Fig. S5E), suggesting its potential roles in migration.
Fig. 5.
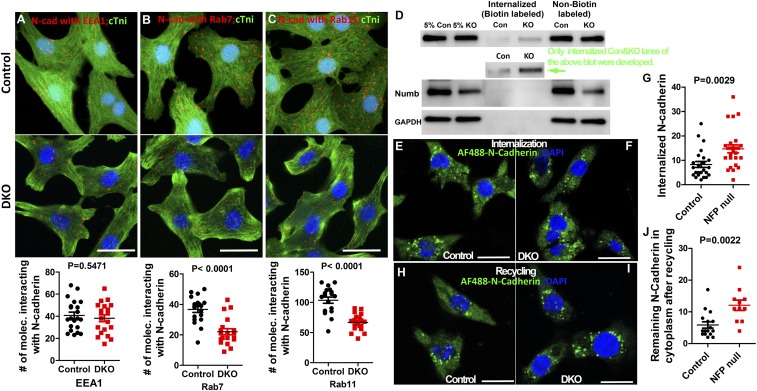
Numb is localized to diverse endocytic organelles and might be required for endosome biogenesis and transition. (A) mCherry:Numb can be detected by antibody for Flag or for mCherry using samples from brain or heart of mCherry:NumbKI. (B–E) mCherry:Numb interacts with EEA1, Rab7, and Rab11 in cardiomyocytes based on the DPL assay. cTni is a marker for cardiomyocytes. Each dot in B–D represents a colocalization of mCherry:Numb and the protein of interest. The number of colocalizations in each cell was quantified in E. (F and G) Protein levels of EEA1, clathrin, RAB11, RAB7, and RAB5 were determined and compared between control and MDKO hearts at E13.5 via Western blot. (Scale bars in B–D: 10 μm.)
To determine the subcellular localization of Numb in cardiomyocytes, cardiomyocytes from mCherry:NumbKI were ex vivo-cultured and costained with mCherry and endosomal or lysosomal markers and then 3D-imaged. We found that Numb colocalized with EEA1-labeled early endosomes, LAMP1-labeled lysosomes, RAB7-labeled late endosomes, and RAB11-labeled recycling endosomes (SI Appendix, Fig. S6 A–D). To further determine that Numb localizes to different endocytic organelles, we applied the Duolink proximity ligation (DPL) assay to detect endogenous protein spatiotemporal interactions in vivo with single-molecule sensitivity. Consistently, Numb colocalizes with markers of diverse endocytic organelles and mainly localizes to the recycling endosome compared with other endocytic organelles (Fig. 5 B–E). This localization pattern suggests that Numb might be involved in protein endocytosis, recycling, and degradation. We then examined whether deletion of NFPs would affect the biogenesis of endosomes or the transition of early endosomes to late endosomes or recycling endosomes. Different from a previous study showing that MDKO hearts displayed more EEA1 based on immunostaining (57), we used the Western blot to determine the amount of EEA1 in whole hearts and found that MDKO hearts contain significantly reduced levels of EEA1, which was further confirmed by immunostaining (Fig. 5 F and G and SI Appendix, Fig. S6 E and F), suggesting that NFPs might play roles in early endosome biogenesis. We also examined the protein levels of RAB5, a marker for early endosomes, and RAB7, a marker for late endosomes, and found that RAB7 is significantly reduced (Fig. 5 F and G). These results suggest that NFPs might modulate signaling pathways and biological processes through endocytosis.
NFPs Regulate the Transition of N-Cadherin from Early Endosome to Recycling Endosome and Its Trafficking to the Membrane.
EEA1 is a RAB5 effector protein that is required for fusion of early and late endosomes, for sorting of early endosomes, and is a docking factor required for sorting and budding of recycling vesicles (66). The reduction in membrane-localized N-cadherin and reduced EEA1 in MDKO hearts suggest that NFPs might play a role in the balance of internalization and recycling of N-cadherin. We examined whether NFPs regulate N-cadherin internalization and trafficking using cultured cardiomyocytes. Cardiomyocytes from p0-2 hearts (ROSA26CreERT2/+; Nbfl/fl; NlD/D) were isolated and cultured. Twenty-four hours later, hydroxytamoxifen or vehicle was added to induce deletion of Numb. We first examined the protein levels of Numb and N-cadherin at different time points. Numb was reduced 72 h after Cre induction, but N-cadherin levels were not reduced significantly until 96 h (SI Appendix, Fig. S7A). We applied the DPL assay to detect endogenous protein spatiotemporal interactions and found that Numb and N-cadherin colocalize in the membrane and cytoplasm (SI Appendix, Fig. S7 B–D). We then determined whether NFPs are required for N-cadherin endocytosis and recycling in vivo. We found that the quantity of N-cadherin localized to early endosomes in NFPs-null cells is not significantly different from the control cardiomyocytes based on the DPL assay, suggesting that the reduction in early endosomes in the MDKO is not a limiting factor for N-cadherin endocytosis (Fig. 6A). However, the quantity of N-cadherin localized to late and recycling endosomes is significantly reduced compared with the control (Fig. 6 B and C), suggesting a defect in transition from early endosomes to late and recycling endosomes in DKO cardiomyocytes. To further determine whether NFPs play roles in N-cadherin internalization and trafficking to the membrane, control and NFP-null cardiomyocytes (72 h after Cre induction) were used in the biotin–IP-based internalization assay. In this assay, membrane N-cadherin was labeled by sulfo-NHS-SS-biotin for 30 min at 4°, and the labeled N-cadherin was allowed to internalize for 20 min at 37°. The remaining biotin on the cell surface was dissociated from proteins as per the protocol, and the internalized biotin-labeled N-cadherin proteins were pulled down via the NeutrAvidin agarose resins and were detected by N-cadherin antibody via Western blot. We found that the levels of N-cadherin after biotin removal (non-biotin-labeled) in control and NFPs-null cells were not significantly different, while the levels of internalized biotin-labeled N-cadherin in the NFP null cells were higher than those of the control heart (Fig. 6D). We also examined N-cadherin internalization using the Alexa Fluor 488-conjugated N-cadherin antibody (Fig. 6 E–G) and consistently there was more internalized N-cadherin in NFP null cardiomyocytes based on immunostaining (Fig. 6 E–G), suggesting that there is not an endocytosis defect but likely a recycling defect for N-cadherin. To determine if NFPs play a role in N-cadherin recycling, internalized Alexa Fluor 488-conjugated N-cadherin was allowed to traffic back to the membrane, and the remaining N-cadherin in the cytoplasm was quantified. We found that more N-cadherin remained in the cytoplasm in NFP null cells (Fig. 6 H–J), suggesting that NFPs play roles in N-cadherin trafficking to the membrane. The decreased N-cadherin recycling to the cell membrane leads to accumulation of N-cadherin in the cytoplasm, which might trigger N-cadherin degradation, leading to the reduced N-cadherin protein level in MDKO hearts.
Fig. 6.
NFPs regulate early endosome transition to recycling endosomes, and N-cadherin trafficking to membrane. (A–C) N-cadherin interacts with EEA1, Rab7, and Rab11 in control and DKO cardiomyocytes based on the DPL assay. Each dot in A–C represents a colocalization between N-cadherin and the protein of interest. (D–J) Seventy-two hours after Cre induction, control and NFP-null cardiomyocytes were used for antibody-based or biotin–IP-based internalization and recycling assays. The Alexa Fluor 488-conjugated N-cadherin or biotin-labeled N-cadherin inside the cytoplasm was detected by confocal imaging or Western blot, respectively, and the amount of labeled N-cadherin in control and knockout was quantified. There is more internalized N-cadherin in the DKO cardiomyocyte based on immunostaining (E–G) and Western blot (D). (H–J) Internalized N-cadherin was allowed to traffic back to the membrane, and the remaining N-cadherin in the cytoplasm was detected. We found that there was more labeled N-cadherin in NFP null cells. (Scale bars in A–C, E, F, H, and I: 10 μm.)
N-Cadherin Overexpression Rescues the Defects of Cellular Orientation and Trabeculation in MDKO Hearts.
To further study the NFPs’ regulation of cellular orientation and trabeculation through N-cadherin, we generated N-cadherin transgenic lines to determine whether N-cadherin overexpression can rescue these defects in the MDKO hearts. The transgenic lines, in which chicken N-cadherin cDNA (cN-cadherin) is driven by the αMHC promoter (Fig. 7A and SI Appendix, Fig. S8 A and B) as previously described (67), show expression of cN-cadherin in the heart based on Western blot (Fig. 7B), which is further confirmed by the cN-cadherin–specific antibody staining (Fig. 7 C and D). We then determined whether cN-cadherin overexpression could rescue the defects in MDKO hearts. We took advantage of the mosaic expression of cN-cadherin in the early heart at E10.5, as cN-cadherin driven by the αMHC promoter was expressed in a few cells at that early stage. We found that cN-cadherin–expressing cardiomyocytes localize to both trabecular and compact zones in control hearts at E10.5 with 25.2% (n = 708 from 3 hearts) of cN-cadherin–expressing cells localized in the trabeculae (Fig. 7C). In contrast, in the MDKO, cN-cadherin–expressing cardiomyocytes predominantly localize to the trabecular zone with 78.3% (n = 578 from 3 hearts) of them being localized in the trabeculae (Fig. 7D). We then examined whether cN-cadherin overexpression will rescue the trabeculation defect in the MDKO at E12.5, a stage in which many cells express cN-cadherin (Fig. 7 L and N). We found that trabecular thickness was reduced and that trabecular density was increased compared with the MDKO, but the trabeculation defect was not fully rescued compared with the control, possibly due to the delayed expression of cN-cadherin expression driven by αMHC promoter (Fig. 7 E–J).
Fig. 7.
N-cadherin overexpression rescues the defects of cellular orientation and trabeculation in MDKO hearts. (A and B) Chick N-cadherin (cN-cadherin) driven by the cardiomyocyte-specific promoter αMHC transgenic lines were generated. All 3-founder lines express cN-cadherin based on Western blot with cN-cadherin–specific antibody. (C and D) cN-cadherin is expressed in cardiomyocytes in both compact and trabecular zones in the control hearts, but cells expressing cN-cadherin predominantly localize to the trabecular zone MDKO hearts. (E–J) MDKO hearts display thicker trabeculae than controls, and cN-cadherin expression reduces the thickness of trabeculae in MDKO. (K–O) Cells in control display a spindle shape, while cells in MDKO display a round shape. cN-cadherin expression in the MDKO rescues the cell shape. (Scale bars in C and D: 20 μm and in E–L and K–N: 40 μm.)
We examined whether cN-cadherin overexpression can rescue this cellular orientation defect in MDKO hearts. The trabecular cardiomyocytes are more mature (68) and display a spindle shape aligning parallel to the trabecula in the control (Fig. 7 K and O), while in the MDKO the cells are round (Figs. 1 G and H and 7 M and O) and display a cellular orientation defect as described in Fig. 1 and SI Appendix, Fig. S1. The control cells expressing cN-cadherin in the trabecular zone display a spindle shape similar to the wild-type cardiomyocytes (Fig. 7 K and L). In MDKO hearts, some of the cN-cadherin–expressing cells were restored to the spindle shape and aligned parallel to trabecula, compared with the cardiomyocytes in MDKO (Fig. 7 M–O). These results indicate that cN-cadherin expression at least partially rescues the cell shape and cellular orientation defect in MDKO hearts.
Discussion
In this study, we found that the patterns of OCD and cellular orientation and transmural growth are abnormal during trabecular morphogenesis in MDKO hearts. With the newly generated mCherry:NumbKI, we found that Numb is expressed in diverse endocytic organelles and mainly in recycling endosomes. In the MDKO hearts, the level of N-cadherin is reduced and its subcellular localization is altered. The cellular orientation and trabeculation defects in the MDKO hearts were at least partially rescued by transgenic cN-cadherin overexpression. Further studies showed that NFPs regulate N-cadherin membrane localization by controlling its recycling to the plasma membrane, suggesting a mechanism that NFPs modulate trabecular and ventricular wall morphogenesis through N-cadherin.
Cardiomyocyte Transmural Migration Contributes to Trabecular Growth.
Recent studies have shown that the cardiomyocytes in the monolayer myocardium undergo perpendicular OCD, which can send a daughter cell into the cardiac jelly to initiate trabeculation (12, 16, 18). Another mechanism of trabecular initiation is directional migration, in which some cardiomyocytes undergo cytoskeleton rearrangement, become elongated, orient perpendicularly to the heart wall, and eventually migrate into the cardiac jelly (16, 18). In zebrafish, the trabeculae are initiated by apical constriction-mediated directional migration but not OCD (15, 58). After trabecular initiation, the myocardium will consist of multiple layers of cardiomyocytes, wherein the endocardial cells sprout and attach to the cardiomyocytes of the compact zone and separate the cardiomyocytes to form trabeculae (11). After trabecular formation, the next step for trabecular morphogenesis will be trabecular growth, and our sparse lineage tracing reveals that the transmural growth distance of cells examined at E12.5 is tripled for both transmural clones and trabecular clones compared with the clones that are examined at E10.5. The invasion depth in the control clones is significantly larger than those of the iDKO clones (Fig. 3). The shorter invasion depth of iDKO clones is not due to less proliferation, as the iDKO clones have larger cell numbers than the control clones. These results suggest that NFPs might be involved in the transmural migration of cardiomyocytes and the transmural growth of trabeculae toward the heart lumen during trabecular and ventricular wall morphogenesis.
N-Cadherin Protein Level and Subcellular Localization Are Critical for Trabecular and Ventricular Wall Morphogenesis.
Previous work has shown that N-cadherin is required to establish cell–cell associations in embryonic cardiomyocytes (49) and for cardiomyocyte migration during trabecular morphogenesis (16, 53). During trabecular initiation in zebrafish, N-cadherin is expressed along the lateral sides of embryonic cardiomyocytes, and N-cadherin accumulates to the basal domain of cells that are extruded from the compact zone, allowing tight adhesion between the compact and trabecular zones (13). In our study, N-cadherin in the mouse displays a similar subcellular localization to the lateral domain of adjacent cardiomyocytes in the compact zone, but in cardiomyocytes that display perpendicular orientation N-cadherin localizes to the basal domain in addition to the lateral domain. After trabecular initiation, N-cadherin in cardiomyocytes at late stages such as E11.5 localizes to the cell membrane evenly. In the chicken heart, cardiomyocyte transmural migration is coincident with up-regulation of N-cadherin (53). A lower level of N-cadherin is required for the dissociation of cells from epithelial tissue, while a higher level will promote cell migration and invasion after dissociation from the epithelial tissue (69). Another study showed that N-cadherin expression modulates cell polarity and impacts speed and directionality of migrating cells in vitro (70) and of tumor cell (71) and neural crest cells (72). We found that N-cadherin–null cardiomyocytes in a mosaic heart display fewer cellular protrusions and are less migratory (16), suggesting its role in cellular orientation and cell migration. In this study, the reduction of N-cadherin protein level and membrane localization resulted in the round shape of cardiomyocytes, abnormal cellular orientation, and trabeculation defects in the MDKO hearts. N-cadherin overexpression might promote cardiomyocyte translocation to trabeculae in the MDKO, consistent with previous studies that N-cadherin induces changes in fibroblastic phenotype, rendering the cells more motile and invasive (73, 74). cN-cadherin overexpression at least partially rescues the defects of cellular orientation and trabeculation in the MDKO. So, it is likely that NFPs working through N-cadherin in the early embryonic stages might be required to establish the cytoskeleton and subsequently to establish cell shape, cell polarity, orientation, and migration during cardiac morphogenesis.
NFPs Modulate Different Biological Processes by Regulating Different Downstream Targets via Endocytosis.
NFPs are essential for cardiac morphogenesis, and the MDKO hearts display a variety of morphogenetic defects. However, the key downstream target or signaling pathway altered and responsible for each defect in the MDKO has not been determined. The localization of Numb to endocytic organelles and lysosomes suggests that Numb might be involved in protein endocytosis, recycling, and degradation and that Numb might function as a homeostatic sensor, regulating signal attenuation, termination and maintenance in response to different biological process cues. Previous work showed that NFPs are involved in clustering of early endosomes in cultured cells (75). We found that cardiomyocytes of MDKO hearts have fewer early endosomes identified by whole-heart Western blot and EEA1 labeling, and that N-cadherin fails to translocate from early endosomes to recycling endosomes, suggesting that NFPs play roles in endosome biogenesis and in the transition from early endosomes to recycling endosomes. These results suggest a possibility that NFPs act at multiple steps in endocytosis.
In summary, the trabecular and ventricular wall morphogenetic defects in MDKO hearts (SI Appendix, Fig. S9A) are caused by abnormal cellular orientation, OCD, and transmural growth. This is at least partially a consequence of reduced N-cadherin protein and membrane localization in the MDKO cardiomyocytes (SI Appendix, Fig. S9 A and B). The cellular orientation might determine the directionality of cellular migration, mitotic spindle orientation, and cellular organization during trabecular morphogenesis. We propose a model that NFPs regulate N-cadherin trafficking to the plasma membrane and that the deletion of NFPs attenuates the transition from early endosomes to recycling endosomes, disturbing the balance of N-cadherin distribution between the cytoplasm and the plasma membrane (SI Appendix, Fig. S9C). However, the mechanism by which NFPs regulate EEA1 protein levels, the mechanism by which NFPs regulate recycling, and whether cellular orientation contributes to trabecular compaction will require further investigation.
Materials and Methods
Mouse and Cell Lines.
Mouse strains Numbfl/fl&Numblikefl/fl (65, 76), Cdh2fl/fl (49), Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP) (mTmG) (53), Rosa26CreERT2 (63), and R26R-Confetti (64) were purchased from The Jackson Laboratory. Robert Schwartz, University of Houston, Houston, TX, provided Nkx2.5Cre/+ mice (41). The aMHC-cN-cadherin transgene was injected into fertilized oocytes as previously described (67). Embryos harvested at around noon on embryonic day 9 were counted as E9.5, and those harvested at around 6 PM were counted as E9.75. All animal experiments are approved by the Institutional Animal Care and Use Committee at Albany Medical College and performed according to the Guide for the Care and Use of Laboratory Animals (77).
Various experimental protocols were applied in this study, and most of the experiments were briefly introduced in the text to make the content easier to be understood. The details of each protocol are available in SI Appendix.
Statistics.
Data are shown as mean ± SD. An unpaired, 2-tailed Student’s t test and χ2 test as specified were used for statistical comparison. A χ2 test was used to compare the cellular orientation patterns, division patterns, or clonal patterns between control and knockout. A P value of 0.05 or less was considered statistically significant.
Supplementary Material
Acknowledgments
We thank the M.W. laboratory members for scientific discussion. This work is supported by American Heart Association (AHA) Grant 13SDG16920099; National Heart, Lung, and Blood Institute Grants R01HL121700 to M.W. and R01HL049426 to H.A.S.; and AHA Grant 19POST34410093 to L.M.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904684116/-/DCSupplemental.
References
- 1.Olson E. N., A decade of discoveries in cardiac biology. Nat. Med. 10, 467–474 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Manasek F. J., Embryonic development of the heart. I. A light and electron microscopic study of myocardial development in the early chick embryo. J. Morphol. 125, 329–365 (1968). [DOI] [PubMed] [Google Scholar]
- 3.Van Mierop L. H., Embryology of the univentricular heart. Herz 4, 78–85 (1979). [PubMed] [Google Scholar]
- 4.Sedmera D., Pexieder T., Vuillemin M., Thompson R. P., Anderson R. H., Developmental patterning of the myocardium. Anat. Rec. 258, 319–337 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Icardo J. M., Fernandez-Terán A., Morphologic study of ventricular trabeculation in the embryonic chick heart. Acta Anat. (Basel) 130, 264–274 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Sedmera D., Thomas P. S., Trabeculation in the embryonic heart. Bioessays 18, 607 (1996). [DOI] [PubMed] [Google Scholar]
- 7.D’Amato G., et al. , Sequential Notch activation regulates ventricular chamber development. Nat. Cell Biol. 18, 7–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenni R., Rojas J., Oechslin E., Isolated noncompaction of the myocardium. N. Engl. J. Med. 340, 966–967 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Breckenridge R. A., Anderson R. H., Elliott P. M., Isolated left ventricular non-compaction: The case for abnormal myocardial development. Cardiol. Young 17, 124–129 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Weiford B. C., Subbarao V. D., Mulhern K. M., Noncompaction of the ventricular myocardium. Circulation 109, 2965–2971 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Del Monte-Nieto G., et al. , Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature 557, 439–445 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Passer D., van de Vrugt A., Atmanli A., Domian I. J., Atypical protein kinase C-dependent polarized cell division is required for myocardial trabeculation. Cell Rep. 14, 1662–1672 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherian A. V., Fukuda R., Augustine S. M., Maischein H. M., Stainier D. Y., N-cadherin relocalization during cardiac trabeculation. Proc. Natl. Acad. Sci. U.S.A. 113, 7569–7574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staudt D. W., et al. , High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development 141, 585–593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., et al. , A dual role for ErbB2 signaling in cardiac trabeculation. Development 137, 3867–3875 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., et al. , Single-cell lineage tracing reveals that oriented cell division contributes to trabecular morphogenesis and regional specification. Cell Rep. 15, 158–170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian X., et al. , Identification of a hybrid myocardial zone in the mammalian heart after birth. Nat. Commun. 8, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M., Mechanisms of trabecular formation and specification during cardiogenesis. Pediatr. Cardiol. 39, 1082–1089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H., et al. , BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219–2231 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura T., Shepherd S., Ackerman L., Jan L. Y., Jan Y. N., numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Rhyu M. S., Jan L. Y., Jan Y. N., Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Knoblich J. A., Mechanisms of asymmetric stem cell division. Cell 132, 583–597 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Spana E. P., Doe C. Q., Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Wu M., et al. , Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couturier L., Vodovar N., Schweisguth F., Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 14, 131–139 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Betschinger J., Mechtler K., Knoblich J. A., The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326–330 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Petersen P. H., Zou K., Krauss S., Zhong W., Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat. Neurosci. 7, 803–811 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Li H. S., et al. , Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 40, 1105–1118 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Petersen P. H., Zou K., Hwang J. K., Jan Y. N., Zhong W., Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Frise E., Knoblich J. A., Younger-Shepherd S., Jan L. Y., Jan Y. N., The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. U.S.A. 93, 11925–11932 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C., et al. , Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development 141, 281–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M., Li J., Numb family proteins: Novel players in cardiac morphogenesis and cardiac progenitor cell differentiation. Biomol. Concepts 6, 137–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon C., et al. , Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat. Cell Biol. 13, 1244–1251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shenje L. T., et al. , Precardiac deletion of Numb and Numblike reveals renewal of cardiac progenitors. eLife 3, e02164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong W., Feder J. N., Jiang M. M., Jan L. Y., Jan Y. N., Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17, 43–53 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Verdi J. M., et al. , Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 1134–1145 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Han Z., Bodmer R., Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development 130, 3039–3051 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Niikura Y., et al. , Zebrafish numb homologue: Phylogenetic evolution and involvement in regulation of left-right asymmetry. Mech. Dev. 123, 407–414 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Cottage C. T., et al. , Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ. Res. 106, 891–901 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu M., Meng F., Has the cardiac stem cell controversy settled down? Sci. China Life Sci. 57, 949–950 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Moses K. A., DeMayo F., Braun R. M., Reecy J. L., Schwartz R. J., Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31, 176–180 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Yang J., et al. , Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart. Cardiovasc. Res. 96, 276–285 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Castanon I., González-Gaitán M., Oriented cell division in vertebrate embryogenesis. Curr. Opin. Cell Biol. 23, 697–704 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Wei Y., Mikawa T., Formation of the avian primitive streak from spatially restricted blastoderm: Evidence for polarized cell division in the elongating streak. Development 127, 87–96 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Wu M., et al. , Epicardial spindle orientation controls cell entry into the myocardium. Dev. Cell 19, 114–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baena-López L. A., Baonza A., García-Bellido A., The orientation of cell divisions determines the shape of Drosophila organs. Curr. Biol. 15, 1640–1644 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Karner C. M., et al. , Wnt9b signaling regulates planar cell polarity and kidney tubule morphogenesis. Nat. Genet. 41, 793–799 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wheelock M. J., Johnson K. R., Cadherin-mediated cellular signaling. Curr. Opin. Cell Biol. 15, 509–514 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Kostetskii I., et al. , Induced deletion of the N-cadherin gene in the heart leads to dissolution of the intercalated disc structure. Circ. Res. 96, 346–354 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Li J., et al. , Cardiac-specific loss of N-cadherin leads to alteration in connexins with conduction slowing and arrhythmogenesis. Circ. Res. 97, 474–481 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Radice G. L., et al. , Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181, 64–78 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Ong L. L., Kim N., Mima T., Cohen-Gould L., Mikawa T., Trabecular myocytes of the embryonic heart require N-cadherin for migratory unit identity. Dev. Biol. 193, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Jiao K., et al. , An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 17, 2362–2367 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaikh Qureshi W. M., et al. , Imaging cleared embryonic and postnatal hearts at single-cell resolution. J. Vis. Exp., e54303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J., et al. , CDC42 is required for epicardial and pro-epicardial development by mediating FGF receptor trafficking to the plasma membrane. Development 144, 1635–1647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirai M., et al. , Adaptor proteins NUMB and NUMBL promote cell cycle withdrawal by targeting ERBB2 for degradation. J. Clin. Invest. 127, 569–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiménez-Amilburu V., et al. , In Vivo visualization of cardiomyocyte apicobasal polarity reveals epithelial to mesenchymal-like transition during cardiac trabeculation. Cell Rep. 17, 2687–2699 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Meilhac S. M., et al. , A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development 130, 3877–3889 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Meilhac S. M., Esner M., Kerszberg M., Moss J. E., Buckingham M. E., Oriented clonal cell growth in the developing mouse myocardium underlies cardiac morphogenesis. J. Cell Biol. 164, 97–109 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikawa T., Cohen-Gould L., Fischman D. A., Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus. III: Polyclonal origin of adjacent ventricular myocytes. Dev. Dyn. 195, 133–141 (1992). [DOI] [PubMed] [Google Scholar]
- 62.Mikawa T., Borisov A., Brown A. M., Fischman D. A., Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus: I. Formation of the ventricular myocardium. Dev. Dyn. 193, 11–23 (1992). [DOI] [PubMed] [Google Scholar]
- 63.Badea T. C., Wang Y., Nathans J., A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23, 2314–2322 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livet J., et al. , Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Zilian O., et al. , Multiple roles of mouse Numb in tuning developmental cell fates. Curr. Biol. 11, 494–501 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Mishra A., Eathiraj S., Corvera S., Lambright D. G., Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1). Proc. Natl. Acad. Sci. U.S.A. 107, 10866–10871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo Y., et al. , Rescuing the N-cadherin knockout by cardiac-specific expression of N- or E-cadherin. Development 128, 459–469 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Miao L., et al. , Notch signaling regulates Hey2 expression in a spatiotemporal dependent manner during cardiac morphogenesis and trabecular specification. Sci. Rep. 8, 2678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Péglion F., Etienne-Manneville S., N-cadherin expression level as a critical indicator of invasion in non-epithelial tumors. Cell Adhes. Migr. 6, 327–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Camand E., Peglion F., Osmani N., Sanson M., Etienne-Manneville S., N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J. Cell Sci. 125, 844–857 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Su Y., et al. , N-cadherin haploinsufficiency increases survival in a mouse model of pancreatic cancer. Oncogene 31, 4484–4489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu X., et al. , N-cadherin and Cx43alpha1 gap junctions modulates mouse neural crest cell motility via distinct pathways. Cell Commun. Adhes. 8, 321–324 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Derycke L. D., Bracke M. E., N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int. J. Dev. Biol. 48, 463–476 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Nieman M. T., Prudoff R. S., Johnson K. R., Wheelock M. J., N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 147, 631–644 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao X., et al. , Numb regulates vesicular docking for homotypic fusion of early endosomes via membrane recruitment of Mon1b. Cell Res. 26, 593–612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson A., et al. , Normal hemopoiesis and lymphopoiesis in the combined absence of numb and numblike. J. Immunol. 178, 6746–6751 (2007). [DOI] [PubMed] [Google Scholar]
- 77.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.