Significance
Phosphatidic acid (PA) is a bioactive lipid that functions as an intracellular second messenger, enabling cells to integrate and react to diverse extracellular cues. Because of PA’s strong and pleiotropic effects, its generation is tightly controlled spatiotemporally. It remains unknown how cells regulate local PA production at individual organelle membranes to control different downstream signaling events. We describe a chemical, activity-based imaging approach for precisely visualizing, in real time, subcellular localizations of PA signaling initiated by phospholipase D (PLD) enzymes. This toolset revealed distinct locations of PLD signaling elicited by G protein-coupled receptor and receptor tyrosine kinase pathways and holds promise to shed light on physiological and pathological effects of localized PLD signaling and, more generally, intracellular lipid transport pathways.
Keywords: click chemistry, lipid trafficking, phosphatidic acid, phospholipase D, second messengers
Abstract
The fidelity of signal transduction requires spatiotemporal control of the production of signaling agents. Phosphatidic acid (PA) is a pleiotropic lipid second messenger whose modes of action differ based on upstream stimulus, biosynthetic source, and site of production. How cells regulate the local production of PA to effect diverse signaling outcomes remains elusive. Unlike other second messengers, sites of PA biosynthesis cannot be accurately visualized with subcellular precision. Here, we describe a rapid, chemoenzymatic approach for imaging physiological PA production by phospholipase D (PLD) enzymes. Our method capitalizes on the remarkable discovery that bulky, hydrophilic trans-cyclooctene–containing primary alcohols can supplant water as the nucleophile in the PLD active site in a transphosphatidylation reaction of PLD’s lipid substrate, phosphatidylcholine. The resultant trans-cyclooctene–containing lipids are tagged with a fluorogenic tetrazine reagent via a no-rinse, inverse electron-demand Diels–Alder (IEDDA) reaction, enabling their immediate visualization by confocal microscopy in real time. Strikingly, the fluorescent reporter lipids initially produced at the plasma membrane (PM) induced by phorbol ester stimulation of PLD were rapidly internalized via apparent nonvesicular pathways rather than endocytosis, suggesting applications of this activity-based imaging toolset for probing mechanisms of intracellular phospholipid transport. By instead focusing on the initial 10 s of the IEDDA reaction, we precisely pinpointed the subcellular locations of endogenous PLD activity as elicited by physiological agonists of G protein-coupled receptor and receptor tyrosine kinase signaling. These tools hold promise to shed light on both lipid trafficking pathways and physiological and pathological effects of localized PLD signaling.
Localized production of signaling molecules is a key feature of signal transduction pathways. This phenomenon enables a small, privileged set of ions, metabolites, and lipids to function as second messengers that dictate diverse downstream signaling events through not only their molecular identities but also their sites of production. Sensing the precise locations where second messengers are produced or enriched is a challenging task. Typical approaches include small-molecule, protein, or nucleic acid-based fluorescent sensors that use selective binding or covalent reactivity to localize or produce a fluorescent signal where the second messenger is produced (1–4). Whereas these approaches have been revolutionary for sensing signals such as divalent cations, reactive oxygen species, and certain lipid second messengers, they have limitations in many contexts, particularly for targets without selective binding domains or that are produced by multiple different biosynthetic routes.
Phosphatidic acid (PA) is a multifunctional lipid that acts both as a key intermediate in de novo phospholipid biosynthesis and as a lipid second messenger (5). In this latter function, PA is generated by one of several isoforms of phospholipase D (PLD) or diacylglycerol kinase (DGK) enzymes (6, 7). The PLD-mediated pathway is highly clinically relevant, as PLD levels are elevated in several diseases, including many cancers, viral infection, and neurological diseases (8–11), and, consequently, PLD inhibition is emerging as a promising therapeutic strategy to attenuate pathological PLD signaling (12–15). Two PLD isoforms, PLD1 and PLD2, mediate the bulk of their biological effects through the production of PA via hydrolysis of the abundant membrane phospholipid phosphatidylcholine (PC) in several organelle membranes, including the plasma membrane (PM), the Golgi apparatus, endosomes, and lysosomes (6, 11, 16).
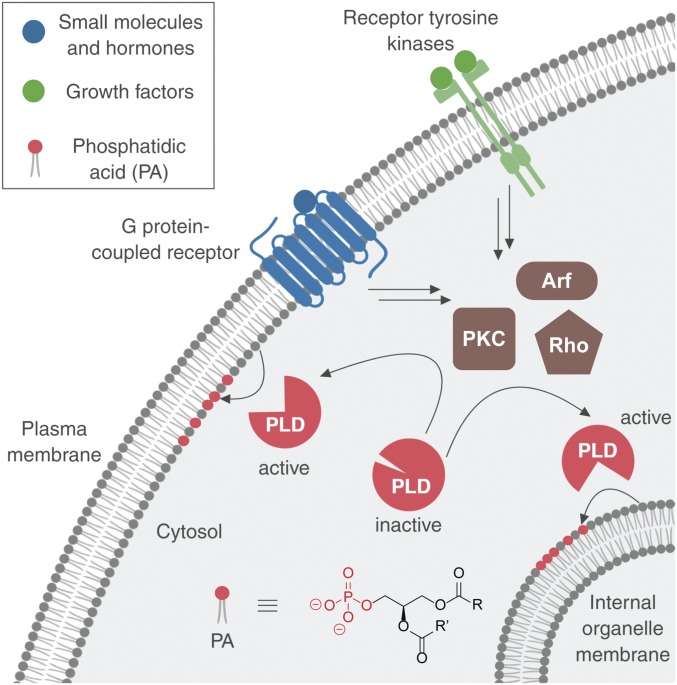
Even though the localizations of PLD1 and PLD2 differ and can change in accordance with their activation, it remains unknown which cellular compartments possess active PLD enzymes during different physiological stimulations (17–21). In particular, PLD enzymes can receive input from 2 different classes of cell-surface receptors, G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs). Both GPCRs and RTKs can stimulate PLDs via activation of several different intermediary, intracellular effectors, including those from the protein kinase C (PKC), Rho GTPase, and Arf GTPase families. Ultimately, downstream of PLD activation, these pathways elicit different physiological effects on cells (Fig. 1) (6, 22–27).
Fig. 1.
Phospholipase D (PLD) enzymes are stimulated by different classes of cell-surface receptors through intracellular effectors, including those from the protein kinase C (PKC), Rho GTPase, and Arf GTPase families, to catalyze phosphatidic acid (PA) formation at different intracellular membrane locations. R and R′ denote the fatty acyl tails.
Despite much effort aimed at understanding PLD signaling, it remains unknown whether activation of PLDs by these different inputs occurs at different subcellular locations. More generally, how cells regulate the local production of PA to effect specific signaling outcomes remains an unsolved mystery, despite the many established effector proteins whose downstream signaling activities depend on PA production (16).
Existing tools for visualizing PA are inadequate for addressing these questions. First, directly tracking fluorescent protein fusions to PLDs and DGKs does not reveal where these enzymes are actively producing PA. A specialized fluorescence lifetime imaging approach, in which cells expressing fluorescently tagged PLDs are dosed with a fluorescent PC analog, reveals changes in enzyme–substrate interactions, which is an indirect measure that can correlate with enzymatic activity (28). Conversely, PA-binding protein domains exhibit only moderate selectivity for PA and, critically, do not distinguish between different biosynthetic sources of PA (29–31). The challenge of distinguishing between pools of PA derived from PLDs and DGKs is highly relevant, given that these enzymes can receive overlapping inputs from upstream signaling molecules including integral membrane receptors, cytosolic enzymes, and small-molecule activators and yet impact distinct downstream targets (6, 7, 11). The deficiencies in these toolsets have prompted us to address the challenge of visualizing a precise subset of PA produced for acute signaling by PLDs.
Here, we report a chemoenzymatic, activity-based imaging method to visualize the precise subcellular locations of PLD activity with high spatiotemporal resolution. Our approach relies on the ability of endogenous PLD enzymes to accept unnatural substrate analogs that are rapidly converted, in sequential enzyme-mediated and bioorthogonal tagging steps, into fluorescent lipids whose subcellular location faithfully reports on the location(s) of active PLD enzymes. The major advances reported here address the key deficiency of earlier versions of this method, namely its poor temporal resolution that, because of trafficking of the fluorescent lipid reporters before imaging, prevented the visualization of the true localizations of PLD activation (32, 33).
These tools enabled us to reveal the precise organelle localizations of PLD activity as elicited by diverse physiological stimuli, including selective activators of GPCR and RTK signaling. We identified striking differences in the locations of PLD activity between different stimulus treatments. Additionally, we observed rapid transport of the fluorescent lipid reporters produced at the PM to the endoplasmic reticulum (ER) and, subsequently, Golgi apparatus, highlighting the need for these time-resolved tools to accurately visualize these dynamic molecules and suggesting additional applications to visualize rapid intracellular phospholipid trafficking occurring on the second-to-minute time scale (34, 35).
Results
Conceptualization of a Real-Time Version of IMPACT.
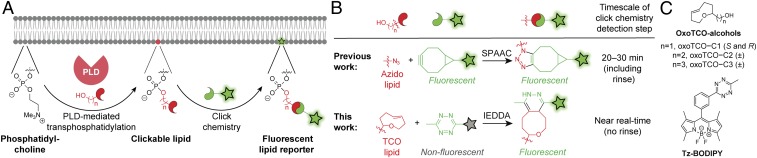
Our approach to visualize sites of PLD-mediated PA production relies on the well-known ability of PLDs to catalyze transphosphatidylation reactions with exogenous small, linear primary alcohols, forming phosphatidyl alcohols as lipid reporters of PLD activity (36, 37). We have previously demonstrated that, when cells are supplied with alkynyl (32, 38) or azido (33) alcohols, their endogenous PLD enzymes convert these bioorthogonally labeled alcohols into the corresponding alkynyl or azido phosphatidyl alcohols, which can be subsequently fluorescently tagged via Cu-catalyzed or strain-promoted azide–alkyne cycloadditions (CuAAC [39, 40] and SPAAC [41, 42]) using appropriately derivatized fluorophores (Fig. 2A). This method, termed IMPACT for Imaging Phospholipase D Activity with Clickable Alcohols via Transphosphatidylation, enabled fluorescent labeling of cellular membranes within live cells, using SPAAC tagging, as a function of their PLD activities (33).
Fig. 2.
Overview of the imaging phospholipase D (PLD) activity with clickable alcohols via transphosphatidylation (IMPACT) method for visualizing PLD activity in live cells. (A) Cartoon schematic of IMPACT. (B) Comparison of established IMPACT method using azido alcohols and strain-promoted azide–alkyne cycloaddition (SPAAC) detection with the advance reported here, real-time (RT) IMPACT, using trans-cyclooctene (TCO)-containing alcohols and inverse electron-demand Diels–Alder (IEDDA) detection using tetrazine (Tz) reagents. (C) Structures of trans-5-oxocene (oxoTCO) alcohols and the fluorogenic Tz–BODIPY conjugate used in this study.
SPAAC-based IMPACT requires 20 to 30 min for the cycloaddition and rinse-out (Fig. 2 B, Top). This low temporal resolution makes it nonideal for ascertaining the precise, subcellular localizations of active PLD enzymes because lipid diffusion and trafficking can occur on more rapid time scales (43–45). In particular, we noted the lack of PM-derived IMPACT fluorescence upon stimulation with phorbol 12-myristate 13-acetate (PMA), which is thought to directly stimulate PKC (23, 33) and result in translocation of PLD1 to the PM (19).
We sought to substantially improve the time resolution of IMPACT by replacing SPAAC with the inverse electron-demand Diels–Alder (IEDDA) reaction of trans-cyclooctene (TCO) and tetrazine (Tz) reagents (46, 47). Two key advantages of this bioorthogonal reaction are its rapid reaction kinetics and the ability to perform no-rinse, real-time imaging of reaction progress using fluorogenic, or “turn-on,” Tz reagents (Fig. 2 B, Bottom) (48). The challenges inherent in using this IEDDA reaction for IMPACT are that (i) TCO groups are substantially larger than azides or terminal alkynes, raising the question of whether endogenous PLD enzymes would accept a bulky, TCO-derivatized primary alcohol as a surrogate for water; and (ii) large, hydrophobic TCO alcohols need to be both efficiently delivered to the cytosolic space, where PLDs reside, and also rinsed out from the cell after transphosphatidylation and before the IEDDA reaction.
Synthesis and Evaluation of TCO-Containing Alcohols as PLD Transphosphatidylation Substrates.
To address these challenges, we first designed and synthesized a series of carbocyclic TCO-containing primary alcohols with different linker lengths and polarities (SI Appendix, Fig. S1 and Schemes S1 and S2). Remarkably, all of these TCO alcohols were transphosphatidylation substrates of both a commercially available PLD from Streptomyces sp. PMF (SI Appendix, Fig. S2) and endogenous mammalian PLDs (SI Appendix, Fig. S3 and Scheme S3) as evaluated by transphosphatidylation followed by in vitro IEDDA tagging of lipid extracts with a Tz-fluorophore conjugate and fluorescence-coupled HPLC analysis. Among these alcohols, those with aliphatic and nonpolar linkers (e.g., ether) were better PLD substrates than those with more polar, bulky linkages (e.g., amide and carbamate). Unfortunately, none of these carbocyclic TCO alcohols were suitable for IMPACT because attempts to carry out both transphosphatidylation and IEDDA tagging within cells resulted in substantial background fluorescence present in cells treated with a pan-PLD inhibitor, FIPI (49), indicating PLD-independent labeling, likely caused by incomplete rinse-out of the lipophilic TCO alcohols (SI Appendix, Fig. S4).
At this point, we became aware of the trans-5-oxocene (oxoTCO; Fig. 2C), a more hydrophilic strained alkene that retains similarly high reactivity toward tetrazine reagents but is more polar than the carbocyclic TCOs because of the endocyclic ether functionality (50). We prepared a group of oxoTCO-containing primary alcohols with 1- to 3-carbon aliphatic linkers: oxoTCO–C1, –C2, and –C3 (Fig. 2C and SI Appendix, Schemes S4 and S5). Because the stereogenic center in oxoTCO–C1 derives from the chiral pool, we prepared both enantiomers of this compound. Excitingly, (S)-oxoTCO–C1 was a substrate of human PLDs, as confirmed by fluorescence-coupled HPLC analysis of IEDDA-tagged lipid extracts from cells stimulated with PMA in the presence of the oxoTCO alcohol (SI Appendix, Fig. S5). LC-MS analysis verified that the labeled species were the expected transphosphatidylation products (SI Appendix, Table S1).
(S)-oxoTCO–C1 Is an Optimal Alcohol Probe for Monitoring PLD Activity.
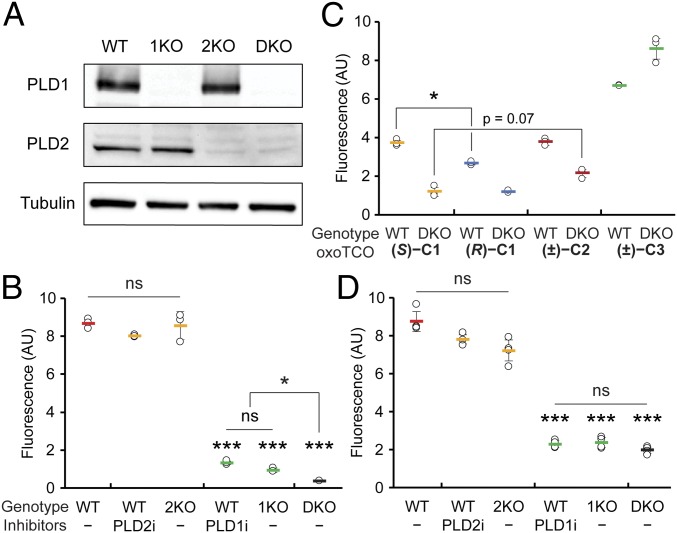
We next assessed the ability of a fluorogenic Tz–BODIPY conjugate (51) to detect the oxoTCO-containing lipids produced by transphosphatidylation within cells. Key tools to establish the specificity of the oxoTCO alcohols as substrates of PLD enzymes were a set of knockout HeLa cell lines for PLD1 and/or PLD2 that we generated using CRISPR/Cas9-mediated mutagenesis (termed 1KO, 2KO, and DKO, for, respectively, PLD1 knockout, PLD2 knockout, and PLD1/2 double knockout). We validated these cell lines by assessing both loss of PLD proteins via Western blot (Fig. 3A) and extent of PMA-stimulated IMPACT labeling using azidopropanol and a cyclooctyne–fluorophore in comparison with wild-type (WT) cells treated with isoform-selective PLD inhibitors (VU0359595 [52] [PLD1i] and VU0364739 [53] [PLD2i]) by flow cytometry (Fig. 3B). The 1KO cells had the same level of IMPACT labeling as WT cells treated with PLD1i, with similar relationships observed between 2KO cells and PLD2i treatment and between DKO cells and FIPI treatment (Fig. 3B and SI Appendix, Fig. S5).
Fig. 3.
oxoTCO alcohols are effective and selective reporters of endogenous human PLD activity. (A and B) Generation and validation of PLD1 knockout (1KO), PLD2 knockout (2KO), or PLD1/2 double knockout (DKO) HeLa cells made by using CRISPR/Cas9-mediated mutagenesis. (A) Knockout was verified by Western blot analysis for PLD1, PLD2, or as a loading control, α-tubulin. (B) The indicated cells were pretreated with the indicated PLD inhibitor (PLD1i, VU0359595; or PLD2i, VU0364739) or DMSO for 30 min, followed by transphosphatidylation with 3-azido-1-propanol (1 mM) under PMA stimulation (100 nM) for 20 min, rinsing with PBS solution, and labeling with a cyclooctyne–BODIPY conjugate (1 µM) for 10 min, rinsing for 20 min, and analysis by flow cytometry. Indicated are mean fluorescence intensities in arbitrary units (AU). (C and D) Evaluation of oxoTCO alcohols as selective probes for endogenous PLD enzymes. Shown are mean fluorescence intensities from flow cytometry experiments wherein IMPACT labeling was performed as in A, except using the indicated oxoTCO alcohol (C) or (S)-oxoTCO–C1 (3 mM, 5 min; D) in place of 3-azido-1-propanol and Tz–BODIPY (0.33 µM, 1 min, no subsequent rinse) in place of cyclooctyne–BODIPY. Statistical significance was assessed by using a 1-way ANOVA followed by Games–Howell post hoc analysis. Asterisks directly above data points denote statistical significance compared with WT without the presence of inhibitors, asterisks above horizontal lines denote statistical significance comparing the 2 indicated data groups, and error bars represent SD (*P < 0.05 and ***P < 0.001; ns, not significant).
To evaluate the panel of oxoTCO alcohols as probes for IMPACT, we performed in-cell transphosphatidylation with PMA stimulation followed by a 1-min IEDDA reaction with Tz–BODIPY and quantification by flow cytometry. Gratifyingly, the oxoTCO–C1 and –C2 were suitable probes for IMPACT in cells (Fig. 3C). (S)-oxoTCO–C1 was the best alcohol probe, approximately 50% more efficient than (R)-oxoTCO–C1. Background labeling, i.e., labeling that occurred even in DKO cells or in the presence of FIPI, trended slightly higher with oxoTCO–C2 and was significantly higher with oxoTCO–C3, whose linkers are more hydrophobic. We thus used (S)-oxoTCO–C1 for all subsequent studies. We note that the optimized IEDDA-based IMPACT protocol does not have as high sensitivity (i.e., signal-to-background) as SPAAC-based IMPACT (Fig. 3 B and D), likely because of differences in PLD-mediated turnover rates of the bulky (S)-oxoTCO–C1 in the transphosphatidylation step compared with the small, linear azidopropanol.
We then ascertained whether (S)-oxoTCO–C1 was a substrate of both PLD isoforms. We compared the extent of PMA-stimulated IMPACT labeling in WT cells to that in the relevant PLD knockout cells or in WT cells treated with isoform-selective PLD inhibitors and observed identical relative labeling efficiencies to those obtained using SPAAC-based IMPACT (Fig. 3 B and D). These experiments clearly indicated that (S)-oxoTCO–C1 was a substrate of PLD1.
The differences in mean fluorescence intensities of cellular IMPACT fluorescence quantified by flow cytometry between PLD2i-treated or 2KO cells and WT cells (and between PLD1i-treated or 1KO cells and DKO cells), however, were subtle or not statistically significant. As to whether (S)-oxoTCO–C1 might be a substrate of PLD2, HPLC analysis of transphosphatidylation products from PLD1i-treated or 1KO cells were suggestive (SI Appendix, Fig. S5). To gain support for this hypothesis, we turned to a gain-of-function approach to test whether (S)-oxoTCO–C1 was a substrate of PLD2. We assessed the level of IMPACT labeling using (S)-oxoTCO–C1 due to PLD2 by overexpressing fluorescently tagged PLD2 in DKO cells. Gratifyingly, we observed IMPACT labeling when overexpressing WT PLD2 but not a catalytically dead variant (K758R; SI Appendix, Fig. S6).
We note that these data do not allow us to establish whether PLD1 and PLD2 are equally efficient in using (S)-oxoTCO–C1 as a transphosphatidylation substrate. Ultimately, whether because of lower PLD2 expression in this cell line, less efficient activation of PLD2 by PMA (18), or (S)-oxoTCO-C1 being a less efficient substrate for PLD2 than for PLD1, in this setting, PLD1 is the major isoform contributing to IMPACT signal.
IMPACT with (S)-oxoTCO–C1 Reveals PM Localization of Active PLD.
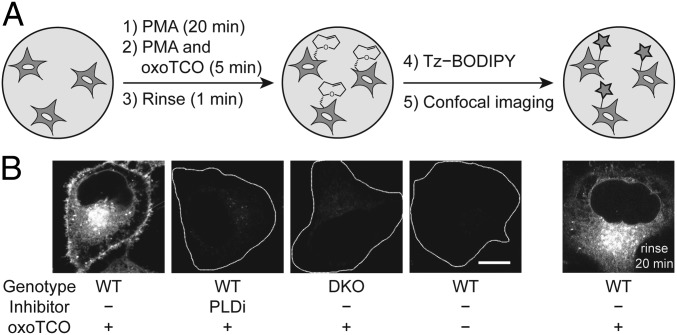
By using confocal microscopy, we evaluated the subcellular localization of IEDDA-based IMPACT using (S)-oxoTCO–C1 labeling with PMA stimulation followed by a 1-min IEDDA reaction with Tz–BODIPY (Fig. 4A). We observed substantial cellular fluorescence, and addition of FIPI during the transphosphatidylation step or the use of DKO cells abrogated the labeling, again demonstrating the specificity of our probes to active PLD enzymes (Fig. 4B). Excitingly, a major portion of the labeling was apparent at the PM, in contrast to SPAAC-based IMPACT using azidopropanol, which requires a 20- to 30-min SPAAC labeling and rinse-out and gives exclusively intracellular labeling. Colocalization analysis using conventional confocal microscopy and superresolution structured illumination microscopy indicated that the IEDDA-based IMPACT predominantly labeled the PM, ER, and Golgi apparatus, with minimal labeling observed of endosomes, lysosomes, and mitochondria (SI Appendix, Figs. S7 and S8).
Fig. 4.
IMPACT with (S)-oxoTCO–C1 reveals the PM localization of active PLD enzymes stimulated with a phorbol ester. (A) Experimental setup: endogenous PLD enzymes were stimulated with PMA (100 nM, 20 min), followed by transphosphatidylation with (S)-oxoTCO–C1 (3 mM, 5 min, in the presence of PMA), rinse (1 min), and IEDDA reaction with Tz–BODIPY (0.33 µM, 1 min), followed by confocal microscopy imaging. (B) Representative images of HeLa cells labeled as described in A with the indicated negative controls. Where indicated, PLDi (the pan-PLD inhibitor FIPI) was applied 30 min before and during the transphosphatidylation step. DKO, PLD1/2 double knockout cells. Far right: cells were rinsed after the IEDDA reaction for 20 min before imaging. White dotted lines indicate cell outlines in the negative controls. (Scale bar: 10 μm.)
We hypothesized that IEDDA-based IMPACT successfully revealed the expected PM localization of PMA-stimulated PLD activity because of the rapid time scale of the labeling: 5 min of (S)-oxoTCO–C1, a quick rinse, a 1-min IEDDA reaction with Tz–BODIPY, and imaging immediately afterward. To test this hypothesis, we performed a chase experiment by incubating the IMPACT-labeled cells at 37 °C for an additional 20 min, followed by imaging, and observed entirely intracellular labeling, similar to the spatial pattern created by SPAAC-based IMPACT (33) (Fig. 4 B, far right). These results suggested that, after 20 min, the BODIPY-labeled phospholipids had been fully internalized.
Real-Time IMPACT Enables Precise Determination of PLD Localization.
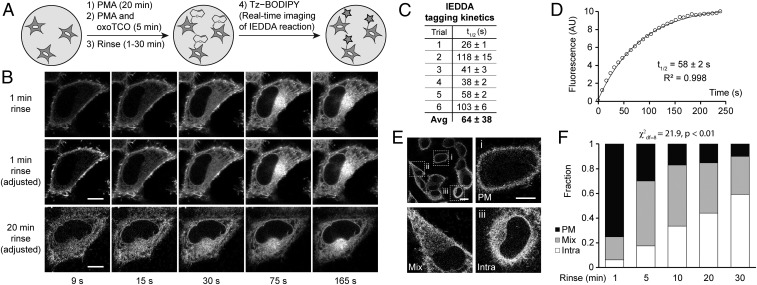
To this point, we had not taken advantage of a key benefit of the TCO–tetrazine IEDDA, namely its fluorogenicity (48, 54). Tz–BODIPY is only very dimly fluorescent and experiences a 1,600-fold increase in brightness upon IEDDA reaction (51). We envisioned that we could adapt this oxoTCO-based IMPACT so that the IEDDA reaction would be monitored in real time on the microscope, allowing us to visualize the localization of fluorescent lipids with no time delay. The optimized protocol, which we termed RT-IMPACT (for real-time IMPACT), involves the identical 5-min (S)-oxoTCO–C1 labeling under PMA stimulation (or, as described later, stimulation with alternate agonists), followed by a quick rinse and time-lapse imaging before and during the IEDDA reaction with Tz–BODIPY (Fig. 5A). With these tools, we would be able to visualize the precise localizations of the TCO-lipid reporters with rapid temporal resolution (i.e., seconds), limited only by microscope acquisition times and the IEDDA’s intrinsic kinetics.
Fig. 5.
Real-time IMPACT enables precise determination of the subcellular localization of active PLD enzymes. (A) The experimental setup for RT-IMPACT is the same as indicated in Fig. 4 except that IEDDA reaction with Tz–BODIPY is monitored in real time by time-lapse confocal microscopy. (B) Representative images from time-lapse RT-IMPACT imaging of HeLa cells. The rinse (chase step) following the transphosphatidylation with (S)-oxoTCO–C1 and before IEDDA tagging was 1 min (Top and Middle) or 20 min (Bottom). (Top) Images are shown with identical intensity settings to facilitate visualization of reaction progress. (Middle and Bottom) Fluorescence intensities of the 9-, 15-, and 30-s time points have been brightened to facilitate visual comparison of localization of the IMPACT signal at different time points. (C and D) Quantification of the kinetics of the IEDDA tagging reaction based on total cellular fluorescence from 6 independent experiments (where the graph represents trial 5, with circles indicating data points, the curve indicating an exponential fit, and R2 indicating the coefficient of determination of the fit) with n = 8 to 16 cells for each trial and average (Avg) reaction half-life (t1/2) indicated as mean ± SD. (E and F) Quantification of the subcellular localizations of RT-IMPACT fluorescent signal at the 9-s time point of the IEDDA reaction as a function of the posttransphosphatidylation chase step, i.e., the rinse time after (S)-oxoTCO–C1 labeling and before IEDDA tagging. Cells were blindly scored as having plasma membrane (PM, black), intracellular (Intra, white), or a combination of PM and intracellular (Mix, gray) IMPACT fluorescence. y axis indicates fraction of cells with the indicated localization. (E, Top Left) Representative zoomed-out image of a population of cells at the 9-s time point from a 20-min rinse time point from F, with examples of PM (i), mix (ii), and intra (iii) localizations indicated in the other panels. (Scale bars: B and E, i–iii, 10 µm; E, Top Left, 20 μm.) Statistical significance was assessed by using a χ2 test for independence, with the χ2 value (df, degrees of freedom) and associated P value indicated. Each bar contains data of 3 to 5 biological replicates with n = 30 to 72 total cells.
Time-lapse RT-IMPACT movies, with frames acquired every 3 s, revealed several observations (Fig. 5B and Movie S1). First, total cellular fluorescence increased over the first minute and then stabilized, indicating that the IEDDA reaction had reached completion during that time period (Fig. 5 B, Top). By measuring total cellular fluorescence over time, we calculated the half-life of IEDDA tagging of (S)-oxoTCO–C1-labeled lipids with Tz–BODIPY (0.33 µM and presumably under pseudo–first-order conditions) to be 64 ± 38 s (Fig. 5 C and D), in line with the reaction’s intrinsically high kinetics (48).
Second, the fluorescence at early time points is almost entirely located at the PM (Fig. 5 B, Middle; and see Movie S1). These data suggest that the bulk of PMA-stimulated PLD activity in HeLa cells is at the PM, with there being differential trafficking rates of PLD-generated PA compared with 2 different types of phosphatidyl alcohols generated via IMPACT, i.e., those whose head group is TCO or BODIPY, which we will address later. Third, when we examined cells that had been rinsed for 20 min before IEDDA and at an early time point of the time-lapse movie after addition of Tz–BODIPY (9 s), we observed a distribution of localizations, which we classified as “PM” for predominantly plasma membrane, “intra” for predominantly intracellular, or “mix” for a mixture of PM and intracellular (Fig. 5 B, Bottom, and Fig. 5E). This distribution of localizations at the early time point of the IEDDA reaction indicates that diffusion of unreacted fluorophore is not rate-limiting compared with IEDDA reaction kinetics. Put another way, the data show that TCO-lipids on intracellular organelle membranes have similar access to Tz–BODIPY compared with those at the PM.
By using this localization classification system (PM, mix, and intra), we performed pulse-chase experiments to examine TCO-lipid trafficking, using as a readout the distribution of localizations at the 9-s time point of a time-lapse movie. Here, the chase was a 37 °C rinse-out after the 5-min transphosphatidylation step and before the real-time IEDDA reaction. As we increased the chase time before IEDDA, the 9-s time point exhibited fewer PM cells and more mix and intra cells (Fig. 5F). We conclude that, even though the TCO-lipid is internalized from the PM, the time scale of this process, with a half-life of ∼10 min (Fig. 5F), is longer than that of the optimized RT-IMPACT protocol. Thus, these data indicate that the localization of fluorescent lipids observed at the early time points of the IEDDA tagging reflects the sites of active PLD enzymes before major trafficking of the lipid reporters.
As well, we observed RT-IMPACT–derived fluorescence at the plasma membrane at the 9-s time point of time-lapse movies in PMA-stimulated DKO cells overexpressing fluorescently tagged PLD1 or PLD2 but not catalytic dead versions of these enzymes (SI Appendix, Fig. S6). These experiments further suggest that RT-IMPACT reveals the expected localizations of the active subsets of these enzymes, which are expected to be at the plasma membrane under PMA stimulation.
Real-Time IMPACT Reveals PM-to-ER Trafficking of Fluorescent Lipid Reporters.
An obvious feature of the RT-IMPACT time-lapse movie is that the fluorescent lipids, which were predominantly at the PM at the early time points (i.e., 9 s), were rapidly internalized, first over the next 15 to 30 s to the ER and then subsequently to the perinuclear Golgi apparatus (Fig. 5 B, Middle). This trafficking from the PM to internal organelle membranes appeared to be unidirectional (Fig. 4 B, far right, and Fig. 5 B, Bottom).
The ability to monitor the trafficking of the labeled lipids from the very moment that they become fluorescently tagged allowed us to gain information about their internalization pathway and its kinetic properties. Notably, the first apparent intracellular destination of the IMPACT-derived fluorescent lipids was the ER, not endosomes, suggesting that they were internalized predominantly not via endocytosis (Fig. 5B and SI Appendix, Figs. S7 and S8). Based on such a localization, we propose that the lipids were internalized by vesicle-independent pathways mediated by lipid transfer proteins that largely operate at ER–PM contact sites (34, 55). These contact sites, or junctions, are assemblies where the 2 organelle membranes are located within 10 to 20 nm of each other to facilitate rapid lipid transport and other signaling processes (55).
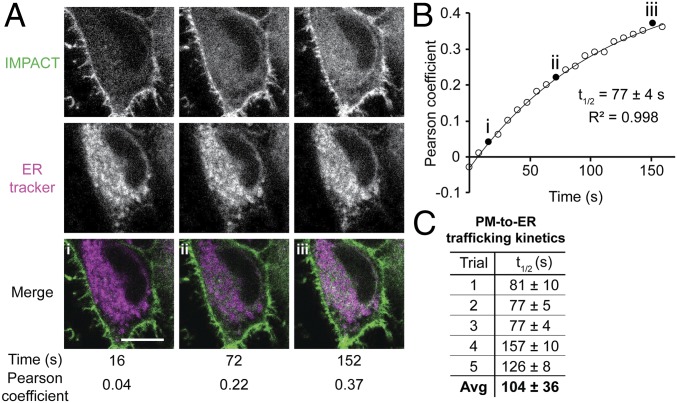
By quantifying the extent of colocalization of the RT-IMPACT labeling with an ER marker over time, we measured the half-life of fluorescent lipid PM-to-ER trafficking, determining it to be 104 ± 36 s (Fig. 6 A and B). We envision that this type of RT-IMPACT–based experiment, i.e., tracking the fate of the BODIPY-labeled lipid, might prove useful for studying lipid trafficking mediated by lipid transfer proteins, suggesting applications of RT-IMPACT entirely separate from applications to visualize locations of PLD signaling. Notably, some lipid transfer proteins interact only with the lipids’ hydrophobic tails and have little preference for head group, suggesting that they may transport the BODIPY-labeled phospholipids similarly to natural glycerophospholipids (56).
Fig. 6.
Real-time IMPACT reveals PM-to-ER trafficking of fluorescent lipids. (A) Representative images of HeLa cells from a 2-color time-lapse movie. HeLa cells were incubated with ER tracker, PMA, and (S)-oxoTCO–C1 for 5 min and rinsed for 1 min before time-lapse imaging. Values of Pearson correlation coefficients of colocalization between IMPACT (green) and ER tracker (magenta) were determined. Colocalization appears as white in merge. (B) Representative exponential fitting of kinetics of PM-to-ER trafficking. Plotted is increase in colocalization (Pearson coefficient) between IMPACT and ER tracker over time. Circles are data points, and the curve indicates an exponential fit. Filled circles (i–iii) correspond to Pearson coefficients for the images shown in A, and R2 indicates the coefficient of determination of the fit. (C) Summary of quantification of PM-to-ER trafficking kinetics from 5 independent experiments (where the graph in B represents trial 3) with n = 8 to 16 cells for each trial and average (Avg) half-life of internalization (t1/2) indicated as mean ± SD. (Scale bar: 10 µm.)
Applying Real-Time IMPACT to Visualize Differences in Active PLD Localization Elicited by Different Stimuli.
Finally, having validated RT-IMPACT, we sought to apply it to evaluate the localizations of PLD signaling activity elicited by different physiological stimuli. We focused on agonists that would specifically activate either GPCR or RTK signaling pathways, which both lead to PLD-mediated PA biosynthesis. In these experiments, localization was determined by examining the 9-s time point after Tz–BODIPY addition of a time-lapse movie of the IEDDA reaction to avoid effects of fluorescent lipid trafficking, and evaluating the fraction of cells with PM, mix, and intra localizations. We varied 2 parameters: the type of stimulus and the amount of time the cells were exposed to the stimulus before the 5-min transphosphatidylation step.
First, to assess GPCR-mediated signaling, we generated HeLa cells stably expressing the M1 muscarinic receptor (M1R). Upon stimulation with its agonist oxotremorine-M (oxo-M), M1R activates Gαq to turn on phospholipase Cβ (PLCβ), which hydrolyzes phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] to generate inositol trisphosphate and diacylglycerol (57, 58), an allosteric activator of PKC. GPCR signaling also activates the RhoA GTPase, and both active PKC and RhoA can stimulate PLD activity downstream of GPCR activation (24).
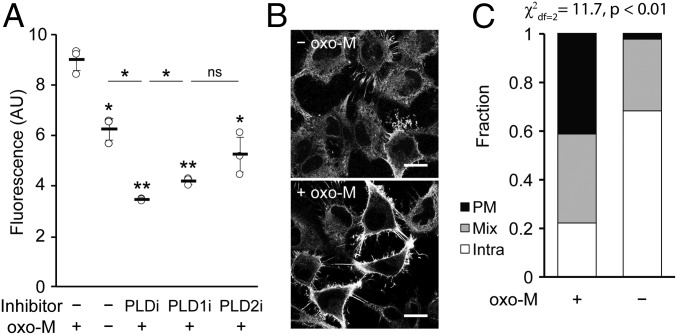
RT-IMPACT revealed that, in the absence of oxo-M stimulation, HeLa cells exhibited a small amount of basal PLD activity, and its localization was intracellular (Fig. 7A, second vs. third sample; Fig. 7 B, Top; Fig. 7C, quantification). Exposure of cells to oxo-M led to an approximate doubling of PLD activity within the first 5 min of oxo-M stimulation (Fig. 7A, first vs. second sample), and treatment with isoform-selective PLD inhibitors revealed that this activity was predominantly the result of PLD1 (Fig. 7A). Prolonged exposure to oxo-M led to attenuation of PLD activity, consistent with the known property of this system to desensitize under sustained oxo-M stimulation (SI Appendix, Fig. S9A) (57, 58). The localization of PLD activity under oxo-M stimulation was nearly evenly split among PM, mix, and intra (Fig. 7 B, Bottom; Fig. 7C, quantification). Because this oxo-M–stimulated activity represented a roughly even mixture of M1R-derived and basal PLD activities (Fig. 7A), we conclude that the bulk of the M1R-stimulated PLD enzymes were located at the PM, either through translocation from intracellular locations or through activation of subsets that were previously located in this membrane.
Fig. 7.
Muscarinic M1 receptor (M1R) activation leads to PLD activity that is predominantly at the plasma membrane. HeLa cells stably expressing M1R were labeled for RT-IMPACT by treatment with the indicated PLD inhibitor or DMSO for 30 min (A only) and then simultaneous treatment of (S)-oxoTCO–C1 in the presence or absence of oxo-M (5 min), rinsing (1 min), and IEDDA reaction with Tz–BODIPY (0.33 µM) for 1 min followed by flow cytometry analysis (A), with mean fluorescence intensity in arbitrary units (AU) indicated, or (B and C) in real time with time-lapse confocal images taken (B) and quantified (C) 9 s after the addition of Tz–BODIPY, using the PM/Mix/Intra rubric described in Fig. 5. In B, the brightness of the –oxo-M image was increased to facilitate comparison of the localization of IMPACT-derived fluorescence in the 2 images. (Scale bars: 20 µm.) For A, statistical significance was assessed by using 1-way ANOVA followed by Games–Howell post hoc analysis. Asterisks directly above data points (*P < 0.05 and **P < 0.01; ns, not significant) denote statistical significance compared with the first sample (−inhibitor, +oxo-M), and error bars represent SD. For C, statistical significance was assessed by using a χ2 test for independence, with the χ2 value (df, degrees of freedom) and associated P value indicated. Each bar contains data of 3 (−oxo-M) or 8 (+oxo-M) biological replicates, with n = 41 (−oxo-M) or 112 (+oxo-M) total cells.
Second, we examined the localization of PLD signaling during RTK activation. Here, we stimulated NIH 3T3 cells with platelet-derived growth factor (PDGF), the agonist for the PDGF receptor (PDGFR), which activates PLDs through PLC-mediated PI(4,5)P2 hydrolysis, but through a different PLC isoform (PLCγ) from GPCR pathways (59–61). PDGFR additionally stimulates PLDs through activation of small Ras superfamily GTPases, including Arf, RhoA, Cdc42, and Rac family GTPases, which can localize to several organelle membranes (62–65).
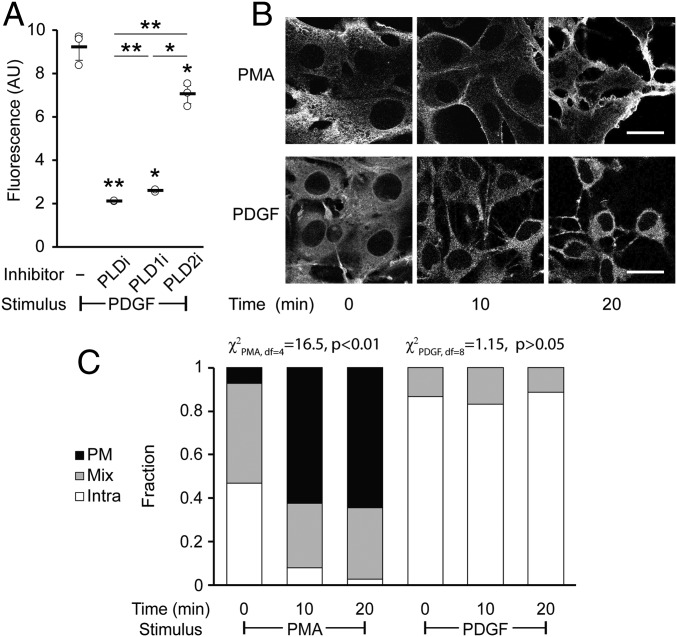
For these experiments, we varied the amount of time that the PDGF stimulus was applied before the RT-IMPACT labeling. We found that PDGF treatment resulted in a robust and sustained stimulation of PLD enzymes over 60 min (SI Appendix, Fig. S10A) and that such activity is caused predominantly by the PLD1 isoform (Fig. 8A). To our surprise, we discovered that the localization of the RT-IMPACT signal was predominantly intracellular for all time points evaluated during the time course of PDGF stimulation (Fig. 8 B and C and SI Appendix, Fig. S10B). As a control, we examined PMA stimulation in this cell line and found that, similar to in HeLa cells, it led to predominantly PM-localized PLD activity after at least 10 min of stimulation before RT-IMPACT (Fig. 8 B and C and SI Appendix, Fig. S10C). Thus, RT-IMPACT labeling suggests that PDGFR signaling leads to activation of predominantly intracellular pools of PLD1, in contrast to M1R signaling or PMA stimulation, which leads to activation of PLD enzymes mostly at the PM.
Fig. 8.
Platelet-derived growth factor (PDGF) receptor activation leads to intracellular PLD activity. NIH 3T3 cells were labeled for RT-IMPACT by treatment with the indicated PLD inhibitor or DMSO for 30 min (A only). Cells were stimulated with PDGF or PMA for the indicated time (0 to 20 min) followed by addition of (S)-oxoTCO–C1 in the continued presence of PDGF or PMA (5 min) and then rinsed (1 min), and the IEDDA reaction with Tz–BODIPY (0.33 µM) was performed for 1 min followed by flow cytometry analysis (A), with mean fluorescence intensity in arbitrary units (AU) indicated, or (B and C) in real time with time-lapse confocal images taken (B) and quantified (C) 9 s after the addition of Tz–BODIPY, using the PM/Mix/Intra rubric described in Fig. 5. (Scale bars: 20 µm.) For A, statistical significance was assessed by using 1-way ANOVA followed by Games–Howell post hoc analysis. Asterisks directly above data points (*P < 0.05 and **P < 0.01) denote statistical significance compared with the first sample (−inhibitor), asterisks above horizontal lines denote statistical significance comparing the 2 indicated samples, and error bars represent SD. For C, statistical significance was assessed by using a χ2 test for independence, with the χ2 value (df, degrees of freedom) and associated P value indicated. Each bar contains data of 3 to 4 biological replicates with n = 58 to 78 total cells for PDGF and 3 to 6 biological replicates, with n = 41 to 95 cells for PMA.
Discussion
Localized production or accumulation of intracellular signaling agents is a key feature of signal transduction pathways. Chemical imaging tools have helped pave the way to understand biological signaling pathways dependent on second messengers (2–4, 66). We initiated this study with the goal of establishing an activity-based imaging tool to enable precise monitoring of the production of the lipid second messenger PA by PLD enzymes. Previously, we had established that PLDs, which physiologically catalyze hydrolysis of phosphatidylcholine to release PA and choline, can accept primary azido alcohols in transphosphatidylation reactions to generate azido phosphatidyl alcohol reporters of PLD activity (33). Subsequent tagging of these azido lipids with cell-permeable cyclooctyne–fluorophore conjugates, followed by rinse-out of excess unreacted fluorophore, enabled fluorescent tagging of intracellular membranes bearing PLD activity, in a method termed IMPACT. The lack of IMPACT labeling at the PM combined with the long labeling and rinse-out times and the knowledge that lipids can diffuse and traffic rapidly throughout the cell led us to consider an alternative approach for precisely determining the subcellular locations of PLD signaling.
Taking advantage of rapid and fluorogenic IEDDA bioorthogonal chemistry, which has proven useful for tagging and imaging highly dynamic molecules such as lipids (67–69), we report here a real-time variant of IMPACT (RT-IMPACT). We have replaced azido alcohols with a bulky, hydrophilic trans-5-oxocene (oxoTCO)-containing primary alcohol (50), which acts as a replacement for the water cosubstrate in the PLD active site. Following this transphosphatidylation step, excess oxoTCO alcohol is quickly rinsed away, and the locations of the oxoTCO-labeled lipids are then visualized within 10 s by using time-lapse monitoring, in real time, of a no-rinse, IEDDA reaction with a fluorogenic tetrazine–BODIPY reagent (Tz–BODIPY).
In developing these tools, we discovered that human PLDs accept a wide group of TCO-containing primary alcohols as transphosphatidylation substrates, revealing a previously unappreciated tolerance of these enzymes for large, bulky side chains on the primary alcohol. However, carbocyclic TCO alcohols and even oxoTCO alcohols with longer aliphatic side chains did not effectively rinse out of cells, likely because of hydrophobicity, even though many were substrates of PLDs. This finding highlights the challenges of using TCO-based probes for metabolic labeling with small precursors, where the physicochemical properties of the bioorthogonal group dominate those of the overall probe, which, in the case of our optimal alcohol, (S)-oxoTCO–C1, is simply the oxoTCO with a hydroxymethyl side chain.
Because the goal of RT-IMPACT was to faithfully report on the localization of PLD signaling activity (i.e., transphosphatidylation of PC to form reporter lipids occurring simultaneously with physiological PC hydrolysis to form PA), a key challenge we faced was to establish that the time scale of RT-IMPACT was faster than that of the trafficking of the reporter lipids to membrane locations other than their site of generation. We found that PMA stimulation led to a strong PM localization of the RT-IMPACT signal, consistent with literature reports that phorbol esters activate PLD enzymes predominantly in this membrane. By monitoring the IEDDA reaction in real time, we directly visualized rapid internalization of BODIPY-lipids on the second time scale to other intracellular locations. In contrast, by performing a chase before the IEDDA tagging and then examining the earliest IEDDA reaction time points, i.e., those before this BODIPY-lipid trafficking, we established that the oxoTCO-labeled lipids were internalized on a slower time scale of 10 to 20 min.
Intriguingly, the pathways for internalization of these phosphatidyl alcohol lipids appeared to be not via classic endocytic routes but instead via vesicle-independent pathways. These routes are mediated by lipid transfer proteins, which shuttle individual lipid molecules between bilayers of different organelles (34, 70, 71). For maximal efficiency of lipid transport, many of these transfer proteins have recently been proposed to operate at membrane contact sites, which are sites of close apposition between 2 organelles where the distance between membranes is minimized, often as little as 10 to 20 nm (55, 72).
Because of these short distances and the rapid kinetics of lipid diffusion and transport, nonvesicular lipid transport at membrane contact sites has proven challenging to directly observe. Even though some lipid transfer proteins have precise lipid preferences based on head group binding, others primarily engage the hydrophobic tails and are thought to transport a wide variety of types of lipids (34, 70, 71, 73). IMPACT-derived fluorescent lipids could be substrates for these latter lipid transfer proteins. An unexpected application of RT-IMPACT is that it may serve as a useful tool for directly visualizing PM-to-ER lipid trafficking via these nonvesicular pathways, which have recently been appreciated to play important roles in lipid homeostasis (55, 70).
To that end, we have investigated the internalization of 2 types of lipids: phosphatidyl alcohols with an oxoTCO head group (TCO-lipids) and those with a fluorescent, BODIPY head group (BODIPY-lipids). Even though both appear to be internalized largely by the same route (i.e., PM to ER), the kinetics of these processes are quite different. These differences could arise from several factors beyond the differences in the membrane surface areas of the PM and ER. Different transfer proteins may mediate their trafficking, or the different chemical properties of the lipids may impact their distribution within a given membrane bilayer, affinity for transfer proteins, and/or relative affinity for origin and destination organelle membranes. A potential future direction is the application of real-time oxoTCO–tetrazine IEDDA reactions for probing the trafficking pathways of lipids labeled with oxoTCO groups by processes other than PLD activity.
Finally, we applied RT-IMPACT to interrogate the spatial control of PLD signaling, taking advantage of its ability to report on the sites of synthesis of phosphatidyl alcohols by active PLD enzymes. As test cases, we examined stimulation of 2 distinct physiological pathways for activation of PLDs: M1R as a prototypical GPCR and PDGFR as a prototypical RTK. Even though the major players in the signaling pathways to activate PLDs have been established, including different PKC isoforms, small GTPases and heterotrimeric G proteins, and lipid cofactors, they can act in parallel and at different locations. Consequently, the precise locations where PA generation occurs remained unknown. Because of different subcellular localizations of the numerous PA-binding effector proteins, the location of PLD-mediated PA synthesis could greatly impact downstream signaling outcomes.
By using RT-IMPACT, we determined that, in the cell types we investigated, the majority of M1R-dependent PLD activity appears to occur at the PM, whereas the bulk of PDGFR-dependent PLD activity appears to occur on intracellular membranes. These results suggest that unique physiological agonists may activate pools of PLD enzymes at different subcellular locations, even when operating in part through common intermediates (e.g., PKC, RhoA, Rac1).
Regarding the PDGF studies, we note that this growth factor stimulates membrane ruffle formation at the plasma membrane in a process that requires PLD-mediated PA production (65), implying that some PDGF-stimulated PLD activity is likely in this membrane. Membrane ruffles are highly dynamic structures that involve major alterations to the actin cytoskeleton, and their effects on rates of phosphatidyl alcohol trafficking are unknown. Thus, it is possible that differences in the localization of IMPACT-derived fluorescent lipids—even at the early, 9-s time point of RT-IMPACT time-lapse movies—could reflect different rates of phosphatidyl alcohol trafficking induced by various stimuli that also activate PLDs.
Interesting future directions will involve dissecting the biological mechanisms and implications of these findings, including differential effects on relevant PA effector proteins, as well as potential roles of choline, the soluble head group liberated from PC by PLD, which has recently been proposed to play signaling roles of its own at the ER (74).
In conclusion, we demonstrate that synthetic oxoTCO-containing primary alcohols function as substrates of endogenous, mammalian PLD enzymes and as metabolic labels for their activity within cells. In conjunction with real-time monitoring of a fluorogenic IEDDA tagging reaction of the resultant oxoTCO-containing lipids, we imaged precise subcellular locations of PLD signaling activity. Compared with IMPACT using azido alcohols and SPAAC-based fluorescent tagging, RT-IMPACT, even though it has a lower extent of labeling, is an activity-based imaging approach that affords temporal resolution of seconds and is thus ideally suited to applications in which the precise subcellular localization of lipid synthesis is critical. As well, continuous monitoring of the trafficking of lipids generated by RT-IMPACT holds promise as a method for interrogating intracellular lipid transport pathways.
Methods
General Materials and Methods.
The 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) was purchased from Echelon Biosciences, and 5-fluoro-2-indolyl deschlorohalopemide (FIPI) was purchased from Cayman Chemical. Phorbol 12-myristate 13-acetate (PMA) and oxotremorine-M were purchased from Santa Cruz Biotechnology. Streptomyces sp. PMF PLD was purchased from Sigma-Aldrich. Recombinant PDGF-BB was purchased from Shenandoah Biotechnology. Methyltetrazine-amine (CAS no. 1345955–28-3) was purchased from Click Chemistry Tools. Azidopropanol was synthesized as reported previously (33). HPLC analysis was performed on a Shimadzu LC-20AR HPLC equipped with an SPD20AV UV/Vis detector, an RF-20A fluorescence detector, and a Phenomenex Luna silica 3-µm, 100-Å, 25-cm × 4.6-mm column. LCMS analysis was performed on an Agilent 6230 electrospray ionization–time-of-flight (ESI-TOF) MS coupled to an Agilent 1260 HPLC equipped with a Zorbax Rx-Sil normal-phase silica column (2.1 × 50 mm, 1.8 µm). Dulbecco’s modified Eagle medium (DMEM), phosphate-buffered saline (PBS) solution, fetal bovine serum (FBS), and 0.05% trypsin-EDTA were purchased from Corning. ER-Tracker Red, CellMask Deep Red Plasma Membrane Stain, and Lipofectamine 2000 were purchased from Thermo Fisher. For all imaging experiments except those in SI Appendix, Fig. S8, confocal imaging was performed on a Zeiss LSM 800 confocal laser scanning microscope equipped with 20× 0.8 NA and 40× 1.4 NA Plan Apochromat objectives; 405-, 488-, 561-, and 640-nm solid-state lasers; and 2 GaAsP PMT detectors by using Zen Blue 2.3 software. For SI Appendix, Fig. S8, superresolution structured illumination microscopy (SR-SIM) was performed on a Zeiss Elyra superresolution microscope, and image reconstruction was performed using default parameters in the Zeiss Zen Black software. All other image analysis was performed by using FIJI/ImageJ. Flow cytometry was performed on a BD Accuri C6 flow cytometer, and analysis was performed by using the BD Accuri C6 analysis software. For measurements of half-life of lipid trafficking, the values of Pearson correlation coefficients were fitted in Origin Pro-8 by monoexponential regressions.
Cell Culture.
HeLa cells and NIH 3T3 cells were grown in DMEM (Dulbecco’s modified Eagle medium) supplemented with 10% FBS and 1% penicillin/streptomycin and were maintained in a 5% CO2, moisture-saturated atmosphere at 37 °C. HEK 293TN cells were grown as described here earlier and additionally supplemented with 1 mM sodium pyruvate. Cell densities were maintained between 105 and 1.6 × 106 cells per milliliter. For cell labeling experiments, all buffers or media were warmed to 37 °C or room temperature before addition to cells unless otherwise noted, and incubations were done at 37 °C unless otherwise specified.
Plasmids.
Plasmids were obtained from the following sources: STIM1-mRFP from Barbara Baird, Cornell University, Ithaca, NY; mCherry-P4M-SidM (a marker of structures positive in phosphatidylinositol 4-phosphate, principally the Golgi apparatus and the plasma membrane) from Addgene (no. 51471); and EEA1-mRFP, OMP25(transmembrane domain)-mCherry, PM-mRFP, and LAMP1-mRFP from Pietro De Camilli, Yale University, New Haven, CT. PLDs were tagged with mScarlet-i by subcloning of mScarlet-i (from pmScarlet-i-C1; Addgene no. 85044) in place of GFP in GFP-PLD1, GFP-PLD1* (K898R), GFP-PLD2, and GFP-PLD2* (K758R; obtained from Mike Frohman, Stony Brook University, Stony Brook, NY) using SnaBI and BspEI.
In Vitro PLD Transphosphatidylation Reactions.
To a 1.5-mL Eppendorf tube was added the desired TCO-alcohol (2 µmol, neat) and DOPC (10 µL of an 8-mg/mL solution in chloroform). The chloroform was evaporated under a stream of nitrogen. The tube was then charged with SDS (1.5 µL of a 50-mM aqueous solution), sodium acetate (3 µL of a 1-M, pH 5.6, aqueous solution), and deionized water (19.5 µL), and the solution was vortexed and centrifuged briefly. Subsequently, calcium chloride (3 µL of a 500-mM aqueous solution) and 2 µL of DI water were added, and the solution was vortexed and centrifuged again. Last, a freshly made solution of Streptomyces sp. PMF PLD (1 U/µL in water, 2 µL) was added, and the mixture was heated to 30 °C for 1.5 h. The mixture was then diluted with PBS solution (70 µL, pH 7.4), methanol (250 µL), chloroform (250 µL), and 20 mM aqueous acetic acid (125 µL). The 2-phase mixture was vortexed for 1 min and then centrifuged for 2 min at 16,000 × g. The organic layer was then transferred to a new Eppendorf tube, and another fresh 250 µL of chloroform was added to the remaining aqueous layer. The mixture was vortexed and centrifuged as before. The 2 organic layers were then combined, dried under a stream of nitrogen, and stored at −80 °C or further tagged via IEDDA click chemistry tagging. The lipid residue stored in a 1.5-mL Eppendorf tube was dissolved in 100 µL of a 2:1 vol/vol chloroform:methanol solution, charged with 2.5 nmol of Tz–BODIPY (1.94 µL, 1.3 mM stock in DMSO), reacted for 5 min at 37 °C, and then filtered (0.45 µm) before HPLC analysis. Samples were analyzed by normal-phase HPLC with a binary gradient elution system whereby solvent A was chloroform:methanol:ammonium hydroxide (95:7:0.5) and solvent B was chloroform:methanol:water:ammonium hydroxide (60:34:5:0.5). Separation was achieved by using linear gradients from 100% A to 100% B over 45 min. The method was as follows: 0–5 min, 0% B; 5–21 min, 0–75% B; 21–22 min, 75–100% B; 22–27 min, 100% B; 27–29 min, 100–0% B; and 29–45 min, 0% B. Detection wavelengths were 495 nm (excitation) and 510 nm (emission).
Detection of Endogenous Mammalian PLD Activity by HPLC.
HeLa cells (n = 500,000) were seeded on a 60-mm dish 24 h before the experiment. On the day of the experiment, the cells were first treated with the indicated PLD inhibitor (PLDi [FIPI], 750 nM; PLD1i [VU0359595], 250 nM; PLD2i [VU0364739], 350 nM) from 1,000× DMSO stock solutions or the appropriate amount of DMSO for 30 min in DMEM (1 mL). PMA (100 µM or 1,000×, stock solution in DMSO) was then added to the DMEM solution to achieve a final concentration of 100 nM, and the cells were incubated for a further 20 min. The media was aspirated and replaced with the desired freshly prepared TCO-alcohol (3–10 mM) in the continued presence of PMA (100 nM) in DMEM (500 µL). The cells were incubated for an additional 20 min at 37 °C (during which the dish was swirled every 5 min to cover the entire surface, as required by the use of a minimal volume of labeling solution) before the treatment buffer was aspirated. The cells were rinsed with DMEM for 3 min at 37 °C, followed by a brief rinse with PBS solution at 4 °C (1 mL). The PBS solution was aspirated, and the cells were then subjected to a modified Bligh–Dyer extraction. In brief, using solutions at 4 °C, 100 µL of cold PBS solution, 125 µL of cold acetic acid (20 mM, aqueous), and cold methanol (250 µL) were added to the aspirated dish. The cells were then scraped off the dish and transferred to 1.5-mL conical tubes. Chloroform (250 µL) was then added to the suspension. The mixture was then vortexed for 1 min and centrifuged at 16,000 × g for 2 min. The organic layer (bottom) was removed and placed in a separate conical tube, and an additional 250 µL chloroform was added to the remaining aqueous layer. The vortexing and centrifugation steps were repeated, and the 2 organic layers were combined, dried under a stream of nitrogen, and stored at −80 °C or tagged by IEDDA with Tz–BODIPY and analyzed by HPLC as described here earlier.
Identification of oxoTCO-Containing Phosphatidyl Alcohols by LC–MS.
HeLa cells were labeled with (S)-oxoTCO–C1 (5 mM) under PMA stimulation as described in the previous section. After isolation of cellular lipidomes, the extracts were derivatized with methyltetrazine-amine (0.01 mg, 10 mg/mL stock in DMSO) for 5 min at room temperature and then filtered (0.45 µm). LC-MS analysis was performed using normal-phase HPLC with a binary gradient elution system whereby solvent A was chloroform:methanol:ammonium hydroxide (85:15:0.5) and solvent B was chloroform:methanol:water:ammonium hydroxide (60:34:5:0.5). Separation was achieved by using a linear gradient from 0 to 100% B over 8.5 min with a flow rate of 0.5 mL/min. Phosphatidyl alcohol species were detected using a dual ESI source operating in positive mode, acquiring in extended dynamic range from m/z 100 to 1,700 at 1 spectrum per second; gas temperature was 325 °C; drying gas, 10 L/min; nebulizer, 20 psig; fragmentor, 300 V.
Generation of PLD1/2 Knockout Cell Lines.
Single and double knockout cells for PLD1 and/or PLD2 (1KO, PLD1 knockout; 2KO, PLD2 knockout; DKO, PLD1/2 double knockout) were generated in HeLa cells using the LentiCRISPR v2 system. The LentiCRIPSR plasmid for PLD1 knockout was made by cloning the sgRNA (GTGAGCCCACAAATAGACGG) into the LentiCRISPRv2-Puro vector (Addgene no. 52961), and the LentiCRISPR plasmid for PLD2 knockout was made by cloning the sgRNA (GGCACCGAAAGATATACCAG) into the LentiCRISPRv2-Blast vector (Addgene no. 83480). The appropriate LentiCRISPR plasmid (7.5 μg) was cotransfected with packaging plasmids (5.6 μg Gag and 1.8 μg Env) into HEK 293TN cells seeded on a 10-cm dish using Lipofectamine 2000. The virus-containing medium was collected 48 h and 72 h after transfection, and cell debris was removed by passing through a 0.45-μm filter. For lentiviral transduction, wild-type HeLa cells seeded on a 6-well plate were treated with transduction medium (freshly prepared by mixing 1.5 mL virus-containing medium, 0.5 mL DMEM media, and polybrene [8 μg/mL]) for 48 h. During transduction, transduction medium was replaced every 12 h. After transduction, cells were placed in regular media for 24 h and then reseeded on a 10-cm dish in media containing 1 μg/mL puromycin (for PLD1 knockout, or 1KO) or 5 μg/mL blasticidin (for PLD2 knockout, or 2KO). The cells were kept in the selective medium for 3 to 5 d until all of the cells on a plate seeded in parallel of nontransduced cells were dead. To eliminate remaining unedited cells in the PLD1 knockout population, the 1KO cells generated as described earlier were labeled with SPAAC-based IMPACT (33) for fluorescence-activated cell sorting (FACS) based on PLD activity. Briefly, cells were treated with azidopropanol (1 mM) and PMA (100 nM) in Tyrode’s-Hepes buffer for 30 min at 37 °C. After 3 rinses with PBS solution, cells were treated with BODIPY-bicyclononyne (1 μM) for 10 min at 37 °C. Cells were rinsed again 3 times with PBS solution and incubated in Tyrode’s-Hepes buffer (135 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 1 mg/mL glucose, 1 mg/mL BSA, 20 mM Hepes, pH 7.4) for 10 min at 37 °C to further rinse out unreacted fluorophore. The cells were then lifted with trypsin, and FACS was used to exclude the minority of cells with residual IMPACT labeling (i.e., the collected cells were those whose IMPACT fluorescence was comparable to negative controls wherein azidopropanol labeling was performed in the presence of PLDi [FIPI]). To generate PLD1/2 double knockout cells (DKO), the sorted PLD1 knockout cells (1KO) cells were transduced with the PLD2 sgRNA-containing LentiCRISPR virus-containing media, followed by blasticidin selection.
IMPACT Labeling with oxoTCO Alcohols for Live-Cell Imaging and Flow Cytometry Analysis.
HeLa cells (n = 150,000) or NIH 3T3 cells (n = 250,000) were seeded on 35-mm glass-bottom imaging dishes (MatTek) for 24 h before experiments. For PDGF stimulation in NIH 3T3 cells, cells were serum-starved by incubation in DMEM + 1% penicillin/streptomycin without serum for 18 h up to and including the transphosphatidylation step of the experiment. Rinses after transphosphatidylation were performed in full media. Where indicated, the cells were then transfected with the indicated plasmid using Lipofectamine 2000 per the manufacturer’s instructions. One day after seeding (or transfection, if any), cells were first treated with the indicated PLD inhibitor or DMSO in media for 30 min. The working concentrations of the stimuli were as follows: PDGF-BB (50 ng/mL), oxo-M (10 μM), and PMA (100 nM). The appropriate stimulus was then added to the media for the indicated period of time (0 to 60 min). Subsequently, freshly prepared (S)-oxoTCO–C1 (or the indicated oxoTCO alcohol; 3 mM) together with the respective stimulus and PLD inhibitor/DMSO in media (100 µL) were carefully added to cover the central glass well.
As a cautionary note, oxoTCOs are reported to have limited water stability (50). Therefore, all aqueous oxoTCO solutions (e.g., those in DMEM-containing media) were used within 20 min of their generation. For example, we dissolved oxoTCO in 200 µL of DMEM with PLD inhibitor/DMSO and respective stimulus and used it only for 2 consecutive replicates, rather than make a single stock solution at the beginning of the day for an entire day of experiments.
The dish was incubated for 5 min, the treatment media was aspirated, the cells were rinsed with PBS solution (1 mL) briefly, and the cells were then incubated in media (500 µL) for 1 min at 37 °C. The media was replaced with Tz–BODIPY (0.33 µM) in PBS solution (100 µL) for 1 min, aspirated, and replaced with 100 µL Tyrode’s-Hepes buffer. Cells were imaged immediately afterward. Multicolor images were obtained in 2-channel, line-switching mode. Z stacks were taken with 0.45-µm sectioning. For flow cytometry analysis, cells were instead seeded in 24-well plates (125,000 cells per well) and labeled as described here earlier. Following the final aspiration of Tz–BODIPY, cells were lifted with trypsin, transferred to 96-well plates, rinsed twice with cold PBS solution + 0.5% FBS by centrifugation at 500 × g, and analyzed by flow cytometry. At least 10,000 live cells were analyzed for each well, as determined by forward/side scatter analysis.
Real-Time IMPACT Imaging of PLD Activity.
For RT-IMPACT, i.e., time-lapse imaging, cells were labeled with (S)-oxoTCO–C1 and the appropriate stimulus as described in the previous section. Following the transphosphatidylation step and 1-min rinse with media (500 µL), the media was aspirated and replaced with 200 µL Tyrode’s-Hepes buffer onto the center of the glass well. The dish was then immediately put onto the microscope stage, and a time-lapse movie was initiated with Definite Focus engaged to maintain the correct focal plane for the duration of the movie (3-s interval between frames for a total of 3 min). Between the second and third frames of the movie, Tz–BODIPY (1 µM) in PBS solution (100 µL) was added to a final concentration of 0.33 μM. All time-lapse experiments were performed with at least 3 replicates. Image analysis for the localization of the RT-IMPACT signal was performed by exporting the third frame (9-s time point) as image files. All images were blinded, and categorization of the IMPACT fluorescence as PM (predominantly plasma membrane), intra (predominantly intracellular), or mix (combination of plasma membrane and intracellular) was carried out by an individual other than the individual who performed the image acquisition. Each image had ∼15 to 20 cells in view and was treated as a single biological replicate; each condition was repeated for the indicated number of biological replicates in the appropriate figure legend (n = 3–8). The final categorization was assembled into stacked bar graphs for data visualization.
Quantification of PM-to-ER Trafficking of Fluorescently Tagged Lipids.
This experiment was performed in a similar fashion compared with the RT-IMPACT time-lapse imaging described in the previous section, except that ER-Tracker Red and CellMask Deep Red Plasma Membrane Stain were added to the (S)-oxoTCO–C1 solution for the transphosphatidylation step. Following the rinse, IEDDA using Tz–BODIPY was carried out during a time-lapse movie acquisition as described in the previous section, with the following microscope settings: frames were acquired in a 3-channel, 2-track, frame-switching mode (track 1, 488 nm and 640 nm; track 2, 561 nm) at an interval of 8 s per time point (4 min total length of acquisition). The Pearson correlation coefficients between the 488-nm (IMPACT) and 561-nm (ER Tracker Red) channels were calculated for each time point and plotted against time, and a monoexponential regression fit was generated in Origin Pro-8 to give half-life (t1/2) of trafficking kinetics from the PM to the ER.
Statistics and Reproducibility.
All imaging experiments show representative images from experiments performed in at least 3 biological replicates on different days. Exact numbers of replicate experiments and sample sizes are provided in each figure legend. For experiments involving quantification of comparisons between more than 2 independent groups (Figs. 3 B–D, 7A, and 8A and SI Appendix, Figs. S9 A and B and S10 A and C), significance was calculated by using 1-way ANOVA followed by Games–Howell post hoc test (for samples of unequal variance). For experiments involving quantification of comparisons between groups with fractional data (i.e., fraction of PM, intra, and mix cells, which, by definition, sum to 1; Figs. 5F, 7C, and 8C and SI Appendix, Figs. S9C and S10B), significance was calculated by using a χ2 test for independence.
Supplementary Material
Acknowledgments
J.M.B. acknowledges support from the Arnold and Mabel Beckman Foundation (Beckman Young Investigator Award), the National Science Foundation (CAREER CHE-1749919), and the Alfred P. Sloan Foundation (Sloan Research Fellowship). D.L. and R.T. were supported by Cornell Fellowships, R.T. was additionally supported by a Funai Overseas Fellowship, and T.W.B. was supported by an NSF graduate research fellowship (DGE-1650441). This work made use of the Cornell University NMR Facility, which is supported in part by the NSF (CHE-1531632), and the Cornell University Biotechnology Resource Center Imaging Facility, which is supported in part by the NSF (DBI-1428922). We thank Joseph Fox and William Lambert (University of Delaware) for generously providing samples of oxoTCO reagents and for assisting with and advising on the synthesis of oxoTCO reagents, Alex Brown (Vanderbilt University) for providing isoform-selective PLD1 and PLD2 inhibitors (VU0359595 and VU0364739), Megan Gelsinger for assistance with statistical analysis, the Emr and Fromme laboratories for sharing equipment and reagents, Weizhi Yu for technical assistance, and members of the laboratory of J.M.B. for helpful discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903949116/-/DCSupplemental.
References
- 1.Giepmans B. N. G., Adams S. R., Ellisman M. H., Tsien R. Y., The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Oldach L., Zhang J., Genetically encoded fluorescent biosensors for live-cell visualization of protein phosphorylation. Chem. Biol. 21, 186–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty K., Veetil A. T., Jaffrey S. R., Krishnan Y., Nucleic acid-based nanodevices in biological imaging. Annu. Rev. Biochem. 85, 349–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J., Dodani S. C., Chang C. J., Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat. Chem. 4, 973–984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Su Y., Wang X., “Phosphatidic acid-mediated signaling” in Lipid-Mediated Protein Signaling (Springer Netherlands, Dordrecht, 2013), pp. 159–176. [DOI] [PubMed] [Google Scholar]
- 6.Selvy P. E., Lavieri R. R., Lindsley C. W., Brown H. A., Phospholipase D: Enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shulga Y. V., Topham M. K., Epand R. M., Regulation and functions of diacylglycerol kinases. Chem. Rev. 111, 6186–6208 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Kang D. W., Choi K. Y., Min S., Functional regulation of phospholipase D expression in cancer and inflammation. J. Biol. Chem. 289, 22575–22582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Frohman M. A., Cellular and physiological roles for phospholipase D1 in cancer. J. Biol. Chem. 289, 22567–22574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira T. G., Di Paolo G., Phospholipase D in brain function and Alzheimer’s disease. Biochim. Biophys. Acta. 1801, 799–805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson R. K., Frohman M. A., Physiological and pathophysiological roles for phospholipase D. J. Lipid Res. 56, 2229–2237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott S. A., et al. , Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5, 108–117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruntz R. C., Lindsley C. W., Brown H. A., Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacol. Rev. 66, 1033–1079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown H. A., Thomas P. G., Lindsley C. W., Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat. Rev. Drug Discov. 16, 351–367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frohman M. A., The phospholipase D superfamily as therapeutic targets. Trends Pharmacol. Sci. 36, 137–144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang J.-H., Lee C. S., Hwang D., Ryu S. H., Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lipid Res. 51, 71–81 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Hammond S. M., et al. , Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J. Biol. Chem. 272, 3860–3868 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Colley W. C., et al. , Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr. Biol. 7, 191–201 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Du G., et al. , Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 162, 305–315 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarri E., Pardo R., Fensome-Green A., Cockcroft S., Endogenous phospholipase D2 localizes to the plasma membrane of RBL-2H3 mast cells and can be distinguished from ADP ribosylation factor-stimulated phospholipase D1 activity by its specific sensitivity to oleic acid. Biochem. J. 369, 319–329 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laulagnier K., et al. , PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 572, 11–14 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Antal C. E., Newton A. C., Tuning the signalling output of protein kinase C. Biochem. Soc. Trans. 42, 1477–1483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallegos L. L., Newton A. C., Spatiotemporal dynamics of lipid signaling: Protein kinase C as a paradigm. IUBMB Life 60, 782–789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du G., et al. , Dual requirement for rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Mol. Biol. Cell 11, 4359–4368 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mozzicato S., Joshi B. V., Jacobson K. A., Liang B. T., Role of direct RhoA-phospholipase D1 interaction in mediating adenosine-induced protection from cardiac ischemia. FASEB J. 18, 406–408 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Everett P. B., Senogles S. E., D3 dopamine receptor signals to activation of phospholipase D through a complex with Rho. J. Neurochem. 112, 963–971 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Powner D. J., Wakelam M. J. O., The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS Lett. 531, 62–64 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Hughes W. E., Larijani B., Parker P. J., Detecting protein-phospholipid interactions. Epidermal growth factor-induced activation of phospholipase D1b in situ. J. Biol. Chem. 277, 22974–22979 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Kassas N., et al. , Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 292, 4266–4279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassas N., et al. , Genetically encoded probes for phosphatidic acid. Methods Cell Biol. 108, 445–459 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Zhang F., et al. , Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol. Cell. Biol. 34, 84–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bumpus T. W., Baskin J. M., A chemoenzymatic strategy for imaging cellular phosphatidic acid synthesis. Angew. Chem. Int. Ed. Engl. 55, 13155–13158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bumpus T. W., Baskin J. M., Clickable substrate mimics enable imaging of phospholipase D activity. ACS Cent. Sci. 3, 1070–1077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong L. H., Gatta A. T., Levine T. P., Lipid transfer proteins: The lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20, 85–101 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Yang Y., Lee M., Fairn G. D., Phospholipid subcellular localization and dynamics. J. Biol. Chem. 293, 6230–6240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown H. A., Henage L. G., Preininger A. M., Xiang Y., Exton J. H., Biochemical analysis of phospholipase D. Methods Enzymol. 434, 49–87 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Philip F., Ha E. E., Seeliger M. A., Frohman M. A., Measuring phospholipase D enzymatic activity through biochemical and imaging methods. Methods Enzymol. 583, 309–325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bumpus T. W., Liang F. J., Baskin J. M., Ex uno plura: Differential labeling of phospholipid biosynthetic pathways with a single bioorthogonal alcohol. Biochemistry 57, 226–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B., A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 41, 2596–2599 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Presolski S. I., Hong V. P., Finn M. G., Copper-catalyzed azide-alkyne click chemistry for bioconjugation. Curr. Protoc. Chem. Biol. 3, 153–162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agard N. J., Prescher J. A., Bertozzi C. R., A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Dommerholt J., et al. , Readily accessible bicyclononynes for bioorthogonal labeling and three-dimensional imaging of living cells. Angew. Chem. Int. Ed. Engl. 49, 9422–9425 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., Kusumi A., Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157, 1071–1081 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusumi A., Tsunoyama T. A., Hirosawa K. M., Kasai R. S., Fujiwara T. K., Tracking single molecules at work in living cells. Nat. Chem. Biol. 10, 524–532 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Nicolson G. L., The fluid-mosaic model of membrane structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim Biophys Acta 1838, 1451–1466 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Blackman M. L., Royzen M., Fox J. M., Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc. 130, 13518–13519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devaraj N. K., Weissleder R., Hilderbrand S. A., Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjug. Chem. 19, 2297–2299 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliveira B. L., Guo Z., Bernardes G. J. L., Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev. 46, 4895–4950 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Su W., et al. , 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75, 437–446 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert W. D., et al. , Computationally guided discovery of a reactive, hydrophilic trans-5-oxocene dienophile for bioorthogonal labeling. Org. Biomol. Chem. 15, 6640–6644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson J. C. T., Meimetis L. G., Hilderbrand S. A., Weissleder R., BODIPY-tetrazine derivatives as superbright bioorthogonal turn-on probes. Angew. Chem. Int. Ed. Engl. 52, 6917–6920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis J. A., et al. , Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg. Med. Chem. Lett. 19, 1916–1920 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavieri R. R., et al. , Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: Discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 53, 6706–6719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devaraj N. K., Hilderbrand S., Upadhyay R., Mazitschek R., Weissleder R., Bioorthogonal turn-on probes for imaging small molecules inside living cells. Angew. Chem. Int. Ed. Engl. 49, 2869–2872 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saheki Y., De Camilli P., Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 86, 659–684 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Schauder C. M., et al. , Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 510, 552–555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Falkenburger B. H., Jensen J. B., Hille B., Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J. Gen. Physiol. 135, 81–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falkenburger B. H., Jensen J. B., Hille B., Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135, 99–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plevin R., Cook S. J., Palmer S., Wakelam M. J., Multiple sources of sn-1,2-diacylglycerol in platelet-derived-growth-factor-stimulated Swiss 3T3 fibroblasts. Evidence for activation of phosphoinositidase C and phosphatidylcholine-specific phospholipase D. Biochem J 279, 559–565 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeo E. J., Kazlauskas A., Exton J. H., Activation of phospholipase C-γ is necessary for stimulation of phospholipase D by platelet-derived growth factor. J. Biol. Chem. 269, 27823–27826 (1994). [PubMed] [Google Scholar]
- 61.van Dijk M. C. M., Muriana F. J. G., de Widt J., Hilkmann H., van Blitterswijk W. J., Involvement of phosphatidylcholine-specific phospholipase C in platelet-derived growth factor-induced activation of the mitogen-activated protein kinase pathway in Rat-1 fibroblasts. J. Biol. Chem. 272, 11011–11016 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Shome K., Rizzo M. A., Vasudevan C., Andresen B., Romero G., The activation of phospholipase D by endothelin-1, angiotensin II, and platelet-derived growth factor in vascular smooth muscle A10 cells is mediated by small G proteins of the ADP-ribosylation factor family. Endocrinology 141, 2200–2208 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Hess J. A., Ross A. H., Qiu R. G., Symons M., Exton J. H., Role of Rho family proteins in phospholipase D activation by growth factors. J. Biol. Chem. 272, 1615–1620 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Pathre P., et al. , Activation of phospholipase D by the small GTPase Sar1p is required to support COPII assembly and ER export. EMBO J. 22, 4059–4069 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanematsu F., et al. , Phosphatidic acid-dependent recruitment and function of the Rac activator DOCK1 during dorsal ruffle formation. J. Biol. Chem. 288, 8092–8100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bumpus T. W., Baskin J. M., Greasing the wheels of lipid biology with chemical tools. Trends Biochem. Sci. 43, 970–983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J., Sˇečkutė J., Cole C. M., Devaraj N. K., Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem. Int. Ed. Engl. 51, 7476–7479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erdmann R. S., et al. , Super-resolution imaging of the Golgi in live cells with a bioorthogonal ceramide probe. Angew. Chem. Int. Ed. Engl. 53, 10242–10246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson A. D., Bewersdorf J., Toomre D., Schepartz A., HIDE probes: A new toolkit for visualizing organelle dynamics, longer and at super-resolution. Biochemistry 56, 5194–5201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiapparino A., Maeda K., Turei D., Saez-Rodriguez J., Gavin A. C., The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling. Prog. Lipid Res. 61, 30–39 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Wong L. H., Cˇopič A., Levine T. P., Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 42, 516–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prinz W. A., Bridging the gap: Membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205, 759–769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reinisch K. M., De Camilli P., SMP-domain proteins at membrane contact sites: Structure and function. Biochim Biophys Acta 1861, 924–927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brailoiu E., et al. , Choline is an intracellular messenger linking extracellular stimuli to IP3-evoked Ca2+ signals through sigma-1 receptors. Cell Rep. 26, 330–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.