Significance
In most animals, offspring sex can be controlled by either genetic or environmental sex determination (GSD or ESD). As an exception, the sex of Daphnia offspring is subject to both GSD and ESD. Normally, female Daphnia offspring are produced in benign conditions, while males are produced in crowded conditions or under hormonal stimuli. However, some females have lost the ability to produce male offspring even in the face of strong hormonal stimuli. Here, we show that the loss of male-producing ability is controlled by a single genetic factor. The putative gene is located downstream of the methyl-farnesoate signaling pathway that controls male induction in response to external stimuli in Daphnia.
Keywords: Daphnia pulex, sex determination, nonmale producing, methyl farnesoate
Abstract
Daphnia normally reproduce by cyclical parthenogenesis, with offspring sex being determined by environmental cues. However, some females have lost the ability to produce males. Our results demonstrate that this loss of male-producing ability is controlled by a dominant allele at a single locus. We identified the locus by comparing whole-genome sequences of 67 nonmale-producing (NMP) and 100 male-producing (MP) clones from 5 Daphnia pulex populations, revealing 132 NMP-linked SNPs and 59 NMP-linked indels within a single 1.1-Mb nonrecombining region on chromosome I. These markers include 7 nonsynonymous mutations, all of which are located within one unannotated protein-coding gene (gene 8960). Within this single gene, all of the marker-linked NMP haplotypes from different populations form a monophyletic clade, suggesting a single origin of the NMP phenotype, with the NMP haplotype originating by introgression from a sister species, Daphnia pulicaria. Methyl farnesoate (MF) is the innate juvenile hormone in daphnids, which induces the production of males and whose inhibition results in female-only production. Gene 8960 is sensitive to treatment by MF in MP clones, but such responsiveness is greatly reduced in NMP clones. Thus, we hypothesize that gene 8960 is located downstream of the MF-signaling pathway in D. pulex, with the NMP phenotype being caused by expression change of gene 8960.
Gynodioecy is a breeding system frequently found in plants, whereby females and hermaphrodites coexist in the same population. Such a system may be an evolutionary intermediate between pure hermaphroditism and dioecy (1–3). Genetic analyses have revealed that gynodioecy is often controlled by cytonuclear interactions between mitochondrial sterility mutations and nuclear restorer genes (4, 5), although a few plant species are thought to experience full nuclear control (6–8). In contrast to plants, very few animal species are known to be gynodioecious (9, 10). However, a form of gynodioecy is found in water flea Daphnia, wherein some individuals are unable to produce males, while others produce both sons and daughters (11–14).
Daphnia normally reproduce by cyclical parthenogenesis, with extended periods of parthenogenesis interspersed with sexual resting-egg production, generally on a yearly cycle (15). Parthenogenetic eggs can develop into either females or males, as sex determination is usually environmentally induced, with emerging evidence also suggesting a genetic component (14, 16–19). Females are produced in benign conditions, while males are more likely to be produced in conditions favoring diapause, such as crowding (15) or short photoperiod (20). Such environmental effects seem to be mediated by maternal hormone stimuli acting on developing oocytes (21). Male production can also be induced artificially by adding the hormone methyl farnesoate (MF) to the culture medium (20).
However, some females have lost the ability to produce males in nature and even in the face of strong hormonal stimuli (12, 13, 18). Crosses between nonmale-producing (NMP) females and males from male-producing (MP) females always generate close to 1:1 ratios of NMP and MP offspring, whereas almost all offspring of MP × MP crosses have MP phenotypes (11, 18), suggesting a dominant allele at a single locus (or several linked loci) underlying the NMP phenotype. Specifically, all NMP clones are hypothesized to be heterozygous Mm (or WZ) at the locus conferring the NMP phenotype, whereas MP clones are mm (or ZZ) homozygotes (18), with the MM (or WW) haplotype being extremely rare, if not absent entirely, as in W/Z sex-determination systems.
An initial effort made by Reisser et al. (14) to identify the locus conferring the NMP trait in Daphnia magna revealed a nonrecombining region spanning a 3.6-Mb segment on linkage group 3, harboring 13 markers and 36 associated SNPs. Furthermore, they provided evidence for a reduced recombination rate in this region, implying a potential supergene in control of the NMP phenotype (14). However, due to the poor quality of the D. magna genome assembly and an insufficient fraction (6%) of the genome sequenced from just a single population, they were unable to identify the genes in this large region likely to be functional determinants of the NMP phenotype.
To better understand the genetic basis of the NMP phenotype in Daphnia, we used the newly published Daphnia pulex genome assembly (22) and generated population-genomic data from 5 D. pulex populations to characterize the nonrecombining region and identify candidate genes. Through whole-genome comparison of 67 NMP clones and 100 MP clones, we identified 132 SNPs and 59 indels unique to all NMP clones, all linked within a 1.1-Mb region, and absent from all MP clones. We further found that one gene contains all 7 of the nonsynonymous SNPs in this entire nonrecombining region. For this candidate gene, NMP haplotypes from different populations form a monophyletic clade, consistent with the expectation for a causal factor with a single origin. This candidate gene is sensitive to treatment by MF in MP clones, but such responsiveness is greatly reduced in NMP clones. Therefore, we predict the product of the candidate gene to be located downstream of the MF signaling pathway.
Results
Markers Specific to NMP Clones in Daphnia pulex.
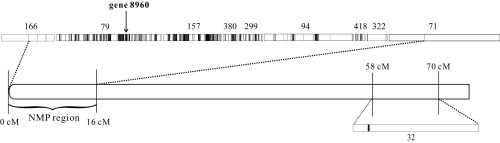
To reveal the difference between male-producing (MP) and nonmale-producing (NMP) clones, we initially relied on whole-genome sequences from 40 MP and 39 NMP females from a specific D. pulex population collected from Kicka Pond, located near Danville, IL (hereafter KAP; ref. 23), and performed genome-wide association mapping using SNPs and indels. We found 1,635 SNPs (from 16 scaffolds) to be present in all NMP clones and absent from all MP clones (Table 1). Of these, 1,429 SNPs (87%) are contained within a region between 0 and 16 centiMorgans (cM) on the genetic map of chromosome I (Fig. 1). A similar pattern is observed for small indel markers, 873 (71%) out of the 1,223 NMP-specific indels are contained within the same region defined by SNP markers (SI Appendix, Fig. S1). All of the SNP marker loci and 69% of the indel loci are heterozygous in NMP clones. This large span of fully linked markers suggests a shared haplotype region across all NMP clones, experiencing little or no recombination. The NMP region spans several scaffolds (Fig. 1), the lengths of which sum to 2.1 Mb, although 80% of the SNP markers and 66% of the indel markers are confined to the middle 2 scaffolds (scaffolds 79 and 157). A few markers are located on scaffold 32, which has been placed in the region between 58 and 70 cM on chromosome I, and may reflect assembly errors. The small remaining fraction (9.5%) of SNP markers located on other chromosomes may simply be false-positive associations resulting from small sample size.
Table 1.
Distribution of NMP-specific markers in the KAP population and markers shared by all populations
| Scaffold | No. of SNPs | No. of indels | Scaffold length (kb) | Chromosome |
| 79 | 582 | 426 | 585 | I |
| 157 | 566 | 153 | 324 | I |
| 94 | 93 | 105 | 498 | I |
| 299 | 82 | 61 | 133 | I |
| 380 | 68 | 11 | 79 | I |
| 418 | 24 | 14 | 56 | I |
| 32 | 50 | 17 | 864 | I |
| 322 | 4 | 44 | 113 | I |
| 71 | 2 | 24 | 619 | I |
| 126 | 27 | 9 | 413 | VI |
| 75 | 41 | 28 | 608 | VII |
| 23 | 40 | 8 | 924 | XI |
| Shared | ||||
| 157 | 79 | 6 | 324 | I |
| 79 | 45 | 51 | 585 | I |
| 94 | 8 | 2 | 498 | I |
For the KAP population, only scaffolds with >20 SNP or indel markers are shown.
Fig. 1.
Distribution of NMP-specific SNPs on chromosome I in the KAP population. Scaffolds (given as numbers) are scaled to their length and vertical black bars indicate NMP-specific SNPs on the scaffolds. Because of potential uncertainties in the genetic map, we only used uniquely mapped scaffolds placed on the chromosomes based on the D. pulex genetic map (22, 46). Gene 8960 is the candidate gene underlying the NMP phenotype.
To explore beyond the SNPs and small indels, we evaluated whether the NMP clones have gained or lost genomic fragments or have unique symbionts compared with MP clones. Because our clones are not treated with antibiotics, if there are symbionts, they should be represented in the raw reads from the genome sequences. To test this, we independently generated composite NMP and MP assemblies by pooling reads from the corresponding clones. Comparison of the 2 assemblies did not reveal any contigs/scaffolds unique to either MP or NMP clones. We also searched for transposable-element (TE) insertions using a preconstructed Daphnia TE library (24), but found no TEs unique to either group. These results suggest that NMP clones do not have unique shared, large genomic insertions/deletions or symbionts with causal contributions to the NMP trait.
Heterozygosity and Linkage Disequilibrium between NMP and MP Clones.
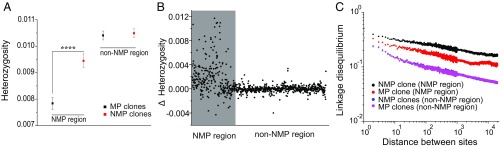
To understand how the NMP region is differentiated between NMP and MP clones, we compared the average levels of heterozygosity between these 2 phenotypes. The NMP clones have a higher level of heterozygosity than MP clones in the NMP region (0.0094 vs. 0.0078, Mann–Whitney U test, P < 0.0001), whereas no difference was observed for regions on the same chromosome outside of the NMP region (0.0105 vs. 0.0104, P = 0.72; Fig. 2 A and B). To evaluate whether excess heterozygosity in NMP clones exists in other chromosomes, we extended the comparison with all 12 chromosomes, but no significant differences were found (SI Appendix, Fig. S2). However, both NMP and MP clones have significantly reduced heterozygosity levels in the NMP region compared to non-NMP regions, ∼25% in the case of the MP clones (Fig. 2A). A 25% reduction is the expectation in the idealized situation when a population has equal numbers of Mm and MM clones, as the effective population size for the M-bearing chromosome would be 75% of that for a normal autosome, a situation that cannot be ruled out.
Fig. 2.
Comparisons of heterozygosity and linkage disequilibrium (LD) on chromosome I within groups of NMP and MP clones. (A) Comparisons of average heterozygosity (mean ± SE; ****P < 0.0001). (B) Difference in average heterozygosity (Δ Heterozygosity = HNMP − HMP) between NMP and MP clones for the NMP and non-NMP regions on chromosome I (defined in Fig. 1; using a 10-kb window size for calculation of average ΔH). X-axis is the summed length of scaffolds (Mb). (C) Measures of LD in NMP and MP clones for the NMP and non-NMP regions; population-level linkage disequilibrium, measured as r2, is calculated by using the maximum likelihood method of Maruki and Lynch (47).
We then compared the profile of linkage disequilibrium (LD), measured as r2, between NMP and MP clones on chromosome I. LD decays more slowly in NMP clones than in MP clones in the NMP region, whereas no difference is found for the non-NMP region (Fig. 2C). In addition, both NMP and MP clones have higher LD in the NMP region compared with non-NMP regions (P < 0.0001, and P < 0.001), reflecting the divergence of the M haplotype.
NMP Clones in Other Populations.
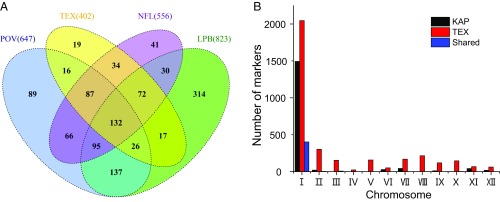
Population-genomic data for 96 clones from each of 4 other D. pulex populations (POV, TEX, NFL, and LPB) permitted a broader examination of the incidence of KAP-based NMP markers, revealing a clear set of marker associations in these populations as well, with a small fraction of clones (POV: 8; TEX: 23; NFL: 2; LPB: 8) having >41% and >46% of the KAP-specific M-chromosome SNP and indel markers, respectively, and the remaining clones having <6% and <9% of the putative SNP and indel markers. All of the marker-rich clones still alive in the laboratory (POV: 8; TEX: 10; NFL: 2; LPB: 8) were experimentally verified to be NMP clones. We also assayed a random set of 15 clones with <6% of the KAP-based NMP markers from each population, and found all 60 to be capable of male production. These results show that the NMP markers identified from the KAP population, as a group, are strongly associated with the NMP phenotype in other populations. Eighty-two percent of the KAP SNP markers and 41% of the indel markers also appear as unambiguous NMP markers for at least one other population, and many are shared by 2 or more populations, but most are not universally informative (Fig. 3A and SI Appendix, Fig. S3).
Fig. 3.
(A) Venn diagram showing the number of KAP-based SNP markers appearing in populations LPB, NFL, POV, and TEX. Just 132 NMP SNP markers are shared by all NMP clones in all populations. (B) Chromosome-level distribution of NMP markers in the KAP and TEX populations.
To understand if other populations share the same NMP region as KAP, we performed a de novo search for NMP-specific SNP markers for population TEX. For TEX clones still alive in the laboratory and with genome sequence data, we first predicted phenotypes using the KAP-based NMP markers, and experimentally verified all predicted TEX NMP clones (10 in total) along with an additional 15 random TEX clones (without obvious KAP markers). With this retrospective data set, we then performed an independent de novo marker-association study for this population and found a total of 3,532 markers, 2,048 (58%) of which are located on chromosome I (Fig. 3B). The lower percentage of TEX-specific markers on chromosome I in the TEX population compared with KAP is likely just a consequence of a higher number of false-positive associations resulting from a much smaller sample size for NMP clones in TEX compared with KAP (10 vs. 39). Of the 2,048 TEX markers on chromosome I, 1,923 (94%) are contained within the NMP region identified from KAP, although only 402 markers match those from KAP.
In principle, a small number of overlapping markers among all populations should narrow down the causal region associated with the NMP phenotype, as recombination will randomly remove spurious associations in different populations. Thus, to further refine the NMP region, we searched for markers shared across all populations, narrowing the remaining pool to just 132 SNP markers and 59 indel markers, based on the comparison of 67 NMP clones and 100 MP clones across populations (Fig. 3A, SI Appendix, Fig. S3, and Dataset S1). These shared NMP markers are located on the middle 3 scaffolds of the NMP region on chromosome I (Table 1), spanning a 1.1-Mb region (from the first marker on scaffold 79 to the last marker on scaffold 94).
Genes and Mechanism Underlying the NMP Phenotype.
To gain further insight into the genetic basis of the NMP phenotype, we mapped the 132 shared SNP markers and 59 shared indel markers to the D. pulex genome annotation (22). Of the SNP markers, 99 were in intergenic regions, 9 in introns, 5 in 5′ UTRs, 4 in 3′ UTRs, and 15 in coding regions. Of the 15 SNP markers in coding regions, 8 are synonymous, and 7 are nonsynonymous mutations. These 15 SNP markers mapped to 8 genes, one of which (gene 8960) contains all of the markers associated with nonsynonymous substitutions (Table 2 and Dataset S1). We also found 6 indel markers located in the gene region, with 3 in introns, 1 in exons, 1 in 5′ UTRs, and 1 in 3′ UTRs (Dataset S1). Five genes were affected by indels, 3 of which overlapped with those involving SNP markers (genes 8960, 12748, and 8948). Of these 3, only gene 8960 has an indel in coding sequence (a 15-bp deletion in NMP clones), and it also has an indel in the 3′ UTR (Dataset S1). Although this gene is not annotated, RNA-seq data from an MP clone cover the full length (22), confirming that it is an expressed protein-coding gene. Although not found in non-Daphnia species, this gene exists in Daphnia pulicaria (25), Daphnia obtusa (22), and D. magna (26), suggesting that it may be a Daphnia-specific gene, although a more encompassing crustacean clade cannot be ruled out.
Table 2.
NMP markers causing nonsynonymous amino acid (AA) substitutions
| Scaffold | Site | Nucleotide (MP-NMP) | AA (MP-NMP) |
| 79 | 372600 | A→G | N→D |
| 79 | 372617 | A→C | K→A |
| 79 | 372618 | A→G | K→A |
| 79 | 372648 | A→C | T→P |
| 79 | 373001 | T→C | F→S |
| 79 | 373231 | C→T | T→I |
| 79 | 373252 | T→A | F→Y |
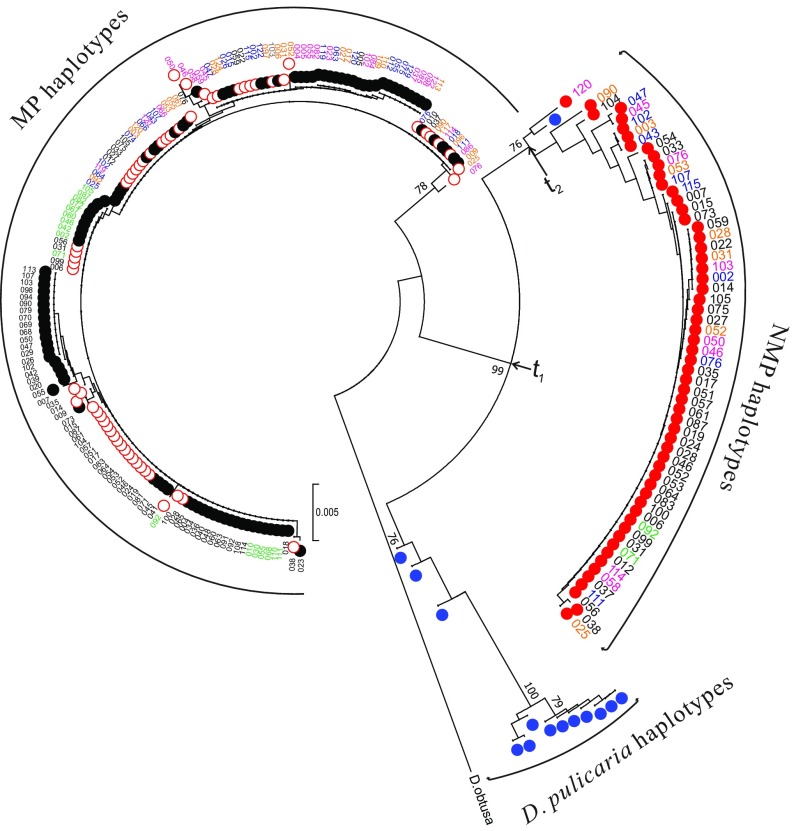
To understand the evolutionary relationships of the allelic variants of this gene in different D. pulex populations, we constructed genealogies using phased haplotypes for gene 8960 from NMP clones. All marker-linked NMP haplotypes from different populations form a monophyletic clade (Fig. 4), confirming a single origin of the NMP phenotype. Moreover, the NMP alleles within this clade are more closely related to those from D. pulicaria than to nonmarker haplotypes from D. pulex, suggesting a D. pulicaria origin of the NMP haplotype (Fig. 4). To determine if other genes in the NMP region (with or without prediscovered SNP or indel markers) show the same phylogeny as gene 8960, we generated phylogenetic trees for all 87 genes in the NMP region. None of these trees cluster NMP marker-linked haplotypes across all populations, i.e., they are all polyphyletic (Dataset S1). Moreover, none of the other marker-containing genes have NMP haplotypes clustering with D. pulicaria, suggesting that either gene 8960 is the only gene with haplotypes from D. pulicaria or that the entire 1.1-Mb NMP region came from D. pulicaria, with subsequent mutation, introgression, and/or recombination within D. pulex breaking down linkage disequilibrium in regions flanking gene 8960.
Fig. 4.
Nucleotide neighbor-joining tree for phased haplotypes of gene 8960. Each NMP clone was phased into 2 sequences, the marker-linked (NMP-associated) haplotype (solid red circle) and the nonmarker haplotype (hollow red circle). MP clones are not phased and have only one sequence (black circle). D. pulicaria clones are marked with blue circles. Clones from each population are color coded by their names (KAP: black; NFL: green; POV: blue; LPB: orange; TEX: pink). The tree is rooted by D. obtusa. Five hundred bootstraps were run, and bootstrap values ≥70% are shown. The tree was constructed using the neighbor-joining method incorporated into MEGA7 (49), and pairwise deletion was applied to deal with gaps and missing data.
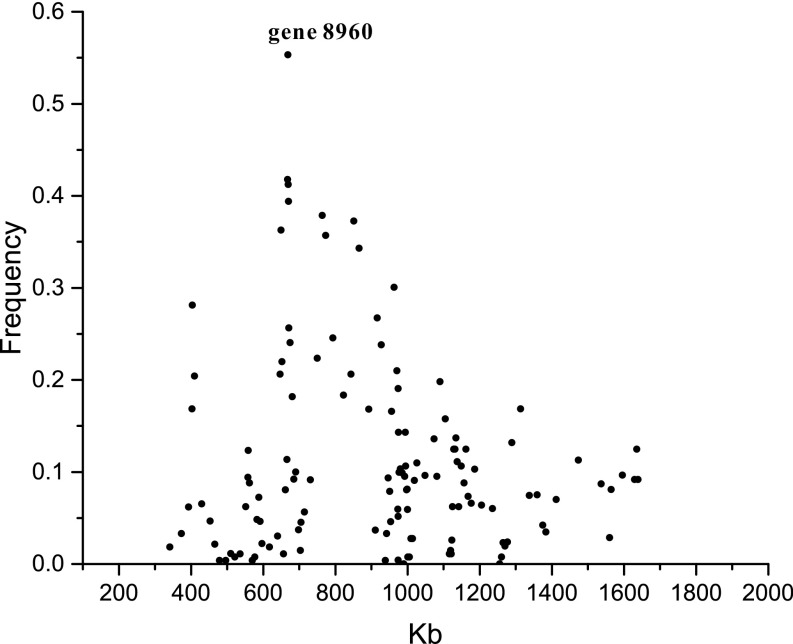
To further evaluate these possibilities, we determined the frequencies of the 132 D. pulex NMP markers in a collection of 14 genomes from D. pulicaria clones evaluated by Xu et al. (25). This analysis shows a gradual decrease of NMP marker frequencies compared with that in gene 8960 in the regions flanking gene 8960 in D. pulicaria (Fig. 5), suggesting that the flanking region also came from D. pulicaria, but that the marker associations with the latter have dissipated over time, due either to recombination since arrival within D. pulex or simple loss of linkage disequilibrium within D. pulicaria with physical distance from the focal locus.
Fig. 5.
Frequency of NMP markers in 14 D. pulicaria clones. The x-axis is the location on chromosome I, and the y-axis is the average frequency of the NMP-specific SNPs within 20-kb windows with step sizes of 10 kb used to shift the window.
Based on the phylogeny of the NMP haplotypes from gene 8960 (Fig. 4), the divergence time (in generations) between the NMP haplotypes and D. pulicaria (t1) can be estimated from the average synonymous nucleotide divergence (ds) between the NMP phased haplotypes and D. pulicaria haplotypes, using ds = 2·μ·t1, where μ = 5.85 × 10−9 per site per generation is the mutation rate obtained from a mutation-accumulation experiment in D. pulex (27). Assuming an average of ∼5 generations per year, the divergence time is estimated to be ∼0.85 Mya. We also estimated the minimum age of the entire NMP haplotype radiation (t2, Fig. 4). Assuming a single origin of the NMP haplotypes, the synonymous nucleotide diversity among NMP haplotypes must have been acquired after the origin, and the base of the NMP haplotype radiation can be estimated from the average synonymous nucleotide diversity between the most distant NMP haplotype (TEX120) and the remaining NMP haplotypes, leading to an estimated minimum age for the NMP haplotype of ∼0.19 My. Thus, we conclude that the NMP-conferring haplotype has been present in D. pulex for at least 0.2 My, with its progenitor allele separating from the remaining D. pulicaria haplotypes ∼0.8 Mya, although it is unknown whether the current NMP trait was present at that time.
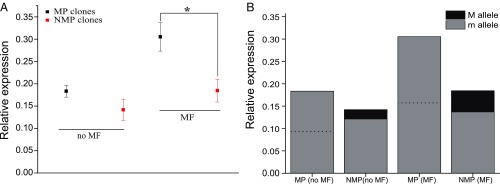
Environmental sex determination in D. pulex is dependent on the synthesis of MF, which can induce the production of male offspring (20, 21). In D. pulex, MF is generated by methylation of farnesoic acid, which is catalyzed by juvenile hormone acid O-methyltransferase (JHAMT) (20). Because the loss of male-producing ability in NMP clones cannot be rescued by adding MF, we hypothesized that the factors conferring the NMP phenotype act downstream of the MF-signaling pathway, i.e., involve the response to MF. To test this hypothesis, we compared the relative expression of gene 8960 in 5 MP and 5 NMP clones with and without MF treatment at the MF-sensitive period for male production (20, 21). Without MF treatment, NMP clones have an expression level not significantly different from that in MP clones (Fig. 6A). However, after treating both groups of clones with MF, gene 8960 showed a significant increase of expression in MP clones, but no significant change in NMP clones (Fig. 6A). Thus, differences in expression levels of this gene in MP and NMP clones in response to MF is due to reduced expression response in NMP clones.
Fig. 6.
Expression levels of gene 8960 relative to α-tubulin. (A) Relative expression (mean ± SE) of gene 8960 in MP and NMP clones with and without MF. (B) Expression of D. pulicaria-derived allele M and D. pulex allele m in NMP clones; dashed lines represent 50% of the relative expression.
To check if genes near gene 8960 are also affected by MF, we quantified the expression level of its nearest flanking genes (genes 8947 and 8972) in MP and NMP clones with and without MF treatment. Gene 8972 does not respond to MF treatment in either MP or NMP clones, while gene 8947 showed significantly increased expression in both MP and NMP clones when treated with MF (SI Appendix, Table S1). This response is consistent with the absence of NMP-specific markers in gene 8947, but also suggests that gene 8947 is influenced by the MF-signaling pathway but likely upstream of gene 8960. Finally, we quantified the expression of 2 additional genes (8948 and 12748) with both NMP-specific SNP and indel markers, neither of which responded to MF treatment (SI Appendix, Table S1).
To further understand how the D. pulex (m) and D. pulicaria derived allele (M) contribute to the expression difference, we quantified allele-specific expression in NMP clones. Without MF, the normalized expression level of D. pulex allele m in NMP clones is not significantly different from the midpoint of m in MP (mm) clones (Fig. 6B). When treated with MF, the expression of D. pulex allele m in NMP clones is also close to 50% of the total expression in MP clones (Fig. 6B). Because MP clones have 2 m alleles, assuming those 2 copies have the same expression level, this indicates that the expression of the m allele is equivalent in MP and NMP backgrounds, and that the NMP phenotype is likely caused by the decreased response of the M allele to MF. Our results suggest that the M allele is under cis-regulation. Consistent with this view, we found 7 NMP SNP markers located within a 1.2-kb region of gene 8960 (4 upstream and 3 downstream) and 1 indel marker located in the 3′ UTR of gene 8960 (Dataset S1), potentially affecting the expression of this gene.
Finally, if fully penetrant, the M allele of gene 8960 is expected to be always cotransmitted with mitochondrial sequences (mtDNA) because both are only maternally transmitted. However, the genealogy of complete mtDNA haplotypes from NMP clones interlaces phylogenetically with those from MP clones and does not associate with any specific population (Fig. 7). This suggests that either Mm individuals occasionally produce functional males or that mm males occasionally transmit mitochondria when mating with Mm females. Moreover, some D. pulicaria clones contain pulex-derived mitochondria, providing further evidence of historical gene flow between these 2 species (28–30). In addition, these observations indicate that mtDNA is not involved in the NMP phenotype, contrary to situations in plants (4, 5).
Fig. 7.
Nucleotide neighbor-joining tree for complete mitochondrial sequences from 5 populations of D. pulex, along with sequences from D. pulicaria isolates. Five hundred bootstraps were performed and bootstrap values ≥70% are shown. MP: male-producing clones (black circles); NMP: nonmale-producing clones (red circles); D. pulicaria sequences are marked with blue circles. Clones from each population are color coded by their names (KAP: black; NFL: green; POV: blue; LPB: orange; TEX: pink).
Discussion
Although the exact molecular mechanism remains to be determined, these population-genomic analyses support the idea that we have located the gene, and a dominant allelic variant of it, that prevents offspring from embarking on a developmental pathway to the male phenotype. Given that 1) only small fraction of NMP-associated markers is shared across all D. pulex populations; 2) these 132 SNP markers and 59 indel markers are confined to a narrow genomic region; and 3) all of the marker-associated amino acid substitutions are concentrated in a single gene with a monophyletic distribution, our results are supportive of a single origin for the nonmale-producing trait in D. pulex.
The NMP Phenotype in Other Daphnia Species.
The genealogy of the M variant of gene 8960 suggests that the NMP haplotype is derived from D. pulicaria, a sister species of D. pulex that also has NMP clones (13). All 7 nonsynonymous mutations in gene 8960 of the NMP D. pulex haplotypes also appear in genome sequences of D. pulicaria. Of these 7 informative sites, 5 harbor the same polymorphisms in D. pulicaria as in NMP D. pulex clones, whereas the remaining 2 exhibit only the NMP marker SNP in D. pulicaria individuals. Because it is unclear whether the D. pulicaria clones from Xu et al. (25) have NMP phenotypes, we are unable to draw conclusions as to which markers may be NMP specific, as opposed to being D. pulicaria specific but not relevant to NMP in this species. A search for the nonsynonymous D. pulex NMP markers in the single genome available for the slightly more distant D. obtusa yielded no such cases. Six of the 7 nonsynonymous NMP marker sites have the MP D. pulex alleles in D. obtusa and one has the NMP allele in D. obtusa.
It is worth noting that this is the second example of an introgression of a dominantly acting genetic factor from D. pulicaria having a dramatic effect on the breeding system of D. pulex. We have previously shown that obligately parthenogenetic (OP) lineages of D. pulex have arisen by the spread of a sex-limited meiosis suppressor derived from D. pulicaria (31). Because this genetic factor prevents meiosis in females, but not in environmentally induced males, when males produced by OP clones backcross to mm sexual (cyclically parthenogenetic, CP) individuals, the resultant progeny are expected to be 50% OP and 50% CP (owing to the segregation of the single dominant factor for OP), and in both cases still capable of producing males. Backcrosses of OP clones to Mm CP clones are still expected to yield 50% OP and 50% CP progeny, but because the meiosis-suppressor resides on a different chromosome than the NMP factor, half of both types of offspring would be expected to be NMP, which implies the production of 25% progeny that are both OP and dead ends with respect to gene flow.
The NMP trait also exists in D. magna (18), a species thought to have diverged from D. pulex 150–200 Mya (32, 33). To understand if the NMP phenotypes in these 2 species share the same mechanism, we searched for D. pulex gene 8960 in the D. magna genome assembly obtained from wfleabase (http://wfleabase.org/), but did not find any homologous region. This may be a result of the poor quality of the D. magna genome assembly. To help avoid this problem, we searched for the coding sequence of D. pulex gene 8960 in the transcriptome assembly of D. magna from 12 environmental stressors (26) and found an expressed ortholog of gene 8960. Although the D. magna orthologous region contains none of the D. pulex NMP-specific nonsynonymous mutations, because it is unclear whether the genotype used for the D. magna transcriptome assembly is an NMP clone, we are unable to tell whether the D. pulex NMP markers apply to D. magna. We also checked RAD-seq read data from MP and NMP D. magna (14), but none mapped to gene 8960, indicating that this gene is not covered by existing RAD-seq. Thus, although 3 species of Daphnia are known to have NMP genotypes, it remains unclear whether they share the same mechanism for the NMP trait.
Mitochondrial Haplotypes of NMP Clones.
Daphnia is an animal with mixed sex determination, with male production determined by both environmental cues and genetic factors (13, 18). In plants, a similar nonmale-producing trait is often controlled by the interaction between a mitochondrial variant and nuclear sex-determining gene (4, 5). However, our results show that D. pulex mitochondrial sequences (mtDNA) from NMP clones are discordant with the allelic genealogy of gene 8960 (Fig. 7), which is completely concordant with the presence of the NMP phenotype. This suggests that the NMP phenotype in D. pulex is under pure nuclear genetic control. The fact that the mtDNA genealogy is polyphyletic with respect to the NMP/MP phenotypes (Fig. 7) suggests either some paternal leakage of mitochondria, incomplete penetrance of the NMP genotype, or multiple introgressions of NMP alleles at gene 8960 from the same clade of D. pulicaria alleles (which would be necessary to account for the monophyly of NMP alleles; Fig. 4).
Maintenance of the NMP Phenotype within Populations.
The NMP mating system in D. pulex is similar to that of gynodioecy in plants, where some females only produce females, and others produce both males and females (1, 3, 34). The usual explanation for such a mating system invokes a single dominant genetic factor controlling the phenotype, with pollen-producing (MP) genotypes being homozygous (mm) and male-sterile (NMP) genotypes being heterozygous (Mm) (1). Consistent with this hypothesis, we found a single, effectively biallelic locus determining the NMP/MP phenotype, with all NMP clones being heterozygous.
Under such a system, an M allele can invade a population made up entirely of mm individuals, by positive selection, provided its carriers (the Mm NMP individuals) have some intrinsic advantage (e.g., avoidance of inbreeding depression; 2, 34, 35). Assuming NMP and MP female Daphnia produce the same number of female offspring parthenogenetically, then very strong inbreeding depression and a high rate of selfing are required for NMP clones to spread in the population (2). If there is no selfing or inbreeding depression, in order for the NMP clones to spread, female offspring production by NMP clones must be more than doubled to compensate for the loss of male-producing ability (2). The most favorable conditions would be fairly high levels of selfing and inbreeding depression and an increased female production by NMP clones (2, 11), and Daphnia do exhibit inbreeding depression (36–38), although the amount of intraclonal mating in natural populations is unknown.
Because half of the genes in sexually produced offspring derive from fathers and mothers, when MP females are rare in the population, at the time of sex all male-derived genes will be concentrated in the small fraction of MP genotypes, endowing them with high relative fitness, thereby increasing the frequency of mm genotypes in the following generation. On the other hand, if there is a surplus of males when the frequency of mm genotypes is high, the NMP (Mm) phenotype may be advantageous owing to the lack of wastage on male production. Under this simplest of models, negative frequency-dependent fitness is expected to lead to an equilibrium frequency of the Mm genotype, the exact frequency depending on the relative fitnesses of the mm and Mm haplotypes before sexual reproduction (11).
Future Plans.
The gene underlying the nonmale-producing trait in D. pulex has 2 alleles, which are homozygous (mm) in MP clones and heterozygous (Mm) in NMP clones. The lower expression level of this locus in NMP clones is largely caused by decreased expression of the M allele, particularly in response to male-inducing hormone. Further confirmation that the NMP trait is determined by just the expression level of gene 8960 can be obtained by decreasing the overall expression of gene 8960 in MP clones, e.g., RNA interference. If the phenotype does not change after expression modification, then the trait may be specified by the protein variant coded by the M allele. Testing this will require regulatory- or coding-region modifications of Mm clones to mm (and ideally MM) by gene transformation and likewise a conversion of mm in MP clones to Mm. There remains the additional, potential issue of comparing noncoding RNAs between the 2 phenotypes, as 75% of the NMP markers are located in noncoding regions.
Methods and Materials
Sample Preparation and Sequencing.
Population samples were collected in the spring of 2013 or 2014 from temporary-pond populations distributed in the Midwest and eastern portions of North America, mostly in the United States (SI Appendix, Table S2 and Fig. S4). As in our prior work (23), to maximize the likelihood that each isolate would represent a hatchling from a unique resting egg, early adults were collected from the water column before reproduction had occurred. We then clonally expanded the individual isolates and sequenced DNA as described in Lynch et al. (23).
Test for Male Reproduction.
Male production was induced by adding the hormone MF. Adult females for each clone were isolated, placed into 50-mL tubes containing 30 mL culture medium and kept at 20 °C with 120,000 cells/mL of the single-celled alga Scenedesmus obliquus as food. Molted adult females were transferred to medium containing MF at a concentration of 400 nM (changed daily) until the next molt. Because oocyte development in female D. pulex starts after each adult molt and eggs are released after the next molting stage, adult females were exposed to MF for the entire period of zygote development, which ensures that the sex of next generation is determined by MF. We examined the sex of juvenile Daphnia from the second molt using a dissection microscope. Males can be visually distinguished from females based on the enlarged antennules and flattened ventral carapace margin. Five adult females were tested for each clone, and clones that did not produce any male offspring were classified as NMP clones. A total of 67 NMP clones and 100 MP clones were assayed in our experiments (SI Appendix, Table S3).
Read Mapping and Screen for NMP Markers.
First, we trimmed adapter sequences from the sequence reads by applying Trimmomatic (version 0.36) (39) with parameters: HEADCROP:3 ILLUMINACLIP:adapter.fa:2:30:10:2 SLIDINGWINDOW:4:15 MINLEN:30. Then, adapter-trimmed sequence reads were mapped to the D. pulex assembly (22) with Novoalign (version 3.02.11) (http://www.novocraft.com/) with the “-r None” option to exclude multiple mapping. SAM files of the mapped reads were converted to BAM files using Samtools (version 1.3.1) (40). After that, we marked duplicates and locally realigned indels in the sequence using GATK (version 3.4–0) (41) and Picard (http://broadinstitute.github.io/picard/). In addition, we clipped overlapping read pairs by applying BamUtil (version 1.0.13) (https://genome.sph.umich.edu/wiki/BamUtil) in the BAM files.
To identify the NMP-specific SNP markers, we first generated mpileup files from the BAM files for each clone using Samtools (40). Then, all of the mpileup files were processed with the proview command of MAPGD (version 0.4.26) (https://github.com/LynchLab/MAPGD) to make a population-level pro file. To obtain the SNP at each site, we applied the high-coverage genotype caller (HGC) (42) to the pro file. We set the minimum coverage required for calling genotypes at 6 and the significance of called genotypes at the 5% level to avoid the false-positive detection of alleles (42).
Comparison of Genome Assembly for NMP and MP Clones.
To make an assembly from NMP clones, we first trimmed the reads from each clone by applying Trimmomatic (39). Then the filtered reads were fed to the assembler SPAdes (version 3.11.1) (43) as independent libraries. We ran the assembler with default settings. After obtaining the raw assembly, we filtered out contigs <1000 bp. We also generated an MP assembly using the same method, and then compared it with the NMP assembly using dnadiff command from MUMmer 3 (44). All of the unaligned fragments from each assembly were collected and tested for their existence in each clone by mapping reads using Novoalign (http://www.novocraft.com/) with the “-r None” option. Then, we obtained coverage for each fragment by applying genomeCoverageBed from Bedtools 2.26.0 (45).
Nucleotide Diversity and Linkage Disequilibrium.
We first generated mpileup files from the BAM files for each clone using Samtools (40). Then, mpileup files from NMP clones and MP clones were processed with the proview command of MAPGD (version 0.4.26) (https://github.com/LynchLab/MAPGD) to make population-level pro files, respectively. We extracted the nucleotide diversity from the pro files and averaged the data within each 10-kb window for each scaffold. Then, scaffolds were placed on the chromosomes based on the PA42 D. pulex genetic map (22, 46). Because of potential uncertainties of the genetic map, we only used uniquely mapped scaffolds and the mean cM for each scaffold was used to determine the locus of scaffold on each chromosome. Population-level linkage disequilibrium was estimated using the maximum likelihood method of Maruki and Lynch (47).
Sequence Analysis.
Genes with NMP markers were phased using fastPHASE 1.4 (48), and those with limited heterozygosity were not phased and removed from further analysis. To construct the phylogeny of mitochondrial sequences, we first obtained mitochondrial sequences from each clone using Samtools consensus-calling pipeline (version 1.3.1) (40). Sequences with >20% gaps were not used. Then, the sequences were aligned using ClustalW from Molecular Evolutionary Genetics Analysis 7 (MEGA7) (49). After that, we used MEGA7 for phylogenetic analysis using neighbor joining with bootstrapping (500 replicates) and pairwise deletion. The same method was also applied to the phylogeny construction of the gene 8960.
RNA Extraction and qRT-PCR.
We kept clones at 20 °C with 120,000 cells/mL of the single-celled alga S. obliquus as food. Twenty adult females from each condition were killed ∼60 h after ovulation. The beginning of the intermolt period (0 h) is determined by the presence of a molted exoskeleton. Clones treated with MF were placed in lake water containing MF at a concentration of 400 nM for 24 h before sacrifice. Total RNA was extracted by RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. Then the quality and concentration of total RNA were checked by NanoDrop and RIN values were obtained by 4200 TapeStation. qRT-PCR was done using SuperScript IV One-Step RT-PCR system following manufacturer’s protocol on the Applied Biosystems ViiA 7 system. Primers used for gene 8960 were: F: TATGTAGTGGGCAGCAACGG, R: GTGTATCCTGCACGAGCCTT; for Alpha tubulin, F: CGATCTGATGTACGCCAAGC, R: CTGCAAGATCTTCACGAGCC. Allele-specific expression was determined by amplicon sequencing at Genomics core, Arizona State University, followed by uniquely mapping the reads to each allele.
Data Accession.
The raw reads for all clones used in this study have been deposited to the NCBI Sequence Read Archive (SRA) under accession PRJNA513203. D. pulicaria sequences are deposited under accession number SRP062107 (25). Reads for the D. magna transcriptomes are deposited under PRJNA284518 (26), and the D. magna genome data can be accessed from NCBI with accession number PRJNA298946. D. obtusa genome can be accessed at the NCBI SRA under accession SAMN06013355.
Supplementary Material
Acknowledgments
We thank Wei-chin Ho for helpful discussions and Ryan Stikeleather, Emily Williams, and Stephan Baehr for technical support. This work is supported by NIH grant R35-GM122566-01 (to M.L.). C.M. was supported by an outgoing mobilities grant: “Immersion Training” from the Laboratoire d’Excellence Centre Méditerranéen de l’Environnement et de la Biodiversité, and C.R.H. acknowledges a Marie Curie Career Integration Grant PCIG13-GA-2013-618961, DamaNMP, from the European Union. The sequence data for the D. magna genome were produced by The Center for Genomics and Bioinformatics at Indiana University and distributed via wFleaBase in collaboration with the Daphnia Genomics Consortium.
Footnotes
The authors declare no conflict of interest.
Data deposition: The raw reads for all clones used in this study have been deposited to the NCBI Sequence Read Archive (SRA) under accession PRJNA513203. D. pulicaria sequences are deposited under accession number SRP062107. Reads for the D. magna transcriptomes are deposited under PRJNA284518, and the D. magna genome data can be accessed from NCBI with accession number PRJNA298946. D. obtusa genome can be accessed at the NCBI SRA under accession SAMN06013355.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903553116/-/DCSupplemental.
References
- 1.Lloyd D. G., The transmission of genes via pollen and ovules in gynodioecious angiosperms. Theor. Popul. Biol. 9, 299–316 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth B., Charlesworth D., A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997 (1978). [Google Scholar]
- 3.Delph L. F., Sex-ratio variation in the gynodioecious shrub Hebe strictissima (Scrophulariaceae). Evolution 44, 134–142 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Delph L. F., Touzet P., Bailey M. F., Merging theory and mechanism in studies of gynodioecy. Trends Ecol. Evol. 22, 17–24 (2007). [DOI] [PubMed] [Google Scholar]
- 5.McCauley D. E., Bailey M. F., Recent advances in the study of gynodioecy: The interface of theory and empiricism. Ann. Bot. 104, 611–620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor H., Charlesworth D., Genetics of male-sterility in gynodioecious Cortaderia (Gramineae). Heredity 63, 373–382 (1989). [Google Scholar]
- 7.Weller S. G., Sakai A. K., The genetic basis of male sterility in Schiedea (Caryophyllaceae), an endemic Hawaiian genus. Heredity 67, 265 (1991). [Google Scholar]
- 8.Spigler R. B., Lewers K. S., Main D. S., Ashman T. L., Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101, 507–517 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Dunn D. F., Gynodioecy in an animal. Nature 253, 528–529 (1975). [DOI] [PubMed] [Google Scholar]
- 10.Schärer L., Tests of sex allocation theory in simultaneously hermaphroditic animals. Evolution 63, 1377–1405 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Innes D. J., Dunbrack R. L., Sex allocation variation in Daphnia pulex. J. Evol. Biol. 6, 559–575 (1993). [Google Scholar]
- 12.Innes D. J., Sexual reproduction of Daphnia pulex in a temporary habitat. Oecologia 111, 53–60 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Tessier A. J., Cáceres C. E., Differentiation in sex investment by clones and populations of Daphnia. Ecol. Lett. 7, 695–703 (2004). [Google Scholar]
- 14.Reisser C. M., et al. , Transition from environmental to partial genetic sex determination in Daphnia through the evolution of a female-determining incipient w chromosome. Mol. Biol. Evol. 34, 575–588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebert P. D., The population biology of Daphnia (Crustacea, Daphnidae). Biol. Rev. Camb. Philos. Soc. 53, 387–426 (1978). [Google Scholar]
- 16.Larsson P., Intraspecific variability in response to stimuli for male and ephippia formation in Daphnia pulex. Hydrobiologia 225, 281–290 (1991). [Google Scholar]
- 17.Yampolsky L. Y., Genetic variation in the sexual reproduction rate within a population of a cyclic parthenogen, Daphnia magna. Evolution 46, 833–837 (1992). [DOI] [PubMed] [Google Scholar]
- 18.Galimov Y., Walser B., Haag C. R., Frequency and inheritance of non-male producing clones in Daphnia magna: Evolution towards sex specialization in a cyclical parthenogen? J. Evol. Biol. 24, 1572–1583 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Roulin A. C., et al. , Local adaptation of sex induction in a facultative sexual crustacean: Insights from QTL mapping and natural populations of Daphnia magna. Mol. Ecol. 22, 3567–3579 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Toyota K., et al. , Methyl farnesoate synthesis is necessary for the environmental sex determination in the water flea Daphnia pulex. J. Insect Physiol. 80, 22–30 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Olmstead A. W., Leblanc G. A., Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J. Exp. Zool. 293, 736–739 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Ye Z., et al. , A new reference genome assembly for the microcrustacean Daphnia pulex. G3: Genes, Genomes. G3 (Bethesda) 7, 1405–1416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch M., et al. , Population genomics of Daphnia pulex. Genetics 206, 315–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X., Tang H., Ye Z., Lynch M., Insertion polymorphisms of mobile genetic elements in sexual and asexual populations of Daphnia pulex. Genome Biol. Evol. 9, 362–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S., et al. , Hybridization and the origin of contagious asexuality in Daphnia pulex. Mol. Biol. Evol. 32, 3215–3225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orsini L., et al. , Daphnia magna transcriptome by RNA-Seq across 12 environmental stressors. Sci. Data 3, 160030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keith N., et al. , High mutational rates of large-scale duplication and deletion in Daphnia pulex. Genome Res. 26, 60–69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omilian A. R., Lynch M., Patterns of intraspecific DNA variation in the Daphnia nuclear genome. Genetics 182, 325–336 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan K. K., et al. , Patterns of genetic architecture for life-history traits and molecular markers in a subdivided species. Evolution 55, 1753–1761 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Crease T. J., Omilian A. R., Costanzo K. S., Taylor D. J., Transcontinental phylogeography of the Daphnia pulex species complex. PLoS One 7, e46620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker A. E., Ackerman M. S., Eads B. D., Xu S., Lynch M., Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl. Acad. Sci. U.S.A. 110, 15740–15745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colbourne J. K., Hebert P. D., The systematics of North American Daphnia (Crustacea: Anomopoda): A molecular phylogenetic approach. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351, 349–360 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Kotov A. A., Taylor D. J., Mesozoic fossils (>145 Mya) suggest the antiquity of the subgenera of Daphnia and their coevolution with chaoborid predators. BMC Evol. Biol. 11, 129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bawa K. S., Evolution of dioecy in flowering plants. Annu. Rev. Ecol. Syst. 11, 15–39 (1980). [Google Scholar]
- 35.Charlesworth D., A further study of the problem of the maintenance of females in gynodioecious species. Heredity 46, 27–39 (1981). [Google Scholar]
- 36.Lynch M., Deng H.-W., Genetic slippage in response to sex. Am. Nat. 144, 242–261 (1994). [Google Scholar]
- 37.Innes D., Genetics of Daphnia obtusa: Genetic load and linkage analysis in a cyclical parthenogen. J. Hered. 80, 6–10 (1989). [Google Scholar]
- 38.Haag C. R., Hottinger J. W., Riek M., Ebert D., Strong inbreeding depression in a Daphnia metapopulation. Evolution 56, 518–526 (2002). [PubMed] [Google Scholar]
- 39.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruki T., Lynch M., Genotype calling from population-genomic sequencing data. G3 (Bethesda) 7, 1393–1404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurtz S., et al. , Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu S., et al. , A male-specific genetic map of the microcrustacean Daphnia pulex based on single sperm whole-genome sequencing. Genetics 201, 31–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruki T., Lynch M., Genome-wide estimation of linkage disequilibrium from population-level high-throughput sequencing data. Genetics 197, 1303–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheet P., Stephens M., A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.