Abstract
The dispersal of anatomically modern human populations out of Africa and across much of the rest of the world around 55 to 50 thousand years before present (ka) is recorded genetically by the multiple hominin groups they met and interbred with along the way, including the Neandertals and Denisovans. The signatures of these introgression events remain preserved in the genomes of modern-day populations, and provide a powerful record of the sequence and timing of these early migrations, with Asia proving a particularly complex area. At least 3 different hominin groups appear to have been involved in Asia, of which only the Denisovans are currently known. Several interbreeding events are inferred to have taken place east of Wallace’s Line, consistent with archaeological evidence of widespread and early hominin presence in the area. However, archaeological and fossil evidence indicates archaic hominins had not spread as far as the Sahul continent (New Guinea, Australia, and Tasmania), where recent genetic evidence remains enigmatic.
Keywords: human evolution, archaic introgression, anthropology, genetics
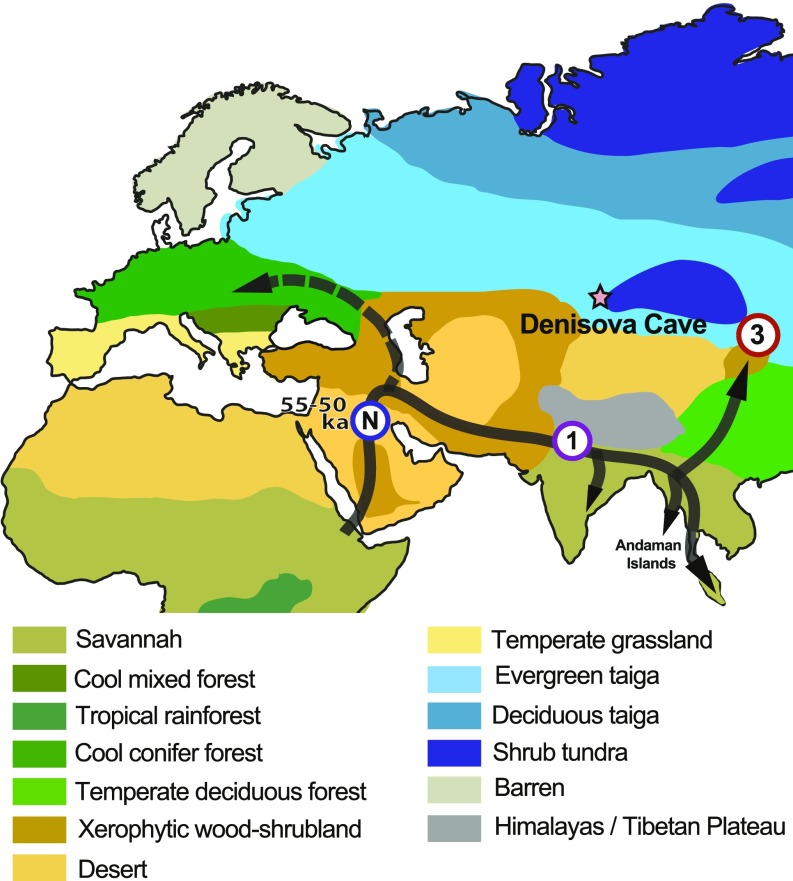
As anatomically modern humans (AMHs) migrated out of Africa and around the rest of the world, they met and interbred with multiple extinct hominin species (1). The traces of genetic input from these past interbreeding events, recorded in the genomes of modern-day populations, have created a powerful tool for recording past human migrations. The first of these events occurred between a small AMH population and Neandertals somewhere in western Eurasia around 55 to 50 thousand years before present (ka), and created a genomic signal of about 2% introgressed Neandertal DNA that is now found in non-African populations (2). The size and ubiquity of the Neandertal genomic signal indicate that the initial AMH population involved was likely small and remained intact long enough for the Neandertal DNA to be effectively mixed among the founding population, which then went on to initiate modern non-African human populations as it spread widely across Eurasia. Consequently, the Neandertal interbreeding event dates the point at which the ancestors of modern-day non-African populations had left Africa, and has been precisely calculated at 55 to 50 ka based on the size of the Neandertal DNA fragments preserved in the genome of an early (45 to 43 ka) AMH specimen from Ust’Ishim, in western Siberia (3, 4). Fig. 1 depicts these events, and infers the migration routes of AMHs by following savannah-like habitats predicted in paleoenvironmental reconstructions at 60 to 50 ka, generated by the BIOME4 CO2 climate model (https://www.paleo.bristol.ac.uk/ummodel/data/bbc_all_triff_rev_dyn/standard_new_plots/bbc_all_triff_rev_dyn_biome4_co2_ann_msy_jav.html). Genomic studies of current Eurasian populations have detected further signals of subsequent Neandertal introgressions, preserved as discrete differences in the frequency distributions of Neandertal DNA (5). Such encounters left localized and far smaller genomic signals in different parts of Eurasia, presumably due to the much larger size of AMH populations involved. For example, East Asian populations appear to carry about 12 to 20% more Neandertal DNA than Europeans (6–8).
Fig. 1.
Proposed route of AMH movement out of Africa, around 60 to 50 ka (1, 4), following areas of savannah-like habitat reconstructed from BIOME4 CO2 climate models (https://www.paleo.bristol.ac.uk/ummodel/data/bbc_all_triff_rev_dyn/standard_new_plots/bbc_all_triff_rev_dyn_biome4_co2_ann_msy_jav.html). Around 55 to 50 ka, a small founding AMH population met and interbred with Neandertals somewhere in western Eurasia (blue circled N), resulting in a Neandertal genomic signal of around 2% that was subsequently distributed globally outside of Africa (1, 2, 4). Sometime after the first event, the AMH population split, with one of the branches leading to the ancestors of Europeans and the other to the common ancestor of South and East Asians, Australo-Papuans, and related populations. Genetic data (33) suggest that as the latter moved across South Asia, it experienced an initial introgression event (purple circled 1) with an unknown hominin (EH1) that was genetically roughly equidistant to Denisovans and Neandertals. The resulting genomic signal (estimated to have originally been 2.6 to 3.4%) is detected in groups as geographically distant as South Asians, Andaman Islanders, and Aboriginal Australians (33), so we have tentatively positioned the event in northern India. In East Asia, a subsequent introgression with a Denisovan group closely related to the Altai specimen also appears to have taken place (red circled 3).
In contrast to the Neandertals, the interbreeding events with other extinct hominin groups, such as the Denisovans, the eastern Eurasian sister group of Neandertals, remain poorly understood, and are potentially far more complex. Our knowledge of the Denisovans remains intimately tied to genetic data retrieved from Denisova Cave in the Altai Mountains in southern Siberia, which, until recently, contained the only known fossil record of this group (9, 10) (Fig. 1). The complete Denisovan genome was reconstructed using DNA from a small distal phalanx found in the cave, and demonstrated the Denisovans were a sister group to Neandertals and diverged around 400 ka (6). In turn, the common ancestor of Denisovans and Neandertals separated from AMHs somewhat earlier, around 600 ka (11). Further Denisovan mitochondrial and nuclear DNA has been retrieved from isolated teeth and sediment samples in the cave (12–15), which Denisovans utilized between at least 200 and 50 ka, and which also records the presence of Neandertals between 140 and 80 ka (16, 17). The 2 groups interacted during the period of temporal overlap, as demonstrated by bone fragments of a first-generation Denisovan-Neandertal hybrid dating to around 90 ka (16, 18). Recently, the fossil evidence for Denisovan range was further extended to the Tibetan Plateau, through ancient protein analysis of a >160-ka mandible (19).
The lack of information about Denisovan morphology, combined with the paucity of fossil evidence beyond central Eurasia, continues to limit studies of both their diversity and range. A recent morphological description of 3 hominin crania uncovered in Xuchang, central China (20), has led to speculations about the likely presence of Denisovans in China, while the extensive diversity of fossil forms in Southeast Asia (SEA) (9, 21–23) potentially hides Denisovans and even more species, as demonstrated by the recent identification of Homo luzonensis in the Philippines (24). For the moment, the Denisovans and relatives remain some of the more elusive members of the recent human family, and genetic studies continue to be the most effective approach to understanding their history. The absence of Denisovan ancestry from both modern-day western Eurasian populations and European Neandertal specimens provides compelling evidence that the group rarely (if ever) ventured into the West. While Neandertals appear to have been widely distributed across western Eurasia, Denisovans appear to have occupied a parallel range across eastern Eurasia. To investigate this issue in more detail, the genomes of human populations around the world are being screened for archaic introgression using new genetic approaches, including methods which are agnostic to the putative archaic source (e.g., refs. 25–27). For example, S* is a commonly used statistic designed to detect genomic regions with an accumulation of variants in strong linkage disequilibrium, which are absent from a reference population that lacks a given archaic introgression of interest (e.g., African populations, for Neandertals or Denisovans) (26). The resulting putatively archaic genomic regions can then be “matched” to known hominin genomes (i.e., Neandertal, Denisovan), with the remainder likely originating from other sources that potentially represent unknown hominin groups.
Denisovan Ancestry in Contemporary AMH Populations
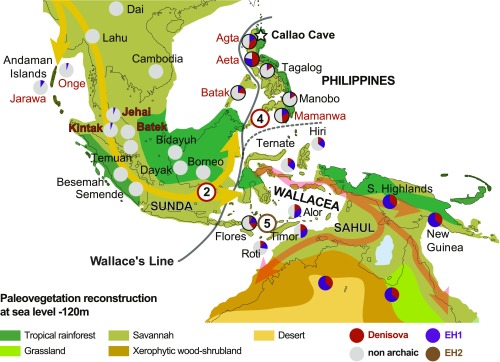
The patterns of Denisovan genetic ancestry in current human populations point to a far more complex and mysterious history of interactions than those of Neandertals. The distribution of Denisovan genomic signals is markedly uneven, with the largest amounts found in New Guinea and Australia, thousands of kilometers away from Siberia (28) and east of Wallace’s Line, one of the world’s most notable geographic barriers for faunal dispersion (29). Wallace’s Line is formed by the strong marine currents running between the Pacific and Indian Oceans through Island Southeast Asia (ISEA), and effectively separates Eurasian placental mammal-dominated ecosystems from the marsupial and reptile-dominated landscapes of Sahul, modern-day Australia and New Guinea (6) (Fig. 2). Early studies reported that the highest genomic proportions of Denisovan ancestry in current populations (∼3 to 6%) are found in Aboriginal Australians and New Guinea Papuan Highlanders (Australo-Papuans), along with Oceanian populations with Papuan-related ancestry (28, 30). These are followed by hunter-gatherer populations in the Philippines (about half the Denisovan content observed in Australo-Papuans), while only trace amounts (<1%) of Denisovan signals have been reported in the genomes of most South and East Asian groups (30, 31) (Fig. 2).
Fig. 2.
Yellow and red arrows indicate the inferred route of AMH movement through ISEA around 50 ka (44) (shown with lowered sea levels), following reconstructed areas of savannah-like habitat as above. Modern-day hunter-gatherer populations with genetic data are shown in red, and farming populations are shown in black. The estimated genomic content of EH1 (purple), Denisovan (red), EH2 (brown), and nonarchaic (gray) in modern-day populations (28, 33, 40, 42) is shown in pie charts, as a relative proportion to that seen in Australo-Papuans (full circles). Gray All populations containing large amounts of Denisovan genomic content are found east of Wallace’s Line. Independent introgression events with Denisovan groups are inferred for both the common ancestor of Australo-Papuan, Philippines, and ISEA populations (red circled 2) and, separately, for the Philippines (red circled 4). The signal for a separate introgression with an unknown hominin on Flores, recorded in genomic data from modern-day populations, remains less secure (brown-circled 5). The precise location of introgression events 2, 4, and 5 currently remains unknown. Pie charts with black borders have estimated hominin proportions.
Further investigation of the Denisovan genomic signals in modern-day human populations using an S* approach have disentangled that there were at least 2 pulses of hominin “Denisovan-like” introgression from very distinct groups, one of which was quite distant from the known Altai Denisovan genome (32). South Asian and Australo-Papuan populations only carry signals from this “distant Denisovan group,” while East Asians appear to have both the “distant” and standard Altai-type Denisovan ancestry components (32).
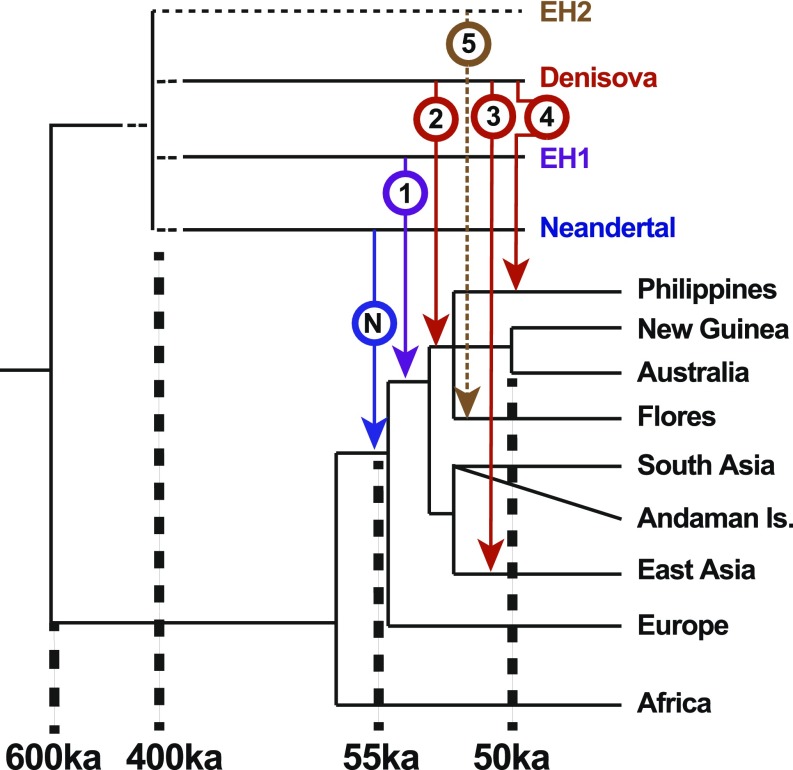
A recent study combining a novel approximate Bayesian computation and deep-learning method (ABC-DL) with extensive demographic modeling clarified that the “distantly related Denisovan” signal is actually so genetically divergent that it is equidistant to both Neandertal and Denisovans, and probably represents an entirely new hominin group (33). As there is currently no obvious suitable taxonomic name, we term this extinct hominin 1 (EH1) (Fig. 3). The ABC-DL study was able to identify that the AMH ancestor of all Asian and Australo-Papuan populations interbred with EH1, resulting in a shared genomic signal of 2.6 to 3.4% EH1 ancestry (purple in Fig. 1). This signal, albeit diluted, can still be detected in modern-day Indian, Andamanese, and East Asian populations (33). Following this initial event, it appears that a second phase of admixture occurred between the ancestors of Australo-Papuans (after they split from other Asian populations) and a Denisovan group more closely related to the sequenced Altai individual, generating an additional ∼1.6% genomic contribution (33) (red-circled 2 in Fig. 2). Previous studies have shown that modern-day ISEA populations have Denisovan genomic content roughly proportional to their Australo-Papuan ancestry (28), suggesting they are derived from this same secondary event (Fig. 2).
Fig. 3.
Recent hominin introgression events in SEA. The phylogenetic tree shows approximate relationships and date estimates for the various events with extinct hominins and modern-day AMH populations. Five inferred hominin introgression events are shown, starting with the Neandertal around 55 to 50 ka (blue-circled N), followed by EH1 (purple-circled 1). The ancestor of Australo-Papuans, Philippines, and other ISEA groups is then inferred to have experienced admixture with Denisovans (red-circled 2), followed by a subsequent event (red-circled 4) detected just in Philippines hunter-gatherer populations. A separate Denisovan introgression is also detected in East Asian populations (red-circled 3). A further contribution from an unknown hominin (EH2) may be recorded in the genomes of modern-day short-statured populations on Flores but remains unclear (brown-circled 5) (42). The phylogenetic relationships among the extinct hominins remains uncertain, but appear to be of roughly similar genetic divergence, occurring around 400 ka (11). We suggest the timing of the above events is constrained between the initial Neandertal introgression at 55 to 50 ka and the colonization of Australia at 50 ka (1, 4).
Interestingly, the ABC-DL study did not detect the separate Denisovan-like introgression signal previously reported in East Asian populations (32), which was also found in a recent genetic survey (34). Such a genetic affinity would certainly be consistent with the geographic proximity of the Altai Denisovan and East Asian populations. It is possible the ancestral EH1 genomic contribution is contributing to this signal, and also the slightly higher Neandertal genomic ancestry reported for these populations (33).
The isolation of Australo-Papuan populations in Sahul around 50 ka (4) appears to have preserved their EH1 and Denisovan genomic content from dilution by admixture with later AMH populations lacking these signals (35). In contrast, the demographic history of SEA seems far more complex. Multiple migrations of mainland Asian farming groups can be detected during the Holocene, including both Austroasiatic and Austronesian speakers (36–40), and appear to have much lower Denisovan ancestry (40). These movements are likely responsible for the low levels of archaic hominin genomic signals surviving in modern-day Asia and western ISEA populations (Fig. 2). Curiously, however, long-isolated SEA hunter-gatherer groups, such as the Andaman Islanders and Malaysian Jehai, also show relatively low amounts of Denisovan ancestry (28, 41). Given they are unlikely to have been impacted by Holocene farming groups, this would suggest that a much earlier phase of admixture with non-Denisovan containing AMH populations also took place. Either way, it appears that the initial introgression event (with EH1) occurred somewhere in South Asia, before the split and geographic spread of the different populations across South and East Asia, ISEA, and Oceania. We have subjectively placed this event in northern India, due to the appearance of the EH1 signal in the South Asian and Andamanese groups and the recent finding of a Denisovan-like mandible in the Tibetan Plateau (19) (Fig. 2).
Denisovan DNA across Wallace’s Line
As noted, the markedly higher amount of Denisovan genomic content observed in modern-day populations across Wallace’s Line (Fig. 2), including many hunter-gatherer groups, generally correlates with the extent of their shared ancestry with the Australo-Papuan populations who first moved through the area (28, 30). Intriguingly however, the Denisovan genomic content of Philippines hunter-gatherer groups is proportionally much higher (28, 41). Initially, this pattern was attributed to dilution of the Denisovan signal in the Australo-Papuan ancestors after their separation from the Philippines groups, presumably by admixture with a population carrying no Denisovan DNA (28). However, this seemed at odds with the Australo-Papuans possessing far more Denisovan DNA than any other population studied, including the Philippines groups, and the geographically widespread relationship between Australo-Papuan and Denisovan genomic content. In fact, a subsequent study of many hunter-gatherer populations across SEA indicated that the higher Denisovan content seen in the Philippines more likely reflects a further independent interbreeding event with Denisovans (41) (Fig. 2). The unique nature of this signal raises the distinct possibility that it originated in the Philippines area, in which case Denisovan populations were indeed present in ISEA. While fossil and archaeological evidence already confirms that early hominin populations had crossed Wallace’s Line [on Flores >1 million years before present, on Philippines >700 ka, and on Sulawesi >120 ka (4, 9)], it is currently unclear whether any of the late-surviving forms in the area might represent Denisovans (4, 10). It is notable that the small fossil hominins on the Philippines [H. luzonensis, reported from 65- to 50-ka deposits in Callao Cave, Luzon (24, 42)] and Flores (Homo floresiensis from Liang Bua Cave) both survive until the estimated arrival of AMHs in the area ∼50 ka, suggesting the latter were involved in their extinction (4, 10). [Reports of earlier AMH presence in the area at Madjedbebe, Australia and Lida Ajer, Sumatra are both questionable (4)].
Recent studies of modern-day human populations in Flores and New Guinea have detected localized genomic signals that could be interpreted as originating from further hominin groups present east of Wallace’s Line (34, 43). Both studies scanned hundreds of human genomes from across Europe, East Asia, ISEA, and Melanesia for putative signatures of archaic introgression, with the source of each introgressed region characterized as either Neandertal, Denisovan, ambiguous (either Neandertal or Denisovan), or unknown. In Flores, analysis of the very short-statured population currently living near Liang Bua Cave reported an enigmatic unknown genomic signature whose source appears to be as divergent from modern-day humans as Neandertals and Denisovans (although the study did not highlight the finding) (43). The unknown genomic component was detected exclusively in Flores, raising the possibility of an additional introgression event with a further extinct hominin (EH2 in Fig. 2), and implies it must also have crossed Wallace’s Line. Interestingly, this signal was not detected in a recent study focused on modern-day New Guinea populations (34), which reported 2 pulses of Denisovan-like introgression as seen in other areas of ISEA. The New Guinea study analyzed just the longest putative hominin-introgressed fragments and suggests that the most recent pulse may have originated in New Guinea as recently as 30 ka (with confidence intervals from 15 to 50 ka). Such a late date for hominin survival seems unlikely, and the finding is in direct conflict with the similar Denisovan genomic content observed in Papuan and Aboriginal Australian populations (28, 30, 31), which separated ∼50 ka (35), and the marked lack of any archaeological evidence for archaic hominin presence in Sahul (4). This extensive discordance means that it will be important to examine introgressed Denisovan DNA signals in geographically widespread Aboriginal Australian populations before concluding there were indeed unique introgression events within New Guinea.
We present what appears to be the most plausible scenario for the current data in Figs. 1 and 2, although the precise location of the introgression events currently remains unknown. For example, the Denisovan introgression with the ancestor of the Australo-Papuan, Philippines, and Flores populations would appear to be on the Sunda Shelf, before the divergence of these groups. We have tentatively placed it near Borneo due to recent studies suggesting the settlement of Sahul occurred via a route that transited through Borneo and Sulawesi, before arriving in Sahul in what is now West Papua (44) (Fig. 2). Alternatively, however, it is possible that the Denisovan introgression might also have taken place east of Wallace’s Line, for example, on Sulawesi (4, 9, 21). Importantly, archaeological evidence of hominin distribution suggests the easternmost point where Denisovan introgression is plausible was Sulawesi, the Philippines, and Flores. Indeed, the western islands in SEA were only separated from mainland SEA by narrow marine gaps during low sea levels in glacial periods (Fig. 2), and the fluctuating extent and nature of land connections during glacial cycles will have played a key role in the original dispersal and subsequent isolation of early hominin groups in the area. However, there remain no clear archaeological signs that archaic hominin migrations extended further east, or as far as the Sahul continent itself. In contrast to this marked absence of evidence, the subsequent arrival and rapid spread of AMHs across New Guinea and Australia around 50 ka is clearly chronicled by archaeological evidence, recording a widespread geographic dispersal across the continent within just a few thousand years, including even remote arid inland areas (4, 45, 46). Megafaunal records are similarly consistent with a late arrival of hominins in Sahul, with no clear extinction phase until 43 to 42 ka (47), well after the widespread presence of AMHs across Australia (35).
Further genetic and archaeological research is required to resolve the many outstanding issues outlined here, but the current genetic evidence suggests that as AMHs first moved through the area, they interbred with EH1 in South Asia, Denisovans in ISEA and the Philippines, and potentially EH2 in Flores. Strikingly, of these hominin groups, only Denisovans are currently known, and even in this case, the evidence stems solely from genomics. The region was clearly occupied by several hominin groups, which probably lived in relative isolation from one another for hundreds of thousands of years, and appear to have contributed unique patterns of ancestry to current populations.
Acknowledgments
We thank Ray Tobler, Fernando Racimo, Chris Turney, Bastien Llamas, Kieren Mitchell, Yassine Souilmi, and Murray Cox for providing useful comments on the manuscript. We also thank Julien Soubrier and Damien Fordham for help in designing the figures. J.C.T. and A.C. are supported by Australian Research Council Discovery Grants and a Laureate Fellowship (A.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Skoglund P., Mathieson I., Ancient genomics of modern humans: The first decade. Annu. Rev. Genomics Hum. Genet. 19, 381–404 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Green R. E., et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Q., et al. , Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connell J. F., et al. , When did Homo sapiens first reach Southeast Asia and Sahul? Proc. Natl. Acad. Sci. U.S.A. 115, 8482–8490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanea F. A., Schraiber J. G., Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol. 3, 39–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer M., et al. , A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prüfer K., et al. , A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wall J. D., et al. , Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics 194, 199–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaifu Y., Archaic hominin populations in Asia before the arrival of modern humans: Their phylogeny and implications for the “Southern Denisovans”. Curr. Anthropol. 58, S418–S433 (2017). [Google Scholar]
- 10.Cooper A., Stringer C. B., Did the Denisovans cross Wallace’s line? Science 342, 321–323 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Prüfer K., et al. , The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause J., et al. , The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawyer S., et al. , Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc. Natl. Acad. Sci. U.S.A. 112, 15696–15700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slon V., et al. , Neandertal and Denisovan DNA from Pleistocene sediments. Science 356, 605–608 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Slon V., et al. , A fourth Denisovan individual. Sci. Adv. 3, e1700186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douka K., et al. , Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova Cave. Nature 565, 640–644 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Jacobs Z., et al. , Timing of archaic hominin occupation of Denisova Cave in southern Siberia. Nature 565, 594–599 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Slon V., et al. , The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561, 113–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F., et al. , A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature 569, 409–412 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Li Z. Y., et al. , Late Pleistocene archaic human crania from Xuchang, China. Science 355, 969–972 (2017). [DOI] [PubMed] [Google Scholar]
- 21.van den Bergh G. D., et al. , Earliest hominin occupation of Sulawesi, Indonesia. Nature 529, 208–211 (2016). [DOI] [PubMed] [Google Scholar]
- 22.van den Bergh G. D., et al. , Homo floresiensis-like fossils from the early Middle Pleistocene of Flores. Nature 534, 245–248 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Sutikna T., et al. , Revised stratigraphy and chronology for Homo floresiensis at Liang Bua in Indonesia. Nature 532, 366–369 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Détroit F., et al. , A new species of Homo from the Late Pleistocene of the Philippines. Nature 568, 181–186 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Plagnol V., Wall J. D., Possible ancestral structure in human populations. PLoS Genet. 2, e105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vernot B., Akey J. M., Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Skov L., et al. , Detecting archaic introgression using an unadmixed outgroup. PLoS Genet. 14, e1007641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich D., et al. , Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 89, 516–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace A. R., On the physical geography of the Malay Archipelago. R. Geogr. Soc. 7, 205–212 (1863). [Google Scholar]
- 30.Qin P., Stoneking M., Denisovan ancestry in East Eurasian and Native American populations. Mol. Biol. Evol. 32, 2665–2674 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Sankararaman S., Mallick S., Patterson N., Reich D., The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 26, 1241–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning S. R., Browning B. L., Zhou Y., Tucci S., Akey J. M., Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell 173, 53–61.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondal M., Bertranpetit J., Lao O., Approximate Bayesian computation with deep learning supports a third archaic introgression in Asia and Oceania. Nat. Commun. 10, 246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs G. S., et al. , Multiple deeply divergent Denisovan ancestries in Papuans. Cell 177, 1010–1021.e32 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Tobler R., et al. , Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia. Nature 544, 180–184 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Abdulla M. A., et al. ; HUGO Pan-Asian SNP Consortium; Indian Genome Variation Consortium , Mapping human genetic diversity in Asia. Science 326, 1541–1545 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Lipson M., et al. , Reconstructing Austronesian population history in Island Southeast Asia. Nat. Commun. 5, 4689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudjashov G., et al. , Complex patterns of admixture across the Indonesian Archipelago. Mol. Biol. Evol. 34, 2439–2452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipson M., et al. , Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McColl H., et al. , The prehistoric peopling of Southeast Asia. Science 361, 88–92 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Jinam T. A., et al. , Discerning the origins of the Negritos, first Sundaland people: Deep divergence and archaic admixture. Genome Biol. Evol. 9, 2013–2022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mijares A. S., et al. , New evidence for a 67,000-year-old human presence at Callao Cave, Luzon, Philippines. J. Hum. Evol. 59, 123–132 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Tucci S., et al. , Evolutionary history and adaptation of a human pygmy population of Flores Island, Indonesia. Science 361, 511–516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kealy S., Louys J., O’Connor S., Least-cost pathway models indicate northern human dispersal from Sunda to Sahul. J. Hum. Evol. 125, 59–70 (2018). [DOI] [PubMed] [Google Scholar]
- 45.McDonald J., et al. , Karnatukul (Serpent’s Glen): A new chronology for the oldest site in Australia’s Western Desert. PLoS One 13, e0202511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamm G., et al. , Cultural innovation and megafauna interaction in the early settlement of arid Australia. Nature 539, 280–283 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Saltré F., et al. , Climate change not to blame for late Quaternary megafauna extinctions in Australia. Nat. Commun. 7, 10511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]