Significance
To address mechanisms by which plants control their associated microorganisms, we designed a screen using a bacterial activity in soil. A detailed analysis of a candidate gene from the screen, CYP71A27, identified an additional root-specific component of synthesis of camalexin, a natural product involved in plant defense against pathogens. Loss of function of CYP71A27 affected not only the microbiota in soil but also interactions with single plant growth-promoting bacteria. The loss of the promoting effect in the mutants could be complemented chemically by adding camalexin to the bacteria. Thus, our study identified an additional role for camalexin in facilitating interaction with microbes in the rhizosphere and an additional gene for its synthesis.
Keywords: plant microbiome function, sulfur containing phytoalexins, Arabidopsis, GWAS
Abstract
Plants in their natural ecosystems interact with numerous microorganisms, but how they influence their microbiota is still elusive. We observed that sulfatase activity in soil, which can be used as a measure of rhizosphere microbial activity, is differently affected by Arabidopsis accessions. Following a genome-wide association analysis of the variation in sulfatase activity we identified a candidate gene encoding an uncharacterized cytochrome P450, CYP71A27. Loss of this gene resulted in 2 different and independent microbiota-specific phenotypes: A lower sulfatase activity in the rhizosphere and a loss of plant growth-promoting effect by Pseudomonas sp. CH267. On the other hand, tolerance to leaf pathogens was not affected, which agreed with prevalent expression of CYP71A27 in the root vasculature. The phenotypes of cyp71A27 mutant were similar to those of cyp71A12 and cyp71A13, known mutants in synthesis of camalexin, a sulfur-containing indolic defense compound. Indeed, the cyp71A27 mutant accumulated less camalexin in the roots upon elicitation with silver nitrate or flagellin. Importantly, addition of camalexin complemented both the sulfatase activity and the loss of plant growth promotion by Pseudomonas sp. CH267. Two alleles of CYP71A27 were identified among Arabidopsis accessions, differing by a substitution of Glu373 by Gln, which correlated with the ability to induce camalexin synthesis and to gain fresh weight in response to Pseudomonas sp. CH267. Thus, CYP71A27 is an additional component in the camalexin synthesis pathway, contributing specifically to the control of plant microbe interactions in the root.
Plant roots in soil are colonized by a large number of diverse bacteria and fungi (1–5). These plant microbiome communities appear to be stable, and their composition is largely controlled by soil characteristics and by plant genotypes (6, 7). It is generally accepted that the way plants communicate with the microorganisms is through metabolites in the root exudates, and several such molecules have recently been identified (8–10). The metabolic composition of root exudates varies among plant genotypes, e.g., Arabidopsis accessions. This provides a possibility to study the links between plant genome and microbiome (11), and a genome-wide association study (GWAS) to investigate how plants control their leaf microbiome revealed variation in taxonomic composition across 196 Arabidopsis accessions (12). The analysis showed that many of the single-nucleotide polymorphisms (SNPs) significantly linked to variation in microbiome composition were localized in genes with a function in defense response, cell wall synthesis, and kinase activity (12). A different approach to assess how plants control their microbiota is the analysis of variation in effects of plant growth-promoting (PGP) bacteria. Arabidopsis accessions showed large difference in changes in fresh weight (FW) and root architecture upon exposure to the rhizobacterium Pseudomonas simiae WCS417r (13). However, although a GWAS led to identification of several candidate genes, they were not further functionally analyzed (13).
We are interested in characterizing how plants control the function of the associated microbial community (2, 14). To understand the mechanisms by which plants shape their microbiota we used the microbial sulfatase activity as a quantitative measure. Bacteria and fungi use sulfatase to cleave sulfate from sulfate esters (15), mineralizing the organic sulfur and playing an important role in plant sulfur nutrition by making it available to plants (16). Sulfatase is commonly used as a measure for biological activity of soil (17) and thus is suitable as a proxy for microbial function in our study. We determined sulfatase activity in rhizosphere soil of 172 Arabidopsis accessions and performed a GWAS. Detailed analysis of one of the candidate genes associated with the variation in microbial sulfatase revealed a so far undescribed component of the camalexin synthesis pathway, CYP71A27 (At4g20240), which is active in the roots. Loss of this P450 enzyme affects not only the sulfatase activity in soil but also the PGP effects of several bacterial strains, and exogenously applied camalexin complemented both effects of the CYP71A27 gene knockout. Our study thus suggests a role for camalexin in plant interaction with beneficial bacteria in the rhizosphere.
Results
Natural Variation in Plant–Microbe Interaction.
To assess how plants influence the activity of the associated microorganisms in soil, we used a genome-wide approach with Arabidopsis accessions. In a pilot experiment we grew 7 Arabidopsis accessions in a soil/sand mix (10% soil) for 2 wk and compared the sulfatase activity in soil after the plants were removed. As control, the activity in soil incubated under the same conditions without plants was measured. A significant variation in sulfatase activity between the different accessions was observed which was always higher than in the control soil (SI Appendix, Fig. S1). Therefore, we extended the analysis to 172 accessions of the core360 collection representing species-wide diversity (18). Measured sulfatase activity varied greatly across the population with more than 20-fold difference between the lowest and the highest activities (SI Appendix, Fig. S1 and Table S1). The data were found to be distributed normally using a chi-square goodness-of-fit test. We therefore performed a genome-wide association mapping using a mixed model algorithm with the web-based platform GWAPP (19). The analysis did not identify any markers significant at the Bonferroni-corrected significance of −log10P of 6.6. Due to the limited power of our association study with only 172 accessions and because we intended to test the candidate genes experimentally, we selected arbitrary P value cutoff of P = 0.0001. This led to a set of 70 SNPs with −log10P values higher than 4 (SI Appendix, Fig. S1 and Table S2). At these loci we inspected the genomic regions of ±20 kBp from the SNP to identify candidate genes possibly affecting the sulfatase activity in the soil (SI Appendix, Table S3). We focused on 2 categories, genes annotated to be connected to sulfur metabolism or to secondary metabolism, and identified 24 candidates from both groups. Next we obtained T-DNA insertion lines for these genes available from the Nottingham Arabidopsis Stock Centre and recovered homozygous mutants by standard PCR genotyping. These mutants were grown in the same way as the accessions in soil/sand mix, and sulfatase activity was measured. From the 15 homozygous lines tested, 6 mutants (40%) showed significant reduction in sulfatase activity (SI Appendix, Table S3), while previous analyses have shown that 10–20% of randomly selected genes might show a significant variation in a given phenotype (20). The significance of the enrichment was determined by a χ2 test to be P = 0.04 or P = 0.09, depending on the dataset used (table S5 in ref. 20). It has to be noted, however, that very different traits are compared, interaction with microbes and nutrient homeostasis, which will have different genetic architectures. Five of the confirmed mutants are associated with secondary metabolism and 1 with sulfur metabolism. For detailed analysis we decided to focus on CYP71A27 since P450 genes are often involved in the biosynthesis of secondary metabolites, which are good candidates for the signals from plants to the soil microbiota (2).
CYP71A27 Is Part of Camalexin Synthesis Network.
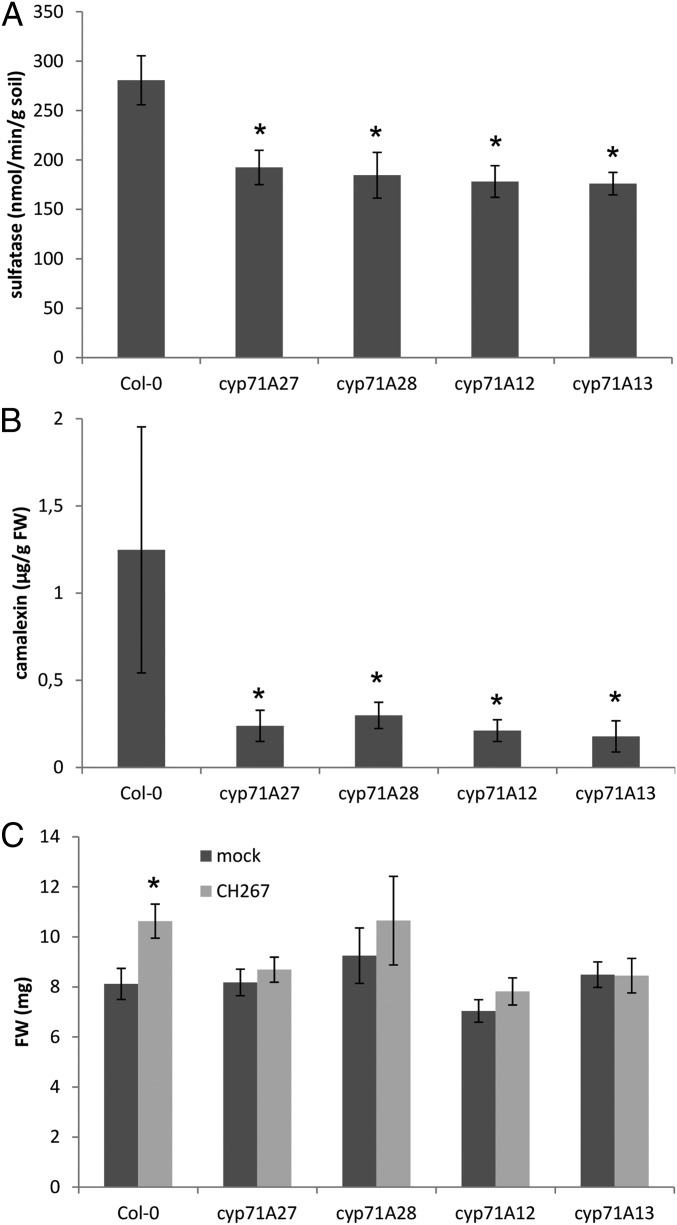
While the function of CYP71A27 was unknown, 2 other members of the cytochrome P450 71A family, CYP71A12 and CYP71A13, participate in synthesis of camalexin, a sulfur containing phytoalexin (21–23). Indeed, the PlantCyc database in the Plant Metabolic Network (https://www.plantcyc.org/) (24) predicts CYP71A27 as well as the product of an adjacent P450 encoding gene CYP71A28 to be part of camalexin synthesis (SI Appendix, Fig. S2). Expression analysis of mutants in these 4 genes revealed that transcript levels for 3 camalexin biosynthesis genes, CYP71A12, CYP71A13, and GST6, were coordinately increased in the roots of the mutants, except when they were missing in the corresponding T-DNA lines (SI Appendix, Fig. S3). On the other hand, the changes in transcript levels of genes involved in glucosinolate synthesis were less prominent, and glucosinolate accumulation was not affected in cyp71A27 and cyp71A28 mutants (SI Appendix, Fig. S4). We therefore asked whether disruption of these genes might have a similar effect on rhizosphere sulfatase activity. Indeed, sulfatase activity was significantly lower in soil from mutants of all 4 P-450s (Fig. 1A). The link to camalexin was confirmed as roots of all 4 mutants grown in soil accumulated less camalexin than WT (Fig. 1B). Thus, it seems that camalexin has a function not only in immunity but also in mediating plant microbiota interactions in the rhizosphere.
Fig. 1.
CYP71A27 is connected to camalexin synthesis. (A) Col-0, cyp71A12, cyp71A13, cyp71A27, and cyp71A28 plants were grown for 2 wk in soil (10%)/sand mixture; afterward, 2 soil samples were taken per plant, and sulfatase activity was measured. Data are presented as means and SE from 10 samples corresponding to 5 independent plants. Asterisks indicate significant differences from the wild-type Col-0 significant at P < 0.05 (Student’s t test). (B) Camalexin accumulation in roots of plants grown in soil (10%)/sand mixture like for sulfatase activity. Data are presented as means and SD from 4 pools of at least 5 independent plants. Asterisks indicate significant differences from Col-0 significant at P < 0.05 (Student’s t test). (C) Col-0 and the 4 mutant lines were grown for 2 wk in presence of Pseudomonas sp. CH267 or 10 µM MgCl2 as mock, and the FW of whole plants was measured. Data are presented as means and SE from at least 20 plants grown on 4 independent plates. Asterisks indicate significant differences between mock and bacterial treatment at P < 0.05 (Student’s t test).
Most mechanistic understanding of plant–microbe interactions has, however, derived from analysis of single bacterial species, pathogenic or PGP (2, 6, 25). Therefore, we tested whether loss of CYP71A27 affects also a different trait, interaction with single bacteria, which is independent from the soil sulfatase assay. Thus, we cultivated the mutants with previously characterized PGP bacterial strain Pseudomonas sp. CH267 (26) on agar plates. Cocultivation with the bacteria resulted in an increased FW of the WT (Fig. 1C). Inactivation of CYP71A27 resulted in loss of the PGP effect, and, similarly, the 3 other mutants did not gain FW after cocultivation with Pseudomonas sp. CH267 (Fig. 1C). Thus, CYP71A27 and possibly also CYP71A28 are parts of the camalexin synthesis network, potentially functioning similarly to the 2 known members of the 71A family, CYP71A12 and CYP71A13 (SI Appendix, Fig. S2). In addition, these results point to a role of camalexin in plant microbiota interactions.
CYP71A27 Is Important for Plant Microbe Interactions.
To investigate whether the observed changes in interaction with Pseudomonas sp. CH267 in cyp71A27 are dependent on special characteristics of the strain or can be generalized, we cultivated the cyp71A27 mutant with 3 additional bacterial strains, 2 well-characterized PGP strains, P. simiae WCS417r (13, 27) and Paraburkholderia phytofirmans PsJN (28), as well as with the root pathogen Burkholderia glumae PG1 (29). Similar to cocultivations with the Pseudomonas sp. CH267, P. simiae WCS417r and P. phytofirmans PsJN only showed PGP effects with WT and not with cyp71A27 (SI Appendix, Fig. S5). On the other hand, the mutant was more susceptible to the growth inhibition by B. glumae (SI Appendix, Fig. S5). We further tested whether the enzyme may be important also for interaction with fungi and incubated the different plant genotypes with the PGP endophytic fungus Serendipita indica (30). The colonization rate, determined by quantification of fungal DNA in roots, was significantly decreased in both cyp71A27 and cyp71A13, taken as a control (SI Appendix, Fig. S5). Thus, the CYP71A27 is important for plant interactions with multiple microbial strains during well-defined cocultivations.
CYP71A27 Is Active in the Roots.
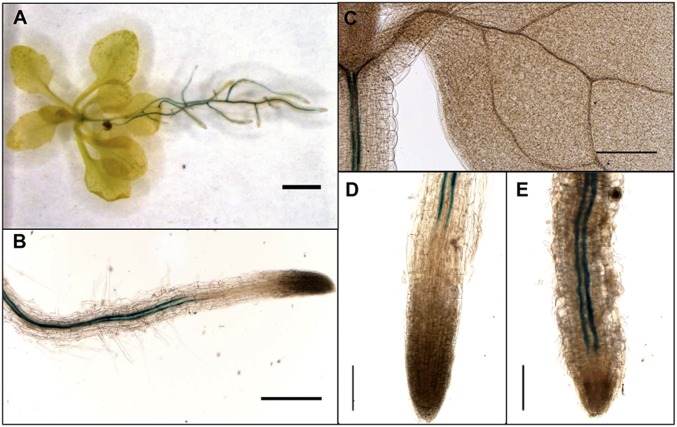
Given the function of the CYP71A27 in plant–microbe interaction in the soil, it is not surprising that the web-based Arabidopsis information portal ARAPORT (31) predicts its expression primarily in the roots (SI Appendix, Fig. S6). To obtain detailed information about tissue-specific expression, Col-0 plants were transformed with the GUS reporter gene uidA under control of the CYP71A27 promoter. Analysis of these lines confirmed an expression in the roots with only a weak staining in the hypocotyl (Fig. 2). In the roots, the staining was observed in the vasculature as 2 strong lines, starting in the differentiation zone of the root, which suggests phloem localization of the transcript (Fig. 2B). The expression pattern was affected by incubation with the Pseudomonas sp. CH267, which after 2 wk resulted in expression extending much closer to the root tip (Fig. 2 D and E). The difference in distance between the root tips and beginning of GUS staining between mock and Pseudomonas sp. CH267-treated plants was significant already after 24 h (SI Appendix, Fig. S7). The bacterial elicitor flagellin (flg22) also triggered the change in spatial expression of CYP71A27 closer to the root tips; on the other hand, no change in spatial pattern was observed by cocultivation with P. simiae WCS417r, which, however, resulted in general decrease in expression levels (SI Appendix, Fig. S7). No expression was observed in mature leaves or reproductive organs. The expression pattern of CYP71A27 is thus consistent with its role in rhizosphere processes. Since CYP71A27 seems to function in the camalexin synthesis pathway, we compared its expression pattern in the roots with CYP71A12. The 2 genes have a very different expression pattern. While CYP71A27 is constitutively expressed, CYP71A12 is not expressed in mock or bacteria-treated roots but strongly induced by flagellin in cortex cells (SI Appendix, Fig. S7).
Fig. 2.
Tissue-specific expression of CYP71A27. GUS staining of transgenic plants expressing CYP71A27pro::GUS. GUS expression in 3-wk-old (A) whole plant, (B) primary root, and (C) leaf. GUS staining of root tips of plants treated for 2 wk with 10 µM MgCl2 as (D) mock or (E) Pseudomonas sp. CH267. (Scale bars: 20 mm in A; 2 mm in B and C; and 1 mm in D and E.) At least 2 plants from 3 independent transgenic lines were stained and analyzed.
To test possible involvement of CYP71A27 in pathogen response in leaves we investigated the response of the cyp71A27 and WT to Botrytis cinerea (32). The lesion area and diameter were not different in cyp71A27 and WT but significantly increased in cyp71A13 (SI Appendix, Fig. S8). Botrytis induced synthesis of camalexin, which accumulated to the same levels in leaves of WT and cyp71A27, while the increased susceptibility of cyp71A13 correlated with low camalexin production (SI Appendix, Fig. S8). Similarly, growth of bacterial pathogen Pseudomonas syringae pv. maculicola, which induces camalexin synthesis without effects on virulence (33), was not affected by loss of CYP71A27 or CYP71A13 (SI Appendix, Fig. S8), and camalexin accumulation in the leaves remained the same in cyp71A27 and WT but was largely abolished in cyp71A13 (SI Appendix, Fig. S8). To further confirm that cyp71A27 is involved in production of camalexin in the roots but not in the leaves we tested whether camalexin induction by abiotic elicitor AgNO3 is compromised in the different organs in the mutants and WT. In accordance with our previous results, camalexin accumulation in AgNO3-treated shoots of cyp71A27 was not different from WT, whereas cyp71A13 possessed less camalexin. On the other hand, in roots of both cyp71A27 and cyp71A13, camalexin concentration was significantly lower than in WT (SI Appendix, Fig. S8). Thus, CYP71A27 does not seem to contribute to camalexin synthesis in the leaves and to plant defense against leaf pathogens but is important for camalexin synthesis and interactions with bacteria and fungi in the roots.
Several CYPs Contribute to Camalexin Production in the Root.
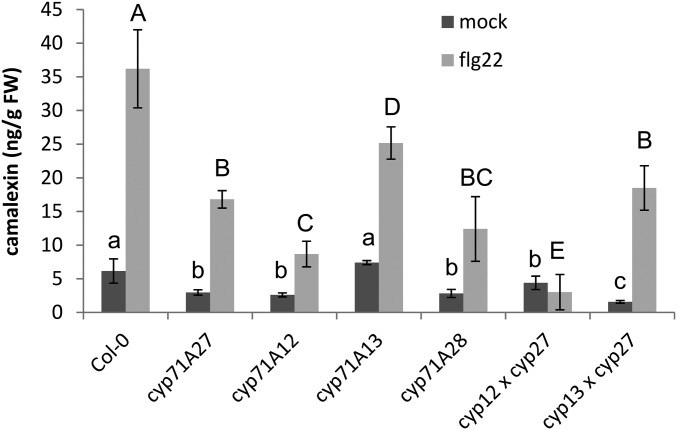
To better position CYP71A27 within the camalexin synthesis pathway, we generated double mutants of cyp71A27 with cyp71A12 and cyp71A13. To compare the contribution of the various CYPs to camalexin synthesis in the roots, the mutants were treated with flagellin. Some camalexin was present already in mock-treated plants, and significantly lower levels than in Col-0 were found in all mutants except cyp71A13, which lacks a main enzyme for camalexin synthesis in the shoots (Fig. 3). The differences were, however, much stronger after the elicitation with flg22, and all mutants produced less camalexin than the WT. Among the single mutants, cyp71A12 showed the lowest camalexin concentration, whereas in cyp71A13 the induction was only slightly less efficient than in WT, similar to the results of mock-treated roots (Fig. 3). Compared with cyp71A27, additional loss of CYP71A12 had an additive effect, but in the double mutant with cyp71A13, camalexin content was not affected compared with cyp71A27. Thus, it seems that the function of CYP71A27 is similar to the function of CYP71A12.
Fig. 3.
Contribution of enzymes of CYP71A family to camalexin synthesis. Roots of 2 1/2-wk-old seedlings were treated with flg22, and camalexin was determined by HPLC. Data are shown as means and SD from 4 independent pools of at least 10 plants. Different letters indicate significant differences at P < 0.05 (Student’s t test).
Camalexin Complements the Loss of CYP71A27.
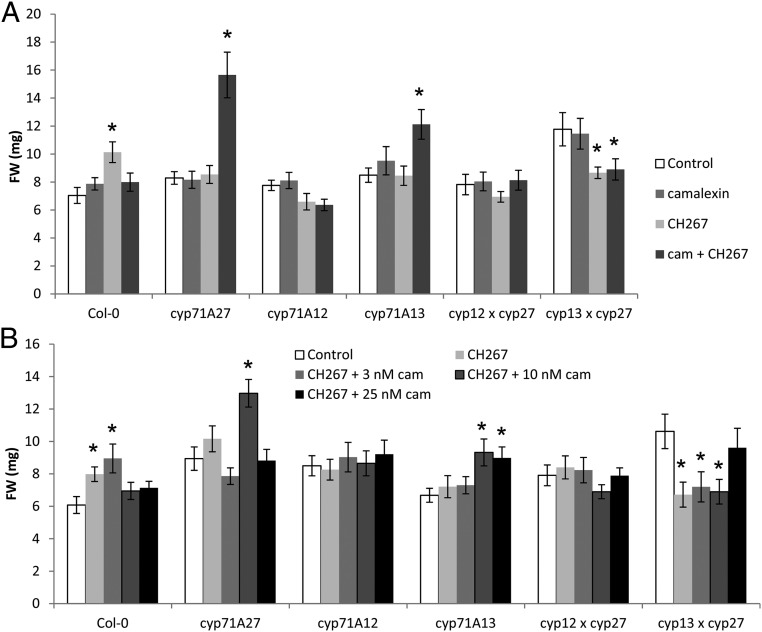
To unequivocally prove that the phenotype of cyp71A27 mutant is due to camalexin, we performed chemical complementation experiments. Col-0 and the single and double mutants in camalexin synthesis were incubated in the presence of 10 nM camalexin, or with Pseudomonas sp. CH267, or with both bacteria and camalexin. This concentration of camalexin corresponds to the difference between cyp71A27 and Col-0 at physiological conditions (Fig. 3) and does not inhibit growth of any genotype (Fig. 4A). As shown before, the cocultivation with Pseudomonas sp. CH267 led to an increase of FW for Col-0 but not for the mutants. When camalexin was added with the bacteria, the PGP effect was restored for cyp71A27 and cyp71A13 but not for cyp71A12 or the double mutants. Interestingly, the growth promotion was lost for Col-0 (Fig. 4A). The cyp71A27 cyp71A13 double mutant was ca. 70% bigger than WT plants and other mutants, and its growth was slightly inhibited with the bacteria irrespective of addition of camalexin. Two-way ANOVA revealed that the FW of the plants depends on genotype (P = 1.3 × 10−10), treatment (P = 0.0065), and genotype:treatment (P = 1.4 × 10−12). Thus, it seems that camalexin not only is important for pathogen defense but has an important function in plant interaction also with beneficial bacteria.
Fig. 4.
Camalexin is important for PGP effect of Pseudomonas sp. CH267. (A) Col-0 and mutants in camalexin synthesis were grown for 2 wk in presence of 10 µM MgCl2 as mock, Pseudomonas sp. CH267, 10 µM camalexin (cam), and both CH267 and camalexin. The FW of the whole plants was determined. Data are presented as means and SE from at least 20 plants grown on 4 independent plates. Asterisks indicate significant differences to mock treatment at P < 0.05 (Student’s t test). Two-way ANOVA revealed that FW of the plants depends on genotype (P = 1.3 × 10−10), treatment (P = 0.0065), and genotype:treatment (P = 1.4 × 10−12). (B) Col-0 and mutants in camalexin synthesis were grown for 2 wk in presence of 10 µM MgCl2 as mock, Pseudomonas sp. CH267, and CH267 supplemented with camalexin (cam) at 3 different concentrations: 3, 10, and 25 µM. The FW of the whole plants was measured. Data are presented as means and SE from at least 20 plants grown on 4 independent plates. Asterisks indicate significant differences to mock treatment at P < 0.05 (Student’s t test). Two-way ANOVA revealed that FW of the plants depends on genotype (P = 5.1 × 10−5) and genotype:treatment (P = 6.4 × 10−5).
It is possible that the total camalexin concentration, resulting from combined plant biosynthesis and external supplementation, was too high in Col-0 and, consequently, the PGP effect of the bacteria was inhibited again. We therefore incubated Col-0 and the mutants with the Pseudomonas sp. CH267 and added camalexin in 3 different concentrations. In Col-0, 3 nM camalexin did not affect the interaction, but the 2 higher concentrations (10 and 25 nM) prevented the PGP effect of the bacteria (Fig. 4B). In cyp71A27 and cyp71A13, however, it was 10 nM camalexin that restored the increase in FW, similar to previous experiments. In cyp71A12 and the double mutants the growth promotion was not restored by camalexin at these concentrations. Here the FW depended on genotype (P = 5.1 × 10−5) and genotype:treatment (P = 6.4 × 10−5) only. Therefore, it seems that there is a specific range of camalexin concentration in the root or the rhizosphere that is necessary for proper interaction with the Pseudomonas sp. CH267 strain. If the concentration is too low or too high, the interaction is disturbed. Thus, in the WT only the smallest camalexin concentration is not inhibitory, whereas in cyp71A27 and cyp71A13, which have a lower capacity for camalexin synthesis, a higher concentration of externally supplied camalexin is necessary to restore the growth promotion. The loss of CYP71A12 could not be complemented by camalexin, which may point to an additional function of this enzyme, e.g., in signaling.
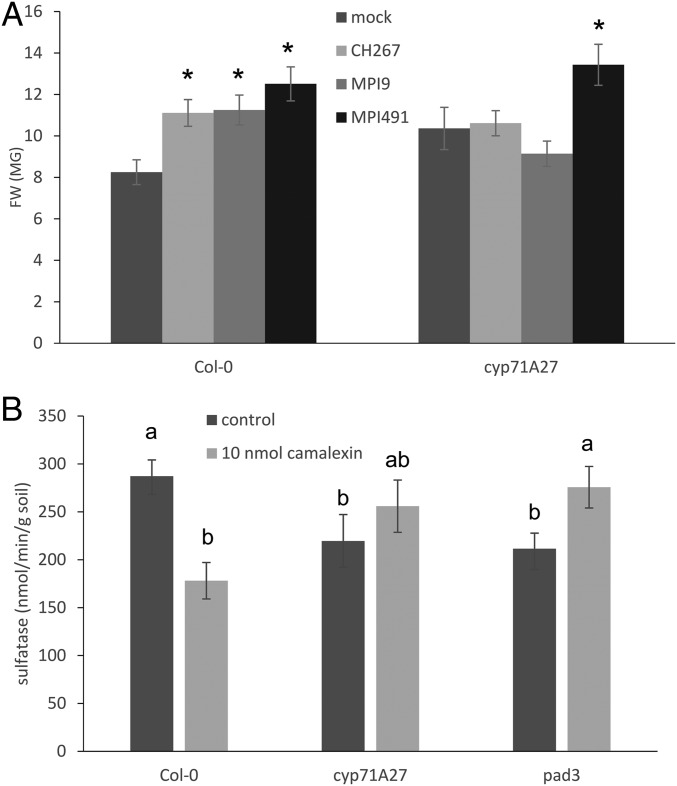
To test the ecological relevance of the observed effect of loss of CYP71A27 on plant microbe interactions we performed cocultivations with bacteria isolated from roots of Arabidopsis grown in the soil used for the initial GWAS (4). Two strains were selected, Pseudomonas sp. MPI9 and Rhizobium sp. MPI491, because they differ in the presence of arylsulfatase (SI Appendix, Fig. S9). Both strains showed a PGP effect with WT Arabidopsis, but only MPI491 (sulfatase negative) displayed PGP also with the cyp71A27 mutant (Fig. 5A). Interestingly, the 2 strains differed also in their sensitivity to camalexin, while at the concentrations used for chemical complementation their growth was not affected; 100 µM camalexin reduced growth of the MPI491 strain but not MPI9 (SI Appendix, Fig. S10). The growth of 5 bacterial strains was not different from WT Col-0, cyp71A27, or cyp71A12 root extracts as carbon source (SI Appendix, Fig. S11). Since the changes in PGP effects of the different strains on the 2 genotypes may be caused by bacterial growth, we quantified bacteria in the roots of Col-0 and cyp71A27 and tested also the effect of addition of 10 nM camalexin. No significant effects on bacterial growth of the model Pseudomonas sp. CH267 or the root isolates MPI9 and MPI491 were observed (SI Appendix, Fig. S12). This suggests that differences in bacterial growth and accumulation are not the drivers of the differences in the PGP effects but points to a metabolic exchange as its basis. Thus, camalexin seems to be affecting individual bacterial strains in Arabidopsis roots/rhizosphere differently.
Fig. 5.
Camalexin is important for sulfatase activity in soil and PGP effect of Arabidopsis root bacteria. (A) Col-0 and cyp71A27 were grown for 2 wk in presence of Pseudomonas sp. CH267 and 2 bacterial strains isolated from Arabidopsis roots MPI9 and MPI491 or 10 µM MgCl2 as mock, and the FW of the whole plants was measured. Data are presented as means and SE from at least 20 plants grown on 4 independent plates. Asterisks indicate significant differences between mock and bacterial treatment at P < 0.05 (Student’s t test). Two-way ANOVA revealed that FW of the plants depends on treatment (P = 7.9 × 10−11) and genotype:treatment (P = 4 × 10−5). (B) Col-0, cyp71A27, and pad3 plants were grown for 2 wk in soil (10%)/sand mixture; afterward, 2 soil samples were taken per plant, and sulfatase activity was measured. Data are presented as means and SE from 10 samples corresponding to 5 independent plants. Different letters indicate significantly different values at P < 0.05 (Student’s t test).
We next tested whether camalexin complements the original phenotype of cyp71A27, lower sulfatase activity in soil. Under control conditions, the activity was indeed lower in cyp71A27 as well as in another mutant in camalexin synthesis, pad3 (34). When 10 nmol camalexin was added to the nutrient solution throughout the 14 d incubation, the activity in soil from both mutants was restored to the WT levels (Fig. 5B). Interestingly, the sulfatase activity in soil from WT Col-0 was reduced after camalexin treatment. This strongly suggests that through differential effects on PGP activity of individual bacterial strains, camalexin is able to modulate microbiome function.
Natural Variation in CYP71A27 Affects Interaction with Bacteria.
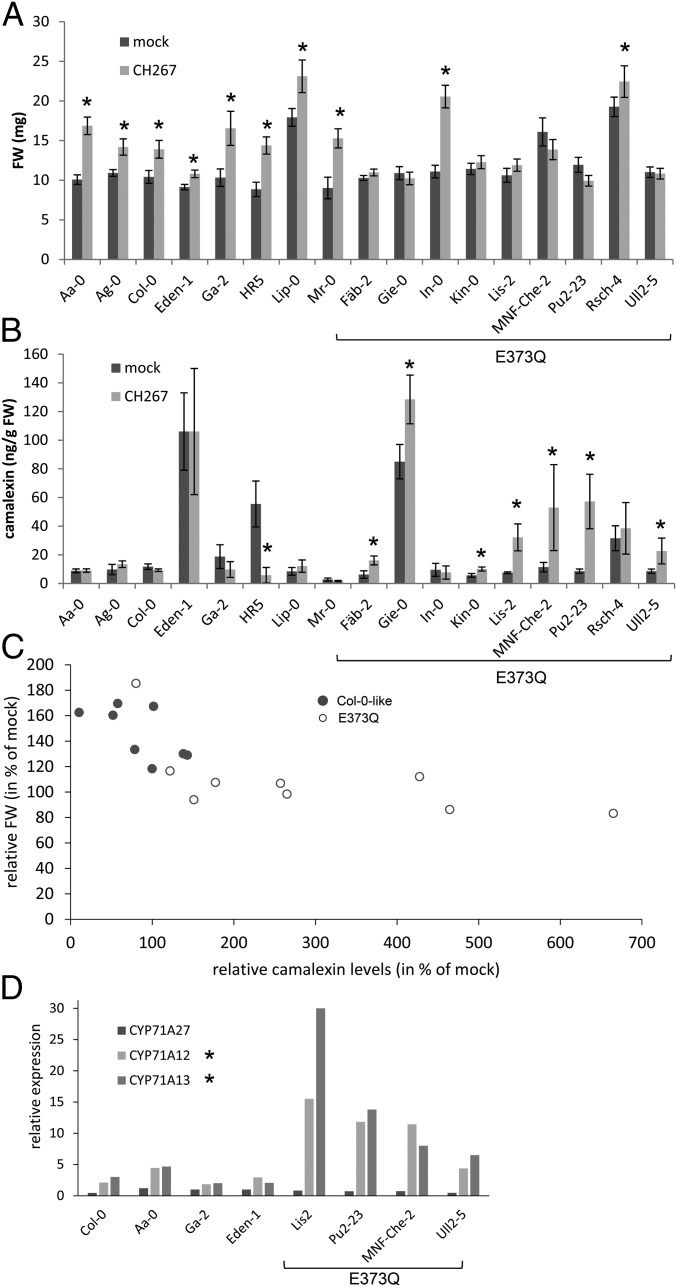
Having established the importance of CYP71A27 for camalexin synthesis and interaction with bacteria, we looked for the gene variation in natural Arabidopsis populations that could explain the different phenotypes in GWAS. We interrogated the Arabidopsis 1001 Genomes database (http://signal.salk.edu/atg1001/3.0/gebrowser.php) for variation in CYP71A27 amino acid sequence among the accessions used for the GWAS. From the 17 nonsynonymous SNPs, only 1 seemed to correlate with low activity of soil sulfatase, the SNP leading to a change of Glu-373 to Gln, albeit at a lower level of significance, P = 0.07. We therefore randomly selected 9 accessions containing the E373Q variation and 8 Col-0–like ones and tested their response to Pseudomonas sp. CH267. Interestingly, while all of the Col-0–like accessions increased FW in presence of the bacteria, 7 from the 9 E373Q harboring accessions did not profit from the PGP effect mediated by the Pseudomonas strain, the exceptions being In-0 and Rsch-4 (Fig. 6A). Three-way ANOVA revealed that FW of the plants depends on ecotype (P = 2 × 10−16), treatment (P = 8.2 × 10−14), haplotype:treatment (P = 4.7 × 10−6), and ecotype:treatment (P = 1.5 × 10−6). Thus, the E373Q variation seems to be responsible for the identification of CYP71A27 in the GWAS analysis and at least partly for the variation in Arabidopsis growth response to Pseudomonas sp. CH267.
Fig. 6.
Natural variation in CYP71A27 affects plant microbe interaction. Eight or nine representative accessions with the 2 CYP71A27 haplotypes were grown for 2 wk in presence of 10 µM MgCl2 as mock treatment or Pseudomonas sp. CH267. (A) The FW of the whole plants was measured. Data are presented as means and SE from at least 20 plants grown on 4 independent plates. Asterisks indicate significant differences between Pseudomonas and mock treatments at P < 0.05 (Student’s t test). Three-way ANOVA revealed that FW of the plants depends on ecotype (P = 2 × 10−16), treatment (P = 8.2 × 10−14), haplotype:treatment (P = 4.7 × 10−6), and ecotype:treatment (P = 1.5 × 10−6). (B) Camalexin content in the roots was determined by HPLC. Data are shown as means and SD from 3 independent pools of at least 10 plants. Asterisks indicate significant differences between Pseudomonas and mock treatments at P < 0.05 (Student’s t test). Kruskal–Wallis test revealed that the camalexin levels depend on ecotype (P = 2.6 × 10−7), treatment (P = 0.28), and haplotype (Col-0-like or E373Q, P = 0.00066). (C) The relative FW after cocultivation with Pseudomonas sp. CH267 was plotted against relative camalexin levels for the different genotypes. (D) RNA was isolated from roots of 4 accessions from each haplotype. Expression of CYP71A27, CYP71A12, and CYP71A13 was determined by qPCR in 3 independent pools of 4 roots for each accession and treatment. Shown are the ratios of transcript levels between roots cocultivated with Pseudomonas sp. CH267 and mock treated roots. The asterisks denote significant differences in the up-regulation levels between the 2 haplotypes at P < 0.05 (Student’s t test).
We then tested whether the different response to the PGP bacteria may correlate with camalexin synthesis. Indeed, while camalexin concentrations in roots varied highly within the 2 groups of accessions, the groups showed differences in the response of camalexin accumulation to the incubation with Pseudomonas sp. CH267 (Fig. 6B). Whereas in the Col-0–like accessions, camalexin levels were not affected or were reduced by the bacteria, the E373Q group showed induction in camalexin contents, except In-0 and Rsch-4. Interestingly, not the steady-state camalexin concentration but rather the ability to increase camalexin synthesis during the incubation correlates with the presence of the E373Q allele and particularly with the growth response to the Pseudomonas sp. CH267. It has to be noted, however, that total camalexin content still influences the PGP effect, as the Col-0–like accession with the highest baseline camalexin level, Eden-1, showed the smallest but still significant increase in FW (Fig. 6).
Camalexin is exuded from roots in response to elicitation with, e.g., flagellin (21). Therefore, we tested whether the variation in root camalexin between the 2 groups affects concentration of camalexin in root exudates. Pseudomonas sp. CH267 and P. simiae WCS417r induced camalexin exudation, but the amount of camalexin in the exudates of accessions with the E373Q allele was strongly increased by each of the strains compared with accessions without this allele (SI Appendix, Fig. S13). Two-way ANOVA confirmed that the camalexin amount depends on haplotype (P = 1.7 × 10−5), treatment (P = 8.2 × 10−5), and haplotype:treatment (P = 0.0059). Thus, in these accessions the elevated root concentration of camalexin correlates with its increased exudation.
Enhanced camalexin synthesis in the accessions with E373Q allele could result from higher activity of the modified enzyme or compensatory effects of other enzymes, e.g., CYP71A12 or CYP71A13. Therefore, we compared the transcript levels of CYP71A12, CYP71A13, and CYP71A27 in roots of plants cocultivated with Pseudomonas sp. CH267 on the agar plates. Whereas CYP71A27 expression was not induced by the bacteria, the CYP71A12 and CYP71A13 transcript levels were significantly higher in all genotypes (Fig. 6D). This induction was, however, much more strongly pronounced in the accessions with the E373Q haplotype, resembling the induction of these genes in the cyp71A27 mutant (SI Appendix, Fig. S3). The increased expression of CYP71A12 and/or CYP71A13 may thus overcompensate the loss or possible inactivation of CYP71A27 and lead to increased camalexin production with effects on the PGP effects of the bacteria. Altogether, this shows that variation in CYP71A27 affects response of both root concentration and exudation of camalexin to bacteria and again points to an important role of camalexin in beneficial plant–microbe interactions.
Discussion
To decipher how plants affect the function of their associated microbial community, sulfatase activity is a suitable quantitative measure since it is absent in plants (16). Indeed, growing diverse Arabidopsis accessions under the same conditions affected strongly the sulfatase activity in soil, which is a strong indication for different shifts in the communities caused by these accessions. The GWAS based on the sulfatase data resulted in 6 candidate genes that potentially affect the soil microbiota confirmed by analysis of T-DNA lines. Since these genes belong to gene families involved in secondary metabolism, they are very good candidates because metabolites excreted from roots in the form of exudates are expected to shape the root microbiome (2, 9, 11). Only a few such metabolites have been shown to have a direct impact on the microbiome, and the candidate genes may thus potentially identify new mechanisms of the interaction. Camalexin, identified in this analysis as associated to the CYP71A27 gene, is an important addition to the still limited number of microbiome shaping metabolites. These include, e.g., scopoletin, a member of a coumarin family of metabolites which are excreted into the rhizosphere to mobilize iron and affect the microbiome composition (8) and aromatic organic acids that cause shifts in the community depending on ability of bacterial strains to metabolize specific acids (9). Camalexin is exuded from Arabidopsis roots in response to pathogens or flagellin (21) but has not been identified in root exudates of sterile grown Arabidopsis previously (11). We detected small amounts of camalexin in exudates of sterile plants and showed that its exudation can be elicited by rhizospheric bacteria (SI Appendix, Fig. S13). Due to this inducibility and its antimicrobial effects on some bacteria, camalexin is thus a good candidate for a plant metabolite capable of shaping the rhizosphere microbial community.
The CYP71A27 has been predicted to be associated with camalexin synthesis by the PlantCyc database. Here we provide multiple lines of evidence that CYP71A27 is indeed a component of camalexin biosynthetic pathway. This includes sequence similarity between CYP71A27 and known camalexin biosynthesis genes CYP71A12 and CYP71A13, comparable phenotypes of cyp71A27 mutant and cyp71A12 and cyp71A13 (Figs. 3 and 4 and SI Appendix, Fig. S8), compensatory induction of camalexin synthesis genes in the cyp71A27 mutant (SI Appendix, Fig. S3), and also the additive effect of the double mutation cyp71A27 cyp71A12 (Fig. 3). The chemical complementation by camalexin of the loss of growth promotion by Pseudomonas sp. CH267 in cyp71A27 (Fig. 4) and of the sulfatase activity (Fig. 5B) then unequivocally placed the gene into the camalexin biosynthetic network. The similarity with the known CYPs suggests that CYP71A27 catalyzes the same reaction(s) as CYP71A12 and CYP71A13, but due to nonoverlapping expression patterns (SI Appendix, Figs. S6 and S7) it is more important for camalexin biosynthesis in the roots. Indeed, while CYP71A12 is strongly inducible by flg22, CYP71A27 is constitutively expressed and therefore most probably responsible for the small amount of camalexin exuded from sterile roots (SI Appendix, Figs. S7 and S13).
The mutant in a gene adjacent to CYP71A27, CYP71A28, also showed similar characteristics: phenotypes similar to the mutants in the other 3 CYPs (Fig. 3), changes in gene expression of the camalexin biosynthetic pathway (SI Appendix, Fig. S4), and slight reduction in camalexin accumulation (Fig. 3). However, due to low expression in comparison with the other genes (SI Appendix, Fig. S6) and the inability to amplify a full coding region it is not possible to draw confident conclusions about the involvement of CYP71A28 in the camalexin pathway. Interestingly, despite some variation among individual experiments, in control conditions and across all experiments the FW of cyp71A27 was ca. 20% greater than FW of the WT Col-0 plants.
Camalexin contributes primarily to Arabidopsis defense against necrotrophic fungal pathogens such as B. cinerea and Alternaria brassicicola (32, 35). Its synthesis is induced by both virulent and avirulent bacteria, such as various strains of P. syringae. Even though some bacterial strains, such as P. syringae pv. maculicola strain ES4326 or P. syringae pv tomato, turned out to be camalexin sensitive, generally, camalexin does not seem to play the key role in resistance against bacterial pathogens (33, 36–39). To date, most investigations of camalexin function were restricted to the leaves (40, 41). The identification of CYP71A27 in our screen thus reveals the importance of extending the analysis of camalexin function to the roots. Several links between root processes and camalexin have been made already. Camalexin has been shown to accumulate in roots infected with an oomycete pathogen (42) and to be exuded from roots in response to flagellin (21). It has also been shown to execute systemic resistance: camalexin synthesis is induced in leaves by a root-colonizing bacterium Pseudomonas fluorescens strain SS101 and is important for resistance against P. syringae pv tomato (38). Genes for camalexin synthesis are also induced in plants treated with the PGP rhizobacterium Paenibacillus polymyxa E681, correlating with increased resistance against B. cinerea in the leaves (43). However, no increase in camalexin accumulation in roots was seen in Col-0 plants treated with Pseudomonas sp. CH267 (Fig. 6B). Remarkably, this was not true for all Arabidopsis accessions. There was a considerable variation in steady-state camalexin levels and in how its accumulation responded to bacteria treatment (Fig. 6). It is not surprising to find natural variation in camalexin content and function. Substantial variations in camalexin steady-state levels and in its responses to B. cinerea or AgNO3 have been measured in leaves of Arabidopsis thaliana accessions before (40, 41, 44). Using quantitative genetics, a substantial overlap between quantitative trait loci for resistance to B. cinerea and camalexin accumulation was revealed (40, 41, 44). The variation seen in our study (Fig. 6) seems to have consequences for the PGP effect of Pseudomonas sp. CH267, however, not necessarily through camalexin concentration but rather through the ability to induce camalexin synthesis and exudation (SI Appendix, Fig. S13). The major difference between the accessions with stable camalexin levels and susceptible to the growth promotion on one hand and those inducing camalexin and not showing a growth response to Pseudomonas sp. CH267 on the other hand seems to be due to allelic variation in CYP71A27. Given the increase in camalexin root content and exudation in the accessions carrying the E373Q allele, it is tempting to speculate that this allele is the more active one. However, E373 is conserved in a number of P450 enzymes of the A71 group, most importantly, the CYP71A12 and CYP71A13. The expression analysis of the 2 groups of accessions suggested that the E373Q allele is linked to a greater induction of the expression of CYP71A12 and CYP71A13 (Fig. 6D) and thus an indirect increase in camalexin synthesis. Whether and how this amino acid change affects the enzyme activity and what is the CYP71A27 exact catalytic function still needs to be determined.
Our results indicate that the microbial response to camalexin is strongly dependent on concentration. For instance, the PGP effect of Pseudomonas sp. CH267 can be abolished both by loss of CYP71A27 function and also by a possible increase in its activity. This is consistent with the results of chemical complementation with varying concentrations of camalexin (Fig. 4B). In Col-0 the lowest concentration of camalexin did not affect the growth promotion, but higher ones abolished the PGP effect, whereas higher camalexin concentrations were necessary for complementation of the biosynthetic mutants cyp71A27 and cyp71A13. It seems therefore that a tightly regulated narrow range of camalexin concentration is necessary to ensure the PGP effect of Pseudomonas sp. CH267. Presumably, too little camalexin, e.g., in cyp71A27 mutant, allows the bacteria to overgrow the plant and suppress the growth promotion or even act antagonistically, which might be the reason for reduction in FW of cyp71A27 cyp71A13 double mutant cultivated with the bacteria. On the other hand, too much camalexin either supplied externally (Fig. 4) or induced by the bacteria (Fig. 6) may inhibit bacterial growth and prevent the growth promotion. However, no significant differences in bacterial titers in the roots were observed between Col-0 and cyp71A27 and in both genotypes with and without camalexin (SI Appendix, Fig. S12). It seems, therefore, that camalexin affects other signaling processes and/or metabolic exchanges necessary for establishing the PGP interaction.
The cocultivations with well-established model as well as newly described PGP bacteria were useful to establish the function of CYP71A27 and camalexin in plant microbe interactions; however, these experiments were independent from the initial screen. Therefore, coming back to the original soil-based experiments, the finding that exogenous camalexin complements sulfatase activity in cyp71A27 and pad3 mutants is important evidence for relevance of our findings with one-to-one interactions in the ecological context (Fig. 5). Interestingly, camalexin reduced sulfatase activity in Col-0 soil, which shows its effect on the global microbiome function and is reminiscent of the reduction in PGP effect of the Pseudomonas sp. CH267 after addition of camalexin (Fig. 4). It seems, therefore, that also in soil, camalexin acts depending on its concentration. The contrasting effect of the 2 strains from the collection of Arabidopsis root bacteria (Fig. 5B), as well as different sensitivity of these strains to camalexin (SI Appendix, Fig. S10), indicates that the changes in sulfatase activity in soil are not caused by a simple increase or decrease in the growth rate of the community but most probably through differential sensitivity of various strains resulting in alterations in community structure. Indeed, camalexin did not directly inhibit the sulfatase activity (SI Appendix, Fig. S14), corroborating this hypothesis. The next step is thus clearly the investigation how camalexin affects the composition of root microbiome in soil. In addition, whether the presence of sulfatase in bacterial genomes is linked to their sensitivity to camalexin, as indicated by the cocultivation experiments, needs to be further investigated with a larger number of strains.
In conclusion, we developed an assay to assess the effect of plant genotype on function of the plant-associated microbiome and used it for GWAS with Arabidopsis accessions to identify candidate genes affecting the rhizosphere microbial community. One candidate gene, CYP71A27, was shown to be an additional component of the camalexin biosynthesis pathway specifically in roots. Using a loss of function mutant in this gene we revealed a function of camalexin in enabling plant growth promotion by rhizobacteria. These results contribute to better understanding of the mechanisms by which plants shape their associated microbiome.
Materials and Methods
Plant Material and Growth Conditions.
For the sulfatase measurements seeds of A. thaliana accessions were surface sterilized with chlorine gas and plated onto plates containing 1/2 strength Murashige Skoog (MS) nutrient solution supplemented with 0.5% sucrose and 0.8% agarose. After 3 d stratification at 4 °C in the dark the plates were incubated vertically in growth chamber (Percival) under long-day conditions (16 h light/8 h dark), 120 μE m−2 s−1, at 22 °C for 9 d. The seedlings were transferred to 40-well (5 × 8) plant trays filled with mix (9:1) of sand and soil, using soil collected at Max Planck Institute for Plant Breeding Research (MPIPZ) Cologne and used previously to characterize Arabidopsis microbiome structure (3). Each tray was planted with Col-0 seedlings for normalization of the data. The trays were further incubated in the growth chambers under the same conditions for 2 wk and were watered with sulfate free Long Ashton nutrient solution. For study of natural variation, 172 accessions (SI Appendix, Table S1) from the core360 collection representing species-wide diversity (18) were analyzed. T-DNA lines disrupting the candidate genes were obtained from the Nottingham Arabidopsis Stock Centre (NASC) and genotyped by PCR (for accession numbers and primers, see SI Appendix, Table S3) to obtain homozygous mutants. The mutants were grown in the same way and were analyzed twice independently.
The camalexin synthesis mutants cyp71A12 (GABI_127H03), cyp71A13 (SALK_105136), pad3 (SALK_026585), and cyp71A12 cyp71A13 (45) were obtained from H. Frerigmann, MPIPZ Cologne, Cologne, Germany. cyp71A27 was crossed with cyp71A12 and cyp71A13 to generate double mutants. Segregating F2 seeds were screened by PCR at 1 locus; plants homozygous for the insertion at this locus were then screened at the second locus. The seeds of CYP71A12::GUS were shared by F. M. Ausubel, Harvard Medical School, Boston, MA.
For metabolite and expression analyses the plants were grown on square Petri dishes filled with 50 mL 1/2 MS media supplemented with 0.5% sucrose and 0.8% agarose, placed vertically in a growth chamber (Percival) under long-day conditions (16 h light/8 h dark), 120 μE m−2 s−1, at 22 °C for 18 d.
Bacterial Strains and Conditions for Cocultivation Experiments.
For cocultivation experiments, 3 previously characterized PGP bacteria, Pseudomonas sp. CH267 (26), obtained from J. R. Dinneny, Stanford University, Stanford, CA; P. simiae WCS417r (27), obtained from C. Pieterse, Utrecht University, Utrecht, The Netherlands; and P. phytofirmans PsJN (28), obtained from A. Zuñiga Sepulveda, Universidad Adolfo Ibañez Peñalolen, Santiago, Chile, were used, as well as a root pathogen B. glumae PG1 (29), obtained from K.-E. Jäger, Heinrich Heine Universität Düsseldorf, Düsseldorf, Germany (SI Appendix, Table S5). In addition, Pseudomonas sp. MPI9 (NCBI Taxonomy ID 1736604) and Rhizobium sp. MPI491 (NCBI Taxonomy ID 1736548), both isolated from field-grown Arabidopsis roots (4) and provided by Prof. P. Schulze-Lefert, MPIPZ Cologne, Cologne, Germany, were used (SI Appendix, Table S5). The bacteria were kept as glycerol stocks and plated freshly before use on LB plates supplemented with corresponding antibiotics. Sulfatase activity was confirmed by enzyme measurements in CH267, MPI9, and PG1 (SI Appendix, Table S5).
Sterile seeds of different A. thaliana genotypes were placed on Petri dishes containing 1/2 MS with 0.5% sucrose and 0.8% agarose. After 3 d stratification the plates were placed vertically into the growth chamber for 5 d. Overnight cultures of the bacteria in LB were grown at 28 °C (CH267, WCS417r, MPI9, and MPI491) or 30 °C (PsJN and PG1). Five milliliters of the cultures were centrifuged for 5 min at 3,200 rpm and washed twice in 3 mL of sterile 10 mM MgCl2. Optical densities at 600 nm of the bacterial suspensions were determined, and the bacteria were diluted into warm (∼42 °C) agar containing Long Ashton medium to final OD = 3.2 × 10−6 (CH267, WCS417r, PsJN, MPI9, and MPI491) or OD = 5 × 10−5 (PG1), and 50 mL were poured into square Petri dishes. For mock treatment, MgCl2 in the same dilution was added. After the plates were hardened, the pregrown plants were transferred onto the plates, using sterile forceps, and incubated for further 14 d in growth chambers as described before.
For quantification of bacteria, roots (ca. 5 mg) were collected in 300 µL 10 mM sterile MgCl2, vortexed for 10 s, and shaken at 500 rpm for additional 10 min. The bacterial suspension was diluted 10,000- and 20,000-fold, and 100 µL were spread on LB plates. The plates (at least 4 per biological replicate) were incubated at 28 °C for 24–40 h, and colonies were counted.
Pathogen Inoculation and Scoring.
Plants were grown for 4 wk under short‐day conditions as described in Liu et al. (46). B. cinerea B05.10 kindly provided by P. Tudszynski, University of Münster, Münster, Germany, was cultured on Sabouraud Maltose agar for ∼2 wk until sporulation. Spores were harvested from agar plates in 4% Sabouraud Maltose Broth and used for spray inoculations of leaves at 2 × 105 spores per mL. For droplet inoculations, 4 5-μL droplets of 5 × 104 spores per mL were applied to single leaves of 4.5‐wk‐old plants (46). For mock treatment, 4% Sabouraud Maltose Broth was used. During infection, plants were kept under sealed hoods at high humidity under short-day conditions (10 h light/14 h dark) at 22 °C. Lesion diameter and area were determined after 3 d in plants inoculated with droplets. Spray-inoculated plants were used for camalexin measurements.
P. syringae pv maculicola ES4326 (Psm) at an OD600 of 0.005 was used to pressure-infiltrate 3 fully expanded rosette leaves of 4.5-wk-old Arabidopsis plants as described in Stahl et al. (47). For camalexin quantification, infiltrated leaves were harvested at 2 d after infiltration. As mock control, 10 mM MgCl2 was used for infiltrations. Six plants were used per treatment, and leaves from 2 plants were pooled per replicate. To assess Psm growth performance, a Psm strain expressing the LUX operon from Photorhabdus luminescens (Psm lux) was pressure-infiltrated into 3 fully expanded rosette leaves of 4.5-wk-old Arabidopsis plants at an OD600 of 0.001. Two days later, discs from infiltrated leaves were used to quantify the luminescence intensity per leaf area (relative light units). Plants were kept at short-day conditions (10 h light/14 h dark) at 20 °C. Leaf discs from 6 plants per genotype (18 replicates) were analyzed.
Fungal Colonization Assays.
S. indica (DSM11827; Deutsche Sammlung von Mikroorganismen und Zellkulturen) was grown at 28 °C in the dark on plates with solid (1.5% agar) complete medium. Fungal spores were obtained as described in Banhara et al. (48). Seven-day-old Arabidopsis seedlings grown at long days (16 h light) on solid 1/2 MS medium were inoculated with 0.3 mL of spore suspension (500,000 spores per mL) in 0.002% Tween20 directly applied to each root. After 14 d incubation in growth chamber at long days the roots were harvested and thoroughly washed to remove fungal hyphae from the surface and pooled from 80 to 100 plants for DNA extraction. DNA was isolated from pools of 200 mg roots, and the colonization rate was determined by qPCR as described in Lahrmann et al. (49) using primers from transcription elongation factor alpha of S. indica (TEF, GenBank AJ249911) and Arabidopsis ubiquitin (UBQ5, At3g62250) (SI Appendix, Table S6).
Generation of Transgenic Plants.
For complementation of cyp71A27, the promoter (2,460 bp upstream from the translation initiation site) and the gene were amplified separately by PCR from Col-0 DNA using primers CYP27PROFOR/CYP27PROREV and CYP27FOR/CYP27REV, respectively, described in SI Appendix, Table S6, so that they overlapped by 270 bp. The PCR fragments were cloned into pENTR/D-TOPO (ThermoFisher) and verified by sequencing. The 2 fragments were joined together by cloning using SpeI (−242 bp from translation initiation) and AscI (pENTR/D-TOPO), and the full promoter–gene construct was inserted to pMDC111 (50) by GATEWAY cloning. The resulting binary plasmid was transformed to Agrobacterium tumefaciens GV3101 (pMP90), and cyp71A27 plants were transformed by the floral dip method. Transgenic plants were selected on 1/2 MS agar media containing 50 mg L−1 hygromycin B. Hygromycin-resistant T2 progenies were tested for their segregation ratio in the T3 generation, and 6 lines with 3:1 ratio of resistant:sensitive seedlings, indicating a single insertion, were selected for further analysis. The 3 lines with expression of CYP71A27 similar to WT showed complementation of both phenotypes, low sulfatase activity and loss of PGP effect of Pseudomonas sp. CH267 proving that all mutant phenotypes are indeed caused by a disruption of the CYP71A27 gene (SI Appendix, Fig. S15).
To express GUS reporter gene under control of CYP71A27 promoter the 2,460-bp promoter fragment in pENTR/D-TOPO was inserted into pGWB3 plasmid (51) by GATEWAY cloning. The binary construct was transformed in Col-0 plants as described above. Transgenic plants were initially selected on 1/2 MS agar media containing 50 mg L−1 kanamycin sulfate, and suitable lines were obtained as described above.
Sulfatase Activity.
After 2 wk of growth the plants were completely bedded out of the tray, and 2 sand/soil samples per well of ca. 1 g were taken with a scoop directly from the area where the root grew, collected in 2-mL Eppendorf tubes, weighed, and frozen in liquid nitrogen. The activity was determined essentially as described in Margesin et al. (52). Sodium acetate (600 µL 0.5 M, pH 5.8) was added to the soil samples, and after vortexing for 5 s and mixing by rotation for 6 min, 37.5 µL toluene were added; 150 µL of 5 mM p-nitrophenyl sulfate were added, vortexed for 5 s, and incubated for 1 h at 37 °C with manual shaking in 10-min intervals. The reactions were stopped by 150 µL 0.5 M CaCl2 and 600 µL 0.5 M NaOH and vortexing for 5 s. The tubes were centrifuged for 10 min at 13,000 rpm, and 850 µL of supernatant were pipetted into semimicro cuvettes. Absorbance at 400 nm was determined with water as blank, and the activity was determined using a standard curve of p-nitrophenyl solutions of different concentrations. Background activity was determined by measuring soil samples incubated at the same conditions without plants and was subsequently subtracted from the activity measured in samples from soil in which the plants grew.
Genome-Wide Association Mapping.
Genome-wide association mapping with sulfatase data from 172 accessions was performed using the GWAPP portal (https://gwas.gmi.oeaw.ac.at/) (19). The accelerated mixed model, which corrects for population structure confounding, was used with 250 k SNP dataset and the default settings. Genomic regions of ±20 kBp from the SNPs with −log10P score > 4 were inspected to determine candidate genes. Selection was performed based on gene annotation, focusing on genes annotated to be connected to sulfur metabolism or to secondary metabolism. T-DNA lines disrupting the candidate genes were obtained from NASC and genotyped by PCR (for accession numbers and primers, see SI Appendix, Table S4) to obtain homozygous mutants.
Camalexin Measurements.
Camalexin was extracted from ca. 20 mg plant material in 100 µL of dimethlysulfoxide (DMSO) for 20 min with shaking, and after centrifugation, 20 µL were injected into a Thermo Scientific Dionex UltiMate 3000 HPLC system with Waters Spherisorb ODS-2 column (250 mm × 4.6 mm, 5 µm). The samples were resolved using a gradient of 0.01% (vol/vol) formic acid (solvent A) and a solvent mixture of 98% (vol/vol) acetonitrile, 2% (vol/vol) water, and 0.01% (vol/vol) formic acid (solvent B). The gradient program was as follows: 97% A for 5 min, 90% A in 5 min, 40% A in 8 min, 20% A in 2 min, 0% A in 20 min and kept at 0% A for 10 min, 100% A in 2.5 min and kept 100% A for 3.5 min, and 97% A in 2 min and kept 97% A for 2 min. Camalexin was detected at an excitation at 318 nm and emission at 368 nm by fluorescence (FLD sensitivity set to 3) as described in Bednarek et al. (53). For the quantification of camalexin, external standards were used ranging from 1 pg to 1 ng per µL. Alternatively, for some experiments, a method using a methanol–water (80:20, vol/vol) extract which was evaporated to dryness before silylation was used to quantify camalexin by GC-MS as described in Hartmann et al. (54).
Camalexin Exudation Experiments.
Six seeds per well were germinated on sterile 100 µm nylon mesh in 12-well plates placed on 1 mL of 1/2 MS medium with 0.5% sucrose. Seedlings were grown at 22 °C at long-day conditions (16 h light) for 6 d. One day before inoculation, the medium was exchanged to 1/2 MS without sucrose. The bacteria were grown overnight at 28 °C and washed 2 times with 10 mM MgCl2. Seven microliters of the cultures diluted to OD600 = 0.0001 were inoculated into each well. The plates were further incubated under the same conditions for 6 d with occasional shaking. The nutrient solution was collected from each well (4 biological replicates per genotype and treatment). The media were centrifuged at maximal speed for 15 min and purified using 1 mL solid phase extraction tubes (Discovery-DSC18) according to manufacturer’s instructions. Samples were eluted with 90% of acetonitrile and 0.1% formic acid, dried in speed vac, and dissolved in 50 µL DMSO. Twenty microliters were injected into HPLC and analyzed as described above.
Expression Analysis.
To determine transcript levels total RNA was isolated by standard phenol/chlorophorm extraction and LiCl precipitation. First-strand cDNA was synthesized from 800 ng of total RNA using QuantiTect Reverse Transcription Kit (Qiagen), which includes a DNase step to remove possible DNA contamination. Quantitative real-time RT-PCR (qPCR) was performed using gene-specific primers (SI Appendix, Table S6) and the fluorescent intercalating dye SYBR Green (Promega) as described in Lee et al. (55). All quantifications were normalized to the TIP41 (AT4G34270) gene. The RT-PCR reactions were performed in duplicate for each of the 3 independent samples.
GUS Activity.
GUS assays were performed in 24- or 6-well plates in 750 µL or 3 mL GUS staining solution (50 mM Na2HPO4 [pH 7.2], 0.2% Triton × 100, 2 mM K-ferrocyanid, 100 mM K-ferricyanid, and 2 mM X-Gluc). After incubation overnight at 37 °C the plants were washed with 10% ethanol for 30 min, followed by 30% ethanol for 1 h and 50% ethanol for 1 h. After these washing steps, the plant samples were stored in 70% ethanol at 4 °C. Photos were made using a Leica DMRB microscope with 5×, 10×, and 20× magnification or a Leica MZ 16 F stereomicroscope. At least 2 plants from 3 independent transgenic lines were analyzed.
To determine the regulation of CYP71A27 and CYP71A12 in roots we analyzed CYP71A27pro::GUS and CYP71A12pro::GUS (21) lines. The seedlings were grown vertically for 7 d on plates containing 1/2 MS medium with 0.5% sucrose. The seedlings were carefully transferred into 1 mL of liquid 1/2 MS without sucrose and kept for 24 h. Pseudomonas sp. CH267 and P. simiae WCS417r were grown overnight, washed 2 times with 10 mM MgCl2, and diluted to OD600 = 0.04. Fifty microliters of bacterial suspensions (final OD600 = 0.002), 20 µM flg22 (final concentration 1 µM), or 10 mM MgCl2 as mock were added to the liquid. After 24 h the medium was substituted with GUS staining solution, and the plates were placed into 37 °C overnight. After washing with ethanol, dilution series photos were made using a Leica DMRB microscope with 10× magnification. At least 2 plants from 3 independent transgenic lines were analyzed.
Photos from whole-plant rhizospheres were made with a Leica MZ 16 F stereomicroscope after 1 wk of cocultivation with corresponding bacteria, as described earlier. For flg22 treatment, 2-wk-old plants were flooded with 1 µM solution on the plate for 5 h (21).
Bacterial Growth Assays.
All bacterial growth assays were conducted in a 48-well plate using a plate reader (Tecan) at 28 °C with shaking. Sulfatase activity of individual bacterial strains was measured by quantifying p-nitrophenyl concentration in the culture supernatant by absorbance at 400 nm, after bacteria were cultivated for 48 h on M9 minimal medium with p-nitrophenyl sulfate (PNPS) as the sole sulfur source. To determine the effect of camalexin on sulfatase activity, the cultures with PNPS were supplemented by camalexin at different concentrations (0, 3 nM, 10 nM, 25 nM, 75 nM, 1 µM, 10 µM, and 100 µM), and the p-nitrophenyl was quantified spectrophotometrically after 48 h. The effect of camalexin on bacterial growth was measured by cultivating individual bacterial strains on 1/2 TSB in the presence of different camalexin concentrations and quantifying the derived growth using the Growthcurver package (56). Growth of bacterial strains on root extracts harvested from different Arabidopsis genotypes was conducted according to the method of Jacoby et al. (14). Arabidopsis plants were grown in shaking liquid culture, then root metabolites were extracted and provided as the sole carbon source in M9 growth medium. Individual bacterial strains were inoculated into media, and bacterial growth was monitored by measuring OD600 values over time.
Statistical Analysis.
The normality of distribution of sulfatase activity was calculated using chi-square goodness-of-fit test in Excel. Significant differences between the WT and the T-DNA lines were analyzed according to the Student’s t test for P < 0.05. On some datasets, 2-way ANOVA analyses with type II sum of squares was performed, followed by a Tukey’s HSD post hoc test, using the R-studio or, alternatively, the Kruskal–Wallis test. χ2 test in Excel was used to test the significance of enrichment of candidate genes by GWAS.
Supplementary Material
Acknowledgments
This research was funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy, EXC-Nummer 2048/1, project 390686111, and within the SPP 2125 DECRyPT. We thank Paul Schulze-Lefert (MPIPZ Cologne), José R. Dinneny (Stanford University), Corné Pieterse (Utrecht University), Ana Zuñiga Sepulveda (Universidad Adolfo Ibañez Peñalolen), and Karl-Erich Jäger (Heinrich Heine Universität Düsseldorf) for the bacterial strains; Henning Frerigmann (MPIPZ Cologne) for Arabidopsis mutants in camalexin synthesis; and Fred M. Ausubel for the CYP71A12::GUS line.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818604116/-/DCSupplemental.
References
- 1.Almario J., et al. , Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl. Acad. Sci. U.S.A. 114, E9403–E9412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacoby R., Peukert M., Succurro A., Koprivova A., Kopriva S., The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant Sci. 8, 1617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulgarelli D., et al. , Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Bai Y., et al. , Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Castrillo G., et al. , Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., Schulze-Lefert P., Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Zgadzaj R., et al. , Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 113, E7996–E8005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stringlis I. A., et al. , MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U.S.A. 115, E5213–E5222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhalnina K., et al. , Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Sasse J., Martinoia E., Northen T., Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Mönchgesang S., et al. , Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data. Sci. Rep. 6, 29033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton M. W., et al. , Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 5, 5320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wintermans P. C., Bakker P. A., Pieterse C. M., Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 90, 623–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby R. P., Martyn A., Kopriva S., Exometabolomic profiling of bacterial strains as cultivated using Arabidopsis root extract as the sole carbon source. Mol. Plant Microbe Interact. 31, 803–813 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Klose S., Moore J. M., Tabatabai M. A., Arylsulfatase activity of microbial biomass in soils as affected by cropping systems. Biol. Fertil. Soils 29, 46–54 (1999). [Google Scholar]
- 16.Kertesz M. A., Fellows E., Schmalenberger A., Rhizobacteria and plant sulfur supply. Adv. Appl. Microbiol. 62, 235–268 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Tejada M., Hernandez M. T., Garcia C., Application of two organic amendments on soil restoration: Effects on the soil biological properties. J. Environ. Qual. 35, 1010–1017 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Platt A., et al. , The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6, e1000843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seren Ü., et al. , GWAPP: A web application for genome-wide association mapping in Arabidopsis. Plant Cell 24, 4793–4805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koprivova A., Harper A. L., Trick M., Bancroft I., Kopriva S., Dissection of the control of anion homeostasis by associative transcriptomics in Brassica napus. Plant Physiol. 166, 442–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millet Y. A., et al. , Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nafisi M., et al. , Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19, 2039–2052 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glawischnig E., Camalexin. Phytochemistry 68, 401–406 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Schläpfer P., et al. , Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173, 2041–2059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lugtenberg B., Kamilova F., Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Haney C. H., Samuel B. S., Bush J., Ausubel F. M., Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 1, 15051 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pieterse C. M., van Wees S. C., Hoffland E., van Pelt J. A., van Loon L. C., Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8, 1225–1237 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poupin M. J., Timmermann T., Vega A., Zuñiga A., González B., Effects of the plant growth-promoting bacterium Burkholderia phytofirmans PsJN throughout the life cycle of Arabidopsis thaliana. PLoS One 8, e69435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao R., et al. , Genome-wide RNA sequencing analysis of quorum sensing-controlled regulons in the plant-associated Burkholderia glumae PG1 strain. Appl. Environ. Microbiol. 81, 7993–8007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahrmann U., et al. , Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol. 207, 841–857 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Krishnakumar V., et al. , Araport: The Arabidopsis information portal. Nucleic Acids Res. 43, D1003–D1009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrari S., Plotnikova J. M., De Lorenzo G., Ausubel F. M., Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35, 193–205 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Glazebrook J., Ausubel F. M., Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 91, 8955–8959 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou N., Tootle T. L., Glazebrook J., Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11, 2419–2428 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomma B. P., Nelissen I., Eggermont K., Broekaert W. F., Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19, 163–171 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Bednarek P. Chemical warfare or modulators of defence responses—The function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15, 407–414 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Rogers E. E., Glazebrook J., Ausubel F. M., Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol. Plant Microbe Interact. 9, 748–757 (1996). [DOI] [PubMed] [Google Scholar]
- 38.van de Mortel J. E., et al. , Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 160, 2173–2188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su T., et al. , Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23, 364–380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kliebenstein D. J., Rowe H. C., Denby K. J., Secondary metabolites influence Arabidopsis/Botrytis interactions: Variation in host production and pathogen sensitivity. Plant J. 44, 25–36 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Corwin J. A., et al. , The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 12, e1005789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bednarek P., Schneider B., Svatos A., Oldham N. J., Hahlbrock K., Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 138, 1058–1070 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon Y. S., et al. , Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16, 122–135 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Rowe H. C., Kliebenstein D. J., Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180, 2237–2250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller T. M., et al. , TRANSCRIPTION ACTIVATOR-LIKE EFFECTOR NUCLEASE-mediated generation and metabolic analysis of camalexin-deficient cyp71a12 cyp71a13 double knockout lines. Plant Physiol. 168, 849–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S., Ziegler J., Zeier J., Birkenbihl R. P., Somssich I. E., Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ. 40, 2189–2206 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Stahl E., et al. , Regulatory and functional aspects of indolic metabolism in plant systemic acquired resistance. Mol. Plant 9, 662–681 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Banhara A., Ding Y., Kühner R., Zuccaro A., Parniske M., Colonization of root cells and plant growth promotion by Piriformospora indica occurs independently of plant common symbiosis genes. Front. Plant Sci. 6, 667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahrmann U., et al. , Host-related metabolic cues affect colonization strategies of a root endophyte. Proc. Natl. Acad. Sci. U.S.A. 110, 13965–13970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curtis M. D., Grossniklaus U., A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakagawa T., et al. , Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Margesin R., Minerbi S., Schinner F., Long-term monitoring of soil microbiological activities in two forest sites in South Tyrol in the Italian alps. Microbes Environ. 29, 277–285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bednarek P., et al. , Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192, 713–726 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Hartmann M., et al. , Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Lee B. R., Koprivova A., Kopriva S., The key enzyme of sulfate assimilation, adenosine 5′-phosphosulfate reductase, is regulated by HY5 in Arabidopsis. Plant J. 67, 1042–1054 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Sprouffske K., Wagner A., Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17, 172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.