Significance
Protein kinases are exceptionally important and effective targets in human diseases, but a significant proportion of the human kinome remains uncharacterized and consequently unexplored as potential therapeutic targets. Here, we describe a kinase not annotated in the original human kinome, C19orf35/PEAK3, which shares structural similarities with PEAK1/Sgk269 and Pragmin/SgK223 pseudokinases. Our work reveals the cellular pool of PEAK3 binding partners and focuses on cellular phenotypes resulting from the PEAK3-dependent inhibition of CrkII, an adaptor protein that controls cell migration. Most importantly, we investigate how the structure and dimerization of the PEAK3 pseudokinase domain regulates its behavior, which highlights potential avenues for pharmacological regulation of PEAK3 function.
Keywords: protein kinase, pseudokinase, NKF3 family, CrkII, motility
Abstract
Members of the New Kinase Family 3 (NKF3), PEAK1/SgK269 and Pragmin/SgK223 pseudokinases, have emerged as important regulators of cell motility and cancer progression. Here, we demonstrate that C19orf35 (PEAK3), a newly identified member of the NKF3 family, is a kinase-like protein evolutionarily conserved across mammals and birds and a regulator of cell motility. In contrast to its family members, which promote cell elongation when overexpressed in cells, PEAK3 overexpression does not have an elongating effect on cell shape but instead is associated with loss of actin filaments. Through an unbiased search for PEAK3 binding partners, we identified several regulators of cell motility, including the adaptor protein CrkII. We show that by binding to CrkII, PEAK3 prevents the formation of CrkII-dependent membrane ruffling. This function of PEAK3 is reliant upon its dimerization, which is mediated through a split helical dimerization domain conserved among all NKF3 family members. Disruption of the conserved DFG motif in the PEAK3 pseudokinase domain also interferes with its ability to dimerize and subsequently bind CrkII, suggesting that the conformation of the pseudokinase domain might play an important role in PEAK3 signaling. Hence, our data identify PEAK3 as an NKF3 family member with a unique role in cell motility driven by dimerization of its pseudokinase domain.
Protein kinases are exceptionally effective therapeutic targets and account for a substantial portion of drug-discovery research. However, the number of newly discovered pathological mutations or translocations in known pro-oncogenic kinases has been decreasing, underscoring the need to turn our attention to understudied protein kinases. To this day, a substantial portion of the human protein kinome, first introduced by Manning et al. in 2002 (1), has remained in the dark, and the physiological functions of many protein kinases are unknown. Pseudokinases, representing approximately one-tenth of all of kinases, embody the darkest matter in the human kinome (1). Their characteristic feature is the presence of active-site mutations that, in most cases, result in loss of catalytic activity. As such, pseudokinases signal primarily through mechanisms based on protein–protein interactions, such as protein scaffolding and allosteric activation, and over the last decade there has been exponential progress in our understanding of these functions (2). The divergence of pseudokinases from their active counterparts, both in structure and function, raises an interesting possibility that there may still remain unidentified kinases, which were missed during the original assembly of the kinome due to significant departure from the canonical sequence motifs and which may be of therapeutic interest. In this paper we characterize a kinase not included in the original human kinome, chromosome 19 Open Reading Frame 35 (C19orf35), as a banner case for discovery of new functions in the human kinome.
C19orf 35, which was only recently annotated as PEAK3, is a new human kinase-like protein in the New Kinase Family 3 (NKF3) that contains 2 known pseudokinases: pseudopodium enriched atypical kinase 1 (PEAK1, also called Sugen kinase [SgK] 269) and Pragmin (PEAK2/SgK223) (1, 3). PEAK1 and Pragmin are large (1,746 and 1,406 residues, respectively) signaling scaffolds comprised of long N-terminal regions with no predicted domain structure followed by protein kinase-like domains at their C termini. The pseudokinase domains of both PEAK1 and Pragmin are atypical and carry multiple substitutions in the canonical consensus motifs critical for the catalysis of phosphorylation. These substitutions include mutation of the DFG motif (to NFS in Pragmin and to NFL in PEAK1), lack of the conserved glutamate in the αC helix, and a degenerate glycine-rich loop. Likely as a consequence of these changes, both kinases were demonstrated to lack the ability to bind nucleotides (4). Consistent with these data, the recent crystal structures of both PEAK1 and Pragmin revealed nucleotide-binding pockets with significantly distorted architectures, which render them incompatible with ATP binding (3, 5, 6). Collectively, these observations support the categorization of PEAK1 and Pragmin as pseudokinases.
Functional studies on PEAK1 and Pragmin have underscored their common role in the regulation of cell morphology and migration. Pragmin was originally described as an effector of Rnd2, a Rho family GTPase expressed primarily in neurons, that participates in negative regulation of neurite outgrowth by activating RhoA (7). Contradictory to this function, overexpression of Pragmin in cultured epithelial cells leads to an elongated cell morphology and promotes cell migration (8–10). Similar to that observed for Pragmin, exogenous expression of PEAK1 is associated with cell elongation and increased cell migration, and knockdown of the protein dramatically reduces cell motility (11–13). The cellular phenotypes induced by both PEAK1 and Pragmin are largely consistent with the hypothesis that they act as late-stage mediators of EGFR signaling, which switch signaling output of EGFR activation from promitogenic to promigratory (14). Upon EGF stimulation, PEAK1 serves as a scaffold for Grb2-independent signaling complexes that enhance cell motility by promoting the phosphorylation of key focal adhesion proteins, including p130Cas, Crk, and Paxillin (11). PEAK1 is also able to recruit Pragmin to these Grb2-independent complexes, where it enhances phosphorylation of JAK1 to induce STAT3-dependent changes in cell morphology (8, 14).
The complex roles of PEAK1 and Pragmin in cell migration pathways likely underlie their oncogenic potential and involvement in the development of a number of tumors (8, 11, 15–20). In recent years, Pragmin has emerged as an important player in cancer progression that is necessary for Src-mediated invasion of colon carcinoma cells, NOTCH-dependent tumorigenesis, and the development of pancreatic cancer through JAK1/STAT3 signaling (8, 16, 17). Furthermore, genetic ablation of Pragmin in human esophageal carcinoma cells was shown to promote tumor growth, emphasizing the critical role it plays in the advancement of cancer (17). PEAK1 has also been shown to promote cancer progression and is overexpressed in subsets of pancreatic, breast, and colon cancers (11, 17, 18, 20).
PEAK3 diverges from PEAK1 and Pragmin in that it has a significantly shorter N-terminal domain and a less conserved kinase-like domain. Using sequence analysis across evolution, we show that, although the NKF3 family emerges in early metazoans (e.g., Placozoa [Trichoplax] and sponges), PEAK3 does not appear in evolution until much later (in some reptilian lineages) and is evolutionarily conserved in birds and mammals. We present bioinformatic and biochemical evidence that PEAK3 shares characteristic topological features with its NKF3 family members, including a split helical dimerization (SHED) domain that supports its homodimerization. Using an unbiased approach to characterize PEAK3 function in cells, we found that one of the direct binding partners of PEAK3 is the focal adhesion protein CrkII. By binding to CrkII in a dimerization-dependent manner, PEAK3 antagonizes CrkII signaling, exerting an opposite effect on cell morphology to PEAK1 and Pragmin.
Results
PEAK3 Is a Distinct Member of the NKF3 Family of Atypical Protein Kinases.
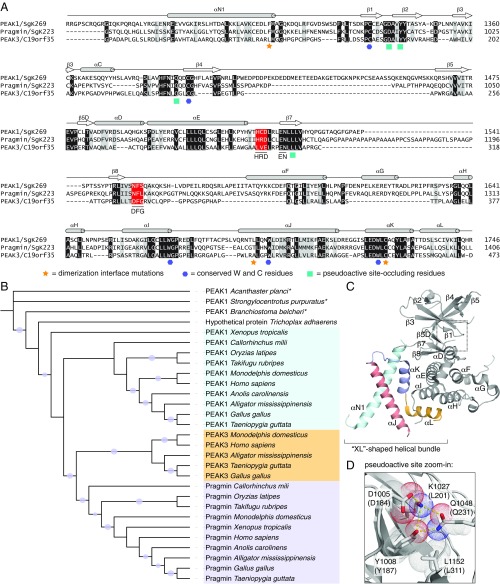
We searched for distant homologs of protein kinases in the human proteome using the sensitive sequence comparison algorithm Fold and Function Assignment System (FFAS) (21), previously used by us to discover novel kinase-like proteins in humans [SELO and FAM69/DIA1 families (22, 23)] and in bacteria and fungi [COTH (24)]. The only novel protein with a significant FFAS score found that was not previously identified as kinase-like was a single uncharacterized protein, denoted as Chromosome 19 Open Reading Frame 35 (C19orf35). C19orf35 has the closest similarity to the PEAK1/SgK269 and Pragmin/SgK223 pseudokinases that together constitute the NKF3 family. Recently, Lecointre et al. (3) reported the identity of C19orf35 as a new member of the NKF3 family, and C19orf35 was annotated in the Universal Protein Resource (Uniprot) as PEAK3. For consistency, from this point on, we refer to C19orf35 as PEAK3. Using PEAK1 or Pragmin as Blast queries results in only partial alignments covering fragments of the PEAK3 kinase-like domain. However, the FFAS method is capable of detecting similarity over the entire length of the putative kinase-like domain (Fig. 1A). The kinase-like domain of human PEAK3 shares only ∼26% sequence identity with human PEAK1 and Pragmin due to low-complexity regions (LCRs), which introduce long gaps in the pairwise alignments. This is most likely why the PEAK3 sequence, although present in the Uniprot database since 2004, was not annotated as kinase-like until recently. Even now, standard protein domain annotation tools (CD-Search or Pfam HMM) do not report kinase similarity for PEAK3.
Fig. 1.
Identification of PEAK3 (C19orf35) as a homolog of PEAK1 and Pragmin. (A) Protein sequence alignment for the kinase domains of human PEAK1/SgK269, Pragmin/SgK223, and PEAK3/C19orf35 and their corresponding secondary structure elements, which are denoted based on the Pragmin structure (PDB ID code 5VE6). Shading for conserved residues (black), conservative mutations (gray), and canonical sequence motifs of active kinases (red) are marked. Residues involved in dimerization, conserved W and C residues, and those shown to occlude the pseudoactive site in Pragmin are marked by the indicated symbols. (B) Phylogenetic tree (PhyML) for selected NKF3 kinases. Branches with bootstrap values better than 70% are marked with circles; an asterisk (*) represents predicted proteins. (C) Cartoon representation of the crystal structure of the Pragmin SHED domain/pseudokinase module (PDB ID code 5VE6). (D) Zoomed-in view of the pseudoactive site in Pragmin depicting residues that occlude the canonical nucleotide-binding pocket. Top numbering is for Pragmin residues (PDB ID code 5VE6), bottom in parentheses for predicted corresponding residues in PEAK3. The Pragmin D184 residue was modeled in the active site based on the other structure of Pragmin (PDB ID code 6EWX).
LCRs, defined as areas of protein sequences with biased amino acid composition (25), located within the kinase-like domain, are a distinct feature of NKF3 proteins. These regions are typically implicated in mediating protein–protein interactions (26). In NKF3 pseudokinases, the LCRs correlate with flexible regions, as judged by missing coordinates corresponding to these sequences in the structures of PEAK1 and Pragmin. One example is the PAPAPAPA motif in Pragmin that is located between the HRD and NFL motifs, where the NFL motif corresponds to the DFG motif in active kinases (SI Appendix, Fig. S1A). The majority of the LCRs in PEAK1, Pragmin, and PEAK3 diverge in sequence from one another and are located in different regions. PEAK3 stands out by having the largest portion of its kinase-like domain sequence (more than 20%) denoted as LCRs. Some of these motifs are relatively well-conserved in evolution, such as the PPGPPGSPGP motif that is immediately downstream of the DFG motif in PEAK3 (SI Appendix, Fig. S1B).
Evolutionary Conservation of PEAK3 and Its Kinase-Like Domain.
The NKF3 family likely appeared at the origin of Metazoa, which is indicated by the presence of homologs in sponges (Amphimedon) and placozoans (Trichoplax). Most invertebrates (e.g., cnidarians, echinoderms) contain a single member of the family, although in some lineages (e.g., insects and nematodes), the NKF3 family has seemingly been lost. The division into the PEAK1 and Pragmin subfamilies likely occurred between the emergence of chordates (the lancelet Branchiostoma has a single family member) and jawed vertebrates (Gnathostomata have both PEAK1 and Pragmin). The PEAK3 subfamily diverges from the Pragmin branch likely during the evolution of reptiles (e.g., it is found in crocodilians) and is present in birds and mammals (Fig. 1B). Interestingly, PEAK3 is missing in some reptile species, such as snakes and lizards. The early members of the NKF3 family (i.e., proteins from sponges and placozoans) exhibit high sequence and structure conservation with those found in more complex metazoans (e.g., vertebrates), suggesting evolutionarily conserved functions of these proteins.
The kinase-like domain of PEAK3 is highly conserved in evolution and carries unique sequence alterations in key catalytic motifs found in active kinases compared with PEAK1 and Pragmin. Interestingly, these alterations within the putative active site of PEAK3 vary markedly between species (Fig. 1A and SI Appendix, Fig. S1). Mammalian PEAK1 and Pragmin do not possess a conserved DFG motif. However, PEAK1 and Pragmin have intact HxD motifs: HRD in PEAK1 and HCD in Pragmin. In contrast, mammalian PEAK3 contains a conserved DFG motif, while the HxD motif is replaced by LxE. These unique features are not conserved in avian PEAK3 homologs, which have an NFF or SFF sequence in place of DFG, more closely resembling sequences present in PEAK1 (NFS) and in Pragmin (NFL). In PEAK1 and Pragmin, these motifs are almost perfectly conserved irrespective of a species. While HxD is also not conserved in avian PEAK3, the catalytic aspartate (contained within the HxD motif) is present within the QGD sequence that replaces the HxD motif.
In all species, PEAK3 has a conserved EN motif, corresponding to the EN sequence located in PKA at positions 170 to 171 that coordinates divalent cations (27). This feature is also present in PEAK1 and Pragmin (Fig. 1A and SI Appendix, Fig. S1). The catalytic lysine, equivalent to K72 in PKA, is conserved in PEAK3 (K204 in PEAK3), but the glutamate (E91 in PKA) with which the catalytic lysine forms a salt bridge in active kinases is missing in PEAK3. In PEAK1 and Pragmin, the catalytic lysine is also present but is “hijacked” by interactions with 3 residues collectively termed the “inhibitory triad” that, in addition to other conserved residues, occlude the nucleotide binding pocket and prevent binding of ATP (Fig. 1 C and D) (3, 5, 6). These residues are well-conserved in PEAK3 and are represented by D184, Y187, L201, Q231, and L311 (Fig. 1 A and D). Collectively, the extent of mutations in the key catalytic motifs strongly indicates that although PEAK3 diverges from other members of the NKF3 family, it still falls into the category of pseudokinases defined by Manning et al. (1) as kinases that lack one or more of the canonical catalytic sequence motifs. Hence, all NKF3 kinases carry pseudokinase characteristics and seem to derive from a common NKF3 pseudokinase ancestor in early Metazoans, as indicated by the sequence variability in the NKF3 proteins and reconstruction of the ancestral NKF3 sequence [Ancescon method (28)].
PEAK3 Interactome.
With no prior insights into the function of PEAK3, we took an unbiased approach to identify its interacting partners using immunoprecipitation followed by mass spectrometry (IP/MS). Due to the current lack of a suitable antibody for detection of endogenous PEAK3, our analysis was conducted using a 3xFLAG-tagged human PEAK3 transiently expressed in HEK293T cells. Identified proteins that coimmunoprecipitated with PEAK3-3xFLAG can be categorized into several subgroups: (i) CrkII and CrkL, highly homologous adaptor proteins that regulate cell proliferation, adhesion, and cytoskeletal integrity downstream from receptor tyrosine kinases and integrins (29, 30); (ii) 14-3-3 scaffold proteins (β, γ, η, and τ), which play diverse roles in signaling, including regulation of cell motility, survival, and intracellular protein trafficking (31); (iii) guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins for Rho family of small GTPases, including ASAP1 that participates in actin cytoskeletal dynamics and cell movement (32, 33); and (iv) several proteins connected to the regulation of cell death and survival, including SIAH1 ubiquitin ligase (34), DRAK1 (35), and another 14-3-3 scaffold protein, 14-3-3σ, also known as SFN (36) (SI Appendix, Table S1). Collectively, this analysis suggests that PEAK3 plays a role in a number of functions involved in cell proliferation, survival, and motility. The function of PEAK3 in cell motility, in particular, was underscored by the abundance of motility regulators in the immunoprecipitates.
PEAK3 Interacts with CrkII via a Proline-Rich Motif/SH3 Domain Interaction.
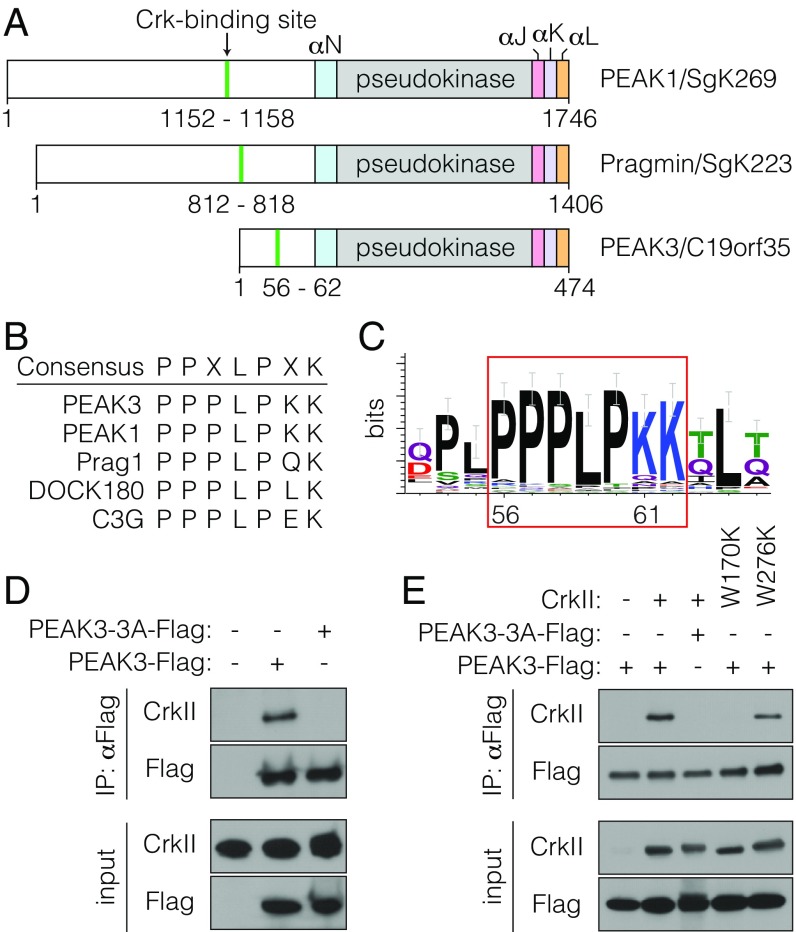
Given the documented roles of NKF3 family members in the regulation of cellular motility (8–13), we focused our functional studies of PEAK3 on the adaptor protein CrkII, which was one of the most abundant PEAK3-interacting proteins identified by the IP/MS analysis measured as a high-confidence score via the comparative proteomic analysis software suite (ComPASS) (37) (SI Appendix, Table S1). Primary sequence analysis of the N-terminal domain of PEAK3 revealed the presence of a putative CrkII-binding site, PPPLPK, located 71 residues upstream from the predicted kinase domain. This site closely resembles the consensus sequence present in known binding partners of CrkII, including the GEF proteins DOCK180 and C3G (Fig. 2 A and B) (11, 38, 39). This putative CrkII-binding motif in PEAK3 is highly conserved across evolution, suggesting a potential importance in PEAK3 function (Fig. 2C). Interestingly, this sequence is also present in PEAK1 and Pragmin, and PEAK1 was previously shown to coimmunoprecipitate with CrkII (11).
Fig. 2.
PEAK3 binds CrkII. (A) Schematic representation of PEAK1, Pragmin, and PEAK3 domain structure. The locations of the CrkII-binding sites and helical regions within the SHED domain are highlighted. (B) Consensus sequence of CrkII-binding sites in selected proteins. (C) Sequence logo depicting conservation of the CrkII-binding site in PEAK3 homologs. (D and E) Coimmunoprecipitation of FLAG-tagged wild-type PEAK3 or a CrkII-binding mutant (PEAK3-3A) transiently expressed in HEK293 cells with either endogenous CrkII (D) or transiently expressed untagged CrkII variants carrying mutations in the SH3 domains (W170K in SH3N and W276K in SH3C) (E). Protein levels were detected with the indicated antibodies by Western blot. All coimmunoprecipitation data are representative of at least 3 independent experiments.
Using coimmunoprecipitation, we verified that transiently expressed PEAK3 in HEK293 cells is indeed able to bind both endogenously and exogenously expressed CrkII (Fig. 2D and SI Appendix, Fig. S2). Mutation of residues in the predicted CrkII-binding site (P56A, L59A, and P60A in PPPLPK) completely abolished the ability of mutant PEAK3 (PEAK3-3A) to coimmunoprecipitate with CrkII (Fig. 2D). CrkII is composed of an SH2 domain followed by 2 SH3 domains, termed N-terminal SH3 domain (SH3N) and C-terminal SH3 domain (SH3C). The inability of the PEAK3-3A mutant to bind CrkII suggested that PEAK3 interacts specifically with the SH3N in CrkII, which has previously been shown to engage similar proline-rich motifs in other CrkII-binding partners (38–40). Indeed, mutation of the SH3N (CrkII-W170K) but not of the SH3C domain (CrkII-W276K) rendered CrkII unable to bind PEAK3 (Fig. 2E).
PEAK3 Antagonizes CrkII-Induced Changes in Cellular Morphology.
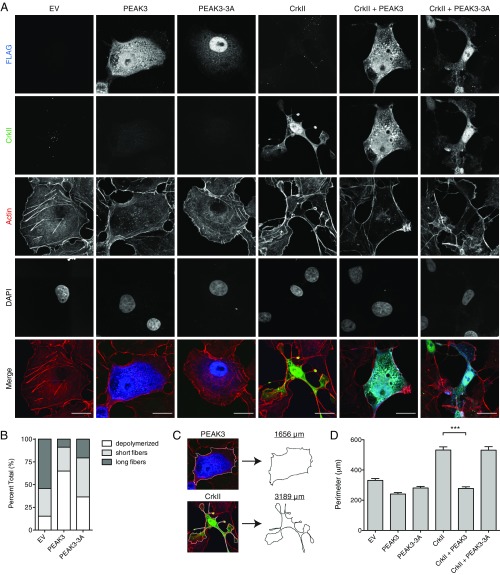
One well-characterized role of PEAK1 and Pragmin is the regulation of cell morphology and migration. Both PEAK1 and Pragmin localize to actin filaments and focal adhesions, induce cell elongation, and promote cell migration when transiently expressed in cells (8–13, 20). In contrast, we found that PEAK3 distributes diffusely throughout the cytoplasm of COS-7 cells and U2OS cells upon transient transfection and had little to no effect on overall cell shape compared with vector-transfected control cells (Fig. 3A and SI Appendix, Fig. S3). However, while the majority of control cells retained prominent actin filaments that traversed the cell (41, 42), cells overexpressing PEAK3 exhibited notably fewer stress fibers and possessed shorter, less-organized actin filaments, mirroring the phenotype typically observed in cells in which the CrkII gene is knocked down (Fig. 3B) (43).
Fig. 3.
PEAK3 prevents CrkII-dependent membrane ruffling and lamellipodia-like extensions. (A) Confocal microscopy imaging of COS-7 cells transiently cotransfected with a FLAG-tagged wild-type or a CrkII-binding deficient mutant of PEAK3 (PEAK3-3A) with either an empty vector or untagged CrkII. CrkII was detected with anti-CrkII antibody (green), PEAK3 with anti-FLAG antibody (blue) and F-actin with Alexa Fluor 647-conjugated phalloidin (red). (Scale bars, 20 μm.) (B) Relative percentage of different actin fiber phenotypes measured in COS-7 cells transiently transfected with PEAK3 and stained with Alexa Fluor 647-conjugated phalloidin (n = 60 cells per group). Cells were blindly scored and binned based on the extent of visible actin fibers within the cytosol: (i) prominent fibers that traversed over 50% of the cell, (ii) short stochastic fibers, and (iii) no significant amount of polymerized actin. (C) Schematic illustrating perimeter calculation in representative cells from A. (D) Average perimeter of COS-7 cells expressing either wild-type or a CrkII-binding deficient (PEAK3-3A) variants of PEAK3 with an empty vector or untagged CrkII, quantified as described in Materials and Methods. Data represent the mean ± SEM of 3 independent experiments (n = 20 cells in each experiment), ***P < 0.001.
Endogenous CrkII participates in actin polymerization and positively regulates cell motility (41, 43–47). Exogenously expressed CrkII localizes to the cell cortex where it induces a spindle-shaped cell morphology with notable membrane extensions resembling lamellipodia or polarized membrane ruffles (29, 47–51). These morphological changes can be visualized robustly in COS-7 and U2OS cells (43, 47, 52, 53), hence we used these cell lines to measure the functional consequences of PEAK3 overexpression on CrkII-dependent effects on cell morphology. Remarkably, cells coexpressing PEAK3 and CrkII had markedly fewer membrane extensions and largely did not adopt a CrkII-dependent morphology (Fig. 3A and SI Appendix, Fig. S4A). In contrast, the PEAK3-3A mutant, which does not interact with CrkII, was unable to interfere with the CrkII-dependent phenotype (Fig. 3A and SI Appendix, Fig. S4A). To quantitatively compare these differences in cellular phenotypes, we developed a metric in which the effect of CrkII on cell morphology is measured as an increase in cell perimeter (Fig. 3C). While CrkII overexpression alone significantly increases cell perimeter, there is no change in cell perimeter when CrkII is coexpressed with wild-type PEAK3 (Fig. 3D and SI Appendix, Fig. S4B). PEAK3-3A mutant has no effect on CrkII-dependent increase in cell perimeter, supporting a conclusion that PEAK3 negatively regulates CrkII as a result of their direct interaction.
Negative Regulation of CrkII by PEAK3 Requires the C-Terminal Domain.
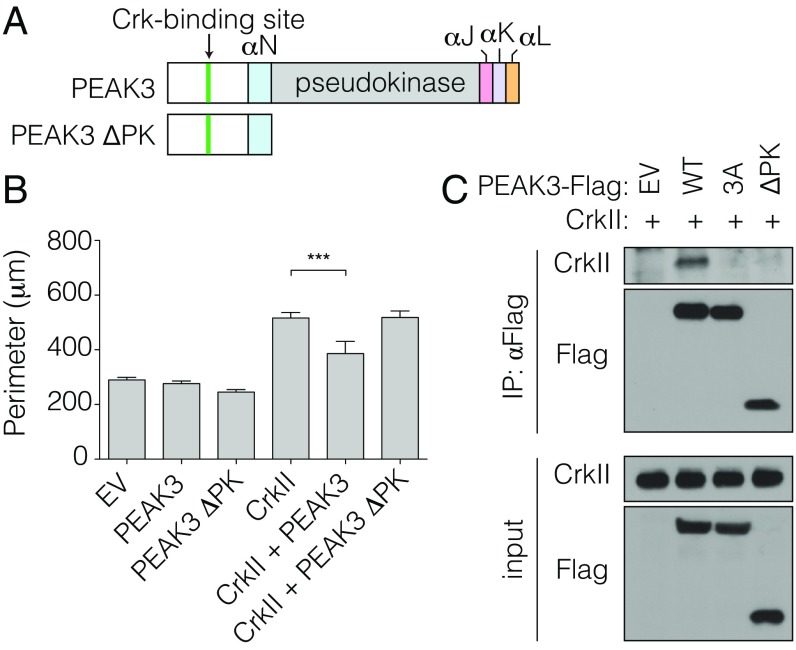
The PEAK3 protein can be arbitrarily subdivided into 2 distinct domains: the N-terminal domain that contains the CrkII-binding site and the C-terminal domain that contains the pseudokinase domain. We exogenously expressed the N-terminal domain of PEAK3 to determine if it alone is sufficient for the negative regulation of CrkII. Unexpectedly, a construct containing only the N-terminal domain and missing the C-terminally located pseudokinase domain (PEAK3 ΔPK) did not antagonize CrkII function (Fig. 4 A and B and SI Appendix, Fig. S5). Even more surprisingly, the PEAK3 ΔPK mutant was also unable to interact with CrkII, despite the presence of the intact CrkII-binding motif (Fig. 4C). Hence, while the CrkII-binding motif is necessary to support binding of PEAK3 to CrkII, it is not sufficient for CrkII binding and inhibition, demonstrating that the C-terminal domain of PEAK3 plays an essential role in mediating the PEAK3/CrkII interaction.
Fig. 4.
CrkII-binding motif is insufficient for PEAK3-dependent inhibition of CrkII. (A) Schematic representation of used PEAK3 constructs. (B) Average perimeter of COS-7 cells transiently expressing either wild-type or the ΔPK variants of PEAK3 with an empty vector or untagged CrkII. Data represent the mean ± SEM of 3 independent experiments (n = 20 cells in each experiment), ***P < 0.001. (C) Coimmunoprecipitation of FLAG-tagged wild-type and ΔPK PEAK3 variants, transiently expressed in HEK293 cells, with exogenously expressed, untagged CrkII. Protein levels were detected with the indicated antibodies by Western blot. Data are representative of 3 independent experiments.
Predicted SHED Domain in PEAK3.
Recent crystal structures of the PEAK1 and Pragmin C-terminal domains have revealed identical dimer forms composed of the pseudokinase domains interacting through a set of helical bundles, termed the SHED domain (3, 5, 6, 12). The SHED domain is unique to NKF3 proteins and is comprised of the helix immediately N-terminal to the pseudokinase domain (αN helix) and 3 helices C-terminal to the pseudokinase domain (αJ, αK, αL helices) that form an “XL”-shaped helical bundle [lettering of helices reflect Pragmin nomenclature (6); PEAK1 helices are off-set by one letter from the αJ helix (5)] (3, 5, 6).
In PEAK3, the highest sequence similarity with PEAK1 and Pragmin, apart from the pseudoactive site, falls within the regions corresponding to the SHED domain. In the 3 C-terminal α-helices, sequence identity between PEAK3 and Pragmin is 34% and 32% between PEAK3 and PEAK1 (SI Appendix, Figs. S1 and S6). The most striking conserved sequence motifs include the tryptophan residue-containing motifs. These motifs include the EDWLCC sequence in the αK helix, WGP in the loop preceding the αJ helix, and WL in the αJ helix (Fig. 1 A and C and SI Appendix, Fig. S1). In the PEAK1 and Pragmin crystal structures, these conserved motifs are involved in the interactions between the pseudokinase domain and the SHED domain (3, 5, 6). PEAK3 has remarkably well-conserved sequences in these regions, indicating that the SHED domain is likely present in PEAK3 adopting a similar structure to the ones in PEAK1 and Pragmin (SI Appendix, Fig. S6). Thus, the SHED domain emerges as a unifying structural feature of the NKF3 family of pseudokinases.
PEAK3 Dimerizes via the SHED Domain.
PEAK1 and Pragmin form homo- and hetero-oligomers through 2 distinct dimer interfaces, one involving the SHED domain and another involving the αG helix/A-loop interface (3, 6, 12). Mutation of these interfaces, especially of the hydrophobic interactions between the helices in the SHED domain, impairs the signaling ability of these pseudokinases (5, 6). While the SHED domain in PEAK3 is highly similar to the SHED domains of PEAK1 and Pragmin (Fig. 1A), the αG helix/A-loop interface is not significantly conserved in PEAK3. This led us to hypothesize that PEAK3 too could dimerize via its putative SHED domain and that this might be critical for its interaction with CrkII.
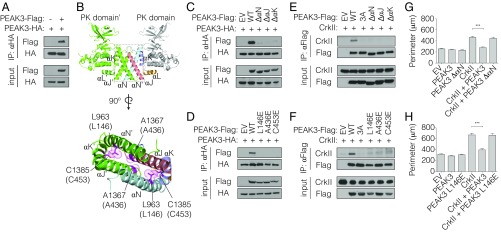
To assess the ability of PEAK3 to homodimerize, we coexpressed HA-tagged and FLAG-tagged wild-type PEAK3 variants in HEK293 cells and assessed their association by coimmunoprecipitation. These 2 differentially tagged PEAK3 constructs robustly coimmunoprecipitated (Fig. 5A and SI Appendix, Fig. S7A). Based on the crystal structures of Pragmin and PEAK1, we designed PEAK3 mutants that carried either individual substitutions of key residues in the predicted dimerization interface or deletion of 1 of the αN, αJ, or αK helices in the SHED domain (Fig. 5B). These mutations/deletions abolished the ability of differentially tagged PEAK3 variants to coimmunoprecipitate (Fig. 5 C and D and SI Appendix, Fig. S7 B and C), while control mutations of residues located within the SHED domain helices but distal from the dimer interface had no effect (SI Appendix, Fig. S7D). These data suggest that PEAK3 self-associates through the SHED domain in a manner analogous to PEAK1 and Pragmin.
Fig. 5.
PEAK3 dimerization via the pseudokinase/SHED module is necessary for binding and inhibition of CrkII. (A) Coimmunoprecipitation of a FLAG-tagged and an HA-tagged variant of wild-type PEAK3, transiently expressed in HEK293 cells. Protein levels were detected with the indicated antibodies by Western blot. (B, Upper) Pragmin SHED/pseudokinase domain (PK) dimer structure (PDB ID code 5VE6). (Lower) Dimerization interface and interactions between helices within the SHED domain. Residues colored in magenta were selected for mutagenesis in PEAK3 based on sequence homology. Top numbering corresponds to Pragmin (PDB ID code 5VE6), bottom in parentheses to PEAK3. (C–F) Coimmunoprecipitation of dimerization-deficient variants of PEAK3 with wild-type PEAK3 (C and D) or with untagged CrkII (E and F), all transiently expressed in HEK293 cells. Proteins levels were detected with the indicated antibodies by Western blot. All coimmunoprecipitation data are representative of 3 independent experiments. (G and H) Average perimeter of COS-7 cells expressing wild-type or dimerization-deficient variants of PEAK3 with an empty vector or untagged CrkII. Data represent the mean ± SEM of 3 independent experiments (n = 20 cells in each experiment), ***P < 0.001.
Dimerization of PEAK3 Is Necessary for CrkII Binding and Its Negative Regulation.
While the C-terminal domain of PEAK3 does not contain a CrkII binding motif, it plays an essential role in CrkII binding and inhibition, as demonstrated by our finding that the construct missing this domain, PEAK3 ΔPK, was unable to engage with and antagonize CrkII-induced membrane ruffling (Fig. 4). To test if this role of the C-terminal domain is linked to its ability to mediate PEAK3 self-association through the SHED domain, we measured how mutations within the SHED domain dimer interface affect CrkII binding. All mutations that compromise PEAK3 dimerization also interfered with the ability of PEAK3 to interact with CrkII (Fig. 5 E and F and SI Appendix, Fig. S8A). Consequently, the PEAK3 dimerization mutants also failed to inhibit the formation of membrane extensions and membrane ruffles caused by CrkII overexpression (Fig. 5 G and H and SI Appendix, Fig. S8 B and C).
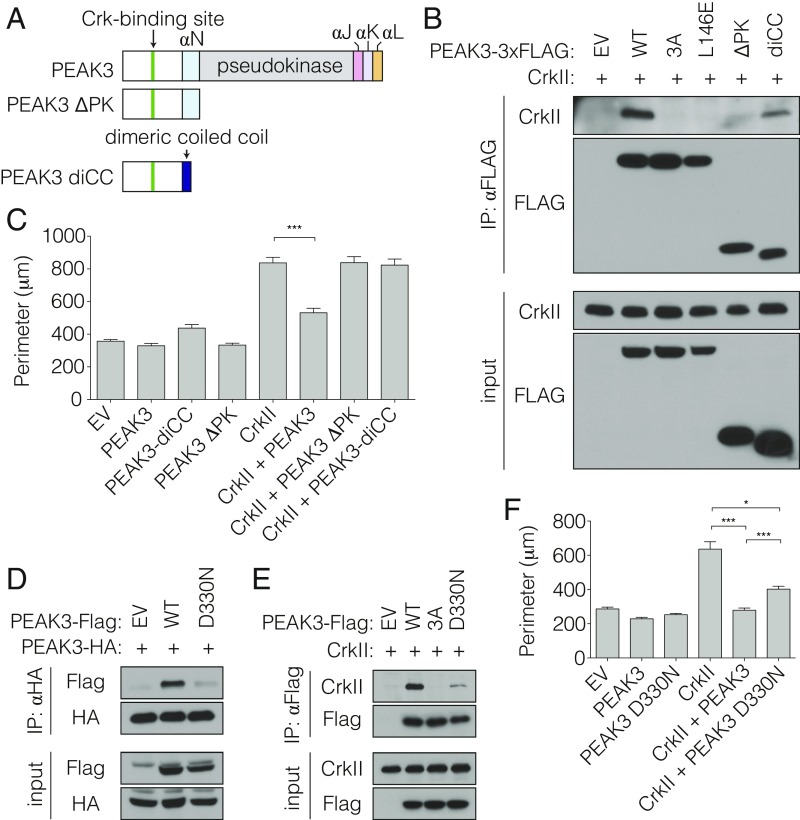
Our data thus far demonstrate that the N-terminal domain of PEAK3 containing the CrkII binding motif is unable to bind to or interfere with CrkII function when not dimerized by the C-terminal domain, which contains the pseudokinase domain. We wondered if the functional effect of the C-terminal domain-mediated dimerization could be mimicked by substitution of the C-terminal domain with an orthogonal domain that drives constitutive PEAK3 dimerization. To test this, we fused the N-terminal region of PEAK3 (residues 1 to 131) containing the CrkII-binding site, but not the SHED domain or the pseudokinase domain, to a constitutively dimeric coiled-coil domain (PEAK3-diCC) (Fig. 6A). While the monomeric N-terminal PEAK3 ΔPK construct is unable to bind CrkII (Fig. 4C), equivalent levels of the immunoprecipitated PEAK3-diCC fusion show notable CrkII binding (Fig. 6B). The ability of this minimal construct to restore CrkII binding underscores the importance of dimerization of the CrkII binding motif as a determinant of interaction between CrkII and PEAK3, and possibly also between CrkII and its other interaction partners. To our knowledge such property in known CrkII-binding proteins has not been described.
Fig. 6.
Role of PEAK3 pseudokinase domain in regulation of CrkII extends beyond dimerization. (A) Schematic representation of used PEAK3 constructs. (B) Coimmunoprecipitation of untagged CrkII with FLAG-tagged dimerization-deficient mutants of PEAK3, transiently expressed in HEK293 cells. Protein levels were detected with the indicated antibodies by Western blot. (C) Average perimeter of COS-7 cells transiently expressing either wild-type or diCC variant of PEAK3 with an empty vector or untagged CrkII. Data represent the mean ± SEM of 3 independent experiments (n = 20 cells in each experiment), ***P < 0.001. (D and E) Coimmunoprecipitation of the PEAK3 D330N variant with wild-type PEAK3 (D) or untagged CrkII (E) transiently expressed in HEK293 cells. Proteins levels were detected with the indicated antibodies by Western blot. All coimmunoprecipitation data are representative of 3 independent experiments. (F) Average perimeter of COS-7 cells transiently expressing either wild-type or the D330N variant of PEAK3 with an empty vector or untagged CrkII. Data represent the mean ± SEM of 3 independent experiments (n = 20 cells in each experiment), *P < 0.05, ***P < 0.001.
Despite the ability to interact with CrkII, PEAK3-diCC did not interfere with CrkII-dependent membrane ruffle formation in cells as observed for the wild-type PEAK3 (Fig. 6C and SI Appendix, Fig. S9). This discrepancy possibly reflects the weaker binding between CrkII and PEAK3-diCC compared with the wild-type PEAK3 (Fig. 6B). Alternatively, or concurrently, the C-terminal pseudokinase/SHED module in PEAK3 might play a role in antagonizing CrkII signaling that extends beyond serving as a dimerization domain for the CrkII binding motif.
Mutation of the DFG Aspartate Impairs CrkII Regulation by PEAK3.
NKF3 kinases have evolved multiple sequence alterations within their pseudokinase domains relative to active kinases, which are predicted to render them catalytically inactive. One notable difference between PEAK3 and PEAK1 and Pragmin is conservation of the DFG motif in PEAK3. The DFG motif is present in a number of human pseudokinases, although its importance for their function is unclear (1). In active kinases, the aspartate residue within the DFG motif plays a critical role in catalysis by coordinating Mg2+ ions (54). In a subset of these kinases, the DFG motif is found to adopt 2 distinct conformations, DFG-in and DFG-out, which are coupled to conformational changes in other regions of the kinase domain (55). The interactions made by the DFG aspartate are key to these changes (56). It is therefore possible that in pseudokinases that conserve the DFG motif, such as PEAK3, the conformation of this motif could be coupled to functional conformational changes in the other parts of the pseudokinase domain.
We examined the importance of the DFG motif in PEAK3’s function as a negative regulator of CrkII by mutating the DFG aspartate to asparagine (PEAK3 D330N) and testing the ability of this mutant to homodimerize, interact with CrkII, and inhibit CrkII-dependent membrane ruffles in cells. Interestingly, PEAK3 D330N did not dimerize as efficiently as wild-type PEAK3 (Fig. 6D). Consistent with our observation that PEAK3 dimerization is necessary for CrkII binding, PEAK3 D330N exhibited markedly impaired binding to CrkII (Fig. 6E) and did not inhibit CrkII-dependent morphological changes to cell shape to the same extent as wild-type PEAK3 (Fig. 6F and SI Appendix, Fig. S10). These data show that the integrity of the DFG motif is essential for PEAK3 function as a negative regulator of CrkII.
Discussion
The PEAK1 and Pragmin pseudokinases have grown in prominence due to the key roles they play in the regulation of cellular motility and oncogenesis (15). Here, we present evidence that PEAK3 is a close homolog of PEAK1 and Pragmin that likely has evaded annotation as a kinase due to the high LCR content within its pseudokinase domain. Despite this difference, PEAK3 retains features that are characteristic of the NKF3 family. These include the residues defined as the inhibitory triad, which occlude the nucleotide-binding pocket. Together with the mutations of several catalytic residues within the kinases domain, these features define PEAK3 as a pseudokinase. Based on sequence analysis and mutagenesis studies, we also demonstrate that PEAK3 self-associates through a conserved SHED domain that flanks its pseudokinase domain, similar to PEAK1 and Pragmin. Hence, the presence of the SHED domain and its role in oligomerization are defining and unique features of NKF3 kinases.
Phylogenetic analysis of the NKF3 family shows that the ancestral NKF3, which likely appeared in the ancestor of Metazoans, was most similar to PEAK1 and was already a pseudokinase. This protein had an NFS motif instead of a DFG, although it retained the HxD motif, which became HCD in human PEAK1, and in some PEAK1 and Pragmin homologs, a canonical HRD motif. It is intriguing that mammalian (but not avian) PEAK3 proteins have apparently reverted to the DFG motif while their HxD drifted into the LxE motif. Thus, at present, our analysis supports a hypothesis that the NKF3 family is an example of a kinase-like family that evolved originally as pseudokinases, similar to examples discussed by Kannan and coworkers (57).
Our unbiased search for PEAK3 binding partners reveals a propensity for PEAK3 to interact with regulators of cell motility, mirroring documented roles of PEAK1 and Pragmin (8–13). While both Pragmin and PEAK1 are known to contain proline-rich CrkII-binding motifs, and PEAK1 was shown to bind CrkII (11), our study describes a clear functional link between CrkII signaling and an NKF3 family member. We show here that PEAK3 regulates CrkII in a manner contrasting that of other NKF3 family members: PEAK3 inhibits CrkII while PEAK1 and Pragmin stimulate promotile signaling in cells and would therefore be expected to potentiate CrkII function. The underlying mechanisms for these differences is unclear but could reflect the distinct domain structure of PEAK3 compared with other NKF3 family members. Both PEAK1 and Pragmin contain large N-terminal regions that likely encode numerous unique functions that are absent in PEAK3. If PEAK1 and Pragmin do indeed promote CrkII signaling, this function may be encoded within their N-terminal domain that is missing in PEAK3.
We further demonstrate that the SHED domain-mediated dimerization of PEAK3 is essential for its ability to bind CrkII. The emerging role of dimerization in binding seems to stem from the necessity to dimerize the CrkII-binding motifs themselves, as efficient CrkII binding can be recapitulated when these sites in PEAK3 are brought together by an orthogonal dimerization module. Since the CrkII binding site in PEAK3 represents the canonical proline-rich motif found in other known CrkII binding partners, we anticipate that these proteins might also regulate their interaction with CrkII through dimerization. Some of the known CrkII interactors, such as PEAK1 and Pragmin, are known to exist as dimers. Interestingly, CrkII and its highly related homolog CrkL, which we also identified as a binding partner of PEAK3 in the IP/MS analysis, have been shown to form dimers (58). Hence, the inherent property of both CrkII and some of its binding partners to oligomerize might be an essential mechanism for regulation of their mutual interactions and signaling.
While necessary for CrkII binding, dimerization of the CrkII binding motif is not sufficient for CrkII inhibition by PEAK3 in the absence of the SHED and pseudokinase domains. These findings suggest the SHED-pseudokinase domain module plays an important role in CrkII inhibition beyond supporting PEAK3 self-association. One such function could be mediating PEAK3 heterodimerization with PEAK1 and Pragmin, which we predict could occur based on sequence similarities within their SHED domains. Since all NKF3 family members have CrkII-binding sites, all possible NFK3 homo- and heterodimers could efficiently bind CrkII in theory. Given the opposing phenotypic outcomes between PEAK3 and other NKF3 members on cell motility, heterodimerization of PEAK3 with PEAK1 or Pragmin could interfere with their positive signaling properties. The outstandingly shorter length of the N-terminal domain in PEAK3 relative to PEAK1 and Pragmin suggests that PEAK3 may have evolved to act as a dominant-negative regulator of PEAK1 and Pragmin, by antagonizing their functions encoded by the N-terminal domains. The regulatory role of PEAK3 for other NKF3 pseudokinases would be consistent with the later appearance of PEAK3 in evolution compared with the other 2 family members. Future studies are needed to parse out the functional relationship of PEAK3 with its family members.
An additional unique feature of PEAK3, which distinguishes it from PEAK1 and Pragmin, is the presence of an intact DFG motif. Our data show that mutation of the DFG motif affects the ability of PEAK3 to homodimerize and subsequently interact with CrkII. The importance of the DFG motif for PEAK3 function is intriguing due to the critical role of this motif in kinase catalysis. While at present we cannot rule out that PEAK3 might be catalytically active, PEAK3 is an unlikely candidate for an active kinase based on its poor conservation of other residues in the putative active site. Rather, the loss-of-function effect of the DFG mutation suggests that it is a resulting conformational change within the pseudoactive site of PEAK3 that influences its dimerization and, consequently, its function. DFG mutations have previously been shown to affect the oligomerization of other kinases and pseudokinases. Notably, mutation of the DFG aspartate (D161N) in the kinase RIPK3 induces oligomerization and assembly of RIPK3 into a multimeric apoptotic complex (59, 60). Other kinase-inactivating mutations, such as those of the β3 lysine or catalytic aspartate, do not affect RIPK3 oligomerization, indicating that the D161N mutation stabilizes a specific conformation of RIPK3 that promotes oligomerization (60). The opposite effect is observed in the pseudokinase MLKL. In MLKL, the DFG motif is replaced by a GFE motif, and mutation of the GFE glutamate (E351K) prevents MLKL oligomerization. This mutation was proposed to stabilize a conformation that is not permissive for the release of the adjacent 4-helix bundle (4HB) domain, which drives oligomerization (61). The SHED domain in NKF3 kinases, composed of helices flanking the pseudokinase domain, maintains close contacts with the pseudokinase domain in the crystal structures of PEAK1 and Pragmin. It is therefore likely that conformational changes within the pseudokinase domain are sensed by the SHED domain and can ultimately affect dimerization.
Potential regulation of PEAK3 dimerization through conformational changes in its pseudokinase domain presents an exciting opportunity for pharmacological modulation of PEAK3 oligomerization, as previously achieved in RIPK3 (60, 62). While RIPK3 inhibitors target the ATP-binding site, this will likely not work for PEAK3, whose nucleotide-binding pocket is predicted to be occluded. However, recent studies on another pseudokinase, TRIB1, point to an important role of protein–protein interactions in regulating the conformation of the pseudoactive site. Like in PEAK3, the nucleotide-binding site in TRIB1 is highly occluded and inaccessible to ligands (63). Binding of the transcription factor C/EBPα to the C-lobe of TRIB1 alters the conformation of the pseudoactive site, including its equivalent DFG motif (SLE in TRIB1), which rotates to a semiactive position upon C/EBPα binding (64). The TRIB1 studies demonstrate that distal binding events can have global effects on the conformation of the pseudoactive site. Further studies can reveal if such interactions exist in PEAK3 and whether they can be leveraged for the pharmacological manipulation of its function.
The therapeutic relevance of targeting PEAK3 remains to be determined, but there is clear therapeutic potential (62). While at present there are no known prevalent disease-associated mutations in PEAK3, PEAK3 mRNA levels are significantly elevated in acute myeloid leukemia (AML) patient samples relative to other cancer types (SI Appendix, Fig. S11) (65, 66). The E3 ubiquitin ligase SIAH1, which is a therapeutic target in AML (34), was identified as a PEAK3 binding partner in our IP/MS analysis. SIAH1 targets for degradation the oncogenic protein FMS-like tyrosine kinase 3 with internal tandem duplication mutation (FLT3-ITD), a mutant FLT3 variant detected in 40% of AML cases (67). It is possible that PEAK3 interferes with this process through direct interaction with SIAH1. CrkL, which too was identified in our IP/MS screen, is one of the downstream effectors of the FLT3 signaling axis that contributes to leukemogenesis (68). Crk family proteins are known to play key roles in cancer invasion and migration by integrating and amplifying extracellular signals (30). Genetic knockdown of CrkII specifically decreases the cell migration and malignant potential of multiple human cancer cells, including lung, breast, and ovarian cancers (30, 43, 69). Mechanistically, inhibition of CrkII leads to reduced or stochastic F-actin networks and reduction in lamellipodia (43, 46, 69), mirroring the morphological changes we observe when PEAK3 is overexpressed in cells. Therefore, pharmacological targeting of PEAK3 could prove useful in AML and potentially additionally types of cancers that are susceptible to inhibitors that target cellular motility pathways.
Materials and Methods
HEK293 cells were lysed and clarified by centrifugation for 10 min at 15,000 rpm. After preclearing with Protein A beads, lysates were incubated with complexed antibody/Protein A beads. Immunoprecipitants were eluted by boiling beads in SDS sample buffer and subjected to analysis by SDS/PAGE and Western blotting. COS-7 cells were fixed, permeabilized, and blocked before incubated with primary and secondary antibodies for visualization of FLAG-tagged PEAK3 constructs, CrkII, and actin. Detailed information is provided in SI Appendix. Additional methods describing the generation of sequence alignments, phylogenetic tree, sequence logos, plasmids, cell culture, IP/MS, Western blotting, and mapping of sequence conservation are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank K. Shokat for critical reading of the manuscript; I. Lucet, R. Daly, and the members of the N.J. laboratory for insightful discussions; D. Larsen and K. Herrington for assistance with confocal imaging; and S. Oakes for providing the wild-type CrkII plasmid. This work was supported by grants from the University of California, San Francisco Program for Breakthrough Biomedical Research (to N.J.); a University of California, San Francisco/National Institute of General Medical Sciences Initiative for Maximizing Student Development fellowship (to M.L.L.); a University of California, San Francisco Genentech Fellowship (to M.L.); NIH Grant U54 CA209891 (to N.J.K.); and Polish National Science Centre grant 2014/15/B/NZ1/03359 (to K.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906360116/-/DCSupplemental.
References
- 1.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S., The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Kung J. E., Jura N., Structural basis for the non-catalytic functions of protein kinases. Structure 24, 7–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lecointre C., et al. , Dimerization of the Pragmin pseudo-kinase regulates protein tyrosine phosphorylation. Structure 26, 545–554.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Murphy J. M., et al. , A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha B. H., Boggon T. J., The crystal structure of pseudokinase PEAK1 (Sugen Kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization. J. Biol. Chem. 293, 1642–1650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel O., et al. , Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association. Nat. Commun. 8, 1157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka H., Katoh H., Negishi M., Pragmin, a novel effector of Rnd2 GTPase, stimulates RhoA activity. J. Biol. Chem. 281, 10355–10364 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Tactacan C. M., et al. , The pseudokinase SgK223 promotes invasion of pancreatic ductal epithelial cells through JAK1/Stat3 signaling. Mol. Cancer 14, 139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senda Y., Murata-Kamiya N., Hatakeyama M., C-terminal Src kinase-mediated EPIYA phosphorylation of Pragmin creates a feed-forward C-terminal Src kinase activation loop that promotes cell motility. Cancer Sci. 107, 972–980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safari F., Murata-Kamiya N., Saito Y., Hatakeyama M., Mammalian Pragmin regulates Src family kinases via the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif that is exploited by bacterial effectors. Proc. Natl. Acad. Sci. U.S.A. 108, 14938–14943 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., et al. , Pseudopodium-enriched atypical kinase 1 regulates the cytoskeleton and cancer progression [corrected]. Proc. Natl. Acad. Sci. U.S.A. 107, 10920–10925 (2010). Correction in: Proc. Natl. Sci. U.S.A.107, 13556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., et al. , Homo- and heterotypic association regulates signaling by the SgK269/PEAK1 and SgK223 pseudokinases. J. Biol. Chem. 291, 21571–21583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bristow J. M., Reno T. A., Jo M., Gonias S. L., Klemke R. L., Dynamic phosphorylation of tyrosine 665 in pseudopodium-enriched atypical kinase 1 (PEAK1) is essential for the regulation of cell migration and focal adhesion turnover. J. Biol. Chem. 288, 123–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y., et al. , Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 499, 166–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Rourke R. L., Daly R. J., The pseudokinases SgK269 and SgK223: A novel oncogenic alliance in human cancer. Cell Adhes. Migr. 12, 524–528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leroy C., et al. , Quantitative phosphoproteomics reveals a cluster of tyrosine kinases that mediates SRC invasive activity in advanced colon carcinoma cells. Cancer Res. 69, 2279–2286 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Weaver K. L., et al. , NACK is an integral component of the Notch transcriptional activation complex and is critical for development and tumorigenesis. Cancer Res. 74, 4741–4751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelber J. A., et al. , KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res. 72, 2554–2564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croucher D. R., et al. , Involvement of Lyn and the atypical kinase SgK269/PEAK1 in a basal breast cancer signaling pathway. Cancer Res. 73, 1969–1980 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Fujimura K., et al. , A hypusine-eIF5A-PEAK1 switch regulates the pathogenesis of pancreatic cancer. Cancer Res. 74, 6671–6681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaroszewski L., Li Z., Cai X.-H., Weber C., Godzik A., FFAS server: Novel features and applications. Nucleic Acids Res. 39, W38–W44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudkiewicz M., Lenart A., Pawłowski K., A novel predicted calcium-regulated kinase family implicated in neurological disorders. PLoS One 8, e66427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudkiewicz M., Szczepińska T., Grynberg M., Pawłowski K., A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PLoS One 7, e32138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen K. B., et al. , Phosphorylation of spore coat proteins by a family of atypical protein kinases. Proc. Natl. Acad. Sci. U.S.A. 113, E3482–E3491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wootton J. C., Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput. Chem. 18, 269–285 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Coletta A., et al. , Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knighton D. R., et al. , Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Cai W., Pei J., Grishin N. V., Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 4, 33 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park T.-J., Curran T., Essential roles of Crk and CrkL in fibroblast structure and motility. Oncogene 33, 5121–5132 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Birge R. B., Kalodimos C., Inagaki F., Tanaka S., Crk and CrkL adaptor proteins: Networks for physiological and pathological signaling. Cell Commun. Signal. 7, 13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu H., Subramanian R. R., Masters S. C., 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Brown M. T., et al. , ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 18, 7038–7051 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen P.-W., et al. , The arf GTPase-activating protein, ASAP1, binds nonmuscle myosin 2A to control remodeling of the actomyosin network. J. Biol. Chem. 291, 7517–7526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krämer O. H., Stauber R. H., Bug G., Hartkamp J., Knauer S. K., SIAH proteins: Critical roles in leukemogenesis. Leukemia 27, 792–802 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Farag A. K., Roh E. J., Death-associated protein kinase (DAPK) family modulators: Current and future therapeutic outcomes. Med. Res. Rev. 39, 349–385 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Lodygin D., Hermeking H., The role of epigenetic inactivation of 14-3-3sigma in human cancer. Cell Res. 15, 237–246 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knudsen B. S., et al. , Affinity and specificity requirements for the first Src homology 3 domain of the Crk proteins. EMBO J. 14, 2191–2198 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiyokawa E., Hashimoto Y., Kurata T., Sugimura H., Matsuda M., Evidence that DOCK180 up-regulates signals from the CrkII-p130(Cas) complex. J. Biol. Chem. 273, 24479–24484 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Lamorte L., Royal I., Naujokas M., Park M., Crk adapter proteins promote an epithelial-mesenchymal-like transition and are required for HGF-mediated cell spreading and breakdown of epithelial adherens junctions. Mol. Biol. Cell 13, 1449–1461 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridley A. J., et al. , Cell migration: Integrating signals from front to back. Science 302, 1704–1709 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Ridley A. J., Rho: Theme and variations. Curr. Biol. 6, 1256–1264 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues S. P., et al. , CrkI and CrkII function as key signaling integrators for migration and invasion of cancer cells. Mol. Cancer Res. 3, 183–194 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Petit V., et al. , Phosphorylation of tyrosine residues 31 and 118 on paxillin regulates cell migration through an association with CRK in NBT-II cells. J. Cell Biol. 148, 957–970 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang D. D., Zhang W., Gunst S. J., The adapter protein CrkII regulates neuronal Wiskott-Aldrich syndrome protein, actin polymerization, and tension development during contractile stimulation of smooth muscle. J. Biol. Chem. 280, 23380–23389 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Nakashima N., et al. , The functional role of CrkII in actin cytoskeleton organization and mitogenesis. J. Biol. Chem. 274, 3001–3008 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Klemke R. L., et al. , CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J. Cell Biol. 140, 961–972 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallés A. M., Beuvin M., Boyer B., Activation of Rac1 by paxillin-Crk-DOCK180 signaling complex is antagonized by Rap1 in migrating NBT-II cells. J. Biol. Chem. 279, 44490–44496 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Kobashigawa Y., et al. , Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat. Struct. Mol. Biol. 14, 503–510 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Saleh T., et al. , Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway. Nat. Chem. Biol. 12, 117–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antoku S., Mayer B. J., Distinct roles for Crk adaptor isoforms in actin reorganization induced by extracellular signals. J. Cell Sci. 122, 4228–4238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kain K. H., Klemke R. L., Inhibition of cell migration by Abl family tyrosine kinases through uncoupling of Crk-CAS complexes. J. Biol. Chem. 276, 16185–16192 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Jacquemet G., et al. , Filopodome mapping identifies p130Cas as a mechanosensitive regulator of filopodia stability. Curr. Biol. 29, 202–216.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huse M., Kuriyan J., The conformational plasticity of protein kinases. Cell 109, 275–282 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Jura N., et al. , Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell 42, 9–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shan Y., et al. , A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc. Natl. Acad. Sci. U.S.A. 106, 139–144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon A., et al. , Tracing the origin and evolution of pseudokinases across the tree of life. Sci. Signal. 12, eaav3810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harkiolaki M., Gilbert R. J. C., Jones E. Y., Feller S. M., The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure 14, 1741–1753 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Newton K., et al. , Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Mandal P., et al. , RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol. Cell 56, 481–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrie E. J., et al. , Conformational switching of the pseudokinase domain promotes human MLKL tetramerization and cell death by necroptosis. Nat. Commun. 9, 2422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kung J. E., Jura N., Prospects for pharmacological targeting of pseudokinases. Nat. Rev. Drug. Discov. 10.1038/s41573-019-0018-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murphy J. M., et al. , Molecular mechanism of CCAAT-enhancer binding protein recruitment by the TRIB1 pseudokinase. Structure 23, 2111–2121 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Jamieson S. A., et al. , Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci. Signal. 11, eaau0597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchwald M., et al. , Ubiquitin conjugase UBCH8 targets active FMS-like tyrosine kinase 3 for proteasomal degradation. Leukemia 24, 1412–1421 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Hartman A. D., et al. , Constitutive c-jun N-terminal kinase activity in acute myeloid leukemia derives from Flt3 and affects survival and proliferation. Exp. Hematol. 34, 1360–1376 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Linghu H., et al. , Involvement of adaptor protein Crk in malignant feature of human ovarian cancer cell line MCAS. Oncogene 25, 3547–3556 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.