The discovery of mutations associated with human genetic disease is an exercise in comparative genomics (see Glossary). Although there are many different strategies and approaches, the central premise is that affected persons harbor a significant excess of pathogenic DNA variants as compared with a group of unaffected persons (controls) that is either clinically defined1 or established by surveying large swaths of the general population.2 The more exclusive the variant is to the disease, the greater its penetrance, the larger its effect size, and the more relevant it becomes to both disease diagnosis and future therapeutic investigation. The most popular approach used by researchers in human genetics is the case–control design, but there are others that can be used to track variants and disease in a family context or that consider the probability of different classes of mutations based on evolutionary patterns of divergence or de novo mutational change.3,4 Although the approaches may be straightforward, the discovery of pathogenic variation and its mechanism of action often is less trivial, and decades of research can be required in order to identify the variants underlying both mendelian and complex genetic traits (see video, available at NEJM.org).
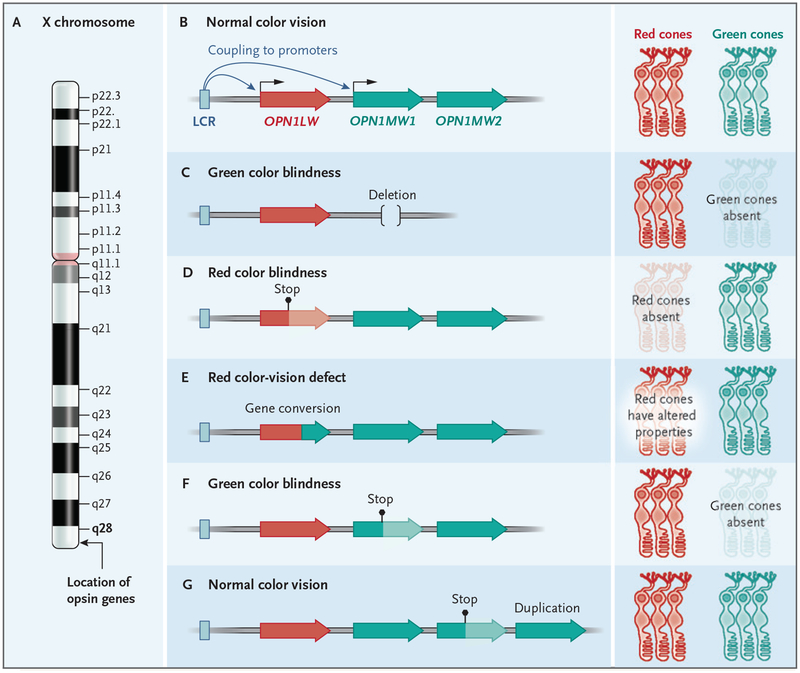
For example, X-linked color blindness is a well-known genetic trait that is commonly observed among males of northern European descent.5 Mutations in redand green-cone color pigment (opsin) genes have long been known to underlie this trait. Remarkably, the prevalence of potentially defective mutations (15.7%) observed among European males at first seemed higher than the observed prevalence of the color-vision defects (8.2%). This discrepancy was partially resolved by the realization that opsin genes were variable in copy number6 and that their expression was restricted to the first red and green opsin genes of a tandem array, the expression of which was under the control of a single locus-control region.7 Irrespective of how many additional copies of the duplicate genes a person carried, what really mattered was only which genes were in the first and second position of the gene cluster (Fig. 1). Thus, only mutations that disrupted, deleted, or created fusion genes in those two critical positions of the tandem array of gene copies would manifest as color-vision defects. Mutations in the other copies of the opsin genes were of little consequence.5
Figure 1. Structure and Expression of the Red–Green Color Blindness Loci.
Genes encoding the red (OPN1LW) and green (OPN1MW1 and OPN1MW2) opsins are organized in a head-to-tail configuration on the X chromosome (Panel A). The locus control region (LCR) couples to the promoter of the red opsin or the first green opsin gene to drive transcription and leads to the formation of either red or green cones in the retina.5 Below the canonical organization (Panel B), five different human mutations are shown, including deletion of the green opsins, leading to green color blindness (Panel C); a stop codon mutation in the red opsin gene, resulting in red color blindness (Panel D); a gene conversion event creating a red–green hybrid gene, resulting in protanomalous color vision (Panel E); a stop codon in the most proximal gene, resulting in green color blindness (Panel F); and a duplication and stop mutation in distal green opsin genes, which have no effect on color vision because distal copy genes are rarely expressed in the retina (Panel G). The sequence structure, regulation, and copy-number variation are key to understanding the genotype–phenotype correlation of this human trait.
There are three key aspects to genetic disease associations: comprehensive variant discovery, accurate allele-frequency determination, and an understanding of the pattern of normal variation and its effect on expression. The normal pattern of genetic variation includes the frequency of de novo mutation, demographic differences, and evolutionary selection at any given locus. An understanding of each of these entities is dependent on advances in genomic technology, including the accurate sequencing and assembly of the genomes of other species. Over the past two decades, there have been biases in our ascertainment of these features driven by technological limitations, funding priorities, and genetic paradigms of disease.
CLASSES OF HUMAN GENETIC VARIATION
Not all classes of mutation occur with equal frequency, nor are they equivalent with respect to their contribution to disease. The spectrum of human genetic variation is wide, ranging from point mutations — such as the substitution of a thymidine nucleotide for an adenine nucleotide in HBB (encoding beta globin), a cause of sickle cell disease — to large chromosomal aneuploidy events involving entire chromosomes, as is the case in trisomy 21 (Down’s syndrome).
Although single-nucleotide variants (SNVs; the most well-characterized type of variant in the human genome8,9) outnumber other types of DNA sequence variants by almost 7 to 1 (Table 1), an analysis of data from the Human Gene Mutation Database (www.hgmd.cf.ac.uk/) indicates that 34% of all disease-causing variation is made up of variants that are larger than a single base-pair substitution — a trend that has been slowly edging upward over the past decade. Almost one third of these variants are classified as gross lesions involving deletions and insertions greater than 20 bp in length. Structural variation is a category that includes copy-number variation and has been used to refer collectively to differences that are at least 50 bp in length between two individual genomes; structural variation also includes insertions, deletions, inversions, and translocations.10,16 Operationally, these variants are distinguished from smaller insertions and deletions (known as indels) on the basis of length, with the latter being between 1 and 49 bp.
Table 1.
Classes of Human Genetic Variation.*
| Class | Size of Variant | No. per Genome† | Size of Region Affected | Percent of Genome |
|---|---|---|---|---|
| bp | Mbp | |||
| Single-nucleotide variants | 1 | 4,000,000–5,000,000 | 4–5 | 0.078 |
| Insertions-deletions | 1–49 | 700,000–800,000 | 3–5 | 0.069 |
| Structural variants | >50 | 23,000–28,000 | 10–12 | 0.19 |
| Inversions | >50 | 153‡ | 23‡ | 0.397 |
| Multi-copy-number variants§ | >1000 | Approximately 500 | 12–15 | 0.232 |
Data are from the 1000 Genomes Project Consortium,8 Sudmant et al.,10–12 Huddleston et al.,13 and Chaisson et al.14,15
The data reflect numbers of mutational events in a diploid human genome (consisting of approximately 5.8 Gbp of euchromatin DNA).
The mean value is shown.
Multi-copy-number variants are a subset of structural variants that have not been completely resolved; they are enriched in segmental duplications but do not include heterochromatic regions of centromeric and acrocentric DNA.
It is now clear that structural variation affecting genes is common and that it contributes sub stantially to disease and disease susceptibility.17–22 Structural variants contribute more base-pair differences between two human haplotypes than any other form of genetic variation (Table 1).10,11 Moreover, as compared with SNVs, large structural variants are 3 times as likely to be associated with a genomewide association signal and more than 30 times as likely to affect the expression of a gene.10,23 This is because larger changes in DNA, such as the deletion or insertion of sequence, are generally more deleterious,24 even in noncoding regions of the genome in which they are more likely than an SNV to add, eliminate, or alter regulatory sequence and lead to gene-expression changes. Of course, when such events intersect the exons of protein-coding genes, they are likely to result in more severely deleterious events, because the loss of entire exons typically disrupts synthesis of the protein. It is not surprising, then, that large structural variants (i.e., >250,000 bp) rarely reach frequencies that exceed 1% in the general population. That said, approximately one quarter of ostensibly unaffected persons carry a structural variant that exceeds 250,000 bp.25
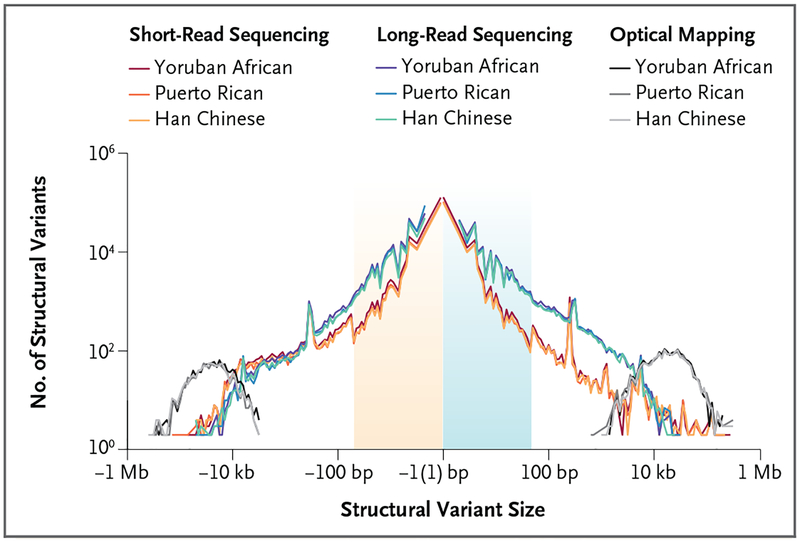
Detection of human genetic variation is imperfect even when deep genome sequence data (i.e., very large numbers of sequence reads covering the area of interest) are available. Structural variants, in particular, have been the most difficult to characterize with the use of short-read DNA-sequencing methods, and so pathogenic alleles have gone undetected.20–22,26 This is because the detection of structural variation with the use of short-read sequencing technology is indirect: it depends on inference. The identification of insertions, deletions, or duplications is based on measurements of read depth or discordances between the DNA sequences obtained from the patient and the reference genome, currently defined as GRCh38.10,12,27–29 With this approach, the reference genome becomes the common benchmark for the discovery of larger variants, when sequence from the new genome shows patterns that are inconsistent with the organization found in the reference genome. Thus, with these indirect assays, the actual sequence of the structural variant is not characterized; rather, the presence of the variant is inferred. Approaches to discovering structural variants are particularly biased by the size and sequence context of the events themselves (Fig. 2). Most notable is the skew against intermediate-size structural variants (<2000 bp), inversions, regions with DNA composition that is GC- or AT-rich, and multi-copy-number variants mapping to duplicated regions.13,14,30 (Multi-copy-number variants are structural variants in which the copy number can range across multiple integer values in the general population.)
Figure 2. Sensitivity of Structural-Variant Detection with the Use of Different Genomic Technologies.
Shown is a comparison of the number of structural variants (gains or losses of DNA shown on a log scale) as a function of variant size among the same three human genomes, detected with the use of different variant discovery technologies. Structural variants, insertions, and deletions were discovered by short-read sequencing, long-read sequencing (e.g., PacBio), and optical mapping technology (e.g., Bionano Genomics). Short-read sequencing shows reduced sensitivity for the detection of structural variants, especially insertions of 50 bp to 2000 bp. Both technologies underperform for sequence resolution of larger multi-copy-number variants (>10,000 bp); optical mapping technology can be used to detected such variants but not to resolve their sequence organization. Adapted from Chaisson et al.14
The fundamental problem is that structural variation is highly enriched within or near repetitive DNA.11,31,32 Repetitive DNA complicates the use of methods that are dependent on mapping, such as structural-variant detection, because sequence reads do not map uniquely but rather map to multiple locations in a genome. The shorter the sequence reads, the greater the fraction of the genome in which mapping becomes ambiguous. Thus, short sequence reads limit our ability to discover and genotype structural variants. Complex patterns of genetic variation and their association with genetic traits, such as red–green color blindness, for example, become difficult to disentangle. Because most of this variation has not yet been discovered or sequence-resolved, some have hypothesized that this cryptic genetic variation may contribute substantially to the “missing heritability” of human disease.33,34
ADDITIONAL AND MORE COMPLETE REFERENCE GENOMES
Given the complexity of human genetic variation, it follows that a single human reference genome is insufficient. Indeed, one of the great surprises after the initial sequencing of the first human genome (which provided the basis of the current human reference genome, GRCh38)35 was the large difference — in content and structure — between the first genome sequenced and additional human genomes.19,36,37 Although most people had accepted the notion that humans would differ from one another by millions of SNVs, the idea that the genetic code of two humans would vary by tens of thousands of larger (>50 bp) insertions, deletions, and inversions took much longer to realize and accept (Table 1). The idea that two humans might in fact might differ in their gene content because of the gain and loss of duplicate genes is still emerging38 but was clearly foreshadowed by the early work on the opsins and color-blindness traits.
This is a critical point, since the reference genome is the benchmark used by clinicians and geneticists to discover genetic variants associated with a disease. Widespread genome structural variation means that any single human haplotype, such as the first human reference genome, may be missing or may have sequence variants, including structural variants, that may or may not be present in the majority of humans. Moreover, complex regions of genetic variation are not particularly well understood outside of humans because nonhuman primate genomes have not been finished to the same standard as the human reference genome and typically carry hundreds of thousands of gaps precisely over these regions of complex genetic variation.39 Because of this missing information, our understanding of tolerance of variation and the extent of conservation in these regions is limited, and the genes therein have been excluded from disease association studies. A more systematic analysis of multiple human genomes — yielding reference genomes for different human populations — is required. There have been several attempts over the years to rapidly identify missing sequence by assembling short-read data from ethnically diverse populations.40,41 Because the sequence is often highly enriched in repetitive DNA, it is not accurately assembled or easily integrated into the current reference genome and, as a result, the associated structural variation is not well-characterized. For this reason, the National Institutes of Health recently issued a request for applications to generate high-quality reference genomes from people of diverse ancestry. Diverse reference genomes would enable variation discovery that is not guided by the human reference genome and would serve as stand-alone independent assemblies for future discovery.
Long-read sequencing technology has allowed us to directly sequence large stretches (ranging from 10,000 bp to 1,000,000 bp) of native DNA. This is particularly advantageous for detecting structural variation, because the long reads provide the necessary context to anchor and sequence-resolve most structural variants regardless of sequence composition. Numerous studies have provided evidence that long reads enhance the detection of structural variants,30,39,42–45 especially those between 50 bp and 2000 bp in length. Direct comparisons have shown that long reads deliver 2.48 times as many structural variants as short reads alone, even at their maximum possible sensitivity. It is estimated that at least 48% of deletions and 83% of insertions are routinely missed by the application of multiple short-read-calling algorithms.14
Long-read sequencing technology provides access to regions that were previously opaque to studies of human genetic variation, including variable-number tandem repeats (VNTRs),30,46 segmental duplications,38 and centromeres.47 This has led to an explosion in the discovery of complex genetic variation. For example, an analysis of 15 human genomes that used long reads resolved approximately 100,000 common structural variants, about half of which were previously unknown.46 Multiple human reference genomes also provided information on the existing human reference genome sequence — enabling the identification of more than 15,000 sites where the sequence is either in error or represents the minor alleles (alleles that are present in the population with a prevalence of <50%). This study also uncovered previously unidentified regulatory sequence, exons, and protein-encoding genes.38,46
Sequence resolution of structural variants has, not surprisingly, improved genotyping of these variants13,46 even when they are “read” by short-read sequencing, allowing for the discovery of new candidates for disease association.20–22
DISEASE VARIANT DISCOVERY
Several studies illustrate the importance of more comprehensive variant discovery (Table 2), especially as it relates to associations between variants and complex genetic diseases.20–22 For 10 years, genomewide association studies of bipolar disorder and schizophrenia have consistently identified a region mapping within the gene CACNA1C (encoding a calcium channel subunit) but have not discovered the disease-associated mutation. Song et al. focused on an intronic 30-bp VNTR and showed that all humans carry an expansion of 100 to 1000 units of this repeat.20 This finding is in contrast to that in the human reference genome, in which it appeared that only two subunits were present, an artifact that was probably caused by a combination of the instability of this repeat and bad luck when preparing it for sequencing as part of the Human Genome Project. Although the expansion is highly variable in length, it is not its size that is associated with the disease but rather the sequence composition and abundance of specific 30-bp repeats that define protective and risk haplotypes. Song et al. showed that composition differences in the sequence (deduced with long-read sequencing) were associated with differences in the expression of CACNA1C in neural cells.20
Table 2.
Genetic Disease and Complex Variants.*
| Disease | Variant or Variants and Location | Gene or Genes | Locus Structure Represented in Human Reference Genome† | Variant Detectable by Whole-Exome Sequencing | Variant Detectable by Whole-Genome Sequencing‡ | Method of Discovery |
|---|---|---|---|---|---|---|
| X-linked dystonia-parkinsonism | SVA insertion, noncoding region21§ | TAF1 | Yes | No | No | Long-read transcript sequencing |
| Bipolar disorder and schizophrenia | VNTR composition, noncoding region20 | CANA1C | No | No | No | Long-read sequencing |
| Schizophrenia | Complex structural variant of C4 genes, coding and noncoding regions48 | C4A, C4B | Yes/No | No | Yes/No | Digital droplet PCR |
| Benign adult familial myoclonic epilepsy | TTTTA expansion, noncoding region22 | SAMD12 | No | No | No | Long-read sequencing |
| Baratela-Scott syndrome | CCG expansion, noncoding region49 | XYLT1 | No | No | Yes | Southern blot and lllumina sequencing |
| Fascioscapulohumeral muscular dystrophy | Macrosatellite D4Z4 contraction and permissive SNVs, coding and noncoding regions26 | FSHD1 | Yes/No | No | Yes/No | Southern blot |
| Amyotrophic lateral sclerosis-frontal temporal dementia | GGGCC repeat expansion, noncoding region50,51 | C90RF72 | No | No | Yes/No | Southern blot, FISH, and repeat-primed PCR |
FISH denotes fluorescence in situ hybridization, PCR polymerase chain reaction, SNV single-nucleotide variant, and VNTR variable-number tandem repeat.
“Yes/No” indicates that the locus structure was incompletely represented in the human reference genome.
“Yes/No” indicates that the variant could be partially detected (depending on the size of the allele — i.e., sequences of the larger alleles are not completely resolved).
SVA (SINE-VNTR-Alu) is a class of retrotransposon found in humans and great apes.
Similarly, linkage analysis in Asian families with benign adult familial myoclonic epilepsy had narrowed the genetic cause of the autosomal dominant disorder to chromosome 8q24 without resolution of the disease-causing variant for more than 20 years. Using long-read sequencing technology, Ishiura and colleagues identified expansions of TTTCA and TTTTA in the intron of the gene SAMD12 in 49 of 51 families.22 The TTTCA expansions, in particular, were exclusive to patients and not found in controls. The authors then searched for expansions of this motif in other genes and identified similar expansions within the introns of different genes — for example, TNRC6A (encoding trinucleotide repeat containing 6A) and RAPGEF2 (encoding Rap guanine nucleotide exchange factor 2) — in the other two families. They concluded that the repeat expansions (irrespective of the gene) cause benign adult familial myoclonic epilepsy, probably through RNA-mediated toxicity mechanisms, which provided a new paradigm for the discovery of pathogenic epilepsy alleles.22
Finally, Aneichyk and colleagues undertook a multilayered genomic analysis to identify the cause of an elusive mendelian neurodegenerative disorder, X-linked dystonia–parkinsonism.21 Using combined long-read transcriptomic and genomic approaches, they identified the likely causal mutation as a retrotransposition mutation within an intron of the gene TAF1. This mutation event, which occurred in a founder haplotype of the island of Panay, Philippines, induced aberrant splicing of pre–messenger RNA. Long-read sequencing confirmed the presence of aberrant splice variants in persons with this rare form of parkinsonism.21 Such examples provide a path forward for identifying the variants involved in a subset of the more than 20% of mendelian disorders for which no pathogenic mutation has yet been discovered.
FUTURE PROSPECTS AND PERSPECTIVES
Technological advances in the past decade have dramatically changed our potential to discover and diagnose disease-causing variation. Despite the many success stories in human genetics, a large fraction of the genetic causes of both rare mendelian and common complex diseases remains unidentified. Although many would argue that this is simply a question of increasing sample sizes to provide increased power, another possibility is that the variant in question has been missed even after “whole”-genome sequencing. In that case, simply sequencing more patient samples with short-read data sets and aligning reads to a single reference genome is a suboptimal approach. Going forward, there are several areas that, if supported, will provide a more comprehensive understanding of the causes of genetic disease.
MULTIPLE HUMAN REFERENCE GENOMES
Given the complexity of genetic variation, it is clear that one reference genome is inadequate to represent human genetic diversity. There is a need to sequence and assemble normal genomes from different human populations but in particular from persons of African descent, the population in which the greatest source of genetic variation lies.52 Long-read DNA sequencing coupled with short-read error correction is leading to the development of dozens of new reference genomes, with more than 50 now in production. It is estimated, on the basis of our current rate of discovery, that sequencing 300 human genomes in such a manner would double the current number of known (at the level of DNA sequence) structural variants, identifying, in theory, the majority of common structural variants (or at least those with an allele frequency of ≥1%).46 Sequence resolution of structural variants has the benefit that such alleles can be better genotyped13,46 in existing short-read data, which in turn permits the identification of new associations in the millions of Illumina genomes that have already been generated.
PHASED GENOMES
Many disease-causing variants map outside of coding regions (Table 2), although their pathogenic effects often influence gene expression and translation. Although exome sequencing incurs less cost and thus permits greater sample size and power, it provides almost no information on regulatory mutations and limits the detection of small structural variants even within coding sequence.53 The discovery of pathogenic mutations associated with noncoding mutations is challenging. However, because structural variants are more likely than SNVs to be deleterious and affect the expression of genes, the systematic discovery of such variants may provide a better foothold for understanding noncoding regulatory mutations and their effect on common and rare genetic disease. Full genome sequencing is critical for the detection of structural variants and, in particular, fully phased long-read genome sequence data are estimated to provide a structural-variant yield that is 2.8 times as high as that obtained by Illumina whole-genome sequencing, as well as to increase the yield by 30% as compared with long-read callers that do not phase.13 We should therefore start thinking of a 6-Gbp (instead of a 3-Gbp) genome, in which both parental haplotypes are fully sequenced and assembled.14,54
ASSEMBLY VERSUS ALIGNMENT
Clinically, there is value in moving away from a model of variant discovery based on aligning fragments of sequence to a reference genome and toward a model in which variants are discovered on the basis of de novo assembly. Incomplete reference genomes can lead to a failure to discover pathogenic alleles as well as to misinterpretation of variants on the basis of mismap-ping. I predict that, within 10 years, it will be possible to clinically sequence and assemble both haplotypes of a patient first, followed by variant discovery by means of a comparison with a reference sequence. This will be particularly valuable for patients with adult-onset diseases (e.g., schizophrenia, Alzheimer’s disease, and Parkinson’s disease), for whom parental DNA may not be available. In such cases, physical phasing of long-read sequencing data with new genomic technologies will allow both parental haplotypes to be resolved and compared with other genomes.14 Such comparisons will facilitate the discovery of private genetic variants. This will require the use of multiple reference genomes and, potentially, graph-based reference genomes (i.e., nonlinear genome representations that build on a reference and capture the haplotypic diversity of a species or a population) as ways of expanding and improving human reference diversity and variant discovery.55,56
SEQUENCING FROM TELOMERE TO TELOMERE
Routine analysis of the human genome with the use of short-read sequence data captures only approximately 85% of the genome and excludes some of the most variation-rich regions, which are thus excluded from tests of association.8 The goal should be simple: complete telomere-totelomere sequence characterization of human chromosomes, including acrocentric, telomeric, centromeric, and segmentally duplicated DNA. Long-read and ultra-long-read sequencing platforms30,38,47 are now providing access to these traditionally opaque regions of human genetic variation.
HIGH-QUALITY COMPARATIVE GENOME SEQUENCING
The interpretation of variants benefits from a fundamental understanding of rates of de novo mutation, conservation of DNA sequence across species, and selection at any given locus. Specific tools57,58 that exploit knowledge of these forces have been helpful in the identification of likely pathogenic variants. However, they require that the pattern of variation be uniformly ascertained both within and between species. Because many stretches of orthologous DNA sequence are historically not well aligned and have rates of mutation that differ by orders of magnitude, it is critical that the same rigor that is applied to the sequencing of additional human genomes also be applied to nonhuman primate, mammalian, and vertebrate genomes.59,60
It is time for a new era of comparative sequencing, with the goal of sequencing the genomes of multiple diverse humans and nonhuman primates to completion. In the short term, this will allow for the development of both the evolutionary and the population frameworks needed to interpret the full spectrum of human genetic variation. Such data will permit the discovery of new disease-causing alleles and the development of new strategies to identify them. In the long term, these genomic advances will provide a template for how to phase, sequence, and assemble patients’ genomes in the clinic, which will become more tenable as the costs of long-read technology decrease and throughput increases.
Supplementary Material
Glossary
- Acrocentric
A term applied to chromosomes in which the short arm is substantially shorter than the long arm. Acrocentric DNA is typically composed of large, highly repetitive DNA encoding ribosomal RNA genes
- Aneuploidy
The occurrence of one or more extra or missing chromosomes, leading to an unbalanced chromosome complement
- Centromere
The point or region on a chromosome to which spindle microtubules attach; it is necessary for meiotic and mitotic segregation. In mammals, the centromere corresponds to the primary constriction of metaphase chromosomes and is typically composed of large tracts of repetitive DNA
- Comparative genomics
A subspecialty of genomics research in which the structure and function of DNA sequence is compared between species or populations of organisms
- Complex genetic disease
A condition caused by the interaction of multiple genes and environmental factors. Examples of complex conditions, which are also called multifactorial diseases, are cancer and heart disease
- Copy-number variation
Variation from one person to another in the number of copies of a particular gene or DNA sequence. The full extent to which copy-number variation contributes to human disease is not yet known
- De novo mutation
Any DNA sequence change that occurs during replication, such as a gene alteration newly occurring in a family as a result of a DNA sequence change in a germ cell or a fertilized egg
- Deletion
A mutation that involves the loss of genetic material. It can be small, involving a single missing DNA base pair, or large, involving a piece of a chromosome
- Exome
All known protein-coding sequences, or exons, in the human genome, constituting approximately 1 to 2% of the 3.2 billion nucleotide base pairs in the human genome
- Exon
The portion of a transcribed gene that encodes amino acids
- Genome
The entire set of genetic instructions found in a cell. In humans, the genome consists of 23 pairs of chromo somes, found in the nucleus, as well as a small chromosome found in the mitochondria of the cell
- Genomewide association study
An approach to the discovery of disease-risk loci that relies on searching the entire genome to identify genetic variants with allele frequencies that are robustly correlated with either disease status or the level of a trait of interest. If the association between the variant and the disease or trait is significant, it is referred to as a genomewide association signal
- Graph-based reference genome
A nonlinear genome representation that builds on a reference sequence, with genetic variants added as edges and invariant sections added as nodes in a graph in order to capture the haplotypic diversity of a species or a population
- GRCh38
A coordinate-based, high-quality representation of the human genome sequence that is highly annotated, accessible to the public, and evaluated by the community. It consists of a composite of different human genomes, rather than being derived from a single person, and mapping of sequence against it is used to discover genetic variation and interpret its effects
- Haplotype
A set of DNA variations, or polymorphisms, that tend to be inherited together. A haplotype can refer to a combination of alleles or to a set of single-nucleotide polymorphisms found on the same chromosome
- Human Genome Project
An international project completed that mapped and sequenced the entire human genome
- Indel
An insertion or deletion mutation between 1 bp and 49 bp in length
- Insertion mutation
A type of mutation involving the addition of genetic material. An insertion mutation can be small, involving a single extra DNA base pair, or large, involving a piece of a chromosome
- Introns
The portions of a gene that are removed (spliced out) before translation to a protein. Introns may contain regulatory information that is critical to appropriate gene expression
- Inversion
A chromosomal segment that has been broken off and reinserted in the same place, but with the genetic sequence in reverse order
- Long-read sequencing
A genetic sequencing platform that relies on long sequence reads (>15,000 bp in length) and is used to discover genetic variants on the basis of sequence assembly and alignment to a reference (e.g., Oxford Nanopore Technologies or Pacific Biosciences)
- Mendelian disease
Genetic disease attributable to variants with large effects on disease status. Because of the high penetrance of such variants, the disease typically cosegregates in a classic mendelian fashion (e.g., dominant or recessive)
- Mutation
A change in a DNA sequence. Germ-line mutations occur in the eggs and sperm and can be passed on to offspring, whereas somatic mutations occur in body cells and are not passed on
- Noncoding DNA sequence
A DNA sequence that does not encode proteins. Noncoding DNA sequences, once referred to as “junk DNA,” account for the majority of genome sequences and are now known to harbor regions that regulate gene expression
- Penetrance
The likelihood that a person carrying a particular genetic variant will have a detectably altered phenotype
- Point mutation
An alteration in DNA sequence caused by a single-nucleotide base change, insertion, or deletion; a base insertion or deletion creates a frameshift
- Private genetic variant
An ultra-rare genetic variant that is typically identified as segregating in only one family in a comparison set
- Read depth
A measure of the number of reads mapping to a genomic interval; a number that is significantly higher or lower than the mean is interpreted as evidence of copy-number variation
- Reference genome
A linear representation of the genome of a species, in which the sequence is ordered and oriented, along with gaps, into chromosomes. Its coordinate system is generally used by the research community for annotation and variant discovery and reporting
- Regulatory sequence
A noncoding DNA sequence that affects the expression of genes
- Retrotransposition
A molecular mechanism responsible for the propagation of mobile elements through RNA intermediates
- Segmental duplication
A duplicated sequence in the genome that occurred during the course of evolution that is typically more than 1000 bp in length and more than 90% identical to the ancestral sequence
- Short-read-calling algorithm
A series of strategies in which the pattern of short-read sequence data mapped to a reference genome is used to infer patterns of structural variation
- Short-read sequencing
The current, dominant approach used to sequence human exomes and genomes. Short stretches of DNA from the patient are sequenced to generate short reads. The original position of each short read is determined by searching for a highly similar or identical DNA sequence in the reference genome sequence
- Single-nucleotide variant
A type of variation affecting a single nucleotide in a DNA sequence, in which the nucleotide (for example, cytosine) is substituted with a different type of nucleotide (for example, thymidine)
- Structural variant
A genetic variant involving the insertion, deletion, duplication, translocation, or inversion of segments of DNA from 50 bp up to millions of base pairs in length
- Telomeric
Pertaining to the terminal portion of a chromosome. In humans, the telomere is characterized by repeating tracts of the TTAGGG motif
- Translocation
The positional change of one or more chromosome segments in cells or gametes without alteration of the normal amount of genetic material
- Variable-number tandem repeat
A copy-number-polymorphic sequence in which the unit of the tandem array is at least 7 bp in length (in contrast to a short tandem repeat, in which the motif size is ≤6 bp in length)
- Whole-genome sequencing
Determination of the primary nucleotide sequence of the entire genome of an organism
Footnotes
References
- 1.Natarajan P, Peloso GM, Zekavat SM, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat Commun 2018; 9: 3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet 2014; 46: 944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeb SS. The molecular basis of variation in human color vision. Clin Genet 2005; 67: 369–77. [DOI] [PubMed] [Google Scholar]
- 6.Neitz M, Neitz J. Numbers and ratios of visual pigment genes for normal red-green color vision. Science 1995; 267: 1013–6. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, Motulsky AG, Deeb SS. Position of a ‘green-red’ hybrid gene in the visual pigment array determines colour-vision phenotype. Nat Genet 1999; 22: 90–3. [DOI] [PubMed] [Google Scholar]
- 8.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015; 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudmant PH, Mallick S, Nelson BJ, et al. Global diversity, population stratification, and selection of human copy-number variation. Science 2015; 349: aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudmant PH, Kitzman JO, Antonacci F, et al. Diversity of human copy number variation and multicopy genes. Science 2010; 330: 641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huddleston J, Chaisson MJP, Steinberg KM, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 2017; 27: 677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaisson MJP, Sanders AD, Zhao X, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 2019; 10:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaisson MJ, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet 2015; 16: 627–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature 2011; 470: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 1998; 14: 417–22. [DOI] [PubMed] [Google Scholar]
- 18.Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 2006; 38: 1038–42. [DOI] [PubMed] [Google Scholar]
- 19.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science 2007; 316: 445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song JHT, Lowe CB, Kingsley DM. Characterization of a human-specific tandem repeat associated with bipolar disorder and schizophrenia. Am J Hum Genet 2018; 103: 421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aneichyk T, Hendriks WT, Yadav R, et al. Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 2018; 172(5): 897–909.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishiura H, Doi K, Mitsui J, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet 2018; 50: 581–90. [DOI] [PubMed] [Google Scholar]
- 23.Chiang C, Scott AJ, Davis JR, et al. The impact of structural variation on human gene expression. Nat Genet 2017; 49: 692–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 2006; 38: 75–81. [DOI] [PubMed] [Google Scholar]
- 25.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemmers RJ, van der Vliet PJ, Klooster R, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 2010; 329: 1650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korbel JO, Urban AE, Affourtit JP, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 2007; 318: 420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handsaker RE, Van Doren V, Berman JR, et al. Large multiallelic copy number variations in humans. Nat Genet 2015; 47: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider VA, Graves-Lindsay T, Howe K, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 2017; 27: 849–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaisson MJ, Huddleston J, Dennis MY, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 2015; 517: 608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharp AJ, Locke DP, McGrath SD, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 2005; 77: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature 2010; 464: 704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009; 461: 747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010; 11: 446–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409: 860–921. [DOI] [PubMed] [Google Scholar]
- 36.Bailey JA, Yavor AM, Viggiano L, et al. Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am J Hum Genet 2002; 70: 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–51. [DOI] [PubMed] [Google Scholar]
- 38.Vollger MR, Dishuck PC, Sorensen M, et al. Long-read sequence and assembly of segmental duplications. Nat Methods 2019; 16: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon D, Huddleston J, Chaisson MJ, et al. Long-read sequence assembly of the gorilla genome. Science 2016; 352: aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman RM, Forman J, Antonescu V, et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat Genet 2019; 51: 30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R, Li Y, Zheng H, et al. Building the sequence map of the human pan-genome. Nat Biotechnol 2010; 28: 57–63. [DOI] [PubMed] [Google Scholar]
- 42.Pendleton M, Sebra R, Pang AW, et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods 2015; 12: 780–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo JS, Rhie A, Kim J, et al. De novo assembly and phasing of a Korean human genome. Nature 2016; 538: 243–7. [DOI] [PubMed] [Google Scholar]
- 44.Shi L, Guo Y, Dong C, et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun 2016; 7: 12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain M, Koren S, Miga KH, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 2018; 36: 338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Audano PA, Sulovari A, Graves-Lindsay TA, et al. Characterizing the major structural variant alleles of the human genome. Cell 2019; 176(3): 663–675.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain M, Olsen HE, Turner DJ, et al. Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol 2018; 36: 321–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekar A, Bialas AR, de Rivera H, et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016; 530: 177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaCroix AJ, Stabley D, Sahraoui R, et al. GGC repeat expansion and exon 1 methylation of XYLT1 is a common pathogenic variant in Baratela-Scott syndrome. Am J Hum Genet 2019; 104: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexa-nucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClellan JM, Lehner T, King MC. Gene discovery for complex traits: lessons from Africa. Cell 2017;171: 261–4. [DOI] [PubMed] [Google Scholar]
- 53.Turner TN, Coe BP, Dickel DE, et al. Genomic patterns of de novo mutation in simplex autism. Cell 2017; 171(3): 710–722. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koren S, Rhie A, Walenz BP, et al. De novo assembly of haplotype-resolved ge nomes with trio binning. Nat Biotechnol 2018. October 22 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen N, Hickey G, Zerbino DR, et al. Building a pan-genome reference for a population. J Comput Biol 2015; 22: 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garrison E, Sirén J, Novak AM, et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol 2018; 36: 875–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013; 9(8): e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kronenberg ZN, Fiddes IT, Gordon D, et al. High-resolution comparative analysis of great ape genomes. Science 2018; 360: eaar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bickhart DM, Rosen BD, Koren S, et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet 2017; 49: 643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.