Abstract
Binge drinking during adolescence increases the risk for neuropsychiatric disorders including alcoholism in adulthood. DNA methylation in post-mitotic neurons is an important epigenetic modification that plays a crucial role in neurodevelopment. We examined the effects of intermittent ethanol exposure during adolescence on adult behavior and whether DNA methylation changes provide a plausible explanation for the lasting effects of this developmental insult. One hour after last adolescent intermittent ethanol (AIE), growth arrest and DNA damage inducible protein 45 (Gadd45a, Gadd45b, and Gadd45g) mRNA expression was increased and DNA methyltransferase (DNMT) activity and Dnmt3b expression was decreased in the amygdala as compared to adolescent intermittent saline (AIS) rats. However, AIE rats 24 hrs after last exposure displayed increased DNMT activity but normalized Gadd45 and Dnmt3b mRNA expression compared to adolescent intermittent saline (AIS) rats. In adulthood, rats exposed to AIE show increased Dnmt3b mRNA expression and DNMT activity, along with decreased Gadd45g mRNA expression in the amygdala. DNA methylation of neuropeptide Y (Npy) and brain-derived neurotrophic factor (Bdnf) exon IV is increased in the AIE adult amygdala compared to AIS adult rats. Treatment with the DNMT inhibitor 5-azacytidine (5-azaC) at adulthood normalizes the AIE-induced DNA hypermethylation of Npy and Bdnf exon IV with concomitant reversal of AIE-induced anxiety-like and alcohol-drinking behaviors. These results suggest that binge-like ethanol exposure during adolescence leads to dysregulation in DNA methylation mechanisms in the amygdala which may contribute to behavioral phenotypes of anxiety and alcohol use in adulthood.
Keywords: Adolescent binge drinking, Alcohol use disorders, Amygdala, Anxiety, DNA methylation/demethylation, Neuropeptide Y, BDNF
1. Introduction
Alcohol consumption during adolescence is a serious health concern due to its deleterious neurodevelopmental consequences and impact on negative affective states (Bekman et al., 2013; Kyzar et al., 2016; McGue et al., 2001; Spear, 2018). Clinical and preclinical studies have shown that binge patterns of drinking during adolescence is hazardous, as intoxicating blood ethanol levels and repeated withdrawals increase the probability for developing alcohol use disorder (AUD) and other psychiatric disorders including anxiety at adulthood (DeWit et al., 2000; Grant and Dawson, 1997; Kokare et al., 2017; Kyzar et al., 2017; Pandey et al., 2015; Pascual et al., 2009). Specifically, early onset of alcohol use during the adolescent period leads to a significant increase in risk for diagnosis of AUD in adulthood (DeWit et al., 2000). Additionally, early age of first alcohol use and later diagnosis of anxiety disorders in adulthood are correlated (Agrawal et al., 2009; Grant et al., 2001), and early onset of anxiety disorders predicts first alcohol use in the general population (Birrell et al., 2015). Preclinical modeling has replicated this phenomenon, as rats exposed to adolescent alcohol exposure show increased alcohol consumption and anxiety-like behavior in adulthood (Alaux-Cantin et al., 2013; Amodeo et al., 2017; Gass et al., 2014; Kokare et al., 2017; Kyzar et al., 2017; Pandey et al., 2015; Pascual et al., 2009). Adult rats previously exposed to adolescent alcohol display molecular alterations in crucial affective brain circuitry such as the amygdala, which is a hub for alcohol drinking and anxiety-like behaviors (Koob and Volkow, 2010) and may explain the persistent behavioral phenotypes seen following binge-like adolescent alcohol consumption (Gilpin et al., 2015; Kokare et al., 2017; Kyzar et al., 2016, 2017; Pandey et al., 2015, 2017).
Epigenetic mechanisms involving histone and DNA chemical modifications are important molecular pathways regulating gene expression during neurodevelopment and are sensitive to early life adversity (Kofink et al., 2013; Labonté et al., 2012; Szyf, 2013). We have shown that adolescent alcohol exposure is associated with condensed chromatin architecture due to deficits in histone acetylation (H3K9/14ac and H3K27ac) and increased histone methylation (H3K9me2 and H3K27me3) in the rat amygdala associated with anxiety-like and alcohol drinking behaviors in adulthood (Kokare et al., 2017; Kyzar et al., 2017; 2019; Pandey et al., 2015). DNA methylation is a relatively stable epigenetic modification that modulates gene expression in response to environmental stimuli (Szyf, 2013). DNA methylation at a given gene promoter is dynamically regulated, and this mark is deposited onto cytosine bases by DNA methyltransferases (DNMTs) (Feng and Fan, 2009; Goll and Bestor, 2005; Razin, 1998). DNMT1 functions as a maintenance methyltransferase, whereas DNMT3a and DNMT3b catalyze the ‘de novo’ methylation of previously unmethylated cytosine residues ( Feng and Fan, 2009; Krishnan et al., 2014). Additionally, DNA demethylation occurs in a process involving base excision repair (BER) mechanisms (Gavin et al., 2013; Wu and Zhang, 2014). On the contrary, the BER pathway is initiated by the ten-eleven translocation (TET) family of enzymes that catalyzes the conversion of methylcytosine to 5-hydroxymethylcytosine (5-hmC), which is ultimately removed through several enzymatic events coordinated by the growth arrest and DNA damage inducible protein 45 (GADD45) family of co-factors (Krishnan et al., 2014; Wu and Zhang, 2014). One of the potential mechanisms by which adolescent alcohol exposure may produce a lasting impact in adulthood could be through changes in DNA methylation and demethylation pathways.
Alcohol alters DNA methylation/demethylation mechanisms in human and rodent brains (Gatta et al., 2017; Guidotti et al., 2013; Manzardo et al., 2012; Ponomarev et al., 2012; Taqi et al., 2011). A single anxiolytic dose of ethanol is capable of inhibiting DNMT activity and modulating Dnmt isoform mRNA levels in the amygdala of adolescent rats (Sakharkar et al., 2014a). Here, we investigated whether adolescent intermittent ethanol (AIE)-induced abnormalities in expression of the enzymes involved in DNA methylation and demethylation in the amygdala during adolescence persist into adulthood, and if abnormal DNA methylation mechanisms may be associated with anxiety-like and alcohol drinking behaviors in adulthood. We probed AIE-induced effects on DNA methylation levels specifically at the promoters of two genes involved in anxiety and alcohol drinking behaviors, the endogenous anxiolytic peptide neuropeptide Y (Npy) and the crucial neurotrophin brain-derived neurotrophic factor (Bdnf). Lower levels of NPY and BDNF in the amygdala have been generally implicated in anxiety and alcohol abuse (Gilpin et al., 2011; Joe et al., 2007; Moonat et al., 2011; Pandey et al., 2017), and AIE treatment leads to an epigenetically-encoded decrease in levels of NPY and BDNF protein and mRNA in the amygdala of adult rats (Kokare et al., 2017; Kyzar et al., 2017; Pandey et al., 2015). However, the role of DNA methylation in the regulation of Npy and Bdnf expression in the amygdala after AIE in adulthood has not been explored. We hypothesize that altered amygdala DNA methylation mechanisms may contribute to increased anxiety-like and alcohol-drinking behaviors after adolescent alcohol exposure in adulthood. We therefore examined the effects of AIE on DNA methylation of the Npy and Bdnf exon IV gene promoters in the amygdala at adulthood. We further examined whether the long-lasting DNA methylation changes in the amygdala and the AIE-induced behavioral changes could be reversed using the DNMT inhibitor 5-azacytidine (5-azaC) in adulthood.
2. Experimental procedures
2.1. Animals and adolescent intermittent ethanol (AIE) exposure
Time-pregnant Sprague-Dawley (SD) rats or rat dams with pups were purchased from Harlan Laboratories (Indianapolis, IN, USA) and maintained on 12:12 hrs light/dark cycle with ad libitum access to food and water. Male pups were weaned at post-natal day (PND) 21 and were group-housed (2 or 3 rats). We used the previously reported paradigm of adolescent intermittent ethanol (AIE) or n-saline (AIS) exposure (Alaux-Cantin et al., 2013; Kokare et al., 2017; Kyzar et al., 2017; Pandey et al., 2015; Pascual et al., 2009). Adolescent male rats received eight intraperitoneal (I.P.) injections of either ethanol (2 g/kg, 20% w/v) or equivalent volume of n-saline during PND 28–41 on alternating 2-days on/off basis. Animals were group-housed throughout all experiments except during alcohol drinking studies. We have previously reported the 2 g/kg dose of ethanol is non-sedative and anxiolytic in adolescent rats (Sakharkar et al., 2014a). All animal experiments followed NIH guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee.
AIS- or AIE-exposed adolescent male rats were sacrificed as previously described (Pandey et al., 2015) after 1 hr (AIE-1hr group; sacrificed on PND 41) and 24 hrs (AIE-24hrs group; sacrificed on PND 42 to determine the effect of alcohol withdrawal) of the last injections along with their respective controls (AIS group). Rats were anesthetized with pentobarbital and decapitated to collect brain tissues for biochemical measurements in the amygdala. Another batch of AIS and AIE male rats were allowed to grow until the adulthood (PND 92) in order to measure baseline differences between adult AIS and AIE rats. Rats were anesthetized with pentobarbital or inhaled isoflurane and then sacrificed for dissection of amygdala tissue and immediately frozen for downstream biochemical measures.
2.2. Effects of 5-azacytidine on behavioral measurements in AIS and AIE adult rats
To examine the effect of DNMT inhibition by 5-azaC on AIS or AIE adult rats, animals were allowed to grow to adulthood prior to treatment with 5-azaC on three consecutive days (PND 92, 93, 94) and were subjected to measurement of anxiety-like behaviors. 5-azaC was dissolved in DMSO to a 5mg/ml concentration and was diluted in normal saline (1:5) to achieve the final concentration of 1 mg/ml. DMSO diluted in n-saline (1:5) was used as a vehicle. 5-azaC (1 mg/kg) or equivalent volumes of vehicle was I.P. injected daily for three consecutive days (each injection 24 hrs apart) and animals were subjected to the light/dark box (LDB) test two hours after the last 5-azaC or vehicle injections. Immediately after the behavioral measurements, rats were anesthetized with pentobarbital, decapitated and the brains were dissected. Amygdala tissues were isolated, quickly frozen and stored at −80 0C prior to processing for biochemical measures as described below.
In another subset of AIS and AIE rats which were allowed to grow to adulthood following adolescent treatment, the baseline differences in ethanol intake at adulthood and the effect of DNMT inhibition by 5-azaC were examined using the two-bottle free-choice paradigm as described previously (Moonat et al., 2013; Pandey et al., 2015; Sakharkar et al., 2014b). AIS and AIE adult rats were single-housed and were habituated to drink water from two bottles for two weeks. Post-training, rats received water in one bottle and increasing concentration of ethanol (3% of ethanol for 3 days from PND 96–98, 7% of ethanol for 3 days from PND 99–101, and 9% of ethanol for 3 days from PND 102–104) in another bottle. The position of the bottles was alternated every day to avoid habit formation for the position of the bottles. All rats received fresh solutions in clean bottles every day in the evening between 5:00 to 6:00 pm, and ethanol and water intake for the previous day (last 24 hrs) was measured (ml/day). After 3 days of 9% ethanol drinking, rats were also concomitantly injected (I.P.) with either vehicle [(AIS+ Vehicle) and (AIE+ Vehicle)] or 5-azaC [1 mg/kg; (AIS+5-azaC) and (AIE+5-azaC)] once daily for four consecutive days (PND 105–108). Rats received vehicle or 5-azaC injections before water and ethanol bottles were given. After four days of vehicle or 5-azaC injection and drinking measurements, rats continued to receive 9% ethanol for additional 3 more days to examine the post-treatment effects on ethanol intake. Alcohol and water intake was measured daily (ml/day) and ethanol-drinking in terms of g/kg/day was calculated for each rat and represented as mean (±SEM). The body weights and total fluid intake (ml/day) were also measured.
2.3. Light/dark box (LDB) exploration test
Anxiety-like behaviors were measured using the LDB exploration test as described previously (Pandey et al., 2008,2015; Sakharkar et al., 2014a). In brief, each rat was conditioned to behavior room for 5 min and was placed into the dark compartment of the LDB apparatus. The activity of the rat was monitored by the computer for the 5-min test session. Time spent in each compartment and ambulation was recorded in each compartment, which was equipped with infrared beam that enabled tracking by computer. Data is represented in terms of mean percent time spent (±SEM) in each compartment and mean total ambulation (±SEM).
2.4. Measurement of DNMT activity
DNMT activity in amygdala was measured using the EpiQuik™ DNA methyltransferases activity/inhibition assay kit (Epigentek; Brooklyn, NY), as described previously (Sakharkar et al., 2014a). Nuclear protein fraction was prepared from the amygdaloid tissues using a nuclear extraction kit (Sigma, St. Louis, MO) and 10 µg of nuclear protein was used for measuring the DNMT activity according to the manufacturer’s protocol. Activity was measured in terms of optical density (O.D.)/mg protein and the results are presented as mean percent of controls (± SEMs).
2.5. Quantitative real-time PCR for mRNA measurements
Quantitative real-time PCR (qRT-PCR) was performed for the analysis of different DNMT isoforms (Dnmt1, Dnmt3a and Dnmt3b), Gadd45a, Gadd45a, Gadd45g, and Tet1 mRNA levels as described previously (Sakharkar et al., 2014a). RNA was isolated from the amygdaloid tissues from the rat brain using TRIZOL reagent (Life Technologies, Grand Island, NY, USA) with subsequent DNA removal using DNA-free™ Kit (Life Technologies). Total RNA was reverse transcribed using High Capacity cDNA Reverse Transcription kit (Life Technologies, Grans Island, NY) or High Capacity Archive Kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green qPCR Master Mix (Agilent Technologies, Santa Clara, CA). Data were analyzed with MxPro software. Primers corresponding to various genes measured are shown in Table 1. PCR conditions included a 10 min 95 0C hold followed by 40 cycles at 95 0C for 30 s, 60 0C for 1 min, and 72 0C for 1 min. The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as an internal control for sample normalization. Target cDNAs for various genes along with Gapdh were analyzed in duplicate for each measurement. The relative expression was determined using 2−ΔΔCT method (Livak and Schmittgen, 2001) after normalization to Gapdh and results are represented as fold change in the mRNA levels.
Table 1.
List of primers used for the measurement of mRNA levels in the amygdala of the rat.
| Gapdh | Forward: ACAAGATGGTGAAGGTCGGTGTGA Reverse: AGCTTCCCATTCTCAGCCTTGACT |
| Dnmt1 | Forward: AAGCCAGCTATGCGACTTGGAAAC Reverse: ACA ACC GTTGGCTTTCTGAGTGAG |
| Dnmt3a | Forward: CACCTACAACAAGCAGCCCATGTA Reverse: AGCCTTGCCAGTGTCACTTTCATC |
| Dnmt3b | Forward: TGTGCAGAGTCCATTGCTGTAGGA Reverse: GCT TCCGCCAATCACCAAGTCAAA |
| Gadd45a | Forward: TGCGAGAACGACATCAACAT Reverse: TCCCGGCAAAAACAAATAAG |
| Gadd45b | Forward: GACAACGCGGTTCAGAAGAT Reverse: TCTTCGTCTATGGCCAGCA |
| Gadd45g | Forward: CATTGCACGAACTTCTGCTG Reverse: ACGCCTGGATCAACGTAAAA |
| Tet1 | Forward: AGAGGGCCAAGATGAAGGAG Reverse: TTTCTGGAAAGCCACCTGAG |
2.6. Measurement of DNA Methylation by MethylMiner™ Assay
Genomic DNA was isolated using DNeasy™ blood and tissue kit according to manufacturer recommendations (Qiagen Inc., Valencia, CA). The DNA was sonicated to 200–500 base-pairs fragments and subjected to the MethylMiner™ methylated DNA enrichment protocol (Life Technologies, Carlsbad, CA) which uses MBD-Biotin molecules to pull down methylated DNA fragments. The methylated DNA complexed with MBD-Biotin was eluted using high salt solution and further cleaned by phenol-chloroform method. Input genomic DNA was used as an internal control to normalize the data. qRT-PCR was performed to amplify the eluted (methylated) and input DNA using SYBR Green qPCR Master Mix (Thermo Scientific) or SYBR Green qPCR SSO Master Mix (BioRad, Hercules, CA, USA) and primers specific to Npy (Forward-5’-AGTAGGTCCAGTAGGTCCAGTAGGT-3’ and Reverse-5’-GAAGCAGTCGAGCAAGGTTTT-3’) and Bdnf exon IV (Forward-5’-GTTCGCTAGGACTGGAAGTGG-3’ and Reverse-5’-CCTCTGCCTCGAAATAGACAC-3’) as mentioned above. The fold changes in DNA methylation were calculated using 2−ΔΔCT method (Livak and Schmittgen, 2001) after normalizing to input and are presented as mean fold changes (±SEM) with respect to control (AIS group).
2.7. DNA methylation and hydroxymethlation by immunoprecipitation assay
Methylated-DNA immunoprecipitation (MeDIP) assay and hydroxymethylated-DNA immunoprecipitation (hMeDIP) assay was performed to measure the DNA methylation and hydroxymethylation levels, respectively, at the Npy gene promoter. DNA was sonicated to 200–500 base-pairs fragments and suspended in 250 μl ChIP dilution buffer. DNA was boiled for 10 min and was immediately placed on ice. Further, DNA was incubated with either 4 μl of 5-methylcytosine monoclonal mouse antibodies (Diagenode, Denville, NJ) or 2 μl of rabbit polyclonal 5-hydroxymethylcytosine antibodies (Active Motif, Carlsbad, CA). Subsequently, antibody-DNA complex was precipitated using Protein A/G PLUS-agarose™ beads (Santa Cruz Biotechnology, Santa Cruz, CA). The beads-antibody-DNA complex was washed using low salt, high salt, lithium chloride and TE buffers. DNA was eluted from beads in elution buffer by heating at 67 0C for 2 hrs and proteinase K digestion of proteins at 37 0C followed by phenol-chloroform purification. Purified DNA was resuspended in water. Genomic DNA that was not subjected to the immunoprecipitation (input) was used as an internal control to normalize the data. qRT-PCR was performed to amplify the precipitated and input DNA using the SYBR Green qPCR Master Mix (Thermo Scientific, Pittsburgh, PA) and Npy primers. Fold changes in DNA methylation and hydroxymethylation levels were calculated after normalizing to input using 2−ΔΔCT method (Livak and Schmittgen, 2001) and results are presented as mean fold changes (±SEM) with respect to control (AIS group).
2.8. Chromatin immunoprecipitation (ChIP) assay for MeCP2 occupancy
Chromatin immunoprecipitation (ChIP) assay was performed as described previously (Kokare et al., 2017; Kyzar et al., 2017) to examine the level of MeCP2 at the promoters of Npy and Bdnf exon IV. Amygdala tissues were homogenized in PBS and briefly fixed in formaldehyde. The fixed tissue homogenate was resuspended in SDS-lysis buffer for 30 min and sonicated to shear the chromatin to 200–500 base-pairs long DNA fragments. Volumes of chromatin from each sample were separated for chromatin immunoprecipitation using an antibody against MeCP2 (ab2828; Abcam, Cambridge, MA, USA) and for quantifying the amount of DNA in different samples before immunoprecipitation (inputs). For immunoprecipitation, chromatin was resuspended in ChIP dilution buffer along with MeCP2 antibody and Protein A/G PLUS-agarose™ beads (Santa Cruz Biotechnology) and incubated overnight at 4 0C. On the following day, chromatin was washed and isolated using Chelex-100 chelating resin (BioRad). qRT-PCR was performed using SYBR Green qPCR SSO Master Mix (BioRad) and primers specific to Npy and Bdnf exon IV as mentioned above. Fold changes in MeCP2 levels were calculated after normalizing to input using 2−ΔΔCT method (Livak and Schmittgen, 2001). Results are presented as mean fold changes (±SEM) with respect to control (AIS group).
2.9. Statistical Analysis
Statistical differences between two and more than two groups were analyzed with SigmaStat (Systat Software Inc., San Jose, CA, USA) using Student’s t-test and one-way analysis of variance (ANOVA), respectively. Two-way repeated measures ANOVA was used to analyze alcohol and total fluid intake. Post hoc analysis for all the ANOVA comparisons was performed using the Tukey’s test. p values less than 0.05 were considered significant.
3. Results
3.1. Effects of AIE on amygdala DNMT activity and DNA methylation/demethylation pathways mRNA levels during adolescence
A schematic of this experiment is provided in Figure 1A. Nuclear DNMT activity was measured in the amygdaloid tissues of AIS, AIE-1hr, and AIE-24hrs animals (Figure 1B). While DNMT activity in the amygdala was decreased (p<0.001) in AIE-1hr rats, it was significantly increased (p<0.001) in AIE-24hrs animals as compared to AIS-exposed animals (Fig. 1B). Dnmt3b mRNA levels were decreased (p<0.05) in the amygdala of AIE-1hr rats, with no significant changes in AIE-24hrs, as compared to AIS-exposed animals (Fig. 1C). No significant differences were observed in mRNA levels of Dnmt1 or Dnmt3a in the amygdala of AIS and AIE (1hr and 24hrs) rats (Fig. 1C). mRNA levels of Gadd45a, Gadd45b and Gadd45g were significantly increased (p<0.05) in the amygdala of AIE-1hr animals, with no significant changes in AIE-24hrs as compared to AIS-exposed animals (Fig. 1D). mRNA levels of Tet1 in the amygdala were not altered by AIE (Fig. 1D). These results indicate that AIE exposure is associated with modifications in the expression of specific enzymes that are involved in DNA methylation and demethylation pathways in the amygdala during the adolescent period.
Figure 1. Effect of adolescent intermittent ethanol (AIE) and n-saline (AIS) exposure on DNMT activity and DNA methylation/demethylation isoform expression in the adolescent amygdala.
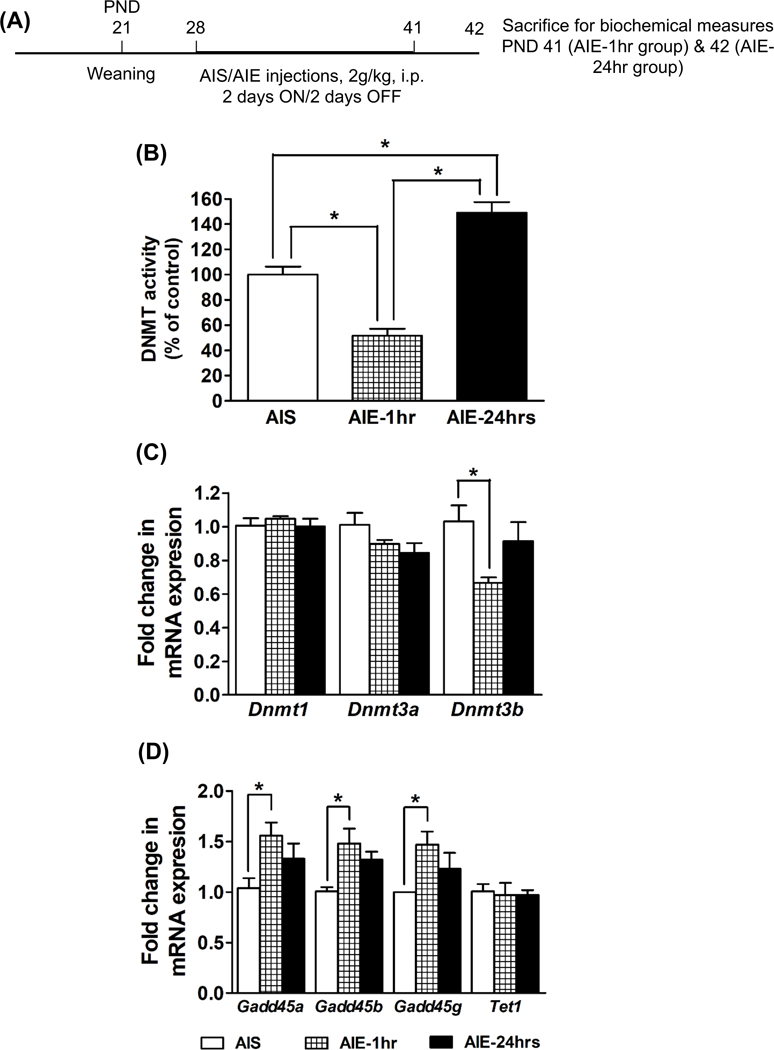
A) Schematic of adolescent alcohol exposure and brain collection for biochemical studies.
B) DNMT activity in the nuclear protein fractions of the amygdala tissues obtained from AIS, AIE (1 hr), and AIE (24 hrs) adolescent rats. Values are the mean percent of control (±SEM) of n=6–7 rats per group. *Significantly different from AIS or AIE-1hr group (p<0.001; ANOVA, F2, 17 = 44.9, p<0.001 followed by post hoc analysis by Tukey’s test).
C) Fold changes in mRNA levels of Dnmt1, Dnmt3a and Dnmt3b in the amygdala of the AIS, AIE (1hr) and AIE (24 hrs) adolescent rats. Values are the mean (±SEM) of n=5 rats in each group. *Significantly different from AIS group (p<0.05; ANOVA, F2, 12 = 4.6, p<0.05 followed by post hoc analysis by Tukey’s test).
D) Fold changes in mRNA levels of Gadd45a, Gadd45b, Gadd45g, and Tet1 in the amygdala of the AIS, AIE (1hr) and AIE (24 hrs) adolescent rats. Values are the mean (±SEM) of n=5 rats in each group. *Significantly different from AIS group (p<0.05; ANOVA, F2, 12 = 4.0, p<0.05 for Gadd45a; p<0.05; ANOVA, F2, 12 = 5.8, p<0.05 for Gadd45b; p<0.05; ANOVA, F2, 12 = 4.0, p<0.05 for Gadd45g followed by post hoc analysis by Tukey’s test).
3.2. Effects of AIE on amygdala DNMT activity and DNA methylation/demethylation pathways mRNA levels in adulthood
A schematic of this experiment is provided in Figure 2A. Nuclear DNMT activity was measured in the amygdala of the AIS and AIE adult rats (Fig. 2B). DNMT activity was significantly higher (p<0.001) in the amygdala of the AIE adult rats as compared to AIS adult rats (Fig. 2B). This is accompanied by the significantly higher (p<0.05) mRNA levels of Dnmt1 and Dnmt3b in the amygdala of AIE adult rats as compared to AIS adult rats (Fig. 2C). Additionally, mRNA levels of Gadd45g were significantly reduced (p<0.05) without changes in the expression of Gadd45a and Gadd45b in the amygdala of AIE adult rats as compared to AIS adult rats (Fig. 2D).
Figure 2. Effect of adolescent intermittent ethanol (AIE) and n-saline (AIS) exposure on DNMT activity and DNA methylation/demethylation isoform expression in the adult amygdala.
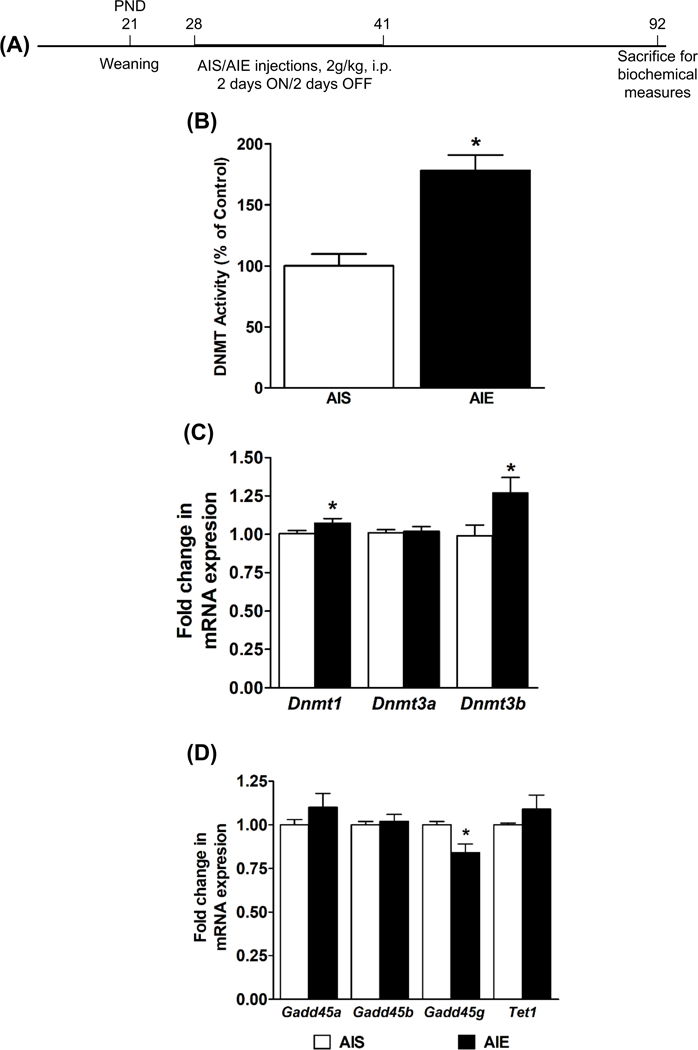
A) Schematic of adolescent alcohol exposure and brain collection in adulthood for biochemical studies.
B) Nuclear DNMT activity in the amygdala of AIS and AIE adult rats. Values are represented as the mean % of control (±SEM) of n=7–8 rats per group. *Significantly different from AIS adult group (p<0.001, student’s t test).
C) Fold changes in mRNA levels of Dnmt1, Dnmt3a and Dnmt3b in the amygdala of the AIS and AIE adult rats. Values are the mean (±SEM) of n=12–13 rats per group. *Significantly different from AIS group (p<0.05; student’s t test).
D) Fold changes in mRNA levels of Gadd45a, Gadd45b, Gadd45g, and Tet1in the amygdala of the AIS and AIE adult rats. Values are the mean (±SEM) of n=8 rats per group. *Significantly different from AIS group (p<0.05; student’s t test).
3.3. Effects of AIE on DNA methylation at Npy and Bdnf exon IV in the amygdala at adulthood
We have previously shown that both NPY and BDNF levels are decreased in the amygdala of AIE-exposed adult rats compared to AIS rats (Kokare et al., 2017; Pandey et al., 2015), but the contribution of DNA methylation levels at the promoter regions of these genes to the decreased expression in adulthood is unknown. Therefore, we used the MethylMiner™ assay to measure methylated DNA levels at the Npy and Bdnf exon IV promoter regions in the adult amygdala. DNA methylation levels were increased at both the Npy promoter (Fig. 3A; p<0.05) and the Bdnf exon IV promoter (Fig. 3B; p<0.05) in the amygdala of AIE-exposed adult rats compared to AIS-exposed adults. DNA methylation recruits repressive co-factors such as methyl-CpG-binding protein 2 (MeCP2) to condense local chromatin and cause transcriptional downregulation (Jones et al., 1998), and MeCP2 is particularly important in maintaining precise levels of synapse-associated gene expression in the brain (Kavalali et al., 2011). Therefore, we used the ChIP assay to investigate whether levels of MeCP2 were also altered at the same promoter regions of Npy and Bdnf exon IV in the amygdala. MeCP2 occupancy was significantly increased at the Npy promoter (Fig. 3C; p<0.05), but not the Bdnf exon IV promoter (Fig. 3D), in the amygdala of AIE adult rats compared to AIS adult rats.
Figure 3. Effect of adolescent intermittent ethanol (AIE) and n-saline (AIS) exposure on DNA methylation of Npy and Bdnf exon IV in the adult amygdala.
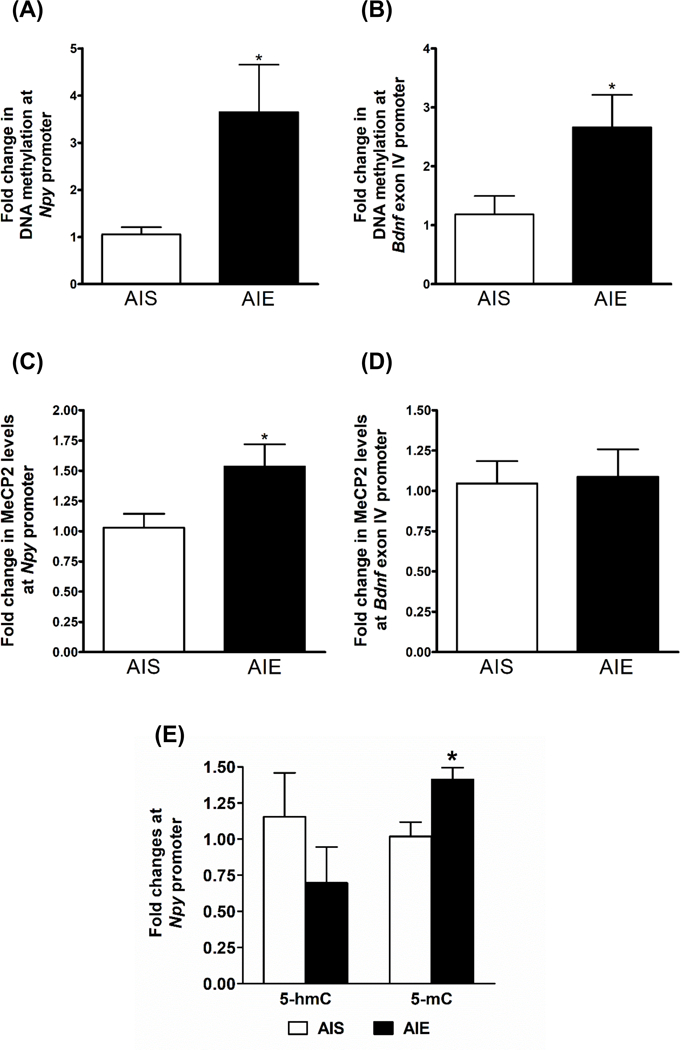
A) Effects of AIE and AIS exposure on DNA methylation levels at the Npy gene promoter in the adult amygdala as measured by the MethylMiner™ assay. Values are represented as the mean (±SEM) of n=6 rats per group. *Significantly different from AIS adult group (p<0.05, student’s t test).
B) Effects of AIE and AIS exposure on DNA methylation levels at the Bdnf exon IV promoter in the adult amygdala as measured by the MethylMiner™ assay. Values are represented as the mean (±SEM) of n=6 rats per group. *Significantly different from AIS adult group (p<0.05, student’s t test).
C) Effects of AIE and AIS exposure on MeCP2 levels at the Npy gene promoter in the adult amygdala as measured by the chromatin immunoprecipitation (ChIP) assay. Values are represented as the mean (±SEM) of n=6 rats per group. *Significantly different from AIS adult group (p<0.05, student’s t test).
D) Effects of AIE and AIS exposure on MeCP2 levels at the Bdnf exon IV promoter in the adult amygdala as measured by the chromatin immunoprecipitation (ChIP) assay. Values are represented as the mean (±SEM) of n=6 rats per group.
E) Effects of AIE and AIS exposure on 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) levels at the neuropeptide Y (Npy) gene promoter in the amygdala as analyzed by the methylated DNA-immunoprecipitation (MeDIP) assay and the hydroxymethylated DNA-immunoprecipitation (hMeDIP). Values are represented as the mean (±SEM) of n=4–5 rats per group. *Significantly different from AIS adult group (p<0.05, student’s t test).
To confirm our DNA methylation findings from the MethylMiner™ assay using a different method, we measured 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) levels at the Npy gene promoter using the MeDIP and hMeDIP assays. Similar to our MethylMiner™ assay findings, 5-mC levels were significantly increased (p<0.05) at the Npy gene promoter, but 5-hmC levels were not significantly altered, in the amygdala of the AIE adult rats as compared to AIS adult rats (Fig. 3E). Taken together, these results indicate that AIE leads to increased DNA methylation around crucial anxiety-related genes such as Npy and Bdnf in the amygdala at adulthood.
3.4. Effects of 5-azaC treatment on AIE-induced anxiety-like behaviors and levels of DNA methylation in adulthood
AIS and AIE adult rats were treated with 5-azaC (1 mg/kg for three consecutive days) to examine the effect of inhibition of DNA methylation on anxiety-like behaviors and on levels of DNA methylation in the Npy and Bdnf gene promoter. A schematic of this experiment is provided in Figure 4A. AIE-adult rats displayed anxiety-like behaviors which were attenuated by 5-azaC treatment (Fig. 4B). AIE adult rats treated with 5-azaC (AIE+ 5-azaC group) spent more time (p<0.001) in the light and less time in the dark compartment (Fig. 4B), as compared to vehicle-treated AIE adult rats (AIE+ Vehicle group). 5-azaC treatment did not alter anxiety measures of AIS adult rats (Fig. 4B). No significant differences were observed in total ambulation among different groups, indicating that the general activity of the rats was not affected by either AIE exposure or 5-azaC treatment.
Figure 4. Effects of 5-azacytidine (5-azaC) treatment in AIS and AIE adult rats on anxiety-like behaviors and DNA methylation mechanisms.
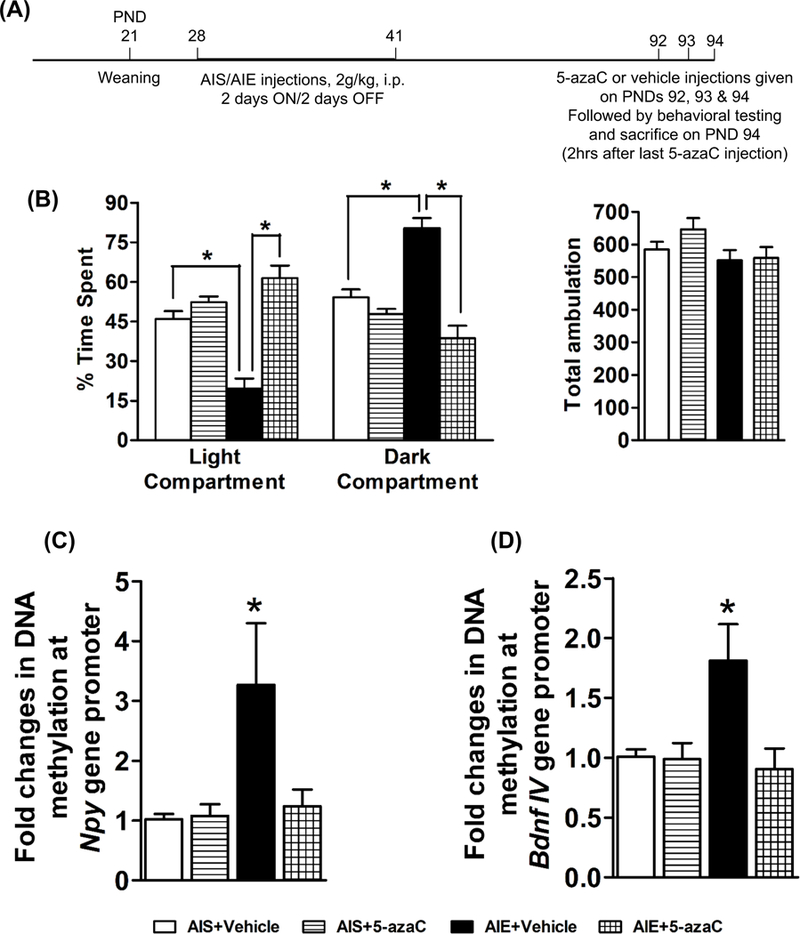
A) Schematic of adolescent alcohol exposure and 5-azaC treatment, behavioral measurements, and brain collection in adulthood for biochemical studies.
B) Effects of 5-azacytidine (5-azaC) treatment in AIS and AIE adult rats on anxiety-like behaviors as analyzed by light dark box (LDB) exploration test. Treatment with 5-azaC (1 mg/kg once daily for three days; 2 hr after last injections) was able to attenuate AIE-induced anxiety-like behaviors at adulthood. Values are represented as the mean (±SEM) of the percent of time spent in each compartment of n=7 rats per group. *Significantly different from respective group [p<0.001, ANOVA (F3, 24 = 25.0, p<0.001) followed by Tukey’s test]. The general activity in terms of total ambulation was not different among the groups.
C) Effects of 5-azacytidine (5-azaC) treatment on DNA methylation levels at the Npy gene promoter in the amygdala as measured by the MethylMiner™ assay. 5-azaC treatment (1 mg/kg once daily for three days; 2 hr after last injections) reduced the AIE exposure induced hypermethylation in the amygdala of the AIE-exposed adult rats. Values are represented as the mean (±SEM) of n=7 rats per group. *Significantly different from AIS-exposed control (AIS+ Vehicle) group [p<0.05, ANOVA (F3, 24 = 4.0, p<0.05) followed by Tukey’s test].
D) Effects of 5-azacytidine (5-azaC) treatment on DNA methylation levels at the Bdnf exon IV promoter in the amygdala as measured by the MethylMiner™ assay. 5-azaC treatment (1 mg/kg once daily for three days; 2 hr after last injections) reduced the AIE exposure induced hypermethylation in the amygdala of the AIE-exposed adult rats. Values are represented as the mean (±SEM) of n=7 rats per group. *Significantly different from AIS-exposed control (AIS+ Vehicle) group [p<0.05, ANOVA (F3, 24 = 5.0, p<0.01) followed by Tukey’s test].
We further examined if DNMT inhibitor treatment could normalize the AIE-induced DNA hypermethylation of Npy and Bdnf exon IV gene promoters. A significant increase (p<0.05) in DNA methylation levels at Npy gene promoter in the amygdala of AIE adult rats using the MethylMiner™ assay (Fig. 4C) was observed as compared to AIS adult rats. Treatment with 5-azaC for three consecutive days (one injection each day; 1 mg/kg) reversed (p<0.05) the AIE-induced DNA hypermethylation at this location in the Npy gene promoter (Fig. 4C). A similar pattern was observed at the Bdnf exon IV promoter (Fig. 4D), where AIE adult rats showed an increase in DNA methylation at baseline (p<0.05) that was normalized by 5-azaC treatment (p<0.05). These results indicate that anxiety-like behaviors and the increased DNA methylation at the Npy and Bdnf exon IV gene promoters associated with AIE exposure are reversible by DNMT inhibitor treatment.
3.5. Effects of 5-azaC treatment on AIE-induced alcohol-drinking behaviors in adulthood
AIS and AIE adult rats were treated with 5-azaC (1 mg/kg for four consecutive days) to examine the effect of inhibition of DNA methylation on alcohol drinking behaviors. A schematic of this experiment is provided in Figure 5A. We observed that the AIE adult rats (AIE+ Vehicle) drank significantly higher (p<0.001) amounts of ethanol as compared to AIS rats (AIS+ Vehicle) in a two-bottle free-choice paradigm (Fig. 5B). To examine if the increased alcohol intake by AIE-adult rats could be reversed by DNMT inhibition, we treated AIS and AIE adult rats with 5-azaC for four consecutive days after three days of 9% ethanol intake (Fig. 5B). Ethanol intake by AIE adult rats treated with 5-azaC (AIE+ 5-azaC) was significantly reduced (p<0.001) after the third injection, as compared to the vehicle-treated AIE adult rats (AIE+ Vehicle). The effect of 5-azaC on the ethanol drinking behavior lasted for one more day following the fourth and last injection of 5-azaC (Fig. 5B) and gradually returned to the levels seen in AIE rats at baseline after two days. However, 5-azaC treatment did not affect the alcohol intake by AIS adult rats (Fig. 5B). There were no significant differences in total fluid intake (ml/day) between the various groups except the AIE+5-azaC group drank significantly higher amount of total fluid on tenth day as compared to AIS+ Vehicle and AIS+5-azaC groups.
Figure 5. Effects of 5-azacytidine (5-azaC) treatment in AIS and AIE adult rats on alcohol-drinking behaviors.
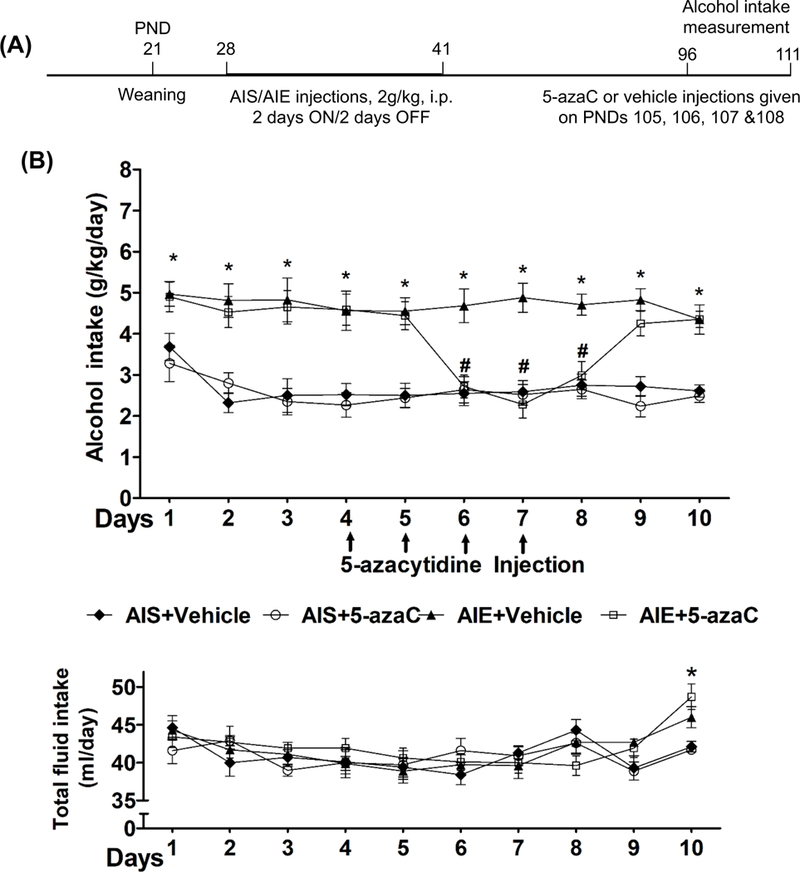
A) Schematic of adolescent alcohol exposure, 5-azaC treatment, alcohol intake measurement in adulthood.
B) Effects of 5-azacytidine (5-azaC) treatment in AIS and AIE adult rats on alcohol-drinking behaviors in two-bottle free-choice paradigm. Line diagrams show daily alcohol and total fluid (water+ethanol) intake and its modulation by 5-azaC treatment in AIS and AIE adult rats. The pattern of ethanol consumption as measured by the two-bottle free choice paradigm indicated that AIE adult rats consumed significantly higher amounts of alcohol (g/kg/day) as compared with AIS adult rats. After 3 days of 9% ethanol intake, AIS and AIE adult rats were treated with either vehicle or 5-azaC (1 mg/kg once daily for 4 days) and consumption of water and 9% ethanol were measured for next seven days. Values are represented as the mean ± SEM (n=7 rats per group) of alcohol intake (g/kg/day) or total fluid intake (ml/day). Two-way repeated measures of ANOVA revealed a significant difference in alcohol intake between various groups overall and daily (Group: F3, 240 = 103.6, p<0.001; Day: F9, 240 =3.7, p<0.001). *Significant difference in alcohol intake was observed between AIS+ Vehicle and AIE+ Vehicle groups (p<0.05–0.001; repeated measures ANOVA followed by post hoc analysis of the group within day comparison using Tukey’s test). # Significant difference in alcohol intake between AIE+ Vehicle and AIE+5-azaC groups (p<0.001; repeated measures ANOVA followed by post hoc analysis of the group within day comparison using Tukey’s test). In addition, there was a significant difference in total fluid intake by days, but not by groups (Day: F9, 240 = 5.5, p<0.001). *Significant difference in total fluid intake was observed between AIE+5-azaC with AIS+Vehicle and AIS+5-azaC groups (p<0.01; repeated measures ANOVA followed by post hoc analysis of the group within day comparison using Tukey’s test).
4. Discussion
In the current study, we demonstrated that adolescent intermittent ethanol exposure produces dysregulation in DNA methylation/demethylation pathways in the amygdala associated with behavioral phenotypes of anxiety and alcohol intake (Fig. 6). Specifically, we found that AIE exposure is associated with an increase in Dnmt3b mRNA expression and DNMT activity, along with a decrease in Gadd45g mRNA expression in the adult amygdala. These changes were also associated with increase in DNA methylation at Npy and Bdnf exon IV, and both of these genes have been highly implicated in AUD and anxiety-like behaviors (Gilpin et al., 2015; Pandey et al., 2017). Additionally, increased DNA methylation at Npy and Bdnf exon IV may contribute to the previously reported downregulation in mRNA and protein levels of NPY and BDNF in the AIE adult amygdala (Kokare et al., 2017; Kyzar et al., 2019; Pandey et al., 2015). Similar to previous studies, AIE produced anxiety-like behaviors (Kokare et al., 2017; Kyzar et al., 2019; Kyzar et al., 2017; Pandey et al., 2015) and increased alcohol intake in adulthood (Alaux-Cantin et al., 2013; Amodeo et al., 2017; Gass et al., 2014; Pandey et al., 2015; Pascual et al., 2009). The data collected here suggest a role for DNA methylation in the observed increased risk for adult alcohol use and anxiety disorders associated with adolescent binge drinking in clinical populations (Birrell et al., 2015; Brown et al., 2008; DeWit et al., 2000; Grant and Dawson, 1997; Grant et al., 2001). Importantly, we found that the observed DNA hypermethylation at Npy and Bdnf and the AIE-induced behavioral changes are reversible using a DNMT inhibitor (5-azaC). DNMT inhibitor treatment in mice has been shown to decrease alcohol intake previously (Warnault et al., 2013). However, in the present study we demonstrated that a behavioral change produced by an early-life environmental insult such as AIE, likely encoded in part through DNA methylation, is pharmacologically reversible using a DNMT inhibitor (Fig. 6).
Figure 6. DNA methylation mechanisms and behavioral consequences of adolescent intermittent ethanol (AIE) exposure during adolescence and in adulthood.
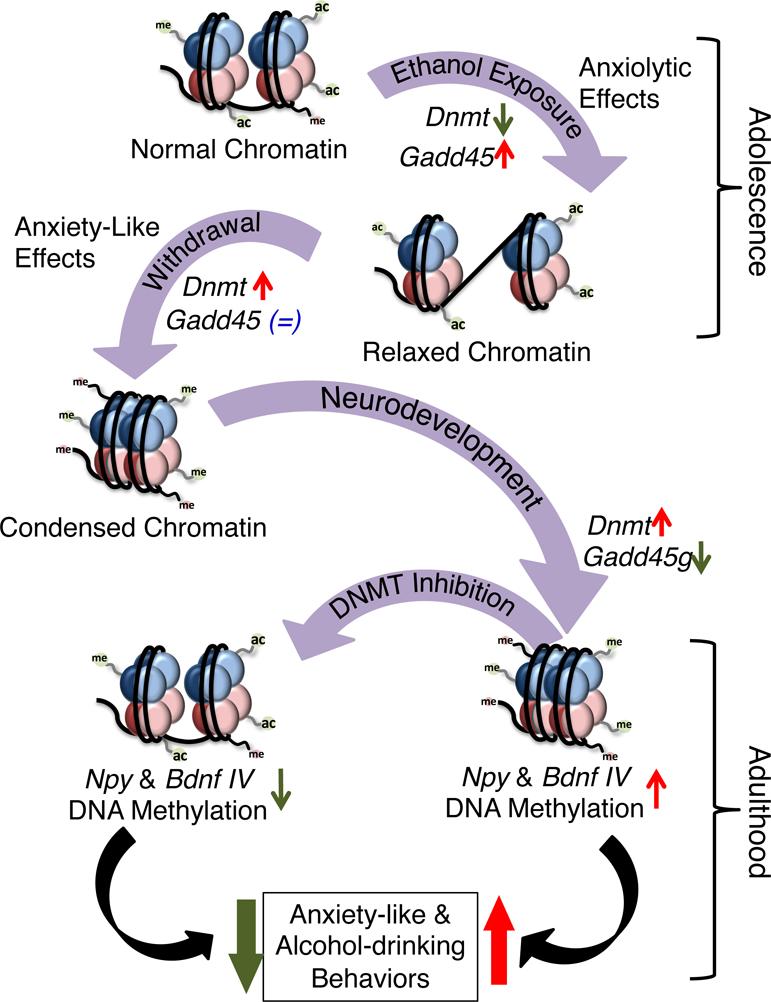
AIE exposure alters DNA methylation mechanisms in the amygdaloid circuitry. Repeated AIE exposure (1 hr after last AIE exposure) leads to a decrease in DNA methyltransferase (DNMT) activity and a concomitant increase in growth arrest and DNA-damage-inducible (Gadd45) family of proteins that mediates active DNA demethylation. Withdrawal (24 hrs) from repeated AIE during adolescence increases DNMT activity in the amygdala. Interestingly, a similar AIE paradigm in adolescent rats produced anxiety-like behaviors (Pandey et al., 2015). During the course of development, this increased DNMT activity is maintained, along with an increase in Dnmt3b mRNA and a reduction in Gadd45g mRNA expression in the amygdala. These changes may produce a DNA hypermethylated state resulting in altered epigenetic dynamics at the Npy and Bdnf exon IV promoter regions and reductions in NPY and BDNF expression (Kokare et al., 2017; Pandey et al., 2015). The treatment with DNMT inhibitor (5-azaC) in adulthood normalizes the AIE-induced DNA hypermethylation at the Npy and Bdnf exon IV promoter regions, along with the AIE-induced anxiety-like behavior and increased alcohol consumption. These results implicate a novel role for DNA methylation/demethylation mechanisms in the amygdala in AIE-induced anxiety-like and alcohol-drinking behaviors at adulthood.
Early-life adversity has been shown to alter DNA methylation in the brain and increase the risk for psychiatric disorders later in life (Khulan et al., 2014; Labonté et al., 2012; Roth et al., 2009; Weaver et al., 2004). For example, early-life stress results in enduring DNA hypermethylation of the Bdnf exon IV promoter region in the adult prefrontal cortex (Roth et al., 2009). Similarly, offspring of low maternal care dams show DNA hypermethylation of the glucocorticoid receptor gene in the hippocampus which is associated with heightened stress responses in adulthood, and this behavior is reversed by inhibition of histone deacetylases (HDACs) (Szyf, 2013; Weaver et al., 2004). DNA methylation and HDACs are known to interact (Jones et al., 1998; Kavalali et al., 2011), and we have previously observed that the HDAC2 isoform is increased in the AIE adult amygdala (Pandey et al., 2015). Along similar lines, previously we observed deficits in H3K9/14ac at the promoter regions of both Npy and Bdnf exon IV, where we observe DNA hypermethylation in the amygdala following AIE in adulthood (Kokare et al., 2017; Pandey et al., 2015). In this context, the present study increases our knowledge of the dynamic interaction between DNA methylation and histone modifications in the amygdala after AIE in adulthood. Additionally, MeCP2 occupancy was found to be increased at the Npy promoter, but unaltered at the Bdnf exon IV promoter, in the AIE adult amygdala in this study. This may reflect the context-dependent transcriptional regulation of MeCP2 (Chahrour et al., 2008), as DNA hypermethylation may recruit this protein to some genomic loci but not others. Notably, mice harboring a mutation in Mecp2 consume less alcohol than wild-type mice (Repunte-Canonigo et al., 2014), and fetal alcohol exposure leads to increased Mecp2 mRNA expression in the pituitary (Gangisetty et al., 2015). Future studies should further probe the interaction between histone acetylation, DNA methylation, and DNA-binding proteins such as MeCP2 to the behavioral phenotypes produced by AIE.
Prior studies show that ethanol impacts DNA methylation dynamics in various in vivo and in vitro experimental models (Bekdash et al., 2013; Chen et al., 2013; Krishnan et al., 2014). DNA methylation in peripheral blood mononuclear cells of chronic alcoholic patients is associated with a decrease in DNMT3B mRNA expression (Bonsch et al., 2006). Changes in DNMT1 mRNA expression in response to chronic alcohol abuse were observed in the prefrontal cortex of psychotic patients (Guidotti et al., 2013). Whole genome-wide approaches have revealed global DNA hypomethylation in the prefrontal cortex of alcoholic patients (Ponomarev et al., 2012). Recently, we observed DNA hypermethylation of the delta subunit of GABA-A receptors in the human post-mortem cerebellum of alcoholics as compared with control subjects (Gatta et al., 2017). Chronic cocaine exposure or stress has been shown to be associated with the persistent changes in Dnmt3a expression in the nucleus accumbens of the adult rats (LaPlant et al., 2010). We previously reported that a single ethanol exposure (2 g/kg; I.P.) inhibits DNMT activity in the amygdala of adolescent rats at 1 hr after injection (Sakharkar et al., 2014a). In the present study, we observed that 1 hr after the last AIE in adolescent rats, DNMT activity is inhibited and Dnmt3b mRNA levels are reduced in the amygdala. However, 24 hrs following the last AIE when adolescent rats were in a state of withdrawal, DNMT activity was increased. This increased DNMT activity was found to persist into adulthood, possibly as a result of increased Dnmt3b mRNA levels in the adult amygdala. However, mRNA levels of Dnmts are not altered in adolescence rats during withdrawal, suggesting that different mechanisms may be involved in the regulation of DNMT activity at adolescence and adulthood after AIE.
In the present study, we found that ethanol significantly altered Gadd45 isoform mRNA expression in the amygdala, but not Tet1 mRNA expression. One hour following the last AIE during adolescence, all three Gadd45 isoforms were significantly increased in the amygdala compared to AIS rats. These alterations were no longer present at 24 hours during withdrawal, but a significant downregulation of Gadd45g mRNA was noted in the amygdala of AIE adult rats. Notably, a similar trajectory of mRNA expression (increased in AIE-1hr rats, not significant in AIE-24hr rats, and decreased in AIE adult rats) was observed for the lysine-specific demethylase 1 (Lsd1) gene in the amygdala of AIE-exposed rats, suggesting that some genes that respond to repeated ethanol during adolescence may become deficient after prolonged abstinence into adulthood and may contribute to long-lasting AIE-induced phenotypes (Kyzar et al., 2017). GADD45B mRNA expression is altered in the prefrontal cortex of alcoholic psychotic patients (Guidotti et al., 2013). Interestingly, Gadd45b and Gadd45g mRNA expression is decreased in the nucleus accumbens of the high alcohol-consuming C57BL/6J mice compared to low alcohol-consuming DBA/2J mice, and Gadd45b heterozygous mice show higher levels of voluntary alcohol consumption (Gavin et al., 2016). Given the role of GADD45 isoforms in the removal of methyl marks from DNA along with up regulation of DNMT activity, it appears that both DNA methylation and demethylation mechanisms in the amygdala are perturbed by AIE which may be associated with altered chromatin architecture in adulthood (Figure 6).
Interestingly, the current study also revealed that two key genes shown to be involved in the risk for alcoholism, Npy and Bdnf, are affected by a persistent dysregulation in DNA methylation/demethylation pathways as a result of AIE. These genes have been shown to exhibit decreased mRNA and protein expression in the amygdala of AIE adult rats compared to AIS rats (Kokare et al., 2017; Pandey et al., 2015). Extensive studies from multiple groups have implicated NPY and BDNF in anxiety and AUD, typically showing that low levels of NPY and BDNF are associated with heightened anxiety and increased alcohol consumption (Gavin et al., 2016; Gilpin et al., 2011, 2015; Heilig, 1995; Joe et al., 2007; Kokare et al., 2017; Kyzar et al., 2017; Moonat et al., 2011, 2013; Pandey et al., 2004,2005,2006, 2008, 2015; Rimondini et al., 2005; Sakharkar et al., 2012, 2014b; Zhang et al., 2010). Previously, we observed that histone acetylation (H3K9/14ac) is an important epigenetic modification involved in Npy gene regulation in the amygdala in both alcohol-preferring (P) rats (Sakharkar et al., 2014b) and AIE adult rats (Kokare et al., 2017). Similarly, we have previously shown that histone acetylation is decreased at the Bdnf gene in the amygdala of P rats and AIE adult rats and have additionally shown that repressive H3K9me2 is increased at the Bdnf exon IV promoter in the AIE adult amygdala (Kyzar et al., 2017; Moonat et al., 2011, 2013; Pandey et al., 2015). It is likely that genes other than Bdnf and Npy are altered by DNA methylation dynamics after AIE exposure, and future studies should aim to discover other methylated sites involved in AIE-induced adult psychopathology. These findings extend our overall knowledge of altered epigenetic processes in the amygdala following AIE exposure (Kyzar et al., 2016). We have previously shown that the HDAC2 isoform and LSD1 are altered in the AIE adult amygdala, leading to globally decreased histone acetylation and increased repressive H3K9me2 (Kyzar et al., 2017; Pandey et al., 2015). Treatment with trichostatin A (TSA), HDAC inhibitor was able to attenuate AIE-induced anxiety-like and alcohol drinking behaviors in rats during adulthood (Pandey et al., 2015). Here, we demonstrate that anxiety-like and alcohol drinking behaviors observed after AIE are also accompanied by an increase in DNMT activity and Dnmt3b mRNA expression in adult rat amygdala that lead to a concomitant increase in DNA methylation at crucial anxiety-related genes such as Npy and Bdnf. Taken together, these results indicate that AIE may alter chromatin conformation in the adult amygdala characterized by increased histone acetylation, methylation, and DNA methylation specifically at the promoter regions of genes involved in anxiety-like and alcohol drinking behaviors (Fig. 6). Interestingly, prior studies have demonstrated the potential use of epigenetic drugs to treat AUD in preclinical models. For example, HDAC and DNMT inhibitors have been shown to ameliorate alcohol intake and other alcohol-related behaviors in animal models of alcohol abuse (Sakharkar et al., 2012, 2014b; Warnault et al., 2013). Here, 5-azaC was shown to attenuate AIE-induced anxiety-like and alcohol drinking behaviors and the increased DNA methylation seen at the Npy and Bdnf exon IV promoters in the amygdala. Additionally, the dose and repeated administration of 5-azaC in this study is in line with previous studies investigating the behavioral effects of DNMT inhibition (Sales et al., 2011; Warnault et al., 2013), but it is possible that side effects could alter behavioral effects seen here. However, it is important to note that 5-azaC treatment did not affect total fluid intake in the 2-bottle choice paradigm and had no effect on anxiety measures in AIS animals as well as total ambulation among groups in LDB test. Further consideration should be given to the DNMT isoform specificity of 5-azaC. The crystal structure of the bacterial DNMT, which is closely related to mammalian DNMT1, in complex with a derivative of 5-azaC has been elucidated and shows that 5-azaC likely blocks the enzymatic activity of DNMT1 (Sheikhnejad et al., 1999). While no such structure exists for DNMT3a or DNMT3b, the ability of these enzymes to methylated non-CpG sites suggest that they may be even more sensitive to the inhibitory effects of 5-azaC on DNA methylation (Christman, 2002). Therefore, determining the effects of specific DNMT isoforms and their alteration by 5-azaC remains an important future direction of research.
There are limitations to the conclusions of the current study. Firstly, we chose to model adolescent binge drinking using i.p. injections to standardize exposure paradigm, as previous studies have noted that alcohol-induced DNA methylation is dose-dependent (Cervera-Juanes et al., 2017; Sakharkar et al., 2014a). It is possible that other modes of AIE exposure including vapor chambers and self-administration may lead to differing effects of DNA methylation in both adolescence and adulthood. Nonetheless, our AIE findings of decreased BDNF expression in the adult rat amygdala translate to humans as shown by decreased BDNF expression in the postmortem amygdala of early onset AUD subjects compared with control subjects (Bohnsack et al., 2019). The current study focused on male rats as previous studies establishing the AIE phenotype were conducted in males (Kokare et al., 2017; Pandey et al., 2015), but future studies should incorporate female animals to determine sex-independent epigenetic effects of adolescent alcohol exposure (Becker et al., 2016).
Taken together, the results of the current study clearly indicate that changes in epigenetic processes including DNA methylation during development after AIE are possibly associated with altered chromatin architecture in the emotional circuitry of the amygdala, contributing to anxiety-like and alcohol drinking behaviors in adulthood (Fig 6). These behaviors may be related to AIE-induced DNA hypermethylation of the crucial anxiety-related genes Npy and Bdnf in the amygdala at adulthood. The behavioral phenotypes and the increase in DNA methylation at Npy and Bdnf exon IV in the amygdala after AIE in adulthood are attenuated by treatment with DNMT inhibitors. Therefore, the present study highlights the therapeutic potential of DNMT inhibitors for AIE-induced anxiety and alcohol use disorders later in adulthood (Fig. 6).
Highlights.
Adolescent intermittent ethanol (AIE) increases adult anxiety and alcohol drinking
AIE increased DMNT function in the amygdala in adulthood
AIE increased DNA methylation at Npy and Bdnf exon IV in the adult amygdala
5-azacytidine treatment normalizes AIE-induced anxiety and alcohol consumption
5-azacytidine treatment restores AIE-induced DNA hypermethylation at Npy and Bdnf
Acknowledgements
This work was supported by National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants U01AA-019971, U24AA-024605 [Neurobiology of adolescent Drinking in Adulthood (NADIA) project], RO1AA-010005, and P50AA-022538 (Center for Alcohol Research in Epigenetics), and by the Department of Veterans Affairs (Merit Review Grant, I01BX000143; Senior Research Career Scientist award) to S.C.P. Additional funding included the NIAAA grant F30AA-024948 to E.J.K., and the Department of Veterans Affairs grant [Merit Review Grant Career Development Award (CDA-2) (IK2BX001650)] to D.P.G. Funders had no further role in the study design; collection, analysis, and interpretation of the data; writing of the report; or decision to submit the paper for publication. Current address of A.J.S. is: Department of Biotechnology, Savitribai Phule Pune University, Ganeshkhind, Pune 411 007, India.
Footnotes
Conflict of interest
Authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Agrawal A, Sartor CE, Lynskey MT, Grant JD, Pergadia ML, Grucza R, Bucholz KK, Nelson EC, Madden PA, Martin NG, Heath AC, 2009. Evidence for an interaction between age at first drink and genetic influences on DSM-IV alcohol dependence symptoms. Alcohol Clin Exp Res 33, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux-Cantin S, Warnault V, Legastelois R, Botia B, Pierrefiche O, Vilpoux C, Naassila M, 2013. Alcohol intoxications during adolescence increase motivation for alcohol in adult rats and induce neuroadaptations in the nucleus accumbens. Neuropharmacology 67, 521–531. [DOI] [PubMed] [Google Scholar]
- Amodeo LR, Kneiber D, Wills DN, Ehlers CL, 2017. Alcohol drinking during adolescence increases consumptive responses to alcohol in adulthood in Wistar rats. Alcohol 59, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JB, Prendergast BJ, Liang JW, 2016. Female rats are not more variable than male rats: a meta-analysis of neuroscience studies. Biol Sex Differ 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK, 2013. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in beta-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res 37, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekman NM, Winward JL, Lau LL, Wagner CC, Brown SA, 2013. The impact of adolescent binge drinking and sustained abstinence on affective state. Alcohol Clin Exp Res 37, 1432–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell L, Newton NC, Teesson M, Tonks Z, Slade T, 2015. Anxiety disorders and first alcohol use in the general population. Findings from a nationally representative sample. J Anxiety Disord 31, 108–113. [DOI] [PubMed] [Google Scholar]
- Bohnsack JP, Teppen T, Kyzar EJ, Dzitoyeva S, Pandey SC, 2019. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Translational Psychiatry 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S, 2006. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm (Vienna) 113, 1299–1304. [DOI] [PubMed] [Google Scholar]
- Brown SA, McGue M, Maggs J, Schulenberg J, Hingson R, Swartzwelder S, Martin C, Chung T, Tapert SF, Sher K, Winters KC, Lowman C, Murphy S, 2008. A developmental perspective on alcohol and youths 16 to 20 years of age. Pediatrics 121 Suppl 4, S290–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera-Juanes R, Wilhelm LJ, Park B, Grant KA, Ferguson B, 2017. Alcohol-dose-dependent DNA methylation and expression in the nucleus accumbens identifies coordinated regulation of synaptic genes. Transl Psychiatry 7, e994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY, 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Zhou FC, 2013. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS One 8, e60503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman JK, 2002. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495. [DOI] [PubMed] [Google Scholar]
- DeWit DJ, Adlaf EM, Offord DR, Ogborne AC, 2000. Age at first alcohol use: a risk factor for the development of alcohol disorders. Am J Psychiatry 157, 745–750. [DOI] [PubMed] [Google Scholar]
- Feng J, Fan G, 2009. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol 89, 67–84. [DOI] [PubMed] [Google Scholar]
- Gangisetty O, Wynne O, Jabbar S, Nasello C, Sarkar DK, 2015. Fetal Alcohol Exposure Reduces Dopamine Receptor D2 and Increases Pituitary Weight and Prolactin Production via Epigenetic Mechanisms. PLoS One 10, e0140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JT, Glen WB Jr., McGonigal JT, Trantham-Davidson H, Lopez MF, Randall PK, Yaxley R, Floresco SB, Chandler LJ, 2014. Adolescent alcohol exposure reduces behavioral flexibility, promotes disinhibition, and increases resistance to extinction of ethanol self-administration in adulthood. Neuropsychopharmacology 39, 2570–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta E, Auta J, Gavin DP, Bhaumik DK, Grayson DR, Pandey SC, Guidotti A, 2017. Emerging Role of One-Carbon Metabolism and DNA Methylation Enrichment on delta-Containing GABAA Receptor Expression in the Cerebellum of Subjects with Alcohol Use Disorders (AUD). Int J Neuropsychopharmacol 20, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Chase KA, Sharma RP, 2013. Active DNA demethylation in post-mitotic neurons: a reason for optimism. Neuropharmacology 75, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Kusumo H, Zhang H, Guidotti A, Pandey SC, 2016. Role of Growth Arrest and DNA Damage-Inducible, Beta in Alcohol-Drinking Behaviors. Alcohol Clin Exp Res 40, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin NW, Herman MA, Roberto M, 2015. The central amygdala as an integrative hub for anxiety and alcohol use disorders. Biol Psychiatry 77, 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin NW, Misra K, Herman MA, Cruz MT, Koob GF, Roberto M, 2011. Neuropeptide Y opposes alcohol effects on gamma-aminobutyric acid release in amygdala and blocks the transition to alcohol dependence. Biol Psychiatry 69, 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH, 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74, 481–514. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, 1997. Age at onset of alcohol use and its association with DSM-IV alcohol abuse and dependence: results from the National Longitudinal Alcohol Epidemiologic Survey. J Subst Abuse 9, 103–110. [DOI] [PubMed] [Google Scholar]
- Grant BF, Stinson FS, Harford TC, 2001. Age at onset of alcohol use and DSM-IV alcohol abuse and dependence: a 12-year follow-up. J Subst Abuse 13, 493–504. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR, 2013. DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcohol Clin Exp Res 37, 417–424. [DOI] [PubMed] [Google Scholar]
- Heilig M, 1995. Antisense inhibition of neuropeptide Y (NPY)-Y1 receptor expression blocks the anxiolytic-like action of NPY in amygdala and paradoxically increases feeding. Regul Pept 59, 201–205. [DOI] [PubMed] [Google Scholar]
- Joe KH, Kim YK, Kim TS, Roh SW, Choi SW, Kim YB, Lee HJ, Kim DJ, 2007. Decreased plasma brain-derived neurotrophic factor levels in patients with alcohol dependence. Alcohol Clin Exp Res 31, 1833–1838. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP, 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kavalali ET, Nelson ED, Monteggia LM, 2011. Role of MeCP2, DNA methylation, and HDACs in regulating synapse function. J Neurodev Disord 3, 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khulan B, Manning JR, Dunbar DR, Seckl JR, Raikkonen K, Eriksson JG, Drake AJ, 2014. Epigenomic profiling of men exposed to early-life stress reveals DNA methylation differences in association with current mental state. Transl Psychiatry 4, e448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofink D, Boks MP, Timmers HT, Kas MJ, 2013. Epigenetic dynamics in psychiatric disorders: environmental programming of neurodevelopmental processes. Neurosci Biobehav Rev 37, 831–845. [DOI] [PubMed] [Google Scholar]
- Kokare DM, Kyzar EJ, Zhang H, Sakharkar AJ, Pandey SC, 2017. Adolescent Alcohol Exposure-Induced Changes in Alpha-Melanocyte Stimulating Hormone and Neuropeptide Y Pathways via Histone Acetylation in the Brain During Adulthood. Int J Neuropsychopharmacol 20, 758–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND, 2010. Neurocircuitry of addiction. Neuropsychopharmacol 35, 217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TD, Pandey SC, 2014. The epigenetic landscape of alcoholism. Int Rev Neurobiol 115, 75–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Floreani C, Teppen TL, Pandey SC, 2016. Adolescent Alcohol Exposure: Burden of Epigenetic Reprogramming, Synaptic Remodeling, and Adult Psychopathology. Front Neurosci 10, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Zhang H, Pandey SC, 2019. Adolescent Alcohol Exposure Epigenetically Suppresses Amygdala Arc Enhancer RNA Expression to Confer Adult Anxiety Susceptibility. Biol Psychiatry 85, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Zhang H, Sakharkar AJ, Pandey SC, 2017. Adolescent alcohol exposure alters lysine demethylase 1 (LSD1) expression and histone methylation in the amygdala during adulthood. Addict Biol 22, 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, Mechawar N, Szyf M, Meaney MJ, Turecki G, 2012. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry 69, 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ, 2010. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci 13, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Manzardo AM, Henkhaus RS, Butler MG, 2012. Global DNA promoter methylation in frontal cortex of alcoholics and controls. Gene 498, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Iacono WG, Legrand LN, Malone S, Elkins I, 2001. Origins and consequences of age at first drink. I. Associations with substance-use disorders, disinhibitory behavior and psychopathology, and P3 amplitude. Alcohol Clin Exp Res 25, 1156–1165. [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Pandey SC, 2011. The role of amygdaloid brain-derived neurotrophic factor, activity-regulated cytoskeleton-associated protein and dendritic spines in anxiety and alcoholism. Addict Biol 16, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC, 2013. Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol Psychiatry 73, 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Kyzar EJ, Zhang H, 2017. Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology 122, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Roy A, Zhang H, Xu T, 2004. Partial deletion of the cAMP response element-binding protein gene promotes alcohol-drinking behaviors. J Neurosci 24, 5022–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Sakharkar AJ, Tang L, Zhang H, 2015. Potential role of adolescent alcohol exposure-induced amygdaloid histone modifications in anxiety and alcohol intake during adulthood. Neurobiol Dis 82, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A, 2008. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci 28, 3729–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Misra K, 2006. Central and medial amygdaloid brain-derived neurotrophic factor signaling plays a critical role in alcohol-drinking and anxiety-like behaviors. J Neurosci 26, 8320–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Xu T, 2005. Deficits in amygdaloid cAMP-responsive element-binding protein signaling play a role in genetic predisposition to anxiety and alcoholism. J Clin Invest 115, 2762–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Boix J, Felipo V, Guerri C, 2009. Repeated alcohol administration during adolescence causes changes in the mesolimbic dopaminergic and glutamatergic systems and promotes alcohol intake in the adult rat. J Neurochem 108, 920–931. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD, 2012. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, 1998. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repunte-Canonigo V, Chen J, Lefebvre C, Kawamura T, Kreifeldt M, Basson O, Roberts AJ, Sanna PP, 2014. MeCP2 regulates ethanol sensitivity and intake. Addict Biol 19, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimondini R, Thorsell A, Heilig M, 2005. Suppression of ethanol self-administration by the neuropeptide Y (NPY) Y2 receptor antagonist BIIE0246: evidence for sensitization in rats with a history of dependence. Neurosci Lett 375, 129–133. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD, 2009. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry 65, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Tang L, Zhang H, Chen Y, Grayson DR, Pandey SC, 2014a. Effects of acute ethanol exposure on anxiety measures and epigenetic modifiers in the extended amygdala of adolescent rats. Int J Neuropsychopharmacol 17, 2057–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Baxstrom K, Shi G, Moonat S, Pandey SC, 2014b. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: a role in anxiety-like and alcohol-drinking behaviours. Int J Neuropsychopharmacol 17, 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC, 2012. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res 36, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales AJ, Biojone C, Terceti MS, Guimaraes FS, Gomes MV, Joca SR, 2011. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br J Pharmacol 164, 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikhnejad G, Brank A, Christman JK, Goddard A, Alvarez E, Ford H Jr., Marquez VE, Marasco CJ, Sufrin JR, O’Gara M, Cheng X, 1999. Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine. J Mol Biol 285, 2021–2034. [DOI] [PubMed] [Google Scholar]
- Spear LP, 2018. Effects of adolescent alcohol consumption on the brain and behaviour. Nat Rev Neurosci 19, 197–214. [DOI] [PubMed] [Google Scholar]
- Szyf M, 2013. DNA methylation, behavior and early life adversity. J Genet Genomics 40, 331–338. [DOI] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Sheedy D, Harper C, Alkass K, Druid H, Wentzel P, Nyberg F, Yakovleva T, Bakalkin G, 2011. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol 16, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D, 2013. Chromatin remodeling--a novel strategy to control excessive alcohol drinking. Transl Psychiatry 3, e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ, 2004. Epigenetic programming by maternal behavior. Nat Neurosci 7, 847–854. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y, 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sakharkar AJ, Shi G, Ugale R, Prakash A, Pandey SC, 2010. Neuropeptide Y signaling in the central nucleus of amygdala regulates alcohol-drinking and anxiety-like behaviors of alcohol-preferring rats. Alcohol Clin Exp Res 34, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]


