Abstract
Cancer development is a complex process influenced by inherited and acquired molecular and cellular alterations. Prevention is the holy grail of cancer eradication, but making this a reality will take a fundamental rethinking based on deep understanding of premalignant biology. Although the seminal multi-step, genetic model of human tumorigenesis was defined in the colorectal adenoma-carcinoma sequence three decades ago, only a handful of sporadic adenomas analyzed by next-generation sequencing (NGS) have been reported, in striking contrast to the wealth of NGS data in cancers. Recent precancer advances include: germline mutation biology driving precision prevention – RANK-L effects on luminal progenitors leading to international trial in BRCA1 carriers, first real signal of potential benefit of early detection research in pancreatic neoplasia, breakthrough trial of combination chemoprevention in familial adenomatous polyposis, and germline/microbiota interactions in intestinal oncogenesis, including in mouse model of Lynch syndrome. Other developments include: novel (e.g., copy number alterations) regulators and imaging of immunosuppressive microenvironments; immune prevention (from HPV to cancer vaccines); mutational signatures; premalignant mutational accumulation in aging, established in clonal hematopoiesis, with recent deep NGS suggesting a more generalized phenomenon; and single-cell analyses of DCIS and Barrett’s esophagus identifying importance of genetic heterogeneity. The challenges are substantial, including hefty computational and data prices for unprecedented whole-genome, single-cell resolution, with population scale data estimates in the exabyte range (>1018 bytes), requiring new computational frameworks. To accelerate the prevention of cancer, this field needs a large-scale, longitudinal national effort, leveraging diverse disciplines, technologies, and models to develop an integrated multi-omics and immunity precancer atlas (PCA), to interrogate and target events that drive oncogenesis.
Keywords: premalignancy, neoplasia, biology, precision medicine, cancer prevention
Introduction
In early 2016, President Obama announced the creation of the National Cancer Moonshot initiative – a commitment to eradicate cancer by investing in dramatically advancing progress within diverse fields of cancer research, including prevention and early detection (1). Historically, the rate-limiting step for developing and implementing precision approaches has been our relatively limited understanding of precancer biology in contrast to the extensive study of advanced disease. For example, The Cancer Genome Atlas (TCGA), which includes volumes of omics data from >11,000 patients across 33 tumor types, has transformed our understanding of cancer biology, identified hundreds of driver mutations that alter hallmark pathways, and become a tremendous national resource for continuing discovery, including catalyzing the development of novel computational tools to analyze mutational signatures and single-cell NGS. Recent studies are redefining the spectrum and biology of neoplasias linked to various hereditary cancer predisposition syndromes (2,3). The influence of mitochondrial genetics and biology on cancer predisposition and development is a critical, and until recently, understudied field (4–7). Finally, a diverse array of engineered preclinical models, technologies, and disciplines are now being leveraged to probe early oncogenesis. Large-scale longitudinal and systematic mapping is critical to develop an integrated omics and immunity precancer atlas (PCA), allowing dissection of the sequential molecular and cellular events that promote oncogenesis leading to novel prevention and interception strategies (8–10) (Figure 1).
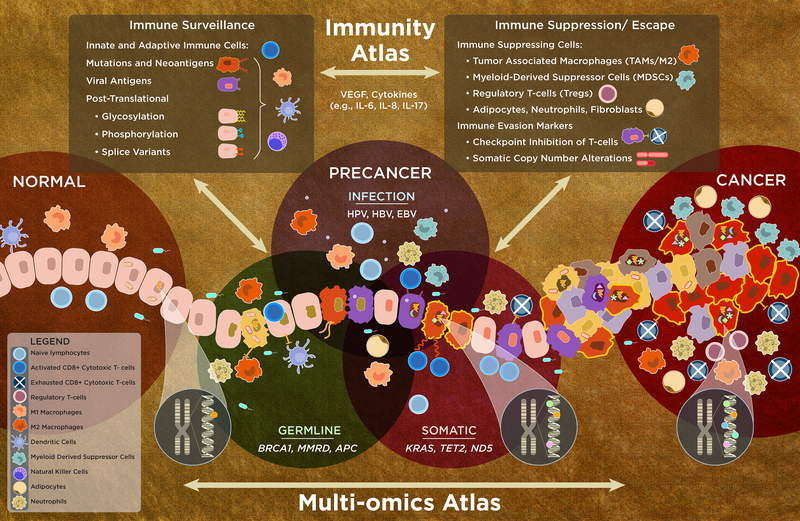
Figure 1.
An integrated multi-omics and immunity precancer atlas (PCA). Inherited and acquired molecular alterations and their interaction with the local microenvironment/immune system influence oncogenic progression to invasive carcinoma. Normal cells (light orange, far left) that have a germline mutation (green nuclei) acquire somatic mutations (dark orange) or molecular alterations due to viral infection (purple). These all can potentially have an altered cell state with dysregulated molecular pathways that result in the loss of cell growth control and other hallmarks of cancer (228), which can then result in the development of advanced precancers (multicolored, subclonal diversity/heterogeneity) that immediately precede invasive cancer (red). Molecular alterations, depicted by symbols in the nucleus, represent mutations, SNPs, or epigenetic modifications. The accumulation of mutations (e.g., UV, ABOBEC) during life creates signatures (shown by the chromosomal insets and colored dots in gradient from cancer to normal). Somatic mutations in nuclear and mitochondrial genes, mutational signatures, other omic events, and germline changes/interactions are key drivers of oncogenesis. The mutational signatures, often identified in cancers, may predict early events, now extending to the precancer state (depicted by the orange/red cells with bold, yellow border). These omic alterations interact (bidirectionally) with the complex tissue microenvironment, including immune cells, stromal cells (adipocytes and fibroblasts), and other events, which promote oncogenesis and invasion/escape; microbiota can prevent or promote oncogenesis (see text). The continuum between the immune state modulated by cytokines and growth factors includes immune surveillance, composed of the antigenic repertoire, adaptive and immune cells (upper left box), and immune suppression/escape (upper right box) along with vascular and endothelial cells (not shown) that can lead to immune escape – another hallmark of cancer. The elucidation of the integrated multi-omics and immunity PCA will continue to evolve in complexity (e.g., recent SCNA finding), requiring continuous updating in this fast moving field.
Translating Inherited Cancer Risk into Precision Prevention
Germline nuclear genetics
The genetics of various hereditary forms of cancer risk have been studied extensively and long been used to aid our understanding of sporadic neoplasia. Our knowledge about the biology of germline mutations in certain cancer genes (e.g., BRCA1) is much better understood than in the somatic setting and has historically provided tremendous insight into fundamental neoplastic processes since the mechanisms of the neoplasia in these mutation carriers directly reflect the biology of the mutated gene itself. For example, study of the biology of tumors that develop in individuals carrying high-penetrance, predisposition mutations has led to paradigm-changing therapy and is beginning to drive prevention and early detection research, as evidenced by PARP inhibitors, one of the most compelling forms of precision cancer therapy in various forms of BRCA1/2-associated cancers (11), shown to delay mammary tumor development in Brca1-deficient mice (12). Understanding familial adenomatous polyposis (FAP) biology, characterized by germline APC mutations and chromosomal instability, led to a recently reported breakthrough trial of combination chemoprevention targeting the convergence of Wnt and EGFR signaling and COX-2 (13) in this devastating syndrome. In contrast, although invasive lobular cancers occur excessively in women who carry germline mutations in BRCA2 or CDH1, there is no reported excess of lobular carcinoma in situ (LCIS) in these rare families (14), illustrating the complexity of precancer types, biology, and patterns in carriers of different high-penetrance germline mutations.
Germline genetic heterogeneity, single phenotype/genetic disorders caused by mutations in several inherited genes, is relatively common. A striking example is pheochromocytoma, which can be caused by a number of germline mutations, including in RET, VHL, FH, IDH1, NF1, and certain SDH nuclear genes (SDHA, SDHB, SDHC, and SDHD), which encode mitochondrial complex II genes (15). The SDH complex functions in mitochondrial energy generation, links the Krebs cycle and the electron transport chain (ETC), and plays a key role in oxygen sensing and tumor suppression. Germline SDHD variants alter PTEN function, a novel mechanism of thyroid pathogenesis in Cowden syndrome (16). Recent large series of pediatric cancer have identified germline mutations in predisposition genes in only about 10% of pediatric cancer patients (17), comparable to the prevalence in adult cancer patients (18). The fundamental question of why certain germline mutations in cancer susceptibility genes predispose to a particular spectrum and magnitude of neoplasias is largely unknown. Although some genes have organ-specific functions (e.g., mutations leading to hepatic overload cause liver cancer) (19), most mutations have a broad range of functions (e.g., mismatch repair (MMR)). It is unclear why MMR inactivation predisposes more to colorectal cancer (CRC) rather than generalized systemic cancer risk or why specific MMR gene mutations are associated with different cancer patterns (20).
Mouse models have begun to unravel some of the key mechanisms of intestinal neoplastic transformation in the germline mutation setting. The first major study involved germline/microbiota interaction in a mouse model of Lynch syndrome (MSH2-deficient intestinal carcinogenesis) and demonstrated that butyrate, generated by gut microbiota from dietary carbohydrates, can act as an oncometabolite (21). Interestingly, butyrate can have opposing effects in different colon models functioning as a tumor-suppressive metabolite with energetic and epigenetic functions, which likely reflect the different germline backgrounds, e.g., reduced dietary butyrate markedly decreased CRC development in MMR-deficient, but not MMR-proficient mice. Building on this germline microbiome/CRC prevention work, a recent study demonstrated that celecoxib (a COX-2 inhibitor known to reduce intestinal adenoma burden) induces alterations in the gut microbiome and metabolome in APCMin/+ mice, a model of FAP (22). Specifically, celecoxib increased gut Coriobacteriaceae, which suppressed production of oncogenic metabolites (e.g., glycine and serine). In this Min mouse model, innate and adaptive sources of IL-17 drive colon tumorigenic response to enterotoxigenic Bacteroides fragilis (ETBF); and anti-IL-17 monoclonal antibody (mAb) and T-regulatory (Treg) cell depletion suppressed tumorigenesis at the micro-adenoma stage (23). Emerging data suggest a link between ETBF, inflammatory bowel disease (IBD), and CRC. ETBF toxin-triggered colon tumorigenesis is characterized by a specific immune signature (combining IL-17-driven colitis and altered myeloid differentiation into MDSC) (24). Germline influence on bacterial translocation also involves GWAS-identified laminin nuclear and mtDNA variants (21,25–28).
Studies in BRCA1/2 carriers are providing novel insights into the high penetrance for breast and ovarian cancer. Data suggest that BRCA1/2 germline mutations are driving oncogenesis via combined effects of compromised DNA repair capability and changes in the endocrine system in the organ at risk, leading to breast and ovary cancers (29). Somatic loss of the BRCA wild-type allele is required to provoke genomic instability and tumorigenesis. Other studies have suggested this tissue specificity may be linked to inhibition of estrogen receptor (ER)-α transcription activation by BRCA1 ubiquitin ligase (30). Although most breast cancers arising in BRCA1-mutation carriers are ER-negative, tamoxifen use appears to be associated with a reduction in risk, particularly of contralateral breast cancer (31), likely due to female hormone effects in the early ontogeny or stromal-/ myeloid-derived suppressor cell (MDSC) (32,33) estrogen signaling. These insights suggest new prevention strategies that exploit hormonal dysregulation in BRCA1/2 carriers.
Elegant studies of luminal progenitor biology in BRCA1-mutation carriers (34,35) have led to a transformative potential to prevent/delay BRCA1-associated breast cancer, a disease for which the best current option is prophylactic mastectomy. A highly proliferative subset of luminal progenitor cells that give rise to basal-like breast cancer, constitutively express RANK and are hyper-responsive to RANK-L, a key mediator of progestin-driven mammary carcinogenesis. Furthermore, RANK+ BRCA1-deficient/mutant progenitor cells are more susceptible to DNA damage and aberrant downstream NFkB activation than RANK- mammary progenitor cells (36). Blocking RANK-L signaling in several Brca1-deficient/mutated mouse models markedly inhibits mammary tumorigenesis (34). RANK-L/RANK signaling (37) also can influence immune surveillance/evasion via innate and adaptive immune responses (38,39). RANK-L produced by Tregs promotes mammary cancer and T-cell tolerance to intestinal bacteria (39). Recently, a second RANK-L receptor, LGR4, has been implicated in the regulation of multiple developmental pathways (40). Serum levels of osteoprotegerin (OPG), the endogenous RANK-L inhibitor, are significantly lower in BRCA1 carriers (vs controls), premenopausal women, and are inversely associated with breast cancer risk (41,42). Certain germline RANK SNPs (TNFRSF11A) have been associated with increased RANK expression and breast cancer risk in BRCA1 carriers (35). Denosumab (a RANK-L mAb inhibitor FDA-approved for the treatment of both osteoporosis and bone metastases) blocked progesterone-induced proliferation in BRCA1-mutant organoids. In small pilot window studies of BRCA1 carriers, denosumab reduced breast epithelial cell proliferation and progenitor cells clonogenic potential (34,35). Furthermore, denosumab has a well-established safety record and was recently shown to significantly delay (by 50%) the time to first fracture in postmenopausal ER+ breast cancer (43). Theoretically, RANK-L inhibition would work best to prevent/delay tumor onset for premenopausal BRCA1-mutation carriers since risk-reducing salpingo-oophorectomy has recently been shown to be ineffective in this setting (44) and RANK-L is a progesterone-responsive gene. Progesterone receptor modulators prevent mammary tumorigenesis in Brca-mutant mice but are limited by toxicity for clinical prevention. Based on these data, denosumab is being developed for a large-scale international breast cancer prevention trial in BRCA-mutation carriers (34,35).
Pancreatic cancer early detection research with magnetic resonance and endoscopic ultrasound in germline high-risk individuals produced the first real signal of potential benefit in this setting (45). Unselected pancreatic cancer patients have a very high (>15% in a clinic-based cohort) prevalence of germline mutations (mostly BRCA1/2), even higher among Ashkenazi Jewish individuals (46). Pancreatic precursor lesions in people with high-penetrance germline mutations have a higher malignant potential (than other pancreas high-risk groups) (47,48). These data recently led some centers to recommend germline testing for all new pancreatic cancer patients. Precedent for such an approach already exists in ovarian cancer, where NCCN guidelines recommend germline analysis of BRCA1/2 in all new patients (49). The development of new molecular imaging techniques to detect high-grade pancreatic intraepithelial neoplasia (PanIN-3) may further improve prevention and early detection of this fatal disease (50–52).
Universal tumor testing for MMR deficiency, a paradigm-changing approach for identifying inherited cancer risk, has become standard practice for all newly diagnosed CRCs, recently extended to endometrial cancers (53,54) to identify Lynch syndrome probands. This benefits both the patient and at-risk family members for intensive and early screening and potentially aspirin and/or other NSAID prevention (54–56). The recent profound activity of immune checkpoint blockade to treat MMR-deficient cancers (57) has added to the enthusiasm for this approach (see below). A similar approach is under study in lung cancer patients: the ~1% with tumor EGFR T790M mutation at diagnosis have a high risk of carrying germline T790M mutations (58). These families appear to have a different lung tumor biology, supporting precision prevention (with T790M inhibitors) and early detection research (59). This highlights the substantial overlap between somatic driver mutations and germline predisposing mutations (19). Recent data reveal that pathogenic genetic variants identified within many cancer types are of germline origin in 10–15% of unselected childhood and adult cancers. As NGS becomes more widely used and germline-somatic relationships comprehensively mapped, shared and distinct oncogenic events can be integrated into the PCA and assessed for preventive targeting (18,19,60,61).
Immunologic mechanisms, including cancer vaccines, may also be key to realizing precision prevention in certain types of neoplasia characterized by immunogenic antigen production, as seen in various inherited settings (Table 1). Cancers that arise in Lynch syndrome with inherited DNA MMR gene defects display a high-level of microsatellite instability (MSI-H) and widespread accumulation of somatic frameshift mutations. This results in very large numbers of neoantigens/mutations (62) that make the tumor appear more foreign to the host immune system, underlying the success of immune checkpoint inhibitors and serving as a model of sporadic MSI-H tumors (57,63). These breakthrough advances using immunotherapy to treat MMR-deficient/MSI-H cancers along with early immunosurveillance (T-cells specific to MSI-related neoantigens (64)) in “healthy” Lynch syndrome carriers have generated interest in developing cancer vaccines as immune prevention targeting predictable frame-shift mutation-derived peptides. DNA damage response plays an important role in innate immunity, activates inflammatory cytokines, and induces the expression of immune-receptor ligands on damaged cells. As such, inhibitors of DNA damage response signaling may, in fact, attenuate the immune response following DNA damage (65). A specific mutational signature associated with both germline and somatic BRCA1 or 2 mutations (66) has been observed in breast, ovarian, pancreatic, gastric, and esophageal cancers (even those without BRCA1 or 2 mutations) (67–72) in association with markers of immunity in subsets of the former three cancers (73), suggesting a potential role for immune-based prevention against such cancers. In contrast, a recent seminal study revealed an increasingly complex interplay between chromosomal abnormalities and immunity. High-level arm- and chromosome-somatic copy number alterations (SCNAs), which can drive precancer/cancer progression, produced immune evasion. Consistent with gene-dosage imbalance rather than a specific gene, SCNAs drove an immunosuppressive microenvironment with immune depletion that opposed immune response (74), ), even in the setting of high mutational/ neoantigen burden as seen in mismatch repair-deficient tumors (57,63) revealing an increasingly complex interplay between chromosomal abnormalities and immunity.
Table 1:
Immunologic features of three distinct neoplastic pathways.
| Germline mechanism of hypermutability | Characteristics of associated cancers | Loci of somatic hypermutations in invasive cancers | Evidence of immunogenic phenotype | Immunologic considerations in premalignant biology |
|---|---|---|---|---|
| DNA mismatch repair gene mutations: Lynch syndrome (LS); biallelic mismatch repair deficiency (BMMR-D) | Gl, gyn, and other cancers with high-level microsatellite instability (MSI); pediatric-onset Gl cancers, gliomas, and lymphomas in BMMR-D | Mono- and dinucleotide repeat microsatellites in non-coding and coding DNA; certain “hotspot” microsatellites within driver genes, e.g., TGFBR2 and BAX in LS; POLE or POLD1 in BMMR-D | MSI-induced frameshift neopeptides/neoantigens found in associated cancers; early success of PD-1 antibodies | Not systematically studied; MSI-H (presumably a surrogate for neoantigen load) seen in various premalignant states (adenomas, intestinal crypt foci, IPMNs, DCIS, etc.) |
| BRCA1/2 mutations | Breast, ovarian, pancreatic, prostate, and other cancers with homologous recombination deficiency | Specific signatures of tandem duplications and/or deletions | BRCA1/2-associated ovarian cancers demonstrate infiltrating lymphocytes and increased expression of genes related to immune-mediated cytotoxicity | Not well studied; loss of wild-type BRCA1/2 allele is seen in some premalignant lesions, but BRCA1/2 haploinsufficiency leads to increased expression of EGFR and genomic instability in non-neoplastic breast epithelium cells prior to loss of wild-type BRCA function |
| APOBEC3A/B chimeric deletion polymorphism | Modestly increased risk of breast (and possibly other) cancers with somatic hypermutation; increased stability of chimeric APOBEC3A/B mRNA leads to increased APOBEC3A activity correlating with germline copy number | TCA and TCG trinucleotide sequences, including certain “hotspots” within driver genes, e.g., PIK3CA | Cancers show upregulation of genes associated with immune activation, cytokine response, and lymphocytic infiltration; penetrance of immune-activating effects appears to be high | Not well studied; APOBEC3A/B-related mutational patterns seen in sporadic (i.e., not associated with germline polymorphism) pre-invasive bladder carcinoma and cervical intraepithelial neoplasia |
This schematic outlines the rationale for investigating the immunobiology of premalignancy in three distinct inherited neoplastic pathways: mismatch repair deficiency, BRCA1/2 mutations, and APOBEC3A/B-mediated neoplasia. Cancers associated with all three pathways are associated with distinct, predictable forms of somatic hypermutability and have various features suggesting an immunogenic phenotype. In each case, however, an in-depth understanding of premalignant biology will be key to devising strategies for primary cancer prevention.
Genome-wide association studies (GWAS) have contributed to expanding catalogs of implicated genes and pathways for many complex human diseases and are beginning to shed light on shared and unique etiological and pathological disease components. A key challenge is that many GWAS-identified loci are not near known coding or regulatory regions, making determining the underlying mechanisms and functions related to the associations difficult. Linking susceptibility variants to their respective causative genes and cell-specific regulatory elements thus remains a high priority in order to realize the potential of association studies to advance understanding of disease biology, etiology, and prevention. Their ability to identify novel cancer genes/pathways (through functional follow-up studies) underlying the observed risk is now being exploited for future drug development or repositioning (75). While the low penetrance of most GWAS-identified risk loci has limited translation to clinical trials, the combined effects of such SNPs are being used to create robust polygenic risk scores (PRS) (76), which may be useful in developing personalized risk estimates. Combining large-scale GWAS findings across cancer types (breast, ovarian, and prostate) and using fine-mapping pathway analysis and PRS discovered cross-cancer risk loci with the potential to shed new light on the shared biology underlying these hormone-related cancers (77). A recent GWAS led to the identification of miR-3662 at the 6q23.3 locus shown in mechanistic studies (when overexpressed) to inhibit NFkB signaling and leukemogenesis (78). Certain SNPs (e.g., involving APOBEC3), underlying cancer (breast and bladder) risk are linked to hypermutability and immune activation (79–81) and intestinal barrier function (discussed above). Future possibilities of drug development or repositioning, possibly aided by studies in relevant animal models, include GWAS-identified loci at 1p31.3 (82) and 6q25 (83), which helped identify cancer prevention relevant drug targets IL-17 (key immune/microbiome target) and ESR1 (tamoxifen target). A GWAS-identified locus at 6p23 (84), associated with breast cancer risk in BRCA2 carriers, is located near the SIRT5 gene, a drug target under active study (85).
Recent findings confirm the occurrence of widespread genetic regulation of immune and host defense pathways overlapping disease loci and involving not only gene expression but also splicing and epigenetic modifications, including CRC precursors (86). The results suggest the convergence of independent regulatory layers for cell-specific function, and used independent measurements to yield robust biological validity to mapped traits for associations between genetic variants and intermediate phenotypes, providing powerful approaches to annotate the putative consequence of disease associations. The biological resolution of this approach was further increased by assessing by evaluating context-specific events in studies probing multiple primary cell types, in different conditions, and extending analyses beyond gene expression to histone modification or methylation greatly enhancing the functional and mechanistic interpretation of genetic associations.
Mitochondrial Biology and Genetics in Cancer Predisposition
Mitochondrial DNA (mtDNA) with its mutations and polymorphisms is a relatively underappreciated field in cancer research. Most mitochondrial proteins are nuclear encoded. Human mtDNA is maternally inherited and exists as a circular, double-stranded genome encoding for 37 mitochondrial genes: 22 transfer RNAs, 2 mitochondrial ribosomal RNAs and 13 protein subunits of the ETC complexes (with the exception of Complex II, which is nuclear encoded) and ATP synthase (mtOXPHOS proteins), essential for respiration. There are several mtDNA copies per mitochondrion and hundreds of mitochondria per cell (87). Generally, neoplastic cells possess functional mitochondria and retain the ability to conduct oxidative phosphorylation. In fact, targeted depletion of mitochondrial DNA generally reduces tumorigenic potential in vivo (88). While it has long been known that somatic mtDNA alterations are frequently acquired during oncogenesis (89), recent intriguing germline data indicate that mtDNA variants influence multiple innate mitochondrial functions, including reactive oxygen species (ROS) production and redox control, signal transduction and epigenome systems, autophagy, apoptosis, and immunity (90). mtDNA has a very high mutation rate, over an order of magnitude higher than the somatic nuclear genes. Furthermore, mtDNA genes are intimately linked with ~2000 nuclear genes encoding proteins that function within mitochondria, which can produce nuclear inheritance of mitochondrial disease. The most common germline mtDNA mutations in neoplasia occur in the non-gene encoding region. Inherited high-penetrance of deleterious missense alterations in mtDNA genes, such as ND6 and COI, which code for subunits of OXPHOS complexes I and IV, have been associated with risks of various cancers. For example, oncocytomas tend to have mutations in one of the seven mtDNA coded polypeptides of respiratory complex I (91), while missense mutations in the mtDNA complex IV (cytochrome c oxidase) subunit COI gene are commonly found in prostate cancer, and certain African mtDNA lineages harbor COI gene variants that may contribute to cancer risk among African Americans (4,92). MtDNA variants have been associated with ovarian (93), bladder (94), breast (95), endometrial (96), and HPV-infection and cervical cancers (97), among multiple other cancers. Mitochondrial mutations are frequent in Barrett’s metaplasia without dysplasia (98). The progression of nonalcoholic fatty liver disease to the liver precancer nonalcoholic steatohepatitis (NASH) has been shown to be associated with a mtDNA SNP in the Mtatp8 gene (a subunit of OXPHOS complex V, ATP synthase). This variant has profound effects on hepatic lipid and acylcarnitine metabolism and susceptibility to diet-induced (e.g., high-fat Western diet) NASH (99). Generating mouse models of these and other deleterious missense mt germline mutations, e.g., COI nt 6589T>C V421A (100) and ND6 nucleotide G13997A P25L (101), will be critical to probe mitochondrial biology.
Deleterious alterations in mtDNA are inherently heteroplasmic (harboring a mixture of mutant and wild-type mtDNA) with high levels of these severe mutations being lethal. Since mtDNA transmission during mitosis is the result of stochastic distribution into daughter cells, milder mtDNA polymorphisms can shift to become predominantly enriched within individual cells and closer to pure mutant (homoplasmy), resulting in neoplastic transformation. The importance of this phenomenon for cancer predisposition has been demonstrated in a pedigree in which a mtDNA complex I ND5 m.12425delA frameshift mutation was transmitted through the maternal germline at lower heteroplasmy levels (5–10% mutant) and was thus masked due to the preponderance of WT mtDNAs. Thus, while the transmission of the mutant mtDNA in this pedigree was phenotypically silent, the chance increase of the mutant mtDNA in somatic cells caused neoplastic transformation and seemingly sporadic cancer (102). In contrast to Mendelian genetics, mtDNA heteroplasmic genotype is continuously changing during successive cytokinesis to generate cells with varying oncogenic potential (92,103). Widespread heterogeneity has been reported in the mtDNA of normal human cells. Furthermore, the frequency of heteroplasmic variants among different tissues of the same individual vary considerably indicating that individuals (and perhaps even a single cell) are characterized by a complex mixture of related genotypes rather than a single genotype. Mechanistic study of heteroplasmy (shifting percentages of WT and mutant mtDNAs) regulation will yield novel prevention insights. Because of its high mutation rate, human mtDNA is highly polymorphic, harboring functional variants that can be beneficial or deleterious depending on environmental context. Because of the strict maternal inheritance of the mtDNA, sequencing mtDNAs from global indigenous populations permitted reconstructing the origins and ancient migrations of women (initially from Africa) (90). A subset of the mtDNA variants cause subtle changes in OXPHOS, which in turn could modulate a wide range of context-dependent cellular functions of adaptive relevance, including inflammatory, stress, autophagy, and oncogenic responses to diet and other factors (104,105). Those functional mtDNA variants that were beneficial (adaptive) in a particular environment increased in number and gave rise to descendant mtDNAs, which share the founders’ beneficial variant creating a regional group of related mtDNA haplotypes known as a haplogroup. A result of migration has been that previously adaptive mtDNA haplogroups have become maladaptive and predisposed to a wide range of common diseases, including diabetes, obesity, aging and cancer (90). Dietary or caloric restriction slows the development of many age related diseases, including cancers, although the mechanisms involved are complex and context-dependent, such as autophagy (106) and translational regulation involving mitochondrial (and ribosomal) genes/proteins/components (107). Autophagy is essential for BRAF V600E-driven melanoma and lung tumor development in GEMMs by overcoming senescence; deletion of Atg7 inhibits tumorigenesis, likely via a mitochondrial mechanism (108).
A recent murine study highlighted the importance of mtDNA genetic background in tumorigenesis by examining PyMT transgenic mice (inherently predisposed to developing mammary tumors) with identical nuclear genomes but varying mtDNA backgrounds. The mtDNA background influenced both mammary tumor latency and progression (109). In normal mice, the mitochondrial-targeted catalase transgene (mtCAT) reduces somatic mtDNA mutations (110) and when crossed with PyMT, the incidence of mammary cancer was greatly reduced (111). Consistent with cancer predisposition, mechanistic studies demonstrated the profound influence of subtle changes in mtDNA haplotype/variation on obesity and aging – two common cancer risk factors (112). Mitochondria may also be intimately involved with T-cell tumor surveillance. T-effector cells are more glycolytic while Treg cells are more oxidative. Within neoplasia, glucose is converted to lactate (which promotes inflammation, angiogenesis and tumorigenesis), thus inhibiting T-effector function. By contrast, the Treg cells can metabolize lactate by OXPHOS (113), further inhibiting T-effector cell immune rejection (114). Germline mtDNA ND6 P25L-mutant mice harboring a mild mtDNA complex I gene mutation have reduced Treg cells (113,114), suggesting that tumor immune rejection might be enhanced by mild complex I inhibitors such as metformin, whose effectiveness should be increased in neoplastic cells with partial OXPHOS dysfunction (115,116). Mitochondria can also influence the inflammasome, innate immunity, IL-1β, and NFkB inflammatory pathways (7,117).
Adding to the tissue- and geographic-specific context of mitochondrial effects (118), emerging data also suggest a complex interplay between the nucleus-cytosol and mitochondria. Murine models with germline mutations in the nuclear gene SUV3, which encodes for a mtRNA helicase, have marked somatic mtDNA instability, hypermutability, shortened lifespan, and various cancers – a unique model to study mitochondrial genomic instability in cancer predisposition. Clinical relevance was shown by reduced SUV3 expression in two independent cohorts of human breast cancer (5). Mutations in nuclear DNA genes influencing transformation include some of the same targets/mechanisms affected by mtDNA, including TETs, succinate, fumarate, NRF2, and α-ketoglutarate dioxygenases (119,120) – all important in cancer risk. Nuclear BRCA1 has been found in the mitochondria, where its function is unclear (121,122). Germline BRCA1 mutation reprograms breast epithelial cell metabolism towards mitochondrial-dependent biosynthesis and increased risk of oncogenesis. Metformin studies in BRCA1 haploinsufficiency-driven oncogenesis support potential prevention approaches in BRCA1 carriers: inhibition of complex I and restriction of mitochondrial-dependent biosynthetic intermediates (123) may open a new avenue for “starvation” strategies; and regulating mitochondrial-nuclear communication and modulating the epigenetic landscape (targeting histone acetylation) in genomically unstable precancerous cells (124), might guide the development of new metabolomic-epigenetic strategies. As with nuclear GWAS, certain mtDNA alterations modify (lower) risks of breast cancer in germline BRCA2 mutations (125). Future GWAS integrating nuclear and mitochondrial studies will provide a more full germline landscape.
Big Genomics Data of Premalignant Somatic Tissues
The collection and analyses of NGS big data are beginning to provide biological insights into prevention/early detection in the context of studies characterizing somatic genomic alterations. It is worth noting that, in addition to comprehensive analyses of “big genomics data,” there are few recent studies that have also examined cancer microbiomes (reviewed in (104,126–128)), transcriptomes (129–131) and epigenomes (reviewed in (132,133)). In addition to big data generated from somatic sequencing efforts, GWAS has involved hundreds of thousands of cancer patients across most organ sites identifying ~3000 cancer-related genetic associations (recently reviewed (134,135)) and has been studied in some precancers – Barrett’s esophagus (BE) (136,137), colorectal adenomas (26), ductal carcinoma in situ (DCIS) (138) and hematologic premalignancies (below).
The genome of a malignancy can be examined as an archeological record bearing the cumulative imprints of all mutational processes that have been operative throughout the cellular lineage between the fertilized egg and cancer (139). Each mutational process leaves a characteristic imprint, termed mutational signature, which can change over time, and almost all mutational signatures detected in a cancer genome have been imprinted during the precursor phase of a cancer cell (139). Examination of the cancer genomes from >12,000 patients has revealed more than 30 known distinct mutational signatures (66,139), including those related to environmental exposures, such as UV-light, aflatoxin, and tobacco (cancer.sanger.ac.uk/cosmic/signatures). Some of these signatures have already been used for identifying the presence of specific carcinogens, including aristolochic acid, one of the most potent known human carcinogens – a chemical present in certain plants still in use even today – to global risks of urologic and hepatic cancers (140). It will be important for the PCA to study mutational signatures of premalignant and normal aged tissues, perhaps guided by the many cancer signatures, to provide a lens into mutational precancer patterns. However, it is important to note that approximately half of the currently known cancer signatures have unknown etiology and ongoing efforts have started exposing experimental systems to known carcinogens in an attempt to reproduce/identify them (141,142).
Another set of widespread and extensively studied endogenous mutational signatures are the ones attributed to ectopic activity of the APOBEC family of deaminases (66,143). Recent examination across more than 10,000 specimens from 36 distinct cancer types revealed that these signatures are found in more than 30% of cancer samples and account for approximately 15% of all somatic mutations across these cancers. The activity of these APOBEC mutational signatures is especially strong in bladder and cervical cancer where they account for more than 75% of all somatic mutations in each of these cancer types (143). In cancers of the cervix and oropharynx, these APOBEC mutational signatures are predominately triggered early by HPV infection (143,144). It has been speculated that the APOBEC mutational signatures have been imprinted during the precancer phases of these cancers (143,144). In lung and most other cancer types, APOBEC signatures are believed to be late events and are found in subclonal expansions and intra-tumor heterogeneity (145). Recent studies using mutational signatures have also demonstrated that the presence of certain germline variants can affect the accumulation of somatic mutations due to environmental exposures. For example, disruptive germline polymorphisms in MC1R, contributor to phosphorylation of DNA repair proteins, have been associated with increased number of somatic mutations due to a UV-light-related mutational signature and a higher risk for developing skin cancer (146).
Multiple mutational signatures reflect failure of different DNA repair pathways. A mutational signature reflecting the accumulation of unrepaired ROS, mainly 8-oxoguanine, has been attributed to failure of base excision repair (BER) due to defects in MUTYH, and has been identified in colorectal cancers and adenomas arising in individuals with pathogenic germline MUTYH mutations (147). Additionally, a failure of transcription-coupled nucleotide excision repair (NER) due to somatic mutations in ERCC2 in bladder (including preinvasive) neoplasia has been shown to exhibit a specific mutational signature (148). Germline defects in other NER genes can cause Xeroderma pigmentosum (XP), a rare autosomal recessive genetic disorder associated with high risk of UV-associated skin cancer due to faster accumulation of UV associated DNA damage and mutational signatures (149). UV-induced non-melanoma lesions can be reduced using bacterial DNA repair enzymes (150) or nicotinamide, which can prevent UV-induced immune suppression and enhance DNA repair (151).
In addition to mutational signatures related to failure of DNA repair mechanisms and ones due to endogenous/exogenous exposures, large-scale genomics studies have also identified mutational signatures responsible for the unavoidable background mutation rate in somatic cells. Notably, two mutational signatures (which are not correlated and have different frequencies in different tissues) have been found to act as endogenous mutational clocks, characterized by accumulating somatic mutations within all normal somatic cells of the human body with the progression of age (152,153). One of these mutational signatures has been attributed to spontaneous deamination of 5-methylcytosine in the context of CpG (its rate of “ticking” appears to be influenced by cellular division), while the etiology of the second clock-like signature remains unknown. Interestingly, a recent study demonstrated the increased rate of one mutational clock to be mechanistically linked to tobacco smoking (148,154). The somatic mutation loads in single-cell lineages provide information about an individual’s lifetime history of mutagenic exposure and the impact of intrinsic factors on mutagenesis. Expanding this study to precancers, more cell types, and larger populations would further refine estimates of the rates of somatic changes in human genomes. Understanding the contributions of environmental and endogenous mutagenic processes to somatic mutation loads is fundamental to develop preventive strategies (155).
Analyses of omics data from precursors are beginning to emerge. Despite their cross-sectional, precancer/cancer pair designs, and relatively small sample sizes, these emerging data suggest that many precancers share genomic alterations with their respective invasive cancers, including ductal and lobular breast cancer (156,157), pancreas (158), non-melanoma skin (159,160), melanoma (161), lung adenocarcinoma (162) and colorectal neoplasia (147). From an NGS, genomics, transcriptomics (see below) and big data perspective, BE is the best-studied epithelial precancer (67,163–166), including three recent GWAS (136,137,167) and a post-GWAS analysis reporting some CDKN2A SNPs associated with reduced EAC risk (168). Somatic tissue studies of Barrett’s/esophageal adenocarcinoma (EAC) pairs revealed that most recurrently mutated genes in EAC were remarkably similar to the matched precancer, only TP53 and SMAD4 were associated with advanced neoplasia (165). Intra-tumor genomic heterogeneity, with some contribution of aberrant methylation, drives neoplastic progression in this setting (see longitudinal section below) and most cancers (169).
Two recent large-scale NGS of mtDNA of cancer (total > 2,000 human cancers, 30 tumor types) identified a unique heavy strand-specific C > T transitions and mutational signature. More importantly, this cancer mitochondrial missense mutational signature was considered neutral (analogous to passenger mutations in nuclear DNA) and did not compromise the function of the mitochondria (170). One of these studies further refined the mtDNA mutational map by requiring that mutations also be detectable in matched transcriptome sequencing (RNA-seq) data from the same tumors (171). Although DNA/RNA allelic ratios generally were consistent, some mutations in mt-tRNAs displayed strong allelic imbalances caused by accumulation of unprocessed tRNA precursors, indicative of impaired tRNA folding and maturation, which underlie a range of diseases. Both studies found a selective pressure against deleterious coding mutations affecting oxidative phosphorylation, indicating that tumors require functional mitochondria. Unexpectedly, known dominant mutagens, such as cigarette smoke or UV light, had a negligible effect on mtDNA mutations. Another recent study has reported significant correlations between mtRNA-Seq and mtDNA copy number, with some important exceptions (e.g. MT-ND5 and MT-ND6) (172). Clonal expansion of mtDNA mutations can result in mitochondrial dysfunction, such as decreased ETC enzyme activity and impaired cellular respiration. NGS of mtDNA of oncocytomas, which are rare benign tumors of epithelial cells defined by excessive amount of mitochondria, has identified a pathogenic mutation signature that compromises the overall function of the mitochondria, proposed to serve as a metabolic barrier for these benign tumors, and perhaps precancers, to progress to more malignant tumors (173).
In addition to the studies above, a systematic approach to classify cancers using transcription profiles at both bulk tumors and single-cell resolution have been well described (174–177). These profiles not only provide molecular basis for classifying cancers with shared transcriptional programs across different cancers but also characterize heterogeneity that exists within individual precancers (178) and tumors. Whole transcriptome profiling using RNAseq, pathway enrichment, and functional assays of BE found novel cell-cell interactions between dysplastic and normal epithelial cells (which often coexist in vivo) in the microenvironment that can dramatically suppress dysplastic cell behavior. These effects are distinct from the stromal and immune cell microenvironment effects on precancer. Differential gene expression revealed TGFβ, EGF, and Wnt as key pathways associated with the differential transcriptional profiles observed in co- vs. mono-culture (166). Single-cell approaches will allow analysis of different subpopulations of cells, including the highly variable epithelial cell motility as well as enumerating the immune cell infiltrates, stromal cells, and other microenvironment components surrounding the neoplasias (166,177). Comparing gene expression-based subtypes defined in tumors with those in their precursor lesion will provide insights that can inform which precursors may progress into malignant disease. Further, oncogenic pathways and developmental- and immune-based gene expression signatures can be used for “pathway/phenotype”-based molecular characterization (179–181). More recently, a novel analytic approach to define oncogenic states and produce functional maps of cancer has been established. This serves as a framework for combining experimental and computational strategies to deconvolve oncogenic pathways/signatures derived from oncogene activation into transcriptional components that can be used to determine oncogenic states. By mapping precancers and tumors onto distinct oncogenic states, the resulting functional map can be used to characterize how these states relate to various omics features, including NGS mutations, copy number alterations, gene and protein expression, gene dependencies, and biological phenotypes; and to predict which interventions are more likely to have a significant effect (182). This approach was used to effectively map cancers with altered KRAS/MAPK pathways into divergent functional states. Studies in pancreatic oncogenesis highlight the need for big data approaches to interpret neoplastic complexity (including KRAS mutation subtypes and Hippo pathway interactions), profound effects on cell metabolism, DNA repair, immunity, mitochondrial biology, and distinct precursor pathways (183). Mutant Kras in pancreatic acinar cells induces expression of ICAM-1 to attract macrophages and drive PanIN development: direct early cooperative mechanism between a driver mutation and inflammatory environment (184). Even B-cells can initiate pancreatic tumorigenesis (185). These maps can be generalized to consider gene networks and interactions (186), including the contributions of the germline and to study the close interplay with the immune microenvironment. Integration of the results from functional genomics studies described above to the functional maps of oncogenic states will provide insight into the cellular contexts in which genomic alterations contribute to malignant transformation (187).
Epigenetics
Previous work has yielded only a limited big data perspective of the neoplastic epigenome, primarily in hematologic neoplasia, where chromatin modifiers are among the most frequently mutated in cancer in general (133,188). Most studies have focused on performing functional analysis on a few genes in a limited number of samples and are reviewed below. Widespread epigenetic field defects have been observed in apparently normal breast tissue located adjacent to breast cancer (189) and also associated with inflammation-related cancers, such as H. pylori-induced neoplasia, in which NGS has revealed more cancer pathway-related genes affected by DNA methylation than by genetic alterations (190). The ten eleven translocation (TET) enzymes oxidize 5-methylcytosines (5mCs) and promote locus-specific reversal of DNA methylation (191).
An epigenetic mitotic-like clock was developed using a novel approach based on an underlying mathematical model. A key feature underlying the construction of this clock is the focus on Polycomb group target promoter CpGs, which are unmethylated in many different fetal tissue types, thus allowing defining a proper ground state from which to then assess deviations in aged tissue. By correlating the tick rate predictions of this model to the rate of stem cell divisions in normal tissue, as well as to an mRNA expression-based mitotic index in cancer tissue, this model approximates a mitotic-like clock. The epigenetic mitotic clock-like signature exhibits a consistent, universal pattern of acceleration in cancer in normal epithelial cells exposed to a major carcinogen. The epigenetic clonal mosaicism is maximal before cancer emerges. Unlike the recently identified mutational clock-like signatures discussed above, this epigenetic clock is based on clinical DCIS and lung CIS progression to cancer and normal at-risk tissue; a concrete example of a molecular mitotic-like clock that predicts universal acceleration in precancer (192). Smoking was associated with an increased rate of this mutational clock. Another approach to ITH analyzed DNA methylation patterns at two genomic loci that were assumed to have no role in gene regulation, in contrast to driver methylation changes. Methylation at such neutral loci were unlikely to be under selective pressure and therefore, could serve as a “molecular clock” to measure mitotic divisions based on the higher error rate of DNA methylation maintenance relative to the error rate of DNA polymerase (193). This molecular clock analysis revealed highly heterogeneous tumors, suggesting that the tumors had not undergone any recent clonal expansion.
Aberrations of the epigenetic modulator TET2 are one of the first alterations in several hematologic premalignancies (TET1 and TET3 are rare in hematologic neoplasias). TET2 mutations are found in premalignant hematopoietic stem cells (HSCs), including of myeloproliferative neoplasms (MPNs) and myelodysplastic syndrome (MDS), and are frequently observed in aged healthy individuals (194) with propensity to transform (see clonal hematopoiesis below). Disruption of TET2 in mouse models increases HSC proliferation and clonal expansion, prone to additional oncogenic events, which are generally required for malignant transformation (191). Mouse models have addressed the functional relevance of co-occurring alterations and found that Tet2 disruption with Asxl, Ezh2, or Jak2 V617F results in MDS or MPN phenotypes. Recurrent dominant point mutations in IDH1 and IDH2 appear to be early events in glioblastoma (affecting a common glial precursor cell population) and hematologic neoplasias that lead to loss of TET activity and other epigenetic changes (195,196). In addition, TET2, IDH1, and IDH2 mutations are frequently observed in lymphoma precursors (197–199), and the frequency of TET loss-of-function (which drives hematologic transformation) in these settings supports testing IDH inhibitors and/or TET activators. For instance, ascorbic acid acts as a cofactor for the α-ketoglutarate-dependent dioxygenases and can affect DNA methylation in embryonic stem cells and mouse embryonic fibroblasts in a manner that is dependent on TET2. TET modulators (200), can enhance antigen presentation and increase IL-6 production by macrophages (201), affect regulatory T-cells (Tregs) (202), and alter expression of endogenous retroviruses, cancer testis antigens, and stem cell antigens in premalignant lesions resulting in enhanced immunogenicity (203).
Another epigenetic mechanism found to be important in premalignant biology involves RNA editing by ADAR enzymes, which results in adenosine-to-inosine conversion of RNA thereby inducing virtual adenosine-to-guanine mutations since inosine bears molecular resemblance to guanine (204). Depending on whether the editing events occur in coding regions or 3’ UTRs, ADAR-mediated editing of mRNAs can result in post-transcriptional protein coding mutations or altered susceptibility to microRNAs (205). Recent data suggest that germline variation involving RNA editing ADAR genes may influence cancer (ovarian) susceptibility (206). ADAR1 editase activity has been implicated in the oncogenic transformation of premalignant progenitors that harbor clonal self-renewal, survival, and cell cycle-altering mutations (207,208), such as in hepatocellular carcinoma precursors, where aberrant RNA editing of AZIN1 has been found to be a key oncogenic driver (207,209). Transgene expression of APOBEC-1 causes dysplasia and cancer in mouse and rabbit livers likely due to RNA editing of NAT1 (210). Study of ADAR1 regulation of APOBEC3 in neoplasia will be critical, potentially suppressing hypermutation and immunity (211). Finally, inflammatory cytokine networks and JAK2/STAT signaling activate ADAR1 during relapse/progression in leukemia stem cell renewal, linking RNA editing to the development of innate immunity and potential preventive activity (212).
Emerging data also suggest that some premalignant lesions may progress to cancer via a fundamental epigenetic reprogramming. Epigenetic defects may be a common mechanism linking genetic mutations to cancer phenotypes, although the details on how they are linked are just beginning to be explored. Indeed, reprogramming of the epigenome to a progenitor-like state may be required for driver mutations to induce tumorigenesis (213). The role of BRAF mutations in benign nevi is a major historical conundrum in the melanoma field (214). In the BRAF V600E zebrafish model of melanoma, deletion of p53 promotes the nevus-to-melanoma transformation, but melanomas remain surprisingly infrequent considering that all of the cells bear both the oncogene and tumor suppressor loss (215) – a feature that replicates the phenomenon of “field defect” in human tumors. Two recent studies using preclinical models addressed this issue. Work with BRAF V600E/p53-null zebrafish now suggests that initiation of malignant transformation within such a “cancerized field” requires fundamental epigenetic reprogramming of these premalignant cells into an embryonic state via transcription factor-mediated reactivation of genes typically expressed only in neural crest progenitor cells (213). This reprogramming involves binding of multiple transcription factors and generation of “superenhancer” regions. New engineered human models, including epigenetic mechanistic studies, suggest a key role of p15 loss in promoting BRAF V600E-mutant benign melanocytic nevi transformation to melanoma (216). Similarly, mouse model research recently demonstrated that basal cell carcinomas, known to be driven by oncogenic signaling in the hedgehog pathway, only originate from stem cells located in very specific areas of the murine epidermis, rather than from more committed progenitor cells (217). Like the zebrafish model, this study provides evidence that the earliest stages of tumorigenesis are characterized by reprogramming to a more embryonic cell state. Such data suggest that tumor-initiating cells can be identified – and potentially targeted for early destruction – through their ability to reactivate an “embryonic” epigenetic state and highlights the importance of studying premalignant cells and model systems to better understand when epigenomic changes arise and how stable they are over time.
The Power of Immunology and Biochemistry to Facilitate Cancer Prevention
Immune oncology
The integration of multiple omics analysis platforms with immune-informatics analysis can be the foundation of a more effective framework for precision prevention (218). There is now a wealth of evidence from both animal models and cancer patients of how the immune system can survey and recognize peptides encoded by certain genetic mutations when such peptides are presented on the surface of the cancer cell bound to MHC-Class I and Class II molecules. For example, RAS mutations, which are key oncogenic drivers in a wide array of cancers, may also be targets of immunosurveillance since T-cells specific to mutated RAS peptides have been found in cancer patients (219) and may be a viable target for immune approaches to treatment and even prevention (220). Proof-of-principle studies of vaccine targeting mutant Kras (with Treg depletion) in a pancreas mouse model induced CD8+ T-cells specific for the Kras mutation and showed preventive efficacy in the early PanIN setting (221). In addition to predicted mutations in well-known oncogenes, cancer cells and their precursors can harbor tens to hundreds of random mutations throughout their genome. Elucidation of the mutated precancer repertoire will allow for efforts aimed at determining which mutated genes produce peptides that can bind MHC molecules and be presented to the immune system (222–224) as potential targets for immunosurveillance and vaccination.
Vaccine-based approaches hold particular promise since they are a form of precision prevention with few side effects. Furthermore, vaccines (e.g., to HPV) could provide long-term protection from cancer development after only one or two treatments unlike prevention drugs that must be taken for many years, challenging an individual’s compliance and/or will to endure accompanying toxicities (9,225). Evidence for immune surveillance has been reported in healthy people and associated with lowered lifetime cancer risk. Childhood febrile viral infections have been associated with reduced cancer risk. Recent mouse model data found that influenza virus infection elicited protective antibodies and T-cells specific for host antigens also expressed on some tumors (226). These data suggest that infection-induced immunity and immune memory could provide long-term immune surveillance of cancer and have important implications for vaccine targets. T-cells are likely the main effector cells in preventing all forms of cancer. The immune system has the ability to recognize precancers and generate immune responses to potentially intercept and prevent cancer (47,64,227) and avoiding immune elimination is a hallmark of cancer (228). We must learn how to both strengthen T-cell immunity – either through immunization, drugs, or engineering – and concurrently overcome a hostile dynamic tumor microenvironment that prevents T-cell activation and infiltration into early neoplasia. The latter involves multiple factors, for example, metabolic reprogramming of the microenvironment by the high utilization of extracellular glucose and glutamine results in extracellular lactate which attenuate dendritic and T-cell activation, stimulate macrophage polarization to an M2 state, induces VEGF secretion by stromal cells and activates NF-kB. The microenvironment can in turn have profound effects on the metabolism of neoplastic cells (88). Emerging data suggest that the microenvironment barriers develop early in precursor lesions but are likely qualitatively different from more established cancer-associated barriers. The progressive accumulation of somatic changes that lead to neoplasia also co-opt neighboring vascular, neuronal, and other normal cells to support/promote oncogenesis. Critical to vaccine development, therefore, is the identification of potent immune enhancers/adjuvants that can specifically target one or more innate pathways (229) and alter the developing inflammation that promotes immune suppression in favor of a neoplastic response (230). Experience with therapeutic cancer vaccines shows that targeting a single antigen or a single mutated peptide invariably leads to outgrowth of cancer cells that have lost that mutation. This may happen in the precancer setting as well, requiring a vaccine that elicits a polyclonal and polyspecific immune response. Trial endpoints could include T-cell receptor sequencing to look at clonality and clone expansion, liquid biopsy approaches to detect low levels of the identified mutations or mutational load, and novel techniques to image immune response (231) and high-grade pre-invasive neoplasia (PanIN-3) (52), or depending on risk, even cancer incidence.
In a clinical feasibility trial in advanced adenoma patients, lack of immune response to a MUC1 cancer vaccine correlated with increased levels of circulating MDSCs responsible for inhibiting adaptive immunity (232), suggesting that these may be useful biomarkers to identify individuals unlikely to benefit from preventive cancer vaccines. For those deemed unlikely to respond to the vaccine alone, research into other immunomodulatory drugs that could help overcome such immune resistance will be critical. Metformin, for example, has been shown to enhance T-cell immunity and immune memory, influence the microbiome in mouse models, by various mechanisms including involving mitochondrial biology and RANK-L inhibition (see above) (37,233–235). Furthermore, recent prospective cohort data suggest that aspirin prevention of CRC is related to its effects on T-cell immunity (236). Adenomas have been reported to have a highly inflammatory microenvironment (237), which varied by histology and location in a recent large microbiota/adenoma study (238). Two recent NGS of tumor from patients with IBD (colitis)-associated CRC were compared with sporadic CRC. The comparisons suggested that colitis-associated CRCs have a distinct mutational profile associated with cell-to-cell signaling, cell adhesion, and epigenetic regulators/chromatin modifiers, all of which may be linked with the inflammatory mediators of IBD (239,240). IDH1 mutation (extremely rare in sporadic CRC) was found only in Crohn’s disease. Extension of NGS to include epigenomic and microbiome profiles in IBD dysplasia has great potential for cancer prevention and early detection research.
Microbiota-immune interactions are an increasingly important and challenging field. Each human body contains at least 40 trillion microorganisms that populate complex ecosystems called microbiomes; 99% of microbiota reside in the gut microbiome and certain bacteria can influence oncogenesis and immune interventions via complex microenvironment interplay. Germline/microbiota interactions effects are discussed above. Intestinal barrier function is regulated by inflammatory cytokines such as IL-1β and IL-18 (241), autophagy (242), and microbiota-accessible carbohydrates (which affect gut mucous layer and microbiota special organization) (243). Thymic stromal lymphopoietin (TSLP) is a cytokine expressed mainly by epithelial cells at barrier surfaces (skin, gut, and lung). Short-course calcipotriol, a topical TSLP inducer FDA approved for psoriasis, suppressed skin cancer development in genetically engineered mouse models (GEMMs) in a TSLP-dependent, long-lasting manner consistent with an immune memory response. A randomized clinical trial of this agent showed a marked reduction in actinic keratosis number mediated by specific induction CD4+ T-cell adaptive immunity (244). Non-specific innate immune activation by imiquimod is also active in actinic keratosis patients (245). Potential immune and/or microbiota prevention factors include lifestyle (104), metformin (234), antibiotics, diet, and microbial reprogramming (246,247). It has been recently shown that gut microbes modulate whole host immune and hormonal factors impacting the fate of distant precancers toward malignancy or regression, for example, by stimulating host immune cells to prevent dietary and genetic predisposition to mammary cancer in mice. This raises the possibility that the tumor microenvironment interacts with broader systemic microbial-immune networks (248). Caloric restriction is the most consistently effective cancer preventive approach in virtually every mouse model/tumor type tested. Recent data, including from the mutant Kras lung mouse model, indicate that caloric restriction or its mimetics (e.g., over-the counter hydroxycitrate) elicits autophagy, which improves immunosurveillance via Treg depletion and prevents malignant transformation (249).
Analyses of NGS genomic data are critical to develop vaccines that target specific epitopes derived from mutations, copy number alterations or other variants common to precancers. However, this direct strategy is especially challenging given the large number of alterations, the low penetrance of driver mutations, the so-called “long tail” problem (low frequency mutations) (250), and the fact that the corresponding mutant peptides do not always lead to effective antigen presentation and response (251). Recent work demonstrates the ability to utilized mass spectrometry (MS)-based analysis to identify attractive target antigen candidates from a native human melanoma tissue, which were subsequently narrowed down using somatic mutation information and subsequent immunogenic assays in mice (252). This approach, coupled with NGS genomic data in precursor lesions, will help better nominate strong candidate antigens for vaccinations. Computational methodologies can also help to identify suitable antigens from a large number of candidates, for example, by using existing resources and databases that catalog potential antigens (253). Prediction of peptide binding affinity to HLA I and II has also been developed using machine-learning classification approaches (254,255). Computational studies have focused on neoantigen-epitope prediction algorithms and have shown that only a very small proportion of predicted neo-epitopes are actually presented on MHC class I as targets of endogenous T-cell responses (57,256,257). Whole-exome sequencing has identified higher antigen load was predictive of overall lymphocytic infiltration, tumor-infiltrating lymphocytes (TILs), memory T-cells, survival in colorectal cancers (258). Using the NGS genomic data from precancers, we will use two strategies to nominate candidate antigens: 1) use the mutation calling algorithms to identify the most frequently occurring neoantigens and 2) for the low frequency events, utilize functional maps described above to identify complementary neoantigens that associate with oncogenic states (182). These filtered lists of neoantigens will then be used to predict strong epitope candidates in silico using algorithms that employ either Artificial Neural Networks (ANN) or Support Vector Machines (SVM). These analytic pipelines can be used generate lists of the most likely vaccine candidates based on stabilized peptide p–MHC-I binding affinity (256), an approach that has already produced promising recent results in mice (259). Attractive candidate neoantigens from this analysis will be used for systematic testing in vivo and for vaccine generation and characterization.
Developing cancer prevention vaccines will require in-depth insight into the molecular events that drive premalignant biology, building off of groundbreaking biochemical studies. A recent analysis found somatic mutations that disrupt beta-2 microglobulin (B2M; a component of the class I MHC complex) protein-protein interactions, with a striking enrichment for mutations at protein interaction interfaces involving B2M’s binding partners (260). It has been shown that disruption of B2M can minimize immunogenicity of human embryonic stem cells (i.e., foreign human cells with possible regenerative benefit but that cause immune reactions) (261). It is conceivable that such mechanisms may be employed by precancers and cancers to escape immune surveillance (262,263).
Biochemistry: Understanding the molecular basis of neoplasia
Recent TCGA and related studies have demonstrated that a large number of genetic and epigenetic factors, such as chromatin modifiers and remodelers, are highly mutated in a large number of solid tumors and in hematological malignancies (264). Recurrent mutations in genes that encode regulators of chromatin structure and function highlight the central role that aberrant epigenetic regulation plays in the pathogenesis of these neoplasms. Deciphering the molecular mechanisms for how alterations in epigenetic modifiers, specifically histone and DNA methylases and demethylases, drive hematopoietic transformation could provide new avenues for developing novel targeted epigenetic prevention for hematological neoplasia and could also inform future studies in solid tumors. Many such protein complexes – including the mixed-lineage leukemia (MLL) family (188), the polycomb complexes PRC1 and PRC2, which contain EZH2, ASXL1 and BAP1 (265), and the SWI/SNF chromatin remodeling complex (266) – contain genes that are frequently mutated in human cancers (264) but were initially identified in simple model systems, such as Drosophila and yeast, emphasizing the importance of including studies of model organisms in any large-scale efforts in cancer prevention. While genomic deletions and nonsense, frameshift and splice site mutations that introduce a premature stop codon or alter protein structure can be obvious loss of function events, missense mutations can be hard to classify unless they alter enzymatic function or disrupt protein-protein interactions within large functional protein complexes.
For example, a large number of hematological malignancies harbor translocations of the N-terminal region of MLL1 to diverse fusion partners that share very little sequence or functional similarity. To understand how these diverse MLL translocations result in leukemogenesis, biochemical and enzymological studies were essential. First, MLL and its yeast homologue SET1 were shown to be present in a complex named COMPASS (Complex of Proteins Associated with Set1) and to function as histone H3K4 methylases (267). Second, AFF4, itself a fusion partner of MLL in leukemia, was found to be a common factor among all purified MLL translocations (268). Third, ELL, one of the frequent translocation partners of MLL in leukemia, was found to function as an RNA Pol II elongation factor that increased the catalytic rate of transcription elongation by RNA Pol II by suppressing transient pausing (269). Finally, it was discovered that many MLL translocation partners are found in association with ELL and the positive transcription elongation factor (P-TEFβ), within a complex named the Super Elongation Complex (SEC) (266,270,271). The translocation of MLL into SEC is involved in the misrecruitment of SEC to MLL target genes, perturbing transcription elongation checkpoints at these loci and resulting in leukemia (271). Recent study of MLL-induced leukemogenesis highlights the role of deregulated histone methylation in tumorigenesis (272).
Another example of the importance of biochemistry is deciphering the molecular role of an observed genetic link of EZH2 in cancer. EZH2 encodes the catalytic subunit of the polycomb repressive complex 2 (PRC2) responsible for methylating lysine 27 of histone 3 (H3K27). Trimethylation at this site is associated with closed chromatin and silencing of neighboring gene expression. In neoplasia, EZH2 can influence T-cell biology (273) and function as either an oncogene or a tumor suppressor gene depending on the cellular context, e.g., EZH is sufficient to transform lung cells in transgenic mouse models overexpressing EZH (274), and loss-of-function EZH2 mutations occur in MDS and chronic myelomonocytic leukemia (CMML) (275). In germinal center diffuse large B-cell lymphomas, recurrent mutations essentially of only one codon (Y641) create a protein with reduced affinity for unmethylated H3K27 but highly increased affinity for mono-methylated H3K27, resulting in higher levels of H3K27 trimethylation overall. In contrast, pre-AML syndromes like MDS and CMML do not develop Y641 mutations but instead recurrently develop nonsense, frameshift, and other loss-of-function mutations in EZH2 resulting in low levels of H3K27 trimethylation (276). These distinctions have important clinical implications for EZH2 inhibitor development. It is possible that EZH2 inhibition will mimic malignancy-associated, loss-of-function EZH2 mutations in normal myeloid cells leading to dysregulated growth or differentiation in these cells, highlighting the need for future context-dependent studies.
SWI/SNF also known as the BAF complex is also a critical regulator of nucleosome remodeling conserved from yeast to humans. Recent biochemical investigation, combined with bioinformatic assessments have demonstrated widespread genomic alterations that occur across the members of the complex in 19.6% of all human tumors reported in 44 studies (277). In synovial sarcoma, SS18-SSX oncogenic fusion that results from a fusion of 78 amino acids of SSX to the SS18 subunit of BAF complex was shown to distrupts binding of BAF47, tumor suppressive member of the complex, leading to reversible dysregulated growth (278). In liver, genetic suppression of SWI/SNF complex member ARID1B was shown to overcome oncogene-induced scenscence and lead to liver neoplastic progression (279). While these studies suggest a newly emerging role for SWI/SNF in tumorigenesis, better delineating the role of SWI/SNF complexes in precancers will also be important. Further, prior studies demonstrate antagonistic relationship between the SWI/SNF and PRC2 complex in mediating oncogenic transformation (280).
A transformative example of biochemistry’s importance in premalignant biology involves the discovery of recurrent mutations in IDH1 and IDH2 in glioblastoma, AML, and their precursor cells. Such mutations were found through broad sequencing efforts (281) although their role at the molecular level was not clear until the advent of modern metabolomics profiling (282), which found that mutant IDH enzymes convert the normal intracellular metabolite alpha-ketoglutarate into 2-hydroxyglutarate (2-HG). 2-HG is a competitive inhibitor of a large class of dioxygenase enzymes that utilize alpha-ketoglutarate, and accumulates to very high levels in IDH-mutated cancers, potently inhibiting many important intracellular dioxygenases, including the TET family, prolyl hydroxylases, and several histone demethylases (283–285). Thus, biochemistry and metabolomics have illustrated how 2-HG contributes to carcinogenesis in a hitherto unprecedented way by acting as a novel “oncometabolite” generated by somatic IDH1/IDH2 mutations that can potentially serve as targets for both cancer prevention and therapy, including vaccines (286).
Recent biochemical approaches have also focused on the significance of metabolism and its link to epigenetic factors, such as the TET family in the regulation of cell-lineage specification and the development of cancer (188). These discoveries are only a few examples among a large number of biochemical approaches in neoplastic cancer studies and are the testimony to the power of biochemistry in understanding neoplasia and the design of its targeted prevention, for example, by highlighting the importance of epigenetic regulation. High-information-content mass spectrometry to profile global histone modifications in human cancers (287), when combined with the DNA sequencing data, can be used to identify novel variants that can drive epigenetic changes that can lead to oncogenic transformation. Chromatin-IP technology combined with NGS sequencing (CHIP-seq) can provide systematic information regarding the architecture of the chromatin cell states of cancers. Recent technological development has demonstrated that CHIP-seq can be carried out in human tissues including tumors (288). Interestingly, examination of the chromatin landscape was able to fully distinguish the normal vs. cancers. These results suggest the possibility of gaining additional insights into precancers by systematic assessment of chromatin states using key histone acetylation and methylation patterns, superenhancers, as well as TET, SWI/SNF, and PRC2 complex, all of which are critical for chromatin regulation.
The study of cancer metabolism is not only shedding light on tumorigenesis carcinogenesis but is also revealing new principles of how the biochemistry of anabolic metabolism is orchestrated to support normal cell growth and function. While most of the studies in this context have been focused on alterations in the metabolism of glucose and glutamine (see above Immune Oncology), neoplastic cells utilize many other nutrients, including sulfur–containing amino acids cysteine and methionine, essential fatty acids, choline, trace metals, and vitamins. We are only beginning to understand the extent to which these nutrients contribute to tumorigenesis. Finally, the contribution of a broad spectrum of metabolites produced by the body’s microbiota, to tumor initiation and progression is only beginning to be elucidated.
It is essential to incorporate detailed biochemical and enzymological studies on purified protein complexes to decipher the precise, context-dependent function of chromatin and other epigenetic modifiers and somatic mutations in cancer development and progression (268). This will also allow the profiles to be cross-referenced with the landscapes in primary tumors, as well as of the corresponding transcriptomic data to identify critical epigenetic changes that are necessary for malignant transformation. The context-dependent, complex roles of EZH2 mutations and PRC2 and SWI/SNF complexes in chromatin regulation in normal development and neoplasia require further study, especially in precancers. Finally, biochemical field of metabolic alterations in neoplasia continues to uncover new connections between nutrient utilization and tumorigenic state, critical to precancer progression.
Single-Cell Analyses
The natural history of precancers is heavily influenced by the heterogeneity of neoplastic cells and tissue microenvironment. Single-cell RNA or DNA sequencing technologies can be specifically leveraged to unravel the complex cellular interactions within these lesions that cannot be addressed by assaying bulk tissue (289,290). In the case of mRNA profiles, downstream analyses can characterize known populations and novel subpopulations of cells and assess how these populations change in abundance as disease progresses or regresses. These data also can be used to more accurately infer important disease-associated gene regulatory and immune cell networks (291) because the gene expression variability has not been averaged across all sampled cells as in bulk tissue. In addition, single-cell sequencing can reveal and monitor lesion heterogeneity in somatic alterations and dissect complex clonal dynamics among epithelial cells sampled at different geographic locations and over time to complement existing multi-region bulk sequencing approaches (292). These data will provide a high-resolution picture of cell types present in precancers and their surrounding microenvironment and the transcriptional programs active within each cell type that drive disease progression.
However, several technical limitations need to be overcome to realize the full potential of single-cell sequencing of precancers. These lesions are relatively small and frequently only diagnosed in formalin-fixed, paraffin-embedded (FFPE) tissues previously precluding comprehensive genome sequencing studies using current methods (293). Furthermore, information regarding the location of neoplastic cells with particular mutations within a given lesion is especially important for early lesions, as this often defines the boundary between preinvasive and invasive neoplasia. Therefore, the development and application of methods that allow assessing the genetic and phenotypic features in situ using intact FFPE tissue samples is especially critical for the improved understanding of preinvasive lesions. Several technologies enable copy number alteration and gene expression analyses at the single-cell level from FFPE slides. These include FISH and immuno-FISH (combination of FISH with immunofluorescence) (294,295), mRNA in situ hybridization, in situ PCR, and STAR-FISH (296–298). The application of immuno-FISH for the analysis of cellular phenotypic heterogeneity and genetic features revealed extensive intratumor diversity in DCIS and clear expansion of minor subclones in DCIS to dominant clones in invasive ductal carcinoma (295). A shortcoming of these methods is the limited set of markers that can be assessed on a single slide and the need for a priori knowledge of the changes to be analyzed.
Single-cell methods are beginning to be applied to premalignancy, including sequencing on both fresh/frozen and FFPE has been applied to an epithelial precancer site, DCIS and associated invasive breast cancers, and included massively parallel single-cell sequencing for copy number analysis (178). This proof-of-principle analysis established technical feasibility and demonstrated intra-lesion genetic heterogeneity at DCIS suggesting complex and distinct evolutionary processes involved in early DCIS to subclonal selection in invasive disease. Multicolor FISH to study clonal evolution at single-cell resolution in BE (see below) found extensive genetic diversity in progressors (299). A whole-exome single-cell sequencing method was developed to assess genetic heterogeneity and tested on a premalignant JAK2-negative myeloproliferative neoplasm (essential thrombocythemia) patient (300). Such profiling of a different myeloproliferative neoplasm myelofibrosis revealed substantial heterogeneity in cytokine production (301). Importantly, however, current single-cell sequencing approaches have important technological limitations. First, the methods available are labor- and cost-intensive. Second, it is currently not possible to obtain accurate detailed copy number and mutational data from the same cell, given that some whole genome amplification methods yield templates optimal for copy number analysis, whereas others are optimal for mutation profiling. Therefore, efforts are required for the development of less labor-intensive and more cost-effective methods for sequencing approaches and clonal lineage tracing, which are essential for a detailed analysis of the evolutionary paths of in situ disease and its progression to invasive cancer. Two more technologies, FISSEQ (fluorescent in situ sequencing) (290,302) and “spatial transcriptomics” (303), allow for complete transcriptome analysis of single cells in intact tissue sections. Recently, single-cell techniques have been developed to study chromatin maps/signatures and epigenetic heterogeneity in neoplasia (304,305) and the microbiome (306), including imaging of host-microbiota interactions (243,307).
Cost reduction and advances in cell sequencing (and cfDNA technology) could theoretically allow temporal monitoring of blood and epithelial premalignancies on a population scale (308–310). Periodic single-cell DNA sequencing of multiple cells from an individual will be invaluable for cancer prevention as it will allow one to assess the overall baseline accumulation of somatic mutations over time in a person, to survey and monitor multiple different endogenous processes and exogenous exposures through the use of mutational signatures, and to reveal the existence of premalignant clones and clonal evolution over time (311–313). The unprecedented resolution of sequencing single cells comes with a hefty computational and data price. Monitoring even a single individual will require multiple sequencing of one’s genome every year resulting in several terabytes of data per person. As such, population scale examinations will generate millions of whole-genome sequences resulting in exabyte scale data (>1018 bytes) that will require a new generation of computational infrastructure (314) and novel computational frameworks (e.g., to take into account their relatively low signal resolution when compared with traditional bulk tissue sequencing). More than ever, the rate-limiting step will be data analysis.
Liquid Biopsies for Early Detection and Intervention
Alterations in precancers and localized tumors may have their greatest impact in early detection of cancer. By virtue of the clonal nature of tumor cells, somatic changes are present in many copies that are continuously released and can be detected in the blood as cell-free (cf) circulating tumor DNA (315). In cancer patients, alterations in cfDNA can be detected using new sequencing and bioinformatic approaches. Such alterations can be difficult to detect as they often represent a minute fraction (<1%) of cfDNA. A variety of both targeted and whole-genome approaches have been developed to detect such alterations in cfDNA (316,317). These have been used for early detection of recurrence in colorectal cancer, pancreatic cancer, neuroblastoma, hematopoietic malignancies, and others (318–320). Of importance to early detection research, cfDNA was recently detected in plasma and urine in patients with premalignant lung and bladder disease (321,322), however, somatic mutations in cfDNA among individuals without any cancer diagnosis poses serious challenges for the development of ctDNA screening tests (323). Importantly, cfDNA detection of pancreatic cancer recurrence appears to precede radiographic detection of recurrence by over six months in some cases, providing a larger window for potential intervention in this challenging disease (319).
Much work remains to be done in improving methodologies for detection of circulating tumor DNA, both in terms of increasing the sensitivity and preventing false positives. A potential confounding issue is the detection of clonal alterations that arise in blood cells of healthy individuals that may be associated with aging and clonal hematopoiesis (see below). Distinguishing between alterations in genes associated with MDS/AML versus solid tumors may be one way to overcome this issue. Additionally, even when molecular alterations are identified, determining the tissue of origin of the incipient neoplastic lesion can be extremely difficult and complex. Combining sequencing of cfDNA with epigenetics markers (324,325), mutational signatures, imaging and mathematical modeling can be used for pinpointing the most likely tissue in which the clone originated. Analysis of other blood/fluid components, such as exosomes, platelets (326), and circulating tumor cells (315), may help increase the sensitivity of detection (327). With these approaches, it is possible to imagine a time when individuals at high risk of developing cancer due to either genetic or environmental risk factors (e.g., individuals with inherited gene mutations at risk for breast, ovary, pancreas, colorectal or other cancers, or heavy smokers or obesity at risk for several cancers) could be serially monitored using a blood-based test. A potential advantage of this approach would be the relative ease of compliance compared with other more invasive screening technologies. Ultimately, the specific methodology would be determined by practical considerations such as cost, sensitivity, specificity, and robustness of the assays, but these approaches may forever change screening for cancers that are currently incurable unless diagnosed at an early stage.
Longitudinal Analysis of Premalignancies
It is likely that synchronous precancer/cancer pair studies will not always accurately reflect the temporal clonal evolution underlying neoplastic transformation, although new analytical methods clonal/subclonal structures from NGS of a single sample. To fully appreciate such mechanisms, systematic longitudinal analyses of malignant cells will be essential. To date, such analyses of epithelial premalignancies, with the exception of BE, have been extremely limited, with reports of a relatively small number of patients with squamous lung premalignancy (328,329). DNA- and RNA-seq of these lesions have identified molecular alterations in both epithelial cell signaling pathways and immune cell pathways that associate with progression of these premalignant lesions over time. The largest longitudinal study of BE assessed genome-wide somatic chromosomal alterations (SCA) using SNP arrays over time and space. Non-progressors largely maintained stable genomes, in contrast to high levels of SCA, increased diversity, and chromosomal instability, were associated with progression to EAC (330). For some patients the transition to malignancy evolves rapidly through a genome-doubling event or chromosomal catastrophe termed chromothripsis; recently shown drive profound immune suppressivon (74,330). Another recent Barrett’s study of clonal evolution at single-cell resolution using multicolor FISH found that baseline genetic diversity predicts progression (to cancer) and remains in stable dynamic equilibrium over time, suggesting that clonal make up and evolutionary trajectory of the lesion is predetermined from the outset (299). Clonal expansions were rare, often involving p16. Importantly, this work has established the feasibility and model of such study in epithelial premalignancy.
Focusing on premalignancies of the blood has several advantages, including the ease of repeatedly acquiring neoplastic cells to study their clonal evolution, discoveries that inform single cell sequencing studies in epithelial neoplasia. For example, study of myeloproliferative syndromes (198) provides the only direct data that somatic mutation order (JAK2 and TET2) can greatly influence disease features. The overall malignant transformation rate for clonal hematopoiesis, monoclonal B-cell lymphocytosis (MBL), and monoclonal gammopathy of undetermined significance (MGUS) is about 1–2% per year, but individual risk is highly variable. Comprehensive single-cell and cfDNA omics studies will play a key role in improving our understanding of disease pathogenesis. We now have the ability to monitor hundreds of individual cells, thus overcoming bulk-cell/tissue limitations and allowing precise study of intraclonal and microenvironment architecture and crosstalk in the process and timing of transformation. The complexity and importance of follow-up is highlighted by recent findings in pancreas precancers, which can exhibit significant clonal heterogeneity/diversity that surprisingly decreases during transformation (331).
Clonal hematopoiesis was only recently characterized (332,333) by somatic mutations in genes (mutant clones of mostly single driver mutations) similar to the mutational spectrum seen in MDS (notably DNMT3A, JAK2, TET2, and ASXL1) that increased markedly with age in the general population (334–336). Nearly 40% of clonal hematopoiesis individuals with unexplained cytopenias harbor detectable mutations (197,336,337), many with clones having more than one driver mutation and higher risk of transformation to MDS/AML and all-cause mortality (197,336–338). Cooperating mutations also can be identified during periods of clonally skewed hematopoiesis in sporadic and hereditary settings that precede myeloid transformation. A recent mouse study showed that DNMT3A haploinsufficiency transforms FLT-mutant myeloproliferative disease into AML (339). Examples of hereditary mutated transcription factors that predispose to hematologic neoplasia include mutations in CEBPA, RUNX1, ETV6, and PAX5 (340). The frequent co-occurrence of germline GATA2 and somatic ASXL1 events (341) and germline SNPs associated with somatic mutations of JAK2 have uncovered several targetable cooperative mutations (342) driving premalignant progression (340). A recent GWAS identified germline variants, which predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms (342). Four genes (JAK2, SH2B3, CHEK2, and TET2) altered in both inherited and somatic settings, contribute to V617F clonal hematopoeisis and/or MPN development. This study’s identification of a predisposition allele associated with TET2 is intriguing since somatic TET2 mutations are one of the common early events in myeloid precursors, including clonal hematopoiesis and myeloproliferative neoplasms, and can be identified in hematopoietic stem cells, either preceding or following the acquisition of V617F, and the mutational order of these two genes can influence the clinical and biologic behavior of these neoplasms (198). Most patients with clonal hematopoiesis can stably harbor small hematopoietic clones for long periods of time that expand to the point of detectability but are then held in check, possibly involving immune mechanisms (336). Clonal hematopoiesis in aplastic anemia characterized a distinct mutational pattern, including 6pUPD and PIGA, which are thought to be involved in immune escape, possibly involving loss of relevant HLA class I (343). Further, the aging microenvironment promotes outgrowth of clones driven by spliceosome mutations (SF3B1, SRSF2) in clonal hematopoiesis, associated with innate and adaptive immune attrition (344). Murine model findings suggest that aberrant splicing could produce neoantigens, which elicit an immune response. Drugs targeting the inflammasome and innate immune responses implicated in remodeling the microenvironment are potential preventive approaches.
MBL is an asymptomatic expansion of clonal B-cells in the peripheral blood present in roughly 4% of all U.S. individuals over the age of 40 (345). Genetic predisposition to MBL is suggested by the finding that the incidence of MBL is three-fold higher for individuals within familial chronic lymphocytic leukemia (CLL; at least two first-degree relatives with CLL). A large GWAS of CLL families (including 60 relatives with MBLs) as part of the replication study samples found significant germline variant associations in two out of eight regions tested (346). NGS has shown that most mutations and intraclonal heterogeneity found in CLL are already present in MBL years before progression (347,348). Furthermore, longitudinal MBL studies, including those from patients who progressed to CLL, have begun to elucidate the sequence, timing, and impact of subclonal expansion and T-cell exhaustion on malignant transformation (346,349–351). Risks of serious bacterial infections in individuals with MBL are similar to those with CLL (352–354) and linked to MBL transformation (355,356) and solid tumor risk is 3–4 fold higher in MBL and CLL (versus healthy controls), all thought to be due to defects in immune surveillance (353,354). Related work involves a hereditary syndrome of susceptibility to pre–B-cell neoplasia caused by inherited mutations of PAX5. Recent mechanistic data indicated that inherited susceptibility and aberrant immune responses to postnatal infections drives B-cell clonal evolution of premalignant B-cells and transformation to leukemia and lymphoma, by showing that pre-B-ALL was initiated in Pax5 heterozygous mice only when exposed to common infections (357). Antibody responses to primary and secondary antigen challenges are typically inefficient among patients with early stage CLL. Preliminary data have shown that immunological T-cell synapse is defective in individuals with MBL as well (345). Efforts to generate efficient vaccine responses will, therefore, be challenging. Enhancement of cytolytic T-cell function in MBL via vaccine therapy should be a long-term challenge since GVL is highly effective in eradicating leukemic B-cells. Both in vitro and in vivo data suggest that lenalidomide can repair defects in the T-cell immune synapse and reduce Tregs in CLL patients (358,359), an approach currently being tested clinically in MBL. Of note, recent mouse model data suggest that age and inflammatory status of the host microenvironment promotes selection for adaptive oncogenic events in B-cell progenitors (360), analogous to clonal hematopoesis.
Studies of MGUS have provided some of the best evidence that the immune system has the capacity to recognize precursor states (361). Search for shared targets of immune response led to the discovery that T-cells against stem-ness antigens (such as SOX2) are particularly enriched in MGUS (versus MM) (362). Prospective data demonstrate that SOX2 immunity correlate with risk of transformation (227). Prevention strategies include boosting pre-existing T-cell immunity, e.g., with SOX2 vaccines and immune modulatory drugs (363). NGS has highlighted clonal evolution and heterogeneity in longitudinal studies (364,365). Similar to clonal hematopoiesis above, MGUS cells demonstrate clinical dormancy despite NGS suggesting that the majority of genomic alterations found in MM are found in precursor gammopathies (364,365). Interestingly, new humanized models developed to grow precursor cells in vivo indicate that MGUS cells have the capacity for progressive growth, suggesting that the clinical stability/dormancy of these cells is in part mediated by features extrinsic to tumor cells, such as the immune system or bone marrow niche where signals derived from osteoblasts may be important for mediating dormancy of MGUS cells (366). The recent development of new humanized models to grow premalignant cells in vivo should greatly advance the study of clonal evolution and malignant transformation in this setting (367). Inherited genetic variation in specific SNPs increases MGUS predisposition and risk of transforming to multiple myeloma (368). Loci identified also points to a role for chronic antigen-driven stimulation in driving clonal origins and evolution in MM and other B-cell tumors. The risk of MM is particularly increased in some populations such as those with inherited lipid storage disorders such as germline GBA mutations in Gaucher disease (GD), due in part to lysolipid-induced chronic inflammation and genomic instability. GD mouse models, e.g., lysolipid substrate reduction in Gba1-deficient mice decreased the risk of gammopathies, studies already show marked preventive efficacy of targeting the underlying trigger of genomic instability (369,370). This led to the recent discovery that in nearly 25% of all cases of MGUS/MM, the underlying clone may be driven by lipid antigens such as inflammation-associated bioactive lipids (369). Studies to characterize the precise nature of microbial/dietary/endogenous lipid antigens in the setting of lipid-reactive gammopathy will facilitate targeted prevention with pharmacologic/lifestyle changes.
Summary
While a number of interventions are already approved for cancer prevention (245), this is only the tip of the iceberg. Recent paradigm changing advances include early detection research (universal tumor testing, pancreatic imaging), vaccine prevention of cervical neoplasia and combination chemoprevention in FAP. New precision prevention approaches will be needed, including novel trial designs (e.g., involving molecular selection (371,372)) and agents (e.g., denosumab for BRCA1 carriers; (34)). Developing cancer vaccines as potent as polio, diphtheria, and rubella vaccines would protect future generations from developing cancer. The elimination of cancers “before” they develop is within our grasp. As in cervical cancer, where vaccination against HPV has virtually eliminated the disease, the development of cancer vaccines to stimulate T-cells to recognize precancer antigens as foreign will prevent cancer. The interaction between the immune system and neoplasia reflects a fundamental principle that is applicable to all organ/cell types and continues to become more involved. For example, recent study revealed a striking complex interplay between chromosomal abnormalities and immunity, with mutational burden and arm- and chromosome-level SCNAs having opposing immune surveillance by showing that high level somatic copy-number alterations, consistent with a mechanism related to general gene dosage imbalance rather than the action of specific gene, creates an immune suppressive microenvironment with immune depletion/evasion that can oppose an immune response (74), even in the setting of high mutational/ neoantigen burden (e.g., MSI-H/mismatch-repair deficient tumors) (57,63). The observation that precursor conditions can exhibit similar genomic complexity to their malignant counterparts, shown in longitudinal studies of hematologic premalignancies and BE to exist years before progression, highlights the importance of microenvironment crosstalk in the process of malignant transformation. Therefore, further such studies using new technologies (e.g., single-cell approaches) are needed to better interrogate and dissect the detailed interactions of precancer cells with immune cells, and other components of the tumor microenvironment. The collaborative, focused study of nuclear, mitochondrial, epigenomic, proteomic, transcriptomic, metabolomic, and multi-omic signatures/ landscapes can be bioinformatically timed to distinguish cancer initiation and clonal diversity/heterogeneity. Organelle-targeted omics and studies of translational activity (e.g., by ribosome profiling) will also yield valuable biologic insights into mitochondrial function and dysfunction influence on oncogenesis. Establishing a PCA that integrates multi-omics and immunity (Figure 1) will be critical to develop approaches to disrupt the disabling immunosuppressive properties in the tumor microenvironment and to identify immunogenic antigens to design vaccines to activate/reprogram the immune response to detect, prevent, and reject precancers.
Specifically, we need a large-scale, systematic effort to longitudinally map the biology of the premalignancies that will bring together leaders from key disciplines (from biology and biochemistry to mathematics and engineering); leverage the NIH/NCI TCGA, GWAS, and other programs; develop/enhance statewide registries and disease specific cohorts, such as web-based registries for hereditary cancers (e.g., the PROMPT registry, www.promptstudy.info) and the recent NHLBI/ NCI National study of clonal hematopoiesis. Analogous to clonal hematopoiesis, albeit in much smaller subsets of older individuals, ultra-deep NGS of somatic cancer mutations reported a distinct mutational profile (mostly NOTCH1) in normal skin (160), and low-frequency p53 mutations in normal peritoneal fluid and peripheral blood from women without ovarian cancer (327), both likely representing a premalignant mutational background events that accumulates in cancer and aging. The NCI PCA demonstration project, which came from the Cancer Moonshot recommendations, is designed to evaluate current state of the science and provide feedback to NCI leadership for a concerted effort to comprehensively profile these premalignant lesions and provide a blueprint, including application of existing genomic, proteomic, immune, bioinformatics methods to delineate the signals and interactions within and around these lesions to create a three dimensional analysis of neoplastic progression; and development of a national database analogous to the Pan-TCGA (373,374) and pan-GWAS (77) shared databases with links to clinical annotations (1). This would provide an immense national resource for discovery and shed new light on shared precancer biology and new targets for interception.
The PCA project will require expansion of a diverse array of companion technologies and preclinical models in which to study (88,375), develop, and implement strategies for cancer prevention and interception. New technologies (e.g., single-cell sequencing, liquid biopsy) and preclinical models, e.g., GEMMs such as autochthonous mouse models of cancer to dissect the in vivo contribution of stroma-specific factors to tumor progression; CRISPR/Cas9-engineered, immunosuppressive mouse strains (376,377); methods to isolate and inducibly engineer primary premalignant/benign human cells to study very early oncogenesis (216); clinically relevant organoids (to study T-cell immune surveillance) and sequential, inducible activation of cancer-causing driver genes (and associated neoantigens) in a cell lineage-specific manner, e.g., in germline (MSH2-deficient) and sporadic intestinal neoplasia (378–382), will allow study of complex interactions between genetically susceptible host, microenvironment, and microbiota (246,383).
In summary, to fully achieve cancer prevention, we must build teams with multiple areas of expertise from the NIH, academia, Food and Drug Administration, private foundations, philanthropic partners, and industry. The best analogy of assembling such a multi-disciplinary team is the Manhattan Project – one goal, multiple experts.
Acknowledgments
The authors thank Jennifer Beane, PhD, for her input on the single-cell sequencing section and Leona Flores, PhD, for editorial assistance with this article.
Additional Information: AS supported by NIH/NCI 1U01CA214182 and 5U01CA196408. SML supported for this work by NCI P30-CA023100–29. AS and SML co-chair the PremalignanT Cancer Genome Atlas (PreTCGA) Demonstration Project, NCI Blue Ribbon Panel (BRP).
References
- 1.Hudson KL, Collins FS. The 21st Century Cures Act - A View from the NIH. N Engl J Med 2017;376(2):111–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamihara J, Rana HQ, Garber JE. Germline TP53 mutations and the changing landscape of Li-Fraumeni syndrome. Hum Mutat 2014;35(6):654–62. [DOI] [PubMed] [Google Scholar]
- 3.Yurgelun MB, Allen B, Kaldate RR, Bowles KR, Judkins T, Kaushik P, et al. Identification of a Variety of Mutations in Cancer Predisposition Genes in Patients With Suspected Lynch Syndrome. Gastroenterology 2015;149(3):604–13 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold RS, Sun Q, Sun CQ, Richards JC, O’Hearn S, Osunkoya AO, et al. An inherited heteroplasmic mutation in mitochondrial gene COI in a patient with prostate cancer alters reactive oxygen, reactive nitrogen and proliferation. Biomed Res Int 2013;2013:239257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen PL, Chen CF, Chen Y, Guo XE, Huang CK, Shew JY, et al. Mitochondrial genome instability resulting from SUV3 haploinsufficiency leads to tumorigenesis and shortened lifespan. Oncogene 2013;32(9):1193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012;12(10):685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Mol Cell 2016;61(5):667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackburn EH. Cancer interception. Cancer Prev Res (Phila) 2011;4(6):787–92. [DOI] [PubMed] [Google Scholar]
- 9.Finn OJ. Vaccines for cancer prevention: a practical and feasible approach to the cancer epidemic. Cancer Immunol Res 2014;2(8):708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spira A, Disis ML, Schiller JT, Vilar E, Rebbeck TR, Bejar R, et al. Leveraging premalignant biology for immune-based cancer prevention. Proc Natl Acad Sci U S A 2016;113(39):10750–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livraghi L, Garber JE. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med 2015;13:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To C, Kim EH, Royce DB, Williams CR, Collins RM, Risingsong R, et al. The PARP inhibitors, veliparib and olaparib, are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev Res (Phila) 2014;7(7):698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samadder NJ, Neklason DW, Boucher KM, Byrne KR, Kanth P, Samowitz W, et al. Effect of Sulindac and Erlotinib vs Placebo on Duodenal Neoplasia in Familial Adenomatous Polyposis: A Randomized Clinical Trial. JAMA 2016;315(12):1266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benusiglio PR, Malka D, Rouleau E, De Pauw A, Buecher B, Nogues C, et al. CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: a multicentre study. J Med Genet 2013;50(7):486–9. [DOI] [PubMed] [Google Scholar]
- 15.Jochmanova I, Pacak K. Pheochromocytoma: The First Metabolic Endocrine Cancer. Clin Cancer Res 2016;22(20):5001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, He X, Ni Y, Ngeow J, Eng C. Cowden syndrome-associated germline SDHD variants alter PTEN nuclear translocation through SRC-induced PTEN oxidation. Hum Mol Genet 2015;24(1):142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 2015;373(24):2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrader KA, Cheng DT, Joseph V, Prasad M, Walsh M, Zehir A, et al. Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol 2016;2(1):104–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman N Realizing the promise of cancer predisposition genes. Nature 2014;505(7483):302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sijmons RH, Hofstra RM. Review: Clinical aspects of hereditary DNA Mismatch repair gene mutations. DNA Repair (Amst) 2016;38:155–62. [DOI] [PubMed] [Google Scholar]
- 21.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 2014;158(2):288–99. [DOI] [PubMed] [Google Scholar]
- 22.Montrose DC, Zhou XK, McNally EM, Sue E, Yantiss RK, Gross SS, et al. Celecoxib Alters the Intestinal Microbiota and Metabolome in Association with Reducing Polyp Burden. Cancer Prev Res (Phila) 2016;9(9):721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Housseau F, Wu S, Wick EC, Fan H, Wu X, Llosa NJ, et al. Redundant Innate and Adaptive Sources of IL17 Production Drive Colon Tumorigenesis. Cancer Res 2016;76(8):2115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bar F, Bochmann W, Widok A, von Medem K, Pagel R, Hirose M, et al. Mitochondrial gene polymorphisms that protect mice from colitis. Gastroenterology 2013;145(5):1055–63 e3. [DOI] [PubMed] [Google Scholar]
- 26.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 2013;144(4):799–807 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13(11):800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiffin N, Hosking FJ, Farrington SM, Palles C, Dobbins SE, Zgaga L, et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet 2014;23(17):4729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widschwendter M, Rosenthal AN, Philpott S, Rizzuto I, Fraser L, Hayward J, et al. The sex hormone system in carriers of BRCA1/2 mutations: a case-control study. Lancet Oncol 2013;14(12):1226–32. [DOI] [PubMed] [Google Scholar]
- 30.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci U S A 2007;104(14):5794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips KA, Milne RL, Rookus MA, Daly MB, Antoniou AC, Peock S, et al. Tamoxifen and risk of contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. J Clin Oncol 2013;31(25):3091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.den Boon JA, Pyeon D, Wang SS, Horswill M, Schiffman M, Sherman M, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A 2015;112(25):E3255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svoronos N, Perales-Puchalt A, Allegrezza MJ, Rutkowski MR, Payne KK, Tesone AJ, et al. Tumor Cell-Independent Estrogen Signaling Drives Disease Progression through Mobilization of Myeloid-Derived Suppressor Cells. Cancer Discov 2017;7(1):72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan E, Vaillant F, Branstetter D, Pal B, Giner G, Whitehead L, et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med 2016. [DOI] [PubMed] [Google Scholar]
- 35.Sigl V, Owusu-Boaitey K, Joshi PA, Kavirayani A, Wirnsberger G, Novatchkova M, et al. RANKL/RANK control Brca1 mutation-driven mammary tumors. Cell Res 2016;26(7):761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sau A, Lau R, Cabrita MA, Nolan E, Crooks PA, Visvader JE, et al. Persistent Activation of NF-kappaB in BRCA1-Deficient Mammary Progenitors Drives Aberrant Proliferation and Accumulation of DNA Damage. Cell Stem Cell 2016;19(1):52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mai QG, Zhang ZM, Xu S, Lu M, Zhou RP, Zhao L, et al. Metformin stimulates osteoprotegerin and reduces RANKL expression in osteoblasts and ovariectomized rats. J Cell Biochem 2011;112(10):2902–9. [DOI] [PubMed] [Google Scholar]
- 38.Galluzzi L, Buque A, Kroemer G. Prevention of breast cancer by RANKL/RANK blockade. Cell Res 2016;26(7):751–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan W, Zhang W, Strasner A, Grivennikov S, Cheng JQ, Hoffman RM, et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature 2011;470(7335):548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G, et al. LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat Med 2016;22(5):539–46. [DOI] [PubMed] [Google Scholar]
- 41.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 2015;313(13):1347–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widschwendter M, Burnell M, Fraser L, Rosenthal AN, Philpott S, Reisel D, et al. Osteoprotegerin (OPG), The Endogenous Inhibitor of Receptor Activator of NF-kappaB Ligand (RANKL), is Dysregulated in BRCA Mutation Carriers. EBioMedicine 2015;2(10):1331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnant M, Pfeiler G, Dubsky PC, Hubalek M, Greil R, Jakesz R, et al. Adjuvant denosumab in breast cancer (ABCSG-18): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet 2015;386(9992):433–43. [DOI] [PubMed] [Google Scholar]
- 44.Kotsopoulos J, Huzarski T, Gronwald J, Singer CF, Moller P, Lynch HT, et al. Bilateral Oophorectomy and Breast Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. J Natl Cancer Inst 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasen H, Ibrahim I, Ponce CG, Slater EP, Matthai E, Carrato A, et al. Benefit of Surveillance for Pancreatic Cancer in High-Risk Individuals: Outcome of Long-Term Prospective Follow-Up Studies From Three European Expert Centers. J Clin Oncol 2016;34(17):2010–9. [DOI] [PubMed] [Google Scholar]
- 46.Salo-Mullen EE, O’Reilly EM, Kelsen DP, Ashraf AM, Lowery MA, Yu KH, et al. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer 2015;121(24):4382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beatty PL, van der Geest R, Hashash JG, Kimura T, Gutkin D, Brand RE, et al. Immunobiology and immunosurveillance in patients with intraductal papillary mucinous neoplasms (IPMNs), premalignant precursors of pancreatic adenocarcinomas. Cancer Immunol Immunother 2016;65(7):771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potjer TP, Schot I, Langer P, Heverhagen JT, Wasser MN, Slater EP, et al. Variation in precursor lesions of pancreatic cancer among high-risk groups. Clin Cancer Res 2013;19(2):442–9. [DOI] [PubMed] [Google Scholar]
- 49.Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 2012;30(21):2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eser S, Messer M, Eser P, von Werder A, Seidler B, Bajbouj M, et al. In vivo diagnosis of murine pancreatic intraepithelial neoplasia and early-stage pancreatic cancer by molecular imaging. Proc Natl Acad Sci U S A 2011;108(24):9945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Li Y, Cui L, Wang B, Cui W, Li M, et al. Monitoring pancreatic carcinogenesis by the molecular imaging of cathepsin E in vivo using confocal laser endomicroscopy. PLoS One 2014;9(9):e106566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrao EM, Kettunen MI, Rodrigues TB, Dzien P, Wright AJ, Gopinathan A, et al. MRI with hyperpolarised [1–13C]pyruvate detects advanced pancreatic preneoplasia prior to invasive disease in a mouse model. Gut 2016;65(3):465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodfellow PJ, Billingsley CC, Lankes HA, Ali S, Cohn DE, Broaddus RJ, et al. Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers From GOG210: An NRG Oncology and Gynecologic Oncology Group Study. J Clin Oncol 2015;33(36):4301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW, et al. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 2015;110(2):223–62; quiz 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ait Ouakrim D, Dashti SG, Chau R, Buchanan DD, Clendenning M, Rosty C, et al. Aspirin, Ibuprofen, and the Risk of Colorectal Cancer in Lynch Syndrome. J Natl Cancer Inst 2015;107(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burn J, Gerdes AM, Macrae F, Mecklin JP, Moeslein G, Olschwang S, et al. Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet 2011;378(9809):2081–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oxnard GR, Heng JC, Root EJ, Rainville IR, Sable-Hunt AL, Shane-Carson KP, et al. Initial results of a prospective, multicenter trial to study inherited lung cancer risk associated with germline EGFR T7conf90M: INHERIT EGFR. 2015 ASCO Annual Meeting. Volume 33 (suppl; abstr 1505): J Clin Oncol; 2015. [Google Scholar]
- 59.Janne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372(18):1689–99. [DOI] [PubMed] [Google Scholar]
- 60.Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoffel EM, Fearon ER. Germline Sequence Variants and Ovarian Cancer: Known-Knowns and Known-Unknowns. JAMA Oncol 2016;2(4):491–2. [DOI] [PubMed] [Google Scholar]
- 62.Saeterdal I, Bjorheim J, Lislerud K, Gjertsen MK, Bukholm IK, Olsen OC, et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci U S A 2001;98(23):13255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22(4):813–20. [DOI] [PubMed] [Google Scholar]
- 64.Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology 2008;134(4):988–97. [DOI] [PubMed] [Google Scholar]
- 65.Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov 2017;7(1):20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500(7463):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2012;2(10):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nat Commun 2015;6:8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Connor AA, Denroche RE, Jang GH, Timms L, Kalimuthu SN, Selander I, et al. Association of Distinct Mutational Signatures With Correlates of Increased Immune Activity in Pancreatic Ductal Adenocarcinoma. JAMA Oncol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 2016;48(10):1131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smid M, Rodriguez-Gonzalez FG, Sieuwerts AM, Salgado R, Prager-Van der Smissen WJ, Vlugt-Daane MV, et al. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat Commun 2016;7:12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strickland KC, Howitt BE, Shukla SA, Rodig S, Ritterhouse LL, Liu JF, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016;7(12):13587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531(7592):47–52. [DOI] [PubMed] [Google Scholar]
- 74.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355(6322). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yurgelun MB, Chenevix-Trench G, Lippman SM. Translating Germline Cancer Risk into Precision Prevention. Cell in press [DOI] [PubMed] [Google Scholar]
- 76.Kuchenbaecker KB, Lesley McGuffog, Daniel Barrowdale, Andrew Lee, Penny Soucy, Sue Healey, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kar SP, Beesley J, Amin Al Olama A, Michailidou K, Tyrer J, Kote-Jarai Z, et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov 2016;6(9):1052–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maharry SE, Walker CJ, Liyanarachchi S, Mehta S, Patel M, Bainazar MA, et al. Dissection of the Major Hematopoietic Quantitative Trait Locus in Chromosome 6q23.3 Identifies miR-3662 as a Player in Hematopoiesis and Acute Myeloid Leukemia. Cancer Discov 2016;6(9):1036–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cescon DW, Haibe-Kains B, Mak TW. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A 2015;112(9):2841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Middlebrooks CD, Banday AR, Matsuda K, Udquim KI, Onabajo OO, Paquin A, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat Genet 2016;48(11):1330–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wen WX, Soo JS, Kwan PY, Hong E, Khang TF, Mariapun S, et al. Germline APOBEC3B deletion is associated with breast cancer risk in an Asian multi-ethnic cohort and with immune cell presentation. Breast Cancer Res 2016;18(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet 2007;80(2):273–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet 2016;48(4):374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 2013;45(4):353–61, 61e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar S, Lombard DB. Mitochondrial sirtuins and their relationships with metabolic disease and cancer. Antioxid Redox Signal 2015;22(12):1060–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen L, Ge B, Casale FP, Vasquez L, Kwan T, Garrido-Martin D, et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 2016;167(5):1398–414 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DeBalsi KL, Hoff KE, Copeland WC. Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res Rev 2017;33:89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene 2006;25(34):4647–62. [DOI] [PubMed] [Google Scholar]
- 90.Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell 2015;163(1):33–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gasparre G, Hervouet E, de Laplanche E, Demont J, Pennisi LF, Colombel M, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet 2008;17(7):986–95. [DOI] [PubMed] [Google Scholar]
- 92.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A 2005;102(3):719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Permuth-Wey J, Chen YA, Tsai YY, Chen Z, Qu X, Lancaster JM, et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidemiol Biomarkers Prev 2011;20(6):1131–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Williams SB, Ye Y, Huang M, Chang DW, Kamat AM, Pu X, et al. Mitochondrial DNA Content as Risk Factor for Bladder Cancer and Its Association with Mitochondrial DNA Polymorphisms. Cancer Prev Res (Phila) 2015;8(7):607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res 2005;65(17):8028–33. [DOI] [PubMed] [Google Scholar]
- 96.Liu VW, Wang Y, Yang HJ, Tsang PC, Ng TY, Wong LC, et al. Mitochondrial DNA variant 16189T>C is associated with susceptibility to endometrial cancer. Hum Mutat 2003;22(2):173–4. [DOI] [PubMed] [Google Scholar]
- 97.Zhai K, Chang L, Zhang Q, Liu B, Wu Y. Mitochondrial C150T polymorphism increases the risk of cervical cancer and HPV infection. Mitochondrion 2011;11(4):559–63. [DOI] [PubMed] [Google Scholar]
- 98.Lee S, Han MJ, Lee KS, Back SC, Hwang D, Kim HY, et al. Frequent occurrence of mitochondrial DNA mutations in Barrett’s metaplasia without the presence of dysplasia. PLoS One 2012;7(5):e37571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schroder T, Kucharczyk D, Bar F, Pagel R, Derer S, Jendrek ST, et al. Mitochondrial gene polymorphisms alter hepatic cellular energy metabolism and aggravate diet-induced non-alcoholic steatohepatitis. Mol Metab 2016;5(4):283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 2008;319(5865):958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin CS, Sharpley MS, Fan W, Waymire KG, Sadun AA, Carelli V, et al. Mouse mtDNA mutant model of Leber hereditary optic neuropathy. Proc Natl Acad Sci U S A 2012;109(49):20065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gasparre G, Iommarini L, Porcelli AM, Lang M, Ferri GG, Kurelac I, et al. An inherited mitochondrial DNA disruptive mutation shifts to homoplasmy in oncocytic tumor cells. Hum Mutat 2009;30(3):391–6. [DOI] [PubMed] [Google Scholar]
- 103.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008;320(5876):661–4. [DOI] [PubMed] [Google Scholar]
- 104.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 2016;13(12):691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lashinger LM, O’Flanagan CH, Dunlap SM, Rasmussen AJ, Sweeney S, Guo JY, et al. Starving cancer from the outside and inside: separate and combined effects of calorie restriction and autophagy inhibition on Ras-driven tumors. Cancer Metab 2016;4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steffen KK, Dillin A. A Ribosomal Perspective on Proteostasis and Aging. Cell Metab 2016;23(6):1004–12. [DOI] [PubMed] [Google Scholar]
- 108.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov 2015;5(4):410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, et al. Mitochondrial Genetics Regulate Breast Cancer Tumorigenicity and Metastatic Potential. Cancer Res 2015;75(20):4429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 2005;308(5730):1909–11. [DOI] [PubMed] [Google Scholar]
- 111.Goh J, Enns L, Fatemie S, Hopkins H, Morton J, Pettan-Brewer C, et al. Mitochondrial targeted catalase suppresses invasive breast cancer in mice. BMC Cancer 2011;11:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sanchez-Cabo F, Torroja C, Acin-Perez R, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 2016;535(7613):561–5. [DOI] [PubMed] [Google Scholar]
- 113.Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Wang W, et al. Foxp3 reprograms T cell metabolism to function in low glucose high lactate environments. Cell Metab (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, et al. Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab 2014;19(5):795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 2014;508(7494):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vernieri C, Casola S, Foiani M, Pietrantonio F, de Braud F, Longo V. Targeting Cancer Metabolism: Dietary and Pharmacologic Interventions. Cancer Discov 2016;6(12):1315–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dan Dunn J, Alvarez LA, Zhang X, Soldati T. Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox Biol 2015;6:472–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Picard M, Zhang J, Hancock S, Derbeneva O, Golhar R, Golik P, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci U S A 2014;111(38):E4033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frezza C, Zheng L, Folger O, Rajagopalan KN, MacKenzie ED, Jerby L, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 2011;477(7363):225–8. [DOI] [PubMed] [Google Scholar]
- 120.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012;26(12):1326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Coene ED, Hollinshead MS, Waeytens AA, Schelfhout VR, Eechaute WP, Shaw MK, et al. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell 2005;16(2):997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Henderson BR. The BRCA1 Breast Cancer Suppressor: Regulation of Transport, Dynamics, and Function at Multiple Subcellular Locations. Scientifica (Cairo) 2012;2012:796808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cuyas E, Fernandez-Arroyo S, Alarcon T, Lupu R, Joven J, Menendez JA. Germline BRCA1 mutation reprograms breast epithelial cell metabolism towards mitochondrial-dependent biosynthesis: Evidence for metformin-based “starvation” strategies in BRCA1 carriers. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cuyas E, Fernandez-Arroyo S, Joven J, Menendez JA. Metformin targets histone acetylation in cancer-prone epithelial cells. Cell Cycle 2016;15(24):3355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Blein S, Bardel C, Danjean V, McGuffog L, Healey S, Barrowdale D, et al. An original phylogenetic approach identified mitochondrial haplogroup T1a1 as inversely associated with breast cancer risk in BRCA2 mutation carriers. Breast Cancer Res 2015;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garrett WS. Cancer and the microbiota. Science 2015;348(6230):80–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature 2016;535(7610):65–74. [DOI] [PubMed] [Google Scholar]
- 128.Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut 2016;65(12):2035–44. [DOI] [PubMed] [Google Scholar]
- 129.Brunner AL, Li J, Guo X, Sweeney RT, Varma S, Zhu SX, et al. A shared transcriptional program in early breast neoplasias despite genetic and clinical distinctions. Genome Biol 2014;15(5):R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Conway C, Graham JL, Chengot P, Daly C, Chalkley R, Ross L, et al. Elucidating drivers of oral epithelial dysplasia formation and malignant transformation to cancer using RNAseq. Oncotarget 2015;6(37):40186–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marquardt JU, Seo D, Andersen JB, Gillen MC, Kim MS, Conner EA, et al. Sequential transcriptome analysis of human liver cancer indicates late stage acquisition of malignant traits. J Hepatol 2014;60(2):346–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schuyler RP, Merkel A, Raineri E, Altucci L, Vellenga E, Martens JH, et al. Distinct Trends of DNA Methylation Patterning in the Innate and Adaptive Immune Systems. Cell Rep 2016;17(8):2101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stahl M, Kohrman N, Gore SD, Kim TK, Zeidan AM, Prebet T. Epigenetics in Cancer: A Hematological Perspective. PLoS Genet 2016;12(10):e1006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stadler ZK, Vijai J, Thom P, Kirchhoff T, Hansen NA, Kauff ND, et al. Genome-wide association studies of cancer predisposition. Hematol Oncol Clin North Am 2010;24(5):973–96. [DOI] [PubMed] [Google Scholar]
- 135.Rebbeck TR, Sellers TA. The Fruits of the Genomic Revolution. Cancer Epidemiol Biomarkers Prev (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Levine DM, Ek WE, Zhang R, Liu X, Onstad L, Sather C, et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett’s esophagus. Nat Genet 2013;45(12):1487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Su Z, Gay LJ, Strange A, Palles C, Band G, Whiteman DC, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett’s esophagus. Nat Genet 2012;44(10):1131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Petridis C, Brook MN, Shah V, Kohut K, Gorman P, Caneppele M, et al. Genetic predisposition to ductal carcinoma in situ of the breast. Breast Cancer Res 2016;18(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Petljak M, Alexandrov LB. Understanding mutagenesis through delineation of mutational signatures in human cancer. Carcinogenesis 2016;37(6):531–40. [DOI] [PubMed] [Google Scholar]
- 140.Poon SL, Pang ST, McPherson JR, Yu W, Huang KK, Guan P, et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med 2013;5(197):197ra01. [DOI] [PubMed] [Google Scholar]
- 141.Meier B, Cooke SL, Weiss J, Bailly AP, Alexandrov LB, Marshall J, et al. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome Res 2014;24(10):1624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nik-Zainal S, Kucab JE, Morganella S, Glodzik D, Alexandrov LB, Arlt VM, et al. The genome as a record of environmental exposure. Mutagenesis 2015;30(6):763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013;45(9):970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 2014;7(6):1833–41. [DOI] [PubMed] [Google Scholar]
- 145.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov 2015;5(7):704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Robles-Espinoza CD, Roberts ND, Chen S, Leacy FP, Alexandrov LB, Pornputtapong N, et al. Germline MC1R status influences somatic mutation burden in melanoma. Nat Commun 2016;7:12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rashid M, Fischer A, Wilson CH, Tiffen J, Rust AG, Stevens P, et al. Adenoma development in familial adenomatous polyposis and MUTYH-associated polyposis: somatic landscape and driver genes. J Pathol 2016;238(1):98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Tiao G, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 2016;48(6):600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol 2012;132(3 Pt 2):785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yarosh D, Klein J, O’Connor A, Hawk J, Rafal E, Wolf P. Effect of topically applied T4 endonuclease V in liposomes on skin cancer in xeroderma pigmentosum: a randomised study. Xeroderma Pigmentosum Study Group. Lancet 2001;357(9260):926–9. [DOI] [PubMed] [Google Scholar]
- 151.Chen AC, Martin AJ, Choy B, Fernandez-Penas P, Dalziell RA, McKenzie CA, et al. A Phase 3 Randomized Trial of Nicotinamide for Skin-Cancer Chemoprevention. N Engl J Med 2015;373(17):1618–26. [DOI] [PubMed] [Google Scholar]
- 152.Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47(12):1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Podolskiy DI, Lobanov AV, Kryukov GV, Gladyshev VN. Analysis of cancer genomes reveals basic features of human aging and its role in cancer development. Nat Commun 2016;7:12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354(6312):618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Saini N, Roberts SA, Klimczak LJ, Chan K, Grimm SA, Dai S, et al. The Impact of Environmental and Endogenous Damage on Somatic Mutation Load in Human Skin Fibroblasts. PLoS Genet 2016;12(10):e1006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim SY, Jung SH, Kim MS, Baek IP, Lee SH, Kim TM, et al. Genomic differences between pure ductal carcinoma in situ and synchronous ductal carcinoma in situ with invasive breast cancer. Oncotarget 2015;6(10):7597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sakr RA, Schizas M, Carniello JV, Ng CK, Piscuoglio S, Giri D, et al. Targeted capture massively parallel sequencing analysis of LCIS and invasive lobular cancer: Repertoire of somatic genetic alterations and clonal relationships. Mol Oncol 2016;10(2):360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murphy SJ, Hart SN, Lima JF, Kipp BR, Klebig M, Winters JL, et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 2013;145(5):1098–109 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 2016;7:12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348(6237):880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med 2015;373(20):1926–36. [DOI] [PubMed] [Google Scholar]
- 162.Vinayanuwattikun C, Le Calvez-Kelm F, Abedi-Ardekani B, Zaridze D, Mukeria A, Voegele C, et al. Elucidating Genomic Characteristics of Lung Cancer Progression from In Situ to Invasive Adenocarcinoma. Sci Rep 2016;6:31628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O’Donovan M, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet 2015;47(9):1038–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet 2015;47(9):1047–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Weaver JM, Ross-Innes CS, Shannon N, Lynch AG, Forshew T, Barbera M, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet 2014;46(8):837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zeng J, Kelbauskas L, Rezaie A, Lee K, Ueberroth B, Gao W, et al. Transcriptional regulation by normal epithelium of premalignant to malignant progression in Barrett’s esophagus. Sci Rep 2016;6:35227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, et al. Genome-wide association studies in oesophageal adenocarcinoma and Barrett’s oesophagus: a large-scale meta-analysis. Lancet Oncol 2016;17(10):1363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Buas MF, Levine DM, Makar KW, Utsugi H, Onstad L, Li X, et al. Integrative post-genome-wide association analysis of CDKN2A and TP53 SNPs and risk of esophageal adenocarcinoma. Carcinogenesis 2014;35(12):2740–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22(1):105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stewart JB, Alaei-Mahabadi B, Sabarinathan R, Samuelsson T, Gorodkin J, Gustafsson CM, et al. Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers. PLoS Genet 2015;11(6):e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Reznik E, Wang Q, La K, Schultz N, Sander C. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, et al. The Genomic Landscape of Renal Oncocytoma Identifies a Metabolic Barrier to Tumorigenesis. Cell Rep 2015;13(9):1895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017. [DOI] [PubMed] [Google Scholar]
- 175.Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015;148(1):88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DT, Capper D, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 2016;164(5):1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tirosh I, Izar B, Prakadan S, Wadsworth M II, Treacy D, Trombetta J, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352(6284):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Martelotto LG, Baslan T, Kendall J, Geyer FC, Burke KA, Spraggon L, et al. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439(7074):353–7. [DOI] [PubMed] [Google Scholar]
- 180.Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, et al. Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity 2016;44(1):194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim JW, Botvinnik OB, Abudayyeh O, Birger C, Rosenbluh J, Shrestha Y, et al. Characterizing genomic alterations in cancer by complementary functional associations. Nat Biotechnol 2016;34(5):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2016;30(4):355–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liou GY, Doppler H, Necela B, Edenfield B, Zhang L, Dawson DW, et al. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov 2015;5(1):52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Roghanian A, Fraser C, Kleyman M, Chen J. B Cells Promote Pancreatic Tumorigenesis. Cancer Discov 2016;6(3):230–2. [DOI] [PubMed] [Google Scholar]
- 186.Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell 2015;58(4):690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373(8):726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev 2016;30(18):2021–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Teschendorff AE, Gao Y, Jones A, Ruebner M, Beckmann MW, Wachter DL, et al. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat Commun 2016;7:10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yoda Y, Takeshima H, Niwa T, Kim JG, Ando T, Kushima R, et al. Integrated analysis of cancer-related pathways affected by genetic and epigenetic alterations in gastric cancer. Gastric Cancer 2015;18(1):65–76. [DOI] [PubMed] [Google Scholar]
- 191.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 2016;30(7):733–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang Z, Wong A, Kuh D, Paul DS, Rakyan VK, Leslie RD, et al. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol 2016;17(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Siegmund KD, Marjoram P, Woo YJ, Tavare S, Shibata D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci U S A 2009;106(12):4828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012;44(11):1179–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012;483(7390):474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009;174(4):1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood 2015;126(21):2355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med 2015;372(7):601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011;20(1):25–38. [DOI] [PubMed] [Google Scholar]
- 200.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013;500(7461):222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015;525(7569):389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yue X, Trifari S, Aijo T, Tsagaratou A, Pastor WA, Zepeda-Martinez JA, et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med 2016;213(3):377–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ali MA, Matboli M, Tarek M, Reda M, Kamal KM, Nouh M, et al. Epigenetic regulation of immune checkpoints: another target for cancer immunotherapy? Immunotherapy 2017;9(1):99–108. [DOI] [PubMed] [Google Scholar]
- 204.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015;28(4):515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wulff BE, Nishikura K. Modulation of microRNA expression and function by ADARs. Curr Top Microbiol Immunol 2012;353:91–109. [DOI] [PubMed] [Google Scholar]
- 206.Permuth JB, Reid B, Earp M, Chen YA, Monteiro AN, Chen Z, et al. Inherited variants affecting RNA editing may contribute to ovarian cancer susceptibility: results from a large-scale collaboration. Oncotarget 2016;7(45):72381–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 2013;19(2):209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A 2013;110(3):1041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Qi L, Chan TH, Tenen DG, Chen L. RNA editome imbalance in hepatocellular carcinoma. Cancer Res 2014;74(5):1301–6. [DOI] [PubMed] [Google Scholar]
- 210.Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev 1997;11(3):321–33. [DOI] [PubMed] [Google Scholar]
- 211.Zipeto MA, Jiang Q, Melese E, Jamieson CH. RNA rewriting, recoding, and rewiring in human disease. Trends Mol Med 2015;21(9):549–59. [DOI] [PubMed] [Google Scholar]
- 212.O’Connell MA, Mannion NM, Keegan LP. The Epitranscriptome and Innate Immunity. PLoS Genet 2015;11(12):e1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 2016;351(6272):aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kato S, Lippman SM, Flaherty KT, Kurzrock R. The Conundrum of Genetic “Drivers” in Benign Conditions. J Natl Cancer Inst 2016;108(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol 2005;15(3):249–54. [DOI] [PubMed] [Google Scholar]
- 216.McNeal AS, Liu K, Nakhate V, Natale CA, Duperret EK, Capell BC, et al. CDKN2B Loss Promotes Progression from Benign Melanocytic Nevus to Melanoma. Cancer Discov 2015;5(10):1072–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sanchez-Danes A, Hannezo E, Larsimont JC, Liagre M, Youssef KK, Simons BD, et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Nature 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Martin SD, Coukos G, Holt RA, Nelson BH. Targeting the undruggable: immunotherapy meets personalized oncology in the genomic era. Ann Oncol 2015;26(12):2367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gjertsen MK, Gaudernack G. Mutated Ras peptides as vaccines in immunotherapy of cancer. Vox Sang 1998;74 Suppl 2:489–95. [DOI] [PubMed] [Google Scholar]
- 220.June CH. Drugging the Undruggable Ras - Immunotherapy to the Rescue? N Engl J Med 2016;375(23):2286–89. [DOI] [PubMed] [Google Scholar]
- 221.Keenan BP, Saenger Y, Kafrouni MI, Leubner A, Lauer P, Maitra A, et al. A Listeria vaccine and depletion of T-regulatory cells activate immunity against early stage pancreatic intraepithelial neoplasms and prolong survival of mice. Gastroenterology 2014;146(7):1784–94 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med 2014;211(11):2231–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Srivastava PK. Neoepitopes of Cancers: Looking Back, Looking Ahead. Cancer Immunol Res 2015;3(9):969–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang X, Sharma PK, Peter Goedegebuure S, Gillanders WE. Personalized cancer vaccines: Targeting the cancer mutanome. Vaccine 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Finn OJ, Beatty PL. Cancer immunoprevention. Curr Opin Immunol 2016;39:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Iheagwara UK, Beatty PL, Van PT, Ross TM, Minden JS, Finn OJ. Influenza virus infection elicits protective antibodies and T cells specific for host cell antigens also expressed as tumor-associated antigens: a new view of cancer immunosurveillance. Cancer Immunol Res 2014;2(3):263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Dhodapkar MV, Sexton R, Das R, Dhodapkar KM, Zhang L, Sundaram R, et al. Prospective analysis of antigen-specific immunity, stem-cell antigens, and immune checkpoints in monoclonal gammopathy. Blood 2015;126(22):2475–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 229.Riteau N, Sher A. Chitosan: An Adjuvant with an Unanticipated STING. Immunity 2016;44(3):522–4. [DOI] [PubMed] [Google Scholar]
- 230.Balkwill F, Mantovani A. Cancer and inflammation: implications for pharmacology and therapeutics. Clin Pharmacol Ther 2010;87(4):401–6. [DOI] [PubMed] [Google Scholar]
- 231.Rashidian M, Keliher EJ, Bilate AM, Duarte JN, Wojtkiewicz GR, Jacobsen JT, et al. Noninvasive imaging of immune responses. Proc Natl Acad Sci U S A 2015;112(19):6146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kimura T, McKolanis JR, Dzubinski LA, Islam K, Potter DM, Salazar AM, et al. MUC1 vaccine for individuals with advanced adenoma of the colon: a cancer immunoprevention feasibility study. Cancer Prev Res (Phila) 2013;6(1):18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Eikawa S, Nishida M, Mizukami S, Yamazaki C, Nakayama E, Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci U S A 2015;112(6):1809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528(7581):262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Incio J, Suboj P, Chin SM, Vardam-Kaur T, Liu H, Hato T, et al. Metformin Reduces Desmoplasia in Pancreatic Cancer by Reprogramming Stellate Cells and Tumor-Associated Macrophages. PLoS One 2015;10(12):e0141392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cao Y, Nishihara R, Qian ZR, Song M, Mima K, Inamura K, et al. Regular Aspirin Use Associates With Lower Risk of Colorectal Cancers With Low Numbers of Tumor-Infiltrating Lymphocytes. Gastroenterology 2016;151(5):879–92.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.McLean MH, Murray GI, Stewart KN, Norrie G, Mayer C, Hold GL, et al. The inflammatory microenvironment in colorectal neoplasia. PLoS One 2011;6(1):e15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 2016;4(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, et al. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology 2016;150(4):931–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yaeger R, Shah MA, Miller VA, Kelsen JR, Wang K, Heins ZJ, et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology 2016;151(2):278–87 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, et al. Epithelial IL-18 Equilibrium Controls Barrier Function in Colitis. Cell 2015;163(6):1444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Levy J, Cacheux W, Bara MA, L’Hermitte A, Lepage P, Fraudeau M, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol 2015;17(8):1062–73. [DOI] [PubMed] [Google Scholar]
- 243.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, et al. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe 2015;18(4):478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cunningham TJ, Tabacchi M, Eliane JP, Tuchayi SM, Manivasagam S, Mirzaalian H, et al. Randomized trial of calcipotriol combined with 5-fluorouracil for skin cancer precursor immunotherapy. J Clin Invest 2017;127(1):106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Maresso KC, Tsai KY, Brown PH, Szabo E, Lippman S, Hawk ET. Molecular cancer prevention: Current status and future directions. CA Cancer J Clin 2015;65(5):345–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. New England Journal of Medicine 2016;375(24):2369–79. [DOI] [PubMed] [Google Scholar]
- 247.Poutahidis T, Varian BJ, Levkovich T, Lakritz JR, Mirabal S, Kwok C, et al. Dietary microbes modulate transgenerational cancer risk. Cancer Res 2015;75(7):1197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Poutahidis T, Erdman SE. Commensal bacteria modulate the tumor microenvironment. Cancer Lett 2016;380(1):356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell 2016;30(1):147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153(1):17–37. [DOI] [PubMed] [Google Scholar]
- 251.Neefjes J, Ovaa H. A peptide’s perspective on antigen presentation to the immune system. Nat Chem Biol 2013;9(12):769–75. [DOI] [PubMed] [Google Scholar]
- 252.Bassani-Sternberg M, Braunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Olsen L, Johan Kudahl U, Winther O, Brusic V. Literature classification for semi-automated updating of biological knowledgebases. BMC Genomics 2013;14 Suppl 5:S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Backert L, Kohlbacher O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med 2015;7:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 2007;8:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 2015;125(9):3413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 2014;124(3):453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Giannakis M, Mu X, Shukla S, Qian Z, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Reports 2016;15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014;515(7528):577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Engin HB, Kreisberg JF, Carter H. Structure-Based Analysis Reveals Cancer Missense Mutations Target Protein Interaction Interfaces. PLoS One 2016;11(4):e0152929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wang D, Quan Y, Yan Q, Morales JE, Wetsel RA. Targeted Disruption of the beta2-Microglobulin Gene Minimizes the Immunogenicity of Human Embryonic Stem Cells. Stem Cells Transl Med 2015;4(10):1234–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Echterdiek F, Janikovits J, Staffa L, Muller M, Lahrmann B, Fruhschutz M, et al. Low density of FOXP3-positive T cells in normal colonic mucosa is related to the presence of beta2-microglobulin mutations in Lynch syndrome-associated colorectal cancer. Oncoimmunology 2016;5(2):e1075692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375(9):819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505(7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016;352(6290):aad9780. [DOI] [PubMed] [Google Scholar]
- 266.Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 2013;152(1–2):144–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Shilatifard A The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012;81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 2011;25(7):661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 1996;271(5257):1873–6. [DOI] [PubMed] [Google Scholar]
- 270.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 2010;37(3):429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 2010;10(10):721–8. [DOI] [PubMed] [Google Scholar]
- 272.Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, et al. ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia. Cancer Discov 2016;6(7):770–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Christofides A, Karantanos T, Bardhan K, Boussiotis VA. Epigenetic regulation of cancer biology and anti-tumor immunity by EZH2. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li F, et al. Oncogenic Deregulation of EZH2 as an Opportunity for Targeted Therapy in Lung Cancer. Cancer Discov 2016;6(9):1006–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med 2016;22(2):128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia 2014;28(1):44–9. [DOI] [PubMed] [Google Scholar]
- 277.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013;45(6):592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013;153(1):71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tordella L, Khan S, Hohmeyer A, Banito A, Klotz S, Raguz S, et al. SWI/SNF regulates a transcriptional program that induces senescence to prevent liver cancer. Genes Dev 2016;30(19):2187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010;18(4):316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321(5897):1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462(7274):739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 2012;483(7390):484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 2013;27(8):836–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016;27(4):599–608. [DOI] [PubMed] [Google Scholar]
- 287.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet 2013;45(11):1386–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015;47(11):1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov 2016;15(3):204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 2015;16(3):133–45. [DOI] [PubMed] [Google Scholar]
- 291.Mahata B, Zhang X, Kolodziejczyk AA, Proserpio V, Haim-Vilmovsky L, Taylor AE, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep 2014;7(4):1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366(10):883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Navin NE. Cancer genomics: one cell at a time. Genome Biol 2014;15(8):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Almendro V, Kim HJ, Cheng YK, Gonen M, Itzkovitz S, Argani P, et al. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res 2014;74(5):1338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest 2010;120(2):636–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Grundberg I, Kiflemariam S, Mignardi M, Imgenberg-Kreuz J, Edlund K, Micke P, et al. In situ mutation detection and visualization of intratumor heterogeneity for cancer research and diagnostics. Oncotarget 2013;4(12):2407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ikeda S, Takabe K, Inagaki M, Funakoshi N, Suzuki K. Detection of gene point mutation in paraffin sections using in situ loop-mediated isothermal amplification. Pathol Int 2007;57(9):594–9. [DOI] [PubMed] [Google Scholar]
- 298.Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 2015;47(10):1212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Martinez P, Timmer MR, Lau CT, Calpe S, Sancho-Serra Mdel C, Straub D, et al. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat Commun 2016;7:12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 2012;148(5):873–85. [DOI] [PubMed] [Google Scholar]
- 301.Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov 2015;5(3):316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, et al. Highly multiplexed subcellular RNA sequencing in situ. Science 2014;343(6177):1360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353(6294):78–82. [DOI] [PubMed] [Google Scholar]
- 304.Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015;348(6237):910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 2015;33(11):1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Cleary B, Brito IL, Huang K, Gevers D, Shea T, Young S, et al. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat Biotechnol 2015;33(10):1053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Geva-Zatorsky N, Alvarez D, Hudak JE, Reading NC, Erturk-Hasdemir D, Dasgupta S, et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med 2015;21(9):1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32(6):579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Grun D, van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell 2015;163(4):799–810. [DOI] [PubMed] [Google Scholar]
- 310.Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet 2014;10(1):e1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Chen X, Love JC, Navin NE, Pachter L, Stubbington MJ, Svensson V, et al. Single-cell analysis at the threshold. Nat Biotechnol 2016;34(11):1111–18. [DOI] [PubMed] [Google Scholar]
- 312.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res 2015;25(10):1499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol 2015;9(4):749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. Big Data: Astronomical or Genomical? PLoS Biol 2015;13(7):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4(6):650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4(136):136ra68. [DOI] [PubMed] [Google Scholar]
- 317.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4(162):162ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet 2013;45(1):12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8(346):346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Birkenkamp-Demtroder K, Nordentoft I, Christensen E, Hoyer S, Reinert T, Vang S, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70(1):75–82. [DOI] [PubMed] [Google Scholar]
- 322.Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun 2015;6:8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Fernandez-Cuesta L, Perdomo S, Avogbe PH, Leblay N, Delhomme TM, Gaborieau V, et al. Identification of Circulating Tumor DNA for the Early Detection of Small-cell Lung Cancer. EBioMedicine 2016;10:117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wen L, Li J, Guo H, Liu X, Zheng S, Zhang D, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015;25(11):1250–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28(5):666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Krimmel JD, Schmitt MW, Harrell MI, Agnew KJ, Kennedy SR, Emond MJ, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A 2016;113(21):6005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Beane JE, Campbell J, Moy C, Perdomo C, Schaffer M, Mazzilli S, et al. Development of the pre-cancer genome atlas (PCGA) for squamous cell lung carcinoma [abstract]. Cancer Research 2015;75(15(Suppl)):Abstract nr 2878. [Google Scholar]
- 329.Pipinikas CP, Kiropoulos TS, Teixeira VH, Brown JM, Varanou A, Falzon M, et al. Cell migration leads to spatially distinct but clonally related airway cancer precursors. Thorax 2014;69(6):548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Li X, Paulson TG, Galipeau PC, Sanchez CA, Liu K, Kuhner MK, et al. Assessment of Esophageal Adenocarcinoma Risk Using Somatic Chromosome Alterations in Longitudinal Samples in Barrett’s Esophagus. Cancer Prev Res (Phila) 2015;8(9):845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Maddipati R, Stanger BZ. Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer Discov 2015;5(10):1086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371(26):2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20(12):1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 2011;364(26):2496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014;28(2):241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Fernandez-Pol S, Ma L, Ohgami RS, Arber DA. Significance of myelodysplastic syndrome-associated somatic variants in the evaluation of patients with pancytopenia and idiopathic cytopenias of undetermined significance. Mod Pathol 2016. [DOI] [PubMed] [Google Scholar]
- 338.Cargo CA, Rowbotham N, Evans PA, Barrans SL, Bowen DT, Crouch S, et al. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood 2015;126(21):2362–5. [DOI] [PubMed] [Google Scholar]
- 339.Meyer SE, Qin T, Muench DE, Masuda K, Venkatasubramanian M, Orr E, et al. DNMT3A Haploinsufficiency Transforms FLT3ITD Myeloproliferative Disease into a Rapid, Spontaneous, and Fully Penetrant Acute Myeloid Leukemia. Cancer Discov 2016;6(5):501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Churpek JE, Pyrtel K, Kanchi KL, Shao J, Koboldt D, Miller CA, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood 2015;126(22):2484–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Lubking A, Vosberg S, Konstandin NP, Dufour A, Graf A, Krebs S, et al. Young woman with mild bone marrow dysplasia, GATA2 and ASXL1 mutation treated with allogeneic hematopoietic stem cell transplantation. Leuk Res Rep 2015;4(2):72–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Hinds DA, Barnholt KE, Mesa RA, Kiefer AK, Do CB, Eriksson N, et al. Germline variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Ogawa S Clonal hematopoiesis in acquired aplastic anemia. Blood 2016;128(3):337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.McKerrell T, Vassiliou GS. Aging as a driver of leukemogenesis. Sci Transl Med 2015;7(306):306fs38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Parikh SA, Kay NE, Shanafelt TD. Monoclonal B-cell lymphocytosis: update on diagnosis, clinical outcome, and counseling. Clin Adv Hematol Oncol 2013;11(11):720–9. [PubMed] [Google Scholar]
- 346.Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood 2011;117(6):1911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Ojha J, Secreto C, Rabe K, Ayres-Silva J, Tschumper R, Dyke DV, et al. Monoclonal B-cell lymphocytosis is characterized by mutations in CLL putative driver genes and clonal heterogeneity many years before disease progression. Leukemia 2014;28(12):2395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015;526(7574):519–24. [DOI] [PubMed] [Google Scholar]
- 349.Barrio S, Shanafelt TD, Ojha J, Chaffee KG, Secreto C, Kortum KM, et al. Genomic characterization of high-count MBL cases indicates that early detection of driver mutations and subclonal expansion are predictors of adverse clinical outcome. Leukemia 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Goldin LR, Lanasa MC, Slager SL, Cerhan JR, Vachon CM, Strom SS, et al. Common occurrence of monoclonal B-cell lymphocytosis among members of high-risk CLL families. Br J Haematol 2010;151(2):152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Oddsson A, Kristinsson SY, Helgason H, Gudbjartsson DF, Masson G, Sigurdsson A, et al. The germline sequence variant rs2736100_C in TERT associates with myeloproliferative neoplasms. Leukemia 2014;28(6):1371–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015;526(7574):525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Moreira J, Rabe KG, Cerhan JR, Kay NE, Wilson JW, Call TG, et al. Infectious complications among individuals with clinical monoclonal B-cell lymphocytosis (MBL): a cohort study of newly diagnosed cases compared to controls. Leukemia 2013;27(1):136–41. [DOI] [PubMed] [Google Scholar]
- 354.Solomon BM, Chaffee KG, Moreira J, Schwager SM, Cerhan JR, Call TG, et al. Risk of non-hematologic cancer in individuals with high-count monoclonal B-cell lymphocytosis. Leukemia 2016;30(2):331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Anderson LA, Landgren O, Engels EA. Common community acquired infections and subsequent risk of chronic lymphocytic leukaemia. Br J Haematol 2009;147(4):444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Slager SL. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukemia. Nature Communications in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Martin-Lorenzo A, Hauer J, Vicente-Duenas C, Auer F, Gonzalez-Herrero I, Garcia-Ramirez I, et al. Infection Exposure is a Causal Factor in B-cell Precursor Acute Lymphoblastic Leukemia as a Result of Pax5-Inherited Susceptibility. Cancer Discov 2015;5(12):1328–43. [DOI] [PubMed] [Google Scholar]
- 358.Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest 2008;118(7):2427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Shanafelt TD, Ramsay AG, Zent CS, Leis JF, Tun HW, Call TG, et al. Long-term repair of T-cell synapse activity in a phase II trial of chemoimmunotherapy followed by lenalidomide consolidation in previously untreated chronic lymphocytic leukemia (CLL). Blood 2013;121(20):4137–41. [DOI] [PubMed] [Google Scholar]
- 360.Henry CJ, Casas-Selves M, Kim J, Zaberezhnyy V, Aghili L, Daniel AE, et al. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J Clin Invest 2015;125(12):4666–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Dhodapkar MV, Krasovsky J, Osman K, Geller MD. Vigorous premalignancy-specific effector T cell response in the bone marrow of patients with monoclonal gammopathy. J Exp Med 2003;198(11):1753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Spisek R, Kukreja A, Chen LC, Matthews P, Mazumder A, Vesole D, et al. Frequent and specific immunity to the embryonal stem cell-associated antigen SOX2 in patients with monoclonal gammopathy. J Exp Med 2007;204(4):831–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Mateos MV, Hernandez MT, Giraldo P, de la Rubia J, de Arriba F, Corral LL, et al. Lenalidomide plus dexamethasone versus observation in patients with high-risk smouldering multiple myeloma (QuiRedex): long-term follow-up of a randomised, controlled, phase 3 trial. Lancet Oncol 2016;17(8):1127–36. [DOI] [PubMed] [Google Scholar]
- 364.Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia 2014;28(2):384–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Zhao S, Choi M, Heuck C, Mane S, Barlogie B, Lifton RP, et al. Serial exome analysis of disease progression in premalignant gammopathies. Leukemia 2014;28(7):1548–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lawson MA, McDonald MM, Kovacic N, Hua Khoo W, Terry RL, Down J, et al. Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nat Commun 2015;6:8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Das R, Strowig T, Verma R, Koduru S, Hafemann A, Hopf S, et al. Microenvironment-dependent growth of pre-neoplastic and malignant plasma cells in humanized mice. Nature Med 2016;(in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Weinhold N, Johnson DC, Rawstron AC, Forsti A, Doughty C, Vijayakrishnan J, et al. Inherited genetic susceptibility to monoclonal gammopathy of unknown significance. Blood 2014;123(16):2513–7; quiz 93. [DOI] [PubMed] [Google Scholar]
- 369.Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar MV. Clonal Immunoglobulin against Lysolipids in the Origin of Myeloma. N Engl J Med 2016;374(6):555–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Pavlova EV, Archer J, Wang S, Dekker N, Aerts JM, Karlsson S, et al. Inhibition of UDP-glucosylceramide synthase in mice prevents Gaucher disease-associated B-cell malignancy. J Pathol 2015;235(1):113–24. [DOI] [PubMed] [Google Scholar]
- 371.Bauman JE, Grandis J. Oral Cancer Chemoprevention--The End of EPOC, the Beginning of an Epoch of Molecular Selection. JAMA Oncol 2016;2(2):178–9. [DOI] [PubMed] [Google Scholar]
- 372.William WN Jr., Papadimitrakopoulou V, Lee JJ, Mao L, Cohen EE, Lin HY, et al. Erlotinib and the Risk of Oral Cancer: The Erlotinib Prevention of Oral Cancer (EPOC) Randomized Clinical Trial. JAMA Oncol 2016;2(2):209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45(10):1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158(4):929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Mazor T, Pankov A, Song JS, Costello JF. Intratumoral Heterogeneity of the Epigenome. Cancer Cell 2016;29(4):440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Dimitrova N, Gocheva V, Bhutkar A, Resnick R, Jong RM, Miller KM, et al. Stromal Expression of miR-143/145 Promotes Neoangiogenesis in Lung Cancer Development. Cancer Discov 2016;6(2):188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 2014;46:49–55. [DOI] [PubMed] [Google Scholar]
- 378.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015;521(7550):43–7. [DOI] [PubMed] [Google Scholar]
- 379.Keysselt K, Kreutzmann T, Rother K, Kerner C, Krohn K, Przybilla J, et al. Different in vivo and in vitro transformation of intestinal stem cells in mismatch repair deficiency. Oncogene 2016. [DOI] [PubMed] [Google Scholar]
- 380.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 2015;21(3):256–62. [DOI] [PubMed] [Google Scholar]
- 381.Purwada A, Jaiswal MK, Ahn H, Nojima T, Kitamura D, Gaharwar AK, et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 2015;63:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Zumwalde NA, Haag JD, Sharma D, Mirrielees JA, Wilke LG, Gould MN, et al. Analysis of Immune Cells from Human Mammary Ductal Epithelial Organoids Reveals Vdelta2+ T Cells That Efficiently Target Breast Carcinoma Cells in the Presence of Bisphosphonate. Cancer Prev Res (Phila) 2016;9(4):305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.DuPage M, Jacks T. Genetically engineered mouse models of cancer reveal new insights about the antitumor immune response. Curr Opin Immunol 2013;25(2):192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]