Abstract
As one economically important fish in the southeastern Himalayas, the giant devil catfish (Bagarius yarrelli) has been known for its extraordinarily large body size. It can grow up to 2 m, whereas the non-Bagarius sisorids only reach 10–30 cm. Another outstanding characteristic of Bagarius species is the salmonids-like reddish flesh color. Both body size and flesh color are interesting questions in science and also valuable features in aquaculture that worth of deep investigations. Bagarius species therefore are ideal materials for studying body size evolution and color depositions in fish muscles, and also potential organisms for extensive utilization in Asian freshwater aquaculture. In a combination of Illumina and PacBio sequencing technologies, we de novo assembled a 571-Mb genome for the giant devil catfish from a total of 153.4-Gb clean reads. The scaffold and contig N50 values are 3.1 and 1.6 Mb, respectively. This genome assembly was evaluated with 93.4% of Benchmarking Universal Single-Copy Orthologs completeness, 98% of transcripts coverage, and highly homologous with a chromosome-level-based genome of channel catfish (Ictalurus punctatus). We detected that 35.26% of the genome assembly is composed of repetitive elements. Employing homology, de novo, and transcriptome-based annotations, we annotated a total of 19,027 protein-coding genes for further use. In summary, we generated the first high-quality genome assembly of the giant devil catfish, which provides an important genomic resource for its future studies such as the body size and flesh color issues, and also for facilitating the conservation and utilization of this valuable catfish.
Keywords: giant devil catfish, Bagarius yarrelli, whole-genome sequencing, genome assembly, body size, flesh color
Introduction
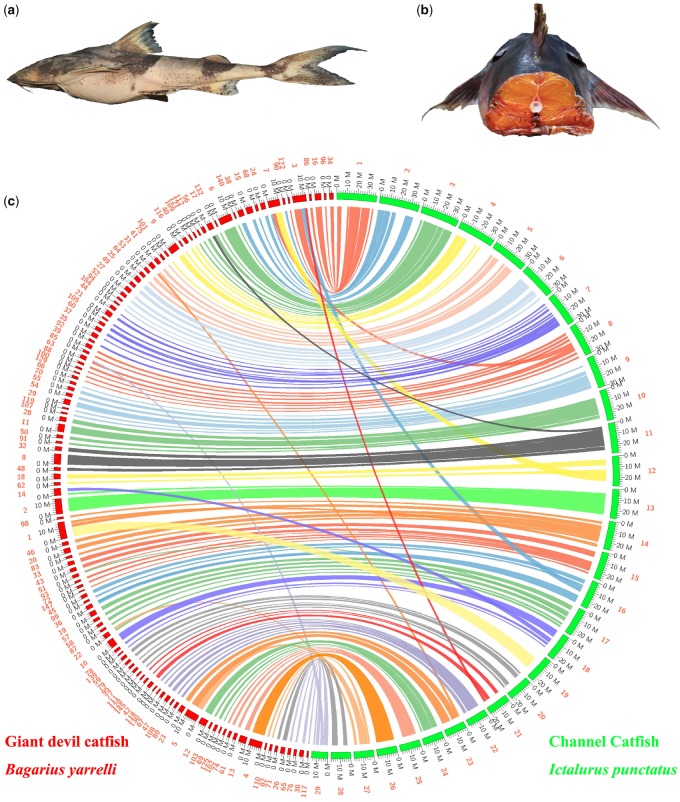
Sisoridae is a group of catfish restrictively resident in the southeastern Himalayas, with more than 200 species in 22 genera (Ng 2015). In general, the members in Sisoridae are small, with standard length usually under 30 cm, and mostly just around 10 cm. However, one exception occurs in the genus Bagarius (supplementary fig. S1, Supplementary Material online, Ng and Jiang 2015); for instance, the giant devil catfish in the Mekong River area, Bagarius yarrelli (fig. 1a), can reach lengths up to 2 m (Allan et al. 2005). The large size of Bagarius is attractive in the fish market, which has led to a decrease of the natural stocks of this genus (Allan et al. 2005). On the other hand, it has also motivated the artificial breeding and aquaculture utilization of B. yarrelli (Xue et al. 2012). The genetic basis of the large size in the genus is largely unknown (Jiang et al. 2019).
Fig. 1.
—Characterization of the giant devil catfish and its genome. (a) A lateral view of the giant devil catfish. (b) A transverse section of the giant devil catfish, demonstrating its unusual reddish flesh color. (c) The collinear relationship between the giant devil catfish (Bagarius yarrelli, assembled in this study) and channel catfish (Ictalurus punctatus, Liu et al. 2016).
Another distinct characteristic of the genus Bagarius (relative to other sisorids and also most other teleosts) is a yellowish to reddish flesh color (fig. 1b). It is similar to that seen in salmonids, and the mechanistic basis is also yet known.
To provide a genome resource for investigations of the body size and flesh color of Bagarius, for sustainable conservation and for the utilization of this economically important fish, here we present the whole-genome assembly, and gene annotation of the giant devil catfish.
Materials and Methods
Sampling, Sequencing, and Genome Size Estimation
A female B. yarrelli with a body length of 52 cm was collected from the main stream of the Lancangjiang (Upper Mekong River) in Yunnan Province of China (supplementary fig. S2, Supplementary Material online). Genomic DNA and total RNA were extracted from muscle tissues of this fish for the whole-genome and transcriptome sequencing. All animal experiments were approved by the Institutional Review Board on Bioethics and Biosafety of the Kunming Institute of Zoology, Chinese Academy of Sciences (Approval ID: 2015-SMKX025).
We employed a combination of two sequencing technologies: Illumina paired-end sequencing and Pacific Bioscience (PacBio) single-molecule real-time sequencing. Six paired-end Illumina sequence libraries, including two short-insert (500 and 800 bp) and four long-insert (2, 5, 10, and 20 kb), were constructed according to the standard protocol from Illumina (San Diego, CA). All these six DNA libraries and one cDNA library were sequenced on an Illumina HiSeq X-Ten platform. Low-quality raw reads (more than 10 Ns or rich in low-quality bases) were filtered using SOAPfilter (SOAP, Li et al. 2009) with optimized parameters (-y -p -g 1 -o clean -M 2 -f 0). The genome size was estimated using the routine 17-mer depth frequency distribution formula: genome size = Kmer number/Kmer depth, where the Kmer number is the total number of 17 k-mer, and Kmer depth indicates the peak frequency that is higher than others (Liu et al. 2013).
De Novo Genome Assembly and Assessment
We generated a de novo assembly that combined both the short reads from the Illumina sequencing and the long reads from the PacBio platform. First, we assembled contigs using these 500- and 800-bp Illumina sequencing data (around 90×) by Platanus (version 1.2.4, Kajitani et al. 2014) with optimized parameters (-k 29 -d 0.3 -t 16 -m 300). Then, we applied DBG2OLC (Ye et al. 2016) to align these contigs against the PacBio reads (about 34×) for construction of consensus contigs. Subsequently, Pilon (version 1.22, Walker et al. 2014) was employed to polish the contigs assembly (Polishing First). Based on the contig assembly, we utilized PacBio reads to construct the initial scaffolds by SSPACE-LongRead (Boetzer and Pirovano 2014). These generated scaffolds were further connected using Illumina long-insert (2, 5, 10, and 20 kb) sequencing data with SSPACE_Standard (Marten et al. 2011). The intrascaffold gaps were then filled using GapCloser (version 1.12, Li et al. 2009) and GapFiller (version 1.10, Nadalin et al. 2012). Again, we applied Pilon to polish the scaffolds after filling the gaps (Polishing Second) and generated the final scaffold assembly of B. yarrelli.
We employed both orthologous gene alignment and transcriptomic data mapping to evaluate our genome assembly. We applied Benchmarking Universal Single-Copy Ortholog (BUSCO, version 2.0, Simao et al. 2015) to align the orthologs of B. yarrelli to a reference gene set of actinopterygii_odb9 (a total of 4,584 orthologs). The transcriptome was first de novo assembled using Trinity (version 2.5.1, Haas et al. 2013) based on the data from the muscle sample, and then mapped to the assembled genome. Furthermore, to compare the scaffold-level genome of B. yarrelli in this study with a reported chromosome-level genome of the channel catfish (Ictalurus punctatus; Liu et al. 2016), we also performed a synteny analysis of these two genome assemblies using Lastz (version 1.02, Harris 2007) with only considering the reliable aligned regions more than 1 Mb in length.
Annotation of Repetitive Sequences
Two traditional methods, de novo and homology-based prediction, were employed to annotate repetitive sequences in the assembled genome. For the de novo prediction, RepeatModeller (Smit and Hubley 2008) and LTR_Finder (version 1.0.6, Xu and Wang 2007) were used to generate a local repeat reference, which was then aligned to our genome assembly for de novo prediction of repeat elements. For the homology-based prediction, the genome was primarily aligned to the RepBase (Jurka et al. 2005) using RepeatMasker (version 4.0.6, Chen 2004) and RepeatProteinMask (Chen 2004). Subsequently, the corresponding outcomes of both the de novo and homology-based predictions were integrated to construct the final nonredundant repeat annotation.
Functional Annotation of Protein-Coding Genes
Three standard strategies, including homology, de novo, and transcriptome-based annotations, were combined to predict a total gene set for B. yarrelli. 1) For the homology annotation, we aligned protein sequences from published genomes of ten vertebrates, including human (Homo sapiens, Shi et al. 2016), zebrafish (Danio rerio, Howe et al. 2013), spotted gar (Lepisosteus oculatus, Braasch et al. 2016), sea lamprey (Petromyzon marinus, Smith et al. 2013), elephant shark (Callorhinchus milii, Venkatesh et al. 2014), medaka (Oryzias latipes, Kasahara et al. 2007), Japanese puffer (Takifugu rubripes, Aparicio et al. 2002), and three recently published catfishes (channel catfish, I. punctatus, Liu et al. 2016; Chinese yellow catfish, Pelteobagrus fulvidraco, Zhang et al. 2018; and Tibetan catfish, Glyptosternum maculatum, Liu et al. 2018), against the B. yarrelli genome using TBlastN (Altschul et al. 1990) with an E-value cutoff of 10−5. Subsequently, GeneWise (version 2.2.0, Birney et al. 2004) was applied to predict the potential gene structure of each alignment. Those low-quality predictions (the predicted genes <150 bp in length) were removed. 2) For the de novo annotation, our genome assembly was first masked to exclude the repetitive elements. Then, we used Augustus (version 3.2.1, Stanke et al. 2006) and GENSCAN (version 1.0, Burge and Karlin 1997) to achieve the de novo predictions with the masked B. yarrelli genome. 3) For the transcriptome annotation, the transcriptome reads obtained after filtering were mapped onto the assembled scaffolds to identify the splice junctions by TopHat (version 2.0.13, Kim et al. 2013) and further integrated into gene structures by Cufflinks (version 2.2.1, Trapnell et al. 2012).
All the gene predictions were integrated to generate consensus gene locations and structures by GLEAN (Elsik et al. 2007). Additionally, we performed functional annotation of these predicted genes by searching several public databases, including SwissProt (Apweiler et al. 2004), Interpro (Hunter et al. 2009), and TrEMBL and KEGG (Kanehisa and Goto 2000).
Clustering of Gene Families
Protein sequences of 13 ray-finned fishes downloaded from Ensemble or NCBI, including spotted gar, zebrafish, Mexican tetra (Astyanax mexicanus), red-bellied piranha (Pygocentrus nattereri), grass carp (Ctenopharyngodon idellus), Atlantic cod (Gadus morhua), Nile tilapia (Oreochromis niloticus), medaka, Japanese puffer, Asian arowana (Scleropages formosus), and three catfishes (channel catfish, Chinese yellow catfish and Tibetan catfish), plus those of B. yarrelli in this study were used to cluster gene families. To eliminate redundant sequences of splicing variations, we retained the longest sequences from unique genomic loci. In addition, any coding sequence <150 bp were discarded from each data set as they may contain unreliable gene predictions. These retained coding sequences were matched at the amino acid level by performing all-to-all BlastP with an E-value cutoff of 10−5. The generated similarities among all the protein sequences were then applied to cluster gene families, which was implemented in OrthoMCL (Li et al. 2003) with the parameter of “-inflation 1.5.”
Phylogenetic Analysis
We carried out a phylogenetic analysis using the single-copy orthologs of the 14 species, identified from gene family clustering, in order to confirm the phylogenetic position of B. yarrelli. This concatenated data set was primarily aligned by MUSCLE (version 3.7, Edgar 2004) and then imported to reconstruct maximum likelihood (ML) tree in PhyML (version 3.0, Guindon and Gascuel 2003) and Bayes inference (BI) tree in MrBayes (version 3.2.2, Ronquist et al. 2012) with HKY85 substitution model. Spotted gar, the only nonteleost species, was used as the outgroup.
Results and Discussion
Genome Assembly and Assessment
A total of 53.9- and 79.3-Gb sequencing reads were generated from these two short-insert (500 and 800 bp) and four long-insert (2, 5, 10, and 20 kb) libraries, respectively. For the PacBio sequencing, a total of 20.2-Gb reads were obtained with an average length of 6.43 kb (supplementary table S1, Supplementary Material online). For the transcriptome sequencing, a total of 8.6-Gb raw reads were generated. For the genome estimation of B. yarrelli, the total 17 kmer number is 47,305,441,040 and the Kmer depth was 79. According to the 17-mer depth frequency distribution formula, the estimated genome size of B. yarrelli was calculated to be 599 Mb.
After the first round of polishing to the contig assembly, we obtained a primary contig assembly with a total size of 572 Mb and a contig N50 of 1.51 Mb. After the second round of polishing to the scaffold assembly, the final assembled genome size reached 571 Mb (95.3% of the estimated genome size), with a scaffold N50 of 3.1 Mb and a contig N50 up to 1.6 Mb (supplementary table S2, Supplementary Material online). The final assembled genome (571 Mb, see statistics in table 1) accounted for 95.3% of the estimated genome size (599 Mb).
Table 1.
Statistics of the Assembled Genome of Bagarius yarrelli
| Scaffold |
Contig |
|||
|---|---|---|---|---|
| Size (bp) | Number | Size (bp) | Number | |
| N90 | 577,296 | 203 | 265,790 | 410 |
| N80 | 1,256,925 | 134 | 591,605 | 266 |
| N70 | 1,848,885 | 97 | 869,246 | 189 |
| N60 | 2,397,639 | 71 | 1,267,632 | 134 |
| N50 | 3,129,371 | 50 | 1,599,318 | 94 |
| N40 | 3,833,100 | 33 | 2,151,192 | 64 |
| N30 | 4,879,671 | 26 | 2,814,748 | 40 |
| N20 | 7,417,786 | 10 | 3,309,792 | 21 |
| N10 | 12,005,657 | 7 | 5,512,941 | 171 |
| Longest | 15,823,707 | 13,429,274 | ||
| Total size | 570,806,968 | 569,629,338 | ||
| Total number (>100 bp) | 541 | 1,002 | ||
| Total number (>2,000 bp) | 541 | 1,002 | ||
| Total number (>10,000 bp) | 537 | 991 | ||
| Total length (>100 bp) | 570,806,968 | 569,629,338 | ||
| Total length (>2,000 bp) | 570,806,968 | 569,629,338 | ||
| Total length (>10,000 bp) | 570,776,638 | 569,557,739 | ||
The results of BUSCO alignment showed that our final assembly contains 4,279 complete BUSCOs (93.4%), of which 4,107 were single-copy, whereas 172 were duplicated (supplementary table S3, Supplementary Material online). Using assessment of transcripts mapping, we found over 98% of the transcripts were covered within the B. yarrelli genome regions (supplementary table S4, Supplementary Material online). Both BUSCO alignment and transcriptomic mapping suggest that our current genome assembly of B. yarrelli is characterized by high quality, completeness, and accuracy. Furthermore, based on the sequence similarities, we observed that most of the scaffolds, as well as most regions in each scaffold of B. yarrelli could align with the chromosome regions of I. punctatus (fig. 1c). These results suggest a highly similar sequence arrangement between these two catfish species and also indicate a high completeness of our B. yarrelli assembly. Interestingly, only a very few examined scaffolds of B. yarrelli (about 8%) presented multiple linear links with the chromosomes of I. punctatus (fig. 1c), suggesting a few chromosomal rearrangements may have occurred between these two catfishes.
Genome Annotation and Gene Family Clusters
We determined that 35.26% of the genome assembly is composed of repetitive elements (see more details in supplementary tables S5 and S6, Supplementary Material online). Employing homology, de novo, and transcriptome-based annotations, we predict a total of 19,027 protein-coding genes. Among them, the average gene length, average coding region length, average exon length, and average exon number are 16,505 bp, 1,724 bp, 178 bp, and 9.58, respectively (supplementary table S7, Supplementary Material online). Through matches to SwissProt, InterPro, TrEMBL, and KEGG, a total of 17,740 (93.24%) predicated genes were assigned biological functions (supplementary table S8, Supplementary Material online).
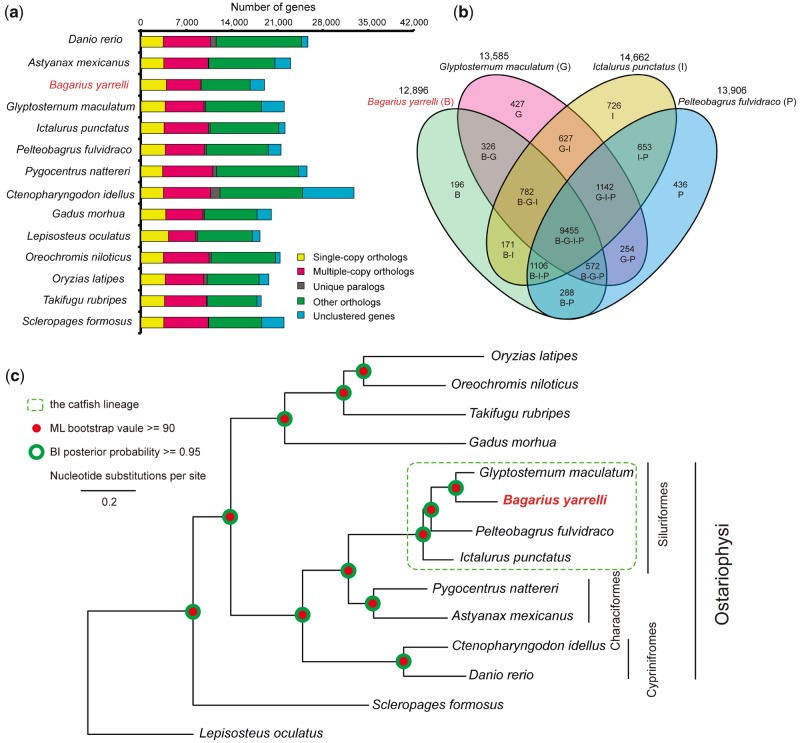
Using the protein sequences of 14 ray-finned fishes, we identified a total of 21,253 gene family clusters (supplementary table S9, Supplementary Material online). For B. yarrelli, 16,807 protein-coding genes grouped with other fishes, clustered into 12,896 gene families. The remaining 2,220 genes did not cluster with the other species. A total of 2,283 clusters were single-copy ortholog families (fig. 2a). A total of 9,455 gene families were shared among the four catfish species. Interestingly, more gene families of B. yarrelli were exclusively shared with G. maculatum (326) than with I. punctatus (171) and P. fulvidraco (288, see more details in fig. 2b).
Fig. 2.
—Gene family clustering and phylogenetic analyses. (a) Gene family clustering of Bagarius yarrelli and other 13 ray-finned fishes. (b) The numbers of clustering gene families among B. yarrelli and other three catfishes. (c) Phylogenetic relationships of B. yarrelli and other 13 ray-finned fishes.
Phylogenetic Position of B. yarrelli
The phylogenetic tree was obtained based on a supermatrix nucleotide data set with 108,820 sites. Both the ML and Bayes inference analyses revealed a consistent phylogenetic topology with robust supporting values (fig. 2c). A relationship of (Cyprinifromes, (Characiformes, Siluriformes)) was found within the Ostariophysi lineage, which corroborates the latest phylogeny of major ray-finned fish lineages based on transcriptomic and genomic data (Hughes et al. 2018). The monophyly of the four catfishes (in the Order Siluriformes) was supported. The Asian catfish species (B. yarrelli, G. maculatum, and P. fulvidraco) were monophyletic relative to the one American species (I. punctatus), which agrees with the “Big Asia” clade, proposed by Sullivan et al. (2006) based on two gene sequences. Bagariusyarrelli was sister to G. maculatum, which is consistent with current taxonomy (they are both in the Sisoridae family, Ng and Jiang, 2015), and also supported by the higher similarity in the shared gene families (as reported above, see in fig. 2b).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Yunnan Provincial Science and Technology Program (Nos. 2016AB024 and 2016FA044), National Natural Science Foundation of China (Nos. 31372182, 31672282, and U1702233), and Youth Innovation Promotion Association of the Chinese Academy of Sciences (to W.J.).
Author Contributions
W.J., Ju.Y., and Q.S. conceived the project. W.J., K.Y., X.W., X.P., Y.Z., S.L., Ji.Y., and C.S. prepared the fish samples. Yu.L., Ya.L., C.B., X.Y., J.L., and X.Z performed data analysis. W.J. and Yu.L. wrote the article. Q.S., Ju.Y., and L.C. revised the article. All authors have read and approved the final manuscript.
Data deposition: All the raw sequence data are available via GenBank under the SRA accessions SRX5413128 (for the muscle transcriptome) and SRX5406122–SRX5406135 (for the whole genome). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession VCAZ00000000 with a BioProject ID of PRJNA523489. The version assembled and described in this article is version VCAZ01000000. The annotation files have been deposited in GitHub (https://github.com/YunyunLv1987/Bagarius_yarrelli_encoding_genes).
Literature Cited
- Allan JD, et al. 2005. Overfishing of inland waters. Bioscience 55(12):1041–1050. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Aparicio S, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 397:1301–1310. [DOI] [PubMed] [Google Scholar]
- Apweiler R, et al. 2004. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32:D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W.. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15:211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 48(4):427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S.. 1997. Prediction of complete gene structures in human genomic DNA1. J Mol Biol. 268(1):78–94. [DOI] [PubMed] [Google Scholar]
- Chen N. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 5:4.10.11–14.10.14. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, et al. 2007. Creating a honey bee consensus gene set. Genome Biol. 8(1):R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignmnet of genomic DNA. State College (PA: ): Pennsylvania State University. [Google Scholar]
- Howe K, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496(7446):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes LC, et al. 2018. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc Natl Acad Sci U S A. 115(24):6249–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, et al. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res. 37(Database issue):D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Guo Y, Yang K, Shi Q, Yang J.. 2019. Insights into body size evolution: a comparative transcriptome study on three species of Asian Sisoridae catfish. Int J Mol Sci. 20(4):944.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1-4):462–467. [DOI] [PubMed] [Google Scholar]
- Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447(7145):714–719. [DOI] [PubMed] [Google Scholar]
- Kim D, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967. [DOI] [PubMed] [Google Scholar]
- Liu B, et al. 2013. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant Boil. 35:62–67. [Google Scholar]
- Liu H, et al. 2018. Draft genome of Glyptosternon maculatum, an endemic fish from Tibet Plateau. GigaScience 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. 2016. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. 7:11757.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten B, Christiaan VH, Hans JJ, Derek B, Walter P.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Nadalin F, Vezzi F, Policriti A.. 2012. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics 13(S14):S8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH. 2015. Phylogenetic systematics of the Asian catfish family Sisoridae (Actinopterygii: Siluriformes). Ichthyol Explor Fres. 26:97–157. [Google Scholar]
- Ng HH, Jiang W.. 2015. Intrafamilial relationships of the Asian hillstream catfish family Sisoridae (Teleostei: Siluriformes) inferred from nuclear and mitochondrial DNA sequences. Ichthyol Explor Fres. 26:229–240. [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, et al. 2016. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun. 7:12065.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R.. 2008. RepeatModeler Open-1.0 [cited 2018 Dec 4]. Available from: http://www.repeatmasker.org.
- Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 45(4):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JP, Lundberg JG, Hardman M.. 2006. A phylogenetic analysis of the major groups of catfishes (Teleostei: Siluriformes) using rag1 and rag2 nuclear gene sequences. Mol Phylogenet Evol. 41:632–662. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505(7482):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H.. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server issue):W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue CJ, et al. 2012. Preliminary studies on artificial propagation and embryonic development of Bagarius yarrelli. J Hydroecol. 33:54–56. [Google Scholar]
- Ye C, Hill CM, Wu S, Ruan J, Ma Z.. 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep. 6:31900.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, et al. 2018. Whole-genome sequencing of Chinese yellow catfish provides a valuable genetic resource for high-throughput identification of toxin genes. Toxins 10(12):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.