Abstract
Chromosomes located in the nucleus form discrete units of genetic material composed of DNA and protein complexes. The genetic information is encoded in linear DNA sequences, but its interpretation requires an understanding of three-dimensional (3D) structure of the chromosome, in which distant DNA sequences can be juxtaposed by highly condensed chromatin packing in the space of nucleus to precisely control gene expression. Recent technological innovations in exploring higher-order chromatin structure have uncovered organizational principles of the 3D genome and its various biological implications. Very recently, it has been reported that large-scale genomic variations may disrupt higher-order chromatin organization and as a consequence, greatly contribute to disease-specific gene regulation for a range of human diseases. Here, we review recent developments in studying the effect of structural variation in gene regulation, and the detection and the interpretation of structural variations in the context of 3D chromatin structure.
Keywords: 3D chromatin structure, gene regulation, Hi-C, structural variation, topologically associating domain
INTRODUCTION
The mammalian genome needs to be tightly packaged into the nucleus in order to fit its relatively large size to the limited space inside nucleus. Thus, there exist long-lasting questions on how the genome is organized into a three-dimensional (3D) structure to be folded into the nucleus and its consequential effect on the genome function. In the last decade, the development of chromosome conformation capture (3C) technology and its variations have revolutionized the analysis of the 3D genome organization at high-resolution compared to imaging based methods and uncovered basic principles underlying chromatin folding (Dekker et al., 2002; Dostie et al., 2006; Lieberman-Aiden et al., 2009; Rao et al., 2014; Simonis et al., 2006; Zhao et al., 2006). These ‘C’ technologies first fragmentize chromatin by restriction enzyme digestion and ligate intra-molecules to convert spatially proximal DNA fragments into a unique DNA ligation product. As a result, the ligation frequency can be an indicator of the spatial distance between two genomic loci, regardless of their linear genomic distance (Dekker et al., 2002). The 3C method detects ligation products one at a time by polymerase chain reaction (PCR) amplification using locus-specific primers as a measurement of one-to-one interactions. Systematic detection of chromatin interactions at increasing scales and resolutions was enabled by developments of various 3C-based methods in conjunction with genome-wide high-throughput approaches. Both 4C (circular chromosome conformation capture/chromosome conformation capture-on-chip) (Simonis et al., 2006; Splinter et al., 2012; van de Werken et al., 2012; Zhao et al., 2006) and 5C (3C-carbon copy) (Dostie et al., 2006) methods begin with 3C templates. The 4C methods capture one-to-all chromatin interactions by detecting the ligation frequencies between one locus (a bait region) and all other genomic loci using inverse PCR that uses primers for the bait sequence to amplify its ligation partners. In contrast to 4C, 5C uses multiplexed ligation-mediated amplification to quantify all potential interactions between the targeted genomic loci of the 3C library to detect many-to-many interactions. The development of Hi-C (high-throughput chromosome conformation capture) technology, combined with high throughput sequencing technologies and biotin mark at ligation junctions, allowed the capture of all-to-all chromatin contacts in a genome-wide and unbiased manner (Lieberman-Aiden et al., 2009; Rao et al., 2014). The unprecedented resolution and comprehensive view of 3D genome maps enabled by Hi-C has revolutionized the characterization of higher-order chromatin structure (Dixon et al., 2012; Guo et al., 2015; Jin et al., 2013; Lieberman-Aiden et al., 2009; Rao et al., 2014; 2017; Schmitt et al., 2016; Vian et al., 2018).
These ‘C’-based technological innovations not only uncover the principles of higher-order chromatin structure, but also reveal that 3D genome is tightly coupled with other nuclear processes including cellular differentiation and reprogramming (Dixon et al., 2015; Krijger et al., 2016; Siersbaek et al., 2017), DNA replication (Pope et al., 2014), and X chromosome inactivation (Crane et al., 2015; Engreitz et al., 2013; Giorgetti et al., 2016). Especially, the interplay between 3D chromatin structure and transcription has a critical role in determining the cell fate, orchestrated by multiple chromatin regulators, specific transcription factors, and long non-coding RNAs (Chen et al., 2016; Chong et al., 2018; de Wit et al., 2013; Stadhouders et al., 2018; 2019). In this aspect, recent studies highlighted the disorganization of 3D chromatin structure as a cause of aberrant gene regulation mechanisms in various human diseases (Flavahan et al., 2016; Franke et al., 2016; Hnisz et al., 2016; Lupianez et al., 2015). In addition to the epigenetic alterations, the disorganized 3D genome often involves large scale genomic variations including deletions, inversions, duplications, and translocations (Lupianez et al., 2015). Although only handful of studies have systematically investigated the relationship between large scale genomic variations and 3D chromatin structure (Chakraborty and Ay, 2017; Dixon et al., 2018; Weischenfeldt et al., 2017), the field is rapidly growing due to its significant biological implications. Here, we introduce recent studies on the basic components of the 3D genome that mediates the effect of large scale genomic variations in gene regulation and the ways in which we can precisely detect and interpret these variations in the context of 3D chromatin structure.
TOPOLOGICALLY ASSOCIATING DOMAINS, BUILDING BLOCKS OF 3D GENOME
Extensive analyses of genome-wide chromatin contact maps have uncovered that the genome is hierarchically organized into multiple layers in the nucleus, from chromatin loops that connect distant DNA fragments such as enhancer and promoter, to larger chromosomal domains known as topologically associating domains (TADs) and compartmentalized structures in the chromosome. In order to understand the effect of large-scale genomic variations on 3D chromatin structure, we first briefly discuss recent discoveries regarding the basic principles of higher-order chromatin organization.
Compartmentalization of interphase chromosomes
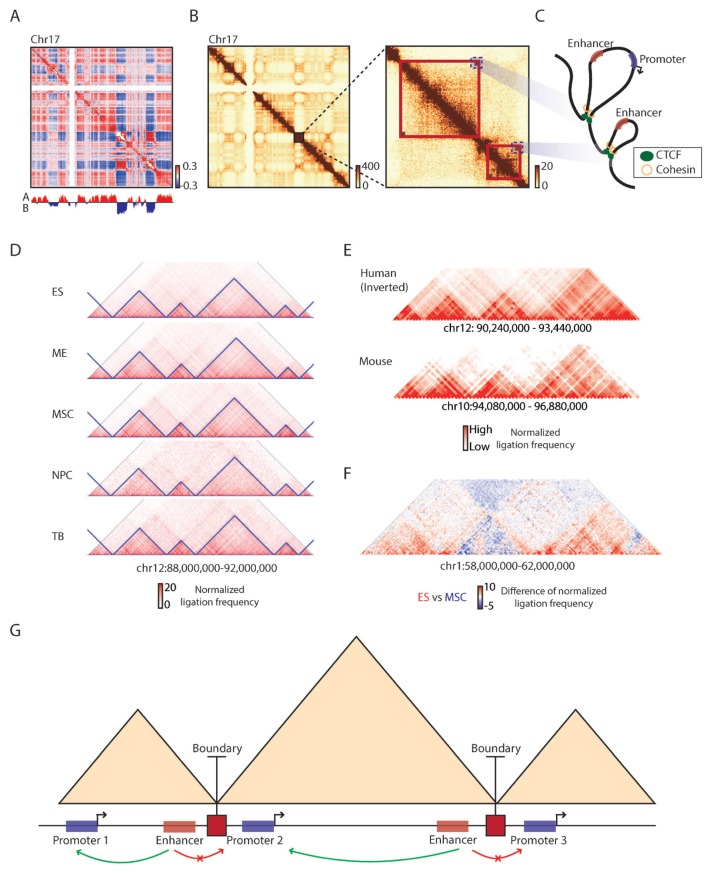
The first Hi-C study generated genome-wide chromatin contact maps at Megabase (Mb) resolution and demonstrated a plaid pattern of 3D chromatin structure of interphase nucleus in the mammalian genome (Fig. 1A), indicating that the mammalian genome is spatially compartmentalized into two parts, labelled compartments A and B (Lieberman-Aiden et al., 2009). Compartment A/B regions are of multi-Mb scale and the regions in the same compartments tend to be spatially proximal compared to the regions in different compartments (Lieberman-Aiden et al., 2009). This spatial segregation of chromatin is highly associated with various nuclear structures. For example, compartment A is often found in the interior nuclear space and euchromatin regions, while compartment B is highly concordant with nuclear lamina associated domains and heterochromatin regions (Pombo and Dillon, 2015; van Steensel and Belmont, 2017). The spatial segregation of chromatin is also dynamically reorganized during cellular differentiation and between cell-types, concordant with cell-type specific gene expression patterns (Dixon et al., 2015; Schmitt et al., 2016), suggesting a close relationship between 3D chromatin structure and gene regulation.
Fig. 1. Principles of topologically associating domains.
(A) A correlation matrix of Hi-C chromatin contact map of chromosome 17 showing plaid compartment A/B pattern (Pearson correlation coefficient, red: positive, blue: negative) with its first eigenvector (PC1 values) obtained from principal component analysis (PCA) analysis (bottom track, red: compartment A, blue: compartment B). (B) 500 kb resolution chromatin contact map of the same chromosome (left), along with a zoomed-in 40 kb resolution contact map showing structures of topologically associating domains (right, red boxes). Dark red color in contact maps indicates higher raw ligation frequency. (C) A schematic showing chromatin loops in the corresponding TAD regions (blue dotted boxes in Fig. 1B). Red boxes: enhancers, Blue boxes: promoters, Green circles: CTCF, Yellow rings: cohesin. (D) Conservation of TAD boundaries during cellular differentiation. ES: embryonic stem cells, ME: mesendoderm cells, MSC: mesenchymal stem cells, NPC: neural progenitor cells, TB: trophoblast-like cells. The blue triangles represent TADs. Red color in contact maps indicates higher normalized ligation frequency. (E) Conservation of TAD boundaries between human and mouse genomes. Red color in contact maps indicates higher normalized ligation frequency. (F) TAD-wise interaction changes observed between ES and MSC. Red color indicates higher normalized ligation frequency in the ES than the MSC and blue color indicates higher frequency in the MSC than ES. (G) A schematic showing enhancer-promoter interactions constricted by TAD boundaries (red squares). Red boxes: enhancers, Blue boxes: promoters, Green arrow arc: enhancer-promoter interactions, Red arrow arc: no enhancer-promoter interactions.
TADs
High-resolution chromatin contact maps obtained from recent Hi-C studies have revealed sub-Mb scale domains in which fragments located in the same domain have a higher number of chromatin contacts compared to the number of interactions between fragments located in different domains. These distinct units are now referred to as TADs (Fig. 1B) (Dixon et al., 2012; Nora et al., 2012). Although the mechanisms underlying TAD formation are yet to be clarified, the most promising model is the loop extrusion procedure that involves dimerization of CTCF proteins located in TAD boundaries and stabilization of loops by cohesin proteins demarcating TAD formation (Fig. 1C) (Rao et al., 2014; 2017; Sanborn et al., 2015; Schwarzer et al., 2017; Vian et al., 2018). A set of studies strongly support that TAD is a basic unit of 3D chromatin structure. First, TAD boundaries are well conserved during cellular differentiation (Dixon et al., 2015) (Fig. 1D) and even between species (Dixon et al., 2012) (Fig. 1E). Second, chromatin interactions are dynamically reorganized during cellular differentiation, albeit the interaction changes occur in a TAD-wise manner, where the pattern of either increase or decrease in chromatin contacts is similar across the fragments in the same TAD (Fig. 1F). Lastly, long-range enhancer-promoter interactions are restricted by TAD boundaries (Fig. 1G). Mammalian genomes, especially the human genome, can be uniquely characterized by the enrichment of cis-regulatory sequences in noncoding regions (Consortium, 2012; Leung et al., 2015; Roadmap Epigenomics et al., 2015), raising one fundamental question of how these noncoding regulatory sequences can regulate non-adjacent genes over large genomic distances (Claussnitzer et al., 2015; Lettice et al., 2003; Smemo et al., 2014; Uslu et al., 2014). Although enhancer-promoter interactions do not necessarily always trigger transcription and the mobility of cis-regulatory sequences within the same TAD increases upon activation (Gu et al., 2018), TADs act as a distinct unit that facilitates the communication between enhancers and their target genes, and the chromatin contacts across TADs are insulated to prevent the establishment of unwanted enhancer-promoter interactions. In this regard, disruption of TADs may perturb enhancer-promoter interactions that are involved in various human diseases as examined in many congenital malformations (Franke et al., 2016; Lupianez et al., 2015).
THE IMPACT OF STRUCTURAL VARIATIONS IN TAD ALTERATION
Since proper folding of chromatin structure is crucial in gene regulation, disruption of TAD boundaries or disorganization of inter-TAD structures can cause aberrant gene expression by exposing genes to inappropriate regulatory elements. The fact that mutations in the genes that encode structural proteins such as CTCF and cohesin have been frequently linked to human diseases and developmental abnormalities supports the notion of a close link between chromatin folding and disease development (Hnisz et al., 2016; Katainen et al., 2015). Recently, two mechanisms have been proposed on how TAD alteration contributes to abnormal gene expression (Valton and Dekker, 2016). One mechanism locally disrupts domains by deleting or dysregulating TAD boundaries, leading to fusion of two adjacent TADs (Flavahan et al., 2016; Hnisz et al., 2016). Another mechanism breaks existing TADs and the resulting TAD fragments form new combinations, creating new TADs, without directly affecting TAD boundaries (Groschel et al., 2014; Northcott et al., 2014). Large scale genomic variations known as structural variations (SV), including duplications, deletions, inversions, and translocations are often involved in TAD boundary disruptions or disorganization of inter-TAD structures.
Structural variations at TAD boundary
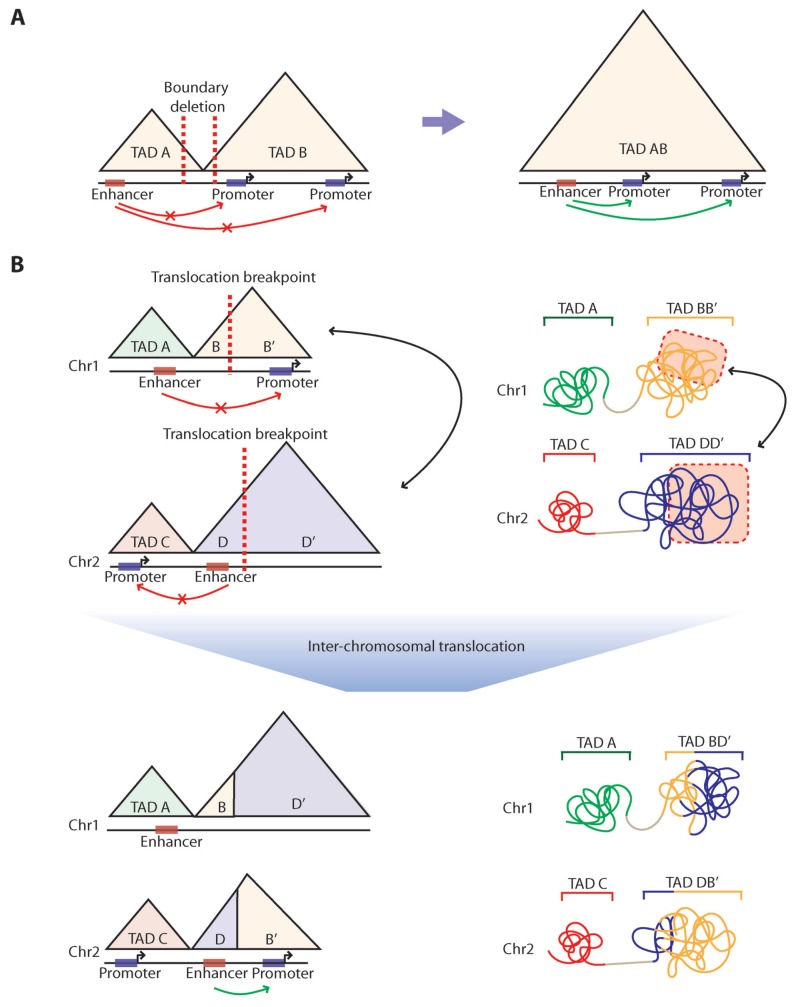
Genomic deletion spanning TAD boundaries or inhibition of structural protein binding sites can result in fusion of neighboring TADs, a phenomenon known as TAD fusion (Fig. 2A), while duplication spanning TAD boundaries can cause formation of neo-TADs (Franke et al., 2016; Hnisz et al., 2016; Lupianez et al., 2015). TAD fusion rewires the spatial distance between enhancers and promoters that were originally located in two different TADs, which can postulate abnormal gene activation. For example, recurrent deletion of the boundary sequences which results in up-regulation of prominent proto-oncogenes via TAD fusion was identified in T cell acute lymphoblastic leukemia (T-ALL) patient samples (Hnisz et al., 2016). This is an evidence for oncogene activation resulting from cis-regulatory element reorganization via TAD fusion, which is known as ‘enhancer hijacking’.
Fig. 2. Structural variations induce TAD alteration.
(A) A schematic of TAD fusion caused by boundary deletion (red dotted line) and the resulting effect of enhancer hijacking. (B) A schematic of TAD shuffling between two TADs in chromosomes 1 and 2 as shown in one-dimensional linear genomic sequence (top left) and 3D chromatin structure (top right), and the effect of enhancer hijacking after the inter-chromosomal translocation in perspective of the one-dimensional linear genomic sequence (bottom left) and 3D chromatin structure (bottom right). Red dotted lines: translocation breakpoints, Black arrow: results of shuffled sub-regions of TADs and 3D chromatin structure, Red boxes: enhancers, Blue boxes: promoters.
Inter-TAD structural variations and TAD shuffling
SVs between TADs (deletions, inversions, translocations, and duplications spanning multiple TADs) can rearrange sub-regions of TADs to create new TADs, through a phenomenon known as neo-TAD formation or TAD shuffling. For example, a balanced translocation or inversion relocates sub-regions of two TADs to one other and deletion of the region spanning multiple TADs may generate shuffled TADs by joining sub-regions of two distant TADs (Fig. 2B). Although further in-depth investigations are required to generalize the effect of TAD alteration on enhancer-promoter interactions, several studies have clearly shown that TAD shuffling caused by inter-TAD SVs can be critical in oncogenesis, as observed in acute myeloid leukemia (AML) patients where relocation of enhancer by inversion in chromosome 3 activates EVI1 proto-oncogene (Groschel et al., 2014). Another form of enhancer hijacking caused by TAD shuffling was also reported; GFI1 family oncogenes were activated in medulloblastoma, where the oncogenes show increased contacts with active enhancers caused by various somatic SV in inter-TAD regions (Northcott et al., 2014). TAD shuffling can also act as a gene repression mechanism. As shown in branchiooculofacial syndrome (BOFS), 89 Mb scale inversion on chromosome 6 leads to enhancer disconnection, altering the expression of a gene involved in pathogenesis (Laugsch et al., 2019). TAD fusion or TAD shuffling driven by SVs may provide a new insight to understand many abnormal gene expression patterns in various human diseases.
DETECTION AND INTERPRETATION OF STRUCTURAL VARIATIONS IN THE CONTEXT OF 3D GENOME
Aberrant gene expression resulted from SV-driven TAD alteration emphasizes the importance of accurate detection and interpretation of large-scale genomic rearrangements in various human diseases. However, despite the significant contributions made by the current usage of whole-genome sequencing (WGS) based approaches, they were limited in precise detection and interpretation of SVs due to sample purity, repeat sequences, transposons, complex form of genomic rearrangements, and limited coverage of sequencing depth and read length (Alkan et al., 2011; Campbell et al., 2008; Rausch et al., 2012; Tattini et al., 2015; Zhang et al., 2018). These limitations led to greater discrepancies in the detected SVs between different calling algorithms (Kosugi et al., 2019). In this aspect, additional information of the 3D architecture of the genome can be employed as a new strategy to precisely detect and interpret SVs. Indeed, recent studies have demonstrated the effectiveness of utilizing 3D chromatin contact maps in characterizing large scale SVs, particularly in cancer cell lines and primary tumor samples (Dixon et al., 2018; Harewood et al., 2017; Rickman et al., 2012).
Detection of SVs with chromatin contact map
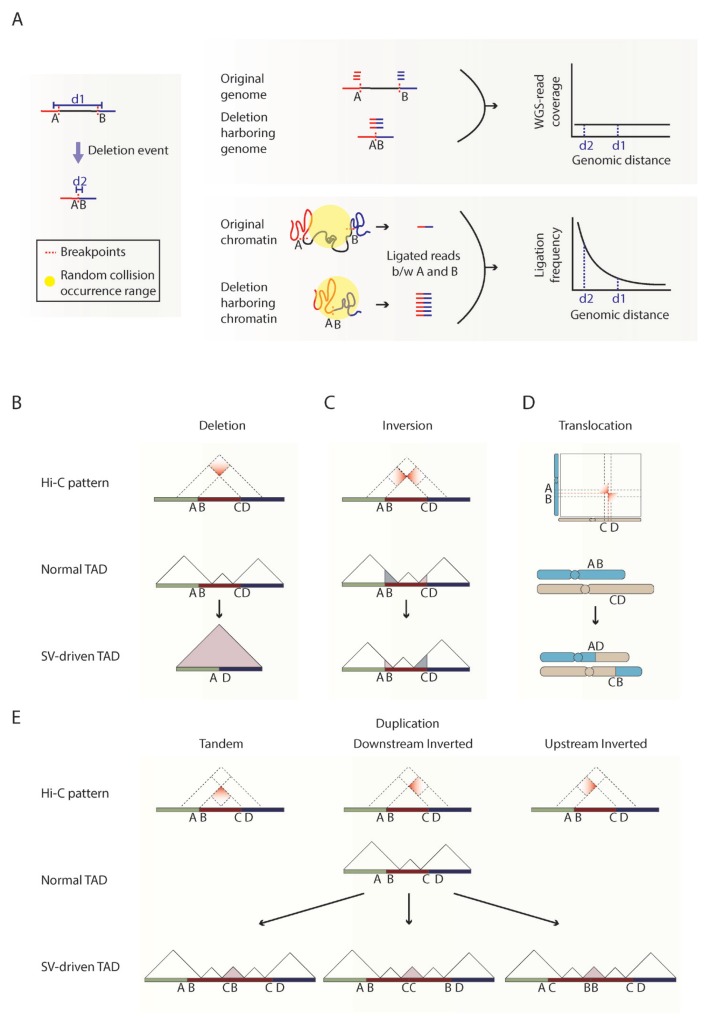
In principle, WGS methods use the number of reads spanning the breakpoint of each SV for detection. However, the number of reads supporting the SV is limited by the read coverage at the breakpoint and the fraction of clones carrying the corresponding SV. Therefore, the detection efficiency of SVs is highly affected and often limited by sequencing depth and allele frequencies (Fig. 3A top right). In contrast, in chromatin contact maps, the reads spanning the breakpoints are not the only evidence for the SV, but also the reads corresponding to ligated DNA fragments located nearby the SV breakpoint can further support the presence of the SV (Fig. 3A bottom right). Due to random collisions in the crowded nucleus, the frequencies of ligated DNA fragments exponentially decay with increasing genomic distance (Fig. 3A bottom right). Therefore, ligated reads located nearby the breakpoints of the SV are detected at an exponentially greater level due to shorter genomic distance after rearrangement of the genome by the SV (denoted ‘d2’ in Fig. 3A left and bottom right), compared to a much lower number of ligated reads in the absence of SV (denoted ‘d1’ in Fig. 3A left and bottom right). Unlike chromatin contact maps, the read coverage of WGS does not reflect any difference according to the change in genomic distance between A and B (denoted in Fig. 3A top right). Therefore, chromatin contact maps are very sensitive to the detection of such large-scale SVs regardless of the original linear genomic distances, which is a great merit for detecting SVs in low purity samples, such as human tumor tissues.
Fig. 3. Precise interpretation of structural variations in the context of 3D genome.
(A) A schematic showing large scale deletion (red dotted lines) changing the distance between two genomic regions A and B from d1 to d2 (left). WGS read coverage in original and deletion-harboring genome and the resulting plot of read coverage against genomic distance (top). A schematic showing regions A (red line) and B (blue line) in 3D genome space for original and deletion-harboring chromatin and the resulting plot of ligation frequency against genomic distance (bottom). (B–E) Categorization of expected gradient patterns in chromatin contact maps (top), normal TAD structure (middle), and the changes in TAD structure (bottom) for deletion (B), inversion (C), translocation (D), and duplications (E).
Interpretation of SV in the context of chromatin contact map
In addition to the importance of detecting SVs, it is also crucial to correctly interpret the type of SVs such as inversion, duplication, deletion, translocation, and other complex form of SVs in order to understand the effect of SVs in gene regulation. WGS, however, uses mapped orientation of reads at breakpoints to determine the type of SVs. Therefore, if the read lengths cannot cover the whole rearranged region or in case of complex SVs, the exact type of SV cannot be clearly distinguished using only mapped read orientation information (Alkan et al., 2011; Soylev et al., 2019). For example, balanced inversion and duplication events containing inverted segments are difficult to be classified solely by read mapping orientation (Soylev et al., 2019). In this regard, chromatin contact maps are highly useful, since different types of SVs produce unique chromatin contact signature (gradient pattern), where the number of chromatin interactions is the highest at the breakpoint and gradually decreases with increasing distance from the breakpoint (Figs. 3B–3E). The direction of the gradient pattern reflects the orientation of the rearranged segments and thereby indicates the type of the corresponding SV. For example, deletion produces new chromatin contact signals between upstream regions of the start breakpoint and the downstream regions of the end breakpoint, generating new chromatin contacts at the upper part of the breakpoint coordinates (Fig. 3B). Inversion produces gradient patterns at both left and right sides of the breakpoint coordinates, which is known as the ‘butterfly’ shape (Fig. 3C) (Harewood et al., 2017). In case of translocations, reciprocal and nonreciprocal translocations can be determined by the presence of either gradient patterns at both sides, similar to those of inversion, or gradient pattern only at one side, respectively (Fig. 3D). In case of duplications, the gradient patterns can be classified into three sub-types (Fig. 3E). Tandem duplications without inverted fragment produce gradient pattern at the bottom of the breakpoint coordinates. In contrast, duplication with inverted fragment produces gradient pattern at either left or right side of the breakpoint coordinates according to the position of inverted duplicated DNA fragment.
Due to its high sensitivity in detecting large-scale SVs and precise interpretation of SV orientations, several computational algorithms have been developed to identify SVs based on chromatin contact maps and combined approaches using multiple platforms including Hi-C and WGS have been proposed to improve the detection power for SVs (Chaisson et al., 2019; Dixon et al., 2018; Harewood et al., 2017; Jacobson et al., 2019; Rickman et al., 2012).
Chromosome-wide complex form of genomic rearrangement
Recent cancer genomic studies have shown superposition of multiple large-scale SVs that can affect the entire chromosome, contributing to oncogenesis or cancer progression (Notta et al., 2016). However, SV detection with WGS can include many false positive SVs due to inaccurate mapping of short sequenced reads, and thus, requires elaborate and laborious manual curation. Therefore, it is challenging to identify complex forms of SV using WGS. Here, we address this issue by describing how chromatin contact information can be used to identify and interpret complex forms of SVs.
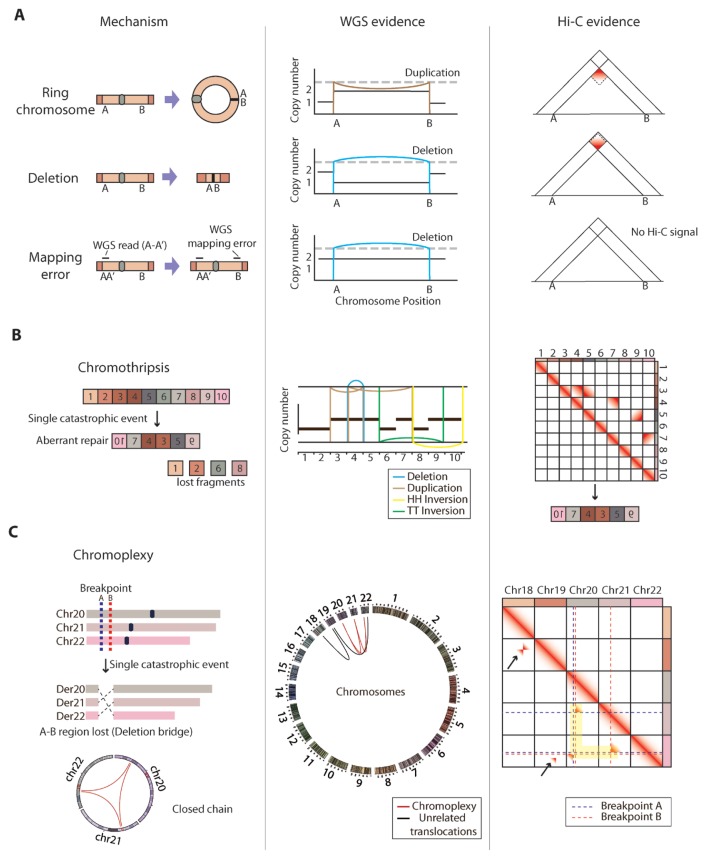
First, chromatin contact maps can provide a direct evidence for the existence of chromosome-wide SV, which can greatly reduce the number of false positive SVs. For example, the detection of paired-end reads spanning A and B regions in WGS can be the result of three possible cases (Fig. 4A left). It can be a result of a ring chromosome formation, which is recognized as an extremely large-scale duplication event. Meanwhile, it can be a result of an extremely large-scale deletion event. Lastly, it could be a simple mapping error of the paired-end reads due to the similarity between A′ and B, resulting in false positive SV calling. Indeed, the orientation of mapped reads is not sufficient to distinguish the three given cases (Fig. 4A middle), requiring additional information such as copy number variation profiles. However, chromatin contact maps show unique gradient patterns for each case (Fig. 4A right). Gradient pattern showing increased chromatin contact signals at the bottom of the breakpoint indicates an actual rearrangement of the chromosome structure, while gradient pattern at the top of the breakpoint validates the occurrence of an extremely large-scale deletion event. In case of the mapping error, we would simply not observe any gradient pattern in the chromatin contact map.
Fig. 4. Detection of complex structural variations with chromatin contact maps.
(A) Schematics showing the three possible cases generating paired-end reads spanning A and B regions in WGS: ring-shaped chromosome formation, very large deletion, and WGS mapping error (left). Shown are the corresponding DNA alteration patterns based on WGS data (middle) and gradient patterns in Hi-C chromatin interaction matrix that can distinguish the three cases (right). (B) Schematics showing the mechanism (left), WGS-based genomic rearrangement patterns (middle), and Hi-C chromatin interaction matrix with distinct gradient patterns that can be used to deduce rearranged genomic order (right) in chromothripsis. Color of the genomic rearrangement patterns (middle) indicates the types of the called SVs. (C) Schematics showing the mechanism (blue/red dotted lines are breakpoints) and circos plot showing the closed chain translocation pattern (red arc lines) of the chromoplexy event (left). Der: derivative chromosomes. A circos plot of all chromosomes shows both chromoplexy (red arc lines) and unrelated translocations (black arc lines) produced from WGS data (middle). Shown is Hi-C chromatin interaction matrix with gradient patterns aligned in breakpoint coordinate lines that can easily distinguish translocations involved in chromoplexy (highlighted in yellow) from unrelated translocations (black arrows) (left).
Second, chromatin contact maps can provide linkage information between the chromosomal segments that were shattered by a complex genomic rearrangement, allowing reconstruction of aberrant genome to understand the effect of complex forms of SVs. For example, chromothripsis is one of the most dramatic types of chromosome-wide genomic rearrangement (Stephens et al., 2011), in which a whole chromosome is shattered into multiple pieces by a single catastrophic event, followed by derivative chromosome formation via aberrant repair process of the broken fragments, resulting in massive rearrangements and loss of the fragments (Fig. 4B left). Due to the complexity of the genomic rearrangement, one-dimensional genomic alteration diagram generated by WGS data can mislead the interpretation of rearranged chromatin structure (Fig. 4B middle). However, the chromatin contact map provides linkage information between aberrantly repaired adjacent fragments, since chromothripsis is caused by a single event that generates fragments sharing no more than two breakpoints, a feature clearly seen in chromatin contact maps (Fig. 4B right). Therefore, reconstruction of derivative chromosome can be enabled by the linkage information obtained from chromatin contact maps (Burton et al., 2013), which in turn allows accurate prediction of the effect of chromothripsis in genome function.
Lastly, chromatin contact maps can also be used to intuitively detect complex forms of SVs. For example, chromoplexy is another form of dramatic chromosome-wide rearrangement where multiple chromosomes are cleaved and joined together through a single catastrophic event (Fig. 4C left) (Baca et al., 2013). The provided example shows chromoplexy between three chromosomes where the points A and B are the breakpoints. Though the feature of chromoplexy is well characterized from the distinct ‘closed-chain’ patterns in the circos plot, it is hard to recognize chromoplexy in presence of other unrelated translocation events located in close proximity (black lines in Fig. 4C middle). However, chromatin contact map generates unique signatures where the breakpoints (blue and red dotted lines in Fig. 4C right) of translocations involved in chromoplexy are aligned into a line as they share breakpoint coordinates (highlighted in yellow in Fig. 4C right), while unrelated translocations are not joined at the alignment lines (black arrows in Fig. 4C right). This unique feature can be utilized to more intuitively understand complex rearrangements of the chromosomes.
FUTURE PERSPECTIVES
SV are important in generating genomic diversity between individuals (Chaisson et al., 2019; Levy-Sakin et al., 2019) and are involved in disease-specific gene regulation mechanisms. Thus, precise detection and interpretation of large-scale genomic rearrangements are crucial, but WGS-based conventional methods are limited in solving this problem. As an alternative or supplementary approach, a set of recent studies strongly suggest that the utilization of 3D genome structure is highly efficient in detecting and interpreting large-scale SVs (Chaisson et al., 2019; Dixon et al., 2018; Harewood et al., 2017; Jacobson et al., 2019; Rickman et al., 2012). Nevertheless, several challenges remain. The first major demerit of the SV identification based on chromatin contact map is the resolution; small size SVs that do not significantly change the chromatin interactions cannot be detected in the chromatin contact map. Currently, Hi-C based chromatin contact maps are very useful in detecting large-scale SVs, but it is limited in detecting SVs within 1 Mb scale, since the gradient patterns generated by SVs are not strong enough compared to the contact frequencies between fragments in close genomic distance in the original chromatin contact maps. Further, the resolution of the chromatin contact map is generally tens of kilobases, which is too large to determine the exact breakpoint of the SVs. Development of new computational methods such as deep learning algorithms is required to precisely detect smaller SVs and compute exact breakpoints solely based on Hi-C result. Second, the inability to predict the effect of SVs in the context of 3D genome hinders the interpretation of the functional consequence of SVs. Due to the limited knowledge of non-coding regulatory regions in the genome, it is difficult to predict the pathogenicity of each SV in the context of higher-order chromatin structure. Although new bioinformatics approaches are being attempted to address this challenge (Weischenfeldt et al., 2017), the complex SVs make it more difficult to predict their effects. Thus, integration of SV-driven 3D chromatin structure and basic principles of gene regulation mechanisms is essential to predict the pathogenicity of each SV.
To conclude, indeed, there are many hurdles in applying this new strategy for detection and interpretation of SVs, but the development of new computational methods and integrative approaches will result in a very powerful tool to comprehensively understand the complex rearrangement of genome driven by SVs in both normal and disease context, beyond the one-dimensional genome.
ACKNOWLEDGMENTS
This work was funded by the Ministry of Science, ICT, and Future Planning through the National Research Foundation in Republic of Korea (2017R1C1B2008838) and Korean Ministry of Health and Welfare (HI17C0328).
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Alkan C., Coe B.P., Eichler E.E. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca S.C., Prandi D., Lawrence M.S., Mosquera J.M., Romanel A., Drier Y., Park K., Kitabayashi N., MacDonald T.Y., Ghandi M., et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P.J., Stephens P.J., Pleasance E.D., O’Meara S., Li H., Santarius T., Stebbings L.A., Leroy C., Edkins S., Hardy C., et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L., et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A., Ay F. Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics. 2017;34:338–345. doi: 10.1093/bioinformatics/btx664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.K., Blanco M., Jackson C., Aznauryan E., Ollikainen N., Surka C., Chow A., Cerase A., McDonel P., Guttman M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361 doi: 10.1126/science.aar2555. pii: eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V., et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Bouwman B.A., Zhu Y., Klous P., Splinter E., Verstegen M.J., Krijger P.H., Festuccia N., Nora E.P., Welling M., et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W., et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Xu J., Dileep V., Zhan Y., Song F., Le V.T., Yardimci G.G., Chakraborty A., Bann D.V., Wang Y., et al. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet. 2018;50:1388–1398. doi: 10.1038/s41588-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S., et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suva M.L., Bernstein B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M., Ibrahim D.M., Andrey G., Schwarzer W., Heinrich V., Schopflin R., Kraft K., Kempfer R., Jerkovic I., Chan W.L., et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- Giorgetti L., Lajoie B.R., Carter A.C., Attia M., Zhan Y., Xu J., Chen C.J., Kaplan N., Chang H.Y., Heard E., et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel S., Sanders M.A., Hoogenboezem R., de Wit E., Bouwman B.A.M., Erpelinck C., van der Velden V.H.J., Havermans M., Avellino R., van Lom K., et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Gu B., Swigut T., Spencley A., Bauer M.R., Chung M., Meyer T., Wysocka J. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science. 2018;359:1050–1055. doi: 10.1126/science.aao3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y., et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L., Kishore K., Eldridge M.D., Wingett S., Pearson D., Schoenfelder S., Collins V.P., Fraser P. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 2017;18:125. doi: 10.1186/s13059-017-1253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Weintraub A.S., Day D.S., Valton A.L., Bak R.O., Li C.H., Goldmann J., Lajoie B.R., Fan Z.P., Sigova A.A., et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson E.C., Grand R.S., Perry J.K., Vickers M.H., Olins A.L., Olins D.E., O’Sullivan J.M. Hi-C detects novel structural variants in HL-60 and HL-60/S4 cell lines. Genomics. 2019 doi: 10.1016/j.ygeno.2019.05.009. pii: S0888-7543(18)30670-0. [DOI] [PubMed] [Google Scholar]
- Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R., Dave K., Pitkanen E., Palin K., Kivioja T., Valimaki N., Gylfe A.E., Ristolainen H., Hanninen U.A., Cajuso T., et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- Kosugi S., Momozawa Y., Liu X., Terao C., Kubo M., Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger P.H., Di Stefano B., de Wit E., Limone F., van Oevelen C., de Laat W., Graf T. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugsch M., Bartusel M., Rehimi R., Alirzayeva H., Karaolidou A., Crispatzu G., Zentis P., Nikolic M., Bleckwehl T., Kolovos P., et al. Modeling the pathological long-range regulatory effects of human structural variation with patient-specific hiPSCs. Cell Stem Cell. 2019;24:736–752.e12. doi: 10.1016/j.stem.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Leung D., Jung I., Rajagopal N., Schmitt A., Selvaraj S., Lee A.Y., Yen C.A., Lin S., Lin Y., Qiu Y., et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Sakin M., Pastor S., Mostovoy Y., Li L., Leung A.K.Y., McCaffrey J., Young E., Lam E.T., Hastie A.R., Wong K.H.Y., et al. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019;10:1025. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R., et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott P.A., Lee C., Zichner T., Stutz A.M., Erkek S., Kawauchi D., Shih D.J., Hovestadt V., Zapatka M., Sturm D., et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., Chan-Seng-Yue M., Lemire M., Li Y., Wilson G.W., Connor A.A., Denroche R.E., Liang S.B., Brown A.M., Kim J.C., et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A., Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K., et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D., et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman D.S., Soong T.D., Moss B., Mosquera J.M., Dlabal J., Terry S., MacDonald T.Y., Tripodi J., Bunting K., Najfeld V., et al. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci U S A. 2012;109:9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J., et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L., et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N.A., Huber W., Haering C.H., Mirny L., et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R., Madsen J.G.S., Javierre B.M., Nielsen R., Bagge E.K., Cairns J., Wingett S.W., Traynor S., Spivakov M., Fraser P., et al. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol Cell. 2017;66:420–435.e5. doi: 10.1016/j.molcel.2017.04.010. [DOI] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gomez-Marin C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F., et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylev A., Le T., Amini H., Alkan C., Hormozdiari F. Discovery of tandem and interspersed segmental duplications using high throughput sequencing. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz237. pii: btz237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., de Wit E., van de Werken H.J., Klous P., de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Stadhouders R., Filion G.J., Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- Stadhouders R., Vidal E., Serra F., Di Stefano B., Le Dily F., Quilez J., Gomez A., Collombet S., Berenguer C., Cuartero Y., et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattini L., D’Aurizio R., Magi A. Detection of genomic structural variants from next-generation sequencing data. Front Bioeng Biotechnol. 2015;3:92. doi: 10.3389/fbioe.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslu V.V., Petretich M., Ruf S., Langenfeld K., Fonseca N.A., Marioni J.C., Spitz F. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat Genet. 2014;46:753–758. doi: 10.1038/ng.2971. [DOI] [PubMed] [Google Scholar]
- Valton A.L., Dekker J. TAD disruption as oncogenic driver. Curr Opin Genet Dev. 2016;36:34–40. doi: 10.1016/j.gde.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken H.J., Landan G., Holwerda S.J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B.A., et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Belmont A.S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L., Pekowska A., Rao S.S.P., Kieffer-Kwon K.R., Jung S., Baranello L., Huang S.C., El Khattabi L., Dose M., Pruett N., et al. The energetics and physiological impact of cohesin extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J., Dubash T., Drainas A.P., Mardin B.R., Chen Y., Stutz A.M., Waszak S.M., Bosco G., Halvorsen A.R., Raeder B., et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet. 2017;49:65–74. doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang L., Kucherlapati M., Chen F., Hadjipanayis A., Pantazi A., Bristow C.A., Lee E.A., Mahadeshwar H.S., Tang J., et al. A pan-cancer compendium of genes deregulated by somatic genomic rearrangement across more than 1,400 cases. Cell Rep. 2018;24:515–527. doi: 10.1016/j.celrep.2018.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjolinder M., Gondor A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]