Abstract
TSPAN12, a member of the tetraspanin family, has been highly connected with the pathogenesis of cancer. Its biological function, however, especially in ovarian cancer (OC), has not been well elucidated. In this study, The Cancer Genome Atlas (TCGA) dataset analysis revealed that upregulation of TSPAN12 gene expression was significantly correlated with patient survival, suggesting that TSPAN12 might be a potential prognostic marker for OC. Further exploration showed that TSPAN12 overexpression accelerated proliferation and colony formation of OVCAR3 and SKOV3 OC cells. Knockdown of TSPAN12 expression in A2780 and SKOV3 cells decreased both proliferation and colony formation. Western blot analysis showed that several cyclins and cyclin-dependent kinases (CDK) (e.g., Cyclin A2, Cyclin D1, Cyclin E2, CDK2, and CDK4) were significantly involved in the regulation of cell cycle downstream of TSPAN12. Moreover, TSPAN12 accelerated mitotic progression by controlling cell cycle. Thus, our data demonstrated that TSPAN12 could be a novel molecular target for the treatment of OC.
Keywords: cell cycle, ovarian cancer, proliferation, TSPAN12
INTRODUCTION
As a member of the tetraspanin family, Tetraspanin 12 (TSPAN12) physically binds to a large number of partners, including immunoglobulin proteins, integrins, growth factors, and other tetraspanin family members (Bailey et al., 2011; Hemler, 2003). The most broadly-known clinical significance of TSPAN12 is its association with familial exudative vitreoretinopathy (Junge et al., 2009). The correlation of tetraspanin proteins (e.g., CD151, NET-6, CD82, and CD9) and cancer progression has been frequently reported (Lafleur et al., 2009; Wang et al., 2011). These data suggest that TSPAN12 may also be correlated with carcinogenesis.
Ovarian cancer (OC) is among the top five of the most lethal gynecological malignancies (Thibault et al., 2014). Mounting evidence has revealed the clinicopathological and molecular mechanisms of OC. Multiple potential causative genes, such as TP53, BRCA1/2, BRAF, KRAS, ERBB2, CDK12, RAD50, ATM, ATR, and PI3K (Banerjee and Kaye, 2013) have been highly associated with the pathogenesis of OC, underlining the molecular heterogeneity of this disease. Much effort has been made in the clinical treatment of OC, albeit with little effect (Spinosa and Kanduc, 2013). Thus, the identification of additional factors may be needed to understand the malignancy of OC.
Cancer cells obtain a growth advantage through uncontrolled cell proliferation (Hanahan and Weinberg, 2011), which may be caused by mutations that help them adapt to the microenvironment through selective pressure. Uncontrolled cell proliferation underlies tumor progression, including tumor initiation and metastasis (Feitelson et al., 2015). Targeted therapy against uncontrolled cancer cell growth could become an effective treatment for cancer patients (Feitelson et al., 2015).
In this study, we investigated how TSPAN12 regulates OC cell proliferation both in vitro and in vivo and found that it contributed to tumor proliferation and poor prognosis in this disease through cyclin and cyclin-dependent kinase (CDK) pathways. Together, these findings suggest that TSPAN12 may be a novel target for OC therapy.
MATERIALS AND METHODS
Cell culture and reagents
Human OC cell lines SKOV3, A2780, and OVCAR3 were obtained from the American Type Culture Collection (ATCC, USA) and maintained as previously described (Li et al., 2019; Yang et al., 2019). CDK inhibitor (#S1524, AT7519) was purchased from Selleck (China).
Establishment of stable cell lines
Lenti-viral vectors (#EX-A6476-Lv203-GS) from GeneCopoeia (China) were used to overexpress TSPAN12 in OVCAR3 and SKOV3 cells according to standard protocol. TSPAN12 gene knockout was performed in A2780 and SKOV3 cells using the lentiCRISPRv2 system from Feng Zhang’s Lab (#52961, #12260, #8454; Addgene, USA) following the protocol (Sanjana et al., 2014; Shalem et al., 2014). Knockout was verified by Sanger sequencing following polymerase chain reaction (PCR) amplification and TA cloning (pMD19 Vector, #3271; Clontech, USA) using primers 5′-AACGTAGTGCACATGGGATCA-3′ (forward) and 5′-CCATACCTCATCCGGTACAGC-3′ (reverse). Specific sgRNA sequences (forward: 5′-GTGGGGATGTTAGGATATTG-3′; reverse: 5′-CAATATCCTAACATCCCCAC-3′) were designed using the online tool (http://chopchop.cbu.uib.no/) (Bailey et al., 2011; Hemler, 2003; Spinosa and Kanduc, 2013). Stable cell clones were selected with puromycin following lentiviral transduction. Overexpression and gene knockout were confirmed by real-time PCR and western blotting.
Data acquisition and analysis
Three expression profile datasets (TCGA_OV_exp_HiSeqV2_PANCAN-2015-02-24; TCGA_OV_exp_G4502A_07_ 3-2015-02-24; TCGA_OV_exp_u133a-2015-02-24) were downloaded from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/). Overall survival was evaluated by Kaplan–Meier analysis using the log-rank method after extraction of available clinical data by online Kaplan–Meier Plotter tool (http://kmplot.com/analysis/index.php?p=service&cancer=ovar).
RNA extraction and real-time PCR
Total RNA was extracted using Trizol following the manufacturer’s protocol (Invitrogen, China). cDNA was reverse transcribed with random primers using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher, China). Real-time PCR was used to quantify mRNA expression using the LightCycler® 480 SYBR Green I Master (Roche, China). β-Actin was used as the control gene. Gene specific primers are:
TSPAN12: 5′-CCAGAGAAGATTCCGTGAAGTG-3′ (forward);
5′-GTCCCTCATCCAAGCAGAAAC-3′ (reverse);
CCNA2: 5′-CGCTGGCGGTACTGAAGTC-3′ (forward);
5′-GAGGAACGGTGACATGCTCAT-3′ (reverse);
CCND1: 5′-GCTGCGAAGTGGAAACCATC-3′ (forward);
5′-CCTCCTTCTGCACACATTTGAA-3′ (reverse);
CCNE2: 5′-TCAAGACGAAGTAGCCGTTTAC-3′ (forward);
5′-TGACATCCTGGGTAGTTTTCCTC-3′ (reverse);
CDK2: 5′-CCAGGAGTTACTTCTATGCCTGA-3′ (forward);
5′-TTCATCCAGGGGAGGTACAAC-3′ (reverse);
CDK4: 5′-ATGGCTACCTCTCGATATGAGC-3′ (forward);
5′-CATTGGGGACTCTCACACTCT-3′ (reverse).
Western blotting analysis
Minced frozen tissue and cells were lysed with 100 [Symbol]ml lysis buffer (Cell Signaling Technology, USA) supplemented with proteinase inhibitors (5 mg/ml aprotinin, 1 mg/ml leupeptin, and 10 mM PMSF; Sigma, USA) and phosphatase inhibitor cocktail (Thermo Fisher Scientific, USA). Equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membrane. Blots were incubated with antibodies against TSPAN12 (1:400, anti-rabbit, Av46887; Sigma), Cyclin D1, Cyclin E2, CDK2, CDK4 (1:400, anti-rabbit, Cell Cycle Antibody Sampler kit #9932, #9870; Cell Signaling Technology), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1,000, anti-mouse, 60004-1; Proteintech, USA). Staining was detected using the SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific). GAPDH was used as control for equal protein loading.
OC xenograft mouse models
All animal experiments were conducted with five-week-old female BALB/c nude mice (NCI-Charles River, China) in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animal. The animal protocol was approved by the Institutional Review Board of Harbin Medical University (approval No. HMUIRB20170033). Eight mice were employed to establish subcutaneous xenograft experiments with cells injected on either side flank of mice. Subcutaneous tumor size was measured every three days, and mice were euthanized three weeks after injection. Tumor volume was measured by caliper and calculated as (L × W2) / 2 (mm3) (L, length; W, width). Five orthotopic mice were generated according to previous reports (Yi et al., 2014). Orthotopic mice were euthanized four week after inoculation.
Cell proliferation assay
Cells were first seeded in six-well plates. Then relative cell proliferation were visualized by crystal violet staining (0.5% w/v), and reflected by absorbance of crystal violet dissolved in dimethyl sulfoxide (DMSO), which was recorded at 490 nm with Sunrise microplate reader (Tecan, Switzerland).
Colony formation assay
Cells were seeded in six-well plates. Cell clones were fixed with ice-cold methanol, stained by crystal violet (0.5% w/v) and counted. The colony is defined by containing a minimum of 50 cells.
Cell cycle analysis
Cells were synchronized with serum-free medium for 24 h, and then harvested at time point 0 h, 4 h, 8 h, and 16 h after released in regular cell culture medium containing 10% fetal bovine serum. Then cells were fixed with ethanol and analyzed by flow cytometry with 488 nm excitation laser line following the manufacturer’s protocol of FxCycle™ PI/RNase Staining kit (Thermo Fisher, USA).
Statistical analysis
Overall survival (OS) was determined using Kaplan–Meier analysis and compared via the log-rank test. Differences between control and treatment groups were determined using the Student’s t-test. Repeated measures ANOVA was used to compare cell cycle progression differences between groups. P < 0.05 was considered statistically significant, and “n.s.” indicated a lack of statistical significance.
RESULTS
Over-expression of TSPAN12 is correlated with poor prognosis in OC patients
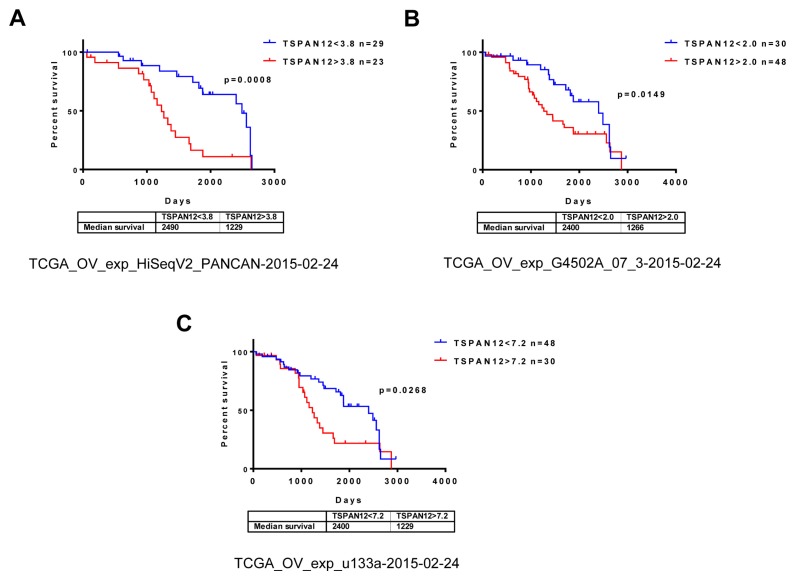
To understand the potential function of TSPAN12 in OC, we extracted the profiling data of three cohorts of human tumor specimens from TCGA including 208 OC patients (Grade 2) in total. Statistical analysis based on open-source data from TCGA database unraveled that high expression of TSPAN12 gene exacerbated patient survival (Fig. 1). These clinical data implicated that TSPAN12 gene may have important role in regulating proliferation of OC.
Fig. 1. TSPAN12 expression is significantly correlated with poor prognosis in OC patients.
(A–C) Three datasets were downloaded from TCGA database. Patients were automatically categorized into two groups with high and low expression level of TSPAN12 gene by the online software. Median survival of each group were shown in days. Overall survival of OC patients from each dataset was analyzed with log-rank method after extraction of available clinical data from online Kaplan–Meier Plotter tool. P < 0.05 was considered significant.
TSPAN12 facilitates proliferation of OC in vitro and in vivo
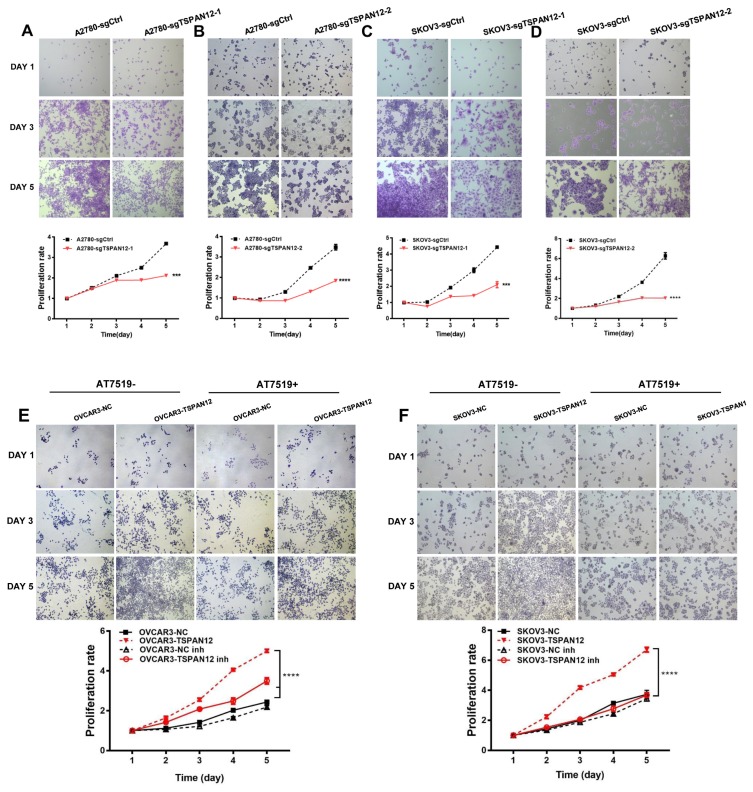
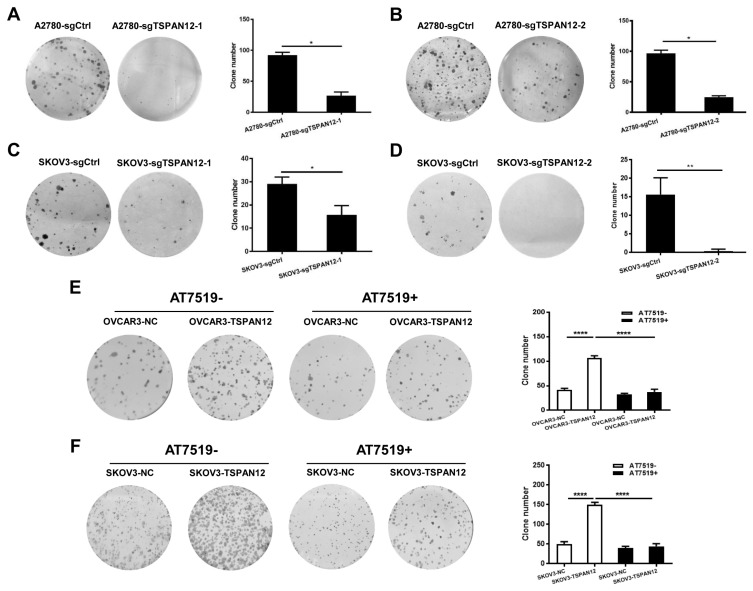
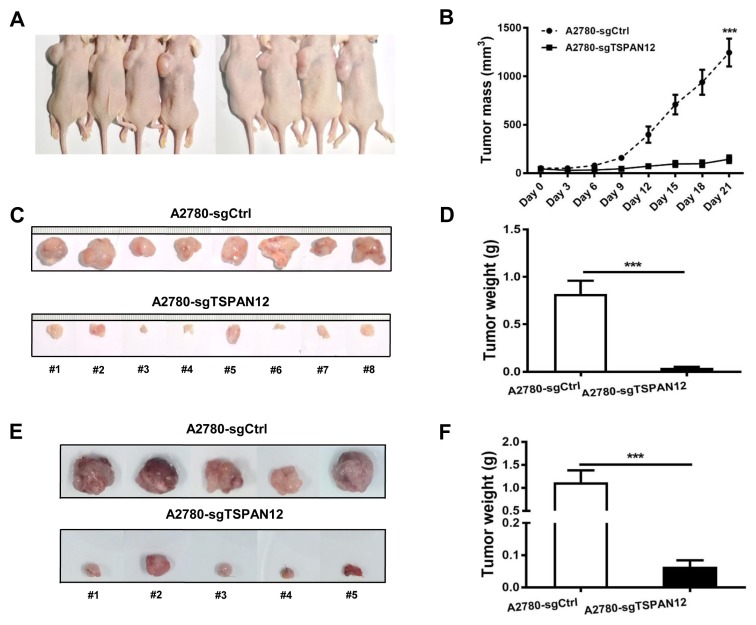
The most common characteristic of cancer cells is their unlimited growth. To understand whether TSPAN12 may involve in the proliferation of OC, we manipulated TSPAN12 gene expression in OC cell lines (Supplementary Fig. S1). In vitro experiments demonstrated that depleted expression of TSPAN12 inhibited the proliferation potential of A2780 and SKOV3 cells (Figs. 2A–2D). In contrast, exogenous expression of TSPAN12 in OVCAR3 and SKOV3 cells showed much higher cell proliferation rate than control cells (Figs. 2E and 2F). Meanwhile, the ability to form clones in A2780 and SKOV3 cells was sharply abrogated following TSPAN12 gene knockout (Figs. 3A–3D), whereas overexpression of TSPAN12 (OVCAR3-TSPAN12 and SKOV3-TSPAN12 cells) resulted in a significantly increased number of colonies compared to control cells (Figs. 3E and 3F). Furthermore, knockout of TSPAN12 in A2780-sgTSPAN12 cells retarded tumor growth both in subcutaneous (Figs. 4A–4D) and orthotopic nude mice (Figs. 4E and 4F). These results indicated that deregulation of TSPAN12 gene could exert crucial functions in cell proliferation of OC both in vitro and in vivo.
Fig. 2. TSPAN12 promotes cell proliferation in OC cells.
(A–D) Stable A2780 and SKOV3 cells with TSPAN12 knockout were seeded in 12-well plates (5 × 103 cells/well). Cells were stained by crystal violet (0.5% w/v) on day 1 to 5, and then the absorbance of crystal violet dissolved in DMSO was recorded at 490 nm. Two individual knockout clones were employed in evaluating the proliferation effect of TSPAN12 knockout in OC cells. (E and F) Overexpression of TSPAN12 on the proliferation of OC cells were examined following the above protocol after treatment with or without CDK inhibitor, AT7519 (Inh, 100 nM) for 72 h at concentration of 100 nM. ***P < 0.001; ****P < 0.0001. Results are representative of three distinct experiments.
Fig. 3. TSPAN12 promotes colony formation in OC cells.
(A–D) Stable A2780 and SKOV3 cells from two single clones with TSPAN12 knockout were seeded in 6-well plates (5 × 102 cells/well) in parallel with control group. Two weeks after inoculation, cell colonies were stained by crystal violet (0.5% w/v), and colony numbers were counted. (E and F) Colony formation assay in OVCAR3 and SKOV3 cells with TSPAN12 overexpression were performed following the above protocol (1 × 103 cells/well for inoculation) after treatment with or without CDK inhibitor, AT7519 (Inh, 100 nM) for 72 h. *P < 0.05; **P < 0.01; ****P < 0.0001. Results are representative of three independent experiments.
Fig. 4. TSPAN12 gene knockout inhibits cell growth in subcutaneous and orthotopic xenograft mouse model.
(A and C) In subcutaneous model, 5 × 106 A2780-sgCtrl cells were subcutaneously inoculated into the left flank area of five-week-old female BALB/c nude mice with A2780-sgTSPAN12 cells into the right flank (mice were numbered from #1 to #8). (B and D) Subcutaneous xenograft tumor mass and tumor weight were measured of A2780-sgTSPAN12 mice and the controls. (E) Orthotopic mice were established by surgically implantation of tumor pieces into mouse ovary after the subcutaneous tumors have reached the diameter of 1 cm. (F) Tumors in orthotopic xenograft mice were extracted and measured after euthanization. ***P < 0.001.
TSPAN12 facilitates cell proliferation by controlling cell cycle in OC cells
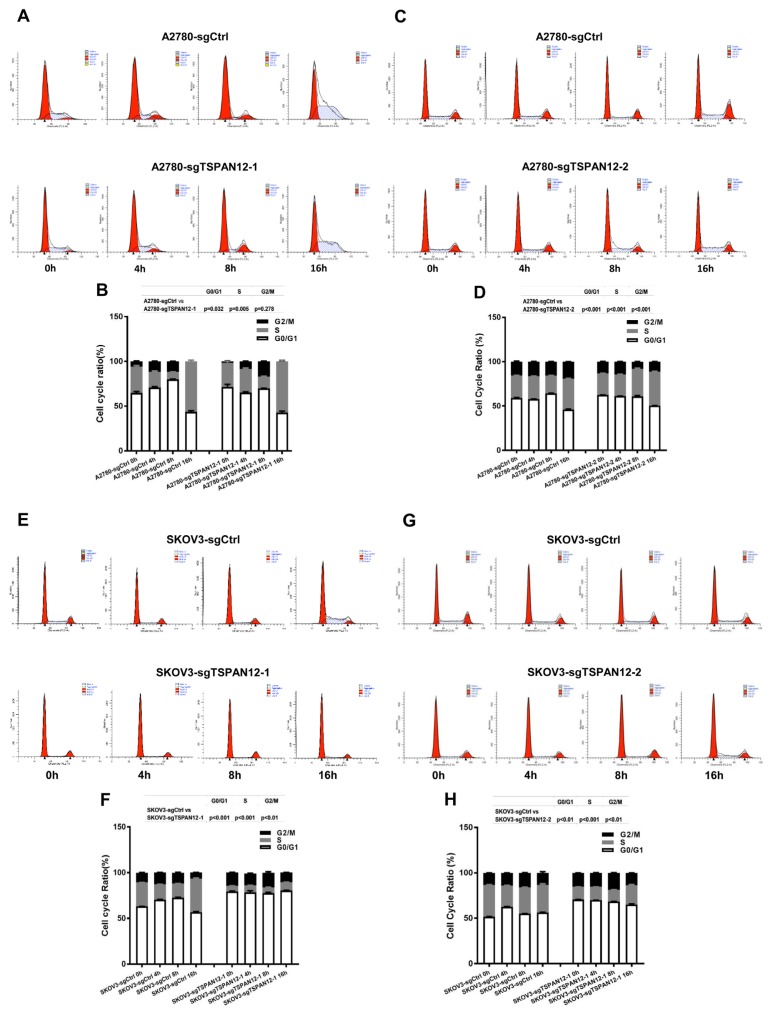
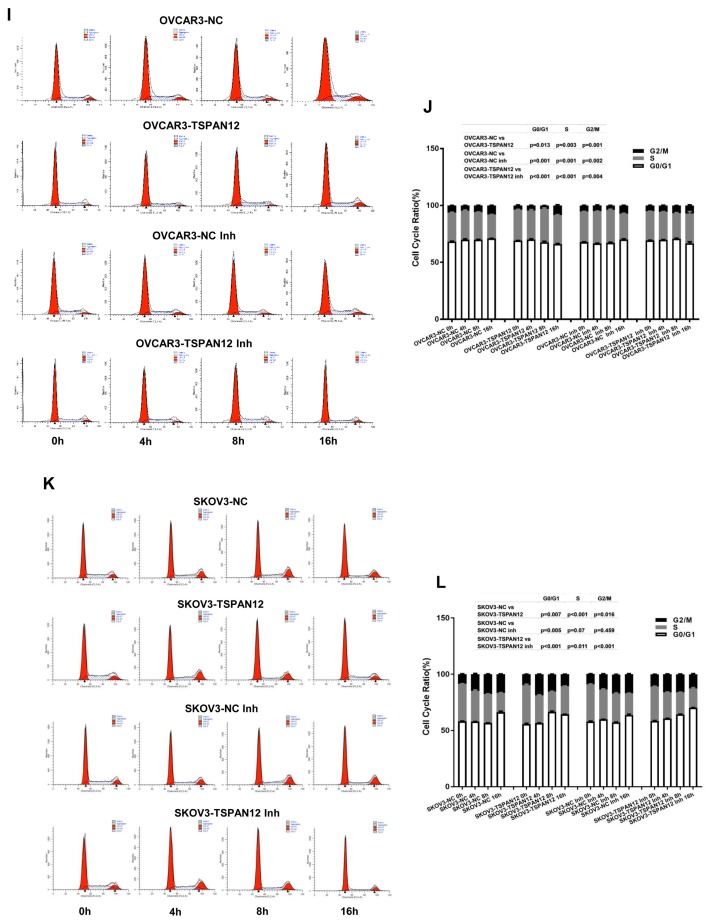
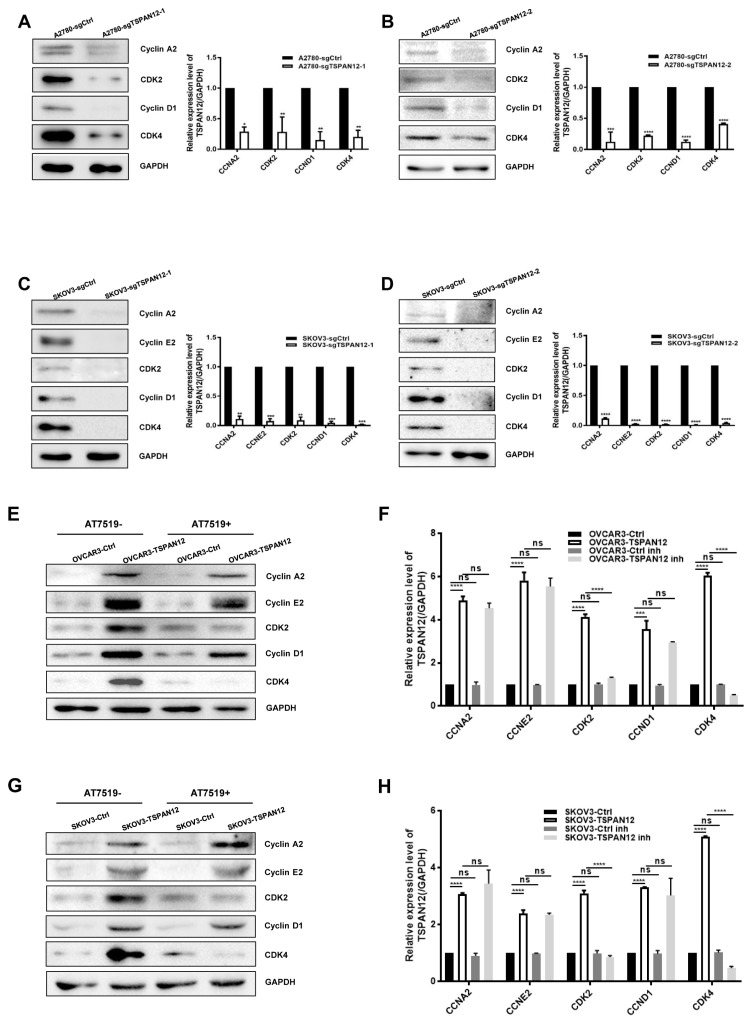
To get further understanding on how TSPAN12 gene may take its role in OC, we measured the cell cycle ratio of stable cell lines with both knockout and overexpression of TSPAN12 by propidium Iodide (PI) staining, and found that attenuation of TSPAN12 expression in A2780 and SKOV3 cells significantly slowed cell cycle progression through G1-S-G2/M (Figs. 5A–5H). Conversely, TSPAN12 overexpression accelerated the cell cycle of OVCAR3 and SKOV3 cells (Figs. 5I–5L). Western blot analysis showed that depletion of TSPAN12 suppressed the expression of these proteins (Figs. 6A–6D), and upregulation of TSPAN12 induced the expression of several cyclin and CDK proteins (Figs. 6E–6H).
Fig. 5. TSPAN12 regulates cell cycle in OC cells.
(A–H) Cells were treated with serum-free medium for 24 h and then collected at time 0 h, 4 h, 8 h, and 16 h after releasing. Cell cycle progression in A2780, SKOV3 with TSPAN12 knockout were examined by flow cytometry analysis after PI staining. (I–L) OVCAR3 and SKOV3 stable cells with TSPAN12 overexpression were first treated with or without CDK inhibitor, AT7519 (Inh, 100 nM) for 72 h (100 nM), and then evaluated by flow cytometry analysis according to the above protocol. P values between groups were listed above each bar chart. Results are representative of three independent experiments.
Fig. 6. Regulation of OC progression by TSPAN12 is highly associated with cyclin and CDK proteins.
(A–D) Stable A2780, SKOV3 cells with TSPAN12 silencing were analyzed by western blot analysis of Cyclin A2, Cyclin E2, Cyclin D1, CDK2, and CDK4. (E-H) OVCAR3 and SKOV3 cells with TSPAN12 overexpression were analyzed by western blot analysis after being treated with or without CDK inhibitor, AT7519 (Inh, 100 nM) for 72 h. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are presented as the mean ± standard deviation (SD) of three independent experiments.
To further confirm that TSPAN12 regulated cell proliferation by controlling the cell cycle, we suppressed CDK activity with AT7519, a specific CDK2 and CDK4 inhibitor (Figs. 6E–6F). We found that cell proliferation (Figs. 2E and 2F) and colony formation (Figs. 3E and 3F) was significantly reversed in the presence of this inhibitor. Meanwhile, cell cycle progression was also compromised following AT7519 treatment in the context of TSPAN12 overexpression (Figs. 5I–5L). These data suggested that TSPAN12 may contribute to cell proliferation by controlling the cell cycle through cyclins and CDKs regulation, which was further supported at the mRNA level by data from the TCGA database analysis (Supplementary Fig. S2) and stable cell lines (Supplementary Figs. S3A–S3E) that showed a positive correlation between TSPAN12 and cyclins or CDKs. Examination of cyclins and CDKs in xenograft tissues at mRNA and protein levels did not demonstrate any universal positive correlations except between CDK2 and TSPAN12 at the mRNA level (Supplementary Figs. S4–S6). These findings have provided preliminary information on the mechanism by which TSPAN12 modulates OC proliferation.
DISCUSSION
In our investigation of the clinical relevance of TSPAN12 in OC, we found that high expression of TSPAN12 was significantly correlated with poor prognosis in this disease. Thus, we hypothesized that TSPAN12 might be an oncogene in OC development.
Retrospective analysis of previous studies demonstrated that TSPAN12 was associated with the progression of breast, lung, and colon cancer (Knoblich et al., 2013; Lafleur et al., 2009; Liu et al., 2017; Otomo et al., 2014; Wang et al., 2011). Downregulation of TSPAN12 expression in human MDA-MB-231 breast cancer cells significantly decreased primary tumor growth, increased tumor apoptosis, and inhibited metastasis (Knoblich et al., 2013). In addition, silencing of TSPAN12 inhibited the growth of non-small cell lung carcinoma cells both in vitro and in vivo (Hu et al., 2018). Moreover, upregulation of TSPAN12 correlated with poor prognosis, late pathologic stage, and chemo-resistance in small cell lung carcinoma (Ye et al., 2017). However, the function of TSPAN12 may not be unilateral but might be tissue-dependent (Knoblich et al., 2013; Ye et al., 2017). For this reason, we evaluated the molecular function of TSPAN12 in OC cells. Based on our findings, we speculate that high expression of TSPAN12 in OC cells provides a strong proliferative advantage, which extends cancer cell survival beyond its normal life span.
In the current study, we performed mechanistic studies to determine how TSPAN12 may regulate the proliferation of OC cells. We identified cell cycle control as the mechanism by which TSPAN12 could contribute to OC progression. Cell cycle control is regulated by a panel of enzymes (i.e., cyclins and CDKs) to maintain accurate DNA replication and chromosome segregation (Bendris et al., 2015). It is widely accepted that cyclin D1 leads to progression through the G1 phase of the cell cycle by activating CDK4 in multiple cancer types (Ewen and Lamb, 2004); cyclin E2-CDK2 is implicated in the transition of human cells from G1 to S (Gudas et al., 1999; Payton and Coats, 2002). Furthermore, cyclin A2 regulates cell cycle progression by interacting with CDK2 during S phase and the G2/M transition (Blanchard, 2000; Pagano et al., 1992). The results of our study indicated that TSPAN12 could trigger the upregulation of a broad spectrum of cyclin and CDK proteins, which promoted cell cycle progression. These results were supported by correlation studies at the mRNA level from tissue samples in the TCGA database and cell lines established in this study. However, we did not find a positive correlation between the cyclins or CDKs and TSPAN12 in xenograft tumors, which may be due to the limited sample size of the xenograft tumors. Therefore, in this study, cyclin and CDK proteins were identified as potential targets for TSPAN12, which have not been evaluated in previous studies focusing on the role of TSPAN12 in carcinogenesis.
Our data demonstrated that TSPAN12 could induce cell proliferation in OC cells, making it a potential therapeutic target in OC. How the interplay between TSPAN12 and cell cycle proteins regulate OC development requires further exploration.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation of China (No. 81602276), foundation of Health Commission of Heilongjiang Province (No. 2016-162), the Program for Changjiang Scholars and Innovative Research Team in University of China (IRT1230). Part of the results are based upon data in the TCGA (https://www.cancer.gov/tcga).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Bailey R.L., Herbert J.M., Khan K., Heath V.L., Bicknell R., Tomlinson M.G. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans. 2011;39:1667–1673. doi: 10.1042/BST20110745. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Kaye S.B. New strategies in the treatment of ovarian cancer: current clinical perspectives and future potential. Clin Cancer Res. 2013;19:961–968. doi: 10.1158/1078-0432.CCR-12-2243. [DOI] [PubMed] [Google Scholar]
- Bendris N., Lemmers B., Blanchard J.M. Cell cycle, cytoskeleton dynamics and beyond: the many functions of cyclins and CDK inhibitors. Cell Cycle. 2015;14:1786–1798. doi: 10.1080/15384101.2014.998085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J.M. Cyclin A2 transcriptional regulation: modulation of cell cycle control at the G1/S transition by peripheral cues. Biochem Pharmacol. 2000;60:1179–1184. doi: 10.1016/S0006-2952(00)00384-1. [DOI] [PubMed] [Google Scholar]
- Ewen M.E., Lamb J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol Med. 2004;10:158–162. doi: 10.1016/j.molmed.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Feitelson M.A., Arzumanyan A., Kulathinal R.J., Blain S.W., Holcombe R.F., Mahajna J., Marino M., Martinez-Chantar M.L., Nawroth R., Sanchez-Garcia I., et al. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin Cancer Biol. 2015;35(Suppl):S25–S54. doi: 10.1016/j.semcancer.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudas J.M., Payton M., Thukral S., Chen E., Bass M., Robinson M.O., Coats S. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol Cell Biol. 1999;19:612–622. doi: 10.1128/MCB.19.1.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hemler M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- Hu Z., Hou D., Wang X., You Z., Cao X. TSPAN12 is overexpressed in NSCLC via p53 inhibition and promotes NSCLC cell growth in vitro and in vivo. Onco Targets Ther. 2018;11:1095–1103. doi: 10.2147/OTT.S155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge H.J., Yang S., Burton J.B., Paes K., Shu X., French D.M., Costa M., Rice D.S., Ye W. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- Knoblich K., Wang H.X., Sharma C., Fletcher A.L., Turley S.J., Hemler M.E. Tetraspanin TSPAN12 regulates tumor growth and metastasis and inhibits beta-catenin degradation. Cell Mol Life Sci. 2013;71:1305–1314. doi: 10.1007/s00018-013-1444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur M.A., Xu D., Hemler M.E. Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol Biol Cell. 2009;20:2030–2040. doi: 10.1091/mbc.e08-11-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chen W., Jin Y., Xue R., Su J., Mu Z., Li J., Jiang S. miR-142–5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Biochem Pharmacol. 2019;161:98–112. doi: 10.1016/j.bcp.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Liu J., Chen C., Li G., Chen D., Zhou Q. Upregulation of TSPAN12 is associated with the colorectal cancer growth and metastasis. Am J Transl Res. 2017;9:812–822. [PMC free article] [PubMed] [Google Scholar]
- Otomo R., Otsubo C., Matsushima-Hibiya Y., Miyazaki M., Tashiro F., Ichikawa H., Kohno T., Ochiya T., Yokota J., Nakagama H., et al. TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc Natl Acad Sci U S A. 2014;111:18691–18696. doi: 10.1073/pnas.1412062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton M., Coats S. Cyclin E2, the cycle continues. Int J Biochem Cell Biol. 2002;34:315–320. doi: 10.1016/S1357-2725(01)00137-6. [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinosa J.P., Kanduc D. Ovarian cancer: designing effective vaccines and specific diagnostic tools. Immunotherapy. 2013;6:35–41. doi: 10.2217/imt.13.144. [DOI] [PubMed] [Google Scholar]
- Thibault B., Castells M., Delord J.P., Couderc B. Ovarian cancer microenvironment: implications for cancer dissemination and chemoresistance acquisition. Cancer Metastasis Rev. 2014;33:17–39. doi: 10.1007/s10555-013-9456-2. [DOI] [PubMed] [Google Scholar]
- Wang H.X., Li Q., Sharma C., Knoblich K., Hemler M.E. Tetraspanin protein contributions to cancer. Biochem Soc Trans. 2011;39:547–552. doi: 10.1042/BST0390547. [DOI] [PubMed] [Google Scholar]
- Yang H.L., Lin R.W., Karuppaiya P., Mathew D.C., Way T.D., Lin H.C., Lee C.C., Hseu Y.C. Induction of autophagic cell death in human ovarian carcinoma cells by Antrodia salmonea through increased reactive oxygen species generation. J Cell Physiol. 2019;234:10747–10760. doi: 10.1002/jcp.27749. [DOI] [PubMed] [Google Scholar]
- Ye M., Wei T., Wang Q., Sun Y., Tang R., Guo L., Zhu W. TSPAN12 promotes chemoresistance and proliferation of SCLC under the regulation of miR-495. Biochem Biophys Res Commun. 2017;486:349–356. doi: 10.1016/j.bbrc.2017.03.044. [DOI] [PubMed] [Google Scholar]
- Yi C., Zhang L., Zhang F., Li L., Ling S., Wang X., Liu X., Liang W. Methodologies for the establishment of an orthotopic transplantation model of ovarian cancer in mice. Front Med. 2014;8:101–105. doi: 10.1007/s11684-014-0315-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.