Abstract
The human intestine harbors an immense, diverse, and critical population of bacteria that has effects on numerous aspects of host physiology, immunity, and disease. Emerging evidence suggests that many of the interactions between the host and the gut microbiota are mediated via the microbial metabolome, or the collection of small-molecule metabolites produced by intestinal bacteria. This review summarizes findings from recent work by focusing on different classes of metabolites produced by the gut microbiota and their effects in modulating host health and disease. These metabolites ultimately serve as a form of communication between the gut microbiome and the host, and a better understanding of this chemical language could potentially lead to novel strategies for treating a wide variety of human disorders.
Gut Microbial Metabolites Mediate Host-Microbiota Interactions
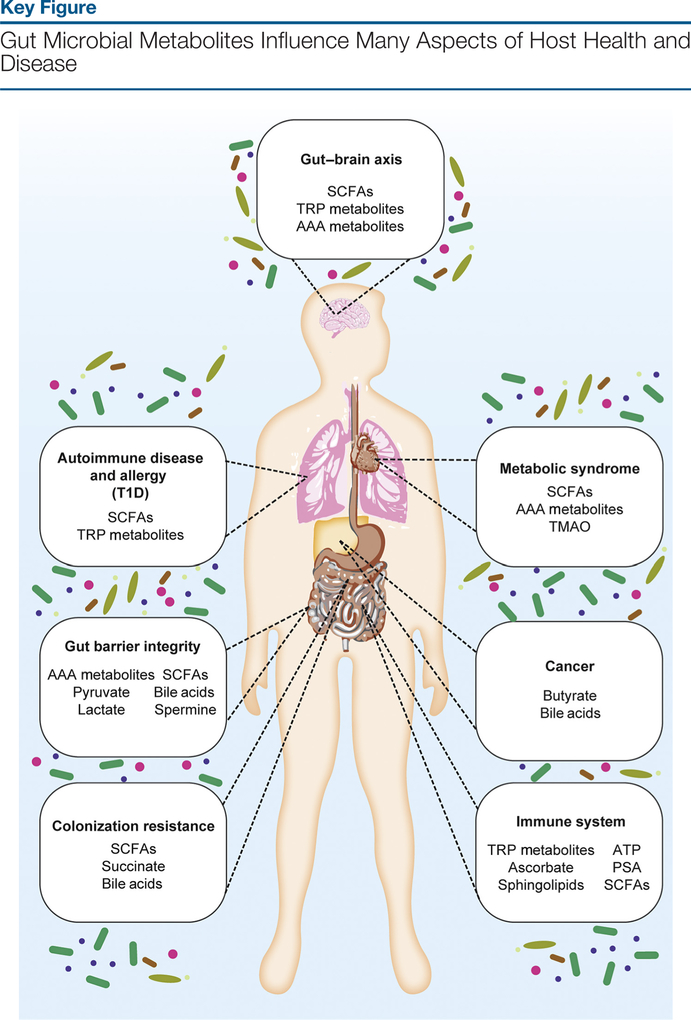
Next-generation sequencing (see Glossary) technologies have catalyzed an expansion of gut microbiome-related research over the past decade. As a consequence, these studies have provided great insight into the composition of our microbiome and how it correlates with different health outcomes for the host. These observations, coupled with large phenotypic differences between germ-free and conventionally raised mice in models of various diseases, including metabolic syndrome (MBS), cancer, and inflammatory bowel disease (IBD), suggest that the microbiota participate in regulating many different aspects of host physiology [1,2]. There is also substantial evidence that suggests that the gut microbiome is critical to the proper development and regulation of our intestinal immune system [3]. Microbial metabolites serve as signals from the gut microbiome that can activate or inhibit endogenous signaling pathways or act as nutrient sources for host cells [1]. These chemical messengers ultimately modulate the intestinal microenvironment to be tolerant or intolerant to specific commensal microbes. Together, these findings highlight the importance of understanding how the gut microbiota exert these effects on the host. This review outlines the recent work carried out in the past several years that contribute to our understanding of how the gut microbiota communicates with us through their production of small-molecule metabolites and how these host-microbe interactions affect a wide range of diseases (Figure 1, Key Figure).
Figure 1.
Small-molecule metabolites that are produced by the gut microbiota can modulate myriad physiological processes in the host, thereby impacting disease outcomes in many inflammatory disorders, including metabolic syndrome, cancer, autoimmune diseases including type 1 diabetes (T1D), allergy, and inflammatory bowel diseases. Furthermore, these metabolites can also influence the gut-brain axis and regulate the immune system and host susceptibility to gastrointestinal infection via colonization resistance. Abbreviations: AAA, aromatic amino acid; PSA, polysaccharide A; SCFAs, short-chain fatty acids; TMAO, trimethylamine N-oxide; TRP, tryptophan.
Short-Chain Fatty Acids (SCFAs) Have Pleiotropic Effects on Host Health and Disease
SCFAs, including acetate, propionate, butyrate, and pentanoate, result from bacterial fermentation of dietary fiber and are among the most abundant microbial metabolites present in the intestinal lumen. SCFAs facilitate host-microbiota communication through several mechanisms:(i) they are readily used as carbon sources for the generation of host endogenous metabolites;(ii) they serve as signaling molecules that activate host G-protein-coupled receptors (GPCRs); and (iii) they affect the expression of host genes through inhibition of histone deacetylases (HDACs) [1–3]. The following section synthesizes recent studies describing the roles of SCFAs in the host-gut microbiota axis.
SCFAs Exhibit Cell Type-Specific Effects on the Host Immune System
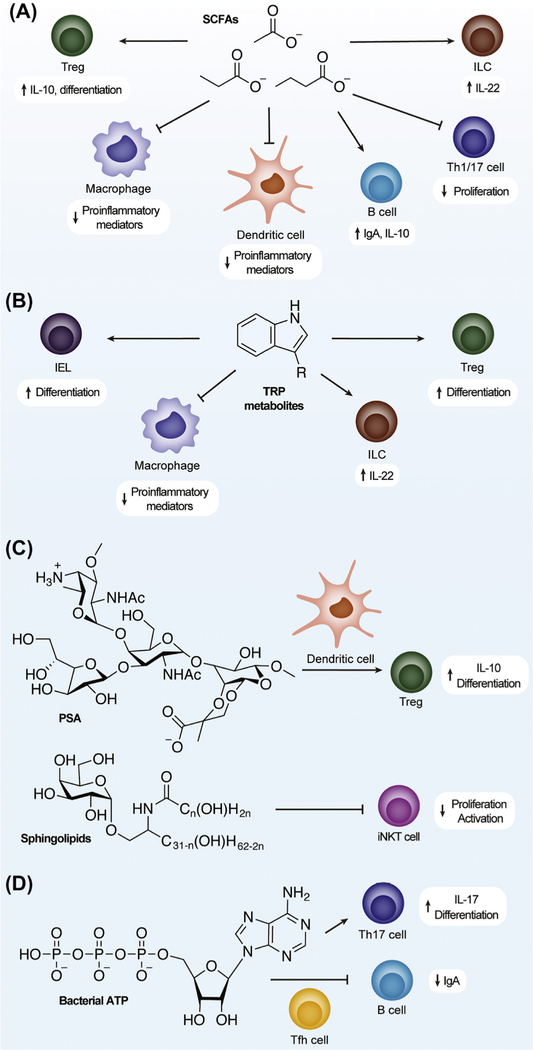
Tolerance of commensal microorganisms relies on a delicate balance of pro- and anti-inflammatory signals, which are regulated by various immune cell types (for a brief orientation on immune cell types referred to in this review, see Table 1). Foxp3+CD4+ regulatory T cells (Tregs) are critical in the downregulation of inflammatory responses in the gut and their differentiation is increased by bacterial SCFAs (Figure 2A), which are mainly produced by Clostridia [4–9]. Atarashi et al. found that a consortium of 17 different Clostridial strains was capable of inducing Foxp3+CD4+ Treg differentiation [9]. Propionate and butyrate produced from Bacteroides thetaiotaomicron were found to increase the differentiation of peripheral, but not thymic, Foxp3+CD4+ Tregs via HDAC inhibition by causing increased acetylation at the conserved noncoding sequence 1 enhancer in the Foxp3 promoter [5,6]. Acetate and propionate cause the proliferation of colonic Foxp3+CD4+ Tregs through GPR43 signaling, which is much more highly expressed in colonic T cells [4]. SCFA-mediated increases in colonic Foxp3+CD4+ Tregs are observed in both the steady state and during infection; however, there is also evidence of acetate increasing Teffector cell populations (T helper cells that mediate adaptive immune responses) during Citrobacter rodentium infections [7]. Butyrate promotes Foxp3+CD4+ Treg differentiation by activation of GPR109A on intestinal macrophages and dendritic cells (DCs), which express interleukin (IL)-10 [8]. Butyrate is also capable of downregulating proinflammatory mediators through the inhibition of HDACs in intestinal macrophages and DCs, which support its role in maintaining a commensal-tolerant environment (Figure 2A) [5,10].
Table 1.
A List of the Immune Cell Types, Including Markers and Functions, Mentioned in This Review
| Cell type | Subset | Function | Marker |
|---|---|---|---|
| T cell | Treg | Suppressor T cells that downregulate inflammatory responses | Foxp3+, IL-10+, CD4+ |
| Th17 | Teffeotor cells that that protect against bacteria and fungi | ROR-γt+, IL-17+, CD4+ | |
| Th1 | Teffector cells that protect against intracellular pathogens | T-bet+, IFN-γ+, CD4+ | |
| Th2 | Teffector cells that protect against extracellular pathogens | GATA3+, IL-4+, IL-13+, IFN-γ+, CD4+ | |
| T follicular helper | Teffector cells critical to the regulation of germinal centers | Bcl6+, PD1+, ICOS+, CXCR5+, CD4+ | |
| Macrophage | Colonic | Tissue-resident innate immune cells that phagocytose and secrete proinflammatory cytokines | CD11b+, CD64+, F4/80+, MHCII+, CXCR1+, CD103− |
| Pancreatic, M1 | Pancreatic innate immune macrophages with a proinflammatory phenotype | F4/80+, CD11b+, CD11c+, CD206− | |
| Pancreatic, M2 | Pancreatic innate immune macrophages with an anti-inflammatory and regulatory phenotype | F4/80+, CD11b+, CD11c−, CD206+ | |
| Adipose, M1 | Adipose innate immune macrophages with a proinflammatory phenotype | CD45+, CD11b+, F4/80+, iNOS+, CD206− | |
| Adipose, M2 | Adipose innate immune macrophages with an anti-inflammatory phenotype | CD45+, CD11b+, F4/80+, iNOS−, CD206+ | |
| Microglia | Tissue-resident macrophages of the central nervous system | CD11b+, CD45Io, CSF1R+(CD115), F4/80+, CD31+ | |
| DC | Splenic | Immune cells responsible for the presentation of antigen to T cells | CD11c+ |
| Colonic, CD103+ | Immune cells responsible for the presentation of antigen to T cells that reside in the colon and upregulate Treg differentiation | CD45+, I-Ab+, CD11c+, CD103+ | |
| Mesenteric lymph node (MLN), CD103+ | DCs that migrate from the intestines to the MLN to upregulate Treg differentiation and promote oral tolerance to food antigens | MHCII+, CD11chi, CD103+ | |
| B cell | Germinal center | Adaptive immune cells that recognize specific antigens to produce antibodies | CD19+ |
| Pancreatic, regulatory | Adaptive immune cells that secrete IL-4 to induce regulatory macrophages, which upregulate Treg induction | CD19+, CD11b−, CD5+, CD1d+, B220+, CD21+, CD24+ | |
| ILC | Group 3 | Regulatory innate immune cells that are critical to intestinal homeostasis via IL-22 secretion | CD45+, CD127+, CD90+, Lin−, IL22+, ROR-γt+ |
| IEL | CD4+ | Intestinal epithelial effector T cells that recognize both autoreactive and foreign antigens | CD45+, CD3+, TCRγδ−, CD8β−, CD4+ |
| CD8+ | CD45+, CD3+, TCRγδ−, CD8β−, CD8α+ | ||
| CD4+ CD8αα+ | Intestinal epithelial tolerogenic effector T cells | CD45+, CD3+, TCRγδ−, CD8β−, CD4+, CD8α+ | |
| NKT | CXCR6+ hepatic NKT | Liver effector T cells with tumor suppressing function | TCRβ+, CD1d−, CXCR6+ |
| Colonic invariant NKT | Effector T cells with both innate and adaptive functions | CD3+, CD1d+ |
Figure 2. Gut Microbial Metabolites Regulate Specific Immune Cell Types.
Microbial metabolites regulate immune responses in the gut by modulating the activities of different immune cell types as indicated. The schematic shows immune cell types that are affected by metabolites: (A) short-chain fatty acids (SCFAs); (B) tryptophan (TRP); (C) polysaccharide A (PSA) and sphingolipids; and (D) bacterial ATP. Abbreviations: IEL, intraepithelial lymphocyte; IL, interleukin; ILC, innate lymphoid cell; iNKT, invariant natural killer T; Tfh, T follicular helper; Th1/17, T helper 1/17; Treg, T regulatory cell.
SCFAs, including pentanoate, also contribute to microbial tolerance by elevating glucose oxidation, which increases regulatory B cells [11]. Pentanoate also suppresses Th17 cell responses by inhibiting HDACs [11]. Alternatively, SCFAs were found to mediate proinflammatory effects by up-regulating B cell metabolism, which increases the systemic production of IgG and IgA to regulate both homeostatic and pathogen-specific immune responses [12]. Collectively, these results suggest that SCFAs can contribute to an anti-inflammatory environment in the gut while simultaneously bolstering host defense against pathobionts and pathogens.
SCFAs Ameliorate Autoimmune Diseases and Allergy
The ability of SCFAs to promote anti-inflammatory responses by increasing Foxp3+CD4+ Treg differentiation ameliorates autoimmune diseases, such as type 1 diabetes (T1D), in which immune homeostasis is disrupted (Figure 1 and Table 2). Nonobese diabetic (NOD) mice, which model T1D, fed acylated starch that is fermented to acetate and butyrate showed expanded Bacteroides in the intestine and decreased disease severity via GPR43 activation by acetate and Foxp3+CD4+ Treg induction by butyrate [13]. Miani et al. also demonstrated that butyrate ameliorates T1D in NOD mice via induction of IL-22 in pancreatic innate lymphoid cells (ILCs), which causes increased expression of β-defensin 14, an antimicrobial peptide that induces pancreatic regulatory macrophages and Foxp3+CD4+ Tregs via stimulation of Toll-like receptor 2 (TLR-2) on IL-4-secreting B cells [14].
Table 2.
Effects of SCFAs in the Host
| Disease | Metabolite | Effect | Target | Refs |
|---|---|---|---|---|
| T1D | Acetate | Suppression of autoreactive T cells, increased IL-22 | Decreased expression of MHC-I molecules in B cells, GPR43 | [13] |
| Butyrate | FOXP3+/IL-10+ Treg expansion, increased IL-22 | [13] | ||
| Butyrate | Pancreatic regulatory macrophage and T cell expansion, IL-22 induction in pancreatic ILCs | [14] | ||
| EAE | Butyrate and propionate | Differentiation to Tregs over Th1/17 cells | [15] | |
| AAD | Propionate | Expansion of DC precursors with an impaired ability to induce Th2 effector cells | GPR41 | [16] |
| Butyrate, propionate, and acetate | IL-10 and TGF-βa-mediated Treg suppressor activity | GPR41 | [17] | |
| Acetate | Increases FOXP3+ Treg | Inhibition of HDAC9 | [18] | |
| Food allergy | Butyrate and acetate | Upregulate CD103+ tolerogenic DCs, which induce Treg differentiation | GPR109A and GPR43 | [19] |
| GVHD | Butyrate | Protection against reactive T cell damage | [20] | |
| MBS | Acetate | Suppression of insulin-mediated fat accumulation in adipocytes | GPR43 | [27] |
| Butyrate, propionate, and acetate | Prevent HFD-induced obesity, stimulate insulin sensitivity, and abrogate hepatic steatosis | Downregulation of PPAR-γ | [28] | |
| Acetate and butyrate | Improved glucose metabolism through increased post-prandial insulin, GLP-1, and fasting PYY | [29] | ||
| Butyrate | Stimulation of IGN | cAMP-dependent IGN gene activation | [30] | |
| Propionate | Stimulation of IGN | GPR41 | [30] | |
| Butyrate | Atherosclerosis prevention and stimulation of fatty acid oxidation | [31] | ||
| Acetate | Appetite suppression | Hypothalamic AMPK inactivation | [32] | |
| Activation of the parasympathetic nervous system to increase glucose-stimulated insulin secretion, hunger, insulin resistance, and hypertriglyceridemia | [33] | |||
| Gut/brain | Butyrate, propionate, and acetate | Increase alpha-synuclein-related inflammation | [35] | |
| Butyrate | Decrease blood-brain barrier permeability | Occludin upregulation | [36] | |
| Butyrate, propionate, and acetate | Microglial maturation and IGN stimulation | GPR41/43 | [37] | |
| Cancer | Butyrate | Suppression of colonic tumorigenesis | HDAC inhibition and GPR109A | [8] |
| Proliferation and transformation of intestinal epithelial stem cells in MSH2−/− mice | [38] | |||
| Attenuation of tumor progression via increased DC recruitment and upregulation of Muc2 and antigen recognition genes | [39] | |||
| Caused hyperbilirubinemia, hepatic inflammation, and upregulation of liver fibrosis and HCC markers in TLR-5 KO mice | [40] | |||
| Colonization resistance | Butyrate | Maintains a hypoxic environment in the gut to prevent Salmonella typhimurium from accessing oxygen | PPAR-γ | [41,42] |
| Propionate | Disrupts pH buffering and destabilizes virulence factors in S. typhimurium | HilD | [43,44] | |
| Butyrate | Promotion of antimicrobial activity against Citrobacter rodentium in intestinal macrophages |
HDAC3 inhibition | [45] |
TGF-β, transforming growth factor beta.
Regulation of the Foxp3+CD4+ Treg/Teffector, cell axis by SCFAs also affects experimental autoimmune encephalomyelitis (EAE) and allergic airway disease (AAD) in mice (Figure 1 and Table 2). SCFAs mitigate EAE by promoting the differentiation of Foxp3+CD4+ Tregs over Th1/Th17 cells via suppression of the c-Jun N-terminal kinase, JNK1, and p38 signaling pathways (Figure 2A) [15]. These metabolites also alleviate AAD through several mechanisms. Propionate treatment causes a GPR41-dependent expansion of macrophage and DC precursors in the lung that have reduced ability to induce Th2 cells [16]. The GPR41 -dependent expansion of these cells and an increase in lung Foxp3+CD4+ Tregs were also observed with increased levels of SCFAs resulting from intestinal helminth infection [17]. Acetate also decreased asthma severity by upregulating Foxp3+CD4+ Treg populations via HDAC9 inhibition, which increases expression of Foxp3 [18]. Butyrate and acetate also suppressed immune responses to oral antigens to protect against food allergy via activation of GPR43 and GPR109a in intestinal epithelial cells (IECs) and CD103+ DCs, respectively [19] (Table 2).
SCFAs Regulate Intestinal Barrier Function and Protect against IBD
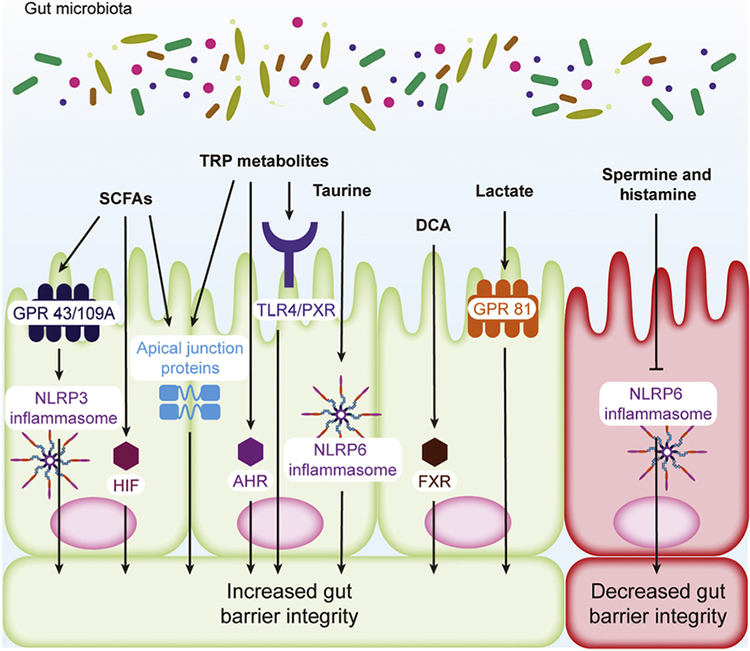
Maintenance of the gut epithelial barrier is critical to immune homeostasis because exposure of the intestinal immune system to colonic contents leads to inflammation. SCFAs increase host defense by enhancing the barrier integrity of the gut epithelium via HDAC inhibition and GPCR activation [20,21]. Clostridia-derived butyrate increases IEC proliferation and apical junctional protein expression, which maintain the epithelial barrier, via HDAC inhibition, and as a consequence mitigates graft-versus-host disease (GVHD) [20]. SCFAs also ameliorate colitis in mouse models by enhancing gut barrier function via GPR43/109A-mediated NOD-like receptor protein 3 (NLRP3) activation (Figure 3), while the NLRP1 inflammasome serves as a negative regulator for SCFA-producing commensals [21,22]. It is important to note here that acetate, propionate, and butyrate activate GPR41/43, while only butyrate is known to activate GPR109A [23,24]. Butyrate also increases gut barrier integrity by stabilizing hypoxia inducible factor (HIF), an important transcription factor that maintains tissue barrier function [25]. Interestingly, butyrate inhibits the proliferation of intestinal epithelial stem cells, which seems counter to maintaining gut barrier integrity, but the colonic crypt prevents these cells from accessing luminal butyrate [26].
Figure 3. Gut Microbial Metabolites Regulate Intestinal Epithelial Barrier Integrity.
Metabolites produced by the gut microbiota, such as short-chain fatty acids (SCFAs), tryptophan (TRP) metabolites, taurine, deoxycholic acid (DCA), lactate, spermine, and histamine, can modulate the barrier function of the intestinal epithelium by regulating receptor expression and/or activation, transcription factor activation, increasing the expression of cytokines that confer barrier protection, and modulating apical junction proteins, which directly regulate epithelial permeability. Abbreviations: AHR, aryl hydrocarbon receptor; FXR, farnesoid X receptor; GPR, G protein receptor; HIF, hypoxia inducible factor; NLRP, NOD-like receptor protein; PXR, pregnane X receptor; TLR4, Toll-like receptor 4.
SCFAs Can Exacerbate or Alleviate Symptoms of MBS
The incidence of MBS, which is characterized by obesity, reduced high-density lipoprotein (HDL) cholesterol, increased blood pressure and triglycerides, heart disease, and type 2 diabetes (T2D), is a growing crisis in many industrialized nations [1]. HDAC inhibition and GPR41/43 activation in adipose and pancreatic tissues provide a mechanism by which SCFAs attenuate pathology in this disease (Figure 1) [27–34]. SCFAs target GPR43 in white adipose tissue to decrease insulin sensitivity and fat accumulation in high-fat diet (HFD)-fed mice [27]. Adipose peroxisome proliferator-activated receptor gamma (PPAR-γ) downregulation by SCFAs attenuates HFD induced obesity and insulin resistance, while hepatic PPAR-γ downregulation limits dysregulated fat accumulation in the liver [28]. These studies describe the targeting of differentially expressed GPR43 and PPAR-γ in the liver and adipose tissue to decrease insulin resistance and fat accumulation in MBS [27,28]. Interestingly, GPR43 promotes inflammatory signals in adipose tissue by stimulating tumor necrosis factor alpha (TNF-α) expression in anti-inflammatory M2 macrophages, but this effect may be due to TNF-α suppressing fat accumulation by regulating insulin signaling in adipose tissues [27,34]. Increased acetate and butyrate levels also result in increased anorectic hormones, such as glucagon-like peptide-1 (GLP-1) and fasting peptide YY (PYY), a peptide hormone that reduces appetite, and alleviates T2D in humans [29]. Elevated SCFA levels from fiber-rich diets cause up-regulation of intestinal gluconeogenesis (IGN), which decreases the glucose and energy dysmetabolism observed in MBS [30]. Butyrate serves as an energy substrate for enterocytes to increase ATP levels and, in turn, cAMP to directly upregulate IGN gene expression, while propionate activates GPR41 in the portal vein to stimulate IGN as a result of a gut-brain signaling [30]. Colonization by Roseburia intestinalis in the intestine increases butyrate levels to attenuate atherosclerosis by stimulating fatty acid oxidation in enterocytes and reducing endotoxemia [31].
In addition to regulating how the body stores and metabolizes fat, SCFAs modify behavior by regulating appetite. Acute acetate administration to HFD-fed mice was shown to decrease food in-take and induce an anorectic phenotype in the hypothalamus, potentially through the downregulation of 5′-AMP-activated kinase AMPK [32]. Alternatively, increased acetate was shown to drive MBS through upregulation of glucose-stimulated insulin secretion, hunger, insulin resistance, and hypertriglyceridemia through the parasympathetic nervous system [33].
SCFAs Regulate the Gut-Brain Axis
In addition to modulating feeding behavior, SCFAs facilitate additional crosstalk between the brain and the intestine. SCFAs are sufficient to increase α-synuclein-related inflammation in Parkinson’s disease models, and gut microbiota transfer from Parkinsonian human subjects to healthy mice replicates motor impairments observed in this neurological disease [35]. Systemic SCFAs are capable of decreasing the blood-brain barrier’s permeability through upregulation of the tight junction protein occludin, demonstrating that the barrier integrity-promoting effects of SCFAs are not limited to the intestinal epithelial barrier [36]. Treating gnotobiotic mice with SCFAs also reverses global microglial maturation defects observed in germ-free mice, indicating that SCFAs regulate immune cells in the central nervous system (CNS) under homeostatic conditions [37].
Butyrate Regulates Tumorigenesis and Cancer Progression
Chronic inflammation in the gut has been associated with increased intestinal tumorigenesis [2]. As summarized above, SCFAs have anti-inflammatory effects in the gut, thereby decreasing the potential for tumor formation. The majority of the studies investigating the roles of SCFAs in intestinal cancer have been largely focused on butyrate. A consensus on butyrate’s role in intestinal carcinogenesis has yet to be reached due to different models demonstrating that butyrate is capable of either suppressing or potentiating colorectal cancer [8,38,39]. Butyrate activates GPR109A in both enterocytes and immune cells, which protects against inflammation and carcinogenesis by upregulating the differentiation of IL-10-producing Foxp3+CD4+ Tregs and stimulating IL-18 production in APCMin/+ mice, which lack the tumor suppressor APC and spontaneously develop intestinal adenomas [8]. In addition, depletion of the gut microbiome with antibiotics followed by treatment with GPR109A agonists exhibited reduced polyp formation in APCMin/+ mice but not in GPR109a−/− APCMin/+ [8]. By contrast, butyrate was shown to drive polyp formation and the transformation of enterocytes in an APCMin/+ MSH2−/− model, which lacks the DNA mismatch repair protein MSH2 and is prone to tumor formation, at lower concentrations but had no effect at higher concentrations associated with HDAC inhibition, demonstrating that lower concentrations of butyrate can drive the proliferation of APCMin/+ MSH2−/− enterocytes [38]. Butyrate was also shown to attenuate tumor progression in HFD-induced intestinal carcinogenesis in dysbiotic K-rasG12Dint mice by increasing DC recruitment and reversing the downregulation of barrier-promoting Muc2 and genes involved in antigen recognition [39]. In the liver, the incorporation of soluble fiber into the diets of TLR-5-deficient mice led to hepatocellular carcinoma (HCC) [40]. In this study, butyrate supplementation alone increased HCC markers but was insufficient for tumorigenesis, suggesting the need for additional tumor-promoting factors [40].
SCFAs Modulate Colonization Resistance against Intestinal Pathogens
Colonization resistance via microbiota-related factors helps to prevent harmful enteric pathogens from colonizing and expanding in the gut. Gut microbiota metabolites can mediate colonization resistance (Figure 1) by directly inhibiting gut pathogens or by indirectly modulating the host environment to make the intestinal landscape less susceptible to infection [3]. Multiple studies have demonstrated that depletion of SCFA-producing commensals drives expansion of Salmonella typhimurium, an enteric pathogen [41,42]. Baumler and coworkers demonstrated that butyrate prevents aerobic expansion of this pathogen by maintaining hypoxia in the gut epithelium that limits S. typhimurium’s access to oxygen [41]. They also demonstrated that oxygen limitation at the epithelium was mediated by butyrate stimulation of PPAR-γ in IECs [42]. Propionate has also been shown to protect against Salmonella infection by the disruption of intracellular pH buffering and destabilization of the S. typhimurium invasion virulence factor HilD, providing an example of the direct effects that SCFAs have on this pathogen [43,44].
In addition to S. typhimurium, butyrate was found to potentiate colonization resistance against C. rodentium and Staphylococcus aureus by promoting antimicrobial activity in intestinal macrophages through the inhibition of HDAC3 [45]. Modulation of the gut barrier also extends to the mucosal layer because dietary fiber depletion promotes the expansion of mucus-degrading bacteria, resulting in enhanced virulence of pathogens such as C. rodentium [46].
In summary, SCFAs are capable of enhancing host health by decreasing inflammation, ameliorating autoimmune diseases and allergy, maintaining the gut barrier, and mediating colonization resistance to enteric pathogens. SCFAs modulate these diseases and physiological processes in different tissues because of their ability to disseminate into the bloodstream, where they can access GPCRs in many tissues or inhibit HDAC activity in various cell types. The ability of SCFAs to modulate MBS and tumorigenesis appear to be tissue and context dependent.
Gut Microbial Tryptophan (TRP) Metabolism Regulates Host Physiology and Immunity
Gut microbial catabolism of TRP produces indole-containing metabolites that regulate the host immune system by activating the aryl hydrocarbon receptor (AHR), a ligand-gated transcription factor that regulates immunity. Stimulation of AHR by TRP metabolites largely upregulates anti-inflammatory responses and helps to maintain host-gut microbiota homeostasis.
Tryptophan Metabolites Mitigate Inflammation and Autoimmune Disease
Through AHR agonist activity, indole-3-lactic acid produced by Lactobacillus reuteri was shown to differentiate CD4+ intraepithelial lymphocytes (IELs) into CD4/CD8 double-positive IELs, which are regulatory cells known to prevent intestinal inflammation (Figure 2B) [47]. Indole-3-acetic acid (IAA) and tryptamine (TRA) were found to reduce macrophage production of proinflammatory mediators, such as TNF-α, IL-1 β, and monocyte chemoattractant protein 1 (MCP-1), and indole-3-aldehyde (I3A) produced by L. reuteri increases the expression of IL-22 by ILCs (Figure 2B) [48,49]. Increased expression of IL-22 by ILCs due to I3A leads to the expansion of pancreatic regulatory macrophages and T cells that provides protection against T1D, similar to SCFAs [14]. Gut microbiota-derived I3A, indole-3-propionic acid (IPA), and indoxyl-3-sulfate (I3S) also regulate T cells and DCs in EAE mouse models via AHR (Table 3) and have been shown to suppress inflammation in the CNS mediated by inhibition of NF-κB in astrocytes (Figure 1) [50].
Table 3.
Effects of Additional Microbial Metabolites on the Host
| Disease | Metabolite | Effect | Target | Refs |
|---|---|---|---|---|
| TRP metabolites | ||||
| EAE | I3A, IPA, and I3S | Suppress inflammation through regulation of type I IFN signaling in astrocytes | AHR | [50] |
| MBS | Indole | Regulation of GLP-1 secretion in colonic L cells; acute exposure leads to GLP-1 release while longer exposures inhibit secretion | Short exposure: K+ channel inhibition Long exposure: NADH dehydrogenase inhibition |
[55] |
| lAA and TRA | Prevent macrophage proinflammatory cytokine production and migration to MCP-1, abrogation of TNF-α and fatty acid-mediated effects in hepatocytes | AHR | [48] | |
| Indole, IAA, and TRA | Improve glucose/insulin dysmetabolism, increase GLP-1 secretion, decrease fasting glucose levels, and decrease hepatic steatosis | AHR | [56] | |
| Gut-brain axis | Serotonin | Serotonin level regulation in colonic epithelial cells | [57] | |
| TRA | Enhancement of serotonergic activity | [58] | ||
| Increased fluid secretion to accelerate intestinal transit | 5-HT4Ra | [59] | ||
| Bile acids | ||||
| Cancer | DCA | Drives HCC, rescued by antibiotic depletion of HFD microbiome | [62] | |
| LCA | Decrease in tumor suppressing CXCR6+ NKTs via downregulation of CXCL16 | [63] | ||
| Colonization resistance | DCA and LCA | Reconstitution of antibiotic-treated mice with known DCA and LCA producers is sufficient to provide resistance against Clostridium difficile | [64] | |
| Enhance the activity of TRP-derived antibiotics | [65] | |||
| Additional | ||||
| MBS | TMAO | Increases cardiovascular risk and platelet aggregation and adhesion to collagen and inhibits reverse cholesterol transport | [71,72] | |
| Insulin resistance | [73] | |||
| Microbial choline depletion and TMAO accumulation exacerbates metabolic disease through inguinal fat accumulation, increased levels of circulating leptin, triglycerides, and free fatty acids | DNA methylation | [31] | ||
| Colonization resistance | Pyruvate and lactate | Induce intestinal CX3CR1+ mononuclear cells to promote antigen-specific immune responses and provide resistance to Salmonella typhimuhum | [74] | |
| Succinate | Nutrient source for the expansion of C. difficile | [75] | ||
| Upregulation of virulence-associated genes in Escherichia coli O157:H7 | Cra | [76] | ||
| Gut-brain axis | 4EPSb | Induces anxious behavior in maternal immune activation mice | [81] | |
5-HT4R, serotonin receptor-4.
4EPS, 4-ethylphenyl sulfate.
Microbially Produced TRP Metabolites Enhance Gut Barrier Function
TRP metabolites also increase intestinal barrier function, as indicated by studies describing the effects of these metabolites in ameliorating mouse colitis models. Card9 is a risk allele of IBD and mice deficient in this gene exhibit increased susceptibility to dextran sodium sulfate (DSS) colitis and gut microbiota with impaired TRP metabolism [51]. This phenotype can be transferred to germ-free mice by transplantation of Card9−/− microbiota and subsequently rescued via colonization with indole metabolite-producing lactobacilli or treatment with AHR agonists [51]. AHR expression in TRP diet-fed mice strongly correlates with the expression of IL-22 and Foxp3 as well, highlighting how multiple classes of microbial metabolites promote commensal tolerance through parallel mechanisms [13,14].
Independent of AHR, IPA was shown to decrease intestinal permeability and inflammation through a pregnane X receptor (PXR)- and TLR-4-mediated pathway [52] (Figure 3). Its levels can be increased through colonization by gut microbiota, including Clostridium sporogenes, which is known to produce this metabolite [53]. Indole is capable of increasing intestinal barrier function by increasing the expression of apical junction proteins (Figure 3) that mediate gut epithelial permeability, which improves pathology associated with DSS colitis [54].
Gut Microbial TRP Metabolites Protect against MBS
Gut microbial TRP metabolites largely show beneficial effects in MBS due to the involvement of AHR in the regulation of anorectic hormone secretion and glucose and insulin-regulated metabolism [55, 56]. Indole has been shown to regulate the release of the anorectic hormone GLP-1, a potential target for treating MBS (Table 3) [55]. Brief exposure to physiological levels of indole in colonic enteroendocrine L cells causes increased release of GLP-1, whereas prolonged exposure suppresses its secretion [55]. AHR activation was shown to decrease fasting glucose levels, improve glucose and insulin dysmetabolism, and increase GLP-1 secretion [56]. Similar to SCFAs, TRP metabolites such as IAA cause a reduction in cytokine-induced lipogenesis in hepatocytes, highlighting how gut microbial metabolites target lipid metabolism to attenuate MBS [48].
TRP Metabolites from the Gut Microbiota Modify Host Neurotransmitter Pools
TRP is actively metabolized by the gut microbiota into neurotransmitters that affect host serotonergic activity [57,58]. Serotonin is also a product of TRP metabolism by the gut microbiota, which has been demonstrated to significantly effect host biosynthesis and levels of this important neurotransmitter (Table 3) [57]. In addition, the neurotransmitter TRA is produced by Clostridial species in the gut via TRP catabolism, and it is hypothesized that sequestration of TRP reduces its bioavailability to the host, thus altering behavior by reducing serotonin biosynthesis [58]. TRA has also been demonstrated to activate the GPCR serotonin receptor-4, which in turn drives fluid secretion in the intestines to accelerate intestinal transit [59].
To summarize, microbial catabolism of TRP produces many metabolites that can serve as ligands for AHR. These metabolites have generally been shown to promote anti-inflammatory signals, maintain the gut barrier, and ameliorate MBS. Some of these metabolites also serve as neurotransmitters, such as serotonin and TRA, which regulate the gut-brain axis.
Regulation of Host Responses by Secondary Bile Acids
Primary bile acids are produced by the host in the liver to solubilize dietary lipids and fat-soluble vitamins in the small intestine. The primary bile acid pool is largely recycled back to the liver, but a small proportion of these bile acids escapes to the large intestine where they are readily deconjugated and further metabolized by the microbiota into secondary bile acids, which have numerous effects on the host. Below we review these effects briefly.
Gut Barrier Integrity
The gut microbiota influences barrier integrity in the intestine by activation of inflammasomes to increase IL-18 levels [60]. Deconjugation of taurine-conjugated primary bile acids by the gut microbiota increases the host taurine pool, which in turn causes activation of the NLRP6 inflammasome, increasing IL-18 secretion and inflammatory responses (Figure 3) [60]. In addition, deoxycholic acid (DCA), a secondary bile acid, was found to positively regulate intestinal crypt regeneration and repair through farnesoid X receptor-mediated downregulation of elevated wound prostaglandin E2 levels, which is critical to maintaining barrier function [61].
Cancer and Colonization Resistance
HFD-induced obesity was found to drive HCC through increased levels of DCA produced by Clostridia and antibiotic depletion of the microbiota helped to prevent tumor formation [62]. Secondary bile acids produced by Clostridium scindens also promote tumorigenesis in the liver through negative regulation of CXCL16, causing a decrease in tumor-suppressing CXCR6+ natural killer T cells (NKTs) [63]. Altered secondary bile acid profiles in dysbiotic microbiomes may also drive liver cancer through cooperative effects with SCFAs [40].
Secondary bile acids known for their carcinogenic effects exhibit beneficial roles in colonization resistance because DCA and lithocholic acid (LCA) inhibit the growth of Clostridium difficile [64] (Figure 1 and Table 3). Reconstitution of antibiotic-treated mice with C. scindens or a consortium of bacterial strains capable of producing secondary bile acids, including DCA and LCA, was shown to restore colonization resistance against this pathogen [64]. Furthermore, DCA and LCA inhibit the growth of C. difficile by enhancing the activity of TRP-derived antibiotics produced by the known DCA and LCA producers C. scindens and Clostridium sordellii [65].
Secondary bile acids are known to drive liver cancer yet also maintain the gut barrier and prevent the colonization of enteric pathogens. The beneficial and deleterious effects of secondary bile acids on the host underscore the need to understand how bacterial metabolites regulate infection and inflammatory diseases on a holistic level. Thus, further studies with various models and the effects of these metabolites with additional metabolites are needed.
Additional Microbial Metabolites That Modulate Host Health and Immune Responses
Membrane Polysaccharide A and Sphingolipids Regulate T Cells
Capsular and membrane components of gut commensals also play a role in maintaining intestinal immune homeostasis. Polysaccharide A (PSA) is a zwitterionic polysaccharide present in the capsule of the gut commensal Bacteroides fragilis and has been known for its immunomodulatory role in inducing IL-10 production in Foxp3+CD4+ Treg cells (Figure 2C). Robust IL-10 production in T cells requires both plasmacytoid DC (PDC) presentation of PSA fragments via MHC-II and PSA activation of TLR-2 on PDCs [66]. Recognition of the PSA-MHC-II complex by T cells was found to induce clonal expansion of CD4+CD45RBlow effector/memory cells, causing an anti-inflammatory response due to a decrease in interferon gamma (IFNγ) and an increase in IL-10 production [67]. In addition, membrane glycosphingolipids from B. fragilis negatively regulate in-variant NKTs (iNKTs) by inhibiting developmental iNKT proliferation and activation to attenuate proinflammatory responses and ameliorate colitis (Figure 2C) [68].
Commensal ATP Levels Mediate Anti-inflammatory Responses
Extracellular ATP in the intestinal lumen is a result of dying host cells, host export, and microbially produced ATP. In the steady state, host ectonucleoside triphosphate diphosphohydrolases (ENTPDases) hydrolyze ATP to control luminal levels, and ENTPDase-knockout mice exhibit increased differentiation of Th17 cells (Figure 2D) in the intestine and more severe inflammatory disease [69]. Commensal-derived ATP also reduced the activity of T follicular helper cells, resulting in diminished microbe-specific IgA secretion by B cells and an increase in commensal outgrowth (Figure 2D) [70].
Gut Microbial Choline Metabolism Negatively Affects Cardiovascular Health
Gut microbial metabolism of choline, phosphatidylcholine, and L-carnitine produces trimethylamine, which is oxidized by the host liver into trimethylamine N-oxide (TMAO). Gut microbe-dependent TMAO levels have been correlated with increased risk of cardiovascular disease (Figure 1 and Table 3) and depend on both L-carnitine and choline metabolism [71]. TMAO was also observed to increase platelet aggregation and adhesion to collagen in vitro and to potentiate thrombosis in vivo [72]. These effects could be replicated by increasing dietary choline and were significantly influenced by gut microbiota composition [72]. Gut microbial L-carnitine metabolism is not limited to cardiovascular health and also correlates with insulin resistance [73]. Effects of bacterial choline catabolism have also been shown to result in the production of TMAO and induction of a choline-deficient state, which in turn aggravates metabolic disorders through regulating DNA methylation [31].
Other Metabolites That Regulate Colonization Resistance in the Intestine
Pyruvate and Lactate
Pyruvate is generated from bacterial fermentation of dietary fiber and further reduced to produce lactate. Bacterial pyruvate and lactate induce small-intestinal CX3CR1 + mononuclear cells to extend dendrites into the intestinal lumen to capture luminal antigens and promote antigen-specific immune responses, which provides resistance to Salmonella infection [74].
Succinate
While some metabolites, such as secondary bile acids, may impede C. difficile infection, others can promote the expansion of this pathogen. One example of such a metabolite is succinate. Antibiotic treatment to cause dysbiosis increases local intestinal succinate levels that C. difficile utilizes as a metabolic source and converts into butyrate [75], causing growth of the bacterium. Increased succinate levels caused by B. thetaiotaomicron also promote enterohemorrhagic Escherichia coli strain O157:H7 pathogenesis through upregulation of the transcription factor Cra, which positively regulates virulence-associated genes [76].
Other Metabolites That Help in Maintaining Gut Barrier Function
Lactate
Microbially produced lactate is also beneficial to the maintenance of the gut barrier because it promotes the differentiation of intestinal stem cells through a GPR81-dependent mechanism [77].
Spermine and Histamine
These metabolites are produced by both the host and the gut microbiota and reduce gut epithelial barrier integrity through inhibition of the NLRP6 inflammasome, which decreases IL-18 levels [60] (Figure 3).
Concluding Remarks and Future Perspectives
Identifying the molecular mechanisms of how the gut microbiota regulates host physiology is critical to understanding the roles of these microbes in our bodies. Microbially derived metabolites produced by the gut microbiota serve as chemical messengers that mediate crosstalk between the microbes and host and can play both beneficial and deleterious roles in human health. The effects of these metabolites have been shown to influence the outcomes of many disorders, including MBS, IBD, cancer, autoimmune diseases, allergy, and neurodegenerative diseases. The gut microbial metabolome has also been shown to modulate colonization resistance to enteric pathogens.
A comprehensive understanding of all gut microbially produced small-molecule metabolites, their molecular targets, and their biological significance remains an important objective in the field (see Outstanding Questions). As a result, the development of new approaches for deciphering this host-gut microbiota crosstalk at the systems level are at the forefront of the field. Combining metagenomic analysis and serum metabolomic profiling has allowed bacteria capable of reductive metabolism of aromatic amino acids such as histidine, valine, leucine, and isoleucine to be correlated with improvements in insulin sensitivity and obesity [73,78–80]. Similarly, comparisons of gnotobiotic mice colonized with commensals either proficient or deficient in reductive aromatic amino acid metabolism translate to significant changes in circulating IPA levels that improve gut barrier function [53]. The effect of circulating metabolites on the host also extends to the gut-brain axis, as 4-ethylphenyl sulfate can induce autism spectrum disorder behaviors [81]. Further, use of experimental models comparing healthy and diseased fecal metabolomes have demonstrated which metabolite classes are important for IBD prevention and how their production may be impaired in individuals suffering from this disease [82]. For example, ascorbate was recently identified as a bioactive microbial metabolite associated with Crohn’s disease and was validated to exhibit suppressive effects on activated effector CD4+ T cells by targeting T cell metabolism [83]. Alternatively, extrathymic Foxp3+CD4+ Treg deficiency leads to increased type 2 responses against commensals and disruption of niche establishment for border-dwelling bacteria during colonization that correlated with systemic changes in lipid and amino acid metabolism in the fecal and serum metabolomes [84].
Outstanding Questions.
Which microorganisms in the gut microbiome produce the metabolites that have effects on the host?
How do host factors such as diet, age, gender, environment, and mental health affect gut microbiome composition and, in turn, the production of microbially derived metabolites?
Of the estimated thousands of unknown microbial metabolites in the gut, which of these metabolites are biologically active, what are their chemical structures, and what effects do they have on host pathways?
How are the biologically active microbial metabolites biosynthesized by the gut microbiota?
Which enzymes are involved in and which metabolites are produced by the sequential action of biosynthetic pathways in different microbes?
What is the interplay between the multiple metabolites produced by different microbes and can we establish a better understanding of the overall effect of these metabolites on host health?
What strategies can be developed to modify the metabolome or manipulate target pathways to treat inflammatory diseases?
Recently, Macpherson and coworkers used isotopic labeling of metabolic precursors to label bacterially produced metabolites in the gut [85,86]. These approaches use stable isotope tracing of nonreplicating 13C-labeled E. coli HA107 in gnotobiotic mice with high-resolution mass spectrometry, which can differentiate the 12C host metabolome to identify the distribution of microbially produced metabolites in different host tissues [86]. A similar approach using 13C-labeled glucose determined that maternal antibodies facilitate the transfer of certain gut microbiota-derived metabolites to the offspring [85].
Alternatively, new approaches have been pioneered to understand the effects of microbial metabolites on the host. Instead of traditional metabolomic approaches, metagenomic data can be bioinformatically mined for operons capable of biosynthesizing certain metabolites [87]. In addition, novel computational methods to analyze existing metagenomic data have enabled the identification of associations between metabolite levels and disease outcomes [88]. Ultimately, these systems-level approaches and novel methods will provide a deeper understanding of host-gut microbiota crosstalk that is mediated by microbial metabolites. Future studies will include the discovery of new microbial metabolites, the identification of bacterial species responsible for metabolite production, and understanding of the individual and systemic effects of these metabolites on host health and disease.
Highlights.
Microbially produced metabolites serve as chemical signals between the gut microbiota and the host and regulate many tissues throughout the body, thereby influencing host physiology.
Gut microbiota metabolites modulate host immune responses and inflammation, thereby influencing host health and disease.
Disorders affected by gut microbial metabolites include metabolic syndrome, inflammatory bowel diseases, cancer, allergy, autoimmune diseases, and neurodegenerative diseases.
The gut microbial metabolome can modulate colonization resistance against intestinal infections due to direct inhibition of enteric pathogens or by improving host defense mechanisms.
Identifying the molecular mechanisms that influence these outcomes is critical to understanding the impact of the gut microbiome and their metabolites on the host.
Understanding the individual and systemic effects of these metabolites is important for deciphering the chemical lexicon of the gut microbiota.
Acknowledgments
We sincerely apologize to those authors whose research we could not include due to space limitations. G.R.N. was supported by a National Institutes of Health Chemistry Biology Interface Training Program (T32GM008500). This work was supported by an Arnold and Mabel Beckman Young Investigator Award and the President’s Council of Cornell Women Affinito-Stewart Grant (both to P.V.C.).
Glossary
- Commensal microbes
microorganisms that cohabitate with the host within tissues and are generally thought to not cause harm to the host.
- Conventionally raised mice
mice that harbor a diverse and largely undefined microbiome.
- Dysbiosis
microbial imbalance or perturbation that is thought to cause host maladaptation.
- Endotoxemia
presence of endotoxin in the bloodstream, which may case hemorrhages, kidney necrosis, or toxic shock.
- Germ-free mice
mice that are raised in a sterile environment and are devoid of any microorganisms.
- Gnotobiotic mice
germ-free mice that have been colonized with a defined microbiota.
- Gut microbiome
trillions of microorganisms that reside in the intestinal lumen, including bacteria, viruses, fungi, parasites, and archaea.
- Hepatic steatosis
accumulation of fat in the liver.
- Inflammasomes
multiprotein oligomers that are responsible for the activation of inflammatory responses.
- Intestinal gluconeogenesis (IGN)
metabolic pathway in the gut that results in glucose production from noncarbohydrate carbon sources.
- Microbial metabolome
collection of small-molecule metabolites that are produced or modified by the gut microbiota.
- Next-generation sequencing
high-throughput DNA sequencing that is processed massively in parallel.
- Pathobiont
commensal microbes that have the potential to lead to disease under certain host physiological states.
References
- 1.Sonnenburg JL and Bäckhed F (2016) Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett WS (2015) Cancer and the microbiota. Science 348, 80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkaid Y and Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157, 121–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith PM et al. (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arpaia N et al. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furusawa Y et al. (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 [DOI] [PubMed] [Google Scholar]
- 7.Park J et al. (2015) Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol 8, 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh N et al. (2014) Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40,128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atarashi K et al. (2013) Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 500, 232–236 [DOI] [PubMed] [Google Scholar]
- 10.Chang PV et al. (2014) The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U. S. A 111, 2247–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luu M et al. (2019) The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun 10 10.1038/s41467-019-08711-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M et al. (2016) Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariño E et al. (2017) Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol 18, 552–562 [DOI] [PubMed] [Google Scholar]
- 14.Miani M et al. (2018) Gut microbiota-stimulated innate lymphoid cells support β-defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab 28,557–572 [DOI] [PubMed] [Google Scholar]
- 15.Haghikia A et al. (2015) Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43, 817–829 [DOI] [PubMed] [Google Scholar]
- 16.Trompette A et al. (2014) Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med 20, 159–166 [DOI] [PubMed] [Google Scholar]
- 17.Zaiss MM et al. (2015) The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43, 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorburn AN et al. (2015) Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun 6 10.1038/ncomms8320 [DOI] [PubMed] [Google Scholar]
- 19.Goverse G et al. (2016) Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 15, 2809–2824 [DOI] [PubMed] [Google Scholar]
- 20.Mathewson ND et al. (2016) Gutmicrobiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol 17, 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macia L et al. (2015) Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun 6 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- 22.Tye H et al. (2018) NLRP1 restricts butyrate producing commensals to exacerbate inflammatory bowel disease. Nat. Commun 9 10.1038/s41467-018-06125-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Poul E et al. (2003) Functional characterization of human receptors for short chain fatty acids and their role in polymorpho-nuclear cell activation. J. Biol. Chem 278, 25481–25489 [DOI] [PubMed] [Google Scholar]
- 24.Taggart AKP et al. (2005) (D)-β-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J. Biol. Chem 280, 26649–26652 [DOI] [PubMed] [Google Scholar]
- 25.Kelly CJ et al. (2015) Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiko GE et al. (2016) The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 165, 1708–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura I et al. (2013) The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun 4 10.1038/ncomms2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Den Besten G et al. (2015) Short-chain fatty acids protect against high-fat diet-induced obesity via a PPAR γ-dependent switch from lipogenesis to fat oxidation. Diabetes 64, 2398–2408 [DOI] [PubMed] [Google Scholar]
- 29.Zhao L et al. (2018) Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 30.De Vadder F et al. (2014) Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96 [DOI] [PubMed] [Google Scholar]
- 31.Romano KA et al. (2017) Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 22, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frost G et al. (2014) The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun 5 10.1038/ncomms4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry RJ et al. (2016) Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 534, 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakajima A et al. (2017) The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS One 12 10.1371/journal.pone.0179696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampson TR et al. (2016) Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braniste V et al. (2014) The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med 6, 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erny D et al. (2015) Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci 18, 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belcheva A et al. (2014) Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299 [DOI] [PubMed] [Google Scholar]
- 39.Schulz MD et al. (2014) High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 514, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh V et al. (2018) Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell 175, 679–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivera-Chávez F et al. (2016) Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byndloss MX et al. (2017) Microbiota-activated PPAR- γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobson A et al. (2018) A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 24, 296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung CC et al. (2013) The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol. Microbiol 87, 1045–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulthess J et al. (2018) The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desai MS et al. (2016) A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cervantes-Barragan L et al. (2017) Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cels. Science 357, 806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan S et al. (2018) Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep 23, 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zelante T et al. (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 [DOI] [PubMed] [Google Scholar]
- 50.Rothhammer V et al. (2016) Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med 22, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamas B et al. (2016) CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med 22, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatesh M et al. (2014) Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dodd D et al. (2017) A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimada Y et al. (2013) Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8 10.1371/journal.pone.0080604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chimerel C et al. (2014) Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep 9, 1202–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natividad JM et al. (2018) Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab 28, 737–749 [DOI] [PubMed] [Google Scholar]
- 57.Yano JM et al. (2015) Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams BB et al. (2014) Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmit-tertryptamine. Cell Host Microbe 16, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattarai Y et al. (2018) Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe 23, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levy M et al. (2015) Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain U et al. (2018) Temporal regulation of the bacterial metabolite deoxycholate during colonic repair is critical for crypt regeneration. Cell Host Microbe 24, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshimoto S et al. (2013) Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 [DOI] [PubMed] [Google Scholar]
- 63.Ma C et al. (2018) Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360 10.1126/science.aan5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buffie CG et al. (2015) Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang JD et al. (2019) Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem. Biol 26, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dasgupta S et al. (2014) Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 15, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson JL et al. (2015) Polysaccharide A from the capsule of Bacteroides fragilis induces clonal CD4+ T cell expansion. J. Biol. Chem 290, 5007–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.An D et al. (2014) Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kusu T et al. (2013) Ecto-nucleoside triphosphate diphosphohydrolase 7 controls Th17 cell responses through regulation of luminal ATP in the small intestine. J. Immunol 190, 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perruzza L et al. (2017) T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing microbiota-derived extracellular ATP. Cell Rep 18, 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koeth RA et al. (2013) Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med 19, 576–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu W et al. (2016) Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujisaka S et al. (2018) Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep 22, 3072–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morita N et al. (2019) GPR31-dependent dendrite protrusion of intestinal CX3CR1+ cells by bacterial metabolites. Nature 566, 110–114 [DOI] [PubMed] [Google Scholar]
- 75.Ferreyra JA et al. (2014) Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Curtis MM et al. (2014) The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee YS et al. (2018) Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe 24, 833–846 [DOI] [PubMed] [Google Scholar]
- 78.Pedersen HK et al. (2016) Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 [DOI] [PubMed] [Google Scholar]
- 79.Liu R et al. (2017) Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med 23, 859–868 [DOI] [PubMed] [Google Scholar]
- 80.Koh A et al. (2018) Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175, 947–961 [DOI] [PubMed] [Google Scholar]
- 81.Hsiao EY et al. (2013) Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franzosa EA et al. (2019) Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol 4, 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang Y-L et al. (2019) A screen of Crohn’s disease-associated microbial metabolites identifies ascorbate as a novel metabolic inhibitor of activated human T cells. Mucosal Immunol 12, 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell C et al. (2018) Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity 48, 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gomez de Agüero M et al. (2016) The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 86.Uchimura Y et al. (2018) Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity 49, 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen LJ et al. (2017) Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanna S et al. (2019) Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet 51, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]