Abstract
Purpose of the review:
The purpose of this review is to examine evaluate similarities and differences in bladder cancer expression subtypes and to understand the clinical implications of the molecular subtyping.
Recent Findings:
Four independent classification systems have been described, and there are broad similarities among the subtyping callers. Two major subtypes have been identified i.e. luminal and basal, with underlying subcategories based on various distinct characteristics. Luminal tumors generally bear a better prognosis and increased survival than basal tumors, although there is subtle variation in prognosis among the different subtypes within the luminal and basal classifications. Clinical subtyping is now commercially-available, although there are limitations to its generalizability and application.
Summary:
Expression subtyping is a new method to personalize bladder cancer management. However, there is probably not sufficient evidence to incorporate use into current standards-of-care. Validation cohorts with clinically meaningful outcomes may further establish the clinical relevance of molecular subtyping of bladder cancer. Additionally, genetic alterations in bladder cancer may ‘color’ the interpretation of individual tumors beyond the expression subtype to truly personalize care for bladder cancer.
INTRODUCTION
Bladder cancer has traditionally been classified as non-muscle invasive bladder cancer (NMIBC) or muscle-invasive bladder cancer (MIBC) according to the invasion depth of the tumor and according to the histopathologic subtype. NMIBC is diagnosed in the majority of patients and has frequent recurrences with low tendency to progress, while MIBC has a high rate of metastasis and a 5-year survival rate of approximately 60% [1]. Both NMIBCs and MIBCs are heterogeneous at the molecular level and their identification can be used to differentiate tumors across multiple intrinsic subtypes that may be used to predict clinical outcome. Expression subtyping is emerging as a significant factor associating with patient outcome and may enhance traditional clinical assessments relying on stage and grade. Expression subtyping and profiling using tools such as Oncotype® are an established method in breast cancer and prostate cancer, with strong clinicopathologic associations. The establishment of molecular subtypes of bladder cancer is poised to bring new opportunities for personalized therapies for the treatment of MIBC and NMIBC.
Molecular subtyping of cancer
The most highly reproducible and most widely applied clinical molecular subtyping tool is used in breast cancer where four different subtypes were identified based on gene expression profiling using cDNA microarray representing over 8,000 genes from 42 different individuals [2]. With breast cancer as a model, subsequent large-scale genomic expression profiling using cDNA microarray or RNAseq have been performed to successfully and reproducibly identify an increasing number of molecular subtypes of bladder cancer, which surprisingly reflect the intrinsic breast cancer subtypes [3–5]. This model was applied to bladder cancer with surprising insights into the relatedness of the two tumor types and clinical outcomes despite their disparate treatment standards clinically.
The Lund taxonomy of bladder cancer
Lindgren et al. at Lund University first defined two expression subtypes of bladder cancer, MS1 and MS2, based on their biologic processes and mutation profiles [3]. The MS1 tumors had activating mutations of FGFR3, PIK3CA while MS2 tumors had significant alterations of TP53, MDM2 and loss of RB1. The same group, after comprehensive analysis of 308 new tumors, extended their bladder cancer molecular classification into five different subtypes [5]. These subtypes were termed urobasal A, genomically unstable, urobasal B, squamous cell cancer (SCC)-like, and a heterogeneous infiltrated class of tumors, based on their distinct expression profiles. These subtypes showed different signatures of genes related to the immune system, cell cycle, cyokeratin and cell adhesion along with different mutation and expression pattern in FGFR3. Urobasal A tumors overexpressed FGFR3, CCND1, TP63, and KRT5 along with expression of RBL2 and ID genes and mainly consisted of NMIBCs. The genomically unstable subtype had TP53 mutations, CCNE and ERBB2 expression, and low PTEN and cytokeratin expression. The SCC-like subtype had high expression of basal keratins, KRT4, KRT6A, KRT6B, KRT6C, KRT14, and KRT16. The urobasal B subtype had TP53 mutations in addition to urobasal A subtype signature. The major drawback of this classification system was that the molecular subtypes were independent of bladder cancer pathology and no clinical correlates were identified at the time.
The UNC classification and development of BASE47
Researchers at University of North Carolina demonstrated that MIBCs can be distinguished into basal and luminal subtypes [6] that can be correlated with prognosis. Damrauer et al. showed that luminal bladder cancers express markers of terminal urothelial differentiation including UPK1B, UPK2, UPK3A, and KRT20 along with PPAR genes. Basal bladder tumors were found to express high levels of genes that typically mark urothelial basal cells including KRT14, KRT5, and KRT6 along with several transcription factors associated with stem cell homeostasis and cancer progression [6, 7]. Basal bladder cancer cells also had overexpression of STAT3 and EGFR which promote tumor progression. Importantly, they showed that basal-like bladder cancers were similar to basal-like and normal-like breast cancer subtypes, whereas luminal bladder cancers had similar genetic signatures as luminal A and luminal B breast cancer subtypes. They also showed that a subset of basal-like bladder tumors were claudin-low and had molecular features similar to claudin-low subtype of breast cancer. These claudin-low bladder tumors were shown to be immune-infiltrated and actively immune suppressed with very high level of PD-L1 expression [8]. The outcome of the study was the development of bladder cancer analysis of subtypes by expression using a 47-gene predictor (BASE47) that could accurately classify bladder cancer into basal-like or luminal tumors. Basal-like tumors as classified by BASE47 had significantly decreased disease-specific survival and overall survival, showing the possible prognostic significance of BASE47.
The TCGA taxonomy
The Cancer Genome Atlas (TCGA) initially identified four distinct molecular subtypes based on their sequencing analysis that included 131 high-grade muscle invasive carcinomas and included clusters I to IV [9]. Cluster I (papillary-like) subtype was enriched with alterations of FGFR3 along with overexpression of HER2 and ESR2, the latter two of which were also overexpressed in cluster II subtypes. These subtypes also had signatures of the luminal subtype described by the UNC group. Cluster III (basal/squamous) was shown to have similar expression signature to that of basal-like breast cancer and SCC of head, neck and lungs. This included characteristic epithelial lineage genes like KRT14, KRT5, KRT6 and EGFR.
In the most recent comprehensive analysis of the full TCGA cohort of 412 MIBC cases, TCGA expanded its taxonomy of urothelial carcinoma from previously reported four subtypes into five subtypes after discovery of 58 significantly mutated genes and multiple recurrent translocations, contributing to formation of fusion genes [10]. Clustering of RNA sequencing data identified luminal-papillary (35%), luminal-infiltrated (19%), luminal (6%), basal-squamous (35%), and neuronal (5%) subtypes. The luminal-papillary cluster had either mutation or overexpression of FGFR3 or FGFR3-TACC3 fusions. The luminal-infiltrated subtype had high expression of smooth muscle and myofibroblast gene signatures and increased expression immune markers such as PD-L1 and PD-1. The luminal subtype had upregulated expression of UPK1A, UPK2, KRT20, and SNX31. The basal-squamous tumor subtype had high expression of CD44, KRT5, KRT6A, KRT14, in addition to squamous differentiation markers, along with TP53 mutations. This subtype also had high expression of immune expression genes. The neuronal subtype showed elevated expression of neuronal differentiation and development genes such as PLEKHG4B, SOX2, MSI1, and GNG4. In addition, this subtype also had alterations in genes in the p53/cell-cycle pathway.
The MDA classification
Choi et al. at MD Anderson Cancer Center (MDA) introduced an additional p53-like subtype harboring wild type p53 gene expression signature along with luminal biomarkers [11], establishing a k=3 classifier, although what this MDA classifier calls basal tumors are recognized as basal or basal/claudin low tumors by the UNC classifier. In contrast, the UNC classifier groups all luminal tumors together, whereas the MDA classifier splits them into p53-like luminal tumors and a separate group of luminal tumors. Basal subtype in the MDA classification, similar to basal-like in UNC classification, had shorter DSS and overall survival rate, and were characterized by squamous differentiation, similar to SCC-like subtype in Lund classification. The p53-like subtype had luminal biomarkers but were more resistant to neoadjuvant chemotherapy (NAC) than other subtypes. Choi et al. also found that chemoresistant tumors were enriched with p53-like subtype after NAC due to expression of an active WT p53 pathway gene signature, which may be a mechanism of resistance against chemotoxicity. They also discovered that basal tumors had high expression of immune infiltration biomarkers.
Interestingly, all these multiple independent groups classified the bladder tumors very similarly (Table 1). The UNC classification (using 3 clusters; two basal subtypes and one luminal subtype) and MDA classifier (also using 3 clusters; two luminal subtypes and one basal subtype) came together to make a 4-cluster classifier described by TCGA, which later was expanded to 5-cluster classifier by addition of neuronal subtype. It is reassuring that the classifiers were consistent across these separate studies, despite difference in bladder cancer samples which confirms their clinicopathologic correlation.
Table 1.
Classification of bladder tumors
| Characteristics | TCGA | MDA | UNC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Luminal-papillary | Luminal-infiltrated | Luminal | Basal-squamous | Neuronal | Luminal | p53-like | Basal | Luminal | Basal | ||
| Luminal characteristics | High expression of FGFR3 | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||
| Low molecular weight keratins (KRT20) | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||
| High urothelial differentiation (FOXA1, GATA3, PPARG) | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||
| High uroplakins (UPK2, UPK1A) | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||||
| High immune infiltrates | ✓✓ | ✓✓ | ✓✓ | ||||||||
| High expression of immune markers (PD1, PDL1) | ✓✓ | ✓✓ | ✓✓ | ||||||||
| Papillary morphology | ✓✓ | ||||||||||
| Basal characteristics | High molecular weight keratins (KRT5, KRT6, KRT14) | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ||||||
| More stem-like phenotype | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||||
| Upregulated EMT markers | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||
| RB1 pathway alteration | ✓✓ | ✓✓ | ✓✓ | ✓✓ | |||||||
| p53 mutations | ✓✓ | ✓✓ | ✓✓ | ||||||||
| p63 activation | ✓✓ | ✓✓ | |||||||||
| High squamous differentiation markers | ✓✓ | ✓✓ | ✓✓ | ||||||||
| High neuronal differentiation | ✓✓ | ||||||||||
| Prognosis | Good | Intermediate | Intermediate | Poor | Poor | Good | Intermediate | Poor | Good | Poor | |
The BOLD taxonomy
In a more recent study, Thiery et al. identified six molecular subtypes (MC1–6, n>100 in each cluster) of bladder cancer based on the analysis of all publicly available datasets annotated as bladder cancer (data available till May 4, 2018) that included 2411 bladder cancer gene expression profiles, calling it bladder cancer subtypes of meta-cohort database (BOLD) taxonomy [12]. Subtype MC1 (neural-like) had similar properties to Lund SCC-like or TCGA neuronal subtypes. MC2 (luminal-like) was similar to NMIBC subtype reported by Hedegaard et al. (described below), and MC3 (papillary-like) was in conjunction with Lund urobasal or TCGA luminal-papillary. MC4 (HER2-like) was similar to Lund genomically unstable or TCGA luminal subtypes, and MC5 (SCC-like) had similarities with Lund SCC-like or MDACC basal or TCGA basal-squamous. Lastly, MC6 (mesenchymal-like) was similar to Lund infiltrated or MDACC p53-like or TCGA luminal-infiltrated.
NMIBC subtypes
Most studies of molecular tumor heterogeneity have either focused on MIBC or have analyzed NMIBC in combination with MIBC, which ascribes NMIBC as having either basal or luminal subtype membership. In a comprehensive study of early-stage bladder cancer, Hedegaard et al. reported three molecular subtypes of NMIBC in a comprehensive multicenter transcriptional analysis of 460 patients [13]. The class 1 subtypes had high expression of early cell-cycle genes while class 2 tumors had high expression of late cell-cycle genes. Class 1 and class 2 tumors had upregulated uroplakins. Class 3 tumors showed expression of CD44, KRT5 and KRT15. Thus, the class 1 and 2 subtypes showed luminal-like characteristics while the class 3 subtype showed basal-like characteristics.
Knowles et al from Leeds University identified two different subtypes of NMIBC based on DNA copy number profile by analyzing 140 primary stage Ta transitional tumors [14]. GS1 subtype consisted of no or few copy number alterations while GS2 consisted of cells with high copy number alterations and high chromosome 9 deletion. Note that these subtypes weren’t defined as expression subtypes but by their genetic alterations. GS2 subtypes also had overexpression of genes related to mTORC1 signaling. The GS2 subtype showed significant overlap with aforementioned class 2 NMIBC Hedegaard subtype.
Clinical implications of molecular subtyping
Radical cystectomy is the standard method of care for the treatment of non-metastatic MIBC and recurrent forms of NMIBC. Cisplatin-based neoadjuvant chemotherapy (NAC) is recommended prior to radical cystectomy with the aim of downstaging MIBC [15] with 15–40% patients responding completely to cisplatin-based chemotherapy [16–19]. Interestingly, the molecular subtypes discovered by the Lund, UNC, and MDACC groups, and TCGA displayed a varied response to platinum-based neoadjuvant chemotherapy and differential overall survival [9, 11, 20]. Whereas it was previously difficult to prospectively identify patients who would benefit from platinum-based neoadjuvant chemotherapy, molecular subtyping of MIBC has opened new doors to potential solutions to this problem. To that end, a commercially-available predictive tool, called Decipher Bladder, was developed to prospectively identify patients who might gain the greatest benefit from platinum-based neoadjuvant chemotherapy [20].
Seiler et al. used a pool of samples retrospectively collected from several institutions to train a single-sample genomic subtyping classifier (GSC) using differential biological characteristics and clinical outcomes, which is the basis for Decipher Bladder. The GSC identified four classes of muscle invasive bladder tumors: basal, basal claudin-low, luminal, and luminal infiltrated, in concordance with previously described classification methods with a high degree of accuracy. Among the four subtypes, patients with basal tumors received the greatest benefit from platinum-based neoadjuvant chemotherapy, which improved 3-year overall survival by almost 30%. This is significant due to the aggressiveness of the basal tumors and the poor overall survival associated with this subtype in the absence of chemotherapy. Interestingly, this marked improvement in overall survival was independent of pathological response, a departure from the existing literature which has consistently shown responder status (at cystectomy) and overall survival to be tightly correlated. A similar but attenuated effect was also seen in patients with basal claudin-low tumors, where NAC prior to radical cystectomy improved 3-year overall survival by almost 15% compared to radical cystectomy alone. Nonetheless, this subset of tumors bore a poor prognosis and should also be considered for novel therapeutic strategies in conjunction with platinum-based NAC [20]. Patients with luminal tumors experienced little additional overall survival benefit from NAC and their prognosis seemed to depend on the amount of immune infiltrate in the tumors. Whereas luminal tumors with no immune infiltrate bore the best prognosis with cystectomy alone, luminal tumors with high immune infiltrates bore poor prognosis irrespective of the treatment strategy [20]. With this in mind it is noteworthy that the PD-L1 inhibitor, azetolizumab, has shown significantly higher response rate in luminal infiltrated tumors [21]. It is possible that molecular subtype may be a useful predictive biomarker for both immune checkpoint therapy and cytotoxic chemotherapy, with non-overlapping groups benefitting from each treatment. Although tantalizing, this is only speculative right now.
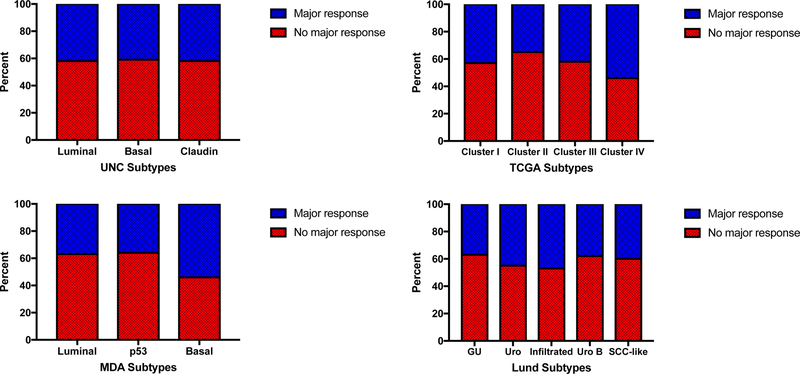
Decipher Bladder offers a novel route to a personalized approach in the treatment of MIBC. It does, however, have pitfalls that may limit its clinical value. Firstly, the Decipher Bladder data shows that patients with non-basal tumors do not receive additional survival benefits from NAC and the marketing literature suggests these patients should undergo radical cystectomy without NAC. However, the does not test associate with major pathologic responses to NAC (Figure 1). In the cohort receiving neoadjuvant chemotherapy (n=200), there was no difference in complete response or downstaging to NMIBC regardless of whether the GSC was used or any of the four academic classifiers. Lack association between subtype and pathologic response was confirmed in an independent study [22]. Clinical downstaging (absent complete pathologic response) appears to be important clinically regarding survival. Indeed, it has been shown that neoadjuvant chemotherapy patients achieving pathological downstaging at the time of cystectomy have an equal survival to those who achieve complete pathological response [23]. Therefore, these patients should not categorically be denied NAC based on the Decipher Bladder test. Secondly, there appears to be some plasticity among the subtype membership after selective therapies such as chemotherapy, and it is not known if other therapies (such as BCG) might also influence subtype membership [11]. Choi et al. observed that chemoresistant tumors that were originally classified as luminal switch subtypes to the p53-like subtype post chemotherapy [11]. This plasticity in tumor subtypes creates an additional pitfall in using molecular subtyping to guide clinical treatment strategies. In addition, it is clear from studies of other genitourinary or non-genitourinary cancers that multiple subtypes or clones may be present in the same tumor, and therefore sampling bias may artificially impact subtype assignment and inadvertently triage a patient into a suboptimal therapy [24–27].
Figure 1: Pathologic response to chemotherapy according to subtype.
Major pathologic response to neoadjuvant chemotherapy, defined as downstaging to NMIBC or pT0 status, was compared between subtypes using the UNC, MDA, TCGA, and Lund systems, from the cohorts in Seiler et al (reference 20). This analysis shows that rate of pathologic response, a well-known discriminator of survival after neoadjuvant chemotherapy is almost identical across subtypes.
Lastly, the conclusion that the test is useful for treatment allocation is based entirely on a survival benefit in the absence of a response correlation. Yet both the TCGA and the data from various intuitions is collected under a waiver of consent for retrospective review. Without consent, patients cannot be contacted directly to collect treatment or survival data. Therefore, data will be missing for all patients who do not have their follow up treatment or survival information documented at the index institution. This is a common situation for patients who undergo cystectomy at tertiary referral centers but ultimately receive chemotherapy and end of life care closer to home. In the absence of robust follow up data, pathologic complete response is an ideal outcome as it is highly correlated to survival [28]. In this context, it is counter-intuitive, and indeed diametrically opposed to level I clinical literature, to believe that molecular subtype associates with differences in survival despite pathologic complete response rates being the major driver of survival and these rates being equal in each subtype [29]. Lastly, the predictive power of this test has yet to be independently validated. Taken together, these caveats underscore that the general treatment guidelines recommended by the Decipher Bladder marketing materials are too broad and presumptive to be an effective clinical tool in guiding therapeutic strategies.
Bladder cancer subtypes are somewhat agnostic to the underlying genetics
It is interesting that expression subtypes are to a large degree agnostic of genetic alterations. While it is clear that FGR3 signaling and mutations play a key role in the luminal subtypes of tumors, FGFR3 is not always mutated in this subtype. Very few genes are enriched in one subtype or another, with the exceptions being TP53, Rb1, EP300, FGFR3, and PPARG. There are data from several groups, including our own, showing that genetic alterations are highly predictive of chemoresponse in bladder cancer to agents such as cisplatin [30] and EGFR inhibitors [31]. Ultimately, clinical implications of such biomarkers may be enhanced by combining the expression subtype and the mutational contexts of each tumor to precipitate a true era of personalized medicine in the treatment of urothelial cancer.
Conclusions
There is high concordance among classification systems according to expression profiling of urothelial cancers. In the absence of chemotherapy, there is broad agreement among the classifiers that basal classified tumors have poorer overall survival compared to luminal tumors. There is association between expression subtype and prognosis. However, the association between pathologic response to chemotherapy and molecular subtype (i.e. predictive biomarker) is not strong enough to warrant exemption from chemotherapy. As such, expression subtype has more utility as a prognostic biomarker than a predictive biomarker.
KEY POINTS:
Robust bladder cancer subtypes are consistently identified across platforms and institutions.
Bladder cancer subtypes are associated with unique clinicopathologic, genomic, and genetic features.
Bladder cancer subtypes may have an association with clinical outcome.
Bladder cancer subtypes do not associate with or predict chemotherapy response, and therefore may not be suitable to use in assigning treatment strategies.
ACKNOWLEDGEMENTS
FINANCIAL SUPPORT
Philip Abbosh: BCAN Young Investigator Award, NCI CA218976, Bucks County Board of Associates.
CONFLICTS OF INTEREST
David J. McConkey: patent pending on Methods of characterizing and treating molecular subsets of muscle-invasive bladder cancer. Research support (Astra-Zeneca); Advisory board (Janssen, Bioclin)
Elizabeth Plimack: Scientific Advisor (BMS, Genentech, Incyte, Janssen, Merck); Data Safety Monitoring (AstraZeneca, Pfizer); CME presentations (AUA, Clinical Care Options, Fox Chase Cancer Center, Medscape, NCCN, Omniprex, PriME Oncology, Research to Practice); Grants for clinical research (Astellas, AstraZeneca, BMS, Genentech, Merck, Peloton, Pfizer [Clovis])
Philip Abbosh: Advisory board (Janssen, Astra Zeneca)
REFERENCES
- 1.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2019. 2019. 69(1): p. 7–34. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, et al. , Molecular portraits of human breast tumours. Nature, 2000. 406: p. 747. [DOI] [PubMed] [Google Scholar]
- 3.Lindgren D, et al. , Combined Gene Expression and Genomic Profiling Define Two Intrinsic Molecular Subtypes of Urothelial Carcinoma and Gene Signatures for Molecular Grading and Outcome. Cancer Research, 2010. 70(9): p. 3463–3472. [DOI] [PubMed] [Google Scholar]
- 4.Dyrskjøt L, et al. , Identifying distinct classes of bladder carcinoma using microarrays. Nature Genetics, 2002. 33: p. 90. [DOI] [PubMed] [Google Scholar]
- 5.Sjödahl G, et al. , A Molecular Taxonomy for Urothelial Carcinoma. Clinical Cancer Research, 2012. 18(12): p. 3377–3386. [DOI] [PubMed] [Google Scholar]
- 6.Damrauer JS, et al. , Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proceedings of the National Academy of Sciences, 2014. 111(8): p. 3110–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi W, et al. , Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nature Reviews Urology, 2014. 11: p. 400. [DOI] [PubMed] [Google Scholar]
- 8.Kardos J, et al. , Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight, 2016. 1(3). [DOI] [PMC free article] [PubMed] [Google Scholar]; **Kardos et al identify a new subtype of tumors, those that are claudin-low which share characteristics with claudin-low breast tumors. These tumors are enriched for specific genetic abberations and are both inflamed and immunosuppressed. Authors propose that these tumors are ideal candidates for immune checkpoint inhibtion.
- 9.The Cancer Genome Atlas Research, N., et al. , Comprehensive molecular characterization of urothelial bladder carcinoma. Nature, 2014. 507: p. 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson AG, et al. , Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell, 2017. 171(3): p. 540–556.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi W, et al. , Identification of Distinct Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer with Different Sensitivities to Frontline Chemotherapy. Cancer Cell, 2014. 25(2): p. 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan TZ, et al. , Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-cohort Analysis of 2411 Tumors. European Urology, 2019. 75(3): p. 423–432. [DOI] [PubMed] [Google Scholar]; **This is the largest cohort of bladder cancers to date to which expresion cluster analysis was performed, identifying 6 subtypes with unique clinical, pathologic, genetic, and genomic features.
- 13.Hedegaard J, et al. , Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell, 2016. 30(1): p. 27–42. [DOI] [PubMed] [Google Scholar]
- 14.Hurst CD, et al. , Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell, 2017. 32(5): p. 701–715.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfred Witjes J, et al. , Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer . European Urology, 2017. 71(3): p. 462–475. [DOI] [PubMed] [Google Scholar]
- 16.Iyer G, et al. , Multicenter Prospective Phase II Trial of Neoadjuvant Dose-Dense Gemcitabine Plus Cisplatin in Patients With Muscle-Invasive Bladder Cancer. 2018. 36(19): p. 1949–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plimack ER, et al. , Accelerated Methotrexate, Vinblastine, Doxorubicin, and Cisplatin Is Safe, Effective, and Efficient Neoadjuvant Treatment for Muscle-Invasive Bladder Cancer: Results of a Multicenter Phase II Study With Molecular Correlates of Response and Toxicity. 2014. 32(18): p. 1895–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choueiri TK, et al. , Neoadjuvant Dose-Dense Methotrexate, Vinblastine, Doxorubicin, and Cisplatin With Pegfilgrastim Support in Muscle-Invasive Urothelial Cancer: Pathologic, Radiologic, and Biomarker Correlates. 2014. 32(18): p. 1889–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah JB, McConkey DJ, and Dinney CPN, New Strategies in Muscle-Invasive Bladder Cancer: On the Road to Personalized Medicine. Clinical Cancer Research, 2011. 17(9): p. 2608–2612. [DOI] [PubMed] [Google Scholar]
- 20.Seiler R, et al. , Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. European Urology, 2017. 72(4): p. 544–554. [DOI] [PubMed] [Google Scholar]; **Seiler et al describe in this publication the genomic subtyping classifer which would later be called Decipher Bladder using discovery and validation cohorts.
- 21.Rosenberg JE, et al. , Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. The Lancet, 2016. 387(10031): p. 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConkey DJ, et al. , A Prognostic Gene Expression Signature in the Molecular Classification of Chemotherapy-naïve Urothelial Cancer is Predictive of Clinical Outcomes from Neoadjuvant Chemotherapy: A Phase 2 Trial of Dose-dense Methotrexate, Vinblastine, Doxorubicin, and Cisplatin with Bevacizumab in Urothelial Cancer. European Urology, 2016. 69(5): p. 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zargar H, et al. , Final Pathological Stage after Neoadjuvant Chemotherapy and Radical Cystectomy for Bladder Cancer—Does pT0 Predict Better Survival than pTa/Tis/T1? 2016. 195(4 Part 1): p. 886–893. [DOI] [PubMed] [Google Scholar]
- 24.Gerlinger M, et al. , Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature Genetics, 2014. 46: p. 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlinger M, et al. , Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. 2012. 366(10): p. 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King H, et al. , Intratumoural evolutionary landscape of high-risk prostate cancer: the PROGENY study of genomic and immune parameters. Annals of Oncology, 2017. 28(10): p. 2472–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Bruin EC, et al. , Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science, 2014. 346(6206): p. 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrelli F, et al. , Correlation of Pathologic Complete Response with Survival After Neoadjuvant Chemotherapy in Bladder Cancer Treated with Cystectomy: A Meta-analysis. European Urology, 2014. 65(2): p. 350–357. [DOI] [PubMed] [Google Scholar]
- 29.Grossman HB, et al. , Neoadjuvant Chemotherapy plus Cystectomy Compared with Cystectomy Alone for Locally Advanced Bladder Cancer. New England Journal of Medicine, 2003. 349(9): p. 859–866. [DOI] [PubMed] [Google Scholar]
- 30.Plimack ER, et al. , Defects in DNA Repair Genes Predict Response to Neoadjuvant Cisplatin-based Chemotherapy in Muscle-invasive Bladder Cancer . European Urology, 2015. 68(6): p. 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhury NJ, et al. , Afatinib Activity in Platinum-Refractory Metastatic Urothelial Carcinoma in Patients With ERBB Alterations. Journal of Clinical Oncology, 2016. 34(18): p. 2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]