Abstract
Most of the studies focused on antimicrobial resistance (AMR) performed in wildlife describe Escherichia coli as the principal indicator of the selective pressure. In the present study, several species of Enterobacterales with a large panel of cephalosporin resistant (CR) genes have been isolated from wildlife in Catalonia. A total of 307 wild animals were examined to determine the prevalence of CR enterobacteria, AMR phenotypes and the presence of common carbapenem and CR genes. The overall prevalence of CR-phenotype was 13% (40/307): 17.3% in wild mammals (18/104) and 11.5% in wild birds (22/191) (p<0.01). Hedgehogs showed the highest prevalence (13.5% of 104) of the mammal specimens, and raptors the highest in bird specimen (7.3% of 191). Although CR E. coli was the most frequently isolated (45%), other CR- Enterobacterales like Klebsiella pneumoniae (20%), Citrobacter freundii (15%), Enterobacter cloacae (5%), Proteus mirabilis (5%), Providencia spp (5%) and Serratia marcescens (2.5%) were also isolated. A high diversity of CR genes was identified among the isolates, with 50% yielding blaCMY-2, 23% blaSHV-12, 20% blaCMY-1 and 18% blaCTX-M-15. Additionally, resistance to carbapenems associated to OXA-48 gene was found. Most of the CR isolates, principally K. pneumoniae and C. freundii, were multi-resistant with co-resistance to fluoroquinolones, tetracycline, sulphonamides and aminoglycosides. This study reports high prevalence of Enterobacterales harbouring a variety of CR genes and OXA-48 mediated-carbapenem resistance, all of them frequently associated to nosocomial human infections, for the first time in wild mammals and wild birds. Implementation of control measures to reduce the impact of anthropogenic pressure in the environment is urgently needed.
Introduction
In the last decades, the prevalence of opportunistic and antimicrobial resistant (AMR) bacteria associated with nosocomial infections has increased in hospital settings. The overuse of antibiotics in human and veterinary medicine have led to the spread of AMR pathogens, becoming a global health problem [1].
Extended-spectrum β-lactamases (ESBLs) and AmpC-type β-lactamases (AmpC) are the most common enzymes that confer resistance to broad-spectrum cephalosporins among members of the family Enterobacterales. These β-lactamases have extensively diversified in response to the clinical use of new generation drugs: cephalosporins, carbapenems and monobactams [2]. There are currently two classification systems for beta-lactamase enzymes. The first one classifies beta-lactamases according to the amino acid sequence [3,4]. The second classification, described by Bush and Jacoby (2010) is based on the functional activity of the enzymes. Within this classification, the group 1 contains cephalosporinases encoded in the chromosome of many Enterobacterales, such as AmpC, CMY, ACT, FOX and MIR. Some variants of these enzymes have also been detected in plasmids. The group 2 serine beta-lactamase represents the largest group with a broad spectrum against penicillins, cephalosporins, and carbapenems. They include the TEM, SHV, CTX, OXA and KPC enzymes. These enzymes are mostly encoded by genes located in plasmids that can be horizontally transferred to different bacteria genera [1]. Finally, the group 3 metallo-beta-lactamases (MBLs) are zinc dependent and include NDM, IMP, VIM and SPM enzymes [5].
Carbapenems are last-line beta-lactam antibiotics with the broadest spectrum of activity. Nowadays, carbapenems are commonly used in hospital settings for the treatment of life-threatening infections caused by Enterobacterales resistant to beta-lactamic drugs, including cephalosporins, monobactams and inhibitors of beta-lactamases. However, the emergence of resistance to carbapenems mediated by the production of carbapenemases has led to limited therapeutic options in human health [6]. The OXA-48 variant of carbapenemases is becoming highly prevalent in human clinical infections [7].
The dissemination of cephalosporin resistance (CR) has been studied widely in Enterobacterales from humans and livestock, whereas studies concerning the environment, including wildlife, are still lacking [2]. In recent years, an important increase of CR Escherichia coli has been reported in different epidemiological settings such as humans, pets, livestock, retail meat and the environment [8–13]. The study of wildlife as sentinel of the AMR environmental contamination has recently acquired more consideration worldwide [14]. However, most of the environmental-wildlife interface studies have been focused on wild birds, as principal AMR disseminators by their migratory routes, with a limited variety of AMR bacteria species described. Isolation of CR-carrying bacteria from wild birds has been globally reported in E. coli [15–20] and less frequently in Klebsiella pneumoniae [21]. All these results confirm the dissemination success of ESBL blaSHV-12 and blaCTX-M variants in wild birds worldwide. More recently, presence of CR E. coli has also been described in wild mammals, but at lower prevalence in comparison with wild birds [22].
In the present study, we report for the first time in Spain, the presence of diverse families of CR-encoding genes in a large variety of Enterobacterales including E. coli, K. pneumoniae, Citrobacter freundii, Enterobacter cloacae, Serratia marcescens and Proteus mirabilis- in wild mammals and wild birds. Furthermore, we describe the presence of carbapemenase resistant E. coli and P. mirabilis associated with the presence of OXA-48 variant in isolates of wildlife origin. These bacteria are frequently found in recurrent and severe urinary tract infections and other nosocomial infections in hospitals of Spain [7,23].
Material and methods
Study population
Wild animals attended at the Wildlife Rehabilitation Centre (WRC) of Torreferrusa (Catalonia, North-East Iberian Peninsula) were analysed between November 2016 and May 2017. This is a public WRC under the direction of the Catalan Wildlife-Service (“Direcció General de Polítiques Ambientals, Departament de Territori i Sostenibilitat of the Generalitat de Catalunya”). Sampling methods and handling protocols of animals were in agreement with the Catalan Wildlife Service who stipulates the management protocols and Ethical Principles according to the Spanish legislation [24]. All animals were examined and tested using cloacal or rectal swabs on arrival at the centre before receiving any pharmacologic or antimicrobial treatment. The most frequent cause of hospitalization was related to anthropogenic origin due to direct persecution (gunshot, poisoning, illegal captivity or traps) to involuntary human induced threats (collisions with vehicles, fences or electric lines and electrocution).
Microbiological analysis
Rectal and cloacal swabs were plated in MacConkey agar supplemented with ceftriaxone (1mg/L). Single colonies growing on the plate were subculture and identified biochemically using API (bioMérieux, Marcy l’Etoile, France) or VITEK 2 (bioMérieux, Spain) systems.
Antimicrobial susceptibility testing
Minimal inhibitory concentration (MIC) was performed using a commercial broth microdilution method (VetMIC GN-mo, SVA, Sweden) for the following antimicrobials: ampicillin (1 to 128 mg/liter), cefotaxime (0.016 to 2 mg/liter), ceftazidime (0.25 to 16 mg/liter), nalidixic acid (1 to 128 mg/liter), ciprofloxacin (0.008 to 1 mg/liter), gentamicin (0.12 to 16 mg/liter), streptomycin (2 to 256 mg/liter), kanamycin (8 to 16 mg/liter), chloramphenicol (2 to 64 mg/liter), florfenicol (4 to 32 mg/liter), trimethoprim (1 to 128 mg/liter), sulfamethoxazole (8 to 1,024 mg/liter), tetracycline (1 to 128 mg/liter), and colistin (0.5 to 4 mg/liter). The E. coli ATCC 25922 was used as control strain. Epidemiological cut-off values (ECOFF) selected were those described by the European Committee on Antimicrobial Susceptibility testing (EUCAST, https://mic.eucast.org/Eucast2/). For the combinations of species-antimicrobial with no cut-off values defined by EUCAST, ECOFF values were obtained from the British Society for Antimicrobial Chemotherapy (BSAC) or the Clinical and Laboratory Standards Institute (CLSI, 2017).
Molecular characterization of antimicrobial resistance genes
The detection of genes coding for ESBLs -blaCTX-M [25], blaTEM [26], blaSHV [27]-, AmpCs -blaCMY-1 [28], blaCMY-2 [29], carbapenemases -blaOXA-48, blaVIM, blaIMP, blaNDM and blaKPC [30]- and colistin-resistance genes mcr1-5 variants [31] was carried out using PCR as previously described (S1 Table).
Sanger DNA sequencing was done for blaTEM, blaSHV, blaCTX-M, and blaOXA PCR products at the Genomic and Bioinformatics Service of the Universitat Autònoma de Barcelona (Spain). Sequences and chromatograms were manually explored to trim bad-quality bases with BioEdit 7.2. Once the assembly of the consensus sequences was done, partial sequences were aligned using Clustal Omega program, and finally blasted against the public database (National Center for Biotechnology Information, NCBI). Allelic variants of the ESBL-resistance genes were determined based on these partial sequences, and AmpC genes were classified according to the CMY-1 and CMY-2 groups.
Statistical analysis
Descriptive analysis was performed under 95% confidence, using SPSS Advanced Models TM 15.0 (SPSS Inc. 233 South Wacker Drive, 11th Floor Chicago, IL 60.606–6412). The Chi-square test or Fisher exact test was used for comparison between proportions when appropriate. Statistically significant results were considered for unadjusted p-value < 0.05.
Results
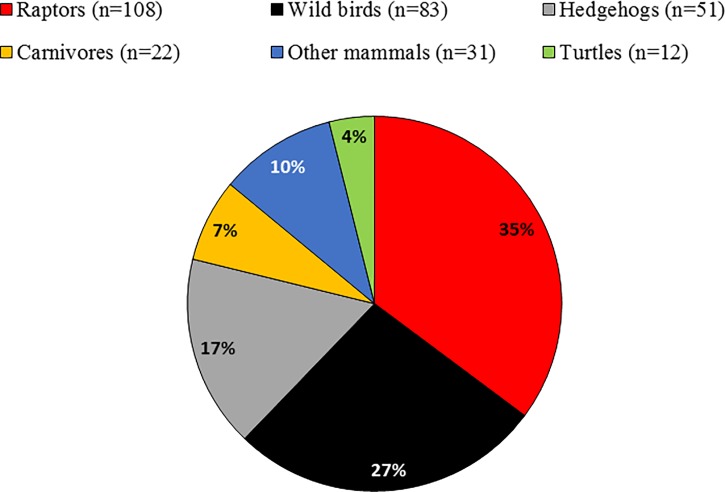
The sample size comprised 307 wild animals belonging to 67 different species grouped as birds (62%), mammals (34%) and reptiles (4%) (Fig 1). Animals came from different regions of Catalonia with a high density of urban areas and pig farming production.
Fig 1. Proportion of wildlife analysed in the study according to the zoological category.
Animal groups: raptors (different species of birds of prey and owls), wild birds (principally passerines and seagulls), insectivorous (European and Algerian hedgehogs), carnivores (mainly mustelids), and other mammals (wild boars and roe deer).
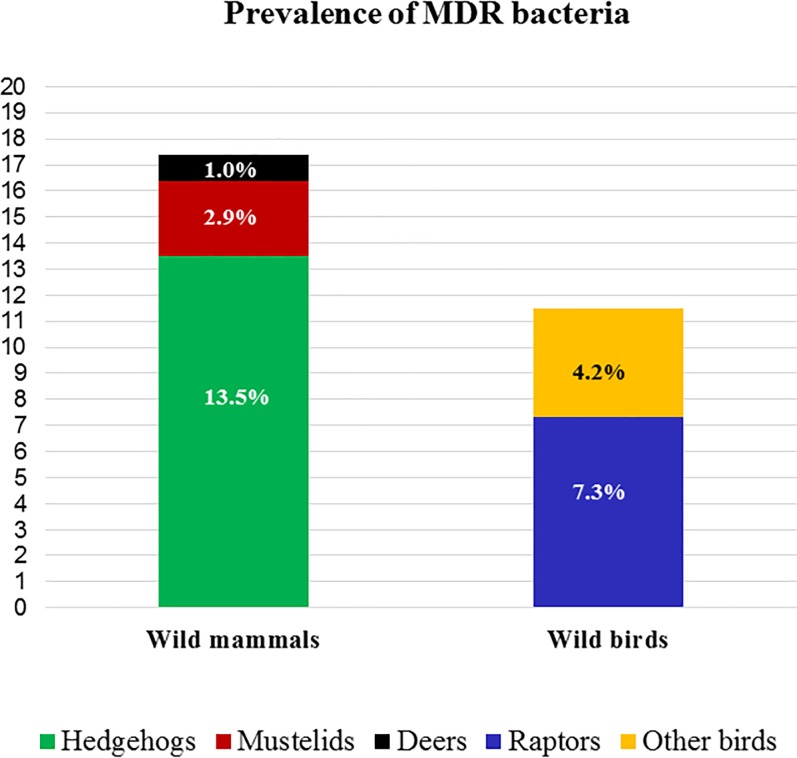
Ceftriaxone resistant isolates were detected in 65 out of the 307 (21%) faecal samples analysed. Of those, 40 harboured ESBL or AmpC-encoding genes, representing an overall prevalence of 13% (Fig 2). The prevalence of CR-carrying isolates was 17.3% in wild mammals (18/104) and 11.5% in wild birds (22/191). Within the mammal group, hedgehogs showed the largest prevalence of resistant isolates in comparison to the total mammal species examined (13.5%, 14/104, p = 0.022). Precisely, 67% of the Algerian (2/3) and 26% of the European (12/47) samples harboured CR-genes. Within the bird group, raptors represented the highest prevalence with 7.3% (14/191) of the total bird specimens [23% (14/60) of the raptor species examined] (Fig 2).
Fig 2. Prevalence of cephalosporin resistant (CR) bacteria in the different wildlife categories.
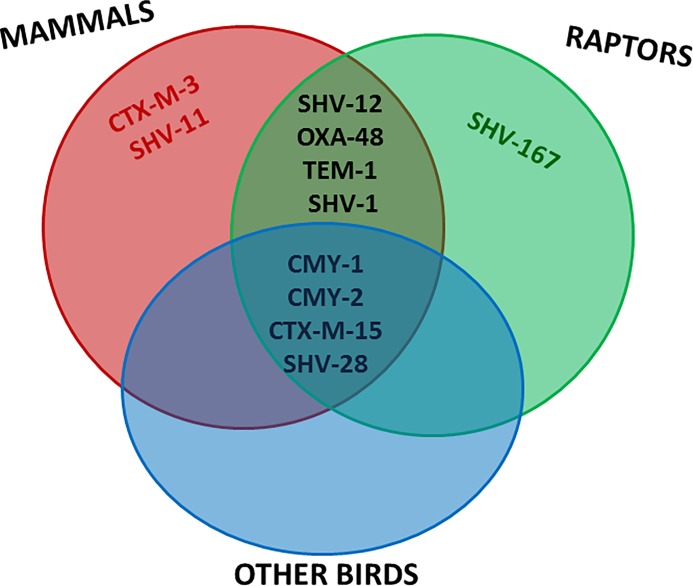
CR isolates belonged to several genera within the Enterobacterales order, with E. coli being detected most frequently (45%). Interestingly, other clinically relevant enterobacteria, including K. pneumoniae (20%), C. freundii (15%), E. cloacae (5%), P. mirabilis (5%), Providencia spp (5%) and S. marcescens (2.5%) were also identified as carriers of CR genes. The proportion of positive samples for AmpC-encoding genes was 65% (26/40) distributed in 27% blaCMY-1 and 38% blaCMY-2 families. Additionally, 65% (26/40) of the isolates exhibited ESBL genes with blaSHV-12 (9/40, 22.5%) and blaCTX-M-15 (6/40, 15%) representing 35% and 23% of the total ESBL respectively. Isolates from 12 animals presented the combination of both, ESBL and AmpC genes. Finally, mammals and raptors shared the largest part of the detected ESBL types, and other minority gene variants, such as blaCTX-M-3 and blaSHV-11 or blaSHV-167 were only detected in mammals or raptors, respectively (Fig 3).
Fig 3. Venn diagram showing the distribution of AMR genes in the different animal groups encountered in this study.
A high genetic diversity of CR encoding genes was observed in all Enterobacterales, with 40% (16/40) of the isolates harbouring 2 to 5 different resistance genes in the same isolate (Table 1). Furthermore, the carbapenemase-encoding gene OXA-48 was detected in E. coli and P. mirabilis isolated from European hedgehog and Barn owl, respectively (Table 1). Other carbapenemase-encoding genes tested were not found.
Table 1. Prevalence and antimicrobial resistance genotypes and phenotypes of beta-lactamase producing Enterobacterales detected in wildlife.
| Scientific name (common name) | Total sample | AMR genes | Bacterial spp | Drug-resistance genes | Resistance phenotype to non-Beta-lactams |
|
|---|---|---|---|---|---|---|
| Mammals (N = 104) | N | n | Prev | |||
| Aetechinus algirus (Algerian hedgehog) | 3 | 2 | 67% | Escherichia coli | CMY-2 | CIP, NAL, KAN, TMP |
| Klebsiella oxytoca | CTX-M-3 | GEN, ST, FF, CF, TET, COL, TMP | ||||
| Erinaceus europeus (European hedgehog) | 47 | 12 | 26% | Escherichia coli | CMY-2 | nd |
| Escherichia coli | CMY-2 | KAN | ||||
| Escherichia coli | CMY-2 | nd | ||||
| Escherichia coli | SHV-12 | STR | ||||
| Escherichia coli | SHV-11,OXA-48 | CIP, NAL, KAN, TET, SUL, TMP | ||||
| Klebsiella pneumoniae | CMY-1,CMY-2, SHV-1,TEM-1,CTX-M-15 | CIP, GEN, ST, KAN, TET, SUL, TMP | ||||
| Klebsiella pneumoniae | SHV-11, TEM-1 | CIP, NAL, GEN, STP, KAN, TET, SUL, TMP | ||||
| Klebsiella pneumoniae | SHV-28 | CIP, NAL, GEN, STR, KAN, TET, COL, SUL, TMP | ||||
| Klebsiella pneumoniae | SHV-12 | CIP, NAL, KAN, TET, SUL, TMP | ||||
| Citrobacter freundii | CMY-2, TEM-1 | CIP, NAL, KAN, TET, TMP | ||||
| Citrobacter freundii | CMY-2, SHV-12 | CIP, NAL, STR, KAN, TET, COL, SUL, TMP | ||||
| Citrobacter freundii | CMY-2 | CIP, NAL | ||||
| Capreolus capreolus (European roe deer) | 2 | 1 | na | Enterobacter cloacae | CMY-2 | SUL |
| Martes foina (Beech marten) | 2 | 1 | na | Citrobacter freundii | CMY-2, SHV-12 | CIP, NAL, GEN, TET, SUL, TMP |
| Meles meles (European badger) | 1 | 1 | na | Escherichia coli | SHV-12 | CIP, NAL, CHL, SUL, TMP |
| Mustela vison (American mink) | 13 | 1 | 8% | Enterobacter cloacae | CMY-2 | SUL |
| PREVALENCE IN MAMMALS | 104 | 18 | 17.3% | |||
| Raptors (n = 108) | N | n | Prev | |||
| Accipiter gentilis (northern goshawk) | 13 | 3 | 23% | Escherichia coli | TEM-1 | COL |
| Escherichia coli | CMY-2 | CIP, NAL | ||||
| Proteus mirabilis | CMY-1, CMY-2, SHV-28, TEM-1 | CIP, NAL, GEN, STR, KAN, TET, SUL, TMP | ||||
| Accipiter nisus (Eurasian sparrowhawk) | 8 | 3 | 38% | Escherichia coli | CMY-1, SHV-1, TEM-1, CTX-M-15 | CIP, NAL, KAN, TET, SUL, TMP |
| Escherichia coli | TEM-1 | CIP, TET, TMP | ||||
| Serratia marcensis | CMY-1, CTX-M-15 | CIP, TET, COL, SUL, TMP | ||||
| Bubo bubo (Eurasian eagle-owl) | 1 | 1 | na | Escherichia coli | CMY-1, SHV-167 | nd |
| Buteo buteo (Common buzzard) | 17 | 2 | 12% | Escherichia coli | SHV-12 | ST, CHL, TET, SUL, TMP |
| Providencia alcalifaciens | SHV-12 | CIP, NAL,GEN,ST,KAN, FF,CHL, TET,SUL, TMP | ||||
| Strix aluco (Tawny owl) | 18 | 3 | 17% | Klebsiella pneumoniae | CMY-2, SHV-28 | STR, SUL, TMP |
| Escherichia coli | CMY-2, SHV-1 | nd | ||||
| Klebsiella pneumoniae | SHV-12, CTX-M15 | CIP | ||||
| Tyto alba (Barn owl) | 3 | 2 | 67% | Escherichia coli | CMY-2 | CIP, NAL, STR, TET |
| Proteus mirabilis | SHV-12,TEM-1, OXA-48 | CIP, NAL, STR, KAN, CF, TET, COL, SUL,TMP | ||||
| Other birds (n = 83) | N | n | Prev | |||
| Carduelis carduelis (European goldfinch) | 12 | 1 | 8% | Citrobacter freundii | CMY-2 | CIP, NAL, GEN, STR, KAN, CHL, TET, SUL, TMP |
| Carduelis choris (European Greenfinch) | 2 | 1 | na | Klebsiella pneumoniae | CMY-1 | CIP, NAL, KAN, FF, CHL, SUL |
| Larus michahellis (Yellow-legged gull) | 7 | 1 | 14% | Escherichia coli | CTX-M-15 | CIP, NAL, GEN, KAN, TET, SUL, TMP |
| Serinus serinus (European serin) | 6 | 1 | 17% | Klebsiella pneumoniae | CMY-1, SHV-28 | CIP, NAL, STR, KAN, TET, SUL, TMP |
| Streptopelia decaocto (Eur. collared dove) | 1 | 1 | na | Citrobacter freundii | CMY-2 | FF, TMP |
| Sylvia melanocephala (Sardinian warbler) | 6 | 2 | 33% | Escherichia coli | CMY-2 | CIP, NAL |
| Providencia spp | CTX-M-15, CMY-1 | CIP, NAL, GEN, STR, KAN, CHL, TET, SUL, TMP | ||||
| Turdus merula (Common blackbird) | 8 | 1 | 13% | Escherichia coli | CMY-2 | CIP, NAL, KAN, TMP |
| PREVALENCE IN BIRDS | 191 | 22 | 11.5% | |||
CIP, Ciprofloxacin; NAL, Nalidixic acid; GEN, Gentamicin; STR, Streptomycin; KAN, Kanamycin; FF, Florfenicol; CHL, Chloramphenicol; TET, Tetracycline; COL, Colistin; SUL, Sulphametoxazole; TMP, Trimethoprim. nd, not detected.
Most of the ESBL/AmpC Enterobacterales isolated (92%), with the exception of E. cloacae, were multiresistant with a common resistance phenotype comprising β-lactams-quinolones-tetracycline-sulfamethoxazole/trimethoprim (Table 1). K. pneumoniae and C. freundii isolates both presented a multi-drug resistance profile including the resistance to aminoglycosides (Table 2). Moreover, 90% of the K. pneumoniae isolates were resistant to ciprofloxacin and sulphametoxazole, 70% to kanamycin, 55% to streptomycin, and 10% to florfenicol. Additionally, 83% of the tested C. freundii isolates exhibited resistance to trimethoprim and nalidixic acid and 67% to tetracycline (Table 2). Although none of the mcr- genes were detected in this study, the colistin resistant phenotype was observed in Klebsiella spp isolated from a European greenfinch and Algerian hedgehog, and in a Providencia spp isolated from a common buzzard.
Table 2. Minimal inhibitory concentration of E. coli, K. pneumoniae and C. freundii isolates of wildlife origin.
Dilution ranges for each antimicrobial are those contained within the white area. Vertical lines indicate epidemiological cut off values (ECOFF) or clinical breakpoints in those cases where ECOFF values have not been described.
| E. coli (n = 18) | |||||||||||||||
| MIC [μg/mL]: | 0.12 | 0.25 | 0.5 | 1 | >2 | 4 | 8 | 16 | >16 | 32 | 64 | >128 | 256 | >512 | R (%) |
| Ampicilin | 3 | 15 | 100 | ||||||||||||
| Cefotaxime | 1 | 1 | 1 | 2 | 12 | 1 | 89 | ||||||||
| Ceftazidime | 2 | 1 | 1 | 7 | 7 | 100 | |||||||||
| Ciprofloxacin | 6 | 2 | 2 | 8 | 67 | ||||||||||
| Nalidixic acid | 2 | 3 | 4 | 9 | 50 | ||||||||||
| Gentamicin | 9 | 8 | 1 | 5.5 | |||||||||||
| Streptomycin | 7 | 3 | 5 | 2 | 1 | 17 | |||||||||
| Kanamycin | 12 | 2 | 4 | 33 | |||||||||||
| Florfenicol | 4 | 12 | 2 | 0 | |||||||||||
| Chloramphenicol | 2 | 8 | 5 | 1 | 2 | 11 | |||||||||
| Tetracycline | 6 | 6 | 1 | 2 | 3 | 33 | |||||||||
| Colistin | 17 | 1 | 5.5 | ||||||||||||
| Sulphametoxazole | 2 | 9 | 1 | 1 | 5 | 28 | |||||||||
| Trimethoprim | 1 | 7 | 2 | 8 | 44 | ||||||||||
| K. pneumoniae (n = 9) | |||||||||||||||
| MIC [μg/mL]: | 0.12 | 0.25 | 0.5 | 1 | >2 | 4 | 8 | 16 | >16 | 32 | 64 | >128 | 256 | >512 | R (%) |
| Ampicilina | 1 | 8 | 100 | ||||||||||||
| Cefotaxime | 9 | 100 | |||||||||||||
| Ceftazidime | 3 | 6 | 100 | ||||||||||||
| Ciprofloxacin | 1 | 8 | 100 | ||||||||||||
| Nalidixic acida | 1 | 2 | 1 | 5 | 88 | ||||||||||
| Gentamicin | 1 | 5 | 3 | 33 | |||||||||||
| Streptomycin | 2 | 2 | 2 | 3 | ND | ||||||||||
| Kanamycina | 2 | 7 | 78 | ||||||||||||
| Florfenicol | 1 | 7 | 1 | ND | |||||||||||
| Chloramphenicola | 1 | 5 | 1 | 1 | 1 | 22 | |||||||||
| Tetracycline | 1 | 1 | 2 | 1 | 4 | 56 | |||||||||
| Colistin | 8 | 1 | 11 | ||||||||||||
| Sulphametoxazoleb | 1 | 8 | 89 | ||||||||||||
| Trimethoprimb | 1 | 2 | 6 | 67 | |||||||||||
| C. freundii (n = 6) | |||||||||||||||
| MIC [μg/mL]: | 0.12 | 0.25 | 0.5 | 1 | >2 | 4 | 8 | 16 | >16 | 32 | 64 | >128 | 256 | >512 | R (%) |
| Ampicilina | 1 | 5 | 100 | ||||||||||||
| Cefotaxime | 1 | 5 | 83 | ||||||||||||
| Ceftazidime | 1 | 1 | 4 | 83 | |||||||||||
| Ciprofloxacinb | 1 | 5 | 0 | ||||||||||||
| Nalidixic acida | 1 | 5 | 83 | ||||||||||||
| Gentamicin | 1 | 1 | 2 | 2 | 33 | ||||||||||
| Streptomycin | 1 | 1 | 2 | 1 | 1 | ND | |||||||||
| Kanamycinb | 3 | 3 | 50 | ||||||||||||
| Florfenicol | 3 | 2 | 1 | ND | |||||||||||
| Chloramphenicola | 1 | 2 | 1 | 1 | 1 | 33 | |||||||||
| Tetracycline | 2 | 1 | 2 | 1 | 67 | ||||||||||
| Colistina | 4 | 1 | 1 | 33 | |||||||||||
| Sulphametoxazoleb | 2 | 1 | 3 | 50 | |||||||||||
| Trimethoprimb | 1 | 5 | 83 | ||||||||||||
EUCAST (ECOFF values WT): AMP ≤8, CTX ≤0.25, CAZ ≤0.5 (≤1 C.freundii), CIP ≤0.064 (≤0.125 K.pneumoniae), NAL ≤16, GEN ≤2, STR≤ 16, KAN ≤8, FFL ≤16, CHL ≤16, TET ≤8, COL ≤2, SMX ≤64, TMP ≤2 (≤8 K. pneumoniae and C. freundii).
aBSAC and
bCLSI clinical break points: CIP ≤1 Enterobacteriaceae, SMX Susceptible ≤256 K. pneumoniae and C. freundii; TMP ≤8 K. pneumoniae and C. freundii. ND, not determined due to lack of ECOFF or clinical breakpoint values available. CLSI: Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
Discussion
This study identifies for the first time a high percentage of wild mammals and wild birds as carriers of potential nosocomial Enterobacterales harbouring diverse ESBL, CMY and OXA-48 genes. Moreover, most of the isolates principally K. pneumoniae and C. freundii, presented a high prevalence of resistance also to fluoroquinolones.
In general, E. coli is the most reported ESBL/pAmpC-producing enterobacteria worldwide, with increasing frequency from animals, food, environmental sources and humans. In recent years, CR- E. coli transmission has been reported in different hosts, demonstrating a close human-animal ESBL/pAmpC gene similarity between livestock (broilers and pigs) and personnel working at the farms [13]. Additionally, similar CR genes have been reported between isolates from the community and those from human clinical settings, sewage water and wild birds [13].
Although ESBL transmission has been studied extensively in Enterobacterales from humans and livestock, data on antimicrobial resistance in the environment is still limited [2]. Moreover, most of the studies related to ESBL-carrying bacteria in wildlife are focused on the wild bird population and mainly restricted to E. coli species [32]. Several studies conducted in E. coli from avian species have identified blaCTX-M-1, blaCTX-M-14, blaCTX-M-15 and blaSHV-12 as the predominant ESBL types circulating in Spain [15, 33–36], Portugal [37], Tunisia [20], The Netherlands [38], Poland [39] and the Czech Republic [40]. Contrarily, in the present study, blaCTX-M-1 and blaCTX-M-14 were not detected in our avian species, but blaSHV-12 and blaCTX-M-15 were the most frequent ESBL types identified not only in E. coli but also in K. pneumoniae and C. freundii isolates. K. pneumoniae has been described in low prevalence (average 1.5%) in wild gulls from different European countries [41–43], including wild migratory birds from Spain, which exhibited blaCTX-M-15 ESBL-producing K. pneumoniae [36]. Interestingly, both blaCTX-M-15 and blaSHV-12 are also currently the most predominant genes in human clinical specimens from community and health care-associated infections in Spain [44,45]. Thus, the human community could potentially be a source of ESBL environmental contamination, through water contaminated with human sewage from urban areas and hospital settings.
In this study, blaCMY-1 group was principally detected in E. coli, K. pneumoniae, Proteus and Providencia spp from avian wildlife, like hawks, owls and small forest birds. Although this is an unusual variant in Spain, the presence in the present study might be explained by those species feeding habits. Raptors are predators occupying the top of the food chain; therefore, they can acquire AMR from a wide variety of preys (mammals, birds, reptiles or scavenging livestock). Moreover, some of these raptors are migratory species, being exposed to different environmental habitats in their migratory movements. In consequence, the role of migratory raptors as disseminators of these AMR traits is a serious concern to be further investigated.
Regarding blaCMY-2, it is the most common CMY type reported worldwide [46]. In this study, blaCMY-2 group was highly detected in E. coli and K. pneumoniae from hedgehogs and wild birds. Plasmid mediated genes can spread easily to other organisms. C. freundii, Enterobacter and Serratia spp in this study presented genes of the CMY-2 family. Since these types of AmpC genes are chromosomally encoded in some of these bacteria species, we cannot conclude the plasmidic nature of such enzymes. However, for epidemiological studies, it is important to report this type of resistance since these Enterobacterales can be involved in severe nosocomial infections and they all presented a MDR profile, except for E. cloacae.
Surprisingly, European hedgehogs represented an important reservoir of ESBL/AmpC-producing E. coli and other Enterobacterales, especially for blaCMY-2 (67%) and blaSHV-12 (25%) in this study. Our results are in agreement with previous studies conducted in Spain reporting low to moderate (1.3%-10%) prevalence of blaCMY-2 and blaSHV-12 E. coli variants in hedgehogs, deer and minks [22,47]. It is important to highlight that hedgehogs are in close contact with humans (home range including gardens), but also with livestock in the countryside, which could explain their acquisition of these AMR types.
Plasmid-mediated colistin resistance by mcr-1 has been reported worldwide in Enterobacterales isolated from humans, livestock, companion animals, food and wildlife [48]. Colistin has been used in veterinary medicine during the last decades for the treatment of gastrointestinal infections in livestock, principally in pigs and poultry [49]. Consequently, livestock is considered the main reservoir of mcr-1 selection and dissemination worldwide. Recent works disclosed the relationship among mcr-1-harbouring E. coli isolates recovered from the environment, pig production and human clinical isolates, demonstrating the rapidly evolving epidemiology of plasmid-mediated colistin-resistant E. coli strains worldwide and the importance of the One Health approach [50,51]. In our study, some Klebsiella and Providencia spp isolates were phenotypically resistant to colistin, but no mcr-associated genes were detected in any of the examined isolates.
Information about carbapenem-resistant Enterobacterales is very scarce in wildlife and has only been reported in avian species [36,52]. In this study, we report the presence of blaOXA-48 in E. coli and P. mirabilis isolates from a European hedgehog and a Barn owl, respectively. The presence of blaOXA-48 in wild mammals and birds in Catalonia is highly indicative of the wide environmental pollution of this variant, commonly reported in hospitals in Spain [53].
To our knowledge, there are no reports in wildlife, especially in wild mammals, describing the presence of ESBL genes in such a variety of Enterobacterales, like Klebsiella spp, Citrobacter spp, Serratia spp, or Enterobacter spp in Spain. C. freundii, is considered an opportunistic pathogen, associated with nosocomial infections, especially in patients who have been hospitalized for a prolonged period of time. In the last years, this bacterium has been classified as an emerging health problem associated to urinary tract infections commonly diagnosed in healthcare settings [54]. E. cloacae has been reported as important opportunistic and multi-resistant pathogen involved in outbreaks of hospital-acquired infections worldwide [55,56], including Spain [57]. ESBL- S. marcescens has also been classified as one of the top ten priority pathogens causing infections in intensive care units [58].
The high prevalence of CR Enterobacterales encountered in this study is really concerning, since wildlife is not directly exposed to any antimicrobial agent. Therefore, faecal contamination of water or soil with MDR bacteria and/or antimicrobial residues can lead to a selection pressure. Wastewaters from urban areas and hospitals have been identified as one of the major sources of AMR environmental contamination [2]. High prevalence of blaSHV-12 but also blaTEM-1 and blaCTX-M-1 alleles have been reported in aquatic environments (urban waters, natural or artificial water reservoirs, seawater or drinking water) in several countries worldwide, likely due to their relatively easy transmission to surface water through waste water treatment plant discharges [2,59]. In our study, wildlife in close contact with urban and farming areas of Catalonia carried a large variety of zoonotic/nosocomial bacteria genetically resistant to β-lactams-quinolones-tetracycline-sulfamethoxazole/trimethoprim-aminoglycosides with similar resistant genes to those found in livestock and clinical settings. However, further studies are needed to assess clonal relatedness among different cephalosporin and carbapenem resistant enterobacteria at the human-animal-environment interface.
Conclusions
This study describes for the first time a high prevalence of Enterobacterales harbouring a large variety of ESBL in addition to carbapenem resistant OXA-48 genes in wild mammals, remarkably in hedgehogs, and wild birds in Catalonia (northeast Spain). AmpC CMY-2 group and the ESBL genes blaSHV-12 and blaCTX-M-15 were the most frequent types identified in E. coli, K. pneumoniae and C. freundii isolates. These results support the concept that wildlife is a good sentinel of AMR environmental contamination and underline the importance of the One Health approach since wildlife can contribute indirectly to the dissemination of resistance genes into other natural environments.
Supporting information
F, sense primer; R, antisense primer; bp, base pairs.
(DOCX)
Acknowledgments
Our grateful thanks to the Torreferrussa WRC staff. A.C. was student of the Master´s Degree in Zoonosis and One Health (UAB). The authors are also grateful to the Centres de Recerca de Catalunya (CERCA) Programme.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The contract of LMG was supported by the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) and the European Social Fund. AV was supported by a PIF grant from the Universitat Autònoma de Barcelona. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–66. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- 2.Liakopoulos A, Mevius D, Ceccarelli D. A Review of SHV Extended-Spectrum β-Lactamases: Neglected Yet Ubiquitous. Front Microbiol. 2016;7:1374. 0.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond. 1980; B289:321–331. [DOI] [PubMed] [Google Scholar]
- 4.Ambler RP, Coulson AFW, Frère JM, Ghuysen JM, Joris B, Forsman M, et al. A standard numbering scheme for the class A-lactamases. Biochem J. 1991;276:269–272. 10.1042/bj2760269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–76. 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25:682–707 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andraseviæ AT, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 17:153–163. 10.1016/S1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- 8.Jouini A, Vinué L, Slama KB, Sáenz Y, Klibi N, Hammami S, et al. Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrob Chemother. 2007;60:1137–1141 10.1093/jac/dkm316 [DOI] [PubMed] [Google Scholar]
- 9.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L, et al. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev. 2010;34:295–316 10.1111/j.1574-6976.2009.00198.x [DOI] [PubMed] [Google Scholar]
- 10.Wieler LH, Ewers C, Guenther S, Walther B, Lübke-Becker A. Methicillin-resistant staphylococci (MRS) and extended spectrum-betalactamases (ESBL)-producing Enterobacteriaceae in companion animals: nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. Int J Med Microbiol. 2011;301:635–641 10.1016/j.ijmm.2011.09.009 [DOI] [PubMed] [Google Scholar]
- 11.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother. 2013;68:60–67 10.1093/jac/dks349 [DOI] [PubMed] [Google Scholar]
- 12.Sallem RB, Gharsa H, Slama KB, Rojo-Bezares B, Estepa V, Porres-Osante N, et al. First detection of CTX-M-1, CMY-2, and QnrB19 resistance mechanisms in fecal Escherichia coli isolates from healthy pets in Tunisia. Vector Borne Zoonotic Dis. 2013; 13:98–102 10.1089/vbz.2012.1047 [DOI] [PubMed] [Google Scholar]
- 13.Dorado-García A, Smid JH, van Pelt W, Bonten MJM, Fluit AC, van den Bunt G, et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J Antimicrob Chemother. 2018;73(2):339–347. 10.1093/jac/dkx397 [DOI] [PubMed] [Google Scholar]
- 14.Huijbers PM, Blaak H, de Jong MC, Graat EA, Vandenbroucke-Grauls CM, de Roda Husman AM. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ Sci Technol. 2015;49(20):11993–2004 10.1021/acs.est.5b02566 [DOI] [PubMed] [Google Scholar]
- 15.Alcalá L, Alonso CA, Simón C, González-Esteban C, Orós J, Rezusta A, et al. Wild Birds, Frequent Carriers of Extended-Spectrum β-Lactamase (ESBL) Producing Escherichia coli of CTX-M and SHV-12 Types. Microb Ecol. 2016;72(4):861–869. 10.1007/s00248-015-0718-0 [DOI] [PubMed] [Google Scholar]
- 16.Hasan B, Laurell K, Rakib MM, Ahlstedt E, Hernandez J, Caceres M, et al. Fecal Carriage of Extended-Spectrum β-Lactamases in Healthy Humans, Poultry, and Wild Birds in León, Nicaragua-A Shared Pool of bla(CTX-M) Genes and Possible Interspecies Clonal Spread of Extended-Spectrum β-Lactamases-Producing Escherichia coli. Microb Drug Resist. 2016;22(8):682–687. Epub 2016 Mar 23. 10.1089/mdr.2015.0323 . [DOI] [PubMed] [Google Scholar]
- 17.Parker D, Sniatynski MK, Mandrusiak D, Rubin JE. Extended-spectrum β-lactamase producing Escherichia coli isolated from wild birds in Saskatoon, Canada. Lett Appl Microbiol. 2016;63(1):11–5. 10.1111/lam.12589 [DOI] [PubMed] [Google Scholar]
- 18.Atterby C, Börjesson S, Ny S, Järhult JD, Byfors S, Bonnedahl J. ESBL-producing Escherichia coli in Swedish gulls-A case of environmental pollution from humans? PLoSOne. 2017;12(12):e0190380 10.1371/journal.pone.0190380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohsin M, Raza S, Schaufler K, Roschanski N, Sarwar F, Semmler T, et al. High Prevalence of CTX-M-15-Type ESBL-Producing E. coli from Migratory Avian Species in Pakistan. Front Microbiol. 2017;8:2476 10.3389/fmicb.2017.02476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben Yahia H, Ben Sallem R, Tayh G, Klibi N, Ben Amor I, Gharsa H, et al. Detection of CTX-M-15 harboring Escherichia coli isolated from wild birds in Tunisia. BMC Microbiol. 2018;18(1):26 10.1186/s12866-018-1163-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raza S, Mohsin M, Madni WA, Sarwar F, Saqib M, Aslam B. First Report of bla(CTX-M-15)-Type ESBL-Producing Klebsiella pneumoniae in Wild Migratory Birds in Pakistan. Ecohealth. 2017;14(1):182–186. 10.1007/s10393-016-1204-y [DOI] [PubMed] [Google Scholar]
- 22.Alonso CA, Alcalá L, Simón C, Torres C. Novel sequence types of extended-spectrum and acquired AmpC beta-lactamase producing Escherichia coli and Escherichia clade V isolated from wild mammals. FEMS Microbiol Ecol. 2017;93(8). 10.1093/femsec/fix097 [DOI] [PubMed] [Google Scholar]
- 23.Cantón R, Loza E, Aznar J, Castillo FJ, Cercenado E, Fraile-Ribot PA, et al. ; SMART-Spain Working Group. Monitoring the antimicrobial susceptibility of Gram-negative organisms involved in intraabdominal and urinary tract infections recovered during the SMART study (Spain, 2016 and 2017). Rev Esp Quimioter. 2019;32(2):145–155 [PMC free article] [PubMed] [Google Scholar]
- 24.Spanish. R.D.1201/2005 of the Ministry of Presidency of Spain (10th October 2005). BOE 21st October 2005. Available: www.boe.es/boe/dias/2005/10/21/pdfs/A34367-34391.pdf. Accessed 2018 Dec 27.
- 25.Miro E, Navarro F, Mirelis B, et al. Prevalence of clinical isolates of Escherichia coli producing inhibitor-resistant b-lactamases at a University Hospital in Barcelona, Spain, over a 3-year period. Antimicrob Agents Chemother. 2002; 46: 3991–4. 10.1128/AAC.46.12.3991-3994.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olesen I, Hasman H, Aarestrup FM. Prevalence of b-lactamases among ampicillin resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb Drug Resist. 2004; 10: 334–40. 10.1089/mdr.2004.10.334 [DOI] [PubMed] [Google Scholar]
- 27.Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol. Lett. 1997;152:163–67. 10.1016/s0378-1097(97)00196-1 [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Kwon Y, Pai H, Kim JW, Cho DT. Survey of Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J.Clin. Microbiol, 1998;36:1446–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. 2005. b-Lactamases among extended-spectrum b-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother. 2005;56:115–121. 10.1093/jac/dki190 [DOI] [PubMed] [Google Scholar]
- 30.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–23. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 31.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes Euro Surveill. 2018; 23: 17–00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther S, Ewers C, Wieler LH. Extended-Spectrum Beta-Lactamases Producing E. coli in Wildlife, yet another Form of Environmental Pollution? Front Microbiol. 2011;2:246 10.3389/fmicb.2011.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simões RR, Poirel L, Da Costa PM, Nordmann P. Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg Infect Dis. 2010;16(1):110–2. 10.3201/eid1601.090896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vergara A, Pitart C, Montalvo T, Roca I, Sabaté S, Hurtado JC, et al. Prevalence of Extended-Spectrum-β-Lactamase- and/or Carbapenemase-Producing Escherichia coli Isolated from Yellow-Legged Gulls from Barcelona, Spain. Antimicrob Agents Chemother. 2017. January 24;61(2). pii: e02071–16. 10.1128/AAC.02071-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alonso CA, Michael GB, Li J, Somalo S, Simón C, Wang Y, et al. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J Antimicrob Chemother. 2017;72(6):1589–1596. 10.1093/jac/dkx024 [DOI] [PubMed] [Google Scholar]
- 36.Oteo J, Mencía A, Bautista V, Pastor N, Lara N, González-González F, et al. Colonization with Enterobacteriaceae-Producing ESBLs, AmpCs, and OXA-48 in Wild Avian Species, Spain 2015–2016. Microb Drug Resist, 2018;24:932–938 10.1089/mdr.2018.0004 [DOI] [PubMed] [Google Scholar]
- 37.Costa D, Poeta P, Sáenz Y, Vinué L, Rojo-Bezares B, Jouini A, et al. Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J Antimicrob Chemother. 2006;58(6):1311–2. 10.1093/jac/dkl415 [DOI] [PubMed] [Google Scholar]
- 38.Veldman K, van Tulden P, Kant A, Testerink J, Mevius D. Characteristics of cefotaxime-resistant Escherichia coli from wild birds in the Netherlands. Appl Environ Microbiol. 2013;79(24):7556–61. 10.1128/AEM.01880-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, et al. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microbiol. 2010;76(24):8126–34. 10.1128/AEM.01446-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dolejská M, Bierosová B, Kohoutová L, Literák I, Cízek A. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J Appl Microbiol. 2009;106(6):1941–50. 10.1111/j.1365-2672.2009.04155.x [DOI] [PubMed] [Google Scholar]
- 41.Bonnedahl J, Stedt J, Waldenström J, Svensson L, Drobni M, Olsen B. Comparison of Extended-Spectrum β-Lactamase (ESBL) CTX-M Genotypes in Franklin Gulls from Canada and Chile. PLoS One. 2015;10(10):e0141315 10.1371/journal.pone.0141315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stedt J, Bonnedahl J, Hernandez J, Waldenström J, McMahon BJ, Tolf C, et al. Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand. 2015;57:74 10.1186/s13028-015-0166-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnedahl J, Hernandez J, Stedt J, Waldenström J, Olsen B, Drobni M. Extended-spectrum β-lactamases in Escherichia coli and Klebsiella pneumoniae in Gulls, Alaska, USA. Emerg Infect Dis. 2014;20(5):897–9. 10.3201/eid2005.130325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Díaz MA, Hernández-Bello JR, Rodríguez-Baño J, Martínez-Martínez L, Calvo J, Blanco J, et al. Spanish Group for Nosocomial Infections (GEIH). Diversity of Escherichia coli strains producing extended-spectrum beta-lactamases in Spain: second nationwide study. J Clin Microbiol. 2010;48(8):2840–5. 10.1128/JCM.02147-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merino I, Shaw E, Horcajada JP, Cercenado E, Mirelis B, Pallarés MA, et al. ITUBRAS-GEIH-SEIMC Group. CTX-M-15-H30Rx-ST131 subclone is one of the main causes of healthcare-associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J Antimicrob Chemother. 2016;71(8):2125–30. 10.1093/jac/dkw133 [DOI] [PubMed] [Google Scholar]
- 46.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22(1):161–82 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cristóvão F, Alonso CA, Igrejas G, Sousa M, Silva V, Pereira JE, et al. Clonal diversity of extended-spectrum beta-lactamase producing Escherichia coli isolates in fecal samples of wild animals. FEMS Microbiol Lett. 2017;364(5). [DOI] [PubMed] [Google Scholar]
- 48.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21:30155 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- 49.EMA. Updated Advice on the Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. 2016. http://www.ema.europa.eu/docs/en_GB/document_library/Press_release/2016/07/WC500211081.pdf. [Google Scholar]
- 50.Guenther S, Falgenhauer L, Semmler T, Imirzalioglu C, Chakraborty T, Roesler U, et al. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J Antimicrob Chemother. 2017;72(5):1289–1292. 10.1093/jac/dkw585 [DOI] [PubMed] [Google Scholar]
- 51.García-Meniño I, García V, Mora A, Díaz-Jiménez D, Flament-Simon SC, Alonso, et al. J. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of mcr-1 ST10 and ST131 E. coli Isolates. Front Microbiol. 2018; 9:2659 10.3389/fmicb.2018.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer J, Schmoger S, Jahn S, Helmuth R, Guerra B. NDM-1 carbapenemase-producing Salmonella enterica subsp. enterica serovar Corvallis isolated from a wild bird in Germany. J Antimicrob Chemother. 2013;68(12):2954–6. 10.1093/jac/dkt260 [DOI] [PubMed] [Google Scholar]
- 53.Aracil-García B, Oteo-Iglesias J, Cuevas-Lobato Ó, Lara-Fuella N, Pérez-Grajera I, Fernández-Romero S, et al. Spanish Group European Antimicrobial Resistance Surveillance network (EARS-Net). Rapid increase in resistance to third generation cephalosporins, imipenem and co-resistance in Klebsiella pneumoniae from isolated from 7,140 blood-cultures (2010–2014) using EARS-Net data in Spain. Enferm Infecc Microbiol Clin. 2017;35(8):480–486 10.1016/j.eimc.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 54.Ranjan KP, Ranjan N. Citrobacter: An emerging health care associated urinary pathogen. Urology Annals. 2013;5(4):313–314. [PMC free article] [PubMed] [Google Scholar]
- 55.Davin-Regli A, Pagès JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Frontiers Microbiol. 2015;6:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-Resistant Enterobacter cloacae Complex Emerging as a Global, Diversifying Threat. Front Microbiol. 2019;10:44 Review 10.3389/fmicb.2019.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernández J, Montero I, Martínez Ó, Fleites A, Poirel L, Nordmann P, Rodicio MR. Dissemination of multiresistant Enterobacter cloacae isolates producing OXA-48 and CTX-M-15 in a Spanish hospital. Int J Antimicrob Agents. 2015; 46(4):469–74. 10.1016/j.ijantimicag.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 58.Dessì A, Puddu M, Testa M, Marcialis MA, Pintus MC, Fanos V. Serratia marcescens infections and outbreaks in neonatal intensive care units. J Chemother. 2009. November;21(5):493–9. 10.1179/joc.2009.21.5.493 [DOI] [PubMed] [Google Scholar]
- 59.Rashid M, Rakib MM, Hasan B. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect Ecol Epidemiol. 2015;5:26712 10.3402/iee.v5.26712 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
F, sense primer; R, antisense primer; bp, base pairs.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.