Abstract

We report the site-specific and covalent bioconjugation of fluorescent polymer chains to proteins in live cells using the HaloTag technology. Polymer chains bearing a Halo-ligand precisely located at their α-chain-end were synthesized in a controlled manner owing to the RAFT polymerization process. They were labeled in lateral position by several organic fluorophores such as AlexaFluor 647. The resulting Halo-ligand polymer probe was finally shown to selectively recognize and label HaloTag proteins present at the membrane of live cells using confocal fluorescence microscopy. Such a polymer bioconjugation approach holds great promises for various applications ranging from cell imaging to cell surface functionalization.
Introduction
Fluorescence microscopy is a powerful technique that is widely used to image living organisms and better understand cell functions and the associated biomolecular processes. This requires engineered fluorescent probes able to specifically label biomolecules such as proteins in their natural environment. Of particular interest is the study of protein dynamics and interactions at the membrane of live cells that often relies on exogenous probes.1 Several parameters have to be taken into account considering that the constraints of live cell imaging differ from those of in vitro studies.2 Techniques relying on the labeling of isolated proteins are indeed hardly transposable as they require the fluorescently labeled protein to be injected after re-purification using methods such as microinjection or electroporation.3 For live cell imaging, less invasive methods are usually preferred, but the challenge is to obtain a selective labeling of the target proteins.
The fluorescent probe needs to adequately fulfill a fine compromise between physicochemical, photophysical, and biological properties. First, it should be water-soluble, should show adequate diffusion properties, and should not be toxic. Moreover, it should be highly bright in the selected wavelength range together with a rather small size and exhibit good photostability (e.g., resistance to photobleaching). In addition, in live cell conditions, the fluorescent probe should promote a very efficient and specific labeling of the target protein.
Recently, we have developed a new family of tunable fluorescent polymer probes that presents many interesting properties.4 These probes are highly water-soluble and biocompatible, highly bright, and resistant to photobleaching.4c The polymer chain size can be controlled owing to the RAFT polymerization process. In addition, we have already shown that such polymer chains can be functionalized at their chain end to promote an oriented conjugation onto native proteins5 or histidine-tagged proteins in vitro via a nitrilotriacetic acid (NTA) ligand.6
Considering these properties, the objective of the present study was to design new fluorescent polymer probes that covalently and selectively label proteins in live cell conditions. We chose the HaloTag technology,7 which is particularly well suited for in vivo labeling.8 Based on the use of a modified haloalkane dehalogenase protein tag (HaloTag), this strategy allows the covalent and site-specific conjugation of synthetic haloalkane ligands (Halo-ligands) onto the protein of interest (fused with the HaloTag). This irreversible and specific coupling has recently drawn much attention for the purification of proteins with high yield and efficiency,9 for the grafting of proteins onto various surfaces,7a,10 and for protein labeling with small-molecule ligands.7a,11 During the preparation of this manuscript, Liu et al. reported on this strategy to engineer cancer cells with glycopolymers for immunotherapy.12 To the best of our knowledge, no fluorescent polymer probes have been developed for the direct observation of HaloTag proteins in live cells by fluorescence microscopy. Our aim was thus to functionalize fluorescent polymer chains with the Halo-ligand and evaluate their ability to specifically and selectively label HaloTag proteins in live cell conditions (Figure 1).
Figure 1.
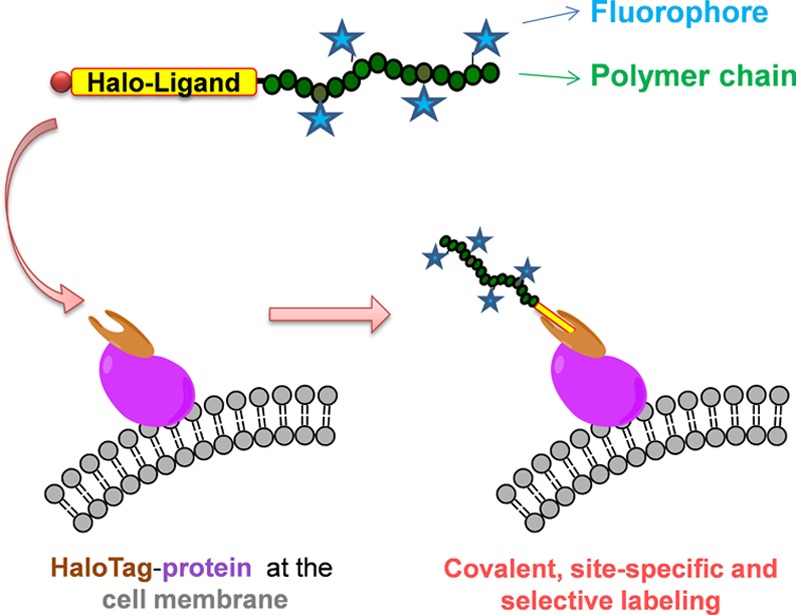
Schematic representation of the conjugation of the fluorescent polymer probe (Halo-ligand–polymer-AF647) with a HaloTag protein expressed at the cell membrane.
Results and Discussion
Halo-ligand-terminated polymer chains were synthesized by RAFT controlled radical polymerization via a new RAFT chain transfer agent (CTA) functionalized with the Halo-ligand (Halo-CTA; Figure 2A). Indeed, we already showed that the use of functionalized CTAs is a highly efficient strategy to introduce entities of interest at the α-chain-end of polymer chains.6,13 An amino-modified Halo-ligand with a tetra(oligoethylene glycol) spacer was first reacted with a precursor RAFT agent (bearing an activated ester function) developed in our laboratory.13b We chose this spacer to better separate the polymer chain bearing the fluorophores and the haloalkane moiety that should efficiently enter and react inside the HaloTag binding pocket of the recombinant protein. The expected Halo-CTA (Figure 2A) was successfully obtained (75% yield after purification) as confirmed by 1H NMR and high-resolution mass spectrometry analyses (see the Supporting Information).
Figure 2.
Structures of the (A) Halo-ligand-functionalized RAFT chain transfer agent (Halo-CTA) and the Halo-ligand chain-end-functionalized (B) homopolymer and (C) reactive copolymer synthesized in this study.
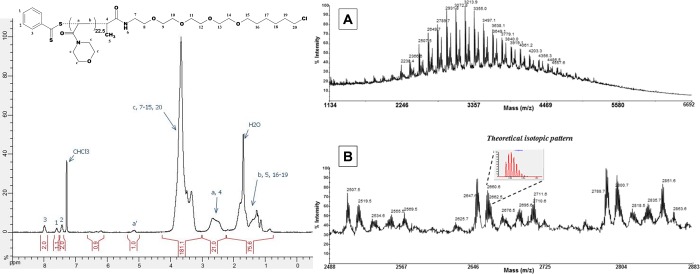
The Halo-CTA was then used to control radical homo- and copolymerizations via the RAFT process. Homopolymerization of N-acryloylmorpholine (NAM) was first performed following a previously optimized procedure.14 A low-molecular-weight poly(N-acryloylmorpholine) (PNAM; Figure 2B) sample was purposely synthesized to precisely investigate chain-end functionalization by 1H NMR and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) analyses (after efficient purification of the polymer sample by precipitation in diethyl ether).
The 1H NMR spectrum (Figure 3, left) exhibited the broad peaks corresponding to NAM repeating units and the expected chains ends. Dithiobenzoate (DB) ω-end-group protons gave three characteristic peaks between 7.3 and 8 ppm and a −CH–S peak at 5.1 ppm (denoted a′). Protons of the Halo-ligand α-end-group were superimposed to the polymer peaks but distinguishable. Oligoethylene glycol and haloalkane protons were superimposed with morpholine protons between 3 and 4 ppm and backbone methylene protons between 1 and 2 ppm. Integration of DB chain-end protons could be used to determine the number-average molecular weight (Mn) of the polymer sample. The NMR Mn value (3700 g mol–1) was in good agreement with the expected one (3560 g mol–1) calculated from eq 1.
Figure 3.
Halo-ligand-functionalized PNAM homopolymer sample (24% monomer conversion, Mn,NMR = 3700 g·mol–1). (Left) 1NMR spectrum in CDCl3. (Right) MALDI-TOF mass spectrum in the positive reflectron mode. (A) Full spectrum. (B) Enlargement showing monoisotopic peaks (inset: theoretical isotopic pattern).
These results were further confirmed by MALDI-TOF MS analysis of the same sample (Figure 3, right). The polymer distribution centered on 3500 m/z and spanning from ca. 1500 to 7000 m/z was composed of a series of peaks spaced by one NAM repetitive unit (ca. 141 m/z). Positive reflectron mode analysis revealed the presence of the expected Halo-ligand-PNAM-DB chains. For instance, for DP = 15, theoretical monoisotopic mass for [C17H33O5NCl(C7H11NO2)15C7H5S2 + Na+], C129H203S2N16O35ClNa: 2658.4 m/z; found: 2658.6 m/z. The isotopic distribution (impacted by the presence of sulfur and chlorine atoms) is in good agreement with the expected one (inset in Figure 3B, right). The additional major populations corresponded to the K+ cationization of the expected product and the usual side products formed inside the MALDI-TOF mass spectrometer during the analysis of dithiobenzoate-terminated chains.15 Indeed, we showed, in a previous study, that fragmentation of the dithiobenzoate ω-chain-end occurs due to the weakness of the carbon–dithiobenzoate C–S bond.16 The main side product was clearly identified as Halo-ligand-PNAM-H chains terminated by a proton. Theoretical monoisotopic mass for [C17H33O5NCl(C7H11NO2)16H + Na+], C129H210N17O37ClNa: 2647.5 m/z; found: 2647.6 m/z.
Altogether, NMR and MALDI-TOF MS analyses demonstrated that the RAFT homopolymerization of NAM with the new Halo-CTA is characterized by good control over molecular weights and chain-end functionalization. Importantly, MALDI-TOF MS analyses demonstrated that the Halo-ligand moiety at the polymer α-chain-end is retained intact after polymerization (and purification).
The Halo-CTA was then used to control the copolymerization of NAM with N-acryloxysuccinimide (NAS) at a 60:40 molar ratio (Figure 2C). We indeed showed that this is an azeotropic composition for this particular co-monomer pair, leading to poly(NAM-stat-NAS) copolymers with regularly distributed activated ester groups along the polymer backbone.17
Polymerization kinetics was followed by 1H NMR (see Methods). A 90% monomer conversion was obtained in less than 2 h (Figure 4, left) with an induction period shorter than 15 min.18 These results were fully consistent with previous copolymerizations performed with other RAFT control agents such as tert-butyl dithiobenzoate.17 NAM and NAS individual conversions were identical at all time points, confirming the azeotropic conditions.
Figure 4.
RAFT copolymerization of NAM and NAS in the presence of the Halo-CTA. (Left) Monomer consumption kinetics. (Right) Evolution of experimental Mn and dispersity (Đ) values with conversion.
Experimental Mn values determined by both 1H NMR and SEC/MALLS analyses of purified polymer samples were in good agreement and also correlated with the calculated Mn values from eq 1 (Figure 4, right). Dispersity remained very low (Đ < 1.1), as expected for well-controlled RAFT polymerization.
In a last synthetic step, fluorescent labeling of polymer chains bearing the Halo-ligand at the α-chain-end (Figure 5D) could be carried out following our previously reported procedure.5 We chose AlexaFluor 647 fluorophore (AF647), which is a very convenient far-red dye for in cellulo studies and especially well adapted to dSTORM super-resolution microscopy.19 A Halo-ligand-poly(NAM-stat-NAS) copolymer sample (Mn = 10 600 g mol–1; Đ = 1.04) was first reacted with amino-modified AF647. The average number of AF647 fluorophores per polymer chain, nf, was controlled by stoichiometry, such coupling reaction leading to a very high yield (>95%) at room temperature. Here, we aimed at four AF647 fluorophores per polymer chain, and the actual value determined experimentally by SEC/UV4a was nf = 4.0, confirming that dye coupling onto the lateral activated esters of the polymer backbone is much faster than the concurrent aminolysis of the dithiobenzoate ω-chain-end.13b After capping the residual activated ester functions of the copolymer with aminoethylmorpholine (AEM), the resulting fluorescent polymer probe (Halo-ligand–polymer-AF647) was purified by dialysis and lyophilized (Mn = 14,400 g mol–1). It is worth noting that this capping procedure also leads to the aminolysis of the polymer dithiobenzoate ω-chain-end that may represent a source of nonspecific interaction in live cells and cytotoxicity.20
Figure 5.
Synthesis of (D) the Halo-ligand–polymer-AF647 fluorescent polymer probe from (C) the well-defined reactive copolymer precursor.
The Halo-ligand–polymer-AF647 probe was finally assessed for its ability to efficiently label HaloTag fusion proteins in live cell conditions. Our objective was to investigate whether our polymer probe leads to (i) a selective labeling of HaloTag proteins (Figure 1) and (ii) the absence of nonspecific cell labeling. As our polymer probe is fluorescent, this study could be performed in a straightforward manner using confocal fluorescence microscopy after a simple incubation of the probe with cells.
As we previously reported that such polymer probes bearing AF647 fluorophores (but without the Halo-ligand) were not cell permeant,5 this study was conducted with HeLa S3 cells engineered21 to express the HaloTag at the cell surface with a GPI-anchor (Halo-GPI cells) (see Methods). Expression of the HaloTag transgene in the Halo-GPI cells was either induced with 50 nM doxycycline (Dox) for 48 h or not induced (negative control) followed by incubation with two types of fluorescent probes. Our Halo-ligand–polymer-AF647 probe was compared with a commercial non-cell permeant Halo-ligand labeled with a molecular dye (HaloTag AlexaFluor 488, Promega; Figure S5).
When the HaloTag expression was Dox-induced, confocal microscopy images showed that the two fluorescent probes nicely labeled the cell surface, however with a much brighter signal for the polymer probe (Figure 6, middle and bottom panels). In addition, still in the presence of the fluorescent polymer probe, the absence of a fluorescent signal when HaloTag expression was not induced (Figure 6, top panels) highlighted the specificity of the interaction. Similar to the commercial probe, the fluorescent polymer probe did not cause a significant background signal in the absence of HaloTag. The probe did not label the cell membrane or other cell compartments in a nonspecific manner.
Figure 6.

Confocal fluorescence microscopy: HeLa S3 cells were incubated for 2 h with the Halo-ligand probes, HaloTag AlexaFluor 488 (0.05 μM), or Halo-ligand–polymer-AF647 probe (0.5 μM), as indicated. HeLa S3 cells were not induced (without doxycycline) (top panels) or induced (with doxycycline) (middle panels) to express the Halo-GPI transgene. Scale bars = 10 μm. Lower panels are zoom-in images of the middle panels.
This study thus shows that the synthesized fluorescent polymer probe bearing a Halo-ligand chain-end very selectively and efficiently recognizes the HaloTag in live cells. A very bright labeling of proteins expressing the HaloTag at the cell membrane was observed by confocal fluorescence microscopy, while there was no nonspecific labeling in the absence of the HaloTag. Such polymer–protein conjugation is covalent and site-specific and ensures that only one polymer chain is bound per protein. This attractive approach thus holds great promise in applications ranging from cell surface functionalization to cell imaging. In particular, Halo-ligand–polymer fluorescent probes may be especially useful in the field of super-resolution light microscopy.
Methods
Materials
N-Acryloyl morpholine (NAM) (Aldrich, 97%) was distilled under reduced pressure (120 °C; 10 mm Hg) to remove the inhibitor. N-Acryloxysuccinimide (NAS) was synthesized as previously described.22 2,2′-Azobis(isobutyronitrile) (AIBN) (Fluka, 98%) was purified by recrystallization from ethanol. 1,4-Dioxane (Acros, 99%) was distilled over LiAlH4 (110 °C). Succinimidoxycarbonylethyl dithiobenzoate (SEDB) was synthesized and purified according to a previously published protocol.13a Trioxane (Acros, 99%), diisopropylethylamine (DIPEA; Sigma-Aldrich, ≥99.5%), 4-(2-aminoethyl)morpholine (AEM; Aldrich, 99%), AlexaFluor 647 cadaverine (AF647, Life Technologies), HaloTag amine (O4) ligand (Halo-ligand-NH2, Promega, France), HaloTag AlexaFluor 488 (Promega), anhydrous dimethylformamide (DMF, Fisher Scientific, 99.99%), pentane (Fisher Scientific), ethyl acetate (Fisher Scientific), silica gel (Sigma-Aldrich), MgSO4 (Sigma-Aldrich), and chloroform (Fisher Scientific) were used as received.
Analytical Techniques
SEC/MALLS
Size exclusion chromatography with multi-angle laser light scattering detection (SEC/MALLS) was performed with a Mixed-C PLgel column (5 μm pore size) and an LC-6A Shimadzu liquid pump (1 mL min–1). Online double detection was provided by a differential refractometer (Waters DRI 410) and a MiniDAWN TREOS three-angle (46°, 90°, 133°) light scattering detector (Wyatt Technologies) operating at 658 nm. Analyses were run by injection of chloroform polymer solutions (3 g L–1, 70 μL). The specific refractive index increment (dn/dc) of the polymers in chloroform (0.130 mL g–1) was previously determined with an NFT-ScanRef interferometer operating at 633 nm. The molecular weight distribution data were obtained via the Wyatt ASTRA SEC/LS software package.
1H NMR
Spectra were acquired at 300 K on a Bruker Ultrashield spectrometer operating at 300.13 MHz.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
Mass spectra were acquired in the reflectron mode using a Voyager-DE PRO (AB Sciex, Framingham, MA) equipped with a nitrogen laser emitting at 337 nm. Ions were accelerated to a 20 kV final potential. Positive ions were detected, and mass spectra were a sum of 300 shots. External mass calibration was provided by a mixture of peptides (Sequazyme standards kit, AB Sciex). Samples were prepared by dissolving polymers in THF at a 10 g L–1concentration. The matrix, 3-indoleacrylic acid (IAA; Sigma-Aldrich, used without further purification), was dissolved in THF at 0.25 M. Matrix and polymer solutions were mixed at a 9:1 (v/v) ratio, and then 1 μL of the mixture was deposited and dried onto the MALDI sample plate.
Synthesis of the Halo-Ligand-Functionalized CTA (Halo-CTA)
To a chloroform solution (0.8 mL) of SEDB (35.2 mg, purity of >93%, 103 μmol, 1.2 equiv), a chloroform solution (1.4 mL) of Halo-ligand-NH2 (86 μmol, 1 equiv) and then DIPEA (30.2 μL, 172 μmol, 2 equiv) was added. The mixture was placed under magnetic stirring at room temperature for 4 h. The orange reaction medium was then diluted with 10 mL of chloroform and the product adsorbed on 6 g of silica gel. After three washes with 20 mL of chloroform and three washes with 20 mL of pentane, the product was recovered with 3 × 50 mL of ethyl acetate. The organic solution was washed with 2 × 250 mL of water, 250 mL of a pH 10 NaOH aqueous solution, and 250 mL of a saturated NaCl aqueous solution, with 2 × 250 mL of deionized water, dried over MgSO4, and evaporated. The final product (red oil) was finally dried under vacuum (yield, 75%).
1H NMR (300.13 MHz, CDCl3; Figure S1): 7.96 (d, 2H, Ar–H), 7.52 (t, 1H, Ar–H), 7.36 (t, 2H, Ar–H), 6.90 (Br, 1H, NH), 4.64 (q, 1H, −CH−), 3.44–3.62 (m, 20H, −CH2–O– and −CH2–Cl), 3.52 (m, 2H), 1.3–1.8 (m, 8H, −CH2−), 1.60 (d, 3H, −CH3).
High-resolution ESI-QTOF mass spectrometry: C24H38ClN05S2; m/z [M + H]+, calcd: 520.1953; found: 520.1935; m/z [M + Na]+, calcd 542.1772; found, 542.1746 (Figure S2).
RAFT Polymerizations
RAFT polymerization protocols were adapted from previously optimized procedures.14,17
Halo-Ligand-PNAM-DB Homopolymers
NAM (1.50 mmol), Halo-CTA (16.3 μmol), AIBN (2.8 μmol), trioxane (0.13 mmol) (internal reference determination of monomer conversion), and dioxane (0.56 mL) were mixed in a Schlenk tube equipped with a magnetic stirrer. The solution was degassed by five freeze–evacuate–thaw cycles before heating at 80 °C under nitrogen in a thermostated oil bath.
Halo-Ligand-P(NAM-stat-NAS) Copolymers
Statistical copolymerization of NAM and NAS monomers followed the same procedure, starting with NAM (2.17 mmol), NAS (1.45 mmol), Halo-CTA (27.8 μmol), AIBN (9.61 μmol), trioxane (0.18 mmol), and dioxane (1.54 mL). Samples were occasionally withdrawn from the polymerization medium via a cannula for analyses.
In both cases, individual monomer conversions were determined by 1H NMR following the decrease in the NAM and NAS vinyl protons in comparison with the trioxane singlet.22 The spectrum acquired at t0 before heating up the reaction mixture allowed us to precisely quantify the monomer/RAFT agent ratio and thus calculate the expected molecular weight of the polymer sample at 100% conversion using eq 1.17
| 1 |
where [NAM]0 and MNAM (141.17 g mol–1) are the initial molar concentration and the molecular weight of the NAM monomer, respectively, [NAS]0 and MNAS (169.13 g mol–1) are the initial molar concentration and the molecular weight of the NAS monomer, respectively, ConvNAM and ConvNAS are the NAM and NAS conversions at time t, respectively, and [CTA]0 and MCTA (520.15 g mol–1) are the initial molar concentration and the molecular weight of the Halo-CTA, respectively.
After the synthesis, polymers were purified by precipitation in diethyl ether and dried under vacuum up to a constant weight.
Synthesis of the Fluorescent Polymer Probe (Halo-Ligand–Polymer-AF647)
Halo-ligand-P(NAM-stat-NAS)-DB (Mn = 10 600 g mol–1, Đ = 1.04; 0.47 μmol, 1 equiv) and AF647 (1.89 μmol, 4 equiv) were dissolved in DMF (0.25 mL). DIPEA (3.77 μmol, 8 equiv) was subsequently added, and the blue solution was stirred for 2 h at room temperature. AEM (56.6 μmol, 3 equiv per NAS unit) was then added, and the reaction was left for another 2 h under stirring at room temperature. The resulting polymer Halo-ligand–polymer-AF647 was purified by dialysis (2 kDa MWCO, SpectraPor) and finally dried by lyophilization.
HaloTag Expressing Cells
A reverse tetracycline (Tet)-dependent transactivator (rtTA) cassette,23 under the control of a bidirectional promoter with ΔLNGFR in the reverse orientation for selection purposes,21a in a lentiviral vector was employed to render cells Tet (and thus Dox)-responsive. Transgene cassettes were genomically integrated in a subsequent manner into HeLa S3 cells by lentiviral delivery, essentially as described.21a Cells with genomic integrations of transgenes, of which the genotype was confirmed by PCR single-locus technology, were cloned by limiting dilution, as described,21b to yield the cell lines used in this study, inducibly expressing HALO-GPI.
The HALO-GPI cassette was created by replacing GFP with the HaloTag in a previously described24 construct with the vitronectin signal sequence at the N-terminus and the C-terminus, the C-terminal portion of human uPAR, which, upon expression, is removed from the protein construct to be replaced by a GPI anchor.
Cell Culture
Hela S3 cells were cultured in DMEM (Thermo Fischer Scientific, Monza, Italy) containing glutamax (1 mM), 5% Tet-System-approved fetal bovine serum (FBS, Takara, Jesi, Italy), 100 U mL–1 penicillin, and 100 mg mL–1 streptomycin. Cells were routinely tested (on a monthly basis) and found to be mycoplasm-free by the use of a standard diagnostic PCR. Expression of transgenes was induced with Dox at the indicated concentrations.
Confocal Fluorescence Microscopy
After incubation with the Halo-ligand probes, cells were fixed in 4% paraformaldehyde for 10 min at room temperature and washed in PBS. Nuclei were stained with Hoechst diluted in PBS for 10 min at room temperature in the dark. After washing in PBS, coverslips were mounted on microscope glass slides with Mowiol. Light microscopy images were acquired at the UltraView spinning disk confocal microscope operated by Volocity software (PerkinElmer), essentially as described.21b A ×63 objective was used. HaloTag AlexaFluor 488 and Halo-ligand–polymer-AF647 were excited with the 488 and 640 nm laser lines, respectively.
Acknowledgments
D.D. acknowledges a Ph.D. grant from the French Ministry of Research and Education. We thank Agnès Crépet (Laboratoire d’Ingénierie des Matériaux Polymères) and the Liquid Chromatography Platform (Institut de Chimie de Lyon) for their expertise and technical support in SEC/MALLS characterization of the polymers. E.v.A.. acknowledges support from the Giovanni Armenise-Harvard Foundation, the Associazione Italiana per la Ricerca sul Cancro (MFAG 13584), and the Ministero della Salute (F-2011-02352852/PE-2011-02352286).
Glossary
Abbreviations
- RAFT
reversible addition–fragmentation chain transfer
- NMR
nuclear magnetic resonance
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- GPI
glycosylphosphatidylinositol
- Dox
doxycycline
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01643.
Halo-CTA characterization, complementary NMR analyses of Halo-ligand poly(NAM-stat-NAS) copolymers and Halo-ligand-polymer-AF647 probe, structure of the HaloTag AlexaFluor 488 (Promega) reference probe (PDF)
Author Present Address
# Department of Chemistry, Loughborough University, UK.
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Marks K. M.; Nolan G. P. Chemical labeling strategies for cell biology. Nat. Methods 2006, 3, 591. 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- a Griffin B. A.; Adams S. R.; Tsien R. Y. Specific Covalent Labeling of Recombinant Protein Molecules Inside Live Cells. Science 1998, 281, 269. 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]; b Crawford R.; Torella J. P.; Aigrain L.; Plochowietz A.; Gryte K.; Uphoff S.; Kapanidis A. N. Long-lived intracellular single-molecule fluorescence using electroporated molecules. Biophys. J. 2013, 105, 2439. 10.1016/j.bpj.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K.; Feramisco J. R. Microinjection and localization of a 130K protein in living fibroblasts: a relationship to actin and fibronectin. Cell 1980, 19, 587. 10.1016/S0092-8674(80)80035-3. [DOI] [PubMed] [Google Scholar]
- a Cepraga C.; Gallavardin T.; Marotte S.; Lanoe P.-H.; Mulatier J.-C.; Lerouge F.; Parola S.; Lindgren M.; Baldeck P. L.; Marvel J.; Maury O.; Monnereau C.; Favier A.; Andraud C.; Leverrier Y.; Charreyre M.-T. Biocompatible Well-Defined Chromophore-Polymer Conjugates for Photodynamic Therapy and Two-Photon Imaging. Polym. Chem. 2013, 4, 61. 10.1039/C2PY20565C. [DOI] [Google Scholar]; b Relogio P.; Bathfield M.; Haftek-Terreau Z.; Beija M.; Favier A.; Giraud-Panis M.-J.; D’Agosto F.; Mandrand B.; Farinha J. P. S.; Charreyre M.-T.; Martinho J. M. G. Biotin-end-functionalized highly fluorescent water-soluble polymers. Polym. Chem. 2013, 4, 2968. 10.1039/c3py00059a. [DOI] [Google Scholar]; c Adjili S.; Favier A.; Fargier G.; Thomas A.; Massin J.; Monier K.; Favard C.; Vanbelle C.; Bruneau S.; Peyriéras N.; Andraud C.; Muriaux D.; Charreyre M.-T. Biocompatible photoresistant far-red emitting, fluorescent polymer probes, with near-infrared two-photon absorption, for living cell and zebrafish embryo imaging. Biomaterials 2015, 46, 70. 10.1016/j.biomaterials.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Duret D.; Haftek-Terreau Z.; Carretier M.; Berki T.; Ladavière C.; Monier K.; Bouvet P.; Marvel J.; Leverrier Y.; Charreyre M. T.; Favier A. Labeling of native proteins with fluorescent RAFT polymer probes: application to the detection of a cell surface protein using flow cytometry. Polym. Chem. 2018, 9, 1857. 10.1039/C7PY02064C. [DOI] [Google Scholar]
- Duret D.; Haftek-Terreau Z.; Carretier M.; Ladavière C.; Charreyre M.-T.; Favier A. Fluorescent RAFT polymers bearing a nitrilotriacetic acid (NTA) ligand at the α-chain-end for the site-specific labeling of histidine-tagged proteins. Polym. Chem. 2017, 8, 1611. 10.1039/C6PY02222G. [DOI] [Google Scholar]
- a Los G. V.; Encell L. P.; McDougall M. G.; Hartzell D. D.; Karassina N.; Zimprich C.; Wood M. G.; Learish R.; Ohana R. F.; Urh M.; Simpson D.; Mendez J.; Zimmerman K.; Otto P.; Vidugiris G.; Zhu J.; Darzins A.; Klaubert D. H.; Bulleit R. F.; Wood K. V. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008, 3, 373. 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]; b England C. G.; Luo H.; Cai W. HaloTag Technology: A Versatile Platform for Biomedical Applications. Bioconjugate Chem. 2015, 26, 975. 10.1021/acs.bioconjchem.5b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng J.-C.; Benink H. A.; McDougall M. G.; Chico-Calero I.; Kung A. L. In Vivo Fluorescent Labeling of Tumor Cells with the HaloTag Technology. Curr. Chem. Genom. 2012, 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Motejadded H.; Kranz B.; Berensmeier S.; Franzreb M.; Altenbuchner J. Expression, One-Step Purification, and Immobilization of HaloTag Fusion Proteins on Chloroalkane-Functionalized Magnetic Beads. Appl. Biochem. Biotechnol. 2010, 162, 2098. 10.1007/s12010-010-8985-1. [DOI] [PubMed] [Google Scholar]; b Locatelli-Hoops S.; Sheen F. C.; Zoubak L.; Gawrisch K.; Yeliseev A. A. Application of HaloTag technology to expression and purification of cannabinoid receptor CB2. Protein Expression Purif. 2013, 89, 62. 10.1016/j.pep.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa I.; Berkovich R.; Alegre-Cebollada J.; Badilla C. L.; Rivas-Pardo J. A.; Taniguchi Y.; Kawakami M.; Fernandez J. M. Nano-mechanics of HaloTag Tethers. J. Am. Chem. Soc. 2013, 135, 12762. 10.1021/ja4056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlag B.; Beutel O.; Janning D.; Czarniak F.; Richter C. P.; Kommnick C.; Göser V.; Kurre R.; Fabiani F.; Erhardt M.; Piehler J.; Hensel M. Single molecule super-resolution imaging of proteins in living Salmonella enterica using self-labelling enzymes. Sci. Rep. 2016, 6, 31601. 10.1038/srep31601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Jiang S.; Liu B.; Yu Y.; Zhao Z.-A.; Wang C.; Liu Z.; Chen G.; Chen H. Take Immune Cells Back on Track: Glycopolymer-Engineered Tumor Cells for Triggering Immune Response. ACS Macro Lett. 2019, 8, 337. 10.1021/acsmacrolett.9b00046. [DOI] [PubMed] [Google Scholar]
- a Adjili S.; Favier A.; Massin J.; Bretonniere Y.; Lacour W.; Lin Y.-C.; Chatre E.; Place C.; Favard C.; Muriaux D.; Andraud C.; Charreyre M.-T. Synthesis of Multifunctional Lipid-Polymer Conjugates: Application to the Elaboration of Bright Far-Red Fluorescent Lipid Probes. RSC Adv. 2014, 4, 15569. 10.1039/C4RA01334D. [DOI] [Google Scholar]; b Bathfield M.; D’Agosto F.; Spitz R.; Charreyre M.-T.; Delair T. Versatile Precursors of Functional RAFT Agents. Application to the Synthesis of Bio-Related End-Functionalized Polymers. J. Am. Chem. Soc. 2006, 128, 2546. 10.1021/ja057481c. [DOI] [PubMed] [Google Scholar]
- Favier A.; Charreyre M. T.; Pichot C. A detailed kinetic study of the RAFT polymerization of a bi-substituted acrylamide derivative: influence of experimental parameters. Polymer 2004, 45, 8661. 10.1016/j.polymer.2004.10.055. [DOI] [Google Scholar]
- Favier A.; Charreyre M. T. Experimental requirements for an efficient control of free-radical polymerizations via the reversible addition-fragmentation chain transfer (RAFT) process. Macromol. Rapid Commun. 2006, 27, 653. 10.1002/marc.200500839. [DOI] [Google Scholar]
- Favier A.; Ladaviere C.; Charreyre M. T.; Pichot C. MALDI-TOF MS investigation of the RAFT polymerization of a water-soluble acrylamide derivative. Macromolecules 2004, 37, 2026. 10.1021/ma030414n. [DOI] [Google Scholar]
- Favier A.; D’Agosto F.; Charreyre M. T.; Pichot C. Synthesis of N-acryloxysuccinimide copolymers by RAFT polymerization, as reactive building blocks with full control of composition and molecular weights. Polymer 2004, 45, 7821. 10.1016/j.polymer.2004.09.042. [DOI] [Google Scholar]
- Favier A.; Charreyre M. T.; Chaumont P.; Pichot C. Study of the RAFT polymerization of a water-soluble bisubstituted acrylamide derivative. 1. Influence of the dithioester structure. Macromolecules 2002, 35, 8271. 10.1021/ma020550c. [DOI] [Google Scholar]
- Heilemann M.; van de Linde S.; Mukherjee A.; Sauer M. Super-Resolution Imaging with Small Organic Fluorophores. Angew. Chem., Int. Ed. 2009, 48, 6903. 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- Chang C.-W.; Bays E.; Tao L.; Alconcel S. N. S.; Maynard H. D. Differences in cytotoxicity of poly(PEGA)s synthesized by reversible addition–fragmentation chain transfer polymerization. Chem. Commun. 2009, 3580. 10.1039/b904456f. [DOI] [PubMed] [Google Scholar]
- a Amendola M.; Venneri M. A.; Biffi A.; Vigna E.; Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat. Biotechnol. 2005, 23, 108. 10.1038/nbt1049. [DOI] [PubMed] [Google Scholar]; b Bakunts A.; Orsi A.; Vitale M.; Cattaneo A.; Lari F.; Tadè L.; Sitia R.; Raimondi A.; Bachi A.; van Anken E. Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. eLife 2017, 6, e27518 10.7554/eLife.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agosto F.; Charreyre M.-T.; Pichot C. Side-Product of N-Acryloyloxysuccinimide Synthesis or Useful New Bifunctional Monomer?. Macromol. Biosci. 2001, 1, 322.. [DOI] [Google Scholar]
- Zhou X.; Vink M.; Klaver B.; Berkhout B.; Das A. T. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 2006, 13, 1382. 10.1038/sj.gt.3302780. [DOI] [PubMed] [Google Scholar]
- Hellriegel C.; Caiolfa V. R.; Corti V.; Sidenius N.; Zamai M. Number and brightness image analysis reveals ATF-induced dimerization kinetics of uPAR in the cell membrane. FASEB J. 2011, 25, 2883. 10.1096/fj.11-181537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.