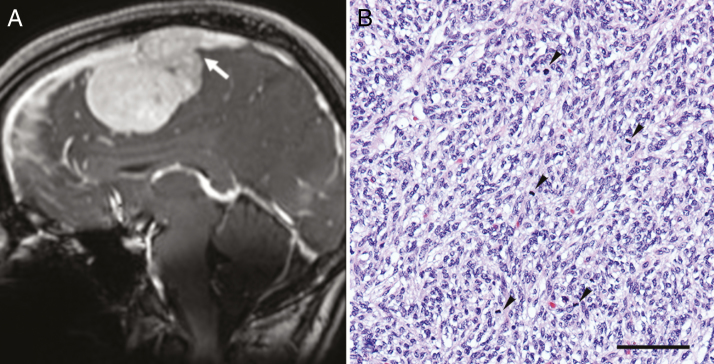
A 22-year-old previously healthy female presented with headache. Her family history was unremarkable. While her physical examination was within normal limits, magnetic resonance imaging revealed a 60 mm homogeneously enhancing, right parafalcine extra-axial, dural-based mass associated with partial occlusion of the anterior superior sagittal sinus (Fig. 1A). She was transferred to our tertiary care center where she underwent a bifrontal craniotomy. Intraoperatively, the tumor was attached to the right falx and invading the superior sagittal sinus. It was resected without incident.
Fig. 1.
(A) Sagittal (coronal) post-contrast T1-weighted image shows avid enhancement in the lesion with invasion of the superior sagittal sinus (arrow). (B) The tumor consists of spindle cells with a vague fascicular-storiform pattern. The nuclei are ovoid and monomorphic, with frequent mitotic figures (arrowheads); hematoxylin and eosin; bar = 100 microns.
Grossly, the tumor was soft and nodular. Histologic sections demonstrated a hypercellular spindle cell neoplasm arranged in sheets and fascicles, with a focal storiform pattern. The nuclei were ovoid and elongated, with mild pleomorphism; mitotic activity was conspicuous (>20 per 10 high power fields [focal depth = 0.52 mm]) (Fig. 1B). Approximately 10% of the sample was necrotic. Rare meningothelial nests were observed within the tumor; it was unclear whether these represented entrapped meningothelial rests or meningothelial differentiation. The tumor was permeated by a prominent capillary-like vasculature that also included occasional “staghorn”-like vessels. Focal dystrophic calcification was also noted. Frank brain invasion was evident with occlusive invasion of the superior sagittal sinus. Immunohistochemistry was diffusely and strongly positive for vimentin, S100, and progesterone receptor, with focal immunoreactivity for epithelial membrane antigen. Signal transducer and activator of transcription 6, CD34, and smooth muscle actin were negative. The Ki-67 proliferation index was focally >20%. A preliminary diagnosis of anaplastic meningioma, World Health Organization (WHO) grade III, was initially rendered. Subsequent molecular classification using DNA methylation was non-contributory, with the methylation profile failing to match any methylation class in the German Cancer Research Center CNS tumor classifier v11b4.
Based on the monomorphic spindle cell morphology, the possibility of a fusion-associated sarcoma was assessed using the Illumina TruSight RNA Fusion Panel, which screens for 507 fusion partners. This revealed a novel MN1-KMT2A fusion gene. The fusion involved exon 1 of MN1 (MN1 proto-oncogene, transcriptional regulator; NCBI Reference Sequence: NM_002430.2) on chromosome 22 and exon 3 of KMT2A (lysine methyltransferase 2A; NM_001197104.1) on chromosome 11. The fusion product was independently verified by PCR and Sanger sequencing. Subsequent copy number variation analysis confirmed MN1 gene instability with both deletions and amplifications (Supplementary Figure 1). Several focal gains and losses were observed involving chromosomes 11 and 22 in copy number variation plots generated from the raw methylation data. This finding correlated with the result of the fusion testing.
Both MN1 and KMT2A function as transcriptional coregulators, and each has been reported to pair with multiple other genes within a subset of hematolymphoid neoplasms.1,2 With respect to CNS tumors, MN1 inactivation has been previously implicated in meningioma due to a balanced MN1-TEL fusion gene.3MN1 has also been reported to partner with BEND2 in a high-grade pediatric neuroepithelial tumor.4 To our knowledge, the MN1-KMT2A fusion gene has not previously been described in any tumor type.
The features of this neoplasm are incompatible with any specific diagnosis listed in the current WHO Classification of CNS Tumors.5 Based on its unique histomorphology, combined with the presence of a novel disease-defining fusion gene, this is presumed to represent a novel entity or perhaps a unique subtype of meningioma; admittedly, the possibility of sarcoma cannot be entirely excluded. Our patient is currently 5 months post-operation. Based on the worrisome histomorphology, she also received a 6-week course of adjuvant radiation therapy. She currently has no radiologic evidence of residual/recurrent disease. Naturally, the prognosis of this tumor cannot be predicted based on a single report; nevertheless, given this tumor’s distinct features, and the increasingly routine application of diagnostic RNA sequencing, it is anticipated additional cases will be recognized in the future.
Funding
No funding sources to declare.
Conflict of interest statement. The authors report no conflict of interest.
Authorship statement
Substantial contributions to the conception of the manuscript: SC, BCD, JC, NZ, FN, SK, GZ, YM, SM, KA, DS, BL, TW, JW
Drafted the manuscript: SC, TW, JW
Revised the manuscript critically for important intellectual content: SC, BCD, JC, NZ, FN, SK, GZ, YM, SM, KA, DS, BL, TW, JW
Approval of the final version: SC, BCD, JC, NZ, FN, SK, GZ, YM, SM, KA, DS, BL, TW, JW
Agreement to be accountable for all aspects of the work: SC, BCD, JC, NZ, FN, SK, GZ, YM, SM, KA, DS, BL, TW, JW
Supplementary Material
References
- 1. Peterson JF, Sukov WR, Pitel BA, et al. . Acute leukemias harboring KMT2A/MLLT10 fusion: a 10-year experience from a single genomics laboratory. Genes Chromosomes Cancer. 2019. doi: 10.1002/gcc.22741. [DOI] [PubMed] [Google Scholar]
- 2. Zerkalenkova E, Lebedeva S, Kazakova A, et al. . Acute myeloid leukemia with t(10;11)(p11-12;q23.3): results of Russian Pediatric AML registration study. Int J Lab Hematol. 2019;41(2):287–292. [DOI] [PubMed] [Google Scholar]
- 3. Lekanne Deprez RH, Riegman PH, Groen NA, et al. . Cloning and characterization of MN1, a gene from chromosome 22q11, which is disrupted by a balanced translocation in a meningioma. Oncogene. 1995;10(8):1521–1528. [PubMed] [Google Scholar]
- 4. Burford A, Mackay A, Popov S, et al. . The ten-year evolutionary trajectory of a highly recurrent paediatric high grade neuroepithelial tumour with MN1:BEND2 fusion. Sci Rep. 2018;8(1):1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. International Agency for Research on Cancer. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon, France: World Health Organization; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.