Abstract
As the field of neuro-oncology makes headway in uncovering the key oncogenic drivers in pediatric glioma, the role of precision diagnostics and therapies continues to rapidly evolve with important implications for the standard of care for clinical management of these patients. Four studies at major academic centers were published in the last year outlining the clinically integrated molecular profiling and targeting of pediatric brain tumors; all 4 demonstrated the feasibility and utility of incorporating sequencing into the care of children with brain tumors, in particular for children and young adults with glioma. Based on synthesis of the data from these studies and others, we provide consensus recommendations for the integration of precision diagnostics and therapeutics into the practice of pediatric neuro-oncology. Our primary consensus recommendation is that next-generation sequencing should be routinely included in the workup of most pediatric gliomas.
Keywords: high-grade glioma, low-grade glioma, next-generation sequencing, pediatric, precision medicine
Within the past decade, considerable progress has been made in uncovering the molecular drivers of pediatric gliomas. Though the cell of origin and evolutionary roadmap of these tumors remain active areas of study, recurrent molecular alterations have been identified through large sequencing datasets and preclinical validation.1–4 Importantly, 4 recent studies demonstrated the feasibility of clinically integrated sequencing and/or targeted therapy in unique prospective cohorts of pediatric brain tumor patients.5–8 These studies generally noted a higher likelihood of sequencing information impacting the management of children with gliomas. Precision medicine approaches are particularly promising for pediatric gliomas as they tend to be more genetically homogeneous and harbor fewer genetic drivers than their adult counterparts, suggesting the presence of distinct oncogenic events critical to tumor development and the potential for a robust, targeted response.5–8 This field is rapidly changing, with potential implications for a new standard of care for young patients with glioma. In this review, we synthesize the data from these and other recent studies and provide consensus recommendations for the integration of precision diagnostics and therapeutics for pediatric gliomas.
Background and Current Treatments/Prognosis
Pediatric High-Grade Glioma and Diffuse Intrinsic Pontine Glioma
Pediatric high-grade glioma (HGG) and diffuse intrinsic pontine glioma (DIPG) are 2 distinct entities often considered together due to similarities in histologic appearance and poor prognosis, with median survival of 12–24 months and less than 12 months, respectively.9–12 HGG is routinely diagnosed by histology that demonstrates a tumor of glial origin with World Health Organization (WHO) grade III or IV features (mitoses, necrosis, and/or microvascular proliferation), whereas DIPG has traditionally been diagnosed by radiographic and clinical features alone. When tissue is obtained, histology shows a diffusely infiltrative glioma with WHO grade frequently varying from II to IV.11,13 A majority of DIPG harbor a point mutation in one of the histone H3 variants (H3.3 or H3.1), which now defines the new pathologic entity of diffuse midline glioma, H3-K27M mutant, associated with a distinct epigenetic phenotype and clinical behavior.1 Mutations in histone chaperone protein alpha-thalassemia/mental retardation syndrome X-linked (ATRX) (31%) and TP53 (54%) are found more frequently in pediatric than in adult glioblastoma (GBM).2 Loss of ATRX has been shown, in pediatric GBM and in other solid tumors, to be associated with a distinct mechanism of telomere elongation called alternative lengthening of telomeres,2,14 as well as with promoting tumor growth and genetic instability.15
Standard of care treatment for HGG in children is not as clearly established as it is in adults. Gross total resection is attempted when feasible and has been shown to improve survival.16 Adjuvant radiotherapy typically follows surgical resection with exceptions made for infants and younger children due to concern for neuro-developmental toxicity.17,18 Unfortunately, DIPG and midline HGG are not amenable to surgical resection, because of their location in essential structures and the infiltrative nature of the tumor. The role of chemotherapy during or after radiation remains unclear. Temozolomide, which is used in the treatment of adult GBM, has not shown a clear benefit in pediatric HGG.19,20 As of yet, no chemotherapeutic regimen has added benefit beyond radiation in the treatment of DIPG.10,21–26 Efforts are being directed toward the use of precision medicine in developing new and innovative chemotherapeutic treatments. A number of clinical trials based on specific molecular targets are ongoing for pediatric HGG and DIPG.
Pediatric Low-Grade Glioma
Pediatric low-grade gliomas (LGGs) include WHO grade I tumors (pilocytic astrocytoma, ganglioglioma, subependymal giant cell astrocytoma [SEGA]) and WHO grade II tumors (pleomorphic xanthoastrocytoma [PXA], diffuse astrocytoma) and are the most common subgroup of CNS tumors found in children.13 These lesions are found throughout the brain and spine, with the most common lesion being cerebellar pilocytic astrocytoma, which is often amenable to complete resection with resultant excellent prognosis.
Gross total resection, when possible, is the recommended first-line therapy for pediatric LGGs and has been associated with an excellent long-term prognosis.27 Unfortunately, many pediatric LGGs are located in eloquent areas of the brain (brainstem, diencephalon, optic pathway), where only a biopsy or partial resection is feasible. While partial resection alone can lead to long-term tumor control in a small subgroup of patients,28 many patients require adjuvant therapy at tumor progression. Radiation is an effective therapy for the majority of pediatric LGGs but is associated with high rates of morbidity, including cognitive deficits,29 endocrinopathies, and secondary malignancies.30 Therefore, it is currently recommended only when all other therapy options have been exhausted, particularly in younger patients (<10 y of age). Given that most children and young adults with LGG have a long overall survival, treatment strategies are now focused on minimization of both acute and chronic morbidity.
Chemotherapy has been shown to be an effective modality for the treatment of pediatric LGG. The most well-established regimen, first studied in the Children’s Oncology Group protocol A9952, involves carboplatin and vincristine, which has an overall tumor control rate (stable disease plus objective radiographic response) of 85% in patients with neurofibromatosis type 1 (NF1) and 67% in non-NF1 patients.31 Unfortunately, this therapy is logistically difficult (weekly intravenous therapy) and immunosuppressive. Alkylating agents such as lomustine (CCNU)32 have proven efficacy in this disease, but their use is limited due to the rare but not negligible risk of alkylator-induced secondary malignancies such as leukemia.33
Several recent genomic studies on archived pediatric LGG specimens have uncovered driving alterations, with the majority located in the receptor tyrosine kinase (RTK)/Ras/Raf/mitogen activated protein kinase (MAPK) pathway.4,34 As discussed below, these studies have sparked a precision medicine revolution for the treatment of these tumors, and it is possible that biologically driven therapy will eventually supplant cytotoxic chemotherapy and radiation therapy for this subgroup of tumors.
Clinically Integrated Molecular Profiling in Pediatric Gliomas
Given the ever-increasing number of therapies targeting specific genetic alterations, it is no surprise that institutions are working to incorporate next-generation sequencing (NGS) into the care of children with brain tumors. Multiple academic centers have reported success in being able to deliver sequencing results in a timely manner with significant implications for both diagnosis and treatment (Supplementary Table 1).5–8
Researchers at UCSF used targeted genome sequencing of 510 genes on a select cohort of 31 pediatric patients enriched for HGG and recurrent tumors.5 Their sequencing infrastructure was able to produce actionable sequencing data in 2–3 weeks and these data had implications for patient care in 81% of cases. Clinical implications included amendment of initial pathologic diagnosis, identification of germline variants necessitating ongoing monitoring for patient and family, and identification of therapeutic targets. Sixty-one percent specifically contained genetic alterations that had potential for clinical interventions.5
Ramkissoon et al (Dana-Farber/Harvard) applied OncoPanel, targeted exome sequencing of 309 genes, and OncoCopy, array comparative genomic hybridization, either alone or in combination to analyze a total of 203 pediatric brain tumors of various subtypes and histologic grades. Of the 146 tumors that were assessed with OncoPanel sequencing, 56% contained alterations that could serve as potential therapeutic targets.6 Additionally, 37 patients who received OncoPanel sequencing had alterations that could be targeted by small-molecule inhibitors currently being investigated in clinical trials, and 8 (22%) of these patients were subsequently enrolled in one of these trials as a consequence of having sequencing results.6
At Seattle Children’s Hospital during a study period ranging from November 2015 to November 2016, all CNS tumors, newly diagnosed or recurrent and without consideration of histologic grade or risk stratification, underwent a universal sequencing protocol. Sequencing was performed on the UW-OncoPlex platform, which detects genetic alterations in 262 cancer-related genes. Eighty-five patients had sufficient tissue for sequencing, of whom 68 (80%) had genetic alterations of some clinical relevance.8 Sequencing revealed disease-defining or -modifying mutations in 57 (67%) patients, and potential therapeutic targets in 44 (52%).8
The University of Michigan conducted a study that specifically sought to evaluate how frequently having access to genetic information would actually result in a demonstrable change in clinical care.7 A group of 50 pediatric patients with brain tumors designated as “high risk” were enrolled. The study used both exome (either whole exome or 1706 gene panel) and transcriptome sequencing with a median turnaround time of 78 days for formalin-fixed paraffin-embedded (FFPE) samples and 49 days for frozen samples.7 Sixty-three percent of sequenced tumors with adequate tissue for analysis revealed alterations with clinical implications and, of these tumors, 53% actually resulted in a change in clinical care—either in the form of treatment decision or amendment of diagnosis. These numbers were even higher when limited to glial tumors, with 85% containing alterations of clinical significance and 70% of these actually affecting therapy.7
Molecular Profiling in Pediatric Gliomas: Current Platforms and Consensus Recommendations
Molecular Tests and Platforms for Profiling Pediatric Gliomas
Many different clinical laboratory methods are used to classify and molecularly profile pediatric gliomas, but there is currently no gold standard. These methods each have their own strengths and weaknesses, as well as different turnaround times, tissue requirements, and costs.
Traditional immunohistochemistry (IHC) is commonly used in the evaluation of pediatric brain tumors. IHC rapidly detects expression of specific proteins in tumors, including those that indicate an underlying mutational status, such as identification of H3F3A or HIST1H3B K27M and mutations of B-Raf serine/threonine kinase (BRAF) V600E.35,36 Similarly, fluorescence in situ hybridization (FISH) may be used to detect single gene fusion or copy events such as the BRAF:KIAA tandem duplication and fusion event or focally amplified genes such as PDGFRA.37,38 Targeted DNA and RNA sequencing (RNA-seq) panels have been developed to simultaneously detect targetable alterations in a defined set of key cancer genes (eg, <50).39,40 While these methods with limited complexity continue to be very useful, the studies described above which analyzed anywhere from 262 to 1706 genes simultaneously have now clearly established the utility of using larger targeted sequencing platforms in the workup of pediatric gliomas which have a diverse set of mutations, copy changes, and rearrangements commonly present.6–9 Due to their breadth, targeted capture-based DNA panels have been shown to identify clinically relevant actionable mutations in a majority of children with glioma, and are currently being performed as either clinical or research tests at several institutions. Sequencing-based testing can offer distinct and confirmatory advantages when added to IHC for recurrent alterations of pediatric glioma, such as for determining clonality of somatic variants and for the isolation of potential rare cases with discrepancy between sequencing and IHC results. Global reduction of H3K27 trimethylation is uniformly present in diffuse midline glioma with H3K27M mutation and is confirmatory of the unique biology of these tumors.41 However, the potential role of H3K27me3 IHC in the workup of pediatric gliomas remains unclear until the significance of K27me3 loss in H3K27 wild-type glioma is further established.
Whole exome and whole genome sequencing have also been used to detect key genetic drivers in tumors. Target coverage is typically lower with these methods, but they have the advantage of identifying alterations which are not easily assessed on more targeted panels. Tumor mutational burden is able to be reliably calculated by whole exome or whole genome sequencing, which may have implications for selection of targeted therapy versus immune checkpoint inhibitors.42 Of note, recent work has shown that tumor mutational burden may be reliably calculated using panels with coverage of as few as 300 genes.43 DNA sequencing of tumor tissue identifies mutations that may be either somatic or germline; therefore, it is important to confirm potential deleterious germline alterations by testing a “normal” tissue sample (eg, peripheral blood, saliva). RNA-seq is well suited for identification of gene fusions, especially those that may be difficult to detect by DNA sequencing.44 Combined DNA and RNA profiling has been used to comprehensively evaluate patients with high-risk pediatric brain tumors.8
Numerous scientific studies have now shown the ability of DNA methylation profiling to aid pediatric brain tumor diagnosis. This method of molecular profiling exhibits exceptional utility in classifying diagnostically challenging high-grade pediatric brain tumors and identifying tumor lineage.45 Additionally, methylation array data can be used for copy number variation analysis to isolate potentially actionable focal amplifications and losses.45 Methylation profiling has been limited to the research setting in the United States, with few laboratories offering this test, but such assays will likely become more readily available as value in other cancer indications increase.
Recommendation for Approach to Molecular Profiling
Recent experience has demonstrated the feasibility and utility of including molecular profiling in the current battery of established molecular and pathologic tests for pediatric gliomas. The pathologic diagnoses themselves have also begun to reflect the impact of key recurrent alterations on the prognosis, clinical course, and choice of therapy by incorporating them into the naming conventions for pediatric tumors in the 2016 revision of the WHO classification of tumors of the central nervous system. Given the formal incorporation of molecular findings into essential classification and the diversity of genetic events required to be detected, our consensus recommendation is for the expanded use of tumor sequencing in clinical management of patients (Table 1). For pediatric gliomas this appears to be best accomplished with an NGS panel to identify relevant genomic drivers. This consensus recommendation reflects the combined experiences with molecular profiling within our groups and aligns with our findings that tumor genomic profiling has the potential to meaningfully impact clinical care in the majority of patients. This recommendation is further driven by a growing list of pathways with targeted therapies available to consider on- and off-trial for the treatment of pediatric gliomas (Supplementary Table 2). The integration of expanded NGS into routine clinical care for glioma patients may require some changes in clinical practice such as ensuring tissue sampling is adequate for both histopathologic and molecular analyses, as well as utilizing tissue preservation protocols in pathology laboratories. Increasingly, the rapid submission of appropriate tumor tissue for NGS evaluation is also required given that most testing requires 2–4 weeks for completion of results, and treatment decisions and clinical trial enrollment often require prompt decision making during or immediately after this period. The potential benefits of incorporating NGS into clinical “standard of care” for pediatric glioma patients are significant as the data can be utilized to refine tumor diagnoses, allow for patient-specific data-driven discussions regarding prognosis, and support therapeutic decision making such as selection of targeted therapies or enrollment into genomically stratified clinical trials.
Table 1.
Summary of recommendation for approach to somatic molecular profiling of pediatric gliomas
| Tumor Type | Molecular Subgroup or Recurrent Alteration | Recommended Expanded Molecular Testing |
|---|---|---|
| LGG at diagnosis | Clinical criteria or molecular confirmation of NF1. | No further testing unless atypical clinical or pathologic features. |
| Does not meet clinical criteria for NF1 | Tumor NGS (DNA cancer-related gene panel, +RNA when available) | |
| OR • Targeted testing for BRAF alterations, including BRAF:KIAA fusion (eg, FISH for BRAF duplication) • BRAF V600E (IHC and/or PCR) • H3K27M mutation (IHC) in midline gliomas • IDH1 R132H mutation (IHC and/or PCR) |
||
| LGG at recurrence or progression | Positive BRAF testing, first relapse. | No further testing |
| Positive BRAF testing, 2+ relapses. | Tumor NGS (DNA cancer-related gene panel, +RNA when available) | |
| Negative BRAF testing. | Tumor NGS (DNA cancer-related gene panel, +RNA when available) | |
| HGG or DIPG at diagnosis or recurrence/progression | • H3K27M mutation (IHC) in midline gliomas • Tumor NGS (DNA cancer-related gene panel, +RNA when available) |
The majority of pediatric LGGs harbor a single driver alteration within a subset of recurrently altered genes (eg, BRAF, FGFR1, NF1) and this alteration may be a point mutation (missense, nonsense), gene fusion, or insertion/deletion. There are 2 general approaches to identifying these genomic drivers in LGGs: (i) sequential or parallel single gene testing or (ii) upfront tumor NGS (cancer-related gene panel). The selection of one approach over another may depend on accessibility to an NGS panel, quantity of tissue, and/or turnaround times for each assay, but NGS clearly offers logistical and clinical advantages. For these tumors, we first recommend clarifying whether the patient meets clinical criteria for a diagnosis of NF1 using National Institutes of Health criteria of at least 2 of the NF1-associated clinical features. For patients with established NF1 and radiographically diagnosed glioma at presentation, expanded molecular profiling may not be required due to the likelihood of NF1 loss or mutation as the sole genetic alteration. However, as co-occurring mutations can be detected in these tumors, advancing with a DNA panel for sequencing is advised if adequate tumor tissue is available, especially in cases with atypical clinical or pathologic features (Table 1).46 For patients who do not meet clinical criteria for NF1, we recommend advancing to expanded NGS-based molecular profiling, particularly if tissue is limited, as all relevant genomic drivers can be evaluated using a single DNA extraction and assay. If, however, access to expanded NGS testing is not possible, then at minimum we recommend targeted testing for BRAF alterations, including BRAF:KIAA fusion (ie, FISH for BRAF duplication) and BRAF V600E, as well as testing for IDH1 R132H (IHC and/or PCR) and histone H3K27M mutations (IHC). In the event that sequential single gene testing returns only negative findings, then advancing to panel-based DNA sequencing is recommended as alterations in FGFR1 (mutation, duplication) or other less common drivers (RAF1, MYB, MYBL1, NTRK, or IDH1 mutations) may be present.46 For pediatric patients with a recurrent or progressive LGG with negative BRAF testing and/or those with positive H3K27M testing, we recommend upfront pursuit of NGS-based molecular profiling to look for tumor drivers. We also recommend this testing for patients with multiply relapsed NF1-related glioma. For recurrent glioma with new biopsy/tissue, we recommend pursuit of repeat NGS-based profiling to clarify clinically relevant molecular changes and tumor evolution in response to prior therapies.
For pediatric patients with DIPG and non-brainstem HGG (including rarer gliomas with high-grade features, such as anaplastic PXA), we recommend IHC testing for H3K27M to quickly clarify diagnosis, as well as tumor sequencing as above. Sequencing may be best performed at a tertiary pediatric brain tumor center with expertise in the field and a sequencing panel from a lab certified by the Clinical Laboratory Improvement Amendments (CLIA), or through send-out sequencing to another CLIA-certified lab with potentially less specific expertise in pediatric brain tumors (ie, academic labs accepting outside cases or private labs with CLIA certification). For centers with a clinical or research platform that includes RNA-seq, we recommend inclusion of RNA-seq to assist with the identification of both established and novel tumor-driving fusions. While RNA-seq is a powerful tool for fusion detection, there is a substantial risk of generating both false negative and false positive calls; consequently, analysis should only be performed at centers with advanced fusion-calling pipelines and computational experience. As an example, sensitivity can be improved with pipelines that allow for fusion detection regardless of breakpoint within gene bodies, such as the CODAC pipeline at the University of Michigan. False positives can be reduced with exclusion of calls that have a low “fusion-quality score” using established metrics (eg, number of spanning reads, alignment quality, repetitiveness of the DNA).47
We also recommend the inclusion of methylation profiling for recurrent or high-grade gliomas when available. Though still an emerging technology, methylation appears to be a promising tool for classification of pediatric brain tumors and may be considered for diagnostically difficult glial tumors and glioneuronal tumors (such as primitive neuroectodermal tumors) as a single platform that is versatile for classification of diverse tumor types. In the future, global or targeted methylation panels may represent important tools in integrated diagnostic workup of pediatric gliomas. Methylation-based subgrouping of medulloblastoma is already seeing use in the clinical realm and this use will presumably increase if CLIA certification is awarded.
Based on our shared published experiences of molecular profiling in pediatric glioma, some important recommendations can be made. All centers generated sequencing results primarily using an Illumina HiSeq platform (2000 or 2500), which is increasingly considered the standard for clinical-grade DNA or RNA sequencing48 and should be considered the current standard for pediatric glioma. While Michigan found that frozen tissue was more often adequate than FFPE (96% vs 72%),7 sequencing from FFPE was found to be feasible in the vast majority of cases among the 4 groups. The gene panels varied in size, but the number of patients with actionable results did not decrease with smaller gene panels down to 262 genes. We performed a comparison of the 4 panels and overlapping genes (Supplementary Table 3). Future panels will need to be adaptive to more recently discovered clinically actionable pathways unique to pediatric glioma (eg, ACVR1, HISTIH3B). As pediatric gliomas may carry potentially actionable focal copy number gains and losses, we further recommend the inclusion of copy number analysis and reporting. This can be performed with NGS data with analysis of interval-placed intergenic probes or concurrent array comparative genomic hybridization analysis.5
In order to capture recurrent and novel rearrangements in actionable genes, we recommend either intronic coverage using a smaller gene panel (Dana-Farber n = 35, Seattle Children’s n = 17, UCSF n = 38) or splice-junction coverage of the entire gene panel paired with tumor RNA-seq (Michigan). Depth of sequencing coverage varied between our centers’ platforms (average coverage ranged 150x–500x). As reported by the UCSF group, for samples with 25% tumor content, >200x coverage provided 99% and 83% sensitivity and 98% and 71% specificity for clonal single nucleotide variants (SNVs) and small indels, respectively.5 We therefore recommend at least 150x average coverage over the entire gene panel.
As is practiced in our collective institutions, we recommend the generation of a short sequencing results report, including SNVs (with variant allele fraction), copy number changes, and fusions deemed to be actionable. The report should utilize variant-specific information (eg, ClinVar, published literature, curated gene-specific resources) and established guidelines for reporting, such as those published by the Association for Molecular Pathology, the American Society of Clinical Oncology, and the College of American Pathologists.49
Germline testing and genetic counseling are being increasingly recognized for their importance in pediatric cancers. In the setting of pediatric gliomas, these should be offered to patients in whom a cancer susceptibility syndrome is suspected, including those with (i) family history of an immediate relative with childhood cancer or exclusive family histories of adult cancer at younger ages, (ii) parents with consanguinity, (iii) history of previous cancer diagnosis, (iv) new clinical diagnosis of germline predisposition syndrome (eg, NF1), (v) features of a predisposition syndrome or multiple unexplained congenital malformations, or (vi) a pathogenic variant in a cancer susceptibility gene detected by tumor sequencing (eg, somatic TP53 mutation).
Targeted Therapies for Pediatric Glioma: Strong Likelihood of Efficacy
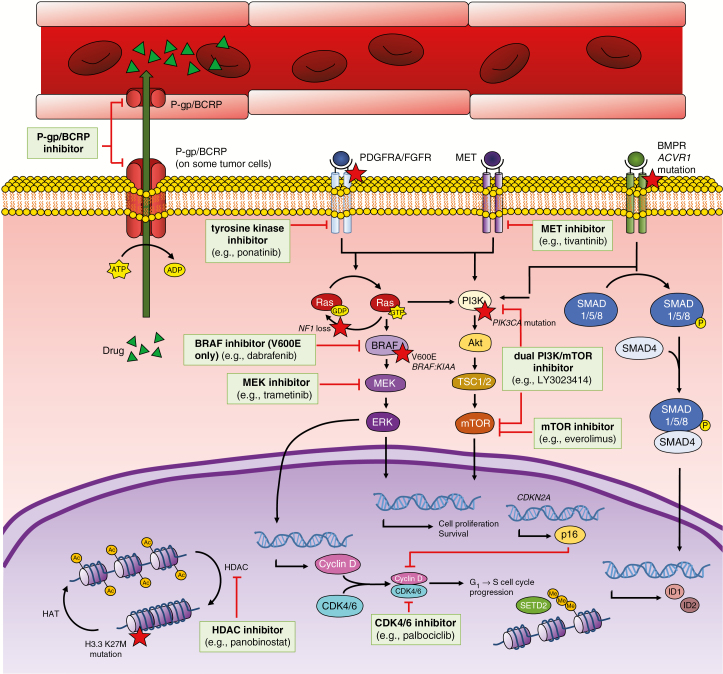
As noted above, pediatric gliomas harbor many of the recurrent genetic alterations found in other human solid tumors (eg, BRAF, FGFR1, PIK3CA) that may drive tumorigenesis. Although the clinical implications of many of these variants have not been firmly established, guidelines have recently been developed for the annotation of tumor variants in a tier-based system.49 With this framework in place, there are a growing number of targeted therapies that are being evaluated in the precision medicine setting for gliomas, with the obvious additional issues of blood–brain barrier (BBB) penetration and drug efflux affecting the potential usefulness of drugs used in other cancers (Fig. 1). To this end, we sought to clarify the pathways and agents that have “strong likelihood of efficacy” and “potential efficacy” in gliomas (Supplementary Table 2).
Fig. 1.
Schematic of key pathways in pediatric glioma, including the frequent efflux of drugs from CNS tissue by efflux proteins such as P-glycoprotein and breast cancer resistance protein
MET = MET receptor tyrosine kinase; BMPR = bone morphogenetic protein receptor; ACVR1 = activin A receptor type 1; Ras = Ras family GTPase; Akt = protein kinase B; SMAD1/5/8 = mothers against decapentaplegic homolog 1/5/8; SMAD4 = mothers against decapentaplegic homolog 4; HAT = histone acetyltransferase; G1 = gap 1 phase; S = synthesis phase; p16 = cyclin-dependent kinase inhibitor 2A (protein); SETD2 = SET domain containing 2; ID1/2 = inhibitor of DNA binding 1/2.
Mutations of Tuberous Sclerosis Complex and Inhibitors of Mechanistic Target of Rapamycin
While the majority of pediatric gliomas are sporadic, a subset arise from germline mutations, such as those related to tuberous sclerosis complex (TSC). TSC results from mutations in the genes TSC1 and TSC2. These mutations are inherited in an autosomal dominant fashion and result in a phenotypically variable syndrome that can affect the skin, heart, lungs, kidneys, eyes, and CNS.50 About 10% of patients with TSC develop slow-growing, low-grade SEGAs.51 SEGAs most frequently arise adjacent to the foramen of Monro and, consequently, are known to cause increased intracranial pressure due to hydrocephalus, as well as seizures.52 The products of TSC1 and TSC2 are hamartin and tuberin, respectively, which form a complex that inhibits downstream mechanistic target of rapamycin (mTOR). Thus, mutations in either of these genes will lead to unchecked activation of mTOR.53 In perhaps the most effective and well-established example of precision medicine therapy for pediatric brain tumors, the mTOR inhibitors sirolimus and everolimus have been used to target the tumor-driving activation of mTOR in the development and progression of SEGAs.54,55 In general, direct clinical comparisons of sirolimus and everolimus are lacking. However, in one cross-sectional review comparing patients treated with either sirolimus or everolimus, tumor responses appeared to be quite similar.56
BRAF V600E Mutation and BRAF and MEK Inhibitors
Alterations in BRAF, such as the V600E mutation, result in dysregulated signaling of extracellular signal-regulated kinase (ERK, also known as MAPK) and glial tumor proliferation. The BRAF V600E mutation is frequently found in multiple different pediatric LGGs, including PXAs (66–78%) and gangliogliomas (49%).4,57,58 While some studies have reported V600E mutations in pilocytic astrocytoma,59 such tumors appear to have increased risk of progression and most likely do not represent classic pilocytic astrocytomas. The majority of studies have shown that V600E and BRAF fusions are mutually exclusive events. Lassaletta et al studied 510 pediatric LGGs (99 with BRAF V600E mutation) and found that patients with BRAF V600E mutant tumors had lower 5-year and 10-year progression-free survival than those with LGGs of wild-type status (5-y PFS, 50.1% mutant vs 72.8% wild type; 10-y PFS, 27% mutant vs 60.2% wild type), despite conventional treatment. When compared with those with BRAF:KIAA fusions, patients with BRAF V600E mutant LGGs showed significantly worse overall survival.58 In a study analyzing the genetic events leading to transformation of pediatric LGG to secondary HGG, Mistry and colleagues found that 39% of secondary HGG patients harbored the BRAF V600E mutation.60 Five of 9 patients identified in the HERBY trial as having “PXA-like” HGGs had BRAF V600E mutations, and 3 of the remaining 4 had an NF1 mutation. All BRAF V600E mutant PXA-like tumors fell into the histologic epithelioid variant of GBM on pathologic re-review.61
BRAF inhibitors prevent the phosphorylation of mitogen-activated protein kinase kinase (MEK; activates MAPK/ERK) and ERK in V600E-mutated cells, preventing signaling required for cellular proliferation. The aforementioned case series analyzed by Lassaletta et al utilized these agents and demonstrated impressive clinical responses, including significant cytoreduction and prolonged survival, in patients with V600E-mutated pediatric glioma58; a case report also described a sustained complete response to vemurafenib.62 Initial findings from a phase I clinical trial of dabrafenib in V600E-mutated pediatric LGG also seem promising, with a 41% overall response rate.63 Importantly, most BRAF inhibitors have poor in vitro activity against cells with non-V600E mutations or wild-type BRAF, and use of BRAF inhibitors in the setting of wild-type BRAF may lead to paradoxical activation of the Raf-MEK-ERK pathway via Ras activation, thus enhancing tumor growth.64 It is therefore of particular importance to determine the status of BRAF with respect to V600E and fusions prior to initiation of therapy to prevent administration to fusion and wild-type patients where use of these inhibitors may be detrimental.
Multiple trials are currently accruing pediatric HGG patients with known V600E mutation to evaluate single agent BRAF inhibitor therapy (NCT01677741, NCT01748149, NCT03220035). Of BRAF inhibitors with FDA approval for any indication, dabrafenib displays reasonable CNS penetration, followed closely by vemurafenib; however, both are substrates of efflux transporters, which may limit their effectiveness in brain tumors.65 Resistance to BRAF inhibitors has been reported to emerge in other cancers due to intrinsic ERK activation via mutation, or by alternative activating BRAF mutations. As such, the addition of a MEK inhibitor (which prevents ERK activation) to a BRAF V600E inhibitor was recently shown to be efficacious in a case series of 3 pediatric patients with HGG.66 A clinical trial is currently accruing to further evaluate BRAF/MEK inhibition as a combination regimen in pediatric HGG (NCT02684058). MEK inhibition has additionally been identified as a targeted therapy for non-V600E BRAF mutations; the MEK inhibitor trametinib has been shown to reduce proliferation of BRAF-mutated cell lines with deficient kinase activity.66
BRAF:KIAA Fusion, NF1 Loss, and MEK Inhibitors
Jones et al first described a tandem duplication at 7q34 which produced an in-frame fusion between KIAA1549 and BRAF. This fusion results in constitutively active kinase activity without the BRAF autoregulatory domain, with subsequent activation of the Ras-MAPK pathway and anchorage-independent growth in NIH3T3 cells.67 The gain at 7q34 is fairly specific for pilocytic astrocytoma; the fusion is found in 59–90% of pilocytic astrocytomas and is used as a diagnostic marker.67,68 Children with NF1, germline loss of neurofibromin 1 (NF1), are predisposed to develop gliomas such as optic pathway low-grade astrocytomas.13 Pathologically, the majority of both NF1-associated and -sporadic optic nerve gliomas are WHO grade I pilocytic astrocytomas. Because of location and ability to diagnose based on radiographic appearance, these tumors are rarely biopsied, thus limiting knowledge of their molecular characteristics.69 NF1 is a classic tumor suppressor, and loss of both copies of NF1 promotes Ras signaling and tumor cell proliferation. Rodriguez et al analyzed 59 patients (both pediatric and adult) with optic pathway gliomas (7 with NF1) and demonstrated that all analyzed tumors were either NF1 associated or had BRAF duplication.69
Monotherapy with a MEK inhibitor has now been established as an effective therapy for NF1-deficient or BRAF:KIAA-driven recurrent or refractory pediatric LGG. A phase II Pediatric Brain Tumor Consortium study examining the use of selumetinib in recurrent pediatric LGG showed impressive efficacy, with 40% of 25 NF1 patients achieving partial response and only 1 patient progressing while on treatment.70 While use of selumetinib has not achieved FDA approval and is thus difficult to obtain off-trial, future trials will likely explore its use for newly diagnosed pediatric LGG. Trametinib and selumetinib both display good CNS penetration. Targeting NF1 loss in HGG with MEK inhibitors shows promise, but will likely require a multipronged approach due to other concurrent alterations. A case report of NF1-associated GBM showed disease regression that was maintained for at least 4 months posttreatment initiation with trametinib.71
Germline Mismatch Repair Deficiency and Checkpoint Inhibitors
Through whole genome sequencing of recurrent GBM, Bouffett et al demonstrated higher rates of mutational burden, including germline mutations associated with cancer predisposition, in pediatric patients compared with adults.42 Cancer predisposition syndromes associated with these tumors include biallelic mismatch repair deficiency (bMMRD) syndrome and Lynch syndrome, caused by germline mutations in mismatch repair genes. Because of their particularly high mutational burden, pediatric HGG in the setting of bMMRD may be responsive to immune checkpoint inhibition, which would allow restoration of antitumor immunity. Restoration of antitumor immunity, allowing for self-destruction of malignant cells by a patient’s own T cells, has shown encouraging results in melanoma, colon, and lung cancers where high mutational burden was demonstrated.
Nivolumab, an anti–programmed cell death 1 directed checkpoint inhibitor, was used as a novel treatment for 2 related pediatric patients with recurrent GBM in the setting of bMMRD. Repeat imaging of the patients demonstrated encouraging results, with both patients returning to school within 9 months of treatment.42 Ongoing studies will hopefully validate this case series. Though targeting germline mismatch repair deficiency via checkpoint inhibition is a promising area of research in other malignancies, early studies in adult and pediatric HGG have shown some concerns for immune-related toxicity in large, unresected tumors.72,73
Other Targeted Therapies: Potential Efficacy and Future Directions
Histone Mutations and Histone Deacetylase Inhibitors
A genetic alteration of particular interest is a group of histone mutations with greater prevalence in pediatric HGG than in any other human cancer. In a hallmark study that sequenced the exomes of 48 pediatric HGG, mutations in H3F3A, the gene that encodes the histone H3.3 variant, were identified in 31% of specimens.2 Histone H3.3 is associated with heterochromatin silencing and is incorporated into nucleosomes in a manner that is replication independent.74 The histone mutations identified in this study lead to amino acid substitutions within the histone tail at positions K27, leading to K27M, and G34, leading to G34R/V.2 The K27M mutation is also seen in HIST1H3B, the gene encoding the histone H3.1 variant, which is only incorporated during DNA synthesis, in contrast to H3.3.74 These single point mutations confer distinct tumor biology in terms of co-occurring mutations, tumor location, age group, and prognosis. Of these mutations, H3F3A G34R/V is associated with the best prognosis, followed by HIST1H3B K27M, and H3F3A K27M is associated with the worst overall survival. Collectively, any one of these histone mutations confers a poorer prognosis than histone wild-type status.1
The K27M mutation is believed to contribute to oncogenesis by interfering with normal posttranslational modifications of the histone H3 protein.75 Histone deacetylase (HDAC) is responsible for the removal of acetyl groups from histone tails, one example of posttranslational modification. HDAC inhibitors induce epigenetic changes, primarily increases in H3 acetylation, which reduce cellular proliferation and viability.76 A study by Grasso et al showed that treatment with the HDAC inhibitor panobinostat restored H3 acetylation of K27M-mutant DIPG cell lines, as well as being efficacious in mouse DIPG xenografts.77 However, a more recent study demonstrated that the activity of HDAC inhibitors is similar among wild-type and K27M-mutated DIPG cell lines and the inhibitors had little to no efficacy in in vivo studies.76
Clinical trials in pediatric HGG have not selected specifically for histone mutations, though a phase I trial of panobinostat is evaluating K27M mutations as a secondary outcome (NCT02717455). Results of clinical trials utilizing HDAC inhibitors in pediatric HGG have had suboptimal outcomes, potentially due to the poor BBB permeability of these agents and/or secondary to efflux transport. However, identification of populations that may specifically benefit from HDAC inhibitor therapy or concurrent therapies to improve BBB penetration may improve outcomes.65 In addition to HDAC inhibitors, a novel therapy, ONC201, is currently in clinical trials for K27M-mutated pediatric glioma (NCT03416530). ONC201 is an imipridone, a novel therapeutic class that antagonizes dopamine receptor D2. ONC201 has demonstrated tumor penetration, and initial studies are reporting promising response rates in both adult and pediatric GBM patients with the K27M mutation.78
PDGFR/FGFR Alterations and Tyrosine Kinase Inhibitors
Recent studies suggest that a large subset (20–30%) of pediatric patients with HGG have mutation and/or amplification of platelet-derived growth factor receptor alpha (PDGFRA), an RTK.3PDGFRA mutations are found in older pediatric patients and are associated with worse prognosis in pediatric HGG as determined by multivariate analysis.79 A recent meta-analysis of over 1000 pediatric high-grade brain tumors showed that PDGFRA alterations tend to co-segregate with H3F3A K27M specifically in tumors located in the pons. PDGFRA amplification is also seen in histone wild-type tumors in conjunction with MET amplification and is associated with an intermediate overall survival compared with other histone wild-type HGG.1 To a lesser extent, alterations in fibroblast growth factor (FGF) and its corresponding receptor, FGFR, are also found in pediatric HGG and may be isolated more frequently with platforms that include RNA sequencing to help isolate fusions.7
Mutations and amplifications of RTKs, such as PDGFRA and PDGFRB, or their ligands, PDGFA and PDGFB, result in activation of both the phosphatidylinositol-3 kinase (PI3K) and Ras/Raf pathways. Multiple tyrosine kinase inhibitors (TKIs), such as dasatinib, pazopanib, ponatinib, and sunitinib, inhibit wild-type PDGFRA at nanomolar concentrations and may be therapeutic options for pediatric patients with PDGFRA mutations. TKIs may also be considered for mutations, fusions, or amplifications of other RTKs such as FGFR. Pazopanib and ponatinib, as well as other investigational agents such as erdafitinib, inhibit wild-type FGFR3 in nanomolar concentrations. The first case report of ponatinib use in a primary pediatric CNS malignancy, resulting in a partial response that was maintained over several months, was recently published. Among the TKIs with current FDA approval, dasatinib and ponatinib appear to have the highest likelihood of achieving good CNS penetration.65
PIK3CA Mutations and PI3K/mTOR Inhibitors
In addition to growth factor receptor alterations, the PI3K/mTOR pathway may also be activated through other somatic variations such as deletion of phosphatase and tensin homolog (PTEN) or mutation of PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha). Activation of this pathway represents an opportunity for additional targeted therapy selection. A study utilizing PedcBioPortal demonstrated PTEN alterations in pediatric HGG and DIPG at a rate of 4.9% overall, 1.9% specifically in the brainstem. Homozygous loss of PTEN was seen in 0.8% of tumors with copy number information available.80 Mutations in the genes encoding subunits of PI3K tend to co-segregate with the HIST1H3B K27M mutation.1
Within the PI3K pathway, mTOR inhibitors function downstream of PI3K by forming a complex with FK506 binding protein 12, which binds to mTOR complex 1 and inhibits pathway signaling. Targeted PI3K inhibitors have also been developed for more upstream inhibition. To date, evaluations of mTOR inhibitors in pediatric HGG have not selected for activating mutations of the PI3K pathway and, similar to HDAC inhibitors, have had suboptimal efficacy despite the adequate BBB penetration of these agents.81 However, utilization of mTOR inhibitors has demonstrated clinical efficacy in other cancers harboring PIK3CA mutations. Dual PI3K and mTOR inhibitors are also being investigated. Targeted utilization of everolimus (mTOR inhibitor) or LY3023414 (dual PI3K and mTOR inhibitor) is currently being investigated in pediatric patients with PI3K-activating mutations (NCT03213678). Another novel dual target therapy, CUDC-907 (HDAC and PI3K inhibitor), has demonstrated preclinical efficacy as a radiosensitizer in pediatric HGG and DIPG, and is currently being evaluated for pediatric patients in a phase I clinical trial (NCT02909777).
CDK4/6 Amplification, CDKN2A Loss, and Cyclin-Dependent Kinase Inhibitors
Genetic alterations in genes encoding cyclin-dependent kinase (CDK) are frequently identified in pediatric HGG. Both alterations in CDK4/6 and loss of CDK inhibitor 2A (CDKN2A) can contribute to dysregulated cellular proliferation.82CDKN2A is inhibitory of CDK signaling, and homozygous loss is exclusively found in non-brainstem pediatric HGG.3 Mistry and colleagues found that 57% of pediatric secondary HGG harbored deletions of CDKN2A.60
Loss of CDKN2A or amplification of CDK4/6 represents an opportunity for targeted treatment with the development of CDK4/6 inhibitors. CDK4/6 inhibitors prevent progression from the G1 phase to the S phase of the cell cycle by inhibiting the phosphorylation of retinoblastoma protein (Rb1), thereby preventing replication.83 Efficacy of CDK4/6 inhibitors therefore is dependent on intact Rb1, and Rb1 loss is a demonstrated mechanism of CDK4/6 inhibitor resistance. This class of agents has been shown to have activity against in vitro and in vivo murine models of GBM.84 Palbociclib demonstrated efficacy in an INK4a/ARF-deficient (CDKN2A loss) murine model of brainstem glioma.82 No reports of CDK4/6 inhibitors in pediatric HGG have been published; however, 2 phase I clinical trials are currently accruing to evaluate the safety and efficacy of these agents in pediatric HGG, though neither of these studies requires that the discussed genetic alterations be present for eligibility (NCT02255461, NCT02644460). Ribociclib has the most promising characteristics for BBB penetration, as it is the only CDK4/6 inhibitor that is not a significant substrate for both efflux transporters, P-glycoprotein and breast cancer resistance protein. Through intrinsic CNS penetration and preclinical data alone, abemaciclib and ribociclib have the most promising characteristics, followed by palbociclib.
Loss of CDKN2A with a BRAF V600E mutation has been identified as a distinct subset of pediatric CNS malignancy.1 The combination of a BRAF inhibitor with a CDK4/6 inhibitor improved survival in a human xenograft model of HGG with BRAF V600E mutation and CDKN2A homozygous loss compared with single agent therapy.85 To date, this combination has not been evaluated in clinical trials in pediatric glioma but may represent a promising therapeutic strategy for patients harboring both mutations.
MET Amplification and MET Inhibitors
Methionine (MET) signaling aberrations are found in many human malignancies, including adult GBM, sarcomas, carcinomas, and cancers of the hematopoietic system. Approximately 3–7% of pediatric GBMs display MET gene amplification.3,86 A recent study found MET fusions in 10% of 53 pediatric GBMs through tumor sequencing.87 MET phosphorylation has the potential to lead to activation of a number of downstream targets that have likewise been implicated in tumorigenesis, including PI3K and Ras/Raf/MEK/ERK.
A targeted strategy to prevent the activation of these downstream targets is the administration of MET inhibitors. MET inhibitors have been shown to reduce Akt and ERK phosphorylation in preclinical HGG models.87 A phase I study of the MET inhibitor tivantinib has been completed in pediatric patients, and, similar to other agents discussed, responses were suboptimal; however, this outcome may be explained by the lack of selection for MET amplification during patient enrollment.88 Of the commercially available MET inhibitors, cabozantinib appears to have the highest BBB penetration.65 Clinical trials are currently accruing to evaluate the impact of targeted MET inhibitor therapy in adult patients, but more studies are needed to determine the clinical impact in pediatric HGG (NCT02978261, NCT02465060).
ROS1, FGFR, and MET Gene Fusions
In considering personalized and targeted therapy for brain cancer, gene fusions may represent a particularly promising area for targeted therapy. As seen in other solid tumors and human cancers, in-frame fusions involving established tumor drivers, such as FGFR intragenic translocations resulting in a fusion protein that causes FGFR dimerization and constitutive activation, are frequently pivotal clonal events that are essential to tumor survival and growth.89 Some of the largest successes in precision medicine involve the targeting of recurrent fusions. As regards adult and pediatric gliomas, targeting fusions are increasingly being studied in vitro and in vivo. Case reports have been published demonstrating the efficacy of targeting of ROS1, FGFR, and MET gene fusions in pediatric HGG.87,90 Bender et al found gene fusions of the MET oncogene in 5 of 53 pediatric GBMs and treated one patient with a PTPRZ1 (protein tyrosine phosphatase, receptor type Z1)–MET fusion-driven GBM with the targeted inhibitor crizotinib, resulting in tumor regression over the next 2 months.87 Multiple TKIs with FDA approval for other indications, such as cabozantinib, as well as many investigational agents, such as brigatinib (ROS1), entrectinib (ROS1/TRK), and larotrectinib (TRK), have demonstrated promise as potential CNS therapies. A prospective study of entrectinib in pediatric solid tumors, including CNS tumors, with ALK, ROS, or TRK fusions is currently accruing (NCT02650401).
Future Directions
Although many of the agents discussed herein have demonstrated in vitro efficacy against pediatric HGG, clinical utility may be significantly limited by their ability to reach the intended site of action. Evaluating drug characteristics can help to predict whether the drug will be able to penetrate the BBB.65 Low molecular weight, high lipophilicity, and low protein binding increase the likelihood that an agent will achieve sufficient CNS concentrations. Agents that are not substrates of efflux transporters (P-glycoprotein and breast cancer resistance protein) are most likely to be maintained in the CSF. These criteria can be applied to determine which agent in a class may be the most beneficial for CNS disease. In addition to poor BBB penetration, unfavorable outcomes from clinical studies may be due to nontargeted patient recruitment to assess efficacy of the agent against the mutation of interest. The National Cancer Institute–Children’s Oncology Group Pediatric MATCH trial (NCT03155620), an umbrella trial for children with treatment-refractory tumors that is currently matching patients with actionable mutations to 9 investigational targeted therapies, with plans to include additional agents in the future, may help to clarify the utility of these targeted therapies for specific mutation types. CNS tumors (and relevant alterations) are eligible for pediatric MATCH, and effective CNS penetration was taken into consideration when deciding which agents had indications for CNS tumors. Future trials should therefore consider both targeted patient selection and BBB penetration of the investigational agent to determine the clinical utility in pediatric glioma. As more institutions incorporate molecular results into the selection of targeted therapies for children with gliomas, we also strongly support institutional and consortium-based clinical programs to prospectively track targeted treatment outcome data with paired tissue and NGS results.
In addition to concerted efforts to devise new treatment strategies, work is being done to develop innovative ways to monitor response to treatment. One promising avenue in this regard is cell-free DNA of tumor origin in the CSF. Previous studies have demonstrated that such DNA can be detected in CSF samples from brain tumor patients,91 and a recent study demonstrated that droplet digital PCR could detect cell-free H3K27M DNA in spinal fluid from patients with midline glioma and detect changes in the amount of this DNA in response to therapy in an in vitro model.92 This could potentially represent a specific and noninvasive method to supplement serial imaging.
In summary, the role of precision diagnostics and therapies in neuro-oncology is rapidly evolving with important implications for the standard of care for management of pediatric gliomas. The clinically integrated molecular profiling and targeting of pediatric brain tumors in recent studies has demonstrated the feasibility and utility of incorporating sequencing into the care of children with brain tumors, particularly children and young adults with glioma. Based on the studies discussed herein and others, there is sufficient justification for our consensus recommendations for the integration of precision diagnostics and therapeutics into the management of pediatric gliomas.
Funding
CK is supported by NIH/NINDS grant K08-NS099427–01, Michigan Medicine Department of Pediatrics Gorman Scholar Award, the University of Michigan Chad Carr Pediatric Brain Tumor Center, the Chad Tough Foundation, and Hyundai Hope on Wheels.
Supplementary Material
Conflict of interest statement
SHR is employed by Foundation Medicine. PB has filed a patent for discovery of the MYB-QKI fusion. The authors have no other disclosures to report.
References
- 1. Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 3. Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang J, Wu G, Miller CP, et al. ; St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kline CN, Joseph NM, Grenert JP, et al. Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol. 2017;19(5):699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramkissoon SH, Bandopadhayay P, Hwang J, et al. Clinical targeted exome-based sequencing in combination with genome-wide copy number profiling: precision medicine analysis of 203 pediatric brain tumors. Neuro Oncol. 2017;19(7):986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koschmann C, Wu Y-M, Kumar-Sinha C, et al. Clinically integrated sequencing alters therapy in children and young adults with high-risk glial brain tumors. JCO Precis Oncol. 2018;(2):1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole BL, Lockwood CM, Stasi S, et al. Year 1 in the molecular era of pediatric brain tumor diagnosis: application of universal clinical targeted sequencing in an unselected cohort of children. JCO Precis Oncol. 2018;(2):1–13. [DOI] [PubMed] [Google Scholar]
- 9. Youmans JR, Winn HR.. Youmans Neurological Surgery. 6th ed Philadelphia, PA: Saunders/Elsevier; 2011. [Google Scholar]
- 10. Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7(3):241–248. [DOI] [PubMed] [Google Scholar]
- 11. Cartmill M, Punt J. Diffuse brain stem glioma. A review of stereotactic biopsies. Childs Nerv Syst. 1999;15(5):235–237; discussion 238. [DOI] [PubMed] [Google Scholar]
- 12. Yoshimura J, Onda K, Tanaka R, Takahashi H. Clinicopathological study of diffuse type brainstem gliomas: analysis of 40 autopsy cases. Neurol Med Chir (Tokyo). 2003;43(8):375–382; discussion 382. [DOI] [PubMed] [Google Scholar]
- 13. Louis DN; International Agency for Research on Cancer, World Health Organization.. WHO Classification of Tumours of the Central Nervous System. Lyon, France: International Agency for Research on Cancer; 2007. [Google Scholar]
- 14. Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koschmann C, Calinescu AA, Nunez FJ, et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8(328):328ra328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wisoff JH, Boyett JM, Berger MS, et al. Current neurosurgical management and the impact of the extent of resection in the treatment of malignant gliomas of childhood: a report of the Children’s Cancer Group trial no. CCG-945. J Neurosurg. 1998;89(1):52–59. [DOI] [PubMed] [Google Scholar]
- 17. Huynh-Le MP, Walker AJ, Burger PC, et al. Management of pediatric intracranial low-grade gliomas: long-term follow-up after radiation therapy. Childs Nerv Syst. 2016;32(8):1425–1430. [DOI] [PubMed] [Google Scholar]
- 18. Armstrong GT. Long-term survivors of childhood central nervous system malignancies: the experience of the Childhood Cancer Survivor Study. Eur J Paediatr Neurol. 2010;14(4):298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cohen KJ, Pollack IF, Zhou T, et al. Temozolomide in the treatment of high-grade gliomas in children: a report from the Children’s Oncology Group. Neuro Oncol. 2011;13(3):317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones C, Karajannis MA, Jones DTW, et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol. 2017;19(2):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sposto R, Ertel IJ, Jenkin RD, et al. The effectiveness of chemotherapy for treatment of high grade astrocytoma in children: results of a randomized trial. A report from the Childrens Cancer Study Group. J Neurooncol. 1989;7(2):165–177. [DOI] [PubMed] [Google Scholar]
- 22. Wolff JE, Driever PH, Erdlenbruch B, et al. Intensive chemotherapy improves survival in pediatric high-grade glioma after gross total resection: results of the HIT-GBM-C protocol. Cancer. 2010;116(3):705–712. [DOI] [PubMed] [Google Scholar]
- 23. Rizzo D, Scalzone M, Ruggiero A, et al. Temozolomide in the treatment of newly diagnosed diffuse brainstem glioma in children: a broken promise? J Chemother. 2015;27(2):106–110. [DOI] [PubMed] [Google Scholar]
- 24. Macy ME, Kieran MW, Chi SN, et al. A pediatric trial of radiation/cetuximab followed by irinotecan/cetuximab in newly diagnosed diffuse pontine gliomas and high-grade astrocytomas: a Pediatric Oncology Experimental Therapeutics Investigators’ Consortium study. Pediatr Blood Cancer. 2017;64(11):5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kilburn LB, Kocak M, Baxter P, et al. A pediatric brain tumor consortium phase II trial of capecitabine rapidly disintegrating tablets with concomitant radiation therapy in children with newly diagnosed diffuse intrinsic pontine gliomas. Pediatr Blood Cancer. 2017;62(2):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Veldhuijzen van Zanten SEM, El-Khouly FE, Jansen MHA, et al. A phase I/II study of gemcitabine during radiotherapy in children with newly diagnosed diffuse intrinsic pontine glioma. J Neurooncol. 2017;135(2):307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tihan T, Ersen A, Qaddoumi I, et al. Pathologic characteristics of pediatric intracranial pilocytic astrocytomas and their impact on outcome in 3 countries: a multi-institutional study. Am J Surg Pathol. 2012;36(1):43–55. [DOI] [PubMed] [Google Scholar]
- 28. Wisoff JH, Sanford RA, Heier LA, et al. Primary neurosurgery for pediatric low-grade gliomas: a prospective multi-institutional study from the Children’s Oncology Group. Neurosurgery. 2011;68(6):1548–1554; discussion 1554‒1555. [DOI] [PubMed] [Google Scholar]
- 29. Merchant TE, Conklin HM, Wu S, Lustig RH, Xiong X. Late effects of conformal radiation therapy for pediatric patients with low-grade glioma: prospective evaluation of cognitive, endocrine, and hearing deficits. J Clin Oncol. 2009;27(22):3691–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merchant TE, Pollack IF, Loeffler JS. Brain tumors across the age spectrum: biology, therapy, and late effects. Semin Radiat Oncol. 2010;20(1):58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ater JL, Xia C, Mazewski CM, et al. Nonrandomized comparison of neurofibromatosis type 1 and non-neurofibromatosis type 1 children who received carboplatin and vincristine for progressive low-grade glioma: a report from the Children’s Oncology Group. Cancer. 2016;122(12):1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prados MD, Edwards MS, Rabbitt J, Lamborn K, Davis RL, Levin VA. Treatment of pediatric low-grade gliomas with a nitrosourea-based multiagent chemotherapy regimen. J Neurooncol. 1997;32(3):235–241. [DOI] [PubMed] [Google Scholar]
- 33. Momota H, Narita Y, Miyakita Y, Shibui S. Secondary hematological malignancies associated with temozolomide in patients with glioma. Neuro Oncol. 2013;15(10):1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122(1):11–19. [DOI] [PubMed] [Google Scholar]
- 36. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tian Y, Rich BE, Vena N, et al. Detection of KIAA1549-BRAF fusion transcripts in formalin-fixed paraffin-embedded pediatric low-grade gliomas. J Mol Diagn. 2011;13(6):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phillips JJ, Aranda D, Ellison DW, et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 2013;23(5):565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nikiforova MN, Wald AI, Melan MA, et al. Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol. 2016;18(3):379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siegfried A, Rousseau A, Maurage CA, et al. EWSR1-PATZ1 gene fusion may define a new glioneuronal tumor entity. Brain Pathol. 2019;29(1):53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bender S, Tang Y, Lindroth AM, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. [DOI] [PubMed] [Google Scholar]
- 42. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 43. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mack SC, Northcott PA. Genomic analysis of childhood brain tumors: methods for genome-wide discovery and precision medicine become mainstream. J Clin Oncol. 2017;35(21):2346–2354. [DOI] [PubMed] [Google Scholar]
- 45. Sturm D, Orr BA, Toprak UH, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnson A, Severson E, Gay L, et al. Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist. 2017;22(12):1478–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DiMario FJ Jr, Sahin M, Ebrahimi-Fakhari D. Tuberous sclerosis complex. Pediatr Clin North Am. 2015;62(3):633–648. [DOI] [PubMed] [Google Scholar]
- 51. Curatolo P, Bombardieri R, Jozwiak S. Tuberous sclerosis. Lancet. 2008;372(9639):657–668. [DOI] [PubMed] [Google Scholar]
- 52. Goh S, Butler W, Thiele EA. Subependymal giant cell tumors in tuberous sclerosis complex. Neurology. 2004;63(8):1457–1461. [DOI] [PubMed] [Google Scholar]
- 53. Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99(21):13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krueger DA, Care MM, Holland K, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363(19):1801–1811. [DOI] [PubMed] [Google Scholar]
- 55. Franz DN, Belousova E, Sparagana S, et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381(9861):125–132. [DOI] [PubMed] [Google Scholar]
- 56. MacKeigan JP, Krueger DA. Differentiating the mTOR inhibitors everolimus and sirolimus in the treatment of tuberous sclerosis complex. Neuro Oncol. 2015;17(12):1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. [DOI] [PubMed] [Google Scholar]
- 58. Lassaletta A, Zapotocky M, Mistry M, et al. Therapeutic and prognostic implications of BRAF V600E in pediatric low-grade gliomas. J Clin Oncol. 2017;35(25):2934–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Horbinski C, Nikiforova MN, Hagenkord JM, Hamilton RL, Pollack IF. Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro Oncol. 2012;14(6):777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mistry M, Zhukova N, Merico D, et al. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol. 2015;33(9):1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mackay A, Burford A, Molinari V, et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 2018;33(5):829–842.e825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson GW, Orr BA, Gajjar A. Complete clinical regression of a BRAF V600E-mutant pediatric glioblastoma multiforme after BRAF inhibitor therapy. BMC Cancer. 2014;14:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kieran MW, Bouffet E, Tabori U, et al. CNS tumoursThe first study of dabrafenib in pediatric patients with BRAF V600–mutant relapsed or refractory low-grade gliomas. Annal Oncol. 2016;27(Suppl 6):LBA19_PR-LBA19_PR. [Google Scholar]
- 64. Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. [DOI] [PubMed] [Google Scholar]
- 65. Marini BL, Benitez LL, Zureick AH, et al. Blood-brain barrier-adapted precision medicine therapy for pediatric brain tumors. Transl Res. 2017;188:27.e1–27.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang J, Yao TW, Hashizume R, et al. Combined BRAFV600E and MEK blockade for BRAFV600E-mutant gliomas. J Neurooncol. 2017;131(3):495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Penman CL, Faulkner C, Lowis SP, Kurian KM. Current understanding of BRAF alterations in diagnosis, prognosis, and therapeutic targeting in pediatric low-grade gliomas. Front Oncol. 2015;5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodriguez FJ, Ligon AH, Horkayne-Szakaly I, et al. BRAF duplications and MAPK pathway activation are frequent in gliomas of the optic nerve proper. J Neuropathol Exp Neurol. 2012;71(9):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fangusaro JR, Onar-Thomas A, Young-Poussaint T, et al. A phase II prospective study of selumetinib in children with recurrent or refractory low-grade glioma (LGG): a Pediatric Brain Tumor Consortium (PBTC) study. J Clin Oncol. 2017;35(15 Suppl):10504. [Google Scholar]
- 71. Ameratunga M, McArthur G, Gan H, Cher L. Prolonged disease control with MEK inhibitor in neurofibromatosis type I-associated glioblastoma. J Clin Pharm Ther. 2016;41(3):357–359. [DOI] [PubMed] [Google Scholar]
- 72. Huang J, Liu F, Liu Z, et al. Immune checkpoint in glioblastoma: promising and challenging. Front Pharmacol. 2017;8:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhu X, McDowell MM, Newman WC, Mason GE, Greene S, Tamber MS. Severe cerebral edema following nivolumab treatment for pediatric glioblastoma: case report. J Neurosurg Pediatr. 2017;19(2):249–253. [DOI] [PubMed] [Google Scholar]
- 74. Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. [DOI] [PubMed] [Google Scholar]
- 75. Lewis PW, Müller MM, Koletsky MS, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hennika T, Hu G, Olaciregui NG, et al. Pre-clinical study of panobinostat in xenograft and genetically engineered murine diffuse intrinsic pontine glioma models. PLoS One. 2017;12(1):e0169485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grasso CS, Tang Y, Truffaux N, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21(6):555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Arrillaga-Romany I, Chi AS, Allen JE, Oster W, Wen PY, Batchelor TT. A phase 2 study of the first imipridone ONC201, a selective DRD2 antagonist for oncology, administered every three weeks in recurrent glioblastoma. Oncotarget. 2017;8(45):79298–79304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koschmann C, Zamler D, MacKay A, et al. Characterizing and targeting PDGFRA alterations in pediatric high-grade glioma. Oncotarget. 2016;7(40):65696–65706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koschmann C, Farooqui Z, Kasaian K, et al. Multi-focal sequencing of a diffuse intrinsic pontine glioma establishes PTEN loss as an early event. NPJ Precis Oncol. 2017;1(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fouladi M, Laningham F, Wu J, et al. Phase I study of everolimus in pediatric patients with refractory solid tumors. J Clin Oncol. 2007;25(30):4806–4812. [DOI] [PubMed] [Google Scholar]
- 82. Barton KL, Misuraca K, Cordero F, et al. PD-0332991, a CDK4/6 inhibitor, significantly prolongs survival in a genetically engineered mouse model of brainstem glioma. PLoS One. 2013;8(10):e77639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–342. [DOI] [PubMed] [Google Scholar]
- 84. Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70(8):3228–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huillard E, Hashizume R, Phillips JJ, et al. Cooperative interactions of BRAFV600E kinase and CDKN2A locus deficiency in pediatric malignant astrocytoma as a basis for rational therapy. Proc Natl Acad Sci U S A. 2012;109(22):8710–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. International Cancer Genome Consortium PedBrain Tumor P. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22(11):1314–1320. [DOI] [PubMed] [Google Scholar]
- 88. Geller JI, Perentesis JP, Liu X, et al. A phase 1 study of the c-Met inhibitor, tivantinib (ARQ197) in children with relapsed or refractory solid tumors: a Children’s Oncology Group study phase 1 and pilot consortium trial (ADVL1111). Pediatr Blood Cancer. 2017;64(11):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–129. [DOI] [PubMed] [Google Scholar]
- 90. Linzey JR, Marini B, McFadden K, et al. Identification and targeting of an FGFR fusion in a pediatric thalamic “central oligodendroglioma”. NPJ Precis Oncol. 2017;1(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang Y, Springer S, Zhang M, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112(31):9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Stallard S, Savelieff MG, Wierzbicki K, et al. CSF H3F3A K27M circulating tumor DNA copy number quantifies tumor growth and in vitro treatment response. Acta Neuropathol Commun. 2018;6(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.