Abstract
The heat shock protein 70 (Hsp70) family of molecular chaperones are crucial for the survival and pathogenicity of the main agent of malaria, Plasmodium falciparum. Hsp70 is central to cellular proteostasis and some of its isoforms are essential for survival of the malaria parasite. In addition, they are also implicated in the development of antimalarial drug resistance. For these reasons, they are thought to be potential drug targets, especially in antimalarial combination therapies. However, their high sequence conservation across species presents a hurdle with respect to their selective targeting. The human genome encodes 17 Hsp70 isoforms while P. falciparum encodes for only 6. The structural architecture of Hsp70s is typically characterized by a highly conserved N-terminal nucleotide-binding domain (NBD) and a less conserved C-terminal substrate-binding domain (SBD). The two domains are connected by a highly conserved linker. In spite of their fairly high sequence conservation, Hsp70s from various species possess unique signature motifs that appear to uniquely influence their function. In addition, their cooperation with co-chaperones further regulates their functional specificity. In the current review, bioinformatics tools were used to identify conserved and unique signature motifs in Hsp70s of P. falciparum versus their human counterparts. We discuss the common and distinctive structure-function features of these proteins. This information is important towards elucidating the prospects of selective targeting of parasite heat shock proteins as part of antimalarial design efforts.
Keywords: Hsp70, Molecular chaperone, Functional specificity, Signature motif, Plasmodium falciparum, Human
General structure-function features of Hsp70
The Hsp70 members are central mediators of cellular proteostasis. They facilitate de-novo protein folding of nascent polypeptides, protein translocation and are implicated in signal transduction (Mayer 2010). Hsp70s thus interact with proteins at virtually every stage of their life cycles, from primary folding through to degradation. Hsp70s are strongly upregulated by physiological stress. Their cooperation with other Hsp members such as Hsp110, Hsp100, Hsp90, Hsp60, Hsp40 and the small Hsps enhances their functional versatility (Mogk et al. 2015). Hsp70s possess a highly conserved N-terminal nucleotide-binding domain (NBD) and a more varied C-terminal substrate-binding domain (SBD), adjoined by a linker (Fig. 1; Mayer 2010). The SBD of Hsp70 is reportedly functionally promiscuous as it allows the chaperone to bind to short degenerate motifs within peptide substrates (Rosenzweig et al. 2017). This technically provides Hsp70 with the capability to bind to virtually all proteins.
Fig. 1.
General structure of Hsp70s. The NBD is comprised of lobes IA (blue), IIA (blue), IB (purple) and IIB (pink). The SBD is constituted by SBD-β (red) and SBD-α subunits (orange) of which the latter is subdivided into helices A–E. The NBD and SBD are adjoined by a highly conserved linker (green)
Generally, Hsp70s of prokaryotic and mammalian family members share at least 50% identity, making Hsp70 one of the most conserved proteins (Yu et al. 2015). Hsp70s are generally ubiquitous, accounting for approximately 1–2% of the cellular proteome (Dhamad et al. 2016). Several organisms express multiple Hsp70s. For instance, while humans express a complement of 17 Hsp70s, the major malaria parasite, Plasmodium falciparum, expresses 6 Hsp70s (Kampinga et al. 2009). The various Hsp70 isoforms localize to different subcellular compartments. Hsp70s particularly play a crucial role in the survival of P. falciparum, since in its development, the parasite cycles between a poikilothermic mosquito vector and a homoeothermic human host. The development of malaria fever at the blood stage is closely linked to the modification of the infected human red blood cell (RBC), making it cytoadherent, thus leading to clinical complications of malaria. Not only does physiological stress upregulate general expression of parasite Hsps, but some of these are exported to the infected RBC (Pryzborski et al. 2015). Amongst the exported proteins are several Hsp40 members and P. falciparum Hsp70-x (PfHsp70-x; Külzer et al. 2012). Hsp40 co-chaperones serve as substrate scanners for Hsp70 and also stimulate the otherwise, functionally rate limiting ATPase activity of the latter (Botha et al. 2011). Consequently, though not essential, PfHsp70-x along with some exported parasite Hsp40s are implicated in augmenting pathogenicity of the malaria parasite (Cobb et al. 2017). Because of their implications in parasite survival, antimalarial drug resistance and pathogenicity, Hsps of P. falciparum are prospective drug targets (Shonhai 2010). Here, we conducted systematic analyses on the structure-function features of Hsp70s from P. falciparum and human systems towards identifying unique features towards selective targeting of parasite Hsp70s towards the development of novel antimalarials.
Subcellular localization and functional features of P. falciparum Hsp70s and their human homologues
P. falciparum expresses 6 Hsp70 members which are located in various subcellular compartments: PfHsp70-1 and PfHsp70-z (cytosol) PfHsp70-2 and PfHsp70-y (ER), PfHsp70-3 (mitochondrium) and PfHsp70-x which occurs in the parasitophorous vacuole and is also exported to the infected RBC (Table 1; Külzer et al. 2012; Shonhai 2014). Hsp70s are classified into two main groups: canonical Hsp70s which structurally resemble E. coli Hsp70 (DnaK), while the larger in size, Hsp110/glucose regulated protein 170 (Grp170) members (Dragovic et al. 2006), constitute the non-canonical Hsp70s. The latter possess chaperone function which is limited largely to suppression of protein aggregation, while the former are efficient protein folders. Apart from their role as chaperones, Hsp110s are thought to serve as nucleotide exchange factors (NEFs) of their canonical Hsp70 counterparts (Dragovic et al. 2006). Structurally, Hsp110s are marked by extended acidic insertions located within their SDB-β and the SBD-α subunits and they possess linker segments that are distinct from those present in canonical Hsp70s (Fig. 2; Oh et al. 1999). PfHsp70-1 (cytosol/nucleus), PfHsp702 (ER) and PfHsp70–3 (mitochondrium) constitute the canonical Hsp70 isoforms of P. falciparum. PfHsp70-z (cytosol) and PfHsp70-y (ER) belong to the Hsp110 and Grp170 families, respectively. On the other hand, humans express 13 Hsp70s, 3 Hsp110s and 1 Grp170. The Hsp70 isoforms carry out specialized protein folding functions within their various subcellular locations (Tables 1 and 2, Kampinga et al. 2009; Shonhai 2014).
Table 1.
Characteristic features of P. falciparum Hsp70s
| PfHsp70 (PlasmoDB accession number) | Size (kDa) | Localization | Stress Inducible (yes/no) | Cellular functions | References |
|---|---|---|---|---|---|
| PfHsp70-1 (PF3D7_0818900) | 74 | Nucleus and cytosol | Yes | Protein folding/translocation/aggregation suppression |
Shonhai et al. 2007 Shonhai et al. 2008 |
| PfHsp70-z (PF3D7_0708800) | 100 | Cytosol | Yes | Predicted NEF of PfHsp70-1; aggregation suppression |
Muralidharan et al. 2012 Zininga et al. 2016 |
| PfHsp70-2 (PF3D7_0917900) | 73 | E.R | Yes | Protein import and folding in the ER, retrograde translocation of proteins for degradation |
Shonhai et al. 2007 Chen et al. 2018 |
| PfHsp70-y (PF3D7_1344200) | 108 | E.R | ND | Thought to be NEF for PfHsp70-2 |
Shonhai et al. 2007 Njunge et al. 2013 |
| PfHsp70-3 (PF3D7_1134000) | 73 | Mitochondrium | ND | Protein translocation into the mitochondrium | Shonhai et al. 2007 |
| PfHsp70-x (PF3D7_0831700) | 76 | P.V and exported to parasite infected RBC | Yes | Protein export and subsequent protein folding of exported proteins in the infected RBC |
Charnaud et al. 2017 Cobb et al. 2017 |
ND not determined
Fig. 2.
Domain architecture of Hsp70 superfamily. The schematic represents canonical and non-canonical Hsp70 structures. The SBDβ is composed of eight sheets while SBDα is constituted by helices A–E (a). Hsp110s, of which PfHsp70-z is a member possess insertions in SBDα and towards the lid segment (b). Grp170 possesses more acidic insertions in SBDα and another leading to the C-terminus (c). The figure was adapted from Oh et al. (1999)
Table 2.
Subcellular localization of human Hsp70s
| Human Hsp70 (accession number) | Size (kDa) | Localization | Stress inducible (yes/no) | Cellular functions | References |
|---|---|---|---|---|---|
| 1. HspA1A/Hsp70-1 (P0DMV8) | 70 | Cytosol, nucleus, cell membrane, extracellular exosomes | Yes | Protein folding, translocation and facilitating degradation of misfolded proteins |
Khalouei et al. 2014 Radons 2016 |
| 2. HspA1B/Hsp70-2 (P0DMV9) | 70 | Cytosol, nucleus, extracellular exosomes | Yes | Protein folding, translocation and facilitating degradation of misfolded proteins | Radons 2016 |
| 3. HspA1L/Hsp70-1L (P34931) | 70 | Cytosol, nucleus | No | Aggregation suppression, protein folding, facilitates spermatogenesis | Zhu et al. 1997 |
| 4. HspA2 (P54652) | 70 | Cytosol, nucleus, cell membrane, extracellular exosomes | Yes | Protein folding and aggregation suppression |
Pigłowski et al. 2007 Redgrove et al. 2012 |
| 5. HspA8/Hsc70 (P11142) | 71 | Cytosol, nucleus, cell membrane, extracellular exosomes | No | Protein folding, translocation, repression of transcriptional activation | Dworniczak and Mirault 1987 |
| 6. HspA14 (Q0VDF9) | 55 | Cytosol, cell membrane | Yes | Component of the ribosome-associated complex (RAC), which is involved in folding or maintaining nascent polypeptides in a folding-competent state | Wu et al. 2011 |
| 7. HspH1 (Q92598) | 97 | Cytosol, nucleus, endocytic vesicle | Yes | Apoptosis suppression, aggregation suppression, NEF | Zappasodi et al. 2015 |
| 8. HspA4L/HspH3 (O95757) | 95 | Cytosol, nucleus | Yes | Elicits humoral immune responses in leukaemia patients | Takahashi et al. 2007 |
| 9. HspA4/HspH2 | 95 | Cytosol, extracellular exosome | N.D | Implicated in spermatogenesis | Held et al. 2011 |
| 10. HspA5/BiP (P11021) | 72 | E.R, extracellular exosomes | No | Protein import and folding in the ER, retrograde translocation of proteins for degradation | Yu et al. 2015 |
| 11. HspA13 (P48723) | 52 | E.R, extracellular exosomes, microsomes | No | Exhibits peptide-independent ATPase activity | Otterson et al. 1994 |
| 12. HspH4/Grp170 (Q9Y4L1) | 111 | E.R | Yes | Aggregation suppression, NEF | Behnke et al. 2015 |
| 13. HSPA9/Grp75 (P38646) | 74 | Mitochondria, nucleus | No | Protein translocation into the mitochondria | Mizzen et al. 1989 |
| 14. HspA7 (P48741) | 40 | Blood microparticles and extracellular exosomes | Yes | N.D | Brocchieri et al. 2008 |
| 15. HspA12B (B7ZLP2) | 76 | Endothelial cells, intracellular, blood plasma | No | N.D |
Han et al. 2003 Radons 2016 |
| 16.HspA6 | 71 | Cytosol, extracellular exosomes | Yes | Protein folding |
Khalouei et al. 2014 Leung et al. 1990 |
| 17.HspA12A | 75 | Intracellular and extracellular exosomes | No | ND |
Han et al. 2003 Radons 2016 |
ND not determined
Structural features of P. falciparum Hsp70s and their human homologues
Multiple sequence alignments between Hsp70s of P. falciparum and their homologues of human origin show high conservation which is enhanced for isoforms that occur in respective subcellular locations (Table 3). For example, the cytosolic homologues (PfHsp70-1 and human HspA1A), as well as the E. R homologues (PfHsp70-2 and human HspA5/Bip) showed high identity scores of 72.23 and 65.18%, respectively. PfHsp70-x which is exported to the RBC by the parasite is highly identical to cytosol localized homologues (PfHsp70-1 and human HspA1A). Mabate et al. (2018) established that PfHsp70-x preferentially binds substrates with asparagine repeat rich regions. Nearly 10% of the malaria parasite proteome is characterized by prion-like repeats and at least 30% of the proteome is characterized by glumatate/asparagine repeat segments (Pallarès et al. 2018). Thus, the substrate preference of PfHsp70-x suggests that it may bind and refold malarial proteins that are exported to the parasite-infected RBC. This may be important for the parasite as nearly 500 of its proteins are thought to be exported to the RBC (Hiller et al. 2004). However, PfHsp70-x was shown to be not essential in P. falciparum lab strain (Charnaud et al. 2017). However, its export to the RBC was shown to correlate with the early stages of parasite development characterized by rapid remodelling of the RBC (Cobb et al. 2017).
Table. 3.
Comparative identities of plasmodial and select human Hsp70s
| PfHsp70-y | PfHsp70-z | PfHsp70-3 | HspA9 | Hsc70 | HspA1A | PfHsp70-1 | PfHsp70-x | PfHsp70-2 | HspA5/BiP | |
|---|---|---|---|---|---|---|---|---|---|---|
| PfHsp70-y | – | 23.28 | 21.04 | 21.34 | 24.17 | 24.36 | 21.22 | 22.55 | 24.88 | 24.68 |
| PfHsp70-z | 23.28 | – | 21.22 | 21.27 | 24.84 | 24.41 | 22.71 | 25.16 | 21.72 | 24.31 |
| PfHsp70-3 | 21.04 | 21.22 | – | 62.22 | 49.92 | 49.92 | 47.44 | 49.06 | 50.00 | 51.21 |
| HspA9 | 21.34 | 21.27 | 62.22 | – | 51.14 | 50.65 | 47.86 | 48.69 | 47.69 | 49.84 |
| Hsc70 | 24.17 | 24.84 | 49.92 | 51.14 | – | 85.80 | 71.83 | 71.00 | 62.04 | 65.43 |
| HspA1A | 24.36 | 24.41 | 49.92 | 50.65 | 85.80 | – | 72.23 | 71.47 | 59.16 | 63.81 |
| PfHsp70-1 | 21.22 | 22.71 | 47.44 | 47.86 | 71.83 | 72.23 | – | 77.61 | 55.89 | 62.32 |
| PfHsp70-x | 22.55 | 25.16 | 49.06 | 48.69 | 71.00 | 71.47 | 77.61 | – | 57.08 | 62.46 |
| PfHsp70-2 | 24.88 | 21.72 | 50.00 | 47.69 | 62.04 | 59.16 | 55.89 | 57.08 | – | 65.18 |
| HspA5/BiP | 24.68 | 24.31 | 51.21 | 49.84 | 65.43 | 63.81 | 62.32 | 62.46 | 65.18 | – |
The percentage identities of the select Hsps were generated after multiple sequence alignments (MSA) of the amino acid sequences of the proteins retrieved from (www.uniprot.org) for human and (www.plasmoDB.org) for P. falciparum proteins, respectively. The MSA were conducted using the BioEdit pairwise tool (Hall et al. 2005)
Multiple sequence alignment of P. falciparum Hsp70s and their human homologues
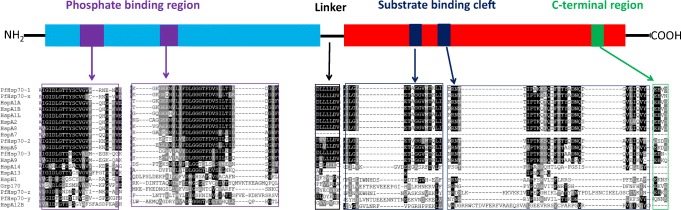
Generally, Hsp70s exhibit higher sequence conservation in the NBDs as compared to the SBDs (Fig. 3). The phosphate-binding region of P. falciparum Hsp70s is more conserved across canonical Hsp70s than it is within the non-canonical Hsp70 family (Hsp110). This could account for the reported differences in the affinities for nucleotide binding and ATP hydrolysis rates between canonical and non-canonical Hsp70s (Zininga et al. 2016). Residues such as Asp10 and Glu175 in subdomain IA, Lys71 in subdomain IB and Asp199 and Thr204 in subdomain IIA, respectively, are highly conserved and act as interaction sites for ADP (Arakawa et al. 2011). The SBDs of non-canonical Hsp70s are less conserved in comparison to those of their canonical counterparts. As such, the two Hsp70 subclasses are reported to exhibit varied substrate preferences (Zininga et al. 2016). Hsp110s and Grp170s are thought to preferentially bind bulky substrates possessing aromatic residues with a higher affinity than their canonical counterparts (Polier et al. 2008, 2010). We further observed that HspA13 possesses a less conserved substrate-binding cleft (SBC) while HspA14 lacks a typical SBC (Fig. 3). HspA13 is an E.R- and microsome-localized protein which could potentially augment the activity of its E.R counterpart HspA5/BiP. Notably, the human Hsp70 complement is not only more expanded in number but is more structurally diverse than that of the parasite, suggesting that the human protein folding system is more versatile than that of the parasite.
Fig. 3.
Comparison of domains of plasmodial and human Hsp70s. PfHsp70-y and PfHsp70-z exhibit unique features in the following segments: phosphate binding region, linker, substrate binding clefts and C-terminal regions. There is however higher sequence conservation within the canonical Hsp70s with HspA7, HspA13 and HspA14 exhibiting the greatest variation
Non-canonical Hsp70s show species-specific variation between the human and P. falciparum homologues. PfHsp70-y and PfHsp70-z exhibit unique features in several regions: the phosphate-binding region, linker, substrate-binding clefts and C-terminal segments. The variations in the phosphate-binding regions of canonical versus non-canonical Hsp70s which transcend across the human and P. falciparum systems may account for the unique nucleotide binding and hydrolysis rates reported in Hsp70 homologues of these species (Table 4). For example, the linker of human Hsp110 is composed of the residues 392EFSVTD396 as compared to residues 422EYECVE427 present in PfHsp70-z (Zininga et al. 2016). Unlike its cytosolic canonical Hsp70 homologues, human HspA14 harbours a less conserved linker (387DSLMIEC392). Since the Hsp70 linker is implicated in allosteric communication (English et al. 2017), human HspA14 may thus coordinate allostery in a unique fashion.
Table 4.
Comparative ATP hydrolysis between cytosolic P. falciparum Hsp70s and other Hsp70s
| Protein | ATP affinity KD value (nM) | ATP hydrolysis Km value (μM) | Reference |
|---|---|---|---|
| PfHsp70–1 | 3.48e-6(a) | 428(a) | Zininga et al. 2016(a) |
| PfHsp70-x | 1.10e-7(b) | 393(c) | Mabate et al. 2018(b); Cockburn et al. 2014(c) |
| PfHsp70-z | 2.50e-5(a) | 283(a) | Zininga et al. 2016(a) |
| Human HspA1A | ND | 85(d) | Bimston et al. 1998(d) |
| E. coli DnaK | 4.13e-7(f) | 20(e) | Liberek et al. 1991(e), Lebepe 2019(f) |
Note: the letters in superscripts depict the various citations of authors that are responsible for the corresponding work reported in the table
ND not determined
Comparative structural features of NBDs of Hsp70s
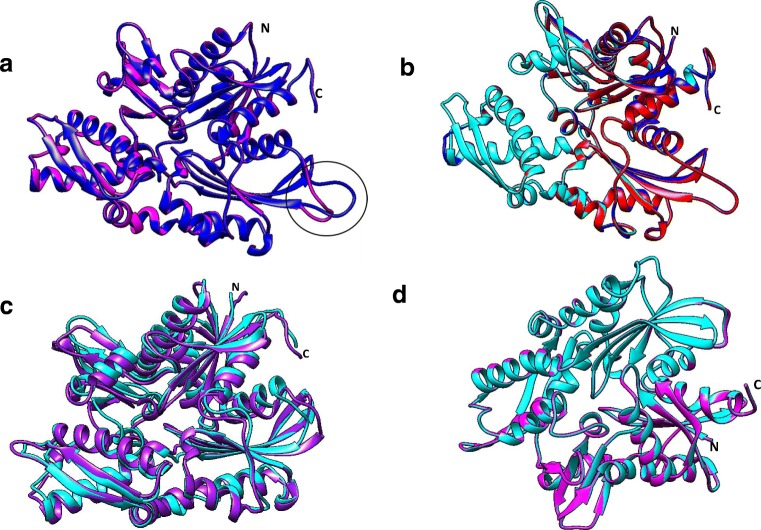
The NBD of Hsp70 is characterized by an ‘actin-like’ fold composed of 4 subdomains, namely IA, IB, IIA and IIB which form two lobes (lobe I and lobe II) (Fig. 1). These in turn form a hydrophobic nucleotide-binding cleft. Notably, canonical Hsp70s generally exhibited the highest predicted structural conservation in NBDs across species in comparison to the Grp170 group which was the most structurally diverse (Fig. 4). However, NBDs of the Hsp110 protein showed high conservation in both humans and P. falciparum with minor variations in a loop connecting the sheet and helical segments in lobe IIA (Fig. 4).
Fig. 4.
Structural comparison of Hsp70 NBDs. Superimposed images of NBDs of cytosolic Hsp110s: PfHsp70-z (blue), human Hsp110 (magenta) (a). Structural variation within the loops in Lobe IIA is encircled. Superimposition of NBDs of cytosolic, canonical Hsp70s shows high structural conservation: PfHsp70-1 (red), HspA1A (blue) and Hsc70 (cyan) (b). The ER Grp170, PfHsp70-y (cyan) versus human Grp70 (purple) NBDs show structural variations (c). Mitochondrial Hsp70s: PfHsp70-3 (cyan) and human Grp75 (magenta) exhibit high structural conservation in their NBDs (d). PfHsp70-2 (cyan) shows minor structural differences from its human ER homologue HspA5 (purple) (e). PfHsp70-x (blue) showed greater structural similarity with human HspA1A (magenta) compared to human HspA7 (cyan) (f). The variation in the loop segments of lobe IIA is encircled. The three-dimensional models were generated using PHYRE2 (Kelley et al. 2015) and models were visualized using Chimera vs1.1 (Pettersen et al. 2004), and were analysed using Matchmaker plugin to generate images
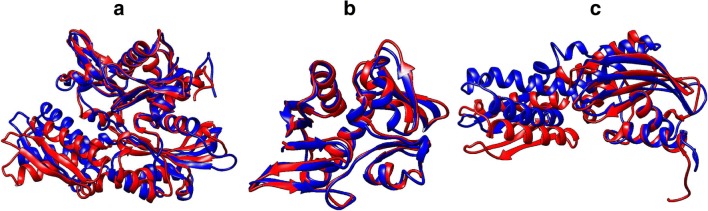
Comparative analysis of the NBDs of the two cytosolic P. falciparum Hsp70s (PfHsp70-1 and PfHsp70-z) showed some degree of divergence between these respective canonical and non-canonical Hsp70 family representatives (Fig. 5). While lobe I showed high structural conservation, there was pronounced variation in lobe II of PfHsp70-1 and PfHsp70-z. This predicted structural difference could account for the varied functional features of the two proteins. For example, Zininga et al. (2016) reported that PfHsp70-z possesses nucleotide independent chaperone activity, in contrast to PfHsp70-1 whose activity is regulated by ATP. In addition, PfHsp70-1 has been shown to exhibit higher nucleotide-binding affinity and ATP hydrolysis rates compared to PfHsp70-z (Zininga et al. 2016). Furthermore, P. falciparum Hsp70s have also been shown to possess much higher ATPase activity as compared to their human counterparts (Table 2).
Fig. 5.
Comparative structural analyses of lobes I and II in NBDs of canonical versus non-canonical Hsp70s. The NBDs of PfHsp70-1 (red) and PfHsp70-z (blue) were superimposed (a); (b). Lobe I of PfHsp70-1 (red) and PfHsp70-z (blue) exhibits minor structural variations. (c). There is more pronounced variation in lobe II of the NBD of PfHsp70-1 and PfHsp70-z. Images were generated using Matchmaker plugin on Chimera 1.11 (Pettersen et al. 2004)
Structural comparisons of SBDs of P. falciparum Hsp70s and their human homologues
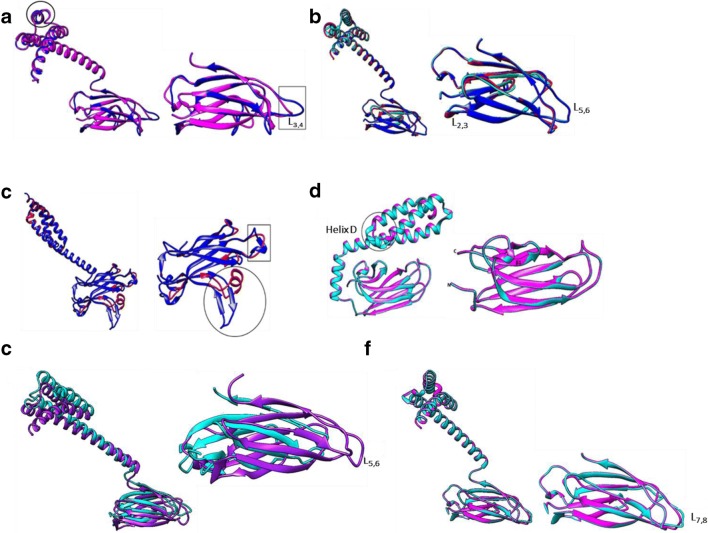
In general, structural analysis of the SBDs of human and P. falciparum Hsp70 homologues showed greater variation within the non-canonical Hsp70s as compared to their canonical counterparts (Fig. 6). PfHsp70-y and its human homologue, Grp170, were shown to possess less conserved SBDs. Grp170 and Hsp110 are also known to exhibit unique substrate preferences compared to canonical Hsp70s (Xu et al. 2012). The substrate-binding residues flanked by the SBD loops L1,2 and L3,4 are thought to be responsible for imparting substrate specificity (Xu et al. 2012). The same residues are conserved in loops L1,2 and L3,4 of the canonical Hsp70s. Variations in these could account for varied substrate preferences between canonical and non-canonical Hsp70 types.
Fig. 6.
Structural comparison of Hsp70 SBDs The Hsp70. SBDs of cytosolic Hsp110s: PfHsp70-z (blue) versus human HspH1 (magenta) (a). b SBDs of cytosolic canonical Hsp70s are more conserved: PfHsp70-1 (red), HspA1A (blue) and Hsc70 (cyan). c The SBDs of E. R Grp170s: PfHsp70-y (red) versus human Grp170 (blue) show structural uniqueness in SBDβ. d Mitochondrial Hsp70s: PfHsp70-3 (cyan) and human Grp75 (magenta) exhibit high structural conservation in their SBDs. e Canonical, parasite ER Hsp70: PfHsp70-2 (cyan) shows minor structural variations from its homologue HspA5 (purple). f Exported parasite protein, PfHsp70-x (cyan) share high structural similarity with human HspA1A (magenta)
It was also interesting to note that in all the Hsp70s, most variations occur in the loop regions of the substrate-binding cleft compared to the helical lid sections. This may suggest that the loops may influence functional specificity of Hsp70 as has been reported recently (Mabate et al. 2018). Furthermore, human Hsp110 (HspH1) and PfHsp70-z exhibited minor variations in the SBD. PfHsp70-z exhibits slightly longer loops located in the substrate-binding cleft as compared to human HspH1 (Fig. 6). While canonical Hsp70s generally show high conservation, human Grp170 exhibits a β-sheet protrusion located within its SBDβ which is fairly distinct from that of its counterpart, PfHsp70-y (Fig. 6). Human Grp75/HspA9 also possesses a helical fold around β5 and β6 segments of the SBC, while PfHsp70-3 has a loop in the same region (Fig. 6). These variations could possibly dictate substrate binding preferences. Notably, the SBD of PfHsp70-2 showed some degree of structural divergence within the SBC region as it seemed to possess shorter loops than its human homologue, HspA2. This could suggest that these ER homologues may possess specialized functions representing possible unique protein folding requirements between parasite and human systems.
Signature motifs of P. falciparum Hsp70s and their human homologues
Despite possessing a highly conserved domain architecture, plasmodial Hsp70s are thought to be tailored for species-specific functional demands (Pryzborski et al. 2015). Outlined below are the unique structural features that delineate P. falciparum Hsp70s and their human counterparts.
P. falciparum Hsp70s possess a valine residue preceding the EEVD motif
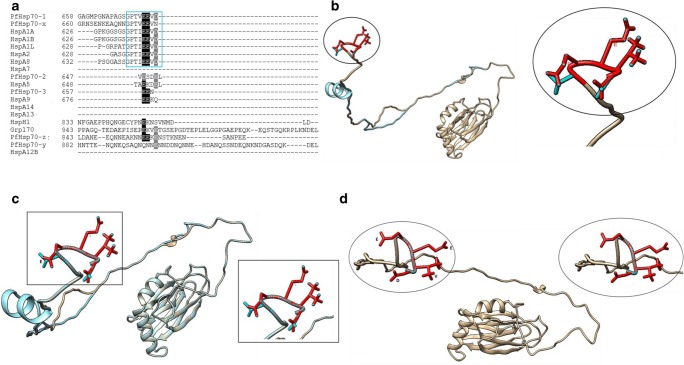
Human and P. falciparum cytosol-localized canonical Hsp70s possess the negatively charged EEVD motif to which Hop, the module that brings Hsp70 and Hsp90 into a functional complex, anchors via its tetratricopeptide repeat (TPR) domains (Zininga et al. 2015). However, unlike their human counterparts, PfHsp70-1 and PfHsp70-x possess a valine instead of an isoleucine residue which precedes the C-terminal EEVD/N motif (Fig. 7). The C-terminal TVEEVD motif of Ssa1 has recently been reported to function as a SUMO-interacting motif (Gong et al. 2018). This suggests that PfHsp70-1 could potentially be involved in SUMOylation. SUMOylation is essential for normal cell function and a potential target of small molecule inhibitors against P. falciparum (Reiter and Matunis 2016).
Fig. 7.
The Hsp70 EEVD/N motif. The C-terminal residues EEVD/EEVN of plasmodial and human Hsp70s PfHsp70-1 and PfHsp70-x possess a valine residue before the EEVD motif, while all human proteins possess isoleucine residues (a). The EEVN (cyan) and EEVD (red) motifs of PfHsp70-x and PfHsp70-1, respectively, show structural conservation (b). The EEVN (cyan) motif and the EEVD motif of Hsc70 show structural conservation (c). Human EEVD motifs exhibit high conservation amongst themselves (HsA1A-red; Hsc70-cyan) (d)
GGMP-repeat motif
Of all the 6 P. falciparum Hsp70 isoforms, PfHsp70-1 is noted for possession of unique GGMP repeat motifs positioned towards its C-terminal in the lid segment. This motif is only present in PfHsp70-1 and absent from other P. falciparum Hsp70 isoforms. In addition, it is absent in human Hsp70 homologues except in the Hsc70 homologue (P11142) which possesses a short 619GGMPGGMP626 motif, representing only two GGMP repeats compared to five GGMP repeats and an additional GGMN segment in PfHsp70-1 (Fig. 8). Thus, the enhanced presence of GGMP repeats in PfHsp70-1 is a distinct feature of this essential protein (Chiang et al. 2009). A closely related motif, the GGAP repeat present in yeast Hsp70 (Ssa1), was recently reported to act as a secondary peptide binding site, hence is thought to regulate the substrate binding specificity of Ssa1 (Gong et al. 2018). In addition, the GGAP motif of Ssa1 was implicated in Hsp40 co-chaperone binding (Gong et al. 2018). Hence, it is possible that the GGMP repeat motifs of PfHsp70-1 could similarly regulate both substrate recognition and co-chaperone binding. In Toxoplasma gondii, the GGMP repeat motif of Hsp70 was reported to be associated with parasite virulence (Lyons and Johnson 1998). Ssa1 ΔGGAP and GGAG mutants were shown to exhibit reduced thermo-tolerance, thus demonstrating the importance of this motif in Hsp70 function (Gong et al. 2018).
Fig. 8.
The GGMP and EKEK repeat motifs on the c-termini of PfHsp70-1 and PfHsp70-z. The C-terminal end of Hsp70s is characterized by sequence divergence and the GGMP repeat motif of PfHsp70-1 is highlighted in green while the EKEK repeat motif of PfHsp70-z is highlighted in red
EKEK-repeat motif of PfHsp70-z
We also identified here a novel 18mer EKEK repeat motif on the C-terminus of PfHsp70-z (Fig. 8). This feature is located between positions 810EKEK--K827 of PfHsp70-z and is one of the features that distinguishes this protein from other Hsp70 homologues (Fig. 8). This highly charged region could potentially play a role in facilitating electrostatic interactions with functional partners. It may also influence the stability of the molecule, since C-terminal segments of Hsp70 have previously been reported to confer stability to the protein (Misra and Ramachandran 2009; Mabate et al. 2018).
TEDWYLEE and Magic motifs
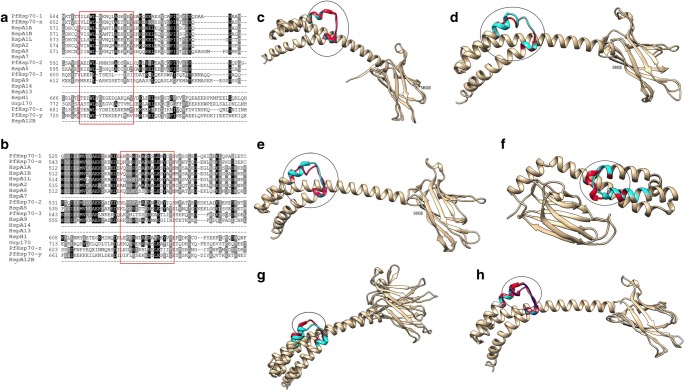
Human and P. falciparum Hsp70s both possess Magic and Tedwlyee motifs which are thought to be involved in substrate or co-chaperone recognition (Easton et al. 2000; Fig. 9). The Magic motifs are more conserved as compared to the TEDWYLEE motifs across the various Hsp70 isoforms (Fig. 9). While cytosolic Hsp70s generally exhibit high structural conservation, the TEDWLYEE motifs of Hsp110s and ER Hsp70s assume distinct structural orientations (Fig. 9). Notably, in spite of PfHsp70-3 and its mitochondrial counterpart, human HspA9 sharing highly conserved SBDs, there is variation within the TEDWLYEE motifs of these proteins. In addition, PfHsp70-z possesses a helical fold within its TEDWLYEE motif that is lacking in its canonical, cytosolic counterpart, PfHsp70-1 (Fig. 9). Compared to human and parasite cytosol localized Hsp70s, PfHsp70-x also harbours a distinct TEDWYLEE motif which is predicted to form a loop around residue, 609L, and a helical fold around residues 613EK614 (Fig. 9). Variation within the TEDWLYEE and Magic motifs of Hsp70 could further account for substrate specificity and may regulate interaction with co-chaperones.
Fig. 9.
Tedwlyee and Magic motifs in P. falciparum and human Hsp70s. The Hsp70 Tedwlyee motifs of plasmodial and human Hsp70s exhibit high sequence variation (a). Magic motifs are highly divergent (b). Both motifs are absent in HspA7, HspA14, HspA13 and HspA12B. Superimposed SBDs of Tedwlyee motifs. Cytosolic Hsp70s; PfHsp70-1 (red), Hsc70 (cyan) and HspA1A (magenta) (c), Tedwlyee motifs of PfHsp70-z (red) and human Hsp110 (blue) (d). PfHsp70-1 (cyan) and PfHsp70-z (magenta) (e), PfHsp70-3 (cyan) and HspA9 (red) f. PfHsp70-2 (red) and human BiP and h. PfHsp70-x (purple), PfHsp70-1 (red), Hsc70 (cyan) and HspA1A all show structural variation (g)
Conclusion
Although plasmodial Hsp70s generally share high sequence and predicted three-dimensional conservation with their human counterparts, there exists some unique features present in some plasmodial Hsp70s. For instance, Tedwlyee, Magic, GGMP and EKEK motifs appear to be amongst the distinctive features setting parasite Hsp70s apart from their human counterparts. Whether these distinctive features would impart sufficient structural variation to allow for selective inhibition of parasite Hsp70 remains to be fully explored. However, a study by Cockburn et al. (2014) identified small molecule inhibitors that seem to selectively target Hsp70 of parasite origin with minimum adverse effects on human Hsp70 function (Cockburn et al. 2014). It is therefore important to conduct experimental studies to validate the roles of the unique motifs described here towards validating their roles and to further explore how they regulate Hsp70 function in malaria parasite versus human systems.
Compliance with ethical standards
Funding
This project was supported through a grant (L1/402/14–1) provided to A.S. by the Deutsche Forchungsgemeinshaft (DFG) under the theme, “German–African Cooperation Projects in Infectiology.” We are grateful to the Department of Science and Technology/National Research Foundation (NRF) of South Africa for providing an equipment grant (UID, 75464) and NRF mobility grant (UID, 92598) awarded to A.S.; T.Z. is a recipient of the NRF Innovation Post-Doctoral fellowship UID, 111989 and African–German Network of Excellence in Science junior researcher grant.
Conflict of interest
Graham Chakafana declares that he has no conflict of interest. Tawanda Zininga declares that he has no conflict of interest. Addmore Shonhai declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biophysics and Structural Biology at Synchrotrons’ edited by Trevor Sewell.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arakawa A, Handa N, Shirouzu M, Yokoyama S. Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci. 2011;20:1367–1379. doi: 10.1002/pro.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Feige MJ, Hendershot LM. BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol. 2015;427(7):1589–1608. doi: 10.1016/j.jmb.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha M, Chiang AN, Needham PG, Stephens LL, Hoppe HC, Külzer S, Przyborski JM, Lingelbach K, Wipf P, Brodsky JL, Shonhai A, Blatch GL. Plasmodium falciparum encodes a single cytosolic type I Hsp40 that functionally interacts with Hsp70 and is upregulated by heat shock. Cell Stress Chaperones. 2011;16:389–401. doi: 10.1007/s12192-010-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri L, de Conway ME, Macario AJ. hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol. 2008;8:19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnaud S, Dixon M, Nie C, Chappell L, Sanders P, Nebl T, Hanssen E, Berriman M, Chan J, Blanch A, Beeson J, Rayner J, Przyborski J, Tilley L, Crabb B, Gilson P. The exported chaperone Hsp70-x supports virulence functions for Plasmodium falciparum blood stage parasites. PLoS One. 2017;12:e0181656. doi: 10.1371/journal.pone.0181656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Murillo-Solano C, Kirkpatrick M, Antoshchenko T, Park T, Pizarro J. Repurposing drugs to target the malaria parasite unfolding protein response. Sci Rep. 2018;8:10333–10350. doi: 10.1038/s41598-018-28608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AN, Valderramos JC, Balachandran R, Chovatiya RJ, Mead BP, Schneider C. Select pyrimidinones inhibit the propagation of the malarial parasite. Plasmodium falciparum. Bioorg Med Chem. 2009;17:1527–1533. doi: 10.1016/j.bmc.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb D, Florentin A, Fierro M, Krakowiak M, Moore J, Muralidharan V. The exported chaperone PfHsp70x is dispensable for the Plasmodium falciparum intraerythrocytic life cycle. mSphere. 2017;2:363–380. doi: 10.1128/mSphere.00363-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IL, Boshoff A, Pesce ER, Blatch GL. Selective modulation of plasmodial Hsp70s by small molecules with antimalarial activity. J Biol Chem. 2014;395:1353–1362. doi: 10.1515/hsz-2014-0138. [DOI] [PubMed] [Google Scholar]
- Dhamad AE, Zhou Z, Zhou J, Du Y. Systematic proteomic identification of the heat shock proteins (Hsp) that interact with estrogen receptor alpha (ERα) and biochemical characterization of the ERα-Hsp70 interaction. PLoS One. 2016;11(8):e0160312. doi: 10.1371/journal.pone.0160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworniczak B, Mirault ME. Structure and expression of a human gene coding for a 71kD heat shock ‘cognate’ protein. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR. The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CA, Sherman W, Meng W, et al. The Hsp70 interdomain linker is a dynamic switch that enables allosteric communication between two structured domains. J Biol Chem. 2017;292(36):14765–14774. doi: 10.1074/jbc.M117.789313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Hu W, Xu L, Wu H, Wu S, et al. The C-terminal GGAP motif of Hsp70 mediates substrate recognition and stress response in yeast. J Biol Chem. 2018;293:17663–17675. doi: 10.1074/jbc.RA118.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- Han Z, Truong QA, Park S, Breslow JL. Two Hsp70 family members expressed in atherosclerotic lesions. Proc Natl Acad Sci U S A. 2003;100:1256–1261. doi: 10.1073/pnas.252764399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held T, Barakat AZ, Mohamed BA, Paprotta I, Meinhardt A, Engel W, Adham I. Heat-shock protein HSPA4 is required for progression of spermatogenesis. Reproduction. 2011;142:133–144. doi: 10.1530/REP-11-0023. [DOI] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. A host targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalouei Sam, Chow Ari M., Brown Ian R. Stress-induced localization of HSPA6 (HSP70B′) and HSPA1A (HSP70-1) proteins to centrioles in human neuronal cells. Cell Stress and Chaperones. 2013;19(3):321–327. doi: 10.1007/s12192-013-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Külzer S, Charnaud S, Dagan T, Riedel J, Mandal P, et al. Plasmodium falciparum-encoded exported Hsp70/Hsp40 chaperone/co-chaperone complexes within the host erythrocyte. Cell Microbiol. 2012;14:1784–1795. doi: 10.1111/j.1462-5822.2012.01840.x. [DOI] [PubMed] [Google Scholar]
- Lebepe C (2019) Comparative analysis of a chimeric Hsp70 of E. coli and Plasmodium falciparum origin relative to its wild type forms. Msc Thesis, University of Venda
- Leung TK, Rajendran MY, Monfries C, Hall C, Lim L. The human heat-shock protein family. Expression of a novel heat-inducible HSP70 (HSP70B’) and isolation of its cDNA and genomic DNA. Biochem J. 1990;267:125–132. doi: 10.1042/bj2670125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons RE, Johnson AM. Gene sequence and transcription differences in 70 kDa heat shock protein correlate with murine virulence of Toxoplasma gondii. Int J Parasitol. 1998;28:1041–1051. doi: 10.1016/s0020-7519(98)00074-5. [DOI] [PubMed] [Google Scholar]
- Mabate B, Zininga T, Ramatsui L, Makumire S, Achilonu I, et al. Structural and biochemical characterization of Plasmodium falciparum Hsp70-x reveals functional versatility of its C-terminal EEVN motif. Proteins. 2018;86(11):1189–1201. doi: 10.1002/prot.25600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Gymnastics of molecular chaperones. Mol Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Misra G, Ramachandran R. Hsp70-1 from Plasmodium falciparum: protein stability, domain analysis and chaperone activity. Biophys Chem. 2009;142:55–64. doi: 10.1016/j.bpc.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Mizzen LA, Chang C, Garrels JI, Welch WJ. Identification, characterization, and purification of two mammalian stress proteins present in mitochondria, grp 75, a member of the hsp 70 family and hsp58, a homolog of the bacterial GroEL protein. J Biol Chem. 1989;264:20664–20675. [PubMed] [Google Scholar]
- Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Oksman A, Pal P, Lindquist S, Goldberg DE. Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun. 2012;3:1310. doi: 10.1038/ncomms2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njunge JM, Ludewig MH, Boshoff A, Pesce ER, Blatch GL. Hsp70s and J proteins of Plasmodium parasites infecting rodents and primates: structure, function, clinical relevance, and drug targets. Curr Pharm Des. 2013;19:387–403. doi: 10.2174/138161213804143734. [DOI] [PubMed] [Google Scholar]
- Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem. 1999;274:15712–15718. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]
- Otterson G.A., Flynn G.C., Kratzke R.A., Coxon A., Johnston P.G., Kaye F.J. Stch encodes the ‘ATPase core’ of a microsomal stress 70 protein. The EMBO Journal. 1994;13(5):1216–1225. doi: 10.1002/j.1460-2075.1994.tb06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallarès I, de Groot NS, Iglesias V, Sant'Anna R, Biosca A, Fernàndez-Busquets X, Ventura S (2018) Discovering putative prion-like proteins in Plasmodium falciparum: a computational and experimental analysis. Front Microbiol 9:1737 [DOI] [PMC free article] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pigłowski W, Nowak R, Krawczyk Z, Ścieglińska D. The structural and functional analysis of the human HSPA2 gene promoter region. Acta Biochim Pol. 2007;54:99–106. [PubMed] [Google Scholar]
- Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Polier S, Hartl FU, Bracher A. Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J Mol Biol. 2010;401:696–707. doi: 10.1016/j.jmb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Pryzborski J, Diehl M, Blatch G. Plasmodial Hsp70s are functionally adapted to the malaria parasite life cycle. Front Mol Biosci. 2015;2:34. doi: 10.3389/fmolb.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radons J. The human Hsp70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgrove KA, Nixon B, Baker M, Hetherington L, Baker G, Liu D, Aitken JR. The molecular chaperone HSPA2 plays a key role in regulating the expression of sperm surface receptors that mediate sperm-egg recognition. PLoS One. 2012;7(11):e50851. doi: 10.1371/journal.pone.0050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter K, Matunis M. Detection of SUMOylation in Plasmodium falciparum. Methods Mol Biol. 2016;1475:283–290. doi: 10.1007/978-1-4939-6358-4_19. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Sekhar A, Nagesh J, Kay LE. Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. eLife. 2017;6:e28030. doi: 10.7554/eLife.28030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonhai A. Plasmodial heat shock proteins: targets for chemotherapy. FEMS Immunol Med Microbiol. 2010;58:61–74. doi: 10.1111/j.1574-695X.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- Shonhai A. Role of Hsp70s in development and pathogenicity of plasmodium species. In: Shonhai A, Blatch G, editors. Heat shock proteins of malaria. New York: Springer; 2014. pp. 47–70. [Google Scholar]
- Shonhai A, Boschoff A, Blatch G. The structural and functional diversity of Hsp70s from Plasmodium falciparum. Protein Sci. 2007;16:1803–1818. doi: 10.1110/ps.072918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonhai Addmore, Botha Melissa, de Beer Tjaart, Boshoff Aileen, Blatch Gregory. Structure-Function Study of a Plasmodium falciparum Hsp70 Using Three Dimensional Modelling and in Vitro Analyses. Protein & Peptide Letters. 2008;15(10):1117–1125. doi: 10.2174/092986608786071067. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Furukawa T, Yano T, Sato N, Takizawa J, Kurasaki T, Abe T, Narita M, Masuko M, Koyama S, Toba K, Takahashi M, Aizawa Y. Identification of an overexpressed gene, HSPA4L, the product of which can provoke prevalent humoral immune responses in leukemia patients. Exp Hematol. 2007;35(7):1091–1099. doi: 10.1016/j.exphem.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Wu CY, Lin CT, Wu MZ, Wu KJ. Induction of HSPA4 and HSPA14 by NBS1 overexpression contributes to NBS1-induced in vitro metastatic and transformation activity. J Biomed Sci. 2011;18:1. doi: 10.1186/1423-0127-18-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Sarbeng EB, Vorvis C, Kumar DP, Zhou L, et al. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J Biol Chem. 2012;287:5661–5672. doi: 10.1074/jbc.M111.275057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Anmin, Li Ping, Tang Ting, Wang Jiancai, Chen Yuan, Liu Li. Roles of Hsp70s in Stress Responses of Microorganisms, Plants, and Animals. BioMed Research International. 2015;2015:1–8. doi: 10.1155/2015/510319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappasodi R, Ruggiero G, Guarnotta C, Tortoreto M, Tringali C, Cavanè A, Cabras A, Castagnoli L, Venerando B, Zaffaroni N, Gianni A, De Braud F, Tripodo C, Pupa S, Di Nicola M. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood. 2015;125:1768–1771. doi: 10.1182/blood-2014-07-590034. [DOI] [PubMed] [Google Scholar]
- Zhu D, Dix DJ, Eddy EM. HSP70-2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes. Development. 1997;124:3007–3014. doi: 10.1242/dev.124.15.3007. [DOI] [PubMed] [Google Scholar]
- Zininga Tawanda, Makumire Stanely, Gitau Grace Wairimu, Njunge James M., Pooe Ofentse Jacob, Klimek Hanna, Scheurr Robina, Raifer Hartmann, Prinsloo Earl, Przyborski Jude M., Hoppe Heinrich, Shonhai Addmore. Plasmodium falciparum Hop (PfHop) Interacts with the Hsp70 Chaperone in a Nucleotide-Dependent Fashion and Exhibits Ligand Selectivity. PLOS ONE. 2015;10(8):e0135326. doi: 10.1371/journal.pone.0135326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zininga T, Achilonu I, Hoppe H, Prinsloo E, Dirr HW, et al. Plasmodium falciparum Hsp70-z, an Hsp110 homologue, exhibits independent chaperone activity and interacts with Hsp70-1 in a nucleotide-dependent fashion. Cell Stress Chaperones. 2016;21:499–513. doi: 10.1007/s12192-016-0678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]