Abstract
Mesenchymal stem cell (MSC) based therapies are currently being evaluated as a putative therapeutic in numerous human clinical trials. Recent reports have established that exosomes mediate much of the therapeutic properties of MSCs. Exosomes are nanovesicles which mediate intercellular communication, transmitting signals between cells which regulate a diverse range of biological processes. MSC-derived exosomes are packaged with numerous types of proteins and RNAs, however, their metabolomic and lipidomic profiles to date have not been well characterized. We previously reported that MSCs, in response to priming culture conditions that mimic the in vivo microenvironmental niche, substantially modulate cellular signaling and significantly increase the secretion of exosomes. Here we report that MSCs exposed to such priming conditions undergo glycolytic reprogramming, which homogenizes MSCs’ metabolomic profile. In addition, we establish that exosomes derive from primed MSCs are packaged with numerous metabolites that have been directly associated with immunomodulation, including M2 macrophage polarization and regulatory T lymphocyte induction.
Keywords: metabolomics, lipidomics, exosomes, mesenchymal stem cells
Graphical Abstract
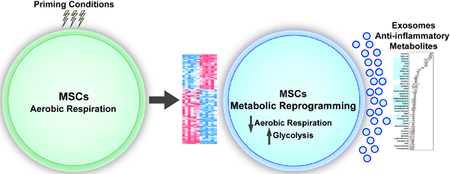
Introduction
In recent years, the therapeutic potential and safety of mesenchymal stem/stromal cells (MSCs) has been investigated preclinically and clinically, in the context of immune regulation and regeneration in a variety of diseases1–3. The therapeutic effects of MSCs on tissue repair and regeneration are mediated by their paracrine activity via various secreted factors, including exosomes4–11. Exosomes are nano-sized vesicles that comprise a robust inter-cellular communication system12,13. There are now over 200 published reports indicating that exosomes are a critical active pharmaceutical ingredient that is responsible for mediating much of the therapeutic effects of MSCs, including their immunomodulatory properties14–17. The application of exosomes isolated and purified from MSC conditioned media holds several potential advantages of the use of cell-based therapies, including increased potency, greater consistency and lower cost. Previously we have reported that exosomes derived from primed MSC possess potent tissue healing properties, and producing a much higher yield of exosomal material, which has important implications for the translation of this technology5. Exosomes transport a variety of factors including proteins and RNAs18. To date, most studies have focused on the RNA, and to a lesser extent, the protein packaged into exosomes derived from MSCs19–21. Consequently, the metabolomic profile of MSC-derived exosomes, remains largely uncharacterized.
Metabolomics uses mass spectrometry to identify and quantify a variety of small molecules that are indicative of the metabolic, and physiological status of cellular subpopulations22. This analytical approach affords the analysis of a large number of samples and characterizes the molecular response of a biological system to any perturbation in its microenvironment23. Metabolomics enables the detection of differential abundance of metabolites between two conditions and adds value to translational studies focusing on elucidating the underlying the dynamic molecular processes within a given system24. We have previously established that pMEX are packaged with biologically active, regenerative proteins4. However, there are few metabolic or lipidomic studies of exosomes reported to date.
Here we report that exosomes derived from primed MSCs are packaged with a variety of metabolites and lipid membrane components. We compare the metabolomic and lipidomic profiles of classically cultured MSC vs primed MSCs demonstrating their metabolic reprogramming under such microenvironmental conditions. We establish that exosomes derived from primed MSCs (pMEX) are packaged with distinct profile of metabolites, the majority of which have been associated with immunomodulation. These findings suggest that further studies establishing the metabolite payload of MSC-derived exosomes may yield insights into their underlying physiological and cell signaling properties.
Materials and Methods
Sample preparation for GC-TOF MS and LC-QTOF MS/MS analysis
Flash frozen cell pellets were ground using a GenoGrinder 2010 (SPEX SamplePrep) for 2 min at 1350 rpm. Cell pellets were then extracted with 225 μl of methanol at −20°C containing an internal standard mixture described previously25, and 750 μL of MTBE (methyl tertiary butyl ether) (Sigma Aldrich) at −20°C containing cholesteryl ester 22:1. Samples were shaken for 6 min at 4°C with an Orbital Mixing Chilling/Heating Plate (Torrey Pines Scientific Instruments, Calsbad, CA). Then 188 μl of LC-MS grade water (Fisher) was added. Samples were vortexed, centrifuged and the upper (non-polar) and bottom (polar) layers were collected (350 μL and 125 μL, respectively) and evaporated to dryness.
The non-polar layer was re-suspended in a methanol:toluene (9:1, v/v) mixture contaning 50 ng/ml CUDA ((12- [[(cyclohexylamino)carbonyl]amino]-dodecanoic acid) (Cayman Chemical, Ann Arbor, MI) vortexed, sonicated for 5 min and centrifuged and prepared for lipidomic analysis. Half the polar layer was resuspended in an acetonitrile:water (4:1, v/v) mixture with 5 μg/ml Val-Try-Val (Sigma) sonicated for 5 min and centrifuged and prepared for HILIC-QTOF MS/MS analysis. The second polar phase aliquots were derivatized with 10 μL of methoxyamine hydro-chloride in pyridine (40 mg/mL) by shaking at 30°C for 90 min followed by trimethylsilylation with 90 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA, Sigma-Aldrich) by shaking at 37°C for 30 min containing C8–C30 fatty acid methyl esters (FAMEs) as internal standards. Derivatized samples were subsequently submitted for analysis by GC-TOF MS (0.5 μl injection). Method blanks and pooled human plasma (BioIVT) were included as quality control samples for all platforms and prepared in same manner.
Chromatographic and mass spectrometric conditions for lipidomic LC-QTOF MS analysis.
For analysis of the non-polar phase, re-suspended samples were injected at 1 μL ESI (+) and 5 μL for ESI(−), onto a Waters Acquity UPLC CSH C18 (100 mm length × 2.1 mm id; 1.7 μm particle size) with a Waters Acquity VanGuard CSH C18 pre-column (5 mm × 2.1 mm id; 1.7 μm particle size) at 65°C coupled to an Agilent 1290 Infinity UHPLC (Agilent Technologies, Santa Clara, CA). For positive mode 10 mM ammonium formate and 0.1% formic acid were used and 10 mM ammonium acetate (Sigma–Aldrich) was used for negative mode. Both positive and negative modes used the same mobile phase composition of (A) 60:40 v/v acetonitrile:water (LC-MS grade) and (B) 90:10 v/v isopropanol:acetonitrile. The gradient started at 0 min with 15% (B), 0–2 min 30% (B), 2–2.5 min 48% (B), 2.5–11 min 82% (B), 11–11.5 min 99% (B), 11.5–12 min 99% (B), 12–12.1 min 15% (B), and 12.1–15 min 15% (B). A flow rate of 0.6 mL/min was used. For data acquisition an Agilent 6550 QTOF with a jet stream electrospray source with the following parameters was used: mass range, m/z 50–1700; capillary voltage, ±3 kV; nozzle voltage, ±1 kV; gas temperature, 200°C; drying gas (nitrogen), 14 L/min; nebulizer gas (nitrogen), 35 psi; sheath gas temperature, 350°C; sheath gas flow (nitrogen), 11 L/min; acquisition rate, 2 spectra/s. For lipid identification, MS/MS spectra were collected at a collision energy of 20 eV with an acquisition rate MS1 of 10 spectra/s (100 ms) and an acquisition rate for MS/MS of 13 spectra/s (77 ms) with 4 precursor ions per cycle.. Mass accuracy was maintained by constant reference ion infusion (purine and HP-0921 in an acetonitrile:water mixture).
Chromatographic and mass spectrometric conditions for polar metabolite HILIC-QTOF MS analysis.
Five microliters of re-suspended sample was injected onto a Waters Acquity UPLC BEH Amide column (150 mm length × 2.1 mm id; 1.7 μm particle size) with an additional Waters Acquity VanGuard BEH Amide pre-column (5 mm × 2.1 mm id; 1.7 μm particle size) maintained at 45°C coupled to an Agilent 1290 Infinity UHPLC. The mobile phases were prepared with 10 mM ammonium formate and 0.125% formic acid (Sigma–Aldrich) in either 100% LC-MS grade water for mobile phase (A) or 95:5 v/v acetonitrile:water for mobile phase (B). Gradient elution was performed from 100% (B) at 0–2 min to 70% (B) at 7.7 min, 40% (B) at 9.5 min, 30% (B) at 10.25 min, 100% (B) at 12.75 min, isocratic until 16.75 min with a column flow of 0.4 mL/min. Spectra were collected using a 5600+ TripleTOF MS (SCIEX, Framingham, MA, USA) using data dependent mode for MS/MS spectra acquisition. Data was collected in ESI(+) mode with the following parameters: m/z 50–1700, curtain gas: 35, ion source gas 1 and 2: 60, temperature: 350°C, ion spray voltage floating: +4.5 kV, declustering potential: 80 V, MS1 accumulation time 100 ms, MS2 accumulation time 50 ms, dependent product ion scan number 8, intensity threshold 1000, active precursor exclusion after 2 spectra for 5 s, collision energy 20 eV with 15 eV collision energy spread. Mass accuracy was maintained through calibration every 10 injections using APCI positive calibration solution delivered using a calibration delivery system.
LC-MS/MS data processing using MS-DIAL and statistics
Both lipidomic and HILIC data processing was performed using MS-DIAL26,13 for deconvolution, peak picking, alignment, and identification. For both data sets, in house m/z and retention time libraries were used in addition to MS/MS spectra databases in msp format as previously described27. Features were reported when present in at least 50% of samples in each group. Statistical analysis was done by first normalizing data using the sum of the knowns, or mTIC normalization, to scale each sample. Peak heights were then submitted using R to DeviumWeb andnormalized further by log transformation and Pareto scaling. ANOVA analysis was performed with FDR correction and post hoc testing.
Chromatographic and mass spectrometric conditions for GC-TOF MS analysis
Primary metabolite data was collected using a Leco Pegasus IV time-of-flight (TOF) MS (Leco Corporation, St. Joseph, MI) coupled to an Agilent 6890 GC (Agilent Technologies, Santa Clara) equipped with a 30 m long 0.25 mm id Rtx-5Sil MS column (0.25 μm film thickness) and a Gerstel MPS2 automatic liner exchange system (Gerstel GMBH & Co. KG, Ruhr, Germany). The chromatographic gradient used a constant flow of 1 ml/min with following gradient: 50°C (1 min), 20°C/min to 330°C, hold 5 min. Mass spectrometry data was collected using 1525 V detector voltage at m/z 85–500 with 17 spectra/s, electron ionization at −70 eV and an ion source temperature of 250°C. QC injections, blanks and pooled human plasma were used for quality assurance throughout the run. Data was processed by ChromaTOF for deconvolution, peak picking, and BinBase28,3 for metabolite identifications.
Results
Primed MSCs Undergo Metabolic Lipidomic Reprogramming
MSCs are typically cultured and expanded in high serum containing media and with atmospheric oxygen tension (~21%)29. Upon administration, MSCs experience a remarkable shift in the microenvironmental niche, including much lower exposure to growth factors, as well as substantially lower oxygen tension (eg 1–5% O2)30. Most published reports to date have focused on elucidating MSC physiology while under classic expansion culture conditions31. Upon serum deprivation and hypoxia, MSC undergo metabolic reprogramming32. We sought to determine how the metabolic reprogramming of MSCs exposed to serum deprivation and hypoxia influenced their metabolomic and lipidomic profiles. MSCs were isolated from human bone aspirates using canonical methods, as previously described5. The resulting cells were verified to express established MSC surface markers via flow cytometry, as previously reported4. At passage 5, expansion media was removed, and cells were washed 3 times with 1X PBS. MSCs either received expansion and were cultured at atmospheric oxygen tensions (~21%), or priming conditions consisting of serum deprivation and culturing under hypoxic conditions, 1% O2. Following 48 hours of incubation, cells were processed for metabolomic and lipidomic profiling using mass spectrometry.
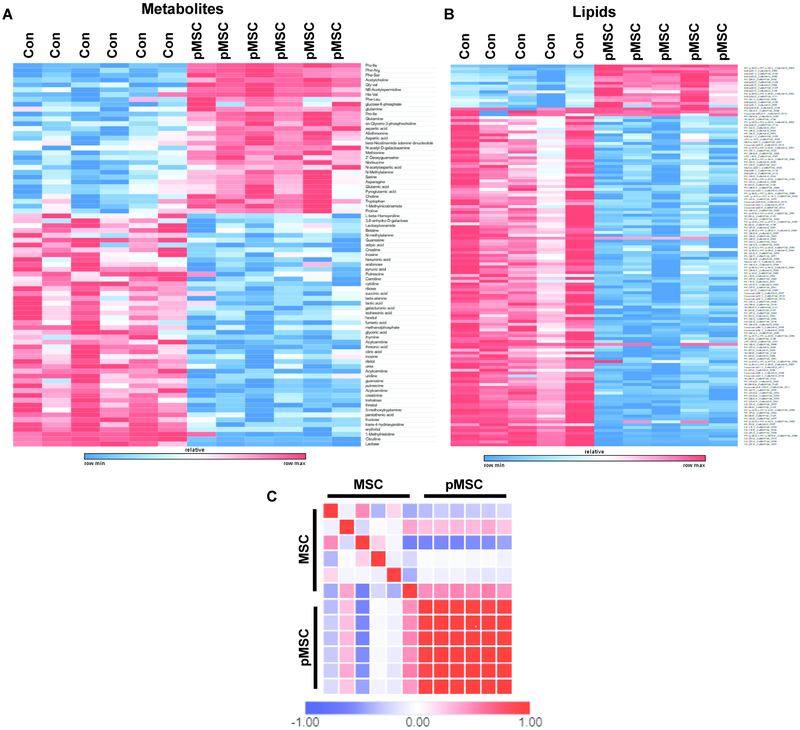
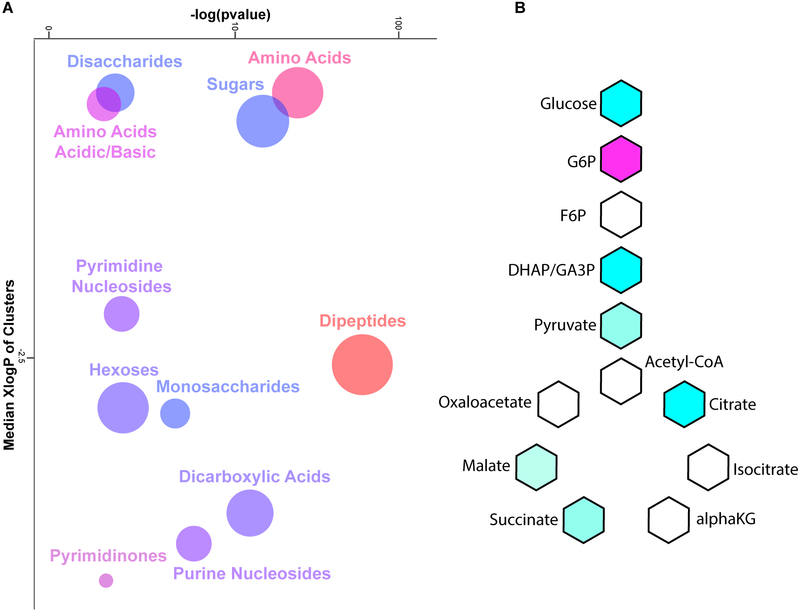
Primed MSCs (pMSC) exposed to hypoxia and serum deprivation resulted in the increased expression of dipeptides, suggesting that hypoxic MSCs increase their pool of free amino acids to fulfill the energetic demands that cannot be sufficiently provided from the glycolytic flux (Fig 1A). Ingenuity Pathway Analysis (IPA) of metabolomics data also determined that pMSC shifted toward a prosurvival metabolic profile (Fig S1). Lipidomics profiling determined that pMSC decreased expression of numerous lipid membrane components, consistent with previous reports of their differential morphology (Fig 1B). Biological replicates of pMSC were also metabolically more similar to each other and displayed significantly less batch to batch variation than MSCs cultured under expansion conditions (Fig 1C). Metabolic examination using the ChemRICH analysis, which creates chemical clusters based on structural similarity33, demonstrated that pMSC modulated expression of numerous nucleosides, biosynthetic and glycolytic associated metabolites, indicating their metabolic reprogramming (Fig 2A). Significant alterations in glycolysis and TCA cycle intermediates in pMSC as compared to MSC, suggest a conversion from aerobic respiration to glycolytic metabolism (Fig 2B). Further IPA analysis established that pMSC metabolites reflected a low carbohydrate environment (Fig S2). Collectively, these data demonstrate that MSCs undergo substantial metabolic reprogramming once they are transitioned from canonical expansion culture conditions, to conditions that mimic the microenviron-mental they experience upon administration.
Figure 1.
Metabolomic and lipidomic profiling of primed MSCs demonstrated significant metabolic reprogramming, increased dipeptide expression, and substantial modulation of lipid membrane components (A, B). Similarity analysis using Pearson’s correlation determined that priming conditions greatly increased the metabolic homogeneity of MSC biological replicates, as compared to classically cultured MSCs (C). N = 6 per condition, significance calculated with T-tests with a multi-testing correction analysis using a false discovery rate of 1%. Similarity matrix was calculated using the Pearson correlation function of the Broad Institute’s Morpheus tool.
Figure 2.
Mass spectrometry metabolic analysis determined that MSCs exposed to hypoxia and serum deprivation significantly modulated amino acid, nucleoside, and carbohydrate metabolism based on chemical ontology analysis using the ChemRICH tool developed by Barupal et al (A). Analysis of metabolites associated with carbohydrate metabolism determined that primed MSCs downregulate several pathways related to aerobic respiration. N = 6 per condition. For ChemRICH analysis enrichment p-values are given by the Kolmogorov–Smirnov-test. For Kreb’s cycle analysis significance calculated with T-tests with a multi-testing correction analysis using a false discovery rate of 1%.
Exosomes Derived from Primed MSCs are Packaged with Metabolites Associated with immunomodulation
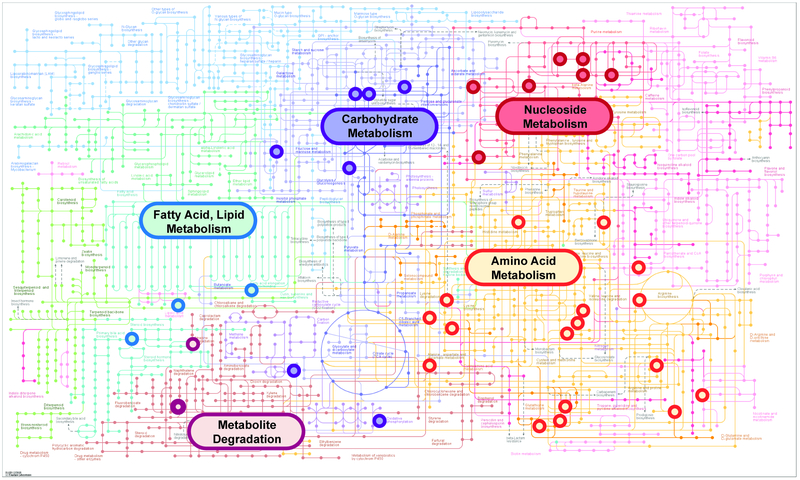
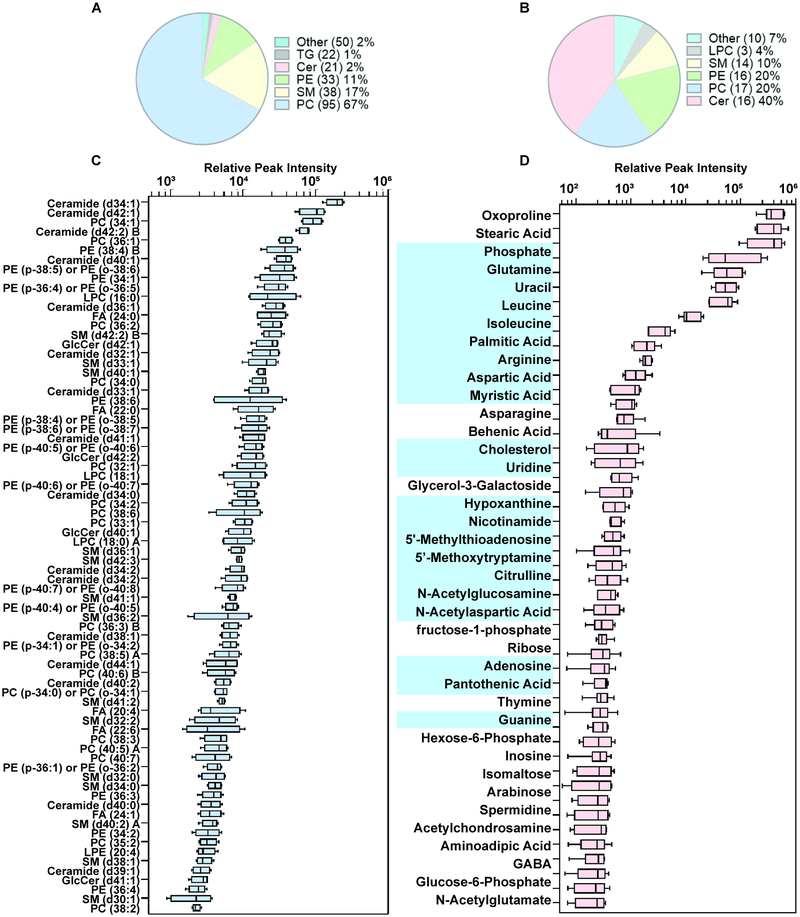
Over 200 studies have now been published establishing the therapeutic properties of MEX in numerous animal models of disease34–36. Several reports have also established the immunomodulatory properties of MEX37–39. Here we demonstrate that exosomes derived from pMSC (pMEX) are packaged with numerous primary metabolites, clustered in metabolic networks associated with amino acids, carbohydrates and nucleosides biosynthetic pathways. (Fig 3). Comparison of the metabolomic profile with our previous proteomics data determined that pMEX are packaged with numerous proteins that correspond to the enzymatic and signaling pathways associated with several of the metabolites detected in pMEX (Fig S3). Using lipidomic analysis we established that pMEX ceramide, lysophosphatidylcholines, and phosphatidyleth anolamine phospholipids, are present at substantially higher ratios as compared to their parental cells of origin (pMSC) (Fig 4A,B). Many of these factors are associated with lipid rafts and ceramide platforms that have been established to be central communication hubs of receptor mediated inter-cellular signaling. In addition, pMEX are packaged with 21 distinct metabolites that have been directly associated with immunoregulation, including: adenosine, arginine, aspartic acid, cholesterol, glutamine, nicotinamide, UDP-N-acetylglucosamine, 5’-deoxy-5’-methylthioadenosine, palmitic acid and isoleucine. Collectively, these metabolites have been directly associated with the modulation of anti-inflammatory responses, M2 macrophage polarization and induction of regulatory T lymphocytes (Fig 4C)40–53. These data establish that pMEX are packaged with numerous metabolites that have been associated with anti-inflammatory and immunoregulatory functions.
Figure 3.
Exosomes derived from MSCs exposed to serum deprivation and hypoxia are packaged with clustered networks of metabolites associated with amino acid, nucleoside, and carbohydrate metabolic pathways. Pathways associations determined using KEGG Metabolic Mapper function using high confidence metabolites with peak intensities significantly above background, n = 6.
Figure 4.
Mass spectrometry based lipidomic analysis established exosomes derived from primed MSCs are packaged with higher fractions of lipid membrane components, ceramide, lysophosphatidylcholines, and phosphatidylethanolamine phospholipids, as compared to pMSCs (A-C). pMEX are packaged with numerous metabolites, including 21 metabolites established to possess immunoregulatory properties (D), using metabolites identified with high confidence above background, n = 6.
Discussion
MSCs and their derived exosomes possess therapeutic potential, but little is currently known about their associated metabolic and lipid membrane profiles54. Historically, metabolites and cellular lipids have been viewed as relatively inert by-products of various cellular activities55–57. However, a growing body of evidence indicates that many metabolites possess functional properties capable of influencing cellular physiology, and immunological properties58–61 which we define as epimetabolites61. Here, we report that priming conditions greatly decreases the variability of MSC batches, on a metabolic level. We also demonstrate for the first time that pMEX are package with numerous metabolites that may underly some of their immunomodulatory properties.
Supplementary Material
Mesenchymal stem cell-based therapies under clinical development.
Exosomes derived from MSCs hold many advantages over cell based therapies.
MSC were primed with in vivo-like priming conditions.
Primed MSCs were metabolically reprogrammed.
Exosomes derived from primed MSCs were packagaed with anti-inflammatory metabolites.
Acknowledgements
This research was supported by funding provided by UC Office of the President’s Multi-campus Research Program Grant (MRP-17–454909), STAIR Grant, STAIR-Plus Grant CTSC Rapid Translational Grant (UL1-TR001860), U2C ES030158 (to OF), T32 Cardio (NIH T32-HL086350), Denny & Jeanene Dickenson Fund, NIH Transformative R01GM099688, NSF GROW 201111600, NIH T32-GM008799, and NSF GRFP 2011116000.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Le Blanc K et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet 371, 1579–1586, doi: 10.1016/S0140-6736(08)60690-X (2008). [DOI] [PubMed] [Google Scholar]
- 2.Le Blanc K & Ringden O Immunomodulation by mesenchymal stem cells and clinical experience. J Intern Med 262, 509–525, doi: 10.1111/j.1365-2796.2007.01844.x (2007). [DOI] [PubMed] [Google Scholar]
- 3.Rosova I, Dao M, Capoccia B, Link D & Nolta JA Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells 26, 2173–2182, doi: 10.1634/stemcells.2007-1104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan O et al. Exosomes Derived from Human Primed Mesenchymal Stem Cells Induce Mitosis and Potentiate Growth Factor Secretion. Stem Cells Dev, doi: 10.1089/scd.2018.0200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson JD et al. Comprehensive Proteomic Analysis of Mesenchymal Stem Cell Exosomes Reveals Modulation of Angiogenesis via Nuclear Factor-KappaB Signaling. Stem Cells 34, 601–613, doi: 10.1002/stem.2298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollock K et al. Human Mesenchymal Stem Cells Genetically Engineered to Overexpress Brain-derived Neurotrophic Factor Improve Outcomes in Huntington’s Disease Mouse Models. Mol Ther 24, 965–977, doi: 10.1038/mt.2016.12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SS et al. Advances in bone marrow stem cell therapy for retinal dysfunction. Prog Retin Eye Res 56, 148–165, doi: 10.1016/j.preteyeres.2016.10.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moisseiev E et al. Protective Effect of Intravitreal Administration of Exosomes Derived from Mesenchymal Stem Cells on Retinal Ischemia. Curr Eye Res 42, 1358–1367, doi: 10.1080/02713683.2017.1319491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fink KD et al. Allele-Specific Reduction of the Mutant Huntingtin Allele Using Transcription Activator-Like Effectors in Human Huntington’s Disease Fibroblasts. Cell Transplant 25, 677–686, doi: 10.3727/096368916X690863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng P et al. Engineered BDNF producing cells as a potential treatment for neurologic disease. Expert Opin Biol Ther 16, 1025–1033, doi: 10.1080/14712598.2016.1183641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velichko S et al. A Novel Nuclear Function for the Interleukin-17 Signaling Adaptor Protein Act1. PLoS One 11, e0163323, doi: 10.1371/journal.pone.0163323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bian S et al. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J Mol Med (Berl) 92, 387–397, doi: 10.1007/s00109-013-1110-5 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Lai RC, Yeo RW, Tan KH & Lim SK Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv 31, 543–551, doi: 10.1016/j.biotechadv.2012.08.008 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Toh WS, Lai RC, Zhang B & Lim SK MSC exosome works through a protein-based mechanism of action. Biochem Soc Trans 46, 843–853, doi: 10.1042/BST20180079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiklander OP et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles 4, 26316, doi: 10.3402/jev.v4.26316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo R et al. BM-MSC-derived exosomes alleviate radiation-induced bone loss by restoring the function of recipient BM-MSCs and activating Wnt/beta-catenin signaling. Stem Cell Res Ther 10, 30, doi: 10.1186/s13287-018-1121-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han YD et al. Co-transplantation of exosomes derived from hypoxia-preconditioned adipose mesenchymal stem cells promotes neovascularization and graft survival in fat grafting. Biochem Biophys Res Commun 497, 305–312, doi: 10.1016/j.bbrc.2018.02.076 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Hurley JH & Odorizzi G Get on the exosome bus with ALIX. Nat Cell Biol 14, 654–655, doi: 10.1038/ncb2530 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Chen TS et al. Mesenchymal stem cell secretes microparticles enriched in premicroRNAs. Nucleic Acids Res 38, 215–224, doi: 10.1093/nar/gkp857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kane NM, Thrasher AJ, Angelini GD & Emanueli C Concise review: MicroRNAs as modulators of stem cells and angiogenesis. Stem Cells 32, 1059–1066, doi: 10.1002/stem.1629 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Momen-Heravi F, Bala S, Bukong T & Szabo G Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine 10, 1517–1527, doi: 10.1016/j.nano.2014.03.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinschen MM, Ivanisevic J, Giera M & Siuzdak G Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol, doi: 10.1038/s41580-019-0108-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marksteiner J, Oberacher H & Humpel C Acyl-Alkyl-Phosphatidlycholines are Decreased in Saliva of Patients with Alzheimer’s Disease as Identified by Targeted Metabolomics. J Alzheimers Dis, doi: 10.3233/JAD-181278 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Manier SK, Keller A, Schaper J & Meyer MR Untargeted metabolomics by high resolution mass spectrometry coupled to normal and reversed phase liquid chromatography as a tool to study the in vitro biotransformation of new psychoactive substances. Sci Rep 9, 2741, doi: 10.1038/s41598-019-39235-w (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Showalter MR et al. Obesogenic diets alter metabolism in mice. PLoS One 13, e0190632, doi: 10.1371/journal.pone.0190632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsugawa H et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 12, 523–526, doi: 10.1038/nmeth.3393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cajka T & Fiehn O LC-MS-Based Lipidomics and Automated Identification of Lipids Using the LipidBlast In-Silico MS/MS Library. Methods Mol Biol 1609, 149–170, doi: 10.1007/978-1-4939-6996-8_14 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Skogerson K, Wohlgemuth G, Barupal DK & Fiehn O The volatile compound BinBase mass spectral database. BMC Bioinformatics 12, 321, doi: 10.1186/1471-2105-12-321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohli N et al. CD271-selected mesenchymal stem cells from adipose tissue enhance cartilage repair and are less angiogenic than plastic adherent mesenchymal stem cells. Sci Rep 9, 3194, doi: 10.1038/s41598-019-39715-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galipeau J et al. International Society for Cellular Therapy perspective on immune functional assays for mesenchymal stromal cells as potency release criterion for advanced phase clinical trials. Cytotherapy 18, 151–159, doi: 10.1016/j.jcyt.2015.11.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gugjoo MB, Amarpal A & Sharma GT Mesenchymal stem cell basic research and applications in dog medicine. J Cell Physiol, doi: 10.1002/jcp.28348 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Luo Z et al. Hypoxia preconditioning promotes bone marrow mesenchymal stem cells survival by inducing HIF-1alpha in injured neuronal cells derived exosomes culture system. Cell Death Dis 10, 134, doi: 10.1038/s41419-019-1410-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barupal DK & Fiehn O Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci Rep 7, 14567, doi: 10.1038/s41598-017-15231-w (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalnapurkar S, Moirangthem RD, Singh S, Limaye L & Kale V Microvesicles Secreted by Nitric Oxide-Primed Mesenchymal Stromal Cells Boost the Engraftment Potential of Hematopoietic Stem Cells. Stem Cells 37, 128–138, doi: 10.1002/stem.2912 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Fujii S et al. Graft-Versus-Host Disease Amelioration by Human Bone Marrow Mesenchymal Stromal/Stem Cell-Derived Extracellular Vesicles Is Associated with Peripheral Preservation of Naive T Cell Populations. Stem Cells 36, 434–445, doi: 10.1002/stem.2759 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Kou X et al. The Fas/Fap-1/Cav-1 complex regulates IL-1RA secretion in mesenchymal stem cells to accelerate wound healing. Sci Transl Med 10, doi: 10.1126/scitranslmed.aai8524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai P et al. A potent immunomodulatory role of exosomes derived from mesenchymal stromal cells in preventing cGVHD. J Hematol Oncol 11, 135, doi: 10.1186/s13045-018-0680-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khatri M, Richardson LA & Meulia T Mesenchymal stem cell-derived extracellular vesicles attenuate influenza virus-induced acute lung injury in a pig model. Stem Cell Res Ther 9, 17, doi: 10.1186/s13287-018-0774-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams AM et al. Mesenchymal Stem Cell-Derived Exosomes Provide Neuroprotection and Improve Long-Term Neurologic Outcomes in a Swine Model of Traumatic Brain Injury and Hemorrhagic Shock. J Neurotrauma 36, 54–60, doi: 10.1089/neu.2018.5711 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Villa-Bellosta R, Hamczyk MR & Andres V Novel phosphate-activated macrophages prevent ectopic calcification by increasing extracellular ATP and pyrophosphate. PLoS One 12, e0174998, doi: 10.1371/journal.pone.0174998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MH & Kim H The Roles of Glutamine in the Intestine and Its Implication in Intestinal Diseases. Int J Mol Sci 18, doi: 10.3390/ijms18051051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmieri EM et al. Pharmacologic or Genetic Targeting of Glutamine Synthetase Skews Macrophages toward an M1-like Phenotype and Inhibits Tumor Metastasis. Cell Rep 20, 1654–1666, doi: 10.1016/j.celrep.2017.07.054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galgani M, De Rosa V, La Cava A & Matarese G Role of Metabolism in the Immunobiology of Regulatory T Cells. J Immunol 197, 2567–2575, doi: 10.4049/jimmunol.1600242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hnia K et al. L-arginine decreases inflammation and modulates the nuclear factor-kappaB/matrix metalloproteinase cascade in mdx muscle fibers. Am J Pathol 172, 1509–1519, doi: 10.2353/ajpath.2008.071009 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeramian A et al. Arginine transport via cationic amino acid transporter 2 plays a critical regulatory role in classical or alternative activation of macrophages. J Immunol 176, 5918–5924 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Yu HR et al. l-Arginine-Dependent Epigenetic Regulation of Interleukin-10, but Not Transforming Growth Factor-beta, Production by Neonatal Regulatory T Lymphocytes. Front Immunol 8, 487, doi: 10.3389/fimmu.2017.00487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rueda CM et al. High density lipoproteins selectively promote the survival of human regulatory T cells. J Lipid Res 58, 1514–1523, doi: 10.1194/jlr.M072835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tall AR & Yvan-Charvet L Cholesterol, inflammation and innate immunity. Nat Rev Immunol 15, 104–116, doi: 10.1038/nri3793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henrich FC et al. Suppressive effects of tumor cell-derived 5’-deoxy-5’-methylthioadenosine on human T cells. Oncoimmunology 5, e1184802, doi: 10.1080/2162402X.2016.1184802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keyel PA et al. Methylthioadenosine reprograms macrophage activation through adenosine receptor stimulation. PLoS One 9, e104210, doi: 10.1371/journal.pone.0104210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasko G & Cronstein B Regulation of inflammation by adenosine. Front Immunol 4, 85, doi: 10.3389/fimmu.2013.00085 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasko G & Pacher P Regulation of macrophage function by adenosine. Arterioscler Thromb Vasc Biol 32, 865–869, doi: 10.1161/ATVBAHA.111.226852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohta A & Sitkovsky M Extracellular adenosine-mediated modulation of regulatory T cells. Front Immunol 5, 304, doi: 10.3389/fimmu.2014.00304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaubey S et al. Early gestational mesenchymal stem cell secretome attenuates experimental bronchopulmonary dysplasia in part via exosome-associated factor TSG-6. Stem Cell Res Ther 9, 173, doi: 10.1186/s13287-018-0903-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Federici M Gut microbiome and microbial metabolites: a new system affecting metabolic disorders. J Endocrinol Invest, doi: 10.1007/s40618-019-01022-9 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Shyh-Chang N & Daley GQ Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab 21, 349–350, doi: 10.1016/j.cmet.2015.02.011 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Berman DE et al. Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat Neurosci 11, 547–554, doi: 10.1038/nn.2100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Passos DF, Bernardes VM, da Silva JLG, Schetinger MRC & Leal DBR Adenosine signaling and adenosine deaminase regulation of immune responses: impact on the immunopathogenesis of HIV infection. Purinergic Signal 14, 309–320, doi: 10.1007/s11302-018-9619-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khayami R et al. Role of adenosine signaling in the pathogenesis of head and neck cancer. J Cell Biochem 119, 7905–7912, doi: 10.1002/jcb.27091 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Dos Santos GG, Hastreiter AA, Sartori T, Borelli P & Fock RA L-Glutamine in vitro Modulates some Immunomodulatory Properties of Bone Marrow Mesenchymal Stem Cells. Stem Cell Rev 13, 482–490, doi: 10.1007/s12015-017-9746-0 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Showalter MR, Cajka T & Fiehn O Epimetabolites: discovering metabolism beyond building and burning. Curr Opin Chem Biol 36, 70–76, doi: 10.1016/j.cbpa.2017.01.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.