Abstract
Invasive lobular carcinoma (ILC) is the second most common histological subtype of breast cancer following invasive ductal carcinoma (IDC). To identify potential genetic drivers of ILC progression, we used NanoString nCounter technology to investigate the DNA copy number (CN) in 70 well-curated primary ILC samples. We confirmed prior observations of frequent amplification of CCND1 (33%), and MYC (17%) in ILC, but additionally identified a substantial subset of ILCs with ESR1 and ERBB2 (19%) amplifications. Of interest, tumors with ESR1 CN gains (14%) and amplification (10%) were more likely to recur compared to those with normal CN. Finally, we observed that MDM4 (MDMX) was amplified in 17% of ILC samples. MDM4 knockdown in TP53 wild-type ILC cell lines caused increased apoptosis, decreased proliferation associated with cell cycle arrest, and concomitant activation of TP53 target genes. Similar effects were seen in TP53 mutant cells, indicting a TP53-independent role for MDM4 in ILC. To conclude, amplification of ESR1 and MDM4 are potential genetic drivers of ILC. These amplifications may represent actionable, targetable tumor dependencies, and thus have potential clinical implications and warrant further study.
Keywords: ILC, ESR1, ERBB2, MDM4, amplification
1. Introduction
Invasive lobular carcinoma (ILC) represents the second most prevalent histological subtype of breast cancer after invasive ductal carcinoma (IDC), accounting for 10–15% of all invasive breast cancer. It is characterized by small, round cells, invading the adjacent stroma in a single-file pattern1. Although ILC differs from IDC in several histological and clinical features, the same therapies are used to treat patients with ILC and IDC, at least in part due to insufficient knowledge of druggable pathways unique to ILC. Numerous studies have reported mutational and copy number (CN) characterization of breast cancer2–5; however, due to low numbers of ILC samples, only limited details are available regarding genomic alterations that drive this unique subtype of breast cancer. These studies have uncovered a number of specific differences in the genetic make-up of ILC compared to IDC, including enrichment of PTEN loss, PIK3CA, FOXA1, ERBB2 and ERBB3 mutations, in addition to the well described ILC genetic hallmark of CDH1 loss6–8. CN analysis using chromosomal comparative genomic hybridization (CGH) revealed gain of chromosomal regions of 1q and loss of 16p9–11. In another study, two of eleven patients with multifocal ILC showed gain in CN at the 11q13.3 locus, which encodes genes such as CCND1, FADD and ORAOV112. Frequent CN amplification at 11q13.3 was further confirmed in a large targeted sequencing study, which additionally revealed high levels of amplification at, 8q24.21 (MYC), 15q26.3 (IGF1R), and 8p11.23 (FGFR1) in ILC6–8. Furthermore, gain of chromosomes 1q and 8q and loss of chromosome 11q were found to be more frequent in the hormone-related subtype, which is characterized by active ER/PR signaling and EMT features13.
A number of studies have recently shown ESR1 CN amplifications, encoding the estrogen receptor (ER), in ILC8,14–18; however, the data are sparse and conflicting, and the clinical relevance remains unclear. While some studies report ESR1 amplifications at a relatively high rate of ~20%, associated with either improved14,15 or worse outcomes16, others have detected a significantly lower ESR1-amplification frequency (<5%)17,18, possibly due to differences in methodology, sample cohorts, and threshold definitions19,20. Intriguingly, Desmedt et al recently reported ESR1 amplification in up to 25% of ILC samples8, a finding that warrants validation in an independent cohort.
To expand upon prior CN analyses in primary ILC, we utilized a highly sensitive NanoString-based approach recently described by us21. Herein, we utilized probes covering 67 genes with known roles in breast cancer progression and therapy resistance, including ESR1, CCND1, MYC, IGF1R, and FGFR1, and comprehensively characterized their CN in 70 primary ILC samples with detailed clinical information. Besides confirming frequent ESR1 CN gain and amplifications, we also show a significant association with higher risk of subsequent recurrence. Finally, we show, amplification of MDM4 (MDMX) in ILC. While MDM4 has primarily been described as a negative regulator of TP5322, our functional studies revealed an essential role in cell proliferation in both a TP53-dependent and independent manner, justifying further studies on the role of MDM4 amplification in ILC.
2. Materials and Methods
2.1. Retrospective cohort of ILC samples, and DNA and RNA isolation
Following review and approval by the Institutional Review Board at the University of Pittsburgh, we obtained formalin-fixed paraffin-embedded (FFPE) sections from primary ER+ ILC cases diagnosed between 1990 and 2011 at UPMC Magee-Women’s Hospital. Samples were macrodissected when tumor cellularity < 40%, as previously described21 . DNA and RNA isolations were performed using the QIAamp DNA FFPE kit (cat#56404) and Qiagen RNeasy FFPE kit (cat#73504) respectively, as per manufacturer’s instructions. Isolated DNA and RNA were quantified using the ThermoFisher Qubit dsDNA S/BR kit and Nanodrop Spectrophotometer (ThermoScientific), respectively.
Recurrence free survival (RFS) was measured as the time between date of diagnosis and date of local, regional, or distant recurrence. The median follow-up of patients in our cohort was 15.9 years. The cohort included 18 cases with recurrences (R; 6 local and 12 distant recurrences), and 52 cases that did not recur (non-recurrence, NR). Additional clinicopathological characteristics are shown in Supplementary Table S1 and further cohort details are described in supplemental text.
2.2. Nanostring copy number (CN) and mRNA expression analysis
A total of 70 samples with sufficient DNA (>150ng) were used for NanoString nCounter-based CN analysis. The development of the NanoString CN library, the overall methodological approach, and extensive quality control (QC) have been recently described by us21. Briefly, we designed 100bp DNA hybridization probes for a total of 67 genes known to be frequently altered in breast cancer. Given prior evidence for ESR1 amplifications in ILC we used 10 probes to cover the ESR1 gene, and 2–3 probes for the other 66 genes. ESR1 CN increase by 35% (CN ≥2.7) and decrease by 50% (CN ≤ 1) were considered gains/amplifications and deletions, respectively. CN calls above ≥10 were considered high amplifications. Given the lower resolution for other genes in the panel, we used one CN cutoff of ≥5 as amplification. Detailed CN calls for all genes in tumor samples and cell lines are provided in Tables S2 and S3, respectively. The Complex Heatmap R package was used to generate oncoprints for CN alterations (CNA). A tile plot was generated in R using the ggplot2 package. To correlate CN levels of genes of interest with expression, several probes covering ESR1, CCND1, ERBB2, MDM4 and MYC were used to determine expression in corresponding RNA samples from the same patients also using the Nanostring nCounter platform as we previously described23.
2.3. Quantitative real time polymerase chain reaction (qRT-PCR)
mRNA from triplicate tumor samples was isolated as per manufacturer’s protocols using Qiagen RNeasy Mini Kit and quantified by NanoDrop. 500 ng of mRNA per sample was converted to cDNA, using 1X iScript (BioRad#1708891) or 1X PrimeScript (Takara #RR036B) as per manufacturer’s protocol. Gene expression was assessed by qRT-PCR using SsoAdvanced™ Universal SYBR® Green Supermix (BioRad#1725274) and primers listed in Table S4, assaying RPLP0 as a housekeeping gene.
2.4. Cell line culture
MCF-7, MCF-10A, MDA-MB-134-VI (MM134), MDA-MB-330 (MM330), and HEK293T cells were obtained from ATCC. Sum44PE cells were purchased from Asterand. BCK4 cells were a generous gift from Britta Jacobsen, PhD (University of Colorado Anschutz Medical Campus) and recently described24, 25. Media formulations for cell culture are outlined in the supplemental text. Cells were periodically confirmed to be mycoplasma negative, authenticated at the University of Arizona Genetics Core by Short Tandem Repeat (STR) profiling and kept in continuous culture for less than 6 months. To assay TP53 status in cell lines, PCR was performed on cDNA (primers listed in Table S4) covering the full length TP53 transcript using Gotaq DNA Polymerase (Promega# M3001) as per manufacturer’s protocol. Sequenced PCR products were blasted against the human reference TP53 sequences (NM_000546).
2.5. Modulation of MDM4 expression
For overexpression studies, cells were reverse transfected with pIRES1neo empty vector or MDM4 cDNA overexpression plasmid (a gift from Wei Gu, PhD; Columbia University) using Lipofectamine™ 3000 (Thermo Fisher Scientific#L3000015), according to the manufacturer’s protocol. Stably transfected cells were selected using 1mg/ml G418. For transient knockdown studies, cells were reverse transfected with ON-TARGETplus Non-targeting Pool (Dharmacon#D-001810-10-05) or SMARTpool ON-TARGETplus Human MDM4 siRNA (Dharmacon#L-006536-02-0005) using Lipofectamine™ RNAiMAX and Opti-MEM® I (Gibco#31985–070) and following the manufacturer’s protocol (Thermo Fisher Scientific#13778–150). For stable knockdown studies, MDM4 shRNA (shMDM4#1 targeting exon 6 and shMDM4#2 targeting the 3’ UTR) and control shRNA (shCTL) in the doxycycline (Dox) inducible GFP-tagged lentiviral vector FH1t (provided by Ygal Haupt, PhD; Peter MacCallum Cancer Center, Australia), along with an additional non targeting Renilla shRNA (shRen; provided by Dr Marco Herold; University of Melbourne, Australia,) were used. shRNA sequences are listed in Table S4. Viruses were generated by transfecting HEK293T cells using third generation packaging systems and polyethylenimine (PEI) (1 μg/mL; Polysciences#23966–2) with a 3:1 ratio of PEI to transfected DNA constructs. shPASHA (targeting human DGCR8) was used to increase titer as previously described26. Virus was collected at the 36-and 48-hour time points, filtered through 0.45 μm filters (Fisher#09-754-21), and treated with 1 μg/mL Polybrene (Sigma#107689–10G) to infect target cells. Cells were infected twice and allowed to recover for three days prior to sorting for GFP positive cells (~40–50%) by FACS. shRNA expression was induced by treatment with 200 ng/ml and 160ng/ml DOX in BCK4 and SUM44PE cells, respectively, with replenishment every 3–4 days.
2.6. Proliferation and Colony formation assay
Cell proliferation was assessed either by FluoReporter™ Blue Fluorometric dsDNA Quantitation Kit (Invitrogen#F2962) or IncuCyte® Live-Cell Imaging System (Essen BioScience) according to manufacturer’s recommendations. Cells were plated in technical replicates (n=4–6) at 15,000 (ILC) or 5,000 (IDC) cells/well in 96-well plates. Data was captured on a PerkinElmer plate reader (Victor). Alternatively, cells were monitored on the IncuCyte Live Cell Analysis Imaging System using integrated confluence as a surrogate for cell number. Relative fold change in growth/confluence was normalized to day 0 data. For colony formation assays, 15,000 cells were seeded in 6 well-plates and treated with DOX next day. After 18 days, cells were fixed with 100% methanol for 10 minutes and stained with 0.5% crystal violet solution in 25% methanol for 10 minutes, followed by gently rinsing with distilled water and drying at room temperature. Plates were imaged on an SZX16 dissecting microscope with a Nikon camera under bright field illumination at 0.8X magnification. Colony confluency fractions were counted using cellSens Dimension (Olympus) software.
2.7. Flow cytometry assays
For cell cycle analysis, cells were collected by trypsinization, diluted to 1 × 106 cells/mL and stained with 20 μg/mL Hoechst 33342 (Life Technologies#H3570) for 30 minutes at 37°C in the dark.
For the apoptosis assay, cells were collected and washed with PBS, and re-suspended in 1X Annexin V binding buffer (BD Biosciences#556454) at a dilution of 1 × 106 cells/mL. 100 μl of cell suspension was stained with Propidium Iodide (PI; BD biosciences#556463) and APC Annexin V (BD biosciences#561012) at room temperature for 15 minutes as per the manufacturer’s protocol. Samples were acquired on a LSR II Flow Cytometer using compensation and analyzed using FACSDiva (BD Biosciences).
2.8. Immunoblotting (IB)
Proteins were extracted using RIPA buffer supplemented with 1X Halt Protease Inhibitor Cocktail. 25–50 μg per sample was run on 9% or 15% gradient SDS-PAGE gels and transferred to PVDF membranes (Millipore#IPFL00010), and blocked using Odyssey PBS blocking buffer (LiCor#927–40000) and incubated with the following antibodies: TP53 (1:500; sc-6243, Santa Cruz Biotechnology), MDM4 (1:500; A300–287A, Bethyl Laboratories), P21 (1:200; sc-397, Santa Cruz Biotechnology), PUMA (1:500; D30C10, Cell Signaling Technology), BAX (1:500; D2E11, Cell Signaling Technology), Tubulin gamma (1:10,000; ab11317, Abcam), ER (1:1000; 6F11, Leica Biosystems) and secondary antibodies anti-mouse 800CW: LiCor#925–32210, anti-rabbit 800CW: LiCor#925–32211. Blots were imaged using the Odyssey Infrared Imaging system (LiCor).
3. Results
3.1. Frequent ESR1 amplifications in ILC and association with increased risk of recurrence
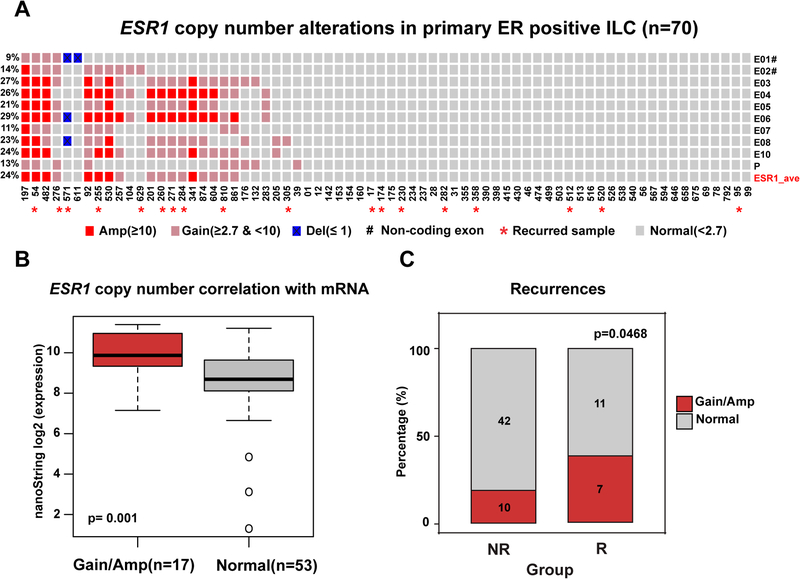
To identify potential genetic drivers of ILC progression, we performed CN analysis on 70 ER+ primary ILC tumors (Table 1) and initially focused on ESR1 CN alterations (Figure 1A). Based on average calls from probes covering the promoter region, two non-coding exons, and seven coding exons, we detected ~24% of samples with ESR1 gains (CN 2.7–10; 10 tumors; 14% of samples) or amplifications (CN ≥10; 7 tumors; 14% of samples). Tumors with elevated ESR1 CN levels showed significantly higher mRNA expression compared to the group with normal CNs (Figure 1B, p value= 0.001). ER histological scores (H-scores) were available for a subset of 53 samples, which were slightly higher in the group with ESR1 gains and amplifications; though did not reach statistical significance (Figure S1A). An analysis on a case-by-case basis shows that samples #197 and #54 (with consistent CNAs across the exons; shown on the very left in Figure 1A) display some of the highest levels of ESR1 mRNA and ER protein expression in our cohort (Supplementary Table S5).
Table 1.
Clinical characterization of ILC primary tumors cohort.
| Patients without recurrence (N=52) | Patients with recurrence (N=18) | p value | |
|---|---|---|---|
| Median age | 60.5 | 60 | 0.37 |
| Pathologic stage | 0.009 | ||
| I | 15(28.8%) | 1(5.6%) | |
| II | 27(51.9%) | 10(55.5%) | |
| III | 6(11.5%) | 6(33.3%) | |
| NA | 4(7.7%) | 1(5.6%) | |
| Pathologic grade | 0.77 | ||
| G1 | 11(21.2%) | 4(22.2%) | |
| G2 | 28(53.8%) | 9(50.0%) | |
| G3 | 3(5.8%) | 2(11.1%) | |
| NA | 10(19.2%) | 3(16.7%) | |
| ER (IHC) | |||
| Positive | 52(100%) | 18(100%) | |
| Average ER H-scorea | 237.6 | 214.1 | 0.24 |
| PR (IHC) | |||
| positive | 47(90.4%) | 17(94.4%) | >0.99 |
| negative | 5(9.6) | 1(5.6%) | |
| Average PR H-scoreb | 170.6 | 112.7 | 0.076 |
| HER2 (IHC) | |||
| Positive | 2(3.8%) | 1(5.6%) | >0.99 |
| Negative | 46(88.5%) | 16(88.9%) |
NA, not available; ER, estrogen receptor; PR, progesterone receptor.
41 samples of non-recurrent tumors and 12 samples of later recurrent tumors were available for ER histological scores.
40 samples of non-recurrent tumors and 11 samples of later recurrent tumors were available for PR histological scores.
Figure 1. Frequent ESR1 copy number alterations in 70 primary ER positive ILC specimens.
A. Oncoprint visualization of ESR1 CN alterations in primary ER positive ILC. Levels of amplification and deletion are color-coded. Each column represents a single sample. Each row indicates the copy number call of the corresponding single exon probe. Frequencies of alterations are indicated to the left side of each row. Samples with subsequent recurrences are annotated with ‘*’ symbol. E01-E10: exons 1–10. Untranslated exons (E1 and E2) are annotated with ‘#’ symbol. P: Promoter probe. ESR1_ave: average copy number call of all probes. B. Box plot comparison of ESR1 mRNA expression measured by NanoString between normal and gain/amplified samples (Wilcoxon rank-sum test). C. Frequency of ESR1 CN amplifications for the non-recurrence (NR) and recurrence groups (R) (Chi-square test, one-side, p=0.0468). Numbers show the count of tumors for each group. Amp: amplification. Del: deletion.
To determine whether ESR1 CN alterations were associated with clinical outcome, we compared groups with (n=18) or without recurrence (n=52) (Table 1). 39% (n=7) of samples from patients that recurred (R) showed ESR1 gain or amplification while only 19% (n=10) of the non-recurrent group (NR) presented with gain or amplification (Figure 1C) (p = 0.047). Recurrence-free survival (RFS) data was also available for 67/70 patients. As expected, CN gains or amplifications showed a trend towards a worse RFS outcome, although this failed to reach statistical significance (Figure S1B, p value= 0.2), potentially due to our limited sample size.
Overall, these results demonstrate that primary ILC exhibit frequent ESR1 gains and amplifications, which are significant enriched in samples with subsequent recurrence, suggesting a potential role for ESR1 CN gains/amplifications in endocrine resistance.
3.2. Enrichment of CCND1, ERBB2, MDM4 and MYC amplifications in ILC
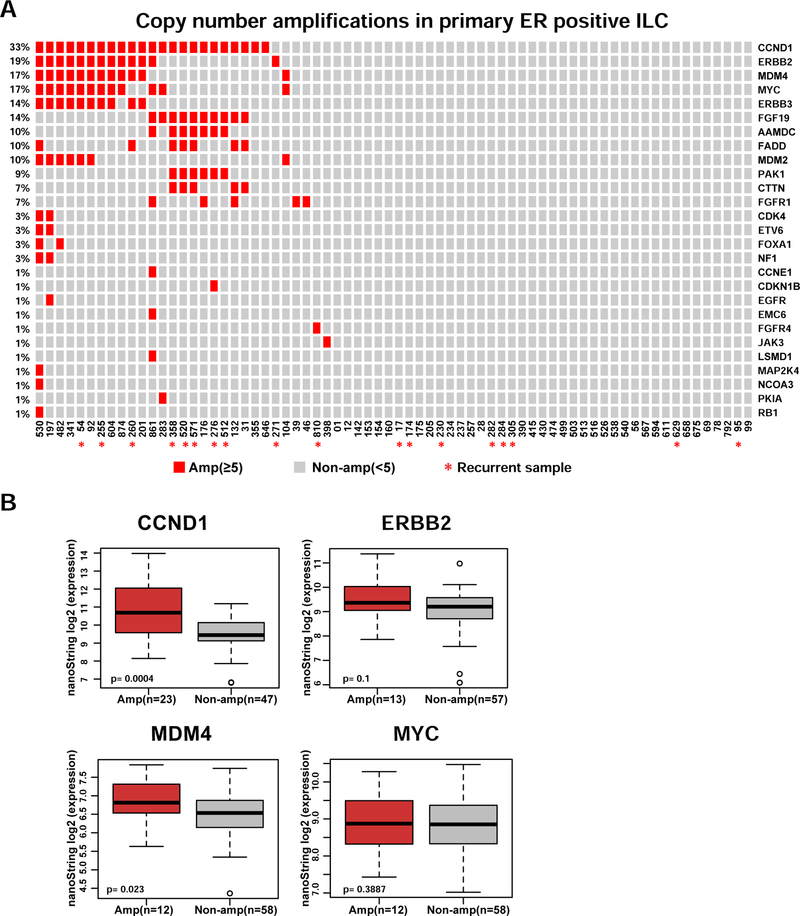
In addition to ESR1, the NanoString library also contained probes covering 66 genes that were previously implicated in breast cancer progression (Table S2, and Material and Methods). The most frequently amplified genes from this CN analysis are shown in Figure 2A. Deletions were rare (Figure S2), with the most frequently deleted gene (NCOR2) being lost in 5 samples. Genes with the highest frequency of amplification included CCND1(33%), ERBB2 (19%), MDM4 (17%), and MYC (17%). The amplification events were significantly correlated with mRNA expression for CCND1, and MDM4, along with a trend for ERBB2 but not MYC (Figure 2B). We observed some associations between amplifications of CCND1, FGF19, FADD, AAMDC, PAK1 and CTTN as was expected given these genes map to the well described 11q13 amplicon27,28. Due to low numbers, we did not perform a formal statistical co-occurrence test.
Figure 2. CCND1, ERBB2, MDM4 and MYC are the most frequently amplified genes in ER positive primary ILC.
A. Oncoprint visualization of CN amplifications by genes (rows) and samples (columns). Frequencies of the amplifications are indicated to the left side of each row. Samples with later recurrence are annotated with ‘*’ symbol. B. Comparison of NanoString mRNA expression of most frequently amplified genes in the ILC cohort between amplified and non-amplified samples (Wilcoxon rank-sum test). Amp: amplification.
In summary, the NanoString analysis confirmed prior observations of frequent amplification of 11q13, including CCND1, but also identified a higher than expected rate of ERBB2 amplification as well as novel MDM4 amplifications.
3.3. Association of MDM4 amplification with TP53 status in primary ILC and ILC cell models
Despite not observing a significant association between MDM4 amplification and recurrence free survival in our cohort (Figure S3), we decided to explore a potential role for MDM4 as a driver of ILC progression due to the high frequency of MDM4 amplifications (12/70). Since MDM4 was initially discovered as a TP53 regulator that possesses significant homology to MDM222, most studies on MDM4 have focused on its TP53-dependent role. In line with this, MDM4 amplification and/or overexpression is more frequent in tumors that retain wild type TP53 (wt TP53)29. We performed sanger sequencing of TP53 in eight tumors with MDM4 amplification, and determined a wt TP53 status in all sequenced samples, in line with the low TP53 mutation frequency seen in primary TCGA ILC tumors (~7%)30.
To further elucidate the role of MDM4 in ILC, we selected a set of ILC cell lines and measured MDM4 CN, RNA expression and protein levels (Figure S4A–C). The IDC cell line MCF-7 harboring a previously described MDM4 amplification31 was included as a positive control and data were normalized to normal breast tissue. In ILC cell lines, we observed MDM4 CN gain in MM134 and BCK4; however, MM134 showed low MDM4 expression compared to the overexpression in BCK4 and the positive control. The TP53 status of ILC cell lines has previously been described32, and also further verified by us using Sanger sequencing. BCK4 cells express relatively high MDM4 and harbor a single nucleotide polymorphism (SNP) in codon 72 of TP53 (RS1042522), a common SNP. MM134 and MM330 cells harbor TP53 E285K and Y220C mutations, respectively, both of which were reported as loss-of-function changes in the IARC TP53 database (http://p53.iarc.fr/). A truncating TP53 frameshift mutation (82_84delinsCA, pE28fsX16) was found in Sum44PE cells, which was also reported in the IARC database. Based on this sequencing information, we selected BCK4 cells as our primary ILC model of functional TP53 in our subsequent studies, while including MM134 and Sum44PE as TP53 mutant models.
3.4. Targeting MDM4 in the wt TP53 breast cancer cell line BCK4 suppresses cell growth by inducing G0/G1 arrest and apoptosis
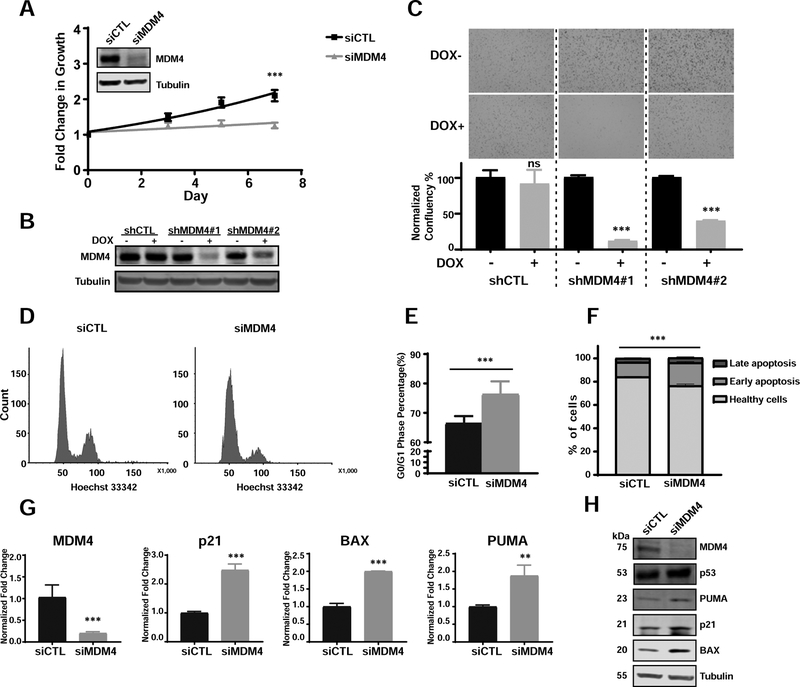
Based on the observation of MDM4 overexpression in BCK4 cells (Figure S4B, C), we performed transient siRNA-mediated MDM4 inhibition studies. We confirmed MDM4 knock-down (KD) at the protein level by immunoblot and observed a significant impact on cell growth/survival over 7 days compared to a control siRNA (Figure 3A). In addition, we tested the effect of MDM4 KD in the wildtype (wt) TP53 IDC cell line MCF7 and observed a similar growth inhibitory effect (Figure S4F). In addition to transient experiments, the ability of MDM4 KD to decrease cell proliferation in BCK4 cells was also seen by lentiviral-mediated doxycycline (DOX)-inducible shRNAs (verified by immunoblot in Figure 3B) in both a growth assay (Figure S5A) and a colony formation assay (Figure 3C). Furthermore, we performed cell cycle and apoptosis assays in consideration of the well-known role of MDM4 in regulating TP53. Cell cycle analysis revealed a reproducible and significantly increased G0/G1 arrest after MDM4 KD for 4 days (Figure 3D, E). In addition to reduced proliferation, we also observed an increase in apoptosis in cells transfected with siMDM4 relative to siCTL, as determined by FACS analysis (Figure 3F).
Figure 3. Targeting MDM4 in p53-functional breast cancer cell line BCK4 suppresses cell growth by inducing G0/G1 arrest and apoptosis.
A. Growth curves of BCK4 cells transiently transfected with siMDM4 vs siCTL (two-way ANOVA, ***p<0.001, n=2). MDM4 transient KD was confirmed by immunoblots shown top left. B. shRNA KD in BCK4 cells confirmed by immunoblot. DOX: doxycycline. C. Representative images from colony formation assays of BCK4 shCTL and shMDM4 cells. Quantifications of normalized confluency are shown below (data represent the average of biological triplicates ±SD; t-tests, ***p<0.001, n=2). D. Representative FACS figures of cell cycle analysis of siCTL and siMDM4 after knockdown for 4 days. E. Percentage of cells in G0/G1 phase in BCK4 siCTL vs siMDM4 after transfection for 4 days (t-tests, data represent the average of 8 biological replicates from three independent experiments ±SD). F. Apoptosis assay profiles in BCK4 cells transfected with siCTL or siMDM4 after 4 days (p value by t-test for total apoptotic cells in siCTL vs siMDM4, ***p<0.001, n=3). G. qPCR of p53 downstream targets in BCK4 cells after MDM4 downregulation (Data represent the average of biological triplicates ±SD; t-tests, **p<0.05, ***p<0.001). H. Immunoblots demonstrating induction of p53 target genes in BCK4 cells after MDM4 downregulation by siRNA.
To understand the downstream effects of MDM4 silencing in BCK4 cells, we assessed RNA and protein levels of TP53 and its known targets (Figure 3G, H). MDM4 siRNA KD in BCK4 cells did not alter TP53 expression at the protein level but did induce the pro-apoptotic Bcl-2 family members PUMA and BAX, and cell cycle inhibitor p21 at both the RNA and protein levels. These results are consistent with previous studies of MDM4 KD in TP53wt cell lines from breast cancer33 and other cancers34. Combined, this data shows that MDM4 expression in the wt TP53 ILC cell line BCK4 contributes to cell cycle progression and that its oncogenic role is predominantly achieved by inhibition of TP53 downstream transactivation.
3.5. MDM4 promotes cell cycle progression in mutant TP53 ILC cell lines
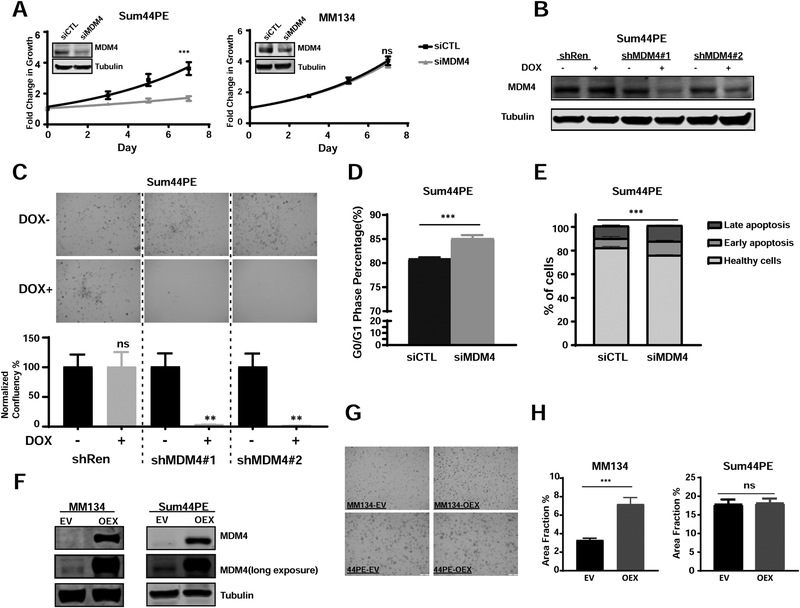
To further investigate if the proliferative role of MDM4 is dependent on functional TP53, we employed MDM4 KD studies in MM330, MM134 and Sum44PE ILC cell lines harboring TP53 mutations. MDM4 depletion suppressed cell growth by more than 50% in the Sum44PE cell line (Figure 4A) in a 7-day growth assay. In addition, we also knocked down MDM4 using dox-inducible shRNAs in Sum44PE cells (Figure 4B) and observed a similar inhibitory effect in both growth (Figure S5), and colony formation assays (Figure 4C). Interestingly, this MDM4 KD induced growth inhibition phenotype was cell line-dependent as we did not observe comparable changes in growth rates of the other two TP53mut ILC cell lines, MM330 (Figure S4D) and MM134 (Figure 4A), while seeing effects in the TP53mut IDC cell line T47D (Figure S4H). We next performed FACS-based cell cycle and apoptosis assays on Sum44PE cells and observed a significant increase in G0/G1 arrest (Figure 4D) and apoptosis (Figure 4E) following MDM4 downregulation by siRNA. As expected, qPCR and immunoblot analyses did not reveal an induction of TP53 downstream targets (Figure S6A), as was seen in the TP53wt cell line BCK4 (see Figure 3G, H), consistent with inactive TP53 signaling.
Figure 4. MDM4 promotes cell cycle progression in mutant TP53 breast cancer cell lines.
A. Growth curves of Sum44PE (left) and MM134 (right) cells transiently transfected with siMDM4 vs siCTL (two-way ANOVA, ***p<0.001, ns: no significance, n=4). MDM4 knockdown was confirmed by immunoblots shown top left. B. shRNA KD in Sum44PE cells was confirmed by immunoblot. C. Representative images from colony formation assay in Sum44PE shCTL and shMDM4 cells. Quantification of normalized confluency are shown below (data represent the average of biological triplicates ±SD; t-tests, ***p<0.001, n=2). D. Percentage of cells in G0/G1 phase in Sum44PE siCTL vs siMDM4 after transfection for 4 days (t-tests, data represent the average of 6 biological replicates from two independent experiments ±SD). E. Apoptosis assay profiles in Sum44PE cells transfected with siCTL or siMDM4 after 4 days (p value by t-test for total apoptotic cells in siCTL vs siMDM4, ***p<0.001, n=2). F. Immunoblot validation of constitutive MDM4 overexpression in MM134 (left) and Sum44PE (right) cells. G. Representative images of MM134 (left) and Sum44PE (right) cells transfected with EV (empty vector) and OEX (MDM4 overexpresssion plasmid) after 18-days growth in a colony formation assay. H. Quantifications by area fraction of all the colonies in MM134 (left) and Sum44PE (right) (t-tests, ***p<0.001, n≥2)
To further confirm the proliferative role of MDM4 in TP53mut ILC cells, we constitutively overexpressed MDM4 in both MM134 and Sum44PE cells (Figure 4F). We detected a mild but reproducible and significant increase in proliferation in a 6-day growth assay for MM134 but not in Sum44PE cells (Figure S6B, C), with similar results from long-term colony formation assays (Figure 4G, H). Consistent with our earlier data (see Figure S6A), we did not observe downregulation of PUMA, p21 and BAX in MDM4-overexpressing cells, (Figure S6D).
Combined, our data suggests that MDM4 KD results in context-dependent suppression of cell proliferation via delaying cell cycle progression and inducing apoptosis through both TP53-dependent and independent mechanisms.
4. Discussion
There is increasing realization that development and progression of ILC is associated with unique pathological, clinical, and molecular features. The goal of further deciphering molecular underpinnings of ILC is to personalize treatment and improve outcome for patients. In our CNA study in ILC, we made a number of novel observations with potential clinical implications, including (1) frequent ESR1 CN gain (14%) or amplification (10%) associated with disease recurrence, (2) frequent ERBB2 amplification (19%), and (3) frequent MDM4 amplifications (17%) and a functional role for MDM4 in ILC. In addition, we were able to confirm previously reported amplifications at 11q13 (CCND1, 33%) and 8q24 (MYC, 17%)7,8,35.
Our CN characterization of ILC revealed frequent ESR1 amplifications in about one quarter of cases (24%), associated with significantly higher mRNA expression. Importantly, ESR1 amplifications were significantly enriched in samples from patients with subsequent recurrences. Our findings are in agreement with the recently reported study detailing ESR1 amplifications in 25% of primary ILC, association with mRNA expression, and enrichment in aggressive ILC tumors by Desmedt et al8. In that study, the alterations were significantly enriched in ILC versus IDC, suggesting a unique role for amplified ESR1 in modulating response to estradiol and endocrine therapy in tumors lacking functional E-cadherin. Collectively, these findings justify further clinical investigation of ILC in the context of endocrine therapies, especially with respect to estradiol-containing therapy regimens as previously described by us and others36–39.
Beyond ESR1, we also observed ERBB2 amplifications in 19% of cases. Given the growing evidence for increased rates of HER2/HER3 mutations in ILC6,8, the finding of higher rates of ERBB2 amplification is of potential interest. Of note, many of these ERBB2 amplified cases were designated as HER2 negative by prior clinical IHC/FISH testing. This discrepancy could result from tissue heterogeneity, but could also reflect low level amplifications detected by sensitive Nanostring technologies that do not cause high level overexpression. Such low-level gene amplifications might reflect genomic instability, and might not be consequential with regard to the biological consequence of overexpression of the gene that has undergone CN changes. An example for complex structural genomic rearrangement in the absence of high level amplification are the recently reported segmental tandem duplications in breast cancer40,41. Additional studies are necessary to further differentiate between CN changes identified with the sensitive Nanostring approach that are associated with true oncogenic driver functions, and those more associated with genetic instability.
In addition to ESR1 and ERBB2, we also observed MDM4 amplifications in 17% of our primary ILC cases. Previous studies have identified varying rates of MDM4 amplification in breast cancer, with studies using fluorescence in situ hybridization (FISH) suggesting a low frequency (<5%)31,33. MDM4 is amplified in about 13% of all TCGA breast cancers and 18% in ILC cases specifically30. MDM4 amplification has previously been implicated in breast cancer metastasis from genomic evolution analyses among paired primary and metastatic tumors. Here MDM4 amplification was shown to be associated with endocrine resistance, while TP53 driver mutations were predominantly early events during metastatic evolution42,43. Subsequent gain of MDM4 in the absence of wt TP53 suggests that MDM4 may have broader functions beyond TP53 transactivation, which is further supported by our current data. Previously, downregulation of MDM4 mRNA using RNAi in vitro and in vivo was shown to attenuate both we TP53 and mutant TP53 breast cancer cell growth33,44; however, little is known about the role of MDM4 specifically in ILC. Our data suggests that in the wt TP53 ILC cell line BCK4, the oncogenic role of MDM4 is likely achieved by inhibition of TP53 transactivation. Conversely, in the ILC cell line Sum44PE, with truncated TP53, MDM4 knockdown led to a reproducible and significant induction of G0/G1 arrest and an increase in apoptotic cell death, further supporting the growing literature that MDM4 has TP53 independent roles. However, using the TP53mut ILC cell line MM134 we observed no effect of MDM4 KD, while we did detect a minor growth advantage in MDM4 overexpressing cells in a colony formation assay. The role of MDM4 mediated proliferation in mutant TP53 cells remains unclear, and our data indicates that this may also be cell line dependent. While studies focusing on the TP53-independent functions of MDM4 are limited, data so far indicated that MDM4 interacts with p21, E2F1, p300, ER and p27 independent of TP5345–47. It will be of interest to understand this mechanism in ILC cells lacking functional E-cadherin.
Our data suggests that MDM4 overexpression in wt TP53 primary ILC tumors may reduce apoptosis, enhance progression through the cell cycle and ultimately contribute to tumor proliferation, implicating it as a novel therapeutic target. The development of small molecules that can target the interaction of TP53 and MDM4 and induce a TP53-dependent cell cycle arrest and/or apoptosis have previously been described48; however, it remains largely unclear whether blocking the binding of MDM proteins to TP53 compromises their TP53-independent functions. While no such agents are currently in clinical development, small molecules specifically targeting the function of MDM4 could potentially hold promise for a subgroup of ILC patients in the future. Given the high frequency of MDM4 CN alterations in ILC reported herein, it will be important to determine the value of such targeted agents and their potential combination with endocrine therapy for ILC patients with MDM4 CNA.
In conclusion, our CNA study of 70 primary ILC specimens has revealed clinically relevant gains and amplifications in ESR1, that are associated with disease recurrence, frequent ERBB2 amplifications often in the absence of clinical IHC scores, and frequent MDM4 amplifications. Further, our functional studies suggest that MDM4 may contribute to ILC progression through both TP53-dependent and independent mechanisms. We believe our findings provide a molecular foundation to further explore the development and progression of ILC, ultimately providing vital insights that may lead to personalized treatment for patients with this understudied subtype of breast cancer.
Supplementary Material
Highlights.
Comprehensive high-resolution copy number alteration study in primary Invasive Lobular Carcinoma (n=70)
Frequent ESR1 CN gain (14%) or amplification (10%) associated with disease recurrence
Frequent ERBB2 amplification (19%) in primary ILC
Frequent MDM4 amplifications (17%) and a functional role for MDM4 in ILC.
These novel findings have potential clinical implications for patients with ILC, an understudied subtype of breast cancer
Acknowledgments:
We acknowledge Dr. Marco Herold (The Walter and Eliza Hall Institute of Medical Research) for kindly providing the FH1tUTG vector, and Dr. Wei Gu (Columbia University) for the gift of pIRES1neo empty vector and MDM4 (cDNA) overexpression plasmid. We are grateful to Britta Jacobsen, PhD (University of Colorado at Denver), for providing the BCK4 cell line.
The work is in part funded by the Third Xiangya Hospital, Central South University, a Shear Family Foundation grant, Susan G. Komen Leadership grants to SO (SAC160073) and AVL (SAC110021), and Breast Cancer Research Foundation grants to SO and AVL. MJS is supported by an NIH Pathway to Independence Award (K99CA193734), and NT is supported by a Department of Defense Breakthrough Fellowship (BC160764). SO and AVL are Hillman Foundation Fellows. This project used the UPMC Hillman Cancer Center and Tissue and Research Pathology/Pitt Biospecimen Core shared resource which is supported in part by award P30CA047904.
Abbreviations:
- (Amp)
Amplification
- (CGH)
chromosomal comparative genomic hybridization
- (CNA)
copy number alteration
- (Del)
deletion
- (DOX)
doxycycline
- (E2)
estradiol
- (ER)
estrogen receptor
- (IDC)
invasive ductal carcinoma
- (ILC)
Invasive lobular carcinoma
- (KD)
knock down
- (RFS)
Recurrence free survival
- (TP53mut)
TP53 mutant
- (TP53wt)
TP53 wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare they have no conflicts of interest with the contents of this article.
References
- 1.Martinez V, Azzopardi JG. Invasive lobular carcinoma of the breast: incidence and variants. Histopathology. 1979;3(6):467–488. [DOI] [PubMed] [Google Scholar]
- 2.Goncalves R, Warner WA, Luo J, Ellis MJ. New concepts in breast cancer genomics and genetics. Breast Cancer Res. 2014;16(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sørile T, Eisen MB, Van De Rijn M, Jeffrey SS, Ress CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø, Pergammenschlkov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. Molecular Characterization of Breast Cancer with High-Resolution Oligonucleotide Comparative Genomic Hybridization Array. Clin Cancer Res. 2009;15(2):441–451. [DOI] [PubMed] [Google Scholar]
- 6.Ross JS, Wang K, Sheehan CE, Boguniewicz AB, Otto G, Downing SR, Sun J, He J, Curran JA, Ali S, Yelensky R, Lipson D, Palmer G, Miller VA, Stephens PJ. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clin Cancer Res. 2013;19(10):2668–2676. [DOI] [PubMed] [Google Scholar]
- 7.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GMK, Factor RE, Collins LC, Allison KH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163(2):506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, Fumagalli D, Brown D, Rothé F, Vincent D, Kheddoumi N, Rouas G, Majjaj S, Brohée S, Van Loo P, Maisonneuve P, Salgado R, Van Brussel T, Lambrechts D, et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol. 2016;34(16):1872–1880. [DOI] [PubMed] [Google Scholar]
- 9.Simpson PT, Reis-Filho JS, Lambros MBK, Jones C, Steele D, Mackay A, Iravani M, Fenwick K, Dexter T, Jones A, Reid L, Da Silva L, Shin SJ, Hardisson D, Ashworth A, Schmitt FC, Palacios J, Lakhani SR. Molecular profiling pleomorphic lobular carcinomas of the breast: Evidence for a common molecular genetic pathway with classic lobular carcinomas. J Pathol. 2008;215(3):231–244. [DOI] [PubMed] [Google Scholar]
- 10.Stange DE, Radlwimmer B, Schubert F, Traub F, Pich A, Toedt G, Mendrzyk F, Lehmann U, Eils R, Kreipe H, Lichter P. High-resolution genomic profiling reveals association of chromosomal aberrations on 1q and 16p with histologic and genetic subgroups of invasive breast cancer. Clin Cancer Res. 2006;12(2):345–352. [DOI] [PubMed] [Google Scholar]
- 11.Nishizaki T, Chew K, Chu L, Isola J, Kallioniemi A, Weidner N, Waldman FM. Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J cancer. 1997;74(5):513–517. http://www.ncbi.nlm.nih.gov/pubmed/9355973. Accessed January 8, 2019. [DOI] [PubMed] [Google Scholar]
- 12.Norton N, Advani PP, Serie DJ, Geiger XJ, Necela BM, Axenfeld BC, Kachergus JM, Feathers RW, Carr JM, Crook JE, Moreno-Aspitia A, Anastasiadis PZ, Perez EA, Thompson EA. Assessment of tumor heterogeneity, as evidenced by gene expression profiles, pathway activation, and gene copy number, in patients with multifocal invasive lobular breast tumors. PLoS One. 2016;11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaut M, Chin SF, Majewski I, Severson TM, Bismeijer T, De Koning L, Peeters JK, Schouten PC, Rueda OM, Bosma AJ, Tarrant F, Fan Y, He B, Xue Z, Mittempergher L, Kluin RJC, Heijmans J, Snel M, Pereira B, et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci Rep. 2016;6(1):18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, Lebeau A, Terracciano L, Al-Kuraya K, Jänicke F, Sauter G, Simon R. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39(5):655–660. [DOI] [PubMed] [Google Scholar]
- 15.Tomita S, Zhang Z, Nakano M, Ibusuki M, Kawazoe T, Yamamoto Y, Iwase H. Estrogen receptor alpha gene ESR1 amplification may predict endocrine therapy responsiveness in breast cancer patients. Cancer Sci. 2009;100(6):1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen KV, Ejlertsen B, Müller S, Møller S, Rasmussen BB, Balslev E, Lænkholm A-V, Christiansen P, Mouridsen HT. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res Treat. 2011;127(2):345–355. [DOI] [PubMed] [Google Scholar]
- 17.Moelans CB, Monsuur HN, de Pinth JH, Radersma RD, de Weger RA, van Diest PJ. ESR1 amplification is rare in breast cancer and is associated with high grade and high proliferation: a multiplex ligation-dependent probe amplification study. Anal Cell Pathol (Amst). 2010;33(1):13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holst F, Stahl P, Hellwinkel O, Dancau AM, Krohn A, Wuth L, Heupel C, Lebeau A, Terracciano L, Al-Kuraya K, Jänicke F, Sauter G, Simon R. Reply to “ESR1 gene amplification in breast cancer: A common phenomenon?” Nat Genet. 2008;40(7):810–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, Gursky A, Siddiqui J, Tomlins SA, Roychowdhury S, Pienta KJ, Kim SY, Roberts JS, Rae JM, Van Poznak CH, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holst F Estrogen receptor alpha gene amplification in breast cancer: 25 years of debate. World J Clin Oncol. 2016;7(2):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basudan A, Priedigkeit N, Hartmaier RJ, Sokol ES, Bahreini A, Watters RJ, Boisen MM, Bhargava R, Weiss KR, Karsten MM, Denkert C, Blohmer J-U, Leone JP, Hamilton RL, Brufsky AM, Elishaev E, Lucas PC, Lee AV, Oesterreich S. Frequent ESR1 and CDK Pathway Copy Number Alterations in Metastatic Breast Cancer. Mol Cancer Res. October 2018:molcanres.0946.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15(19):5349–5357. http://www.ncbi.nlm.nih.gov/pubmed/8895579. Accessed January 12, 2019. [PMC free article] [PubMed] [Google Scholar]
- 23.Gyanchandani R, Lin Y, Lin H-M, Cooper K, Normolle DP, Brufsky A, Fastuca M, Crosson W, Oesterreich S, Davidson NE, Bhargava R, Dabbs DJ, Lee AV. Intratumor Heterogeneity Affects Gene Expression Profile Test Prognostic Risk Stratification in Early Breast Cancer. Clin Cancer Res. 2016;22(21):5362–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jambal P, Badtke MM, Harrell JC, Borges VF, Post MD, Sollender GE, Spillman MA, Horwitz KB, Jacobsen BM. Estrogen switches pure mucinous breast cancer to invasive lobular carcinoma with mucinous features. Breast Cancer Res Treat. 2013;137(2):431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasdemir N, Bossart EA, Li Z, Zhu L, Sikora MJ, Levine KM, Jacobsen BM, Tseng GC, Davidson NE, Oesterreich S. Comprehensive phenotypic characterization human invasive lobular carcinoma cell lines in 2D and 3D cultures. Cancer Res. 2018;78(21):6209–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IAM, Kwon JS, Guan Y, Sinha N, Zuber J. An Optimized microRNA Backbone for Effective Single-Copy RNAi. Cell Rep. 2013;5(6):1704–1713. [DOI] [PubMed] [Google Scholar]
- 27.Lundgren K, Holm K, Nordenskjöld B, Borg Å, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwek SS, Roy R, Zhou H, Climent J, Martinez-Climent JA, Fridlyand J, Albertson DG. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28(17):1892–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Home-The Cancer Genome Atlas-Cancer Genome-TCGA. https://cancergenome.nih.gov/. Accessed January 9, 2019. [Google Scholar]
- 31.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, Helin K, Pelicci PG, Jochemsen AG, Marine J-C. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24(13):5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christgen M, Derksen PWB. Lobular breast cancer: Molecular basis, mouse and cellular models. Breast Cancer Res. 2015;17(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haupt S, Buckley D, Pang JMB, Panimaya J, Paul PJ, Gamell C, Takano EA, Ying Lee Y, Hiddingh S, Rogers TM, Teunisse AFAS, Herold MJ, Marine JC, Fox SB, Jochemsen A, Haupt Y. Targeting Mdmx to treat breast cancers with wild-type p53. Cell Death Dis. 2015;6(7):e1821–e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, Goydos J, Haigh JJ, Haupt Y, Larue L, Jochemsen A, Shi H, Moriceau G, Lo RS, Ghanem G, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012;18(8):1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tajiri R, Inokuchi M, Sawada-Kitamura S, Kawashima H, Nakamura R, Oyama T, Dobashi Y, Ooi A. Clonal profiling of mixed lobular and ductal carcinoma revealed by multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Pathol Int. 2014;64(5):231–236. [DOI] [PubMed] [Google Scholar]
- 36.Song RX, Mor G, Naftolin F, McPherson RA, Song J, Zhang Z, Yue W, Wang J, Santen RJ. Effect of long-term estrogen deprivation on apoptotic responses of breast cancer cells to 17beta-estradiol. J Natl Cancer Inst. 2001;93(22):1714–1723. [DOI] [PubMed] [Google Scholar]
- 37.Ellis MJ, Gao F, Dehdashti F, Jeffe DB, Marcom PK, Carey LA, Dickler MN, Silverman P, Fleming GF, Kommareddy A, Jamalabadi-Majidi S, Crowder R, Siegel BA. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: a phase 2 randomized study. JAMA. 2009;302(7):774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kota K, Brufsky A, Oesterreich S, Lee A. Estradiol as a Targeted, Late-Line Therapy in Metastatic Breast Cancer with Estrogen Receptor Amplification. Cureus. 2017;9(7):e1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, Ding L, Griffith OL, Miller C, Larson D, Fulton RS, Harrison M, Mooney T, McMichael JF, Luo J, et al. Endocrine-Therapy-Resistant ESR1 Variants Revealed by Genomic Characterization of Breast-Cancer-Derived Xenografts. Cell Rep. 2013;4(6):1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menghi F, Barthel FP, Yadav V, Tang M, Ji B, Tang Z, Carter GW, Ruan Y, Scully R, Verhaak RGW, Jonkers J, Liu ET. The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell. 2018;34(2):197–210.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, Ju YS, Smid M, Brinkman AB, Morganella S, Aure MR, Lingjaerde OC, Langerød A, Ringnér M, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullah I, Karthik GM, Alkodsi A, Kjällquist U, Stålhammar G, Lövrot J, Martinez NF, Lagergren J, Hautaniemi S, Hartman J, Bergh J. Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. J Clin Invest. 2018;128(4):1355–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland HK, Lilleng PK, Gundem G, Gerstung M, Pappaemmanuil E, Gazinska P, Bhosle SG, Jones D, Raine K, Mudie L, Latimer C, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell. 2017;32(2):169–184.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda PJ, Buckley D, Raghu D, Pang JMB, Takano EA, Vijayakumaran R, Teunisse AFAS, Posner A, Procter T, Herold MJ, Gamell C, a JC, Fox SB, Jochemsen A, Haupt S, Haupt Y. MDM4 is a rational target for treating breast cancers with mutant p53. J Pathol. 2017;241(5):661–670. [DOI] [PubMed] [Google Scholar]
- 45.Jin Y, Zeng SX, Sun X-X, Lee H, Blattner C, Xiao Z, Lu H. MDMX Promotes Proteasomal Turnover of p21 at G1 and Early S Phases Independently of, but in Cooperation with, MDM2. Mol Cell Biol. 2008;28(4):1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wunderlich M, Ghosh M, Weghorst K, Berberich SJ. MdmX represses E2F1 transactivation. Cell Cycle. 2004;3(4):472–478. [PubMed] [Google Scholar]
- 47.Swetzig WM, Wang J, Das GM. Estrogen receptor alpha mediates the p53-independent overexpression of MDM4/MDMX and MDM2 in human breast cancer. Oncotarget. 2016;7(13):16049–16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Lozano G. Molecular pathways: Targeting Mdm2 and Mdm4 in cancer therapy. Clin Cancer Res. 2013;19(1):34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.