Abstract
The highly polymorphic classical human leukocyte antigen (HLA) genes display strong linkage disequilibrium (LD) that results in conserved multi-locus haplotypes. For unrelated individuals in defined populations, HLA haplotype frequencies can be estimated using the expectation-maximization (EM) method. Haplotypes can also be constructed using HLA allele segregation from nuclear families. It is straightforward to identify many HLA genotyping inconsistencies by visually reviewing HLA allele segregation in family members. It is also possible to identify potential crossover events when two or more children are available in a nuclear family. This process of visual inspection can be unwieldy, and we developed the “HaplObserve” program to standardize the process and automatically build haplotypes using family-based HLA allele segregation. HaplObserve facilitates systematically building haplotypes, and reporting potential crossover events. HLA Haplotype Validator (HLAHapV) is a program originally developed to impute chromosomal phase from genotype data using reference haplotype data. We updated and adapted HLAHapV to systematically compare observed and estimated haplotypes. We also used HLAHapV to identify haplotypes when uninformative HLA genotypes are present in families. Finally, we developed “pould”, an R package that calculates haplotype frequencies, and estimates standard measures of global (locus-level) LD from both observed and estimated haplotypes.
Keywords: 17th International HLA and Immunogenetics Workshop, Next Generation Sequencing, HLA, Haplotype, Linkage Disequilibrium
1. INTRODUCTION
The human leukocyte antigen (HLA) genes are located in the 4 MB major histocompatibility complex (MHC) region on chromosome 6p21.3 [1]. There are three classical (HLA-A, HLA-C and HLA-B) class I and eight classical (HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLADPA1 and HLA-DPB1) class II HLA genes. The HLA system is the most polymorphic and diverse genetic system in the genome [1–3]. Due to the magnitude of the polymorphisms at a given HLA locus, complex allele names are assigned for each gene based on polymorphic nucleotide positions and inferred amino acid variation [4]. These genes are in strong linkage disequilibrium (LD) [5], establishing haplotypes; LD is especially pronounced between physically proximal neighboring genes, such as HLA-C and HLA-B. Although occasional meiotic recombination events are observed, alleles of HLA genes usually segregate as intact chromosomal blocks transmitted from both parents to their offspring [6]. Specific HLA alleles are often observed in distinct haplotypes in different ethnic groups. In allogeneic transplantation, HLA genotype matching correlates significantly with positive outcomes; in addition, certain HLA alleles associate significantly with predisposition to autoimmune diseases [7]. HLA haplotype information elevates the confidence in the assignment of HLA genotypes [8], allows specific alleles to be identified as the factors contributing to disease susceptibility in specific haplotypes, and helps predict match grade in hematopoietic stem cell transplants [9].
The “Study of Haplotypes in Families by NGS HLA” (Family Haplotype) project was organized under the 17th International HLA and Immunogenetics Workshop (IHIW). The advent of next generation sequencing (NGS) technologies allows sequencing nearly entire HLA genes (from 3’-UTR to 5’-UTR) and generates HLA genotypes from a large number of subjects in a cost effective manner. The objective of the 17th IHIW Family Haplotype project was to build a set of HLA haplotypes based on the segregation of HLA alleles generated using NGS-based HLA typing on an international collection of family subjects. To achieve the goal of the project, we developed analytical tools to automatically build haplotypes and calculate critical information, such as haplotype frequencies and standard measures of global (locus-level) LD related to the haplotypes under the “Informatics of Genomic Data” component. In the present report we describe the tools developed as a collaborative product of the 17th IHIW Informatics of Genomic Data component, their functionalities, utility, and limitations for the Family Haplotype project. This manuscript specifically focuses on addressing the algorithmic needs of the software tools and illustrating issues with examples from the dataset collected for the 17th IHIW. The analyses accomplished using these tools are included in a separate report in this issue (Osoegawa et al, manuscript submitted).
2. Materials and Methods
2.1. Family and Subjects
A total of 204 nuclear families consisting of 820 subjects were selected from a clinical database at the Stanford Blood Center’s Histocompatibility, Immunogenetics & Disease Profiling Laboratory (Osoegawa et al. manuscript submitted). Family pedigrees were stored in PED format [10] in the 17th IHIW database [11]. Subjects were de-identified by each participating laboratory, and new IDs were systematically assigned by the 17th IHIW database system. Family haplotypes were built using double-blinded subject IDs. The use of the data from the double-blinded subjects was approved by the Stanford University Institutional Review Board (IRB) eProtocol Title “17th International HLA and Immunogenetics Workshop” eProtocol #: 38899.
2.1.1. Genotype Data
The HLA genotypes for each family were recorded in GL String format [12] in the 17th IHIW database [11]. We used HLA genotypes from the 204 families to develop and validate the tools described in this manuscript. Additionally, we reviewed HLA-DPB1 genotypes for 4949 samples from 1442 nuclear families that were registered for various projects in the 17th IHIW database.
2.2. HaplObserve
The HaplObserve package is written in the Java programming language using Java SE Development kit (JDK – 1.8), and includes data-processing, data-analysis and data-reporting functions. HaplObserve’s data-analysis function generates parental haplotypes from a nuclear family, and was designed to generate haplotypes using HLA genotypes from a nuclear family that consists of two parents and at least one child (sections 2.2.2.1 – 2.2.2.7). The data-processing function accepts genotype and accessory data for multiple families, and selects individual families and subsets of families for analysis (section 2.4). The data-reporting function generates output files for review and downstream analyses (sections 2.2.2.8 and 2.5). HaplObserve is a command line tool, and the software package can be downloaded on a local computer as a zip file (hapl-obs-tools-0.0.1-SNAPSHOT-bin.zip) from the IHIW GitHub repository [A]. HaplObserve requires the Java SE Development kit (JDK – 1.7 or newer). The software installation and execution guide is provided as a Supplemental document (SupplementalHaplObserveInstruction). The detailed software installation document can be obtained from the IHIW GitHub repository [A].
2.2.1. Input file format
Machine-readable input data formats requiring minimal human intervention are key requirements for successful, error-free software execution. The GL String format was developed as a standard software-consumable HLA genotype data format [12], and HLA genotypes were recorded in GL String format in the 17th IHIW database [11]. The advantages using GL Strings have been described previously [11], and this format is being widely adopted in the immunogenetics communtiy for these purpose [9, 13–16]. There are three options as input files for HaplObserve. 1) HaplObserve converts a “master” comma separated value (CSV) file into individual CSV files for each family. The master CSV files contain Labcodes (laboratory-specific IHIW IDs), Family IDs, sample IDs, relationship data, GL String formatted HLA genotypes, and ethnicity/country data (Supplemental Table 1). The ethnicity/country data is used to calculate haplotype frequencies for each ethnicity/country. The individual family CSV files are generated for visual examination of alleles if HaplObserve reports any errors (Supplemental Table 2). 2) HaplObserve is also able to accept individual family CSV files. This option may be convenient for building haplotypes from a single family, and the user would like to organize HLA genotype data in spreadsheet format. 3) Alternatively, the software is capable of accepting HLA genotypes in Histoimmunogenetics Markup Language (HML) formatted HLA genotypes [17], PED file-formatted family pedigree information [10] and CSV-formatted Labcodes and ethnicity/country information (INFO.csv file). The software looks for subjects across all three of these files and combines the information into the master CSV file. This option is convenient for building haplotypes from extended or multi-generation families.
2.2.2. Building haplotypes from a family
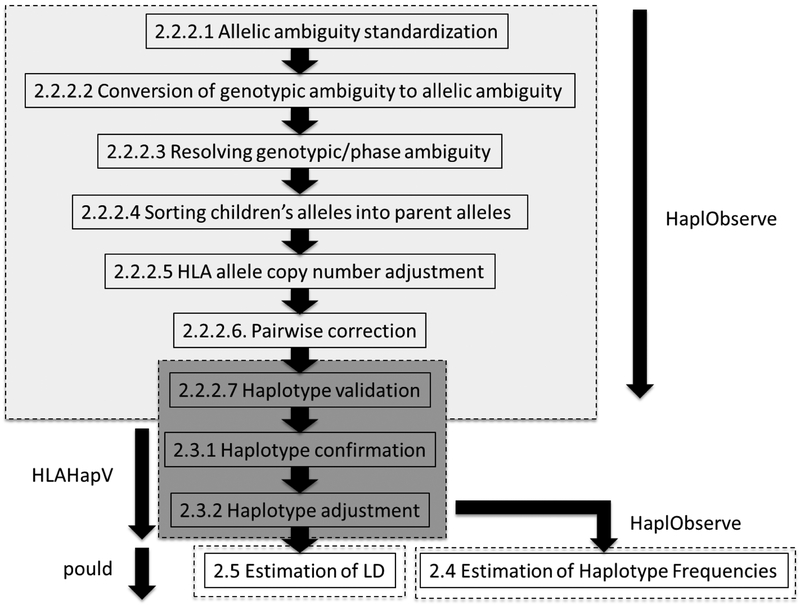
HaplObserve attempts to follow the first seven steps depicted in Figure 1, which are described in sections 2.2.2.1 to 2.2.2.7. Proper handling of ambiguities was one of the most critical elements of building fully phased haplotypes from un-phased genotypes. The first three (sections 2.2.2.1 to 2.2.2.3) focus on describing nature of ambiguities and how to handle different types of ambiguities.
Figure 1:
Figure 1 shows the flow of concepts behind the software suite. Detailed description of each step with the section numbers and titles can be found in Materials and Methods. HaplObserve, HLAHapV and pould were developed and updated to accomplish: [1] building HLA haplotypes from families (light gray highlight: sections 2.2.2.1 – 2.2.2.7); [2] validating the haplotypes (dark gray highlight: 2.2.2.7 – 2.3.2); [3] calculating haplotype frequencies (2.4); [4] estimating measures of LD from the family haplotypes (2.5).
2.2.2.1. Allelic ambiguity standardization
Two types of genotyping ambiguities exist: allelic and genotypic (or “phase”) ambiguities [12]. GL String formatted allelic ambiguity is represented using a slash (/); e.g. HLA-DPB1*04:01:01:01/HLADPB1*04:01:01:02 indicates that these two alleles are not distinguishable using the HLA genotyping method applied [12]. We initially observed frequent differences in the genotyping between parents and offspring for specific alleles. The first group of the differences in the inconsistent alleles were often due to STR or homopolymer length variation, which occurred independently of the NGS HLA genotyping protocols and software applications used in the 17th IHIW. The second group of differences was due to polymorphisms outside of the sequenced region. For example, HLADPB1*13:01:01 and HLA-DPB1*107:01 are often indistinguishable due to a difference in exon1; the NGS HLA genotyping protocols used for the 17th IHIW did not cover exon1 (Table 1) [18]. Similarly, HLA-DPB1*02:01:02 and HLA-DPB1*02:01:19 are not distinguishable when DPB1 exon5 is not sequenced. We treated the alleles in these groups as allelic ambiguity groups (Table 1). HaplObserve addresses these testing limitations by automatically converting alleles in a given STR, homopolymer or unsequenced polymorphism group to an allelic ambiguity string even if an allele has been unambiguously reported.
Table1:
Allelic ambiguities
| Ambiguities | Reason |
|---|---|
| HLA-DPB1*13:01:01/HLA-DPB1*107:01 | Polymorphism outside the sequenced region (Exon 1) |
| HLA-DPB1*04:01:01:01/HLA-DPB1*04:01:01:02 | Short Tandem Repeat |
| HLA-DPB1*02:01:02/HLA-DPB1*02:01:19 | Polymorphism outside the sequenced region (Exon 5) |
| HLA-DQA1*01:01:01:02/HLA-DQA1*01:01:01:03 | Homopolymer |
| HLA-DQA1*01:02:01:01/HLA-DQA1*01:02:01:03/HLA-DQA1*01:02:01:05 | Homopolymer |
| HLA-DQA1*01:02:01:04/HLA-DQA1*01:02:01:06/HLA-DQA1*01:02:01:07 | Homopolymer |
| HLA-DQA1*01:03:01:03/HLA-DQA1*01:03:01:04 | Homopolymer |
| HLA-DQA1*01:03:01:02/HLA-DQA1*01:03:01:06 | Homopolymer |
| HLA-DQA1*02:01:01:01/HLA-DQA1*02:01:01:02 | Homopolymer |
| HLA-DQA1*01:04:01:01/HLA-DQA1*01:04:01:02/HLA-DQA1*01:04:01:04 | Homopolymer |
| HLA-DQA1*05:05:01:01/HLA-DQA1*05:05:01:02/HLA-DQA1*05:05:01:04 | Short Tandem Repeat |
| HLA-DQA1*05:05:01:03/HLADQA1*05:05:01:05/HLA-DQA1*05:05:01:06 | Short Tandem Repeat |
| HLA-DQB1*03:03:02:02/HLA-DQB1*03:03:02:03 | Polymorphism outside the sequenced region |
| HLA-DQB1*05:03:01:01/HLA-DQB1*05:03:01:02 | Polymorphism outside the sequenced region |
| HLA-DRB1*03:01:01:01/HLA-DRB1*03:01:01:02 | Short Tandem Repeat |
| HLA-DRB1*07:01:01:01/HLA-DRB1*07:01:01:02 | Short Tandem Repeat |
| HLA-DRB1*13:01:01:01/HLA-DRB1*13:01:01:02 | Short Tandem Repeat |
| HLA-DRB1*15:01:01:01/HLADRB1*15:01:01:02/HLA-DRB1*15:01:01:03 | Short Tandem Repeat |
| HLA-DRB4*01:03:01:01/HLA-DRB4*01:03:01:03 | Polymorphism outside the sequenced region |
Alleles that could not be distinguished using the methods used for the 17th IHIW are reported as GL String formatted allelic ambiguities listed in the first column. The allelic ambiguities are based on IPDIMGT/HLA Database version 3.25.0. The reasons for the ambiguities are described in the second column.
2.2.2.2. Conversion of genotypic ambiguity to allelic ambiguity
GL String formatted “genotypic” ambiguity is represented using both a pipe (|) and a plus (+) delimiter together to identify alternative genotypes that cannot be distinguished due to an inability of a given typing system to establish phase between detected polymorphisms; for example, the HLADPB1*04:01:01:01+HLA-DPB1*04:02:01:02|HLA-DPB1*105:01+HLA-DPB1*126:01 string indicates that heterozygous genotyping result is either HLA-DPB1*04:01:01:01+HLA-DPB1*04:02:01:02 or HLA-DPB1*105:01+HLA-DPB1*126:01 [12]. Although allelic and genotypic ambiguities result from distinct shortcomings of a given typing system, some HLA genotyping software applications report allelic ambiguities in a genotypic ambiguity format [12]. There is no recommendation on how to express a specific typing within a GL String; neither is there a required order for alleles in a GL String. For example, the GL String HLA-DPB1*04:01:01:01+HLA-DPB1*04:02:01:02|HLADPB1*04:01:01:02+HLA-DPB1*04:02:01:02|HLA-DPB1*105:01+HLA-DPB1*126:01 indicates that the heterozygous genotype is either HLA-DPB1*04:01:01:01+HLA-DPB1*04:02:01:02, HLADPB1*04:01:01:02+HLA-DPB1*04:02:01:02 or HLA-DPB1*105:01+HLA-DPB1*126:01. HLADPB1*04:02:01:02 is shared in the first two allele combinations, indicating that these first two combinations represent allelic ambiguity. Therefore, this genotype ambiguity example can be shortened to HLA-DPB1*04:01:01:01/HLA-DPB1*04:01:01:02+HLA-DPB1*04:02:01:02|HLADPB1*105:01+HLA-DPB1*126:01. Supplemental Table 3 shows more examples of genotypic ambiguity and allelic ambiguity formats. To be consistent, HaplObserve converts genotypic ambiguities to allelic ambiguities when possible (Supplemental Table 3).
2.2.2.3. Resolving genotypic/phase ambiguity
Genotypic ambiguities frequently result when assembling short sequence reads, even if introns are sequenced; this primarily occurs for HLA-DPB1 genotypes. The genotypic ambiguities cannot be converted to the allelic ambiguities (see section 2.2.2.2), because they occur when NGS HLA genotyping software applications are not able to phase exon 2 and exon 3 sequences due to a lack of informative intron 2 SNPs or reference sequences. It is not possible to resolve such phase ambiguities when reviewing a single individual’s genotype. However, it is often possible to identify a single phased allele combination by reviewing all the genotypes in the family and assessing segregation (Table 2A and2B). HaplObserve includes a function to determine a single phased allele combination by reviewing HLA allele segregation prior to building family haplotypes. When phase ambiguity is not resolved due to the lack of informative family members, the software progressively compares each allele name from the first field to the fourth field, and takes the lowest-digit allele name combination as the priority HLA genotype (Table 2C). This approach assumes that the lowest-digit allele name combination is the more common combination, and haplotypes were built using genotypes that included these presumed more common alleles when phased ambiguities were not resolved. This process can be turned off when the end user manually chooses the allele combination, and removes the disapproved allele combination on the basis of their own judgement.
Table 2:
Resolving genotypic ambiguity
| A: Family2A | |||
|---|---|---|---|
| Subject | Relationship | Original | Reduced |
| 767 | child | DPB1*03:01:01+DPB1*05:01:01| DPB1*135:01+DPB1*104:01 |
DPB1*03:01:01+ DPB1*05:01:01 |
| 768 | child | DPB1*03:01:01+DPB1*04:01:01| DPB1*124:01+DPB1*350:01 |
DPB1*03:01:01+ DPB1*04:01:01 |
| 769 | father | DPB1*03:01:01+DPB1*13:01:01| DPB1*03:01:01+DPB1*107:01 |
DPB1*03:01:01+ DPB1*13:01:01/DPB1*107:01 |
| 76A | mother | DPB1*04:01:01:01+DPB1*05:01:01 | DPB1*04:01:01:01+DPB1*05:01:01 |
| B: Family2B | |||
|---|---|---|---|
| Subject | Relationship | Original | Reduced |
| 2B1 | child | DPB1*05:01:01+ DPB1*05:01:01 |
DPB1*05:01:01+ DPB1*05:01:01 |
| 2B2 | child | DPB1*05:01:01+ DPB1*135:01 |
DPB1*05:01:01+ DPB1*135:01 |
| 2B3 | child | DPB1*05:01:01+ DPB1*13:01:01/DPB1*107:01| DPB1*135:01+DPB1*519:01 |
DPB1*05:01:01+ DPB1*13:01:01/DPB1*107:01 |
| 2B4 | father | DPB1*05:01:01+DPB1*135:01 | DPB1*05:01:01+DPB1*135:01 |
| 2B5 | mother | DPB1*05:01:01+ DPB1*13:01:01/DPB1*107:01| DPB1*135:01+DPB1*519:01 |
DPB1*05:01:01+ DPB1*13:01:01/DPB1*107:01 |
| C: | ||||
|---|---|---|---|---|
| Family | Subject | Relationship | Original | Reduced |
| 7457 | 158B | child | DPB1*02:01:02+DPB1*104:01| DPB1*124:01+DPB1*414:01 |
DPB1*02:01:02+DPB1*104:01 |
| 7457 | 158A | mother | DPB1*02:01:02+DPB1*104:01| DPB1*124:01+DPB1*414:01 |
DPB1*02:01:02+DPB1*104:01 |
| 7457 | 1589 | father | DPB1*02:01:02+DPB1*104:01| DPB1*124:01+DPB1*414:01 |
DPB1*02:01:02+DPB1*104:01 |
| 73 | 78B82 | mother | DPB1*04:01:01:01+DPB1*04:02:01:02| DPB1*105:01+DPB1*126:01 |
DPB1*04:01:01:01+DPB1*04:02:01:02 |
| 73 | 78B83 | child | DPB1*04:01:01:01+DPB1*04:02:01:02| DPB1*105:01+DPB1*126:01 |
DPB1*04:01:01:01+DPB1*04:02:01:02 |
| 73 | 78B84 | father | DPB1*04:01:01:01+DPB1*04:02:01:02| DPB1*105:01+DPB1*126:01 |
DPB1*04:01:01:01+DPB1*04:02:01:02 |
| 273 | 1934 | child | DPB1*03:01:01+DPB1*04:01:01:01| DPB1*124:01+DPB1*350:01 |
DPB1*03:01:01+DPB1*04:01:01:01 |
| 273 | 1935 | father | DPB1*03:01:01+DPB1*04:01:01:01| DPB1*124:01+DPB1*350:01 |
DPB1*03:01:01+DPB1*04:01:01:01 |
| 273 | 1936 | mother | DPB1*03:01:01+DPB1*04:01:01:01| DPB1*124:01+DPB1*350:01 |
DPB1*03:01:01+DPB1*04:01:01:01 |
The original HLA genotypes that are obtained from NGS HLA typing software applications are shown in the “Original” column. The prefix “HLA-” is omitted from gene names in this table. Table 2A shows a family (Family2A) in which two children had genotypic ambiguities in their HLA-DPB1 genotypes. These genotype ambiguities are not resolvable when these individual’s HLA genotypes are reviewed. When the father’s (HLA-DPB1*03:01:01+HLA-DPB1*13:01:01/HLA-DPB1*107:01) and mother’s (HLA-DPB1*04:01:01:01+HLA-DPB1*05:01:01) genotypes are reviewed, the only possible genotypes for child1 and child2 are HLA-DPB1*03:01:01+HLA-DPB1*05:01:01 and HLA-DPB1*03:01:01+HLA-DPB1*04:01:01, respectively. Table 2B shows another family (Family2B) in which a child (2B3) and mother (2B5) share genotypic ambiguity. In Family2B, if subject 2B3 were the only child, then it would not be possible to resolve the genotypic ambiguity; the genotypes of children 2B1 and 2B2 made it possible to establish haplotype phase. Table 2C: We identified four families in which all family members had the same ambiguous HLA-DPB1 genotype. Of these four families, Table 2C shows three such families. The fourth family had the same HLA-DPB1 genotype as family 273, and was omitted from the table. This is an uninformative genotype example, in which haplotypes cannot be built using segregation. HaplObserve selected the lowest-digit allele name combination as a default.
2.2.2.4. Sorting children’s alleles
The transmitted HLA alleles of each child are separated into paternal and maternal genotype groups or haplotypes (Supplemental Table 4). The non-transmitted parental alleles are identified for each parent by subtracting the alleles matched with the child. This step produces sets of parental haplotypes for all children.
2.2.2.5. HLA allele copy number adjustment
When a locus is recognized as homozygous, some NGS HLA genotyping software applications report the homozygous allele once (e.g., “HLA-A*02:01:01:01”, which could be interpreted as hemizygous) instead of reporting two identical alleles (e.g., “HLA-A*02:01:01:01+HLA-A*02:01:01:01”). To be consistent, HaplObserve duplicates the potentially hemizygous allele and includes a “+” operator to represent it as truly homozygous for the HLA-A, HLA-C, HLA-B, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1 loci. This rule does not apply for the HLA-DRB3, HLA-DRB4 and HLADRB5 loci, which may be legitimately hemizygous. With a few exceptions, HLA-DRB3, HLA-DRB4 and HLA-DRB5 (HLA-DRB3/4/5) alleles are strongly associated with specific HLA-DRB1 alleles in haplotype groups defined by Andersson [19]. Table 3 shows the DRB haplotype rules. HaplObserve applies these rules to decide the copy numbers of HLA-DRB/3/4/5 alleles and which HLA-DRB3/4/5 alleles are associated with which HLA-DRB1 alleles (Supplemental Table 5). When an HLA-DRB1 and HLA-DRB3/4/5 genotype combination does not follow the Andersson DRB haplotypes, HaplObserve does not force this rule; it leaves the sorted results in step 4 (section 2.2.2.4), and generates a warning message for the user.
Table 3:
Andersson DRB haplotypes
| DRB1 | DRB3 | DRB4 | DRB5 |
|---|---|---|---|
| HLA-DRB1*01 | |||
| HLA-DRB1*08 | |||
| HLA-DRB1*10 | |||
| HLA-DRB1*15 | PRESENT | ||
| HLA-DRB1*16 | PRESENT | ||
| HLA-DRB1*03 | PRESENT | ||
| HLA-DRB1*11 | PRESENT | ||
| HLA-DRB1*12 | PRESENT | ||
| HLA-DRB1*13 | PRESENT | ||
| HLA-DRB1*14 | PRESENT | ||
| HLA-DRB1*04 | PRESENT | ||
| HLA-DRB1*07 | PRESENT | ||
| HLA-DRB1*09 | PRESENT |
PRESENT indicates that HLA-DRB3/4/5 loci are expected based on HLA-DRB1 alleles, while empty cells show the absence of HLA-DRB3/4/5 loci for haplotypes including that DRB1 allele family.
2.2.2.6. Pairwise correction
Table 4A and4B show two families in which both parental and child single locus genotypes are completely identical in a trio. In the case of such an uninformative trio, it is impossible to separate the child HLA genotypes into unique parental genotypes using a segregation strategy. Unlike the DRB haplotypes described in 2.2.2.5, it is difficult to apply specific haplotype rules for the other loci. However, it was possible to find one informative trio that contains distinct genotypes for the specific locus when two or more children are available in the same family (Table 4). When such informative trios are found in a family, HaplObserve builds 2-locus pairwise haplotypes with the neighboring locus alleles from the trio, and deciphers the parental haplotypes. We implemented pairwise correction for HLA-DPA1~HLA-DPB1, HLA-B~HLA-C, and HLA-C~HLA-A.
Table 4A:
Pairwise combination
| Sample | Relationship | HLA-DPA1 | HLA-DPB1 | Informative | HLA-DPA1~ HLA-DPB1 |
|---|---|---|---|---|---|
| 4A1 | child | DPA1*01:03:01:02+ DPA1*01:03:01:04 |
DPB1*04:01:01:01+ DPB1*104:01 |
False | DPA1*01:03:01:04~ DPB1*04:01:01:01+ DPA1*01:03:01:02~ DPB1*104:01 |
| 4A2 | child | DPA1*01:03:01:02+ DPA1*01:03:01:04 |
DPB1*04:01:01:01+ DPB1*104:01 |
False | DPA1*01:03:01:04~ DPB1*04:01:01:01+ DPA1*01:03:01:02~ DPB1*104:01 |
| 4A3 | mother | DPA1*01:03:01:02+ DPA1*01:03:01:04 |
DPB1*04:01:01:01+ DPB1*104:01 |
DPA1*01:03:01:04~ DPB1*04:01:01:01+ DPA1*01:03:01:02~ DPB1*104:01 |
|
| 4A4 | father | DPA1*01:03:01:02+ DPA1*01:03:01:04 |
DPB1*04:01:01:01+ DPB1*104:01 |
DPA1*01:03:01:04~ DPB1*04:01:01:01+ DPA1*01:03:01:02~ DPB1*104:01 |
|
| 4A5 | child | DPA1*01:03:01:04+ DPA1*01:03:01:04 |
DPB1*04:01:01:01+ DPB1*04:01:01:01 |
True |
DPA1*01:03:01:04~ DPB1*04:01:01:01+ DPA1*01:03:01:04~ DPB1*04:01:01:01 |
HLA-DPA1 and HLA-DPB1 columns show the original genotypes presented in GL String format. The prefix “HLA-” is omitted from gene names in this table. “Informative” column indicates whether the child genotypes are informative (True) or not (False) for building haplotypes using allele segregation. Maternal (4A3), paternal (4A4) and two children’s (4A1 and 4A2) genotypes of these loci are completely identical. There are two possible haplotype combinations for mother (4A3), father (4A4) and two children (4A1 and 4A2), thus it is not feasible to separate the children’s HLA alleles into unique parental alleles. These loci for these family members contain uninformative genotypes. However, only one haplotype combination can be built using the third child 4A5’s genotype and the parents’. The haplotypes are shown in bold and underlined letters. Each haplotype for 4A5 is inherited from the parents. When one haplotype is built for the parents, it is possible to build the second haplotype for each parent by subtracting the first haplotype from the parental genotypes.
Table 4B:
Pairwise combination
| Sample | Relationship | HLA-A | HLA-C | Informative | HLA-A~HLA-C |
|---|---|---|---|---|---|
| 3FFE | mother | A*01:01:01:01+ A*24:02:01:01 |
C*08:02:01:02+ C*01:02:01 |
False |
A*01:01:01:01~ C*08:02:01:02+ A*24:02:01:01~ C*01:02:01 |
| 3FFD | father | A*01:01:01:01+ A*24:02:01:01 |
C*04:01:01:06+ C*12:02:02 |
False |
A*01:01:01:01~ C*12:02:02+ A*24:02:01:01~ C*04:01:01:06 |
| 4000 | child | A*01:01:01:01+ A*24:02:01:01 |
C*01:02:01+ C*12:02:02 |
False |
A*01:01:01:01~ C*12:02:02+ A*24:02:01:01~ C*01:02:01 |
| 3FFC | child | A*01:01:01:01+ A*24:02:01:01 |
C*08:02:01:02+ C*04:01:01:06 |
False |
A*01:01:01:01~ C*08:02:01:02+ A*24:02:01:01~ C*04:01:01:06 |
| 3FFF | child | A*01:01:01:01+ A*01:01:01:01 |
C*12:02:02+ C*08:02:01:02 |
True |
A*01:01:01:01~ C*12:02:02+ A*01:01:01:01~ C*08:02:01:02 |
It is not possible to build an unambiguous HLA-A~HLA-C haplotypes from children 4000 and 3FFC, because these two children and their parents have identical HLA-A genotypes. However, it is possible to build a unique haplotype set from a trio with child 3FFF. HaplObserve builds HLA-A~HLA-C haplotypes based on the trio with child 3FFF, and uses the haplotype information to build haplotypes for the remaining two children.
2.2.2.7. Haplotype validation
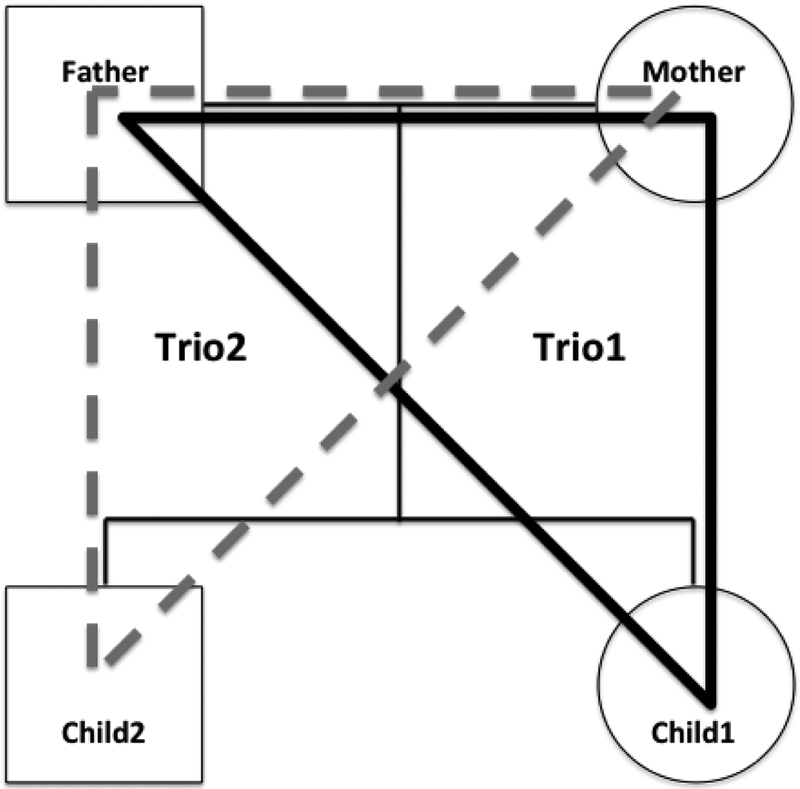
When there are two or more children are available in a family, two or more sets of four haplotypes are independently built based on the number of children (Figure 2), and the set of two paternal haplotypes for a child is compared to the other sets of paternal haplotypes for other children in the same family. Similarly, the set of the mother’s haplotypes for a child is compared to the other sets of mother’s haplotypes for other children. No matter how many children are in the family, the parental haplotypes should be identical unless chromosomal crossover occurred. It is possible to recognize a crossover event in quartet families, but not possible to determine which of the two children inherited the recombinant haplotype. For the families that had more than two children, it is usually possible to identify the parental haplotypes that participated in the crossover events. When the parental haplotypes are consistent for all the children, HaplObserve reports that the haplotype validation is true, indicating no crossover. When HaplObserve reports false in validation in a quartet or larger family, it indicates a possible crossover, which requires critical review of the resulting haplotypes. When only one child is available in a family, the validation result is always false, because there is no haplotype confirmation from additional children, and it is not possible to identify crossover.
Figure 2:
A pedigree of a quartet family is shown with father, mother and two children as indicated with thin solid lines. A set of parental haplotypes is built from Trio1 with father, mother and child1, as shown with a black bold triangle. A second set of parental haplotypes is built independently from Trio2 with father, mother and child2, as shown with a gray bold dotted triangle. The resulting two sets of parental haplotypes are compared for validation; both sets of parental haplotypes are identical if no meiotic crossover event occurred in either of the children. This validation facilitates identification of not only a recombination event but also inconsistent HLA allele segregation within the same haplotype due to HLA typing errors.
2.2.2.8. Output file format
HaplObserve generates CSV files in four formats (Supplemental Tables 6 – 9), three of which include GL String formatted allele-names: 1) “untruncated allele name” phased-haplotypes (Supplemental Table 6), including allelic ambiguities; 2) “unambiguous allele name” phased-haplotypes (Supplemental Table 7), in which ambiguous allele strings are replaced with the allele with the lowest digit in the highest common field; and 3) “two-field allele name” phased haplotypes (Supplemental Table 8), where all allele names have been truncated to 2-fields. HaplOvserve also generates summary CSV file, which contains “Family_ID”, “Sample_ID”, “Relation”, “HLA-A”, “HLA-C”, “HLA-B”, “HLA-DRB345”, “HLA-DRB1”, “HLA-DQA1”, “HLA-DQB1”, “HLA-DPA1”, “HLA-DPB1”, “Validation”, and “Ethnicity/Country” information in separate columns (Supplemental Table 9).
2.3. Updating HLA Haplotype Validator (HLAHapV)
HLAHapV was developed to automate the application of common and well documented (CWD) allele prevalence data [20] and reference haplotype frequency data for assessing haplotypes derived from HLA genotypes and for calculating the likelihood of haplotype combinations [8]. We used HLAHapV to evaluate the haplotypes generated by HaplObserve, using reference haplotype frequency tables generated by the National Marrow Donor Program (NMDP) [21][B]. We improved HLAHapV by enhancing its performance, adding an XML output format (SupplementalHLAHapV) and packaging the program for easy installation. The software accommodates any HLA loci for which reference haplotype frequencies are provided, permitting any properly formatted source data to be used. HLAHapV accepts GL String formatted HLA genotypes as input [8]. The GL Strings can be organized in a tab or comma separated text file or directly imported from Histoimmunogenetics Markup Language (HML) [17] documents generated from the NGS HLA typing software applications. In addition to validating haplotypes derived from HLA genotypes, HLAHapV can also function as a batch-enabled haplotype estimator. Therefore, the software can be potentially used to automate the estimation of haplotype combinations that are often included in a clinical test report for solid organ and hematopoietic stem cell transplantation. The HLAHapV package and its instructions can be obtained from GitHub [C].
2.3.1. Haplotype confirmation
We estimated six-loci (HLA-A~HLA-C~HLA-B~HLA-DRB3/4/5~HLA-DRB1~HLA-DQB1) g group haplotypes [21] using HLAHapV, and compared these with those built using HaplObserve. We were able to correlate almost all of the haplotypes built using HaplObserve with haplotypes estimated using HLAHapV except for the individuals who had rare alleles listed in Table 5.
Table 5:
Rare alleles that could not be used to estimate haplotypes using HLAHapV
| HLA-A*11:199:01 |
| HLA-A*24:284 |
| HLA-B*07:238 |
| HLA-B*40:320 |
| HLA-B*41:18 |
| HLA-B*55:67 |
| HLA-C*06:116N |
| HLA-C*14:18 |
| HLA-DQB1*06:84 |
| HLA-DRB1*08:77 |
| HLA-DRB1*09:21 |
It was not possible to estimate haplotypes from the individuals who had these alleles using HLAHapV, because these alleles did not belong to any g group.
2.3.2. Haplotype adjustment
We visually inspected haplotypes from 204 nuclear families consisting of 820 subjects. When errors were found (Table 6), haplotypes were manually corrected.
2.4. Estimation of Haplotype Frequencies
As noted in section 2.2, HaplObserve can process multi-family data, separate subjects by a specified ethnic group or country, and combine the output data using a single command. We separated data by ethnic group at the subject level, because there are families in which the parents have different ethnic backgrounds. HaplObserve generates a report containing haplotype counts, frequencies, and haplotype rankings within the specified ethnic group. These ranking data allow easy comparison of haplotype frequencies between different ethnic groups. Haplotypes containing missing alleles (“NT”) or alleles that were discordant (denoted as “NoMatch”) are excluded when haplotype counts and frequencies are estimated.
2.5. Estimation of LD
We treated the parents in each family as unrelated individuals for the final haplotype analyses. We developed the “Phased Or Unphased LD” (pould) R package, which accepts either “unambiguous” or “two-field” allele name HaplObserve output file formats. When these files are provided as input, pould calculates D’ [22] and Wn [23], standard measures of global (locus-level) LD, and WLoc1/Loc2 and WLoc2/Loc1, the complementary pair of conditional asymmetric LD (ALD) measures [24, 25]. Pould calculates D’, Wn, WLoc1/Loc2 and WLoc2/Loc1 using phased haplotype information by default or can calculate these measures for EM haplotypes estimated by regenerating the genotypes from the phased HaplObserve output, via an optional argument.
The ALD approach permits the dissection of LD for locus pairs where alleles at the first locus might be in complete LD with alleles at the second locus, but where alleles at the second locus may be in less than complete LD with alleles at the first locus. We also used pould to perform sign tests to compare the measures of LD from phased haplotypes and unphased haplotypes. The pould R package is available for download from GitHub [D].
2.5.1. Pould Data Requirements
Pould requires GL Strings consisting of two tilde (~) delimited HLA haplotypes connected by a plus (+) sign as input, and requires HLA allele names that include complete locus prefixes (e.g., “HLA-A”, “HLA-DRB3”); pould parses these prefixes to identify each locus, but does not perform any additional parsing or validation of HLA allele names. Pould treats unusual allele names (e.g., “HLA DRB1*NoMatch”, “HLA-DPB1*NT”) and truncated versions of allele names (“HLA-A*01”, “HLAA*01:01”, “HLA-A*01:01:01”, etc.) as discrete alleles. The analysis of these unusual allele names or different truncated versions of the same allele name may skew the analytic results, and any inferred parental genotypes that included an allele coded as “NT” were excluded analyses performed with pould.
2.6. Computer operating systems
We tested HaplObserve, HLAHapV and pould on Windows 7, Mac OS X Version 10.9.5 and Linux Red Hat Enterprise Linux Server release 6.4 (version 2.6.32–358.el6.x86_64) operating systems; all of these tools perform as described on these operating systems. The authors welcome comments and feedback from the community on these tools, and will continue to optimize and improve them.
2.7. Software Tools Validation
The software tools and functions described in sections 2.3 - 2.5 were validated iteratively, as the data issues described in each section were encountered and addressed. HaplObserve was developed using a set of 820 subjects in 204 nuclear families. The haplotypes generated for each subject were visually inspected during each development iteration. We used HLAHapV to validate the haplotypes generated by HaplObserve, using CWD prevalence data and the NMDP reference haplotype frequencies to detect potential unexpected HaplObserve-generated haplotypes. Rules we developed to address specific circumstances encountered during this validation process are discussed in section 3.1.
The pould cALD() and LDWrap() functions were validated using the hla.freqs [E] and snp.freqs [26] datasets included with the R asymLD package [F] [25], as well as a publically available HLA dataset (included in the pould package) [27], to ensure that identical LD values were generated by pould’s cALD() and asymLD’s compute.ALD() functions. The LDWrap() function was further validated on the 820 subjects described above, to ensure reporting accuracy.
3. Results
3.1. HaplObserve optimization and validation
As noted in section 2.7, haplotypes for 802 subjects in 204 nuclear families were applied to develop and optimized HaplObserve. As a result of this optimization process, we implemented a set of rules defining locus-specific exceptions associated with specific categories of HLA genotyping results.
We set a “perfect match” requirement for class I genes (aka, “perfect match rule”), but accepted two-field allele name concordance for class II genes (aka, “two-field match rule”), except for HLA-DPA1 and HLA-DPB1 (discussed below). For HLA-DRB3/4/5, HLA-DRB1, HLA-DQA1 and HLA-DQB1 loci, HaplObserve reports ambiguous alleles for children when untruncated HLA allele names are inconsistent but concordant for their two-field allele names. For example, if a child’s allele is reported as HLA-DQA1*01:03:01:02 and the corresponding parental allele as HLA-DQA1*01:03:01:01, then HaplObserve reports the child’s allele as HLA-DQA1*01:03:01:01/HLA-DQA1*01:03:01:02. In these cases, the user needs to decide whether the ambiguities are acceptable or if HLA genotypes should be corrected.
In addition, we implemented a pairwise haplotype correction step. HaplObserve reports errors when inconsistent allele segregation occurs for the HLA-A, HLA-C, HLA-B, HLA-DPA1 and HLA-DPB1 loci. We decided to implement perfect match rules for HLA-DPA1 and HLA-DPB1. We found that all NGS HLA typing software applications had difficulties calling common five HLA-DPA1 alleles, HLADPA1*01:03:01:01, HLA-DPA1*01:03:01:02, HLA-DPA1*01:03:01:03, HLA-DPA1*01:03:01:04 and HLA-DPA1*01:03:01:05, as heterozygous, although the heterozygous genotype options were usually displayed in the applications’ graphical interfaces. In our experience, it was possible to distinguish these five HLA-DPA1 alleles that differ in the fourth-field. It was difficult to build correct HLADPA1~HLA-DPB1 haplotypes using allele segregation if a two-field match rule was applied to HLADPA1 locus, because these five HLA-DPA1 alleles are two-field concordant. Therefore, the two-field concordant rules are not applicable for HLA-DPA1~HLA-DPB1, and HaplObserve requires perfect HLA-DPA1 and HLA-DPB1 genotype matches between a child and its parents.
3.2. Genotypic ambiguity
HaplObserve chooses the lowest-digit allele name combination as the priority HLA genotype if genotypic ambiguity is not resolved due to the lack of informative family members. We investigated whether this process would influence the overall haplotype frequency calculation. We reviewed HLADPB1 genotypes for 4949 samples from 1442 nuclear families that were registered for various projects in the 17th IHIW database (Section 2.1.1). There were 488 families that contained genotypic ambiguity genotypes that could not be converted to be allelic ambiguities for at least one family member. In 484 of 488 families (>99%), HaplObserve identified a single allele combination by reviewing all the genotypes in the family and assessing segregation (Table 2AB), and using pairwise correction (Table 4). The lowest-digit alleles were segregated to the offspring for all the 484 families. We identified only four families that had uninformative HLA-DPB1 genotypes in which all family members had the same ambiguous HLA-DPB1 genotype (Table 2C); in these cases, HaplObserve selected the lowest-digit allele name combination as a default.
3.3. Haplotype adjustment
We also found that in 7 families it was not feasible to automatically assign the correct haplotypes using current HaplObserve. Table 6A shows a family that had uninformative HLA-DQB1 genotypes, in which haplotypes cannot be built using segregation. Table 6B shows a family that had uninformative HLA-B genotypes. Similar to the family in Table 6A, we applied HLAHapV to identify known haplotypes for these uninformative segregation cases. Initially, HLAHapV failed to assign haplotypes with HLA-C*02:10:01:01, and only a single likely HLA-C~HLA-B haplotype (HLA-C*04:01g~HLAB*53:01) was identified. HLA alleles with identical nucleotide sequences of exons 2 and 3 for class I and exon 2 for class II are organized as G groups, while HLA alleles with identical amino acid sequences of these exons are summarized as P groups [28]. HLA alleles were also organized as g (lower case) groups, which are equivalent to P groups including null alleles [8, 29]. HLAHapV uses a g group conversion table [8, 21, 29]. HLA-C*02:10 was renamed from HLA-C*02:02:04 in 2006, thus HLA-C*02:10:01:01 conversion failed, because this allele name change did not follow the g group rule. The NMDP haplotype frequency tables do not contain any haplotypes that include HLA-C*02:10, though they contain haplotypes that include HLA-C*02:02g [21]. We incorporated an exception to the g group conversion rule allowing C*02:10 to be converted to HLA-C*02:02g (Table 6B), in order to achieve consistency with historical NMDP practice when using these frequencies.
Table 6A:
Manually edited HLA-DRB1~HLA-DQB1 haplotypes
| Sample | Relation | HLA-DRB1 | HLA-DQB1 | HLAHapV | Final |
|---|---|---|---|---|---|
| 4584 | child | DRB1*08:04:01+ DRB1*15:03:01:01 |
DQB1*03:19:01+ DQB1*06:02:01 |
DRB1*08:04~DQB1*03:01g+ DRB1*15:03~DQB1*06:02 |
DRB1*08:04:01~DQB1*03:19:01+ DRB1*15:03:01:01~DQB1*06:02:01 |
| 4585 | child | DRB1*15:03:01:01+ DRB1*11:02:01 |
DQB1*03:19:01+ DQB1*06:02:01 |
DRB1*15:03~DQB1*06:02+ DRB1*11:02~DQB1*03:01g |
DRB1*15:03:01:01~DQB1*06:02:01+ DRB1*11:02:01~DQB1*03:19:01 |
| 4582 | father | DRB1*08:04:01+ DRB1*15:03:01:01 |
DQB1*03:19:01+ DQB1*06:02:01 |
DRB1*08:04~DQB1*03:01g+ DRB1*15:03~DQB1*06:02 |
DRB1*08:04:01~DQB1*03:19:01+ DRB1*15:03:01:01~DQB1*06:02:01 |
| 4583 | mother | mother DRB1*15:03:01:01+ DRB1*11:02:01 |
DQB1*03:19:01+ DQB1*06:02:01 |
DRB1*15:03~DQB1*06:02+ DRB1*11:02~DQB1*03:01g |
DRB1*15:03:01:01~DQB1*06:02:01+ DRB1*11:02:01~DQB1*03:19:01 |
Table 6B:
Manually edited HLA-C~HLA-B haplotypes
| Sample | Relation | HLA-C | HLA-B | HapV (g group) | HapV (IHIW) | Final |
|---|---|---|---|---|---|---|
| 769 | child | C*02:10:01:01+ C*04:01:01:01 |
B*15:03:01:02+ B*53:01:01 |
C*02:02g~B*15:03g+ C*04:01g~B*53:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
| 765 | child | C*02:10:01:01+ C*04:01:01:01 |
B*15:03:01:02+ B*53:01:01 |
C*02:02g~B*15:03g+ C*04:01g~B*53:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
| 766 | child | C*02:10:01:01+ C*04:01:01:01 |
B*15:03:01:02+ B*53:01:01 |
C*02:02g~B*15:03g+ C*04:01g~B*53:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
| 76A | child | C*02:10:01:01+ C*07:18 |
B*15:03:01:02+ B*53:01:01 |
C*02:02g~B*15:03g+ C*07:01g~B*53:01 |
C*02:10:01:01~B*15:03:01:02+ C*07:18~B*53:01:01 |
C*02:10:01:01~B*15:03:01:02+ C*07:18~B*53:01:01 |
| 767 | mother | C*02:10:01:01+ C*07:18 |
B*15:03:01:02+ B*53:01:01 |
C*02:02g~B*15:03g+ C*07:01g~B*53:01 |
C*02:10:01:01~B*15:03:01:02+ C*07:18~B*53:01:01 |
C*02:10:01:01~B*15:03:01:02+ C*07:18~B*53:01:01 |
| 768 | father | C*02:10:01:01+ C*04:01:01:01 |
B*15:03:01:02+ B*53:01:01 |
C*02:02g~B*15:03g+ C*04:01g~B*53:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
C*02:10:01:01~B*15:03:01:02+ C*04:01:01:01~B*53:01:01 |
Table 6C and 6D show two families that haplotypes were built inconsistently among children using HaplObserve (Table 6C and 6D). The inconsistencies were not due to chromosomal crossover. The inconsistencies were between HLA-DQB1 and HLA-DPA1~HLA-DPB1 haplotypes. It was not possible to use the NMDP haplotype frequency tables for these families, because they do not contain information about HLA-DPA1 and HLA-DPB1. When inconsistent haplotypes were built within the same family, HaplObserve indicated “false” for validation results. We carefully reviewed the families that contained false validation results, and manually corrected the haplotypes if possible. It is very important to review the validation results for each family.
Table 6C:
Manually edited HLA-DQB1~HLA-DPA1~HLA-DPB1 haplotypes
| Sample | Relation | Likely incorrect | Corrected |
|---|---|---|---|
| 715 | child | DQB1*05:02:01~DPA1*01:03:01:02~DPB1*04:01:01:01+ DQB1*03:02:01~DPA1*01:03:01:02~DPB1*104:01 |
DQB1*05:02:01~DPA1*01:03:01:02~DPB1*104:01+ DQB1*03:02:01~DPA1*01:03:01:02~DPB1*04:01:01:01 |
| 716 | child | DQB1*05:02:01~DPA1*01:03:01:02~DPB1*104:01+ DQB1*03:01:01:02~DPA1*01:03:01:02~DPB1*104:01 |
DQB1*05:02:01~DPA1*01:03:01:02~DPB1*104:01+ DQB1*03:01:01:02~DPA1*01:03:01:02~DPB1*104:01 |
| 717 | father | DQB1*03:02:01~DPA1*01:03:01:02~DPB1*04:01:01:01+ DQB1*03:01:01:02~DPA1*01:03:01:02~DPB1*104:01 |
DQB1*03:02:01~DPA1*01:03:01:02~DPB1*04:01:01:01+ DQB1*03:01:01:02~DPA1*01:03:01:02~DPB1*104:01 |
| 718 | mother | DQB1*05:02:01~DPA1*01:03:01:02~DPB1*104:01+ DQB1*05:01:01:03~ DPA1*01:03:01:02~DPB1*04:01:01:01 |
DQB1*05:02:01~DPA1*01:03:01:02~DPB1*104:01+ DQB1*05:01:01:03~ DPA1*01:03:01:02~DPB1*04:01:01:01 |
Table 6D:
Manually edited HLA-DQB1~HLA-DPA1~HLA-DPB1 haplotypes
| Sample | Relation | Likely incorrect | Corrected |
|---|---|---|---|
| 10C3 | child | DQB1*02:01:01~DPA1*01:03:01:03~DPB1*06:01:01+ DQB1*03:02:01~DPA1*01:03:01:05~DPB1*04:02:01:02 |
DQB1*02:01:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*03:02:01~DPA1*01:03:01:03~DPB1*06:01:01 |
| 10C7 | child | DQB1*02:01:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*04:02:01~DPA1*01:03:01:05~DPB1*04:02:01:02 |
DQB1*02:01:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*04:02:01~DPA1*01:03:01:05~DPB1*04:02:01:02 |
| 10C4 | father | DQB1*02:01:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*03:02:01~DPA1*01:03:01:03~DPB1*06:01:01 |
DQB1*02:01:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*03:02:01~DPA1*01:03:01:03~DPB1*06:01:01 |
| 10C5 | mother | DQB1*04:02:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*03:02:01~DPA1*01:03:01:03~DPB1*06:01:01 |
DQB1*04:02:01~DPA1*01:03:01:05~DPB1*04:02:01:02+ DQB1*03:02:01~DPA1*01:03:01:03~DPB1*06:01:01 |
Table 6A shows a family that had two children, a father and a mother, all of whom had identical HLA-DQB1 genotypes. This is an uninformative genotype example, in which haplotypes cannot be built using segregation. In this case, we applied HLAHapV to review known haplotypes in order to identify the most likely HLA-DRB1~HLA-DQB1 haplotypes [8]. The “HLAHapV” column shows the high priority haplotypes. The “Final” column shows the manually edited haplotypes based on the HLAHapV results. We confirmed the haplotypes by executing HLAHapV using the haplotype frequency tables from the IHIW17 Family Haplotype Project (data not shown). Table 6B shows another uninformative family of three children and both parents, all of whom had identical HLA-B genotypes. Similar to Table 6A, we initially applied HLAHapV using g group haplotype table to identify known haplotypes for these uninformative segregation cases [column “HapV (g group)”]. Later we performed HLAHapV analyses using the accumulated haplotypes of the IHIW17 Family Haplotype Project to identify most likely haplotypes for these uninformative segregation cases [column “HapV (IHIW)”]. Tables 6C and 6D show families for which haplotypes were manually corrected after visual inspection. The HLA-DQB1~HLA-DPA1~HLA-DPB1 haplotype frequency is not available from NMDP.
3.4. Measures of LD from Phased and Unphased haplotypes
We initially validated pould using the test dataset of 838 published DRB1~DQB1 haplotypes for 419 individuals [27] included in the package. We then calculated D’, Wn, WLoc1/Loc2 and WLoc2/Loc1 from both phased haplotypes built from families using HaplObserve and EM-estimated haplotypes built by pould assuming no phase between loci. During the validation of pould, we observed that LD measures for EM-estimated unphased haplotypes were generally higher than those measures for their phased haplotype counterparts. For the test DRB1~DQB1 dataset included in the package, pould built 53 unique EM haplotypes, while the dataset includes 105 unique phased haplotypes. Five EM haplotypes were not found among the phased haplotypes, while 48 phased haplotypes were not found among the EM haplotypes. These 48 phased haplotypes are rare (counts 1–3), indicating that the EM algorithm is returning fewer low frequency haplotypes than are present in the phased data. The EM algorithm thus underestimates the frequency of rare (n<4) haplotypes, thereby overestimating the LD of those 2-locus haplotypes.
To further investigate the phenomena of overestimation of LD for EM estimated haplotypes, we calculated measures of LD for all possible 2-locus phased and unphased haplotypes built from 17th IHIW families using pould. We compared measures of LD values of 2-locus haplotypes for both phased and unphased haplotypes, and performed sign tests of the differences between the LD measures (Table 7). It is apparent that the differences between LD values for the phased and unphased haplotypes trends significantly toward higher LD values for unphased haplotypes, and the number of 2-locus haplotypes trends toward lower numbers of unphased haplotypes than phased. The majority of p-values from the sign test are statistically significant (p < 0.05), except for the family haplotypes from Austria, Egypt and Switzerland. The number of families analyzed for these countries were small (10, 14 and 11 respectively), which is likely the major factor contributing to the non-significant sign test results. This phenomenon is more pronounced where 2-locus LD is weaker, such as for HLA-A~HLA-C, HLA-B~HLA-DRB1 and HLA-DQB1~HLA-DPA1. The weaker LD indicates more haplotype variations or more rare combinations of these loci (not shown).
Table 7:
Sign Test Comparisons of LD Measures in Phased and Unphased Haplotypes
| Ethnicity/Country | D’ | Wn | WLoc1/Loc2 | WLoc2/Loc1 | N_Haplotypes | |
|---|---|---|---|---|---|---|
| African American 2N = 144 |
unphased > phased | 44 | 42 | 45 | 46 | 12 |
| unphased = phased | 5 | 4 | 4 | 4 | 7 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 8.70E-06 | 0.000114 | 2.06E-06 | 4.34E-07 | 3.31E-05 | |
| European American 2N = 424 |
unphased > phased | 48 | 45 | 49 | 47 | 15 |
| unphased = phased | 2 | 2 | 2 | 2 | 5 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 1.31E-08 | 2.06E-06 | 1.82E-09 | 8.07E-08 | 0.001016 | |
| Asian American 2N = 230 |
unphased > phased | 42 | 42 | 43 | 44 | 17 |
| unphased = phased | 2 | 2 | 2 | 2 | 8 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 0.000114 | 0.000114 | 3.31E-05 | 8.70E-06 | 0.006456 | |
| Hispanic 2N = 232 |
unphased > phased | 43 | 45 | 45 | 43 | 16 |
| unphased = phased | 1 | 1 | 2 | 2 | 6 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 3.31E-05 | 2.06E-06 | 2.06E-06 | 3.31E-05 | 0.002667 | |
| ARGENTINA 2N = 348 |
unphased > phased | 43 | 49 | 46 | 46 | 18 |
| unphased = phased | 0 | 0 | 0 | 0 | 2 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 3.31E-05 | 1.82E-09 | 4.34E-07 | 4.34E-07 | 0.014454 | |
| AUSTRIA 2N = 40 |
unphased > phased | 34 | 42 | 37 | 39 | 20 |
| unphased = phased | 7 | 6 | 10 | 10 | 16 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 0.104789 | 0.000114 | 0.014454 | 0.002667 | 0.058064 | |
| CHINA 2N = 932 |
unphased > phased | 15 | 14 | 15 | 15 | 1 |
| unphased = phased | 0 | 0 | 0 | 0 | 1 | |
| locus pairs | 15 | 15 | 15 | 15 | 15 | |
| p-values | 6.10E-05 | 0.000977 | 6.10E-05 | 6.10E-05 | 0.000977 | |
| CZECH REPUBLIC 2N = 336 |
unphased > phased | 48 | 51 | 48 | 49 | 16 |
| unphased = phased | 1 | 1 | 2 | 2 | 7 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 1.31E-08 | 2.05E-11 | 1.31E-08 | 1.82E-09 | 0.002667 | |
| EGYPT 2N = 56 |
unphased > phased | 35 | 33 | 33 | 33 | 21 |
| unphased = phased | 10 | 9 | 10 | 9 | 13 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 0.058064 | 0.177001 | 0.177001 | 0.177001 | 0.104789 | |
| GERMANY 2N = 276 |
unphased > phased | 47 | 49 | 48 | 48 | 11 |
| unphased = phased | 4 | 3 | 4 | 4 | 8 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 8.07E-08 | 1.82E-09 | 1.31E-08 | 1.31E-08 | 8.70E-06 | |
| GREECE 2N = 100 |
unphased > phased | 41 | 44 | 42 | 42 | 16 |
| unphased = phased | 7 | 7 | 7 | 7 | 15 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 0.000355 | 8.70E-06 | 0.000114 | 0.000114 | 0.002667 | |
| ITALY 2N = 268 |
unphased > phased | 21 | 21 | 21 | 21 | 0 |
| unphased = phased | 0 | 0 | 0 | 0 | 0 | |
| locus pairs | 21 | 21 | 21 | 21 | 21 | |
| p-values | 9.54E-07 | 9.54E-07 | 9.54E-07 | 9.54E-07 | 9.54E-07 | |
| KUWAIT 2N = 116 |
unphased > phased | 41 | 44 | 42 | 44 | 13 |
| unphased = phased | 5 | 5 | 6 | 6 | 11 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 0.000355 | 8.70E-06 | 0.000114 | 8.70E-06 | 0.000114 | |
| SWITZERLAND 2N = 44 |
unphased > phased | 36 | 33 | 38 | 36 | 19 |
| unphased = phased | 9 | 9 | 10 | 10 | 18 | |
| locus pairs | 55 | 55 | 55 | 55 | 55 | |
| p-values | 0.030029 | 0.177001 | 0.006456 | 0.030029 | 0.030029 |
The “locus pairs” rows show the total number of locus pairs built for the ethnic group or country. The “unphased > phased” rows indicate the number of locus pair where value of each category shown at the top for unphased haplotypes is larger than that for phased. The “unphased = phased” rows indicate the number of locus pairs where the value of each category for both unphased and phased haplotypes are equal. For example, 44 in the “unphased > phased” row and D’ column means that 44 of 55 locus pairs had higher D’ value for unphased haplotypes; 5 in the “unphased = phased” row and D’ column indicates that 5 of 55 locus pairs had equal value. Sign tests were performed, and p-values are shown for each category. The uncorrected threshold of significance for these p-values is 0.05. Correcting for 14 comparisons in each category [32], the threshold of significance is 3.57E-03; p-values below this threshold are underlined. Correcting for all 70 comparisons in the table, the threshold of significance is 7.14E-04; p-values below this threshold are shown in bold.
4. Discussion
We developed and updated three computational tools for analyzing family-based HLA haplotypes. In developing HaplObserve, we had to make a few assumptions and use knowledge that has been accumulated during the 17th IHIW to account for allele and genotype reporting variation among NGS HLA genotyping software applications. The DRB haplotype adjustment in HaplObserve was essential for building accurate haplotypes, as no NGS HLA genotyping software application currently reports copy numbers of HLA-DRB3/4/5 loci correctly. HaplObserve modifies the copy number of HLADRB3/4/5 loci based on HLA-DRB1 genotypes (Table 3 and Supplemental Table 5). It is, however, the end user’s responsibility to review HLA-DRB3/4/5 copy numbers, since it is impossible to predict all unusual haplotypes that may not follow Andersson’s DRB haplotype rule [19].
The pairwise analysis was especially useful for building HLA-DPA1~HLA-DPB1 haplotype segments (Table 4A). We also implemented a haplotype validation step for cases when two or more children and both parents are present in a family (Figure 2). This validation has been helpful for identifying situations that cannot be explained without chromosomal crossover and/or potential HLA genotyping errors. Historically, HLA genotyping has been focused on returning two-field level allele names, with much less effort given to eliminating ambiguity in the third and fourth fields. HLA genotype errors appear to occur most frequently in the fourth field of HLA-DPA1. We noticed that current NGS HLA genotyping software applications tend to assemble consensus sequences for single HLA-DPA1 alleles. We suggest that there remains room for improvement for the current NGS HLA genotyping systems, as the process of capturing intron sequences is still evolving. HaplObserve does not work unless consistent HLA genotypes are provided within families. In other words, building haplotypes from families using HaplObserve requires clean data sets that have no genotype discordances/dropouts. If even a single locus genotype dropout existed for one of the family members, HaplObserve would either fail to build haplotypes for the family or report “NoMatch” for the locus. This is a consequence of building haplotypes using allele segregation from families. In our experience, the most time-consuming part of building HLA haplotypes from families was to identify and correct the allele name inconsistencies in a family. In contrast, the EM estimation of haplotypes is more tolerant to missing information, and new methods have extended the EM approach to allow the imputation of haplotypes from genotype data including ambiguous genotypes and data of multiple allele name resolutions [30, 31]. Haplotypes have been built from unrelated subjects using EM estimation as another 17th IHIW projects, and the haplotype frequency tables are available from the 17th IHIW website [G].
For the 17th IHIW projects, we applied these tools to build haplotypes for 1017 subjects in 263 families collected from the US clinical laboratories (Osoegawa et al., submitted). Additionally, HaplObserve has been used to build haplotypes for additional 630 families from the various 17th IHIW projects (Osoegawa et al., manuscript in preparation), and the haplotype frequency tables are available from the 17th IHIW website [G].
As the IHIW efforts continue and the community accumulates more high-quality NGS HLA genotype data, the tools described here can be used to enrich and improve our knowledge of HLA haplotypes. After four decades of investigation, we have assessed only a small fraction of the HLA diversity of the human population. Calculating and comparing LD from different populations will enrich our knowledge about HLA haplotypes, and the evolution and migration of human populations. Finally these tools can be used to reproduce multi-locus analysis results and expand our knowledge in the same manner in the future. Computational tools are essential component of reproducible science and enrich our knowledge.
Supplementary Material
Acknowledgements
We thank the Stanford Blood Center for the support and promotion of the 17th IHIW endeavor, Tamara Vayntrub and Susan Twietmeyer for their tremendous administrative support of 17th IHIW efforts. We also thank Kalyan C. Mallempati, Sridevi Gangavarapu and Karl Kornel for their technical support and Jorge R. Oksenberg for providing DNA samples from families, Medhat Askar for organizing 17th IHIW Family Haplotype project and the histocompatibility and immunogenetics community and the International HLA and Immunogenetics Workshop Council for their continued dedication to and support of the International Workshops. The work described here was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) grant R01AI128775 (SM) and National Institute of Neurological Disorders and Stroke (NINDS) grant U19NS095774 (MFV). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIAID, NINDS, NIH or United States Government.
Abbreviations:
- CSV
Comma-Separated Values
- CWD
Common and Well Documented
- GL
Genotype List
- HLA
Human Leukocyte Antigen
- HML
Histoimmunogenetics Markup Language
- IHIW
International HLA and Immunogenetics Workshop
- IMGT
ImMunoGeneTics
- IPD
ImmunoPolymorphism Database
- MIRING
Minimum Information for Reporting Immunogenomic NGS Genotyping
- NGS
Next Generation Sequencing
- NMDP
National Marrow Donor Program
- SNP
Single Nucleotide Polymorphism
- STR
Short Tandem Repeat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interests
The authors declared no conflicting interests.
Online Resources
GitHub HaplObserve Repository, https://github.com/ihiw/haplObserve, accessed December 21, 2018; HaplObserve release versions are available in the /releases directory.
NMDP Haplotype Frequencies Webpage, https://bioinformatics.bethematchclinical.org/hla-resources/haplotype-frequencies/be-the-match-registry-haplotype-frequencies/, accessed December 21, 2018
GitHub HLAHapV Repository, https://github.com/nmdp-bioinformatics/ImmunogeneticDataTools, accessed December 21, 2018
GitHub Pould Repository, https://github.com/IHIW/pould, accessed December 21, 2018
Wilson, C., 2010 Identifying polymorphisms associated with risk for the development of myopericarditis following smallpox vaccine. The Immunology Database and Analysis Portal (ImmPort), Study #26, https://immport.niaid.nih.gov/immportWeb/clinical/study/displayStudyDetails.do?itemList=SDY26, accessed December 26, 2018
CRAN Repository Page for the asymLD package, https://cran.r-project.org/web/packages/asymLD/index.html, accessed December 21, 2018
The 17th IHIW NGS HLA data, http://17ihiw.org/17th-ihiw-ngs-hla-data/, accessed December 26, 2018
References
- [1].Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P et al. : Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 2004;14:1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L et al. : The DNA sequence and analysis of human chromosome 6. Nature 2003;425:805. [DOI] [PubMed] [Google Scholar]
- [3].Shiina T, Hosomichi K, Inoko H, Kulski JK: The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 2009;54:15. [DOI] [PubMed] [Google Scholar]
- [4].Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG: The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 2015;43:D423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Slatkin M: Linkage disequilibrium-understanding the evolutionary past and mapping the medical future. Nat Rev Genet 2008;9:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Choo SY: The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med J 2007;48:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Trowsdale J, Knight JC: Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet 2013;14:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Osoegawa K, Mack SJ, Udell J, Noonan DA, Ozanne S, Trachtenberg E et al. : HLA Haplotype Validator for quality assessments of HLA typing. Hum Immunol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bochtler W, Gragert L, Patel ZI, Robinson J, Steiner D, Hofmann JA et al. : A comparative reference study for the validation of HLA-matching algorithms in the search for allogeneic hematopoietic stem cell donors and cord blood units. HLA 2016;87:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chang CJ, Osoegawa K, Milius RP, Maiers M, Xiao W, Fernandez-Vina M et al. : Collection and storage of HLA NGS genotyping data for the 17th International HLA and Immunogenetics Workshop. Hum Immunol 2018;79:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Milius RP, Mack SJ, Hollenbach JA, Pollack J, Heuer ML, Gragert L et al. : Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens 2013;82:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E et al. : Defining KIR and HLA Class I Genotypes at Highest Resolution via High-Throughput Sequencing. Am J Hum Genet 2016;99:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Geneugelijk K, Wissing J, Koppenaal D, Niemann M, Spierings E: Computational Approaches to Facilitate Epitope-Based HLA Matching in Solid Organ Transplantation. J Immunol Res 2017;2017:9130879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Paunic V, Gragert L, Schneider J, Muller C, Maiers M: Charting improvements in US registry HLA typing ambiguity using a typing resolution score. Hum Immunol 2016;77:542. [DOI] [PubMed] [Google Scholar]
- [16].Sauter J, Schafer C, Schmidt AH: HLA Haplotype Frequency Estimation from Real-Life Data with the Hapl-o-Mat Software. Methods Mol Biol 2018;1802:275. [DOI] [PubMed] [Google Scholar]
- [17].Milius RP, Heuer M, Valiga D, Doroschak KJ, Kennedy CJ, Bolon YT et al. : Histoimmunogenetics Markup Language 1.0: Reporting next generation sequencing-based HLA and KIR genotyping. Hum Immunol 2015;76:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF et al. : High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc Natl Acad Sci U S A 2012;109:8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Andersson G: Evolution of the human HLA-DR region. Front Biosci 1998;3:d739. [DOI] [PubMed] [Google Scholar]
- [20].Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D et al. : Common and well-documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens 2013;81:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gragert L, Madbouly A, Freeman J, Maiers M: Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 2013;74:1313. [DOI] [PubMed] [Google Scholar]
- [22].Hedrick PW: Gametic disequilibrium measures: proceed with caution. Genetics 1987;117:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cramér H: Mathematical Methods of Statistics Princeton University Press; 1999. [Google Scholar]
- [24].Thomson G, Single RM: Conditional asymmetric linkage disequilibrium (ALD): extending the biallelic r2 measure. Genetics 2014;198:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Single RM, Strayer N, Thomson G, Paunic V, Albrecht M, Maiers M: Asymmetric linkage disequilibrium: Tools for assessing multiallelic LD. Hum Immunol 2016;77:288. [DOI] [PubMed] [Google Scholar]
- [26].de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J et al. : A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006;38:1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mack SJ, Udell J, Cohen F, Osoegawa K, Hawbecker SK, Noonan DA et al. : High resolution HLA analysis reveals independent class I haplotypes and amino-acid motifs protective for multiple sclerosis. Genes Immun 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA et al. : Nomenclature for factors of the HLA system, 2010. Tissue Antigens 2010;75:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schmidt AH, Baier D, Solloch UV, Stahr A, Cereb N, Wassmuth R et al. : Estimation of high- resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Hum Immunol 2009;70:895. [DOI] [PubMed] [Google Scholar]
- [30].Madbouly A, Gragert L, Freeman J, Leahy N, Gourraud PA, Hollenbach JA et al. : Validation of statistical imputation of allele-level multilocus phased genotypes from ambiguous HLA assignments. Tissue Antigens 2014;84:285. [DOI] [PubMed] [Google Scholar]
- [31].Schafer C, Schmidt AH, Sauter J: Hapl-o-Mat: open-source software for HLA haplotype frequency estimation from ambiguous and heterogeneous data. BMC Bioinformatics 2017;18:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bland JM, Altman DG: Multiple significance tests: the Bonferroni method. BMJ 1995;310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.