Abstract
Autophagy is a mechanism by which cellular substances are transported to lysosomes for degradation, allowing the basic transformation of cellular components, and providing energy and macromolecular precursors. In cancer, the contradictory role of autophagy in tumor suppression and promotion has been widely acknowledged. Activation and suppression of autophagy have been proposed as cancer therapies, resulting in targeted treatment of cancer by autophagy being considered ambiguous. The dynamic effect of autophagy can also be applied to hepatocellular carcinoma (HCC), a malignant tumor with high incidence and a low survival rate. In this review, we introduce characteristics of different types of autophagy and summarize which genes, non-coding RNAs, and related signaling pathways are involved in autophagy and the regulation of the formation and progress of HCC. More importantly, we discuss the role of autophagy in the treatment of HCC, such as in traditional chemotherapy, molecular targeted drugs, and natural products.
Keywords: Autophagy, hepatocellular carcinoma, signaling pathways, noncoding RNAs, therapy
Introduction
Autophagy is an intracellular catabolic pathway. By removing misfolded proteins, damaged organelles, and lipid droplets, autophagy plays a crucial role in energy balance and cytoplasmic quality control, and promotes liver homeostasis [1,2]. Autophagy is active at a basic level in cells. It may be further upregulated in response to several types of stress that interfere with cell homeostasis, such as low ATP levels, nutrient and growth factor deficiency, hypoxic conditions, endoplasmic reticulum (ER) stress, pathogen entry, or anticancer drugs [3]. The role of autophagy in cancer is important. It is believed that autophagy can prevent the development of cancer. However, once cancer has formed, increased autophagic flux tends to enable cancer cells to survive and grow [4,5]. Accordingly, a major challenge in cancer treatment is should we try to enhance or inactivate autophagy?
Hepatocellular carcinoma (HCC) is ranked as the sixth most common cancer and the third leading cause of cancer death [6]. HCC is one of the leading causes of cancer-related death worldwide and is refractory to nearly all currently available anti-cancer therapies [7,8]. Despite new breakthroughs in treatment and surgery, the 5-year survival rate remains unsatisfactory [9]. In addition, the use of anticancer drugs to treat HCC is limited by the occurrence of primary and acquired drug resistance [10,11]. Increasing numbers of studies have shown that autophagy greatly affects HCC. Autophagy is associated with risk factors for HCC, such as oxidative stress, persistent inflammation, viral infection, metabolic dysfunction, liver alcohol disorders, and fatty liver disease [5,12-14]. Therefore, a comprehensive understanding of the role of autophagy in HCC may be beneficial to develop new diagnostic and therapeutic techniques. From a therapeutic point of view, understanding whether, when, and how autophagy can be used to cure HCC remains a challenge.
In this review, we summarize the characteristics of autophagy and focus on some new research hotspots, such as non-traditional autophagy, secretory autophagy, and selective autophagy. We then summarize which genes, non-coding RNAs, and related signaling pathways are involved in autophagy, and the regulation of the formation and progress of HCC. Finally, we discuss the role of autophagy in the treatment of HCC.
Autophagy
The characteristics of autophagy
Autophagy mainly has three forms: Microautophagy, macroautophagy (referred to hereafter as autophagy), and chaperone-mediated autophagy. Macroautophagy is an evolutionarily conserved metabolic process, including the formation of double membrane vesicles called autophagosomes [15]. In the formation of autophagosomes, a portion of the cytoplasm, which may include organelles, protein aggregates, and lipid droplets, is sequestered in large quantities or selectively [16]. Then, the outer membrane of the autophagosome fuses with a lysosome (forming an autolysosome), which leads to degradation of the enclosed materials along with the inner membrane of the autophagosome. The amino acids and other small molecules produced by autophagic degradation are transported back to the cytoplasm for recycling or energy production. Autophagy is controlled by a series of highly regulated signaling events that occur at a basic level in all cells, and may be induced by different signals and cellular stresses [17]. Macroautophagy is mediated by a group of evolutionarily conserved genes, termed autophagy-related genes (ATGs), which were first found via yeast gene screening [18]. To date, more than 40 ATGs have been identified and their functions have been extensively evaluated. ATGs are involved in the formation, nucleation, expansion, and elongation of autophagic membranes, the binding and fusion with lysosomes, and the degradation of intracapsular products. With a few exceptions, all ATG genes are required for efficient fusion of autophagosomes and lysosomes [19]. The process of autophagy mainly comprises the following processes: (a) The activation of the ULK1 (Unc-51 like autophagy activating kinase 1, also known as ATG1) complex [20]; (b) the activation of the class III PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase) complex, ATG14, Beclin 1, p63, and AMBRA1 (autophagy and beclin 1 regulator 1) [21]; (c) the ATG5ATG12 complex then conjugates with ATG16 to expand the autophagosome membrane and members of the LC3 (microtubule associated protein 1 light chain 3 alpha) and GABARAP (GABA type A receptor-associated protein) families of proteins are conjugated to the lipid phosphatidylethanolamine (PE) and recruited to the membrane [22,23]; (d) ATG4B, in conjunction with ATG7, conjugates LC3-I and PE to form LC3-II [24]; and finally (e) autophagic degradation. Microautophagy involves inward invagination of the lysosomal membrane, which delivers a small portion of the cytoplasm into the lysosomal lumen [25]. Chaperone-mediated autophagy (CMA) is a pathway for protein degradation in intracellular lysosomes. Chaperone-mediated autophagy involves the direct translocation of cytosolic proteins across the lysosomal membrane, which requires protein unfolding via chaperone proteins [26]. In addition to classical autophagy, there are several non-traditional types of autophagy that have received research attention (Figure 1).
Figure 1.
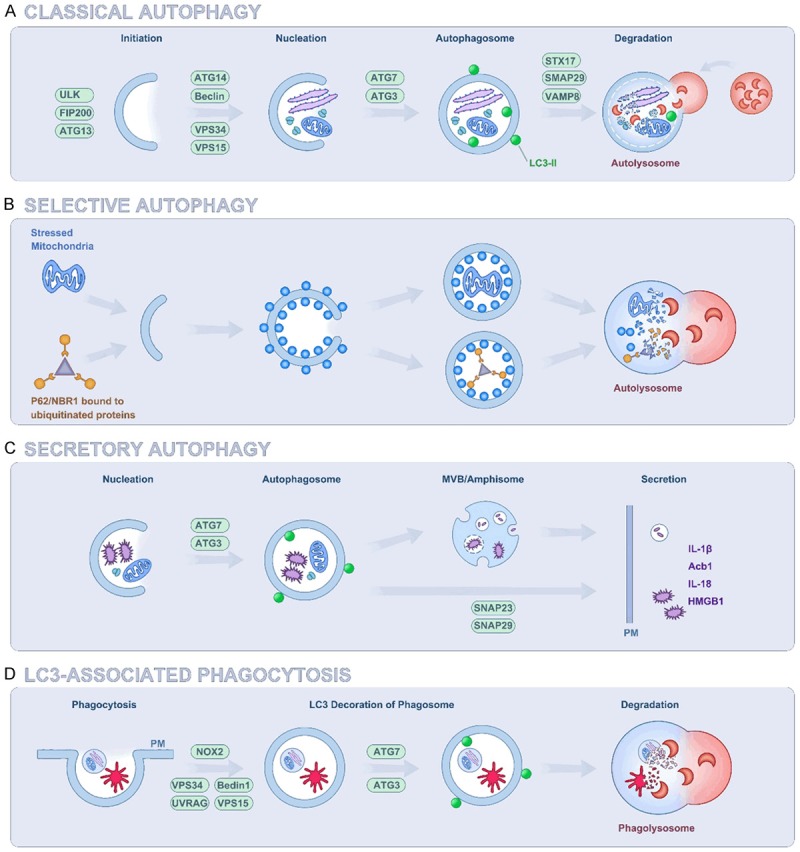
Molecular Mechanisms of different types of autophagy. A. Classic autophagy: Different stimuli cause aggregation and activation of multiple ATGs and other regulatory proteins to construct double membrane autophagosomes. Lipidation of LC3 (LC3-II) is essential for capturing autophagy cargo and stabilizing the autophagosome inner membrane. Then, autophagosomes fuse to lysosomes in a STX17-dependent manner, resulting in lysosomal enzymes degrading the vesicle contents. B. Selective autophagy: Selective autophagy contributes to intracellular homeostasis by modulating the degradation of cytoplasmic substances, such as aggregated proteins, damaged or excessive organelles, and invading pathogens. Its mechanism must ensure efficient identification and isolation of cargo in autophagy. C. Secretory autophagy: ATGs mediate unconventional secretion of multiple proteins lacking the N-terminal signal sequence. First, ATGs promote the formation of LC3+ autophagosome-like intermediates, and the contents encapsulated in the autophagy inner membrane are released extracellularly rather than degraded in lysosomes. Second, the target of secretory autophagy is transferred to the intramembranous space of the LC3+ double-membrane vesicle, which is directly fused to the plasma membrane or fused to the secreted MVB intermediate. Finally, secretory autophagy may involve extracellular release of MVB/Amphisome intermediates. D. LC3-associated phagocytosis: The phagocytosis of pathogens recruits UVRAG and Rubicon (RUBCN), thereby activating the Beclin-1-VPS34 complex. With the participation of ATG3 and ATG7, lysosome fusion and pathogen degradation are accelerated. It is noteworthy that the phagocytic vesicles formed by LAP are single membraned.
Other kinds of autophagy
Non-canonical autophagy is a process that does not require the entire set of ATG proteins, in particular Beclin 1, to form an autophagosome. Scarlatti et al. found that overexpression of BCL2 (BCL2 apoptosis regulator) was unable to reverse the non-canonical autophagy triggered by the polyphenol resveratrol in the breast cancer cell line MCF-7 [27]. Besides, Beclin 1-independent autophagy mediated by the neurotoxin 1-methyl-4-phenylpyridinium is associated with neuronal cell death [28]. Smith et al. found that arsenic trioxide induces a Beclin 1-independent autophagic pathway in ovarian carcinoma cells and implicates SnoN (SKI like proto-oncogene) in promoting arsenic trioxide-mediated autophagic cell survival [29]. These studies suggested that it is feasible to treat cancer by inducing non-canonical autophagy with pro-apoptotic compounds when the function of typical autophagic proteins is impaired. Some ATG genes are involved in the digestion of unwanted extracellular (rather than intracellular) material. One such alternative function of autophagy proteins is LC3-associated phagocytosis (LAP), which is a macroautophagy-like process [30]. During LAP, the phagosomes phagocytose extracellular contents, such as microorganisms or dying cells, which are then transported to lysosomes. Cunha et al. revealed that the anti-tumor effects of LAP impairment require tumor-infiltrating T cells, and are dependent upon STING (stimulator of interferon genes) and the type I interferon response [31]. Muniz-Feliciano et al. found that retinal pigment epithelial cells promote LAP through the expression of RUBCN/Rubicon (RUN domain and cysteine-rich domain containing Beclin 1-interacting protein) and suppress autophagy through the activation of EGFR (epidermal growth factor receptor) [32]. As a pathway related to autophagy, the physiological importance of LAP and its value in tumor research deserve further exploration.
Recent findings indicated that ATGs lack N-terminal signal sequences and are involved in unconventional protein secretion. ATGs may be involved in a process that is significantly different from classical autophagy. In addition to its role in lysosomal degradation, autophagy also controls extracellular secretion. ATGs’ involvement in this process was first discovered in yeast secretion of Acb1 (acyl-CoA-binding protein) [33]. The role of the selective secretion pathway of ATGs regulates the genetic requirements of GRASP55 (Golgi reassembly stacking protein 55) and GRASP65 for Golgi accumulation. This discovery enriched the function of autophagy and demonstrated its involvement in secretion [34,35]. Kimura et al. found that the prototypical cytosolic secretory autophagy cargo, interleukin 1 beta (IL-1β) is recognized by specialized secretory autophagy cargo receptor TRIM16 (tripartite motif containing 16) and that this receptor interacts with the R-SNARE Sec22b to recruit cargo to the LC3-II+ sequestration membranes [36]. Moreover, Adam utilized the autophagy-based involvement in cellular secretion to identify shed proteins associated with autophagy levels in melanoma [37]. Jacob hypothesized that autophagy-dependent secretion of tumor-promoting factors by HNSCC (head and neck squamous cell carcinoma)-associated CAFs (cancer-associated fibroblasts) may explain their role in malignant development [38]. Interestingly, autophagy is also associated with the recent research hotspot, exosomes. Autophagy probably contributes to the decreased exosome release induced by ISGylation (conjugation of proteins to Interferon stimulated gene 15) [39]. Dias demonstrated that PRNP (prion protein) supports CAV1 (Caveolin 1)-suppressed autophagy to protect multivesicular bodies (MVBs) from sequestration into phagophores, thus facilitating exosome secretion. These results indicated that secretory autophagy could affect the microenvironment of the tumor; therefore, further analysis of the cellular mechanisms by which autophagy promotes these different secretory processes remains an important topic for future research.
For a long time, researchers thought that autophagy lacked cargo specificity. However, we now understand that the control of the choice of cargoes is extremely specific. This process is called ‘selective autophagy’, which plays an important role in preventing most mammalian diseases. Liang et al. proposed a spatiotemporal model wherein recruitment of AMPK (AMP-activated protein kinase) in association with components of the VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3) and ATG16 complex to damaged mitochondria regulates selective mitophagy to maintain cancer cell viability [40]. The selectivity of autophagy is achieved by target recognition: Kimura demonstrated that a subset of tripartite motif (TRIM) proteins mediate selective autophagy of key regulators of inflammatory signaling [41]. By removing dangerous cytoplasmic components, selective autophagy protects cells from oxidative and genotoxic stress, which may constitute a tumor suppressor mechanism. In addition, all mechanisms for earmarking cargo must be tightly coordinated with the formation of autophagosomes to ensure final cargo disposal. The mechanisms involved in the recognition of selective autophagy substrates may play an active role in the initiation of autophagy [42,43]. Selective autophagy involves a variety of mechanisms; the ultimate goal is to ensure proper movement of cargo. Substrate phosphorylation is also a common mechanism for targeting selective autophagy, possibly by activating specific kinases that enhance selective autophagy [44]. A more plausible explanation for the tumor-suppressing effect of autophagy may be its role as a selective degradation pathway [45]. Selective autophagy might ensure tumor survival by degrading misfolded proteins and damaged organelles accumulated in genetically unstable tumor cells. Therefore, targeting nonspecific autophagy and its selective forms may prove beneficial in the fight against malignant tumors.
Roles of autophagy in HCC
Autophagy plays a contradictory role in HCC, protecting cells from carcinogenesis at the early stage and promoting tumor progression at the advanced stage [46-49]. This dual role illustrates the complexity of targeting autophagy to treat HCC. Autophagy-related genes, non-coding RNAs, and related signaling pathways (Figure 2) are involved in autophagy and the regulation of the formation and progress of HCC. The exact function of autophagy in HCC has not been fully determined and is controversial. Further in-depth research is required to understand the role of autophagy in the development of HCC.
Figure 2.
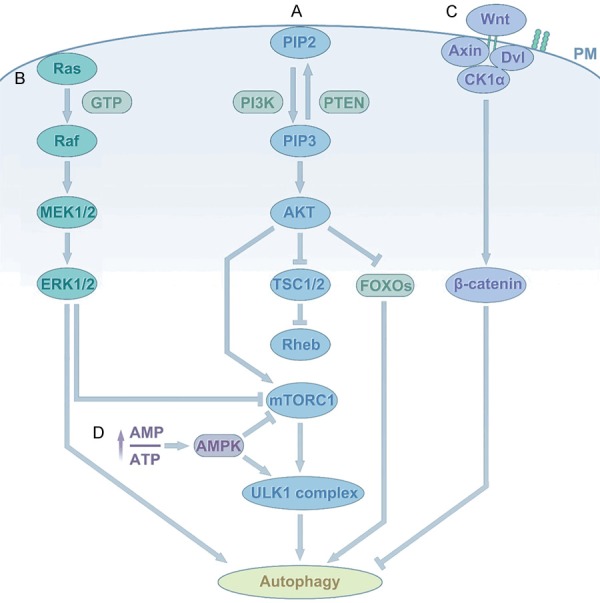
Autophagy signaling in HCC. A. PI3K/AKT/mTOR: Classic autophagy: Binding of growth factors to receptors triggers PI3K, and then activation of PI3K catalyzes the production of PIP3, phosphorylation of PIP3, and activation of Akt serine/threonine kinase. Subsequently, phosphorylation of AKT activates mTORC1, thereby inhibiting autophagy. In addition, activated AKT leads to activation of Rheb. Rheb then activates mTORC1. In addition to direct or indirect activation of mTORC1, active AKT can also directly regulate transcription leading to inhibition of autophagy. B. The RAS/RAF/MEK/ERK pathway: RAS switches from inactive (GDP-bound) to the active (GTP-bound) form. Activated RAS binds and recruits RAF kinase to the cell membrane for RAF dimerization and activation. Subsequently, activated RAF phosphorylates and activates MEK; MEK in turn phosphorylates and activates ERK/MAPK. Finally, phosphorylated ERK directly activates autophagy, or phosphorylates a variety of substrates that trigger autophagy by inhibiting mTORC1. C. The Wnt/β-catenin signaling pathway: This pathway can negatively regulate autophagy. β-catenin is a special target for degradation by autophagy in starvation stress. D. The AMPK signaling pathway: AMPK is an αβγ heterotrimer that is activated by decreasing ATP concentrations and increasing AMP concentrations. It induces autophagy via the activation of ULK1 or the inactivation of mTOR.
Autophagy-related genes
ATG-mediated autophagy has a significant impact on HCC. Deletion of the genes encoding Beclin-1, ATG5, or ATG7 in mice damaged autophagy and promoted the occurrence of spontaneous liver tumors in aged mice [46,50,51]. At the same time, the expression level of Beclin-1 correlated negatively with HCC grading, suggesting that to some extent, Beclin-1 correlates positively with HCC, and the low expression of Beclin-1 in HCC tissues was associated with the recurrence and survival rates [52,53]. In addition, Takamura reported that mice with systemic mosaic deletion of Atg5 and liver-specific Atg7-/- mice develop benign liver adenomas [54]. This result revealed that autophagy has an anti-tumor effect on specific liver cancers and may have a preventative effect on the occurrence and development of HCC. Tian et al. utilized an ATG5 knockdown model to verify that impairing autophagy in hepatocytes would induce oxidative stress and DNA damage, followed by the initiation of hepatocarcinogenesis [55]. Another study showed that p62 is necessary for HCC induction in mice and that its high expression level in non-tumor human liver predicts rapid HCC recurrence after curative ablation [56]. P62 is an ubiquitin-binding autophagy receptor and signaling protein that accumulates in premalignant liver diseases and most HCCs. Ji et al. reported that HuR (human antigen R) functions as a pivotal regulator of autophagosome formation by enhancing the translation of ATG5, ATG12, and ATG16 mRNAs. Augmented expression of HuR and ATGs may participate in the malfunction of autophagy in HCC cells [57]. In addition, UVRAG (UV radiation resistance associated) interacts with BECN1 and PIK3C3, and is a significant regulator of mammalian autophagy. Feng et al. provided in vitro and in vivo evidence that UVRAG ubiquitination at lysine residues 517 and 559 promotes autophagosome maturation and enhances the lysosomal degradation of EGFR, which significantly inhibits HCC cell growth [58]. These reports indicated that ATGs are involved in the progress of HCC and offer insights into autophagy regulation and therapeutic combinations in HCC.
Noncoding RNAs
Noncoding RNAs (ncRNAs), including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), are attracting more attention as potential new drug targets for human diseases. In recent years, the interaction between ncRNAs and autophagy has become a hotspot in the study of HCC. MiRNAs are a class of endogenously expressed, short noncoding RNAs, which regulate gene expression post-transcriptionally [59]. MiRNAs can affect many biological processes, such as cell development, infection, immunity, and carcinogenesis [60]. MiRNAs are involved in various stages of autophagy, including phagophore induction, nucleation and expansion; the maturation of autolysosomes and autophagosomes; and have a regulatory role [61]. MiRNAs are increasingly recognized to play an important role in physiological and pathological processes, including the development and progression of tumors. Many microRNAs are involved in autophagy regulation of HCC [59]. For example, Glycine decarboxylase overexpression inhibited migration and invasion via an increase in cellular autophagy. This effect was reduced by miR-30d-5p transfection [62]. Furthermore, miRNAs regulate autophagy by targeting autophagy-related genes in HCC. Xiu-Tao Fu et al. noted downregulated miR-30a in metastatic HCC, which mediates Beclin 1 and Atg5-dependent autophagy and confers anoikis resistance in HCC cells [63]. In addition, the first reported miRNA, mir-375, with proapoptotic functions, can inhibit autophagy and reduce cell viability in HCC cells by binding directly to ATG7 under hypoxic conditions [64]. In another study, miR-26 family members (miR-26a, miR-26b, and miR-26a/b) could act as potential autophagy inhibitors, making HCC cells sensitive to doxorubicin (Dox) and promoting apoptosis by directly inhibiting the expression of serine/threonine protein kinase ULK1, which is a key promoter of autophagy [65]. Interestingly, Lan et al. were the first to reveal that autophagy selectively regulates miR224 expression through an autophagosome-mediated degradation system. They also found that the off-label use of amiodarone, an antiarrhythmic agent, effectively suppressed HCC tumorigenesis through autophagy-mediated miR224 degradation, both in vitro and in vivo [66]. In general, autophagy and miRNAs are important regulators of HCC development.
Emerging evidence indicates that lncRNAs act as competitive platforms for both miRNAs and mRNAs [67]. LncRNAs are non-coding RNAs longer than 200 nucleotides [68,69]. LncRNAs have a crucial role in various fundamental pathophysiological processes, such as carcinogenesis, that play a regulatory role in the progression of cancer [70,71]. The discovery of lncRNAs provides a new way to regulate genes in almost all essential biological processes, including autophagy. A series of studies have shown that many lncRNAs are abnormally expressed in HCC tissues and participate in their biological behaviors, such as proliferation, apoptosis, metabolism, migration, and invasion [72,73]. In hepatocellular carcinoma, PTEN (phosphatase and tensin homolog) and PLLP (Plasmolipin) interact with miRNA17, miRNA19B and miRNA20A, inhibiting the PI3K-AKT (AKT kinase)-mTOR (mammalian target of rapamycin) signaling pathway to inhibite cell proliferation, migration/invasion as well as induced autophagy and apoptosis [74]. Thereafter, lncRNA HULC was observed to accelerate the development of HCC by inhibiting PTEN through co-operation with the autophagy of miRNA15A, and HULC enhanced the interplay between LC3 and ATG3 [75]. Our research group confirmed that lncRNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in HCC [76]. We also showed that PVT1 (plasmacytoma variant translocation 1) could promote autophagy as a competing endogenous RNA (ceRNA) by targeting miR-365 in HCC [77]. As more and more lncRNAs are identified that regulate autophagy, it will be interesting to see if autophagy also affects the expression of lncRNAs. Given the limitations of the studies conducted to date, we have limited understanding of the underlying mechanisms of regulation between identified lncRNAs and autophagy. Of course, lncRNAs have more complex functions in autophagy regulation, which requires further clarification. Studies also suggest that we may need to classify lncRNAs according to their role in different types of autophagy, to explore the functions of lncRNAs more specifically. Both, lncRNAs and miRNAs are often deregulated in liver cancer, underlining the importance of ncRNAs in hepatocarcinogenic processes [78,79]. Therefore, a joint intervention targeting ncRNAs and autophagy may be a promising therapeutic strategy in HCC.
Signaling pathways of autophagy in HCC
PI3K/AKT/mTOR
The PI3K/Akt/mTOR pathway plays an important role in promoting autophagy and regulates cell growth, survival, metabolism, and apoptosis under physiological conditions, which has great significance for the occurrence and survival of various solid tumors, including HCC [80-83]. As a typical survival pathway, the PI3K/Akt/mTOR pathway plays an increasingly important role in the occurrence of HCC. The PI3Ks, which are divided into three classes (class I (A and B), class II, and class III), are important kinases regulating cell survival, proliferation, and differentiation [84,85]. AKT is a key factor in signaling pathways that regulate autophagy in a variety of ways [86-88]. AKT induces the activation (phosphorylation) of mTORC1, a serine/threonine kinase, using the phosphatase PTEN, which can decrease the level of PI3K and initiate the formation of autophagosomes. Evidence indicates that abnormal activation of the PI3K/Akt/mTOR signaling pathway frequently occurs in HCC [89,90]. Activating or inhibiting this pathway can inhibit or activate autophagy, which has different effects on HCC. Wang et al. demonstrated that overexpressed AFP (alpha-fetoprotein) interacts with PTEN in HCC cells, resulting in activation of PI3K/Akt/mTOR and a reduction in autophagy [91]. Wang applied H2S (hydrogen sulfide) to induce cell autophagy by inhibiting the PI3K/AKT/mTOR signaling pathway in HCC [92]. Moreover, Li et al. verified that IL-37 (anti-inflammatory cytokine interleukin-37) regulates autophagy in HCC via inhibition of the PI3K/AKT/mTOR signaling pathway. In addition, using β-Thujaplicin combined with an autophagy blocker or agonist treatment in HepG2 cells, Zhang found that β-Thujaplicin induced autophagic cell death (ACD), mediated by reactive oxygen species (ROS), which caused inhibition of the Akt-mTOR signaling pathway [93]. In summary, targeting this pathway might result in autophagic cancer cell death, and could be used to treat HCC.
The RAS/RAF/MEK/ERK pathway
This pathway is involved in the induction of apoptosis and autophagic cell death in many cancer cell lines [94-96]. The RAS/RAF (Raf-1 proto-oncogene, serine/threonine kinase)/MEK (MAPK/ERK kinase 1)/ERK (extracellular signal-regulated kinase) pathway plays multiple roles in cell cycle regulation, apoptosis, and differentiation [97]. The mechanism of its involvement in autophagy regulation is extremely complex and sometimes seems to be contradictory. In particular, how it fine regulates autophagy in a specific environment has not yet been clarified. Studies have shown that activation of RAF/MEK/ERK can alter the expression levels of autophagy markers LC3 and SQSTM1 (Sequestosome 1) [98]. At the same time, there is evidence that ERK triggers autophagy by inhibiting mTORC1, which can also affect autophagy by regulating Beclin-1 [99,100]. By contrast, ERK can cause the downregulation of LAMP1 (lysosomal-associated membrane protein 1) and LAMP2, and prevent the binding of autophagosomes to lysosomes, thereby inhibiting the degradation of autophagosomes [101]. Frequent mutations in RAS/RAF/MEK/ERK pathway members are thought to contribute to the development, tumor progression, and metastasis of many solid tumors, including HCC. RAS was the first human oncogene to be identified [102]. To overcome the effects of RAS mutations, only EGFR inhibitors and RAF-MEK-ERK pathway therapies have any effect [103]. RASSF1A (Ras association domain family 1 isoform A) can inhibit HCC by activating autophagy, thus improving the survival rate [104]. Wang et al. reported a novel 2-phenyloxypyrimidine derivative, E5, which induced autophagy via activation of the MAPK (mitogen activated protein kinase)/ERK pathway in HCC cells [105]. Additionally, cytoplasmic sequestration of ERK by binding to PEA-15 (proliferation and apoptosis adaptor protein 15) promotes autophagy [106]. These results indicated that the RAS/RAF/MEK/ERK pathway mediates autophagy and plays a crucial role in HCC.
Wnt/β-catenin signaling pathway
The Wnt signaling pathway is a key signaling pathway that directly determines cell proliferation, cell polarity, and cell fate during embryonic development and in the regulation of tissue homeostasis [107,108]. Mutations in any molecule in the Wnt pathway may cause birth defects, cancer, and a range of other diseases [109]. The Wnt/β-catenin pathway regulates various cell processes, such as initiation, growth, survival, migration, differentiation, and apoptosis in HCC [110-112]. Poor prognosis and disease progression in liver cancer usually involve upregulation of wnt/β-catenin signaling [113]. In addition, this pathway can negatively regulate autophagy. Inhibition of the Wnt/β-catenin pathway leads to the accumulation of autophagy proteins such as LC3-II, ATG7, and Beclin-1 [114,115]. Petherick demonstrated that β-catenin inhibits autophagy and the expression of the autophagy adapter p62, whereas under conditions of nutrient deficiency, β-catenin undergoes proteasome-independent degradation through its interaction with autophagy protein LC3 [116]. However, in terms of autophagy, the relationship between the Wnt pathway and HCC is rarely reported. Lilia’s study revealed that the Wnt/β-catenin pathway inhibitor fh535 and its derivatives (fh535-N) exert anti-tumor effects on hepatoma cells by regulating autophagy activity [117]. Unexpectedly, Wang et al. used Wnt secretion inhibition in HCC to show that autophagy was not successfully induced [118]. Zhang et al. reported the tetrandrine suppresses HCC cell migration via suppression of Wnt/β-catenin signaling, which is regulated by tetrandrine-induced autophagy [119]. Our previous study verified that autophagy could act as an upstream regulatory factor to activate Wnt/β-catenin signaling [120]. These results indicated that the Wnt/β-catenin pathway has a complex relationship with the dynamic effects of autophagy and the development of HCC, indicating that it could represent a new target for liver cancer research.
Other pathways
PERK (pancreatic EIF2-alpha kinase) is a common upstream signaling pathway between autophagy and apoptosis that is induced by endoplasmic reticulum (ER) stress. Under ER stress, PERK is activated and phosphorylates and inactivates EIF2α (eukaryotic translation initiation factor 2 subunit alpha). This results in the selective induction of ATF4 (activating transcription factor 4), which induces autophagy and apoptosis [121]. Autophagy lies downstream of the PERK signaling axis and ultimately leads to tumor cell survival [122]. In addition, hepatocyte growth factor (HGF) and its receptor tyrosine kinase, MET were first discovered in the 1980s because they are highly active in many liver cancers [123,124]. Activated hepatic stellate cells promote the progression of HCC cells after sublethal heat treatment from autophagic survival to proliferation via HGF/c-Met signaling [125]. HGF/c-MET signaling inhibits autophagy by interacting with the PI3K/AKT pathway, while high expression of c-MET is observed in HCC samples [126]. When treated with HGF-MET kinase activity-targeted drugs, tyrosine 1234/1235-dephosphorylated MET activated autophagy in HCC [127]. There are also many pathways involved in the interaction between autophagy and HCC, such as the Hippo pathway [128], EGFR crosstalk [129], and the AMPK pathway [130]; however, all these pathways require further exploration.
Autophagy and HCC therapy
Currently, liver cancer is the second leading cause of cancer-related death worldwide [131,132]. Poor prognosis has lead to a 5-year survival rate of patients with HCC of less than 5% [133]. To date, the most effective treatment for HCC is surgical resection, interventional radiotherapy, or liver transplantation [134]. Therefore, in this section, we will summarize clinical and basic studies that focused on autophagy in the context of HCC treatment (Table 1).
Table 1.
Autophagy and treatment of hepatocellular carcinoma
| Classification | Treatment | Autophagy status | HCC lines | Reference |
|---|---|---|---|---|
| Oxaliplatin | ↑ | Huh-7 | [139,140] | |
| SMMC-7721 | ||||
| HepG2 | ||||
| Epirubicin | ↑ | HepG2 | [142] | |
| Huh7 | ||||
| Conventional chemotherapeutics | Cisplatin | ↑ | SMMC-7721 | [144] |
| HuH-7 | ||||
| HepG2 | ||||
| 5-FU | ↑ | SMMC-7721 | [143] | |
| Hep3B | ||||
| HepG2 | ||||
| Doxorubicin | ↑ | HepG2 | [145,184] | |
| Hep3B | ||||
| Sorafenib | ↑ | SMMC-7721 | [157,185] | |
| HepG2 | ||||
| Huh7 | ||||
| Hep3B | ||||
| Panobinostat | ↓ | Huh7 | [186] | |
| Hep3B | ||||
| HepG2 | ||||
| Molecular Targeting Drugs | Egorafenib | ↑ | HepG2 | [159] |
| Hep3B | ||||
| Bevacizumab | ↑ | SMMC-7721 | [163] | |
| Hep3B | ||||
| Salinomycin | ↓ | HepG2 | [166] | |
| Huh7 | ||||
| β-Thujaplicin | ↑ | HepG2 | [93] | |
| SMMC-7721 | ||||
| HCCLM3 | ||||
| P. bistorta aqueous extract | ↑ | Hep3B | [170] | |
| HepG2 | ||||
| Natural Product | Arenobufagin | ↑ | HepG2 | [171] |
| Hep3B | ||||
| Bel-7402 | ||||
| Baicalin | ↑ | SMMC-7721 | [172] | |
| Oroxylin A | ↑ | HepG2 | [173] |
Note: ↑, means that increase of autophagosomes in HCC; ↓, means that decrease of autophagosomes in HCC.
Conventional chemotherapeutics
In recent years, more and more chemotherapy drugs have been used to treat advanced HCC, often involving combined treatment with a variety of chemotherapy drugs. For example, cisplatin combined with doxorubicin, 5-fluorouracil, and interferon (INF) can significantly improve the survival rate of advanced HCC; however, is prone to chemotherapy resistance [135,136]. Therefore, it is essential to study how HCC resists chemotherapy and develop new drug strategies to overcome chemotherapeutic resistance. Autophagy is a double-edged sword for MDR (multidrug resistance), which occurs after long-term chemotherapy, leading to refractory cancer and recurrence of tumors [137]. Autophagy participates in the development of MDR and protects cancer cells from chemotherapy; however, it also kills MDR cancer cells in which apoptosis pathways are inactive. Oxaliplatin-based chemotherapy has recently been shown to be effective to treat advanced HCC [138]. Studies have shown that Oxaliplatin can activate autophagy in HCC [139,140]. Ren et al. identified EVA1A (transmembrane protein 166, an autophagy-related protein) as a target of miR-125b, and showed that it was upregulated in HCC tissues from Oxaliplatin-resistant patients, suggesting that EVA1A plays a role in resistance to chemotherapy [141]. In general, the response of cancer cells to chemotherapy is usually to increase autophagy. Meanwhile, restraining autophagy makes cancer cells sensitive to anticancer drugs. For example, HSF1 (heat shock transcription factor 1) could upregulate ATG4B expression and enhance epirubicin-induced protective autophagy in HCC [142]. In another study, the inhibition of autophagy using 3-methyladenine or siRNA targeting Beclin-1 increased chemotherapy (cisplatin or 5FU)-induced apoptosis and increased damage to the mitochondrial membrane potential in HCC cells [143]. In addition, Wu et al. demonstrated that autophagy could cause HCC resistance to cisplatin [144]. Moreover, miR-101 inhibited autophagy and synergized with either doxorubicin or fluorouracil to induce apoptosis in HCC [145]. In summary, targeting autophagy is a promising therapeutic strategy to enhance the effects of chemotherapy and improve clinical outcomes in patients with HCC.
Molecular targeting drugs
Sorafenib, a multitargeted kinase inhibitor, has greatly revolutionized the treatment of HCC [146]. Currently, representative phase III trials have shown that Sorafenib significantly improves overall survival in patients with advanced HCC [147]. The role of Sorafenib includes blocking the RAF-MEK-ERK signaling pathway to inhibit the proliferation of cancer cells, and targeting the vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR) to prevent angiogenesis [148]. Although it is now recognized that Sorafenib can activate autophagy and apoptosis, it can also induce autophagy through the ERK/MAPK pathway independently, thus promoting the survival of HCC cells in vivo or in vitro [149-151]. Unfortunately, the long-term value of Sorafenib is limited because of primary and acquired resistance, in which autophagy activation is a factor [152-154]. For example, Lu et al. found that CD24 (a glycoprotein expressed on the surface of most B lymphocytes) could regulate Sorafenib resistance via activating autophagy in HCC [155]. Wu et al. verified that ADRB2 (β-2 adrenergic receptor) signaling promotes HCC progression and Sorafenib resistance by inhibiting autophagic degradation of HIF1α [156]. In another experiment, targeting ATG5/ATG16L1 inhibited autophagy and increased the sensitivity of HCC cells to Sorafenib [157]. In addition, Adriamycin, a traditional chemotherapeutic drug, can be inhibited by Sorafenib, leading to cell progression, increased survival, and reduced autophagy of HCC [158]. In short, Sorafenib can induce autophagy formation and enhance autophagy activity, allowing HCC cells to survive. At the same time, inhibition of autophagy may be an attractive strategy to release the anti-tumor potential of Sorafenib in HCC.
Other targeted drugs also have autophagy activation effects. For instance, Regorafenib may act as an adjunctive therapy for patients with liver cancer. Regorafenib delays the proliferation of HCC by inducing autophagy [159]. Tong found ANXA3 (Annexin A3) inhibition sensitized HCC cells to Regorafenib treatment via suppressing autophagy and activating apoptosis [160]. Concomitantly, autophagy can be induced by EGFR inhibitors [161]. In one study, P57-mediated autophagy promoted the efficacy of Erlotinib/Cetuximab (EGFR inhibitors) in HCC [162]. In addition, Bevacizumab, which targets VEGF, induces autophagy and combined inhibition of autophagy with Bevacizumab can significantly inhibit tumor growth in HCC [163]. Another inhibitor of VEGF, Linifanib, also induces autophagy in HCC cells. After ATG5 and ATG7 were inhibited by pharmacological inhibitors or short interfering RNAs (siRNAs), cell death induced by Linifanib increased significantly [164]. In addition, Wu et al. found that the calcium phosphate nanoparticle system could be further developed for co-delivery of FTY720 (Fingolimod) and a Beclin 1 siRNA to treat HCC, which enhanced the anticancer efficacy of FTY720 [165]. In another study, Klose et al. reported that Salinomycin suppresses the late stages of HCC autophagy [166]. In conclusion, the relationships among small molecule targeted drugs, autophagy and HCC need further research to provide new strategies to treat HCC.
Natural products
Many natural products have been shown to have autophagic effects on the growth and survival of HCC cells. For example, β-Thujaplicin, a natural tropolone derivative, exhibits a variety of biological properties, including antibacterial, antifungal, antiviral, anti-inflammatory, and anticancer potential [167-169]. A study found that β-Thujaplicin might inhibit the growth of HCC cells by inducing autophagy [93]. In another study, P. bistorta (Bistorta officinalis (synonym Persicaria bistorta) aqueous extract (PB) induced autophagy, subsequently triggering caspase-dependent apoptosis in HCC. P. bistorta is used in traditional Chinese medicine owing to its anticancer activities [170]. In addition, Arenobufagin, a natural bufadienolide from toad venom, induces apoptosis and autophagy in human HCC cells through inhibition of the PI3K/AKT/mTOR pathway [171]. Meanwhile, Baicalin has been demonstrated to exert anticancer effects mainly through the induction of tumor cell apoptosis and cell cycle arrest. Baicalin induces autophagic cell death in HCC cells [172]. In addition, Zou et al. reported that Oroxylin A, which is a natural mono-flavonoid extracted from Scutellariae radix, exhibits autophagy-mediated antitumor activity in a dose and time-dependent manner in human HCC cells [173]. Similarly, Berberine, Allicin, Matrine, and Glycyrrhetinic acid are plant-derived molecules that show anti-tumor effects by inducing apoptosis and/or autophagy of HCC cells [174-176]. Natural products have been recognized as a new source of anti-cancer drugs and new adjuvant therapy to improve the efficacy of chemotherapy and to reduce the side effects associated with cancer chemotherapy [177].
Other approaches
Increasing evidence suggests that both autophagy inhibitors and inducers contribute to the response of HCC therapies. Autophagy inhibitors, such as 3-Methyladenine, chloroquine, or knockdown of different ATG genes have been reported to enhance the efficacy of Oxaliplatin, Cisplatin, 5-Fu and Sorafenib in the treatment of HCC [140,143,178]. These studies show that inhibition of autophagy rendered HCC cells susceptibility to chemotherapy-induced apoptosis and cell growth inhibition, identifying autophagy as a sensitizer that can improve the efficacy of conventional chemotherapeutic drugs for HCC. Additionally, Rapamycin, an inhibitor of mTOR, activates autophagy both in vitro and in vivo [179]. Rapamycin showed an anti-tumor effects in therapy for HCC [180]. Another study showed that mTOR inhibition significantly reduced HCC growth and improved survival primarily via antiangiogenic effects [181]. These results indicated that autophagy can be both a promoter and inhibitor of HCC. Currently, this mechanism underlying this paradox is unknown. In addition, the previously mentioned ATGs and non-coding RNAs can also participate in autophagy and the treatment of HCC. For example, Thomas et al. reported that altered expression of autophagy genes was associated with poor prognosis [182]. MiR-375 inhibits autophagy by reducing the expression of ATG7 and impairs the viability of HCC cells under hypoxic conditions [64]. MiRNAs that inhibit autophagy of HCC cells may be developed as therapeutics. Some inhibitors of autophagy-related pathways might also have anti-HCC effects. Recent studies have shown that at an early time point, Sorafenib increases ER stress, which induces the autophagic survival process in HCC cell lines by regulating the JNK/AMPK signaling pathway [183]. Although targeting autophagy in the treatment of HCC is complex, many basic and clinical studies have shown that autophagy can be a potential therapeutic target that could enhance the anticancer potency of both native tumor suppressor mechanisms and chemotherapy.
Conclusion
In general, autophagy plays a double role in HCC. Most studies support the view that autophagy inhibits tumors. Autophagy plays an anti-tumor role in normal hepatocytes by maintaining cell homeostasis. However, once tumors are formed, it will promote the survival of HCC cells in the tumor microenvironment. Autophagy-related genes, non-coding RNAs, and related signaling pathways are involved in autophagy and the regulation of the formation and progress of HCC. In addition, the effects of autophagy on traditional chemotherapy, molecular targeted drugs, and natural products can also be associated with the treatment of HCC. Although it remains a challenge to understand the specific molecular mechanisms of autophagy in different stages of HCC, this understanding will help to develop therapeutic targets and overcome resistance to current therapies.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81472302/No. 81572425/No. 81871983).
Disclosure of conflict of interest
None.
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Allaire M, Rautou PE, Codogno P, Lotersztajn S. Autophagy in liver diseases: time for translation? J Hepatol. 2019;70:985–998. doi: 10.1016/j.jhep.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 7.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 8.Grohmann M, Wiede F, Dodd GT, Gurzov EN, Ooi GJ, Butt T, Rasmiena AA, Kaur S, Gulati T, Goh PK, Treloar AE, Archer S, Brown WA, Muller M, Watt MJ, Ohara O, McLean CA, Tiganis T. Obesity drives STAT-1-dependent NASH and STAT-3-dependent HCC. Cell. 2018;175:1289–1306. e1220. doi: 10.1016/j.cell.2018.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GA, Giulini SM. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006;243:229–235. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asghar U, Meyer T. Are there opportunities for chemotherapy in the treatment of hepatocellular cancer? J Hepatol. 2012;56:686–695. doi: 10.1016/j.jhep.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 13.Yu S, Wang Y, Jing L, Claret FX, Li Q, Tian T, Liang X, Ruan Z, Jiang L, Yao Y, Nan K, Lv Y, Guo H. Autophagy in the “inflammation-carcinogenesis” pathway of liver and HCC immunotherapy. Cancer Lett. 2017;411:82–89. doi: 10.1016/j.canlet.2017.09.049. [DOI] [PubMed] [Google Scholar]
- 14.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 15.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 17.Towers CG, Thorburn A. Therapeutic targeting of autophagy. EBioMedicine. 2016;14:15–23. doi: 10.1016/j.ebiom.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 19.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Banhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolome A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bedard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Benard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Bockler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouche M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Butikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Cena V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Claria J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D’Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Davila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D’Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Duran RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martinez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Faergeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernandez AF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernandez-Lopez A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, Francois A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannage M, Gao FB, Gao F, Gao JX, Garcia Nannig L, Garcia Vescovi E, Garcia-Macia M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gomez-Sanchez R, Goncalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, Gonzalez-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson AB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hebert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernandez A, Hernandez C, Hernandez-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Hoglinger GU, Hohfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jaattela M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jimenez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhasz G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kagedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knaevelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Kohler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovacs AL, Kovacs T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc’h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, Lopez-Otin C, Lopez-Vicario C, Lorente M, Lorenzi PL, Lorincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magarinos M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manie SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Marino G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martin-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martinez-Velazquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Melendez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Rios MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Moller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Munoz-Moreno R, Munoz-Pinedo C, Munz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nurnberger T, O’Donnell VB, O’Donovan T, O’Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O’Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palkova Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstadt H, Pavone F, Pedrozo Z, Pena FJ, Penalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Perez-de la Cruz V, Perez-Perez ME, Perez-Rodriguez D, Perez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muinos FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Poggeler S, Poirot M, Polcic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodriguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roue G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schonenberger MJ, Schonthal AH, Schorderet DF, Schroder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Segui-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninova I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Strom AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Sward K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takats S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thome MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcategui NL, Vaccari T, Vaccaro MI, Vachova L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jaattela M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Munz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarlatti F, Maffei R, Beau I, Ghidoni R, Codogno P. Non-canonical autophagy: an exception or an underestimated form of autophagy? Autophagy. 2008;4:1083–1085. doi: 10.4161/auto.7068. [DOI] [PubMed] [Google Scholar]
- 28.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DM, Patel S, Raffoul F, Haller E, Mills GB, Nanjundan M. Arsenic trioxide induces a beclin-1-independent autophagic pathway via modulation of SnoN/SkiL expression in ovarian carcinoma cells. Cell Death Differ. 2010;17:1867–1881. doi: 10.1038/cdd.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 31.Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, Natarajan S, Turnis ME, Finkelstein D, Opferman JT, Gawad C, Green DR. LC3-associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell. 2018;175:429–441. e416. doi: 10.1016/j.cell.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muniz-Feliciano L, Doggett TA, Zhou Z, Ferguson TA. RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy. 2017;13:2072–2085. doi: 10.1080/15548627.2017.1380124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M, Schekman R. Cell biology. Unconventional secretion, unconventional solutions. Science. 2013;340:559–561. doi: 10.1126/science.1234740. [DOI] [PubMed] [Google Scholar]
- 35.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura T, Jia J, Kumar S, Choi SW, Gu Y, Mudd M, Dupont N, Jiang S, Peters R, Farzam F, Jain A, Lidke KA, Adams CM, Johansen T, Deretic V. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2017;36:42–60. doi: 10.15252/embj.201695081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraya AA, Piao S, Xu X, Zhang G, Herlyn M, Gimotty P, Levine B, Amaravadi RK, Speicher DW. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy. 2015;11:60–74. doi: 10.4161/15548627.2014.984273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.New J, Arnold L, Ananth M, Alvi S, Thornton M, Werner L, Tawfik O, Dai H, Shnayder Y, Kakarala K, Tsue TT, Girod DA, Ding WX, Anant S, Thomas SM. Secretory autophagy in cancer-associated fibroblasts promotes head and neck cancer progression and offers a novel therapeutic target. Cancer Res. 2017;77:6679–6691. doi: 10.1158/0008-5472.CAN-17-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernandez-Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sanchez-Madrid F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. 2016;7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang J, Xu ZX, Ding Z, Lu Y, Yu Q, Werle KD, Zhou G, Park YY, Peng G, Gambello MJ, Mills GB. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat Commun. 2015;6:7926. doi: 10.1038/ncomms8926. [DOI] [PubMed] [Google Scholar]
- 41.Kimura T, Jain A, Choi SW, Mandell MA, Johansen T, Deretic V. TRIM-directed selective autophagy regulates immune activation. Autophagy. 2017;13:989–990. doi: 10.1080/15548627.2016.1154254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, Wilkinson S. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-Phagy and pancreatic ER proteostasis. Dev Cell. 2018;44:217–232. e211. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, Zhu Y, Zhang X, Li L, Zhang L, Sui S, Zhao B, Feng D. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kluge AF, Lagu BR, Maiti P, Jaleel M, Webb M, Malhotra J, Mallat A, Srinivas PA, Thompson JE. Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg Med Chem Lett. 2018;28:2655–2659. doi: 10.1016/j.bmcl.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- 46.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, Jaeschke H, Ding WX. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K, Stevens C, Brunton VG, Langdon WY, Vidal M, Sansom OJ, Dikic I, Wilkinson S, Frame MC. Autophagic targeting of src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol. 2011;14:51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- 50.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D’Orazio M, Skobo T, Bordi M, Rohde M, Dalla Valle L, Helmer-Citterich M, Gretzmeier C, Dengjel J, Fimia GM, Piacentini M, Di Bartolomeo S, Velasco G, Cecconi F. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17:20–30. doi: 10.1038/ncb3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kotsafti A, Farinati F, Cardin R, Cillo U, Nitti D, Bortolami M. Autophagy and apoptosis-related genes in chronic liver disease and hepatocellular carcinoma. BMC Gastroenterol. 2012;12:118. doi: 10.1186/1471-230X-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, Ou JH. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025–1034. doi: 10.1038/cdd.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, Linares JF, Reina-Campos M, Umemura S, Valasek MA, Seki E, Yamaguchi K, Koike K, Itoh Y, Diaz-Meco MT, Moscat J, Karin M. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji E, Kim C, Kang H, Ahn S, Jung M, Hong Y, Tak H, Lee S, Kim W, Lee EK. RNA binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol Cell Biol. 2019;39 doi: 10.1128/MCB.00508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng X, Jia Y, Zhang Y, Ma F, Zhu Y, Hong X, Zhou Q, He R, Zhang H, Jin J, Piao D, Huang H, Li Q, Qiu X, Zhang Z. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy. 2019;15:1130–1149. doi: 10.1080/15548627.2019.1570063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Wang P, Wan L, Xu S, Pang D. The emergence of noncoding RNAs as heracles in autophagy. Autophagy. 2017;13:1004–1024. doi: 10.1080/15548627.2017.1312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhuang H, Wu F, Wei W, Dang Y, Yang B, Ma X, Han F, Li Y. Glycine decarboxylase induces autophagy and is downregulated by miRNA-30d-5p in hepatocellular carcinoma. Cell Death Dis. 2019;10:192. doi: 10.1038/s41419-019-1446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu XT, Shi YH, Zhou J, Peng YF, Liu WR, Shi GM, Gao Q, Wang XY, Song K, Fan J, Ding ZB. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018;412:108–117. doi: 10.1016/j.canlet.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177–187. e178. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 65.Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K, Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. doi: 10.1038/cddis.2016.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy. 2014;10:1687–1689. doi: 10.4161/auto.29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grull MP, Masse E. Mimicry, deception and competition: the life of competing endogenous RNAs. Wiley Interdiscip Rev RNA. 2019;10:e1525. doi: 10.1002/wrna.1525. [DOI] [PubMed] [Google Scholar]
- 68.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 70.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 71.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L, George J, Qiao L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16:165. doi: 10.1186/s12943-017-0734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 74.Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, Hu YC. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 75.Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, Li X, Zheng Q, Pu H, Gui X, Li T, Li J, Jia S, Lu D. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17:94. doi: 10.1186/s12943-018-0843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605–2612. doi: 10.1039/c6mb00114a. [DOI] [PubMed] [Google Scholar]
- 77.Yang L, Peng X, Jin H, Liu J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene. 2019;697:94–102. doi: 10.1016/j.gene.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 78.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gong J, He XX, Tian A. Emerging role of microRNA in hepatocellular carcinoma (review) Oncol Lett. 2015;9:1027–1033. doi: 10.3892/ol.2014.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu M, Guo J, Xia H, Li W, Lu Y, Dong X, Chen Y, Xie X, Fu S, Li M. Alpha-fetoprotein activates AKT/mTOR signaling to promote CXCR4 expression and migration of hepatoma cells. Oncoscience. 2015;2:59–70. doi: 10.18632/oncoscience.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoneda N, Sato Y, Kitao A, Ikeda H, Sawada-Kitamura S, Miyakoshi M, Harada K, Sasaki M, Matsui O, Nakanuma Y. Epidermal growth factor induces cytokeratin 19 expression accompanied by increased growth abilities in human hepatocellular carcinoma. Lab Invest. 2011;91:262–272. doi: 10.1038/labinvest.2010.161. [DOI] [PubMed] [Google Scholar]
- 82.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou Q, Lui VW, Yeo W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011;7:1149–1167. doi: 10.2217/fon.11.95. [DOI] [PubMed] [Google Scholar]
- 84.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 85.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 86.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, Zhao Z, Jung JU. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell. 2016;19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chae YC, Vaira V, Caino MC, Tang HY, Seo JH, Kossenkov AV, Ottobrini L, Martelli C, Lucignani G, Bertolini I, Locatelli M, Bryant KG, Ghosh JC, Lisanti S, Ku B, Bosari S, Languino LR, Speicher DW, Altieri DC. Mitochondrial Akt regulation of hypoxic tumor reprogramming. Cancer Cell. 2016;30:257–272. doi: 10.1016/j.ccell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sahin F, Kannangai R, Adegbola O, Wang J, Su G, Torbenson M. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–8425. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 90.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, Battiston C, Van Laarhoven S, Fiel MI, Di Feo A, Hoshida Y, Yea S, Toffanin S, Ramos A, Martignetti JA, Mazzaferro V, Bruix J, Waxman S, Schwartz M, Meyerson M, Friedman SL, Llovet JM. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. 1983.e1–11. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang S, Zhu M, Wang Q, Hou Y, Li L, Weng H, Zhao Y, Chen D, Ding H, Guo J, Li M. Alpha-fetoprotein inhibits autophagy to promote malignant behaviour in hepatocellular carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death Dis. 2018;9:1027. doi: 10.1038/s41419-018-1036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang SS, Chen YH, Chen N, Wang LJ, Chen DX, Weng HL, Dooley S, Ding HG. Hydrogen sulfide promotes autophagy of hepatocellular carcinoma cells through the PI3K/Akt/mTOR signaling pathway. Cell Death Dis. 2017;8:e2688. doi: 10.1038/cddis.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang G, He J, Ye X, Zhu J, Hu X, Shen M, Ma Y, Mao Z, Song H, Chen F. beta-thujaplicin induces autophagic cell death, apoptosis, and cell cycle arrest through ROS-mediated Akt and p38/ERK MAPK signaling in human hepatocellular carcinoma. Cell Death Dis. 2019;10:255. doi: 10.1038/s41419-019-1492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Balmanno K, Cook SJ. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16:368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- 95.Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 96.Wang SH, Shih YL, Kuo TC, Ko WC, Shih CM. Cadmium toxicity toward autophagy through ROS-activated GSK-3beta in mesangial cells. Toxicol Sci. 2009;108:124–131. doi: 10.1093/toxsci/kfn266. [DOI] [PubMed] [Google Scholar]
- 97.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 98.Kim JH, Hong SK, Wu PK, Richards AL, Jackson WT, Park JI. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res. 2014;327:340–352. doi: 10.1016/j.yexcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang J, Whiteman MW, Lian H, Wang G, Singh A, Huang D, Denmark T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating beclin 1. J Biol Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maddodi N, Huang W, Havighurst T, Kim K, Longley BJ, Setaluri V. Induction of autophagy and inhibition of melanoma growth in vitro and in vivo by hyperactivation of oncogenic BRAF. J Invest Dermatol. 2010;130:1657–1667. doi: 10.1038/jid.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sivaprasad U, Basu A. Inhibition of ERK attenuates autophagy and potentiates tumour necrosis factor-alpha-induced cell death in MCF-7 cells. J Cell Mol Med. 2008;12:1265–1271. doi: 10.1111/j.1582-4934.2008.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 103.Scott AJ, Lieu CH, Messersmith WA. Therapeutic approaches to RAS mutation. Cancer J. 2016;22:165–174. doi: 10.1097/PPO.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li W, Yue F, Dai Y, Shi B, Xu G, Jiang X, Zhou X, Pfeifer GP, Liu L. Suppressor of hepatocellular carcinoma RASSF1A activates autophagy initiation and maturation. Cell Death Differ. 2019;26:1379–1395. doi: 10.1038/s41418-018-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang J, Sun P, Chen Y, Yao H, Wang S. Novel 2-phenyloxypyrimidine derivative induces apoptosis and autophagy via inhibiting PI3K pathway and activating MAPK/ERK signaling in hepatocellular carcinoma cells. Sci Rep. 2018;8:10923. doi: 10.1038/s41598-018-29199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bartholomeusz C, Rosen D, Wei C, Kazansky A, Yamasaki F, Takahashi T, Itamochi H, Kondo S, Liu J, Ueno NT. PEA-15 induces autophagy in human ovarian cancer cells and is associated with prolonged overall survival. Cancer Res. 2008;68:9302–9310. doi: 10.1158/0008-5472.CAN-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 108.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 110.Shanbhogue AK, Prasad SR, Takahashi N, Vikram R, Sahani DV. Recent advances in cytogenetics and molecular biology of adult hepatocellular tumors: implications for imaging and management. Radiology. 2011;258:673–693. doi: 10.1148/radiol.10100376. [DOI] [PubMed] [Google Scholar]
- 111.Wang Z, Sheng YY, Gao XM, Wang CQ, Wang XY, Lu XU, Wei JW, Zhang KL, Dong QZ, Qin LX. Beta-catenin mutation is correlated with a favorable prognosis in patients with hepatocellular carcinoma. Mol Clin Oncol. 2015;3:936–940. doi: 10.3892/mco.2015.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu LJ, Xie SX, Chen YT, Xue JL, Zhang CJ, Zhu F. Aberrant regulation of wnt signaling in hepatocellular carcinoma. World J Gastroenterol. 2016;22:7486–7499. doi: 10.3748/wjg.v22.i33.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Inagawa S, Itabashi M, Adachi S, Kawamoto T, Hori M, Shimazaki J, Yoshimi F, Fukao K. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Clin Cancer Res. 2002;8:450–456. [PubMed] [Google Scholar]
- 114.Su N, Wang P, Li Y. Role of wnt/beta-catenin pathway in inducing autophagy and apoptosis in multiple myeloma cells. Oncol Lett. 2016;12:4623–4629. doi: 10.3892/ol.2016.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin R, Feng J, Dong S, Pan R, Zhuang H, Ding Z. Regulation of autophagy of prostate cancer cells by beta-catenin signaling. Cell Physiol Biochem. 2015;35:926–932. doi: 10.1159/000369749. [DOI] [PubMed] [Google Scholar]
- 116.Petherick KJ, Williams AC, Lane JD, Ordonez-Moran P, Huelsken J, Collard TJ, Smartt HJ, Batson J, Malik K, Paraskeva C, Greenhough A. Autolysosomal beta-catenin degradation regulates wnt-autophagy-p62 crosstalk. EMBO J. 2013;32:1903–1916. doi: 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Turcios L, Chacon E, Garcia C, Eman P, Cornea V, Jiang J, Spear B, Liu C, Watt DS, Marti F, Gedaly R. Autophagic flux modulation by Wnt/beta-catenin pathway inhibition in hepatocellular carcinoma. PLoS One. 2019;14:e0212538. doi: 10.1371/journal.pone.0212538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang W, Xu L, Liu P, Jairam K, Yin Y, Chen K, Sprengers D, Peppelenbosch MP, Pan Q, Smits R. Blocking wnt secretion reduces growth of hepatocellular carcinoma cell lines mostly independent of beta-catenin signaling. Neoplasia. 2016;18:711–723. doi: 10.1016/j.neo.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Z, Liu T, Yu M, Li K, Li W. The plant alkaloid tetrandrine inhibits metastasis via autophagy-dependent wnt/beta-catenin and metastatic tumor antigen 1 signaling in human liver cancer cells. J Exp Clin Cancer Res. 2018;37:7. doi: 10.1186/s13046-018-0678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fan Q, Yang L, Zhang X, Ma Y, Li Y, Dong L, Zong Z, Hua X, Su D, Li H, Liu J. Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and wnt/beta-catenin signaling pathway activation in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2018;37:9. doi: 10.1186/s13046-018-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song S, Tan J, Miao Y, Li M, Zhang Q. Crosstalk of autophagy and apoptosis: involvement of the dual role of autophagy under ER stress. J Cell Physiol. 2017;232:2977–2984. doi: 10.1002/jcp.25785. [DOI] [PubMed] [Google Scholar]
- 122.Song S, Tan J, Miao Y, Zhang Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: involvement of UPR and the core autophagy machinery. J Cell Physiol. 2018;233:3867–3874. doi: 10.1002/jcp.26137. [DOI] [PubMed] [Google Scholar]
- 123.Xie Q, Su Y, Dykema K, Johnson J, Koeman J, De Giorgi V, Huang A, Schlegel R, Essenburg C, Kang L, Iwaya K, Seki S, Khoo SK, Zhang B, Buonaguro F, Marincola FM, Furge K, Vande Woude GF, Shinomiya N. Overexpression of HGF promotes HBV-induced hepatocellular carcinoma progression and is an effective indicator for met-targeting therapy. Genes Cancer. 2013;4:247–260. doi: 10.1177/1947601913501075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu F, Wu L, Zheng S, Ding W, Teng L, Wang Z, Ma Z, Zhao W. The clinical value of hepatocyte growth factor and its receptor-c-met for liver cancer patients with hepatectomy. Dig Liver Dis. 2006;38:490–497. doi: 10.1016/j.dld.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 125.Zhang R, Lin XH, Liu HH, Ma M, Chen J, Chen J, Gao DM, Cui JF, Chen RX. Activated hepatic stellate cells promote progression of post-heat residual hepatocellular carcinoma from autophagic survival to proliferation. Int J Hyperthermia. 2019;36:253–263. doi: 10.1080/02656736.2018.1558459. [DOI] [PubMed] [Google Scholar]
- 126.Tavian D, De Petro G, Benetti A, Portolani N, Giulini SM, Barlati S. u-PA and c-MET mRNA expression is co-ordinately enhanced while hepatocyte growth factor mRNA is down-regulated in human hepatocellular carcinoma. Int J Cancer. 2000;87:644–649. [PubMed] [Google Scholar]
- 127.Huang X, Gan G, Wang X, Xu T, Xie W. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy. 2019;15:1258–1279. doi: 10.1080/15548627.2019.1580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee YA, Noon LA, Akat KM, Ybanez MD, Lee TF, Berres ML, Fujiwara N, Goossens N, Chou HI, Parvin-Nejad FP, Khambu B, Kramer EGM, Gordon R, Pfleger C, Germain D, John GR, Campbell KN, Yue Z, Yin XM, Cuervo AM, Czaja MJ, Fiel MI, Hoshida Y, Friedman SL. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of yap. Nat Commun. 2018;9:4962. doi: 10.1038/s41467-018-07338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Berasain C, Ujue Latasa M, Urtasun R, Goni S, Elizalde M, Garcia-Irigoyen O, Azcona M, Prieto J, Avila MA. Epidermal Growth Factor Receptor (EGFR) crosstalks in liver cancer. Cancers (Basel) 2011;3:2444–2461. doi: 10.3390/cancers3022444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang , Li T, Wang L, Zhang L, Yan R, Li K, Xing S, Wu G, Hu L, Jia W, Lin SC, Dang CV, Song L, Gao P, Zhang H. Hepatocellular carcinoma redirects to ketolysis for progression under nutrition deprivation stress. Cell Res. 2016;26:1112–1130. doi: 10.1038/cr.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 132.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 133.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 134.Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol. 2013;10:34–42. doi: 10.1038/nrgastro.2012.199. [DOI] [PubMed] [Google Scholar]
- 135.Kaseb AO, Shindoh J, Patt YZ, Roses RE, Zimmitti G, Lozano RD, Hassan MM, Hassabo HM, Curley SA, Aloia TA, Abbruzzese JL, Vauthey JN. Modified cisplatin/interferon alpha-2b/doxorubicin/5-fluorouracil (PIAF) chemotherapy in patients with no hepatitis or cirrhosis is associated with improved response rate, resectability, and survival of initially unresectable hepatocellular carcinoma. Cancer. 2013;119:3334–3342. doi: 10.1002/cncr.28209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Deng GL, Zeng S, Shen H. Chemotherapy and target therapy for hepatocellular carcinoma: new advances and challenges. World J Hepatol. 2015;7:787–798. doi: 10.4254/wjh.v7.i5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Petrelli F, Coinu A, Borgonovo K, Cabiddu M, Ghilardi M, Lonati V, Barni S. Oxaliplatin-based chemotherapy: a new option in advanced hepatocellular carcinoma. a systematic review and pooled analysis. Clin Oncol (R Coll Radiol) 2014;26:488–496. doi: 10.1016/j.clon.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 139.Du H, Yang W, Chen L, Shi M, Seewoo V, Wang J, Lin A, Liu Z, Qiu W. Role of autophagy in resistance to oxaliplatin in hepatocellular carcinoma cells. Oncol Rep. 2012;27:143–150. doi: 10.3892/or.2011.1464. [DOI] [PubMed] [Google Scholar]
- 140.Ding ZB, Hui B, Shi YH, Zhou J, Peng YF, Gu CY, Yang H, Shi GM, Ke AW, Wang XY, Song K, Dai Z, Shen YH, Fan J. Autophagy activation in hepatocellular carcinoma contributes to the tolerance of oxaliplatin via reactive oxygen species modulation. Clin Cancer Res. 2011;17:6229–6238. doi: 10.1158/1078-0432.CCR-11-0816. [DOI] [PubMed] [Google Scholar]
- 141.Ren WW, Li DD, Chen X, Li XL, He YP, Guo LH, Liu LN, Sun LP, Zhang XP. MicroRNA-125b reverses oxaliplatin resistance in hepatocellular carcinoma by negatively regulating EVA1A mediated autophagy. Cell Death Dis. 2018;9:547. doi: 10.1038/s41419-018-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang N, Wu Y, Lyu X, Li B, Yan X, Xiong H, Li X, Huang G, Zeng Y, Zhang Y, Lian J, Ni Z, He F. HSF1 upregulates ATG4B expression and enhances epirubicin-induced protective autophagy in hepatocellular carcinoma cells. Cancer Lett. 2017;409:81–90. doi: 10.1016/j.canlet.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 143.Guo XL, Li D, Hu F, Song JR, Zhang SS, Deng WJ, Sun K, Zhao QD, Xie XQ, Song YJ, Wu MC, Wei LX. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–179. doi: 10.1016/j.canlet.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 144.Wu B, Cui J, Yang XM, Liu ZY, Song F, Li L, Jiang JL, Chen ZN. Cytoplasmic fragment of CD147 generated by regulated intramembrane proteolysis contributes to HCC by promoting autophagy. Cell Death Dis. 2017;8:e2925. doi: 10.1038/cddis.2017.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, Liu C, Kabar I, Schmidt HH, Zhang X, Lu M, Cicinnati VR. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590–598. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 146.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, Xu J, Sun Y, Liang H, Liu J, Wang J, Tak WY, Pan H, Burock K, Zou J, Voliotis D, Guan Z. Efficacy and safety of sorafenib in patients in the asia-pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 147.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 148.Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7:3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guidetti A, Carlo-Stella C, Locatelli SL, Malorni W, Pierdominici M, Barbati C, Mortarini R, Devizzi L, Matteucci P, Marchiano A, Lanocita R, Farina L, Dodero A, Tarella C, Di Nicola M, Corradini P, Anichini A, Gianni AM. Phase II study of sorafenib in patients with relapsed or refractory lymphoma. Br J Haematol. 2012;158:108–119. doi: 10.1111/j.1365-2141.2012.09139.x. [DOI] [PubMed] [Google Scholar]
- 150.Fischer TD, Wang JH, Vlada A, Kim JS, Behrns KE. Role of autophagy in differential sensitivity of hepatocarcinoma cells to sorafenib. World J Hepatol. 2014;6:752–758. doi: 10.4254/wjh.v6.i10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, Hosui A, Ishida H, Tatsumi T, Kanto T, Hiramatsu N, Fujita N, Yoshimori T, Hayashi N. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548–557. doi: 10.1002/ijc.26374. [DOI] [PubMed] [Google Scholar]
- 152.Chen KF, Chen HL, Tai WT, Feng WC, Hsu CH, Chen PJ, Cheng AL. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J Pharmacol Exp Ther. 2011;337:155–161. doi: 10.1124/jpet.110.175786. [DOI] [PubMed] [Google Scholar]
- 153.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lu S, Yao Y, Xu G, Zhou C, Zhang Y, Sun J, Jiang R, Shao Q, Chen Y. CD24 regulates sorafenib resistance via activating autophagy in hepatocellular carcinoma. Cell Death Dis. 2018;9:646. doi: 10.1038/s41419-018-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, Li T, Wang CZ, Tan YX, Ding J, Chen Y, Tang L, Guo LN, Tang SH, Yang W, Wang HY. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol. 2016;65:314–324. doi: 10.1016/j.jhep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 157.Zhang K, Chen J, Zhou H, Chen Y, Zhi Y, Zhang B, Chen L, Chu X, Wang R, Zhang C. PU.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 2018;9:312. doi: 10.1038/s41419-018-0344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Manov I, Pollak Y, Broneshter R, Iancu TC. Inhibition of doxorubicin-induced autophagy in hepatocellular carcinoma Hep3B cells by sorafenib--the role of extracellular signal-regulated kinase counteraction. FEBS J. 2011;278:3494–3507. doi: 10.1111/j.1742-4658.2011.08271.x. [DOI] [PubMed] [Google Scholar]
- 159.Han R, Li S. Regorafenib delays the proliferation of hepatocellular carcinoma by inducing autophagy. Pharmazie. 2018;73:218–222. doi: 10.1691/ph.2018.7988. [DOI] [PubMed] [Google Scholar]
- 160.Tong M, Che N, Zhou L, Luk ST, Kau PW, Chai S, Ngan ES, Lo CM, Man K, Ding J, Lee TK, Ma S. Efficacy of annexin A3 blockade in sensitizing hepatocellular carcinoma to sorafenib and regorafenib. J Hepatol. 2018;69:826–839. doi: 10.1016/j.jhep.2018.05.034. [DOI] [PubMed] [Google Scholar]
- 161.Zhang P, Zheng Z, Ling L, Yang X, Zhang N, Wang X, Hu M, Xia Y, Ma Y, Yang H, Wang Y, Liu H. w09, a novel autophagy enhancer, induces autophagy-dependent cell apoptosis via activation of the EGFR-mediated RAS-RAF1-MAP2K-MAPK1/3 pathway. Autophagy. 2017;13:1093–1112. doi: 10.1080/15548627.2017.1319039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li WY, Li Q, Jing L, Wu T, Han LL, Wang Y, Yu SZ, Nan KJ, Guo H. P57-mediated autophagy promotes the efficacy of EGFR inhibitors in hepatocellular carcinoma. Liver Int. 2019;39:147–157. doi: 10.1111/liv.13957. [DOI] [PubMed] [Google Scholar]
- 163.Guo XL, Li D, Sun K, Wang J, Liu Y, Song JR, Zhao QD, Zhang SS, Deng WJ, Zhao X, Wu MC, Wei LX. Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. J Mol Med (Berl) 2013;91:473–483. doi: 10.1007/s00109-012-0966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pan H, Wang Z, Jiang L, Sui X, You L, Shou J, Jing Z, Xie J, Ge W, Cai X, Huang W, Han W. Autophagy inhibition sensitizes hepatocellular carcinoma to the multikinase inhibitor linifanib. Sci Rep. 2014;4:6683. doi: 10.1038/srep06683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu JY, Wang ZX, Zhang G, Lu X, Qiang GH, Hu W, Ji AL, Wu JH, Jiang CP. Targeted co-delivery of beclin 1 siRNA and FTY720 to hepatocellular carcinoma by calcium phosphate nanoparticles for enhanced anticancer efficacy. Int J Nanomedicine. 2018;13:1265–1280. doi: 10.2147/IJN.S156328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Klose J, Stankov MV, Kleine M, Ramackers W, Panayotova-Dimitrova D, Jager MD, Klempnauer J, Winkler M, Bektas H, Behrens GM, Vondran FW. Inhibition of autophagic flux by salinomycin results in anti-cancer effect in hepatocellular carcinoma cells. PLoS One. 2014;9:e95970. doi: 10.1371/journal.pone.0095970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Seo JS, Choi YH, Moon JW, Kim HS, Park SH. Hinokitiol induces DNA demethylation via DNMT1 and UHRF1 inhibition in colon cancer cells. BMC Cell Biol. 2017;18:14. doi: 10.1186/s12860-017-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee YS, Choi KM, Kim W, Jeon YS, Lee YM, Hong JT, Yun YP, Yoo HS. Hinokitiol inhibits cell growth through induction of S-phase arrest and apoptosis in human colon cancer cells and suppresses tumor growth in a mouse xenograft experiment. J Nat Prod. 2013;76:2195–2202. doi: 10.1021/np4005135. [DOI] [PubMed] [Google Scholar]
- 169.Byeon SE, Lee YG, Kim JC, Han JG, Lee HY, Cho JY. Hinokitiol, a natural tropolone derivative, inhibits TNF-alpha production in LPS-activated macrophages via suppression of NF-kappaB. Planta Med. 2008;74:828–833. doi: 10.1055/s-2008-1074548. [DOI] [PubMed] [Google Scholar]
- 170.Liu YH, Weng YP, Lin HY, Tang SW, Chen CJ, Liang CJ, Ku CY, Lin JY. Aqueous extract of polygonum bistorta modulates proteostasis by ROS-induced ER stress in human hepatoma cells. Sci Rep. 2017;7:41437. doi: 10.1038/srep41437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang DM, Liu JS, Deng LJ, Chen MF, Yiu A, Cao HH, Tian HY, Fung KP, Kurihara H, Pan JX, Ye WC. Arenobufagin, a natural bufadienolide from toad venom, induces apoptosis and autophagy in human hepatocellular carcinoma cells through inhibition of PI3K/Akt/mTOR pathway. Carcinogenesis. 2013;34:1331–1342. doi: 10.1093/carcin/bgt060. [DOI] [PubMed] [Google Scholar]
- 172.Zhang X, Tang X, Liu H, Li L, Hou Q, Gao J. Autophagy induced by baicalin involves downregulation of CD147 in SMMC-7721 cells in vitro. Oncol Rep. 2012;27:1128–1134. doi: 10.3892/or.2011.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zou M, Lu N, Hu C, Liu W, Sun Y, Wang X, You Q, Gu C, Xi T, Guo Q. Beclin 1-mediated autophagy in hepatocellular carcinoma cells: implication in anticancer efficiency of oroxylin a via inhibition of mTOR signaling. Cell Signal. 2012;24:1722–1732. doi: 10.1016/j.cellsig.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 174.Hou Q, Tang X, Liu H, Tang J, Yang Y, Jing X, Xiao Q, Wang W, Gou X, Wang Z. Berberine induces cell death in human hepatoma cells in vitro by downregulating CD147. Cancer Sci. 2011;102:1287–1292. doi: 10.1111/j.1349-7006.2011.01933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang L, Gao C, Yao S, Xie B. Blocking autophagic flux enhances matrine-induced apoptosis in human hepatoma cells. Int J Mol Sci. 2013;14:23212–23230. doi: 10.3390/ijms141223212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tang ZH, Li T, Chang LL, Zhu H, Tong YG, Chen XP, Wang YT, Lu JJ. Glycyrrhetinic acid triggers a protective autophagy by activation of extracellular regulated protein kinases in hepatocellular carcinoma cells. J Agric Food Chem. 2014;62:11910–11916. doi: 10.1021/jf503968k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li-Weber M. New therapeutic aspects of flavones: the anticancer properties of scutellaria and its main active constituents wogonin, baicalein and baicalin. Cancer Treat Rev. 2009;35:57–68. doi: 10.1016/j.ctrv.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 178.Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW, Wang XY, Dai Z, Peng YF, Gu CY, Qiu SJ, Fan J. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–1172. doi: 10.4161/auto.7.10.16818. [DOI] [PubMed] [Google Scholar]
- 179.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 180.Ong LC, Song IC, Jin Y, Kee IH, Siew E, Yu S, Thng CH, Huynh H, Chow PK. Effective inhibition of xenografts of hepatocellular carcinoma (HepG2) by rapamycin and bevacizumab in an intrahepatic model. Mol Imaging Biol. 2009;11:334–342. doi: 10.1007/s11307-009-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Semela D, Piguet AC, Kolev M, Schmitter K, Hlushchuk R, Djonov V, Stoupis C, Dufour JF. Vascular remodeling and antitumoral effects of mTOR inhibition in a rat model of hepatocellular carcinoma. J Hepatol. 2007;46:840–848. doi: 10.1016/j.jhep.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 182.Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, Tanaka K, Matsutani T, Iwanami A, Aronow BJ, Manway L, Maira SM, Thorgeirsson SS, Mischel PS, Thomas G, Kozma SC. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med. 2012;4:139ra184. doi: 10.1126/scitranslmed.3003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rodriguez-Hernandez MA, Gonzalez R, de la Rosa AJ, Gallego P, Ordonez R, Navarro-Villaran E, Contreras L, Rodriguez-Arribas M, Gonzalez-Gallego J, Alamo-Martinez JM, Marin-Gomez LM, Del Campo JA, Quiles JL, Fuentes JM, de la Cruz J, Mauriz JL, Padillo FJ, Muntane J. Molecular characterization of autophagic and apoptotic signaling induced by sorafenib in liver cancer cells. J Cell Physiol. 2018;234:692–708. doi: 10.1002/jcp.26855. [DOI] [PubMed] [Google Scholar]
- 184.Kim DG, Jung KH, Lee DG, Yoon JH, Choi KS, Kwon SW, Shen HM, Morgan MJ, Hong SS, Kim YS. 20(S)-ginsenoside Rg3 is a novel inhibitor of autophagy and sensitizes hepatocellular carcinoma to doxorubicin. Oncotarget. 2014;5:4438–4451. doi: 10.18632/oncotarget.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li W, Dong X, He C, Tan G, Li Z, Zhai B, Feng J, Jiang X, Liu C, Jiang H, Sun X. LncRNA SNHG1 contributes to sorafenib resistance by activating the akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2019;38:183. doi: 10.1186/s13046-019-1177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lachenmayer A, Toffanin S, Cabellos L, Alsinet C, Hoshida Y, Villanueva A, Minguez B, Tsai HW, Ward SC, Thung S, Friedman SL, Llovet JM. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J Hepatol. 2012;56:1343–1350. doi: 10.1016/j.jhep.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]


