Abstract
Bifidobacteria are exposed to substantial amounts of dietary β-galactosides. Distinctive preferences for growth on different β-galactosides are observed within Bifidobacterium members, but the basis of these preferences remains unclear. We previously described the first β-(1,6)/(1,3)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04. This enzyme is relatively promiscuous, exhibiting only 5-fold higher efficiency on the preferred β-(1,6)-galactobiose than the β-(1,4) isomer. Here, we characterize the solute-binding protein (Bal6GBP) that governs the specificity of the ABC transporter encoded by the same β-galactoside utilization locus. We observed that although Bal6GBP recognizes both β-(1,6)- and β-(1,4)-galactobiose, Bal6GBP has a 1630-fold higher selectivity for the former, reflected in dramatic differences in growth, with several hours lag on less preferred β-(1,4)- and β-(1,3)-galactobiose. Experiments performed in the presence of varying proportions of β-(1,4)/β-(1,6)-galactobioses indicated that the preferred substrate was preferentially depleted from the culture supernatant. This established that the poor growth on the nonpreferred β-(1,4) was due to inefficient uptake. We solved the structure of Bal6GBP in complex with β-(1,6)-galactobiose at 1.39 Å resolution, revealing the structural basis of this strict selectivity. Moreover, we observed a close evolutionary relationship with the human milk disaccharide lacto-N-biose-binding protein from Bifidobacterium longum, indicating that the recognition of the nonreducing galactosyl is essentially conserved, whereas the adjacent position is diversified to fit different glycosidic linkages and monosaccharide residues. These findings indicate that oligosaccharide uptake has a pivotal role in governing selectivity for distinct growth substrates and have uncovered evolutionary trajectories that shape the diversification of sugar uptake proteins within Bifidobacterium.
Keywords: ABC transporter, surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), crystal structure, actinobacteria, protein evolution, microbiome, probiotic, evolution, enzyme kinetics, bifidobacteria, galactooligosaccharides (GOS), human gut microbiota, human milk oligosaccharides (HMO), prebiotics
Introduction
The human gut microbiota (HGM)4 is increasingly acknowledged as a major contributor to human health as well as a modulator of host metabolism (1) and immune-homeostasis (2). Specific microbiota signatures are associated with serious disease states, including inflammatory disorders (3), obesity, type 2 diabetes (4), and colorectal cancer (5), underscoring the crucial impact of a balanced HGM composition. Diet is a major external factor affecting the composition of the microbiota (6), and oligo- as well as polysaccharides that are nondigestible to the human host are instrumental in shaping this community (7). The abundance and the diversity of dietary glycans have driven the evolution of different microbial strategies to harvest energy from these metabolic resources. Dominant commensals from the Bacteroides genus are considered generalists that target a broad range of glycans via outer membrane–bound extracellular enzymes (7, 8). Other taxonomic groups (e.g. bifidobacteria) typically possess fewer extracellular enzymes compared with Bacteroides. Instead, bifidobacteria rely on effective transport systems, mainly ATP-binding cassette (ABC) transporters, to access oligosaccharides that are either naturally present in diet (9) or produced by hydrolytic enzymes from other HGM members, thereby facilitating cross-feeding (10, 11). Genomes of bifidobacteria typically encode a multitude of oligosaccharide-specific ABC transporters (12), which mediate high-affinity capture of oligosaccharides via their extracellular lipid-anchored solute-binding proteins (SBPs). Uptake of oligosaccharides by these transporters is likely to confer an important advantage in the competitive human gut niche, particularly in the colon, which harbors the highest bacterial density. Despite this key role, our insight into oligosaccharide uptake in the human gut niche remains limited.
β-Galactosides are ubiquitous in the human diet (e.g. in both human and animal milk) (13 and in fruits and vegetables, where they are omnipresent in primary cell wall pectic polysaccharides (14)). Oligomeric β-galactosides of different origin have been shown to selectively promote members of the Bifidobacterium genus that harbor many probiotic health-promoting strains (15). Dietary β-galactosides display source-dependent diversity with respect to degree of polymerization (DP), glycosidic linkages, and the presence of other monosaccharides. β-Galactosides are also considered prime prebiotic dietary supplements (galactooligosaccharides (GOS)) produced by transglycosylation of lactose, partially due to their bifidogenic effect. Notably, commercial GOS preparations typically comprise tens of compounds of DP 2–6 and contain β-(1,3)-, β-(1,4)-, and β-(1,6)-galactosidic linkages (16). Only some of these oligosaccharides are metabolized by bifidobacteria in a species- and strain-specific manner (17–19). Knowledge about β-galactoside preferences of bifidobacteria largely stems from characterization of β-galactosidases (20) or from growth and oligosaccharide consumption profiles (19). The role of oligosaccharide transporters in β-galactooligosaccharide utilization selectivity has received less attention, with two recent studies having addressed the identification and binding properties of β-galactooligosaccharide transport proteins in Bifidobacterium breve (21, 22).
We have previously identified a transcriptionally induced operon during growth of the probiotic strain Bifidobacterium animalis subsp. lactis Bl-04 on GOS (a commercial β-galactoside mixture). This operon encodes a transcriptional regulator, a β-galactosidase of glycoside hydrolase family 42 (BlGH42A), and an ABC transporter (23). The β-galactosidase revealed a broad specificity, hydrolyzing β-(1,6)-, β-(1,3)-, and β-(1,4)-galactosidic bonds with a modest 5-fold preference for β-(1,6)-galactobiose (β6Gal2) as compared with the β-(1,4) isomer (24). Here, we report the molecular characterization of the β-galactoside–specific SBP (Bal6GBP) associated with the ABC transporter. In contrast to the relatively promiscuous β-gal, Bal6GBP displayed selectivity exceeding 3 orders of magnitude for β6Gal2 as compared with the β-(1,4) isomers, which was explained by the structure of Bal6GBP in complex with β6Gal2. The high selectivity of Bal6GBP is reflected in a dramatic difference in growth profiles of B. animalis subsp. lactis Bl-04 showing several hours of lag phase on less preferred substrates. This was shown to be attributed to the inefficient uptake of ligands that are less preferred by the binding protein.
Structural analyses allowed tracking the evolutionary relationship with the lacto-N-biose (human milk-derived) binding protein from Bifidobacterium longum subsp. longum (25), which shares only 26% sequence identity with Bal6GBP. Despite the low primary structure identities, these two binding proteins share a similar architecture of the side chains interacting with the nonreducing galactosyl moiety but diverge in the recognition of the penultimate carbohydrate ring from the nonreducing end.
Altogether, the present study constitutes a unique case enabling the comparison of ligand selectivity of an ABC-associated transport protein and the intracellular glycoside hydrolase mediating uptake and breakdown, respectively, of the same substrates. These findings underscore the leading role of ABC-mediated glycan transport in governing the growth preferences of core gut microbiota members on β-galactosides. This insight paves the way to tailored editing of specific members of the microbiota based on uptake profiles in future interventions to promote health and to alleviate disease.
Results
The ABC transporter–associated binding protein from B. animalis subsp. lactis Bl-04 is highly specific toward β-(1,6)-galactosides
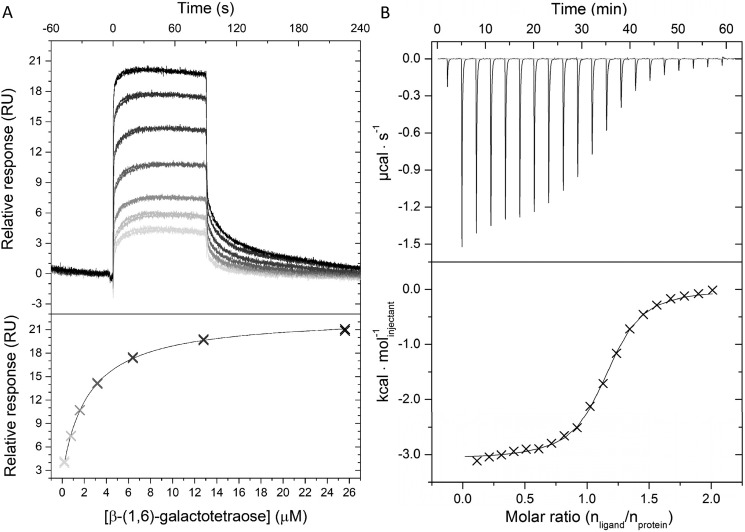
The β-galactoside SBP from B. animalis subsp. lactis Bl-04 (Bal6GBP) was recombinantly produced and purified (yield = 85 mg liter−1 medium). The binding of a panel of di- and oligosaccharides to Bal6GBP was screened using surface plasmon resonance (SPR), which established the specificity toward β-galactopyranosyl–linked ligands (Table S1). The binding affinities and kinetic parameters of the β-galactoside ligands (Fig. 1) were discerned by SPR (Fig. 2A and Table 1). The highest affinity of Bal6GBP was for β6Gal2 (KD ≈ 100 nm), and the selectivity toward this galactosidic linkage was striking as the affinity dropped by 304- and 1630-fold for β-(1,3)-galactobiose (β3Gal2) and β-(1,4)-galactobiose (β4Gal2), respectively. The selectivity was governed by staggering (up to 3 × 104-fold) changes in the association rate constant (kon), whereas the dissociation rate constants (koff) varied modestly with glycosidic linkage. The affinity appeared to decrease with size for β-(1,6)-galactosides with a 26-fold drop for β-(1,6)-galactotetraose (β6Gal4) as compared with β6Gal2. This trend was not valid for the less preferred β-(1,4)-galactosides. The affinity of Bal6GBP was highest at pH 6.5 but varied only modestly in the pH range 5.4–8.2, consistent with the slightly acidic pH prevalent in the colon.
Figure 1.
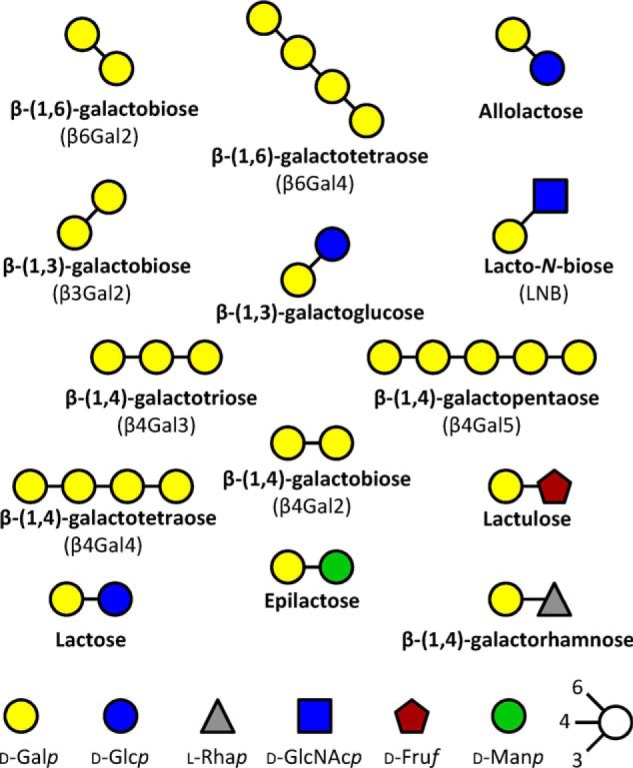
Schematic representation of β-galactosides that display binding to the transport protein Bal6GBP from B. animalis subsp. lactis. The β-galactosides are ordered according to preferred glycosidic bond by Bal6GBP.
Figure 2.
Binding analyses of Bal6GBP. A, binding of β-(1,6)-galactotetraose to Bal6GP as analyzed by surface plasmon resonance at 25 °C and pH 6.5. Shown are blank and reference cell–corrected sensograms (top) and one binding site steady-state analysis (bottom) to seven ligand concentrations (0.2–25.6 μm). B, isothermal titration calorimetry binding analysis of Bal6GBP (246 μm) to β-(1,6)-galactobiose (2.4 mm) at 25 °C and pH 6.5. Top, baseline-adjusted thermogram; bottom, binding isotherm and a one-site binding model fit to the data (solid line).
Table 1.
SPR binding analysis of Bal6GBP to β-galactosides of different glycosidic linkages, DP, and composition
Experiments were performed in at least duplicates.
| Linkage | Ligand | KD | kon | koff | Rmaxa |
|---|---|---|---|---|---|
| m | m−1 s−1 | 10−2 s−1 | RU | ||
| β-(1,6) | β6Gal2 | (9.2 ± 0.5) × 10−8 | (6.91 ± 0.02) × 106 | 7.39 ± 0.02 | 11.7 ± 0.02 |
| β6Gal4 | (2.4 ± 0.1) × 10−6 | (2.12 ± 0.04) × 104 | 1.28 ± 0.02 | 5.6 ± 0.02 | |
| Allolactose | (1.3 ± 0.3) × 10−5 | NDb | ND | 12.1 ± 0.59 | |
| β-(1,3) | β3Gal2 | (2.8 ± 0.2) × 10−5 | (2.48 ± 0.01) × 103 | 4.66 ± 0.01 | 16.2 ± 0.05 |
| Galβ13Glc | (5.7 ± 0.7) × 10−4 | ND | ND | 12.1 ± 0.33 | |
| β-(1,4) | β4Gal2 | (1.5 ± 0.3) × 10−4 | (3.22 ± 0.1) × 102 | 5.97 ± 0.08 | 14.1 ± 0.06 |
| β4Gal3 | (3.4 ± 0.2) × 10−5 | ND | ND | 22.7 ± 0.37 | |
| β4Gal4 | (1.2 ± 0.03) × 10−4 | ND | ND | 25.6 ± 0.13 | |
| β4Gal5 | (1.0 ± 0.1) × 10−4 | (3.20 ± 0.06) × 102 | 1.55 ± 0.02 | 8.5 ± 0.07 | |
| Lactulose | (4.9 ± 0.3) × 10−4 | (2.71 ± 0.06) × 102 | 6.61 ± 0.08 | 8.4 ± 0.10 | |
| Lactose | (7.2 ± 0.4) × 10−4 | (2.00 ± 0.02) × 102 | 6.71 ± 0.03 | 7.5 ± 0.04 | |
| Epilactose | (2.1 ± 0.2) × 10−3 | (6.35 ± 0.06) × 101 | 8.87 ± 0.06 | 7.7 ± 0.03 |
a Maximum binding signal level determined from the fits to the data.
b ND, binding kinetics were not within range for reliable determination.
Isothermal titration calorimetry (ITC) analyses confirmed the binding magnitude and preference of Bal6GBP for β-(1,6)-galactosides (Fig. 2B and Table 2).
Table 2.
Binding energetics of Bal6GBP to β6Gal2 and β6Gal4 at 25 °C and pH 6.5 using ITC
| Ligand | Sites (n) | KD | ΔG | ΔH | −TΔS |
|---|---|---|---|---|---|
| m | kJ mol−1 | kJ mol−1 | kJ mol−1 | ||
| β6Gal2a | 0.61 ± 0.0 | (1.4 ± 0.02) × 10−7 | −39.7 | −59.4 ± 0.7 | 19.8 |
| β6Gal4b | 1.1 ± 0.0 | (4.1 ± 0.2) × 10−6 | −30.7 | −13.2 ± 0.1 | −17.5 |
a Average of three experiments. The deviation of the stoichiometry from 1:1 is likely due to inaccuracy in the ligand concentration, which makes the thermodynamic parameters from this measurement of this ligand less reliable.
b Average of two experiments.
Overall three-dimensional structure of Bal6GBP in complex with the preferred ligand
The crystal structure of Bal6GBP in the free open conformation was solved to a resolution of 1.94 Å by SeMet single anomalous dispersion phasing. Coordinates of the separate domains from the open conformation were used as molecular replacement (MR) models to solve the structure of Bal6GBP in complex with the preferred ligand β6Gal2 to 1.39 Å (Table 3).
Table 3.
Data collection and refinement statistics of the X-ray crystal structure of native Bal6GBP in complex with β-(1,6)-galactobiose and the selenomethionine-labeled protein in open form
| PDB entry 6H0H (complex with β6Gal2) | PDB entry 6Q5G (SeMet open form) | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.000 | 0.979 |
| Resolution range (Å) | 30.21–1.39 (1.44–1.39) | 46.19–1.94 (2.01–1.94) |
| Space group | P 1 21 1 | P 1 21 1 |
| a, b, c (Å) | 56.35, 71.69, 88.71 | 62.94, 69.36, 97.30 |
| β (degrees) | 95.04 | 104.43 |
| Total reflections | 489,576 (19,734) | 378,068 (21,326) |
| Unique reflections | 130,307 (8129) | 57,952 (4289) |
| Multiplicity | 3.8 (2.4) | 6.5 (5.0) |
| Completeness (%) | 91.81 (57.69) | 96.38 (71.74) |
| Mean I/σ(I) | 18.59 (2.39) | 12.69 (2.64) |
| Refinement | ||
| Wilson B-factor (Å2) | 10.84 | 27.65 |
| Rmerge | 0.04525 (0.3927) | 0.09661 (0.3816) |
| Rmeas | 0.05249 (0.4990) | 0.1049 (0.4255) |
| Rpim | 0.02629 (0.3028) | 0.0405 (0.1826) |
| CC½ | 0.999 (0.799) | 0.996 (0.931) |
| CC* | 1.000 (0.943) | 0.999 (0.982) |
| Reflections used in refinement | 129,672 (8129) | 57,952 (4288) |
| Reflections used for Rfree | 6526 (427) | 1078 (80) |
| Rwork | 0.1161 (0.2066) | 0.1588 (0.2408) |
| Rfree | 0.1464 (0.2344) | 0.2035 (0.2848) |
| CC(work) | 0.982 (0.902) | 0.971 (0.935) |
| CC(free) | 0.973 (0.866) | 0.965 (0.809) |
| Number of non-hydrogen atoms | 8134 | 7148 |
| Macromolecules | 6765 | 6444 |
| Ligands | 138 | None |
| Solvent | 1231 | 704 |
| Protein residues | 820 | 832 |
| RMSD (bonds) | 0.007 | 0.010 |
| RMSD (angles) | 0.92 | 0.96 |
| Ramachandran favored (%) | 97.79 | 97.58 |
| Ramachandran allowed (%) | 1.96 | 2.29 |
| Ramachandran outliers (%) | 0.25 | 0.12 |
| Rotamer outliers (%) | 0.86 | 1.97 |
| Clashscore | 1.91 | 2.30 |
| Average B-factor | 16.31 | 33.39 |
| Macromolecules | 13.96 | 32.86 |
| Ligands | 19.93 | None |
| Solvent | 28.78 | 38.32 |
| Number of TLS groups | 14 | None |
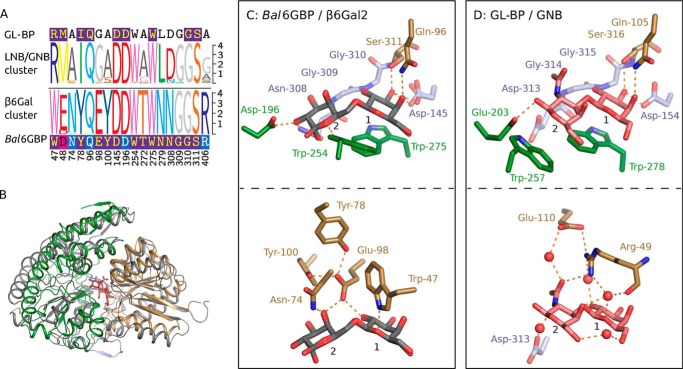
Bal6GBP adopts a classical SBP-fold (cluster B type SBP according to structural classification (26)), which comprises two α/β domains of different sizes interconnected by three hinge regions with the ligand binding site located at the domain interface (Fig. 3A). The smaller domain 1 (residues 1–144 and 311–362) is formed by six α-helices and five centrally positioned β-strands. Domain 2 (residues 149–306 and 374–447) consists of 10 α-helices and six β-strands. The architecture of these two domains is similar to other SBP structures (27). The hinge region that connects the two domains and potentiates domain movement between the open and closed conformation (Fig. S1) consists of two loop regions (residues 145–148 and 307–310) and an anti-parallel β-sheet formed by two short β-strands 363–373 (Fig. 3A). A DALI structure comparison search (28) against the Protein Data Bank (PDB) identified the (galacto/lacto)-N-biose (GNB/LNB)-binding protein (GL-BP) from B. longum subsp. longum ATCC 15697 (PDB code 2Z8D-F; z-score = 42.0; RMSD = 2.8 Å; 26% sequence identity) as the structurally closest related homologue to Bal6GBP (25). The structure of GL-BP was determined in complex with galacto-N-biose (PDB code 2Z8E), lacto-N-biose (PDB code 2Z8D), and lacto-N-tetraose (PDB code 2Z8F).
Figure 3.
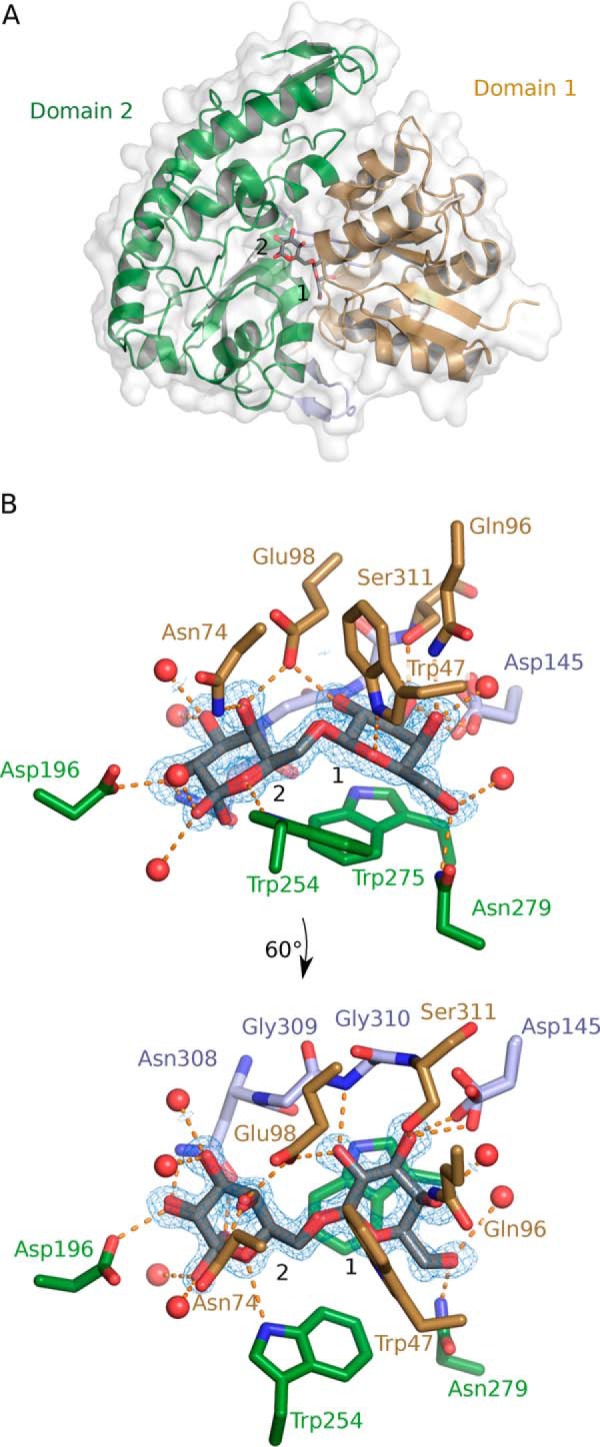
Bal6GBP protein structure in complex with β-(1,6)-galactobiose (β6Gal2). A, overall structure of Bal6GBP with a semitransparent white surface that visualizes the protein surface. Domain 1 (residues 38–144 and 311–362) is shown in brown and domain 2 (residues 149–306 and 374–447) in green, held together by linker regions (residues 145–148, 307–310, and 363–373) in light blue. The nonreducing galactosyl unit of β6Gal2 occupies position 1, which is defined by the aromatic platform provided by Trp-275. B, the residues and waters (red spheres) that directly interact with the bound β6Gal2 are shown, and the dashed orange lines indicate polar interactions. The mFobs − DFcalc difference electron map for β6Gal2 is shown prior to inclusion of the ligand in the model at a contour level of 3σ (blue mesh). The main-chain atoms are omitted for clarity unless they participate in polar interactions. The bottom panel is a view rotated 60° along the x axis.
The crystal obtained for phasing from the SeMet-labeled protein belongs to the same space group as the ligand-bound form, but with a different β angle and slightly different cell lengths (Table 3). The structure of the open form of the protein was determined from the SeMet protein crystals, which revealed high flexibility between domain 1 and domain 2, resulting in an open ligand-free form and a closed ligand-bound form (29). The transition between these two states involves a rotation angle of 32.3° between the domains of Bal6GBP, as estimated with the DynDom server (30) (Fig. S1). Thus, closure of the protein involves a rigid-body movement of domains with changes mainly occurring in the hinge region.
β-(1,6)-Galactobiose-binding site in Bal6GBP
Bal6GBP in complex with β6Gal2 showed a well-defined density of the ligand that primarily assumes the β-configuration. The ligand is almost completely engulfed in the largely solvent-inaccessible binding site (Fig. S2). The nonreducing galactosyl stacks onto Trp-275 from domain 2, which defines position 1. A total of 12 potential hydrogen bonds recognize β6Gal2: three from domain 1, six from domain 2, and three from the hinge region (Table S2 and Fig. 3B). Five of the six water molecules that coordinate the ligand also mediate hydrogen-bonding networks to residues in the binding pocket. All hydroxyl groups of the galactosyl at position 1 are recognized by polar interactions to Gln-96, Glu-98, Asp-145, Asn-279, Gly-310, and Ser-311. This correlates well with the strict requirement for β-galactosyl at position 1 for recognition by Bal6GBP. By contrast, only the C2-OH and C4-OH of the galactosyl at position 2 are recognized by direct polar contacts to Asn-74, Glu-98, Asp-196, and Asn-308. Glucosyl can be accommodated at position 2, albeit with 140-fold reduced affinity for the disaccharide allolactose as compared with β6Gal2 (Table 2), probably due to the recognition of the axial C4-OH in β6Gal2 by Asn-74 and Glu-98, which also interacts with the C2-OH from the reducing end galactosyl unit. Both hemiacetal ring oxygens in β6Gal2 form hydrogen bonds with indole nitrogens of tryptophans (Trp-47 and Trp-254; Fig. 3B).
Transporter preference is reflected in the β-galactobiose isomer growth profiles of B. animalis subsp. lactis Bl-04
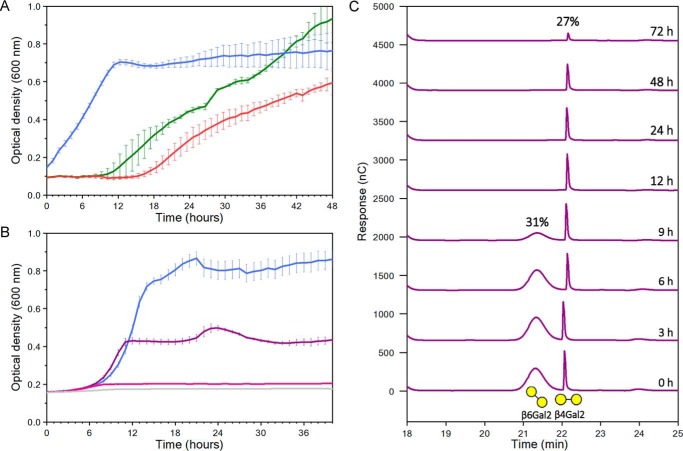
Only two loci were previously observed to be highly up-regulated during growth on galactooligosaccharides, the canonical lactose metabolism locus encoding a LacS transport system and the ABC system characterized in the present study (23), suggesting that this ABC system is the only β-galactoside–specific locus. To evaluate the impact of the preference of Bal6GBP on growth of B. animalis sp. lactis Bl-04, the bacterium was cultivated for 48 h on the β-(1,3), β-(1,4), or β-(1,6) structural isomers of β-galactobiose as a carbon source (Fig. 4A). Starting from the same inoculum, the most rapid growth was on β6Gal2 with no observable lag phase. Conversely, growth on β3Gal2 and β4Gal2 commenced after lag phases of 9 and 15 h, respectively. We carried out another growth experiment to verify whether the growth delay on less preferred ligands was related to inefficient transcriptional response or due to poor uptake. The growth in this case was performed on 0.5% β6Gal2, 0.2% each of β4Gal2 and β6Gal2, or 0.45 and 0.05% of β4Gal2 and β6Gal2, respectively (all w/v) (Fig. 4B). The growth level was proportional to the amount of the preferred β6Gal2 after 40 h, although the final one-point OD measurement before the culture harvest at about 48 h was higher (data not shown) compared with the 40-h sample, consistent with the delayed growth pattern observed on β4Gal2 (Fig. 4A). The reason for using the lower concentration in the 1:1 galactobiose mixture was to have higher sensitivity in the uptake assay for monitoring uptake during the exponential phase. The uptake preference was evaluated by analysis of supernatants from a culture growing on the same equimolar mixture of β4Gal2 and β6Gal2 as in Fig. 4B. The preferred β6Gal2 was depleted after 12 h, whereas consumption of β4Gal2 was only observed after 24 h and was not completed after 72 h of growth (Fig. 4C). These experiments suggest that the uptake preference is a key factor that governs the onset and the extent of growth on both pure substrates and mixtures of those.
Figure 4.
Growth curves and uptake profiles of B. animalis subsp. lactis Bl-04 growing in modified MRS medium supplemented with β-galactobioses. A, growth on 0.5% (w/v) β-(1,6)-galactobiose (β6Gal2, blue), β-(1,3)-galactobiose (β3Gal2, green), or β-(1,4)-galactobiose (β4Gal2, red). B, a different growth experiment on 0.5% (w/v) β6Gal2 (blue), on 0.2% β6Gal2 + 0.2% β4Gal2 (purple), or 0.05% β6Gal2 + 0.45% β4Gal2 (pink). C, uptake analysis of culture supernatants of a static growth experiment in Eppendorf tubes, but otherwise similar to B with the equimolar mixture of β4Gal2 and β6Gal2 (purple) using high-performance anion exchange with peramperometric detection. The experiments in A, B, and C are from different experiments. Experiments in A and B were performed in five replicates, and the data are shown as means ± S.D. (error bars), whereas the data in C are from a single experiment.
Discussion
Transporter proteins as sensitive probes of the evolution of microbial glycan preferences in the gut niche
The evolution of bifidobacteria has been shaped by the acquisition of genes that support the adaptation to the digestive tracts of animals that provide parental care, especially mammals (31, 32). The evolution of unique glycan utilization capabilities by Bifidobacterium members underscores the impact of metabolic specialization as a driving force for species differentiation and niche adaptation (12). Considerable acquisition of carbohydrate transporters, especially of the ABC type, has been proposed in Bifidobacterium (12). The evolution of oligosaccharide transporter specificities to support the adaptation of bifidobacteria to distinct niches in host guts has not received sufficient attention. Transport proteins are suitable probes of the evolution of glycan metabolic preferences due to their superior selectivities as demonstrated in the present study. Biochemical and structural characterization of carbohydrate transporters is key to enable the assignment of metabolic preferences based on genome sequence data. To illustrate this, we performed structural and multiple sequence alignments of Bal6GBP and GL-BP, the homologue conferring uptake of the mucin-derived GNB and the human milk–derived LNB from B. longum sp. longum (25), together with closely related sequences that cluster with these two characterized proteins in the phylogenetic analysis (Fig. S3). The aromatic platform, which is the hallmark of carbohydrate recognition by SBPs at position 1, is strictly conserved in both the primary and tertiary structural alignment together with the four residues recognizing the nonreducing β-galactosyl (Fig. 5, C and D). This striking conservation unveils the evolutionary pressure to retain the functionality of binding subsite 1 of these two proteins despite the divergence of primary structure (26% sequence identity). Another tryptophan (Bal6GBP Trp-254) is also fully conserved in the sequence alignment (Fig. 5A), but this residue serves two different roles at position 2: aromatic stacking onto the N-acetyl hexosamine ring in GL-BP, whereas in Bal6GBP, the indole ring is orthogonal to the galactosyl plane and forms a polar interaction with the hemiacetal oxygen.
Figure 5.
Comparison of the β6Gal and GL-BP proteins. A, multiple-sequence alignment logos of the binding site residues of the β6Gal and GL-BP clusters of bifidobacteria accessed by phylogenetic analysis. Amino acid residues of the logos are colored by amino acid category. The sequences of GL-BP and Bal6GBP are included and are colored according to the conservation as compared with their respective clusters (dark blue, all match; pink, similar; blue, ≥50%). B, structural alignment of Bal6GBP/β6Gal2 (sand, domain 1; green, domain 2; light blue, linker) and GL-BP/GNB (gray). The structural alignment is based on the identical β-galactopyranosyl in position 1 of the binding pockets. C and D, binding sites of Bal6GBP:β6Gal2 and GL-BP:GNB, respectively. The top panels depict the conserved residues, and the bottom panel depicts variant residues (sand, domain 1 residues; green, domain 2 residues; light blue, linker residues).
A key difference is the substitution of Trp-47 in Bal6GBP with Arg-49 in GL-BP. Importantly, Arg-49 recognizes the N-acetyl group unique to GL-BP substrates via direct and water-mediated hydrogen bonds (Fig. 5B). This residue together with Glu-110 also engages the galactosyl at position 1 (ring oxygen and C2-OH) and the C4-OH of the N-acetyl hexoseamine at position 2. This arginine is strictly conserved in close homologs of GL-BP, thereby constituting a specificity signature. Another facet underpinning the divergence of GL-BP toward LNB/GNB recognition entails amino acid changes that reduce the volume of side chains flanking the bulky N-acetyl group to enable its accommodation.
The structure of Bal6GBP also gives an explanation for the extreme preference for the longer β-(1,6)-galactosidic bond that allows higher rotational freedom between the two monosaccharide rings as compared with β4Gal2, with β3Gal2 having intermediate rotational freedom. The rotational flexibility of β6Gal2 allows the positioning the sugar planes at an angle of about 140°, a conformation that is enforced by Glu-98. This residue locks the orientation of the galactosyl at position 2 through a bidentate polar interaction that connects the galactosyl (C4-OH) at position 2 to the counterpart (C2-OH) at position 1 (Fig. 3B). Notably, the position of Glu-98 is fixed by two highly conserved tyrosines (Tyr-78 and Tyr-100) that are from rigid α-helices, which is likely to be important for the high selectivity for β6Gal2. The preference for galactosyl at position 2 is mediated by a polar interaction between the axial C4-OH and Asn-74. A 140-fold weaker affinity is observed when the axial C4-OH is replaced by an equatorial one in allolactose, possibly due to the loss of the hydrogen bond to Asn-74. The glycosidic bond to an axial OH group in β4Gal2 restricts the rotation, rendering this ligand more rigid. Therefore, binding of β4Gal2 is associated with a large penalty due to conformational rearrangements of the protein and a strained conformation of the ligand. Comparing Bal6GBP with the β-galactosyl-(1,4)-lactose–specific homolog from B. breve (21) reveals that the hydrophobic platform and six of seven residues that bind the galactosyl at position 1 are also conserved in the primary structure (cluster 5 in Fig. S3). Strikingly, however, the double tyrosine motif and Asn-74 discussed above are variable, highlighting the sequence divergence underpinning the evolution of transporters for recognition of different β-galactosides.
The selectivity of transport binding proteins correlates to rapid growth and competitiveness on preferred substrates
Dietary glycans are instrumental in shaping the composition of the HGM (7, 8). For bacteria that lack extracellular polysaccharide-degrading enzymes, high-affinity capture and transport of oligosaccharides is crucial to cross-feed on glycan oligomers produced by enzymes of primary degraders. The early dominance of bifidobacteria in the human gut relies partially on their arsenal of ABC transporters that confer the uptake of abundant human milk galactosides (33, 34). The competitiveness of bifidobacteria on preferred substrates for their ABC transporters was recently demonstrated in B. animalis subsp. lactis in co-culture with the human gut symbiont Bacteroides ovatus during growth on raffinose (9). This transporter is highly conserved in adult human gut bifidobacterial species, consistent with the stimulation of bifidobacteria on α-raffinose family α-galactosides from legumes. Similar observations were recently reported in other taxa (e.g. competitive growth of the xylan-degrading symbiont Roseburia intestinalis grown on a preferred ligand for its xylooligosaccharide ABC transporter in a mixed culture with B. ovatus) (35). Most bifidobacteria lack extracellular xylanases but are able to cross-feed on shorter arabinoxylooligosaccharides produced by other primary degraders using an efficient ABC system (10, 11), further supporting the pivotal role of ABC transporters in bifidobacterial adaptation as secondary degraders that efficiently target oligosaccharides.
Bifidobacteria selectively ferment different dietary β-galactosides (e.g. B. animalis subsp. lactis grows well on β-(1,6)-galactosyl lactose, but not on the β-(1,4) version of this trisaccharide (both from bovine milk)) (36). Recent studies have identified an ABC transporter that mediates the uptake of β-(1,6)-galactosyl-lactose from B. breve and characterized the preference of the associated SBP (22, 37). Notably, B. breve was able to grow comparably well on the β-(1,3)-, β-(1,4)-, and β-(1,6)-galactosyl-lactose (GL) isomers. Different ABC transporters appear to mediate this utilization, and the closest homolog of Bal6GBP (WP_003828494.1; amino acid sequence identity 76%) showed the highest affinity for β-(1,6)-GL (KD = 0.357 μm). This protein has a markedly different selectivity as compared with Bal6GBP, based on a decrease in affinity of only 16-fold for β-(1,4)-GL and 460-fold lower affinity for β-(1,3)-GL (22). The shared preference for β-(1,6)-galactosides, but large selectivity differences, are consistent with these SBPs populating different branches in the same clade in the phylogenetic tree (clade 1, Fig. S3). The notable difference in selectivity despite the relatedness of these homologues highlights the need for more structural and biochemical data for reliable assignments of transport proteins from this niche. The second closest homolog (WP_003828303.1; sequence identity 45%) is specific for only β-(1,4)-galactosides with the highest affinity for β4Gal2 (KD = 23.7 nm) followed by β-(1,4)-GL (KD = 10.4 μm) (21). β-(1,6)-Galactoside motifs are present in plant cell wall pectic arabinogalactans (types I and II) and arabinogalactan proteins (type II) (38, 39). We have identified homologs (∼80% identity) of galactan β-(1,3)-galactosidase (40) and an endo-β-(1,6)-galactanase (41) in the Human Microbiome Project database (42), suggesting that β-(1,6)-galactosides are likely to be accessible degradation products from especially type II arabinogalactan in the diet. The about 100-fold lower selectivity of the B. breve closest homolog for the β-(1,6)-galactosides may suggest an adaptation to bovine milk, whereas the strict selectivity of Bal6GBP may suggest an adaptation to different ligands (e.g. from pectin). The selectivity of Bal6GBP for β6Gal2 (1630-fold preference over β4Gal2) demonstrates the role of oligosaccharide transporters in defining metabolic preferences as compared with intracellular enzymes, which may display more promiscuity (24). The preference of Bal6GBP correlates to rapid growth on the preferred ligand, whereas relative delay of several hours was observed on less-preferred ligands (Fig. 4A). The preferential depletion of β6Gal2 before β4Gal2 (Fig. 4C) provides compelling evidence that the large delay in the onset of growth on β4Gal2 is attributable to the inefficient uptake. The impact of the poor uptake is amplified by the lower affinity of the intracellular β-gal. The observed correlation between the selectivity of transport systems and early growth efficiency is likely to be crucial for resource allocation in the competitive human gut niche.
In conclusion, the study gives an unprecedented insight into the selectivity of transport proteins as compared with intracellular enzymes targeting the same substrates and the large impact of this selectivity on bacterial growth patterns. Access to the first structure of a β6Gal2-binding protein uniquely allowed analyses of the structural elements that support the evolutionary divergence of bifidobacterial β-galactoside transport proteins, which is otherwise challenging due to low primary structure conservation. Altogether, these findings promote our understanding of the structural and functional aspect of oligosaccharide transport in prevalent core microbiota.
Experimental procedures
Carbohydrate ligands and chemicals
The carbohydrates tested for binding interactions with Bal6GBP are listed in Table S1 and were 94–99% pure. All other chemicals were of analytical grade.
Cloning, expression, and purification of the galactooligosaccharide-binding protein from B. animalis subsp. lactis Bl-04 (Bal6GBP)
The gene fragment encoding Bal6GBP (locus tag: Balac_0483; GenBankTM: NC_012814.1) without the signal peptide (residues 1–21 as predicted by SignalP 4.0 (43)) was amplified from genomic B. animalis subsp. lactis Bl-04 DNA (kindly provided by DuPont Nutrition & Health, Kantvik Finland) using the sense primer 5′-ATACATATGGCAGCCTGTGGGGGTGGC-3′ and the antisense primer 5′-ACTGGATCCCTACTCCTTCATCGTGAAGCCTTG-3′ with NdeI and BamHI restriction sites highlighted in boldface type and the amplicon was cloned into the same sites of pET28a(+) (Novagen). The resulting recombinant plasmid, pET28 Balac_0483, was verified by full sequencing and transformed into the strains Escherichia coli Rosetta (DE3) and B834 (DE3) (Novagen) to allow production of the unlabeled and seleno-l-methionine–labeled recombinant proteins, respectively. Transformants were grown in lysogeny broth medium with kanamycin (50 μg ml−1) and chloramphenicol (34 μg ml−1) at 30 °C to A600 = 0.5. Expression was induced by isopropyl β-d-1-thiogalactopyranoside (250 μm); cells were harvested after 5 h by centrifugation (10,000 × g, 30 min) and lysed with Bugbuster® (Millipore); and debris was removed by centrifugation (20,000 × g, 2 × 30 min). Clarified lysates were sterile-filtered (0.45 μm) and purified on a HisTrap HP column (GE Healthcare) as described previously (10). Pure fractions were applied onto a HiLoad 16/600 Superdex 75 preparation grade column, and the proteins were eluted during 1.2 column volumes in 20 mm phosphate, 150 mm NaCl, 0.5 mm DTT, pH 6.5, and concentrated by 10-kDa Amicon Ultracentrifugal filters (Millipore). The fractions containing pure Bal6GBP were pooled and dialyzed against 20 mm Tris-HCl, 150 mm NaCl, pH 7.5. The N-terminal hexahistidine tag of Bal6GBP was cleaved using human thrombin (Calbiochem), recovered after passing through a 1-ml HisTrap HP column, and buffer-exchanged into 10 mm MES buffer, pH 6.5. Protein purity was verified by SDS-PAGE, and protein concentration was determined by measuring A280 using the theoretical molar extinction coefficient ϵ280 nm = 93,280 m−1 cm−1 calculated by ProtParam on the EXPASY server (http://web.expasy.org/protparam).
SPR
SPR-binding analysis was performed using a Biacore T100 (GE Healthcare). Bal6GBP (0.1 mg ml−1) in 10 mm sodium acetate, pH 4.0, was immobilized on a CM5 chip by random amine coupling (GE Healthcare) to 2385–3448 response units. Initial binding screening was conducted at a flow rate of 30 μl min−1 and 10 mm carbohydrate ligand at 25 °C with association and dissociation steps of 60 and 150 s, respectively. Subsequently, saccharides that displayed binding were analyzed using 10 concentrations in the range of 0.05–10 KD in at least duplicates. The pH and temperature dependence of β-(1,4)-galactobiose binding to Bal6GBP was analyzed at eight temperatures (15–40 °C) and at nine pH values (3.6–8.2) in 20 mm phosphate citrate buffer. The BIA Evaluation software (GE Healthcare) was used to analyze the data as described previously (10) to determine dissociation constants (KD) as well as association (kon) and dissociation rate (koff) constants where possible.
ITC
Binding thermodynamics of Bal6GBP (8–600 μm) and preferred galactosides were measured at 25 °C in 20 mm phosphate citrate, pH 6.5, using an iTC200 microcalorimeter (MicroCal). The titration was initiated by a 0.3-μl injection followed by 19 2-μl injections of ligand. The concentrations used were 80 μm β6Gal2 injected into 8 μm Bal6GBP and 2.6 (or 2.4 in the duplicate experiment) mm β6Gal4 injected into 222 (or 246 in the dupilcate) μm Bal6GBP. A one-binding site model was fitted to the ITC data after baseline adjustment and correction for the dilution enthalpy using Origin software 7.0552 supplied with the instrument to determine the equilibrium association constant (KA), the stoichiometry of binding (N), and the molar binding enthalpy (ΔH).
Crystallization, data processing, and protein structure determination
Crystallization of Bal6GBP was initially tested with the purified protein in 10 mm MES buffer, pH 6.5, at a concentration of 10 mg ml−1 and 2.5 mm β6Gal2 with a reservoir from the JCSG+ screen (Molecular Dimensions) set up using a Mosquito® liquid-handling robot (TTP Labtech) with 200 nl of protein mixed with a 100-nl reservoir in MRC 3-well plates, and after several months at room temperature (297 K), a crystal was obtained in C12 (reservoir consisting of 10% PEG 1000 and 10% PEG 8000) that diffracted beyond 1.4 Å resolution. Data were collected at the I911-3 beamline at MAX-II (44), and a total of 390 images (195° of data) were collected to a maximum resolution of 1.5 Å on the edge of the square detector, which impacted the completeness of the data in the outer shell (Table 3). The SeMet-labeled protein was crystallized in MRC 2-well plates using an Oryx 8 liquid-handling robot (Douglas Instruments) at a concentration of 20 mg ml−1. Standard screens JCSG+, PACT, and Morpheus (Molecular Dimensions) were initially carried out. Drops consisting of 200 nl of protein and 100 nl of reservoir or 150 nl of each were set up and kept at room temperature (around 297 K). Crystals appeared under several conditions. Data were collected on a crystal obtained with the Morpheus screen, condition E4 (12.5% PEG 1000, 12.5% PEG 3350, 12.5% MDP, 30 mm each di-, tri-, tetra-, and pentaethyleneglycol with 0.1 m MES/imidazole at pH 6.5) at the ESRF on beamline ID29 (45) at a wavelength of 0.979 Å and a temperature of 100 K. A total of 360° were collected over 2400 images to a resolution of 2 Å. For both data sets, the diffraction data were processed and scaled with XDS (46) using XDSAPP (47). Both native and SeMet Bal6GBP crystals belong to space group P21 but are not isomorphous. Phenix.AutoSol (48) found a total of 19 selenium sites with a figure of merit of 0.382, and the density-modified map was used in phenix.autobuild (49) to produce a preliminary model for the SeMet-labeled protein. This model was split into domain 1 and domain 2 and used in molecular replacement with phaser. After automated model building with phenix.autobuild, the model was completed using Coot (50) and phenix.refine (51) and analyzed with Molprobity (52) (see Table 3). The Bal6GBP structures with β6Gal2 (PDB code 6H0H) and in the open SeMet form (PDB code 6Q5G) were submitted to the Protein Data Bank.
Galactobiose uptake preference of B. animalis sp. lactis Bl-04
Growth of Bl-04 was performed as previously described (9). Freeze-dried B. animalis subsp. lactis Bl-04 was grown anaerobically overnight at 37 °C in Bifidus Selective Medium, and a 2% inoculum of the overnight culture was similarly propagated twice in de Man, Rogosa, and Sharpe (MRS) broth supplemented with 0.5 g liter−1 l-cysteine, before 2% (v/v) of this culture was used to inoculate a fresh culture in the modified MRS medium supplemented with either 0.5% (w/v) β3Gal2, β4Gal2, or β6Gal2 in total volumes of 200 μl in a 96-well microtiter plate. In another experiment, the same procedure was used to inoculate 0.5% (w/v) β6Gal2 or mixtures of β6Gal2 and β4Gal2 (0.20% + 0.20% or 0.05% + 0.45%, respectively). Growth was continued for 48 h at 37 °C and monitored at A600 by the 2030 Multilabel Reader VictorTM X4 (PerkinElmer Life Sciences). The growth was performed in five replicates in both cases.
Preferential culture supernatant utilization of the β6Gal2 and β4Gal2 isomers by Bl-04 was analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (Dionex, Sunnyvale, CA) in a time-dependent manner of a growth experiment, performed in Eppendorf tubes, otherwise as described above with a 0.5% (w/v) equimolar mixture of β4Gal2 and β6Gal2, and aliquots were collected for 72 h. An injection volume of 10 μl onto a Carbopac PA200 column and a mobile phase (0.35 ml min−1) of constant 100 mm NaOH and a gradient of sodium acetate of 0–30 min at 0–100 mm and 30–35 min at 100–400 mm was used. Standards and samples were diluted 10× in 100 mm NaOH before analysis, and standards of the two isomers (0.58 mm) were used to identify corresponding peaks in the chromatograms.
Bioinformatic analyses
A BLAST was performed with the native Bal6GBP sequence (GenBankTM accession number ACS45862.1) as the query against all proteins of the Bifidobacterium genus. The 795 resulting protein sequences (length 350–550 amino acids) with maximum scores ≥100 were aligned by MAFFT with default settings (53). From these sequences, 40 and 123 protein sequences clustered together with Bal6GP and GL-BP (the GNB/LNB binder from B. longum sp. longum), respectively, in a neighbor-joining consensus phylogenetic analysis (S3) performed in Geneious version 11.1.5 (54) including bootstrap resampling with 1000 replicates. Amino acid sequence logos of the residues involved in ligand binding were generated by TEXshade (55) for the orthologue groups.
Author contributions
M. C. T., B. S., and M. A. H. conceived the idea of the study. M. C. T. performed all experiments except for the crystallography. F. F. and L. L. L. obtained X-ray crystallography protein structure data and solved the structure. M. C. T., F. F., and M. A. H. drafted the first version of the manuscript, and M. C. T., F. F., B. S., L. L. L., and M. A. H. finalized the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the Carlsberg Foundation for Instrument Grant 2011-01-0598 (to M. A. H.) to fund the isothermal titration calorimeter. We also thank Professor Mads Hartvig Clausen (Department of Chemistry, Technical University of Denmark), Dr. Motomitsu Kitaoka (National Food Research Institute, Tsukuba, Japan), and Assistant Professor Wataru Saburi (Faculty of Agriculture, Hokkaido University, Sapporo, Japan) for generous gifts of oligosaccharides and Professor Tine Rask Licht and technician Bodil Madsen (National Food Institute, Technical University of Denmark) for help regarding bifidobacterial growth. We thank the MAX IV Laboratory and Maria Håkonsson (MAX IV Laboratory, Lund University) for use of the Mosquito robot and help during crystallization and Tobias Tandrup (Department of Chemistry, University of Copenhagen) for doing the Oryx8 crystallization setup. Mickael Blaise (currently at Institut de Recherche en Infectiologie de Montpellier, CNRS) and Tobias Tandrup and Jens-Christian Navarro Poulsen (both from the Department of Chemistry, University of Copenhagen) are thanked for help during data collection. The ESRF and the MAX IV Laboratory are thanked for beamtime.
This study was supported by Independent Research Fund Denmark Natural Sciences (FNU) Research Project 2, Grant 4002-00297 (to M. A. H.) and Large Instrument Grant 272-06-0050 (to B. S.) for the Biacore T100. The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Tables S1–S3 and Figs. S1–S3.
The atomic coordinates and structure factors (codes 6H0H and 6Q5G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HGM
- human gut microbiota
- RMSD
- root mean square deviation
- SBP
- solute-binding protein
- DP
- degree of polymerization
- GOS
- galactooligosaccharide(s)
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- GNB
- galacto-N-biose
- LNB
- lacto-N-biose
- GL-BP
- GNB/LNB-binding protein
- GL
- galactosyl-lactose
- β3Gal2
- β-(1,3)-galactobiose
- β4Gal2
- β-(1,4)-galactobiose
- β6Gal2
- β-(1,6)-galactobiose
- β6Gal4
- β-(1,6)-galactotetraose.
References
- 1. Sonnenburg J. L., and Bäckhed F. (2016) Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furusawa Y., Obata Y., Fukuda S., Endo T. A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N. N., Murakami S., Miyauchi E., Hino S., et al. (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K., Nishida A., Fujimoto T., Fujii M., Shioya M., Imaeda H., Inatomi O., Bamba S., Andoh A., and Sugimoto M. (2016) Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn's disease. Digestion 93, 59–65 10.1159/000441768 [DOI] [PubMed] [Google Scholar]
- 4. Canfora E. E., van der Beek C. M., Hermes G. D. A., Goossens G. H., Jocken J. W. E., Holst J. J., van Eijk H. M., Venema K., Smidt H., Zoetendal E. G., Dejong C. H. C., Lenaerts K., and Blaak E. E. (2017) Supplementation of diet with galacto-oligosaccharides increases bifidobacteria, but not insulin sensitivity, in obese prediabetic individuals. Gastroenterology 153, 87–97.e3 10.1053/j.gastro.2017.03.051 [DOI] [PubMed] [Google Scholar]
- 5. Sivan A., Corrales L., Hubert N., Williams J. B., Aquino-Michaels K., Earley Z. M., Benyamin F. W., Lei Y. M., Jabri B., Alegre M. L., Chang E. B., and Gajewski T. F. (2015) Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., Fischbach M. A., Biddinger S. B., Dutton R. J., and Turnbaugh P. J. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koropatkin N. M., Cameron E. A., and Martens E. C. (2012) How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cockburn D. W., and Koropatkin N. M. (2016) Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 428, 3230–3252 10.1016/j.jmb.2016.06.021 [DOI] [PubMed] [Google Scholar]
- 9. Ejby M., Fredslund F., Andersen J. M., Vujičić Žagar A., Henriksen J. R., Andersen T. L., Svensson B., Slotboom D. J., and Abou Hachem M. (2016) An ATP binding cassette transporter mediates the uptake of α-(1,6)-linked dietary oligosaccharides in Bifidobacterium and correlates with competitive growth on these substrates. J. Biol. Chem. 291, 20220–20231 10.1074/jbc.M116.746529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ejby M., Fredslund F., Vujicic-Zagar A., Svensson B., Slotboom D. J., and Abou Hachem M. (2013) Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp. lactis Bl-04. Mol. Microbiol. 90, 1100–1112 10.1111/mmi.12419 [DOI] [PubMed] [Google Scholar]
- 11. Rogowski A., Briggs J. A., Mortimer J. C., Tryfona T., Terrapon N., Lowe E. C., Baslé A., Morland C., Day A. M., Zheng H. J., Rogers T. E., Thompson P., Hawkins A. R., Yadav M. P., Henrissat B., et al. (2015) Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun. 6, 7481 10.1038/ncomms8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milani C., Turroni F., Duranti S., Lugli G. A., Mancabelli L., Ferrario C., van Sinderen D., and Ventura M. (2016) Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 82, 980–991 10.1128/AEM.03500-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Urashima T., Asakuma S., Leo F., Fukuda K., Messer M., and Oftedal O. T. (2012) The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv. Nutr. 3, 473S–482S 10.3945/an.111.001412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohnen D. (2008) Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 15. O'Callaghan A., and van Sinderen D. (2016) Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7, 925 10.3389/fmicb.2016.00925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Leeuwen S. S., Kuipers B. J. H., Dijkhuizen L., and Kamerling J. P. (2016) Comparative structural characterization of 7 commercial galacto-oligosaccharide (GOS) products. Carbohydr. Res. 425, 48–58 10.1016/j.carres.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 17. Akiyama T., Kimura K., and Hatano H. (2015) Diverse galactooligosaccharides consumption by bifidobacteria: implications of β-galactosidase-LacS operon. Biosci. Biotechnol. Biochem. 79, 664–672 10.1080/09168451.2014.987204 [DOI] [PubMed] [Google Scholar]
- 18. Li W., Wang K., Sun Y., Ye H., Hu B., and Zeng X. (2015) Influences of structures of galactooligosaccharides and fructooligosaccharides on the fermentation in vitro by human intestinal microbiota. J. Funct. Foods 13, 158–168 10.1016/j.jff.2014.12.044 [DOI] [Google Scholar]
- 19. Peacock K. S., Ruhaak L. R., Tsui M. K., Mills D. A., and Lebrilla C. B. (2013) Isomer-specific consumption of galactooligosaccharides by bifidobacterial species. J. Agric. Food Chem. 61, 12612–12619 10.1021/jf403789r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Viborg A. H., Katayama T., Abou Hachem M., Andersen M. C. F., Nishimoto M., Clausen M. H., Urashima T., Svensson B., and Kitaoka M. (2014) Distinct substrate specificities of three glycoside hydrolase family 42 β-galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology 24, 208–216 10.1093/glycob/cwt104 [DOI] [PubMed] [Google Scholar]
- 21. Shigehisa A., Sotoya H., Sato T., Hara T., Matsumoto H., and Matsuki T. (2015) Characterization of a bifidobacterial system that utilizes galacto-oligosaccharides. Microbiology 161, 1463–1470 10.1099/mic.0.000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sotoya H., Shigehisa A., Hara T., Matsumoto H., Hatano H., and Matsuki T. (2017) Identification of genes involved in galactooligosaccharide utilization in Bifidobacterium breve strain YIT 4014(T). Microbiology 163, 1420–1428 10.1099/mic.0.000517 [DOI] [PubMed] [Google Scholar]
- 23. Andersen J. M., Barrangou R., Abou Hachem M., Lahtinen S. J., Goh Y. J., Svensson B., and Klaenhammer T. R. (2013) Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 14, 312–325 10.1186/1471-2164-14-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Viborg A. H., Fredslund F., Katayama T., Nielsen S. K., Svensson B., Kitaoka M., Lo Leggio L., and Abou Hachem M. (2014) A β1–6/β1–3 galactosidase from Bifidobacterium animalis subsp lactis Bl-04 gives insight into sub-specificities of β-galactoside catabolism within Bifidobacterium. Mol. Microbiol. 94, 1024–1040 10.1111/mmi.12815 [DOI] [PubMed] [Google Scholar]
- 25. Suzuki R., Wada J., Katayama T., Fushinobu S., Wakagi T., Shoun H., Sugimoto H., Tanaka A., Kumagai H., Ashida H., Kitaoka M., and Yamamoto K. (2008) Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 283, 13165–13173 10.1074/jbc.M709777200 [DOI] [PubMed] [Google Scholar]
- 26. Berntsson R. P. A., Smits S. H. J., Schmitt L., Slotboom D.-J., and Poolman B. (2010) A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 10.1016/j.febslet.2010.04.043 [DOI] [PubMed] [Google Scholar]
- 27. Fukami-Kobayashi K., Tateno Y., and Nishikawa K. (1999) Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 10.1006/jmbi.1998.2454 [DOI] [PubMed] [Google Scholar]
- 28. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abe K., Sunagawa N., Terada T., Takahashi Y., Arakawa T., Igarashi K., Samejima M., Nakai H., Taguchi H., Nakajima M., and Fushinobu S. (2018) Structural and thermodynamic insights into β-1,2-glucooligosaccharide capture by a solute-binding protein in Listeria innocua. J. Biol. Chem. 293, 8812–8828 10.1074/jbc.RA117.001536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayward S., and Berendsen H. J. C. (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30, 144–154 [DOI] [PubMed] [Google Scholar]
- 31. Milani C., Lugli G. A., Duranti S., Turroni F., Bottacini F., Mangifesta M., Sanchez B., Viappiani A., Mancabelli L., Taminiau B., Delcenserie V., Barrangou R., Margolles A., van Sinderen D., and Ventura M. (2014) Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 80, 6290–6302 10.1128/AEM.02308-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turroni F., van Sinderen D., and Ventura M. (2011) Genomics and ecological overview of the genus Bifidobacterium. Int. J. Food Microbiol. 149, 37–44 10.1016/j.ijfoodmicro.2010.12.010 [DOI] [PubMed] [Google Scholar]
- 33. Garrido D., Kim J. H., German J. B., Raybould H. E., and Mills D. A. (2011) Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6, e17315 10.1371/journal.pone.0017315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsuki T., Yahagi K., Mori H., Matsumoto H., Hara T., Tajima S., Ogawa E., Kodama H., Yamamoto K., Yamada T., Matsumoto S., and Kurokawa K. (2016) A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 7, 11939 10.1038/ncomms11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leth M. L., Ejby M., Workman C., Ewald D. A., Pedersen S. S., Sternberg C., Bahl M. I., Licht T. R., Aachmann F. L., Westereng B., and Abou Hachem M. (2018) Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat. Microbiol. 3, 570–580 10.1038/s41564-018-0132-8 [DOI] [PubMed] [Google Scholar]
- 36. Urashima T., Taufik E., Fukuda K., and Asakuma S. (2013) Recent advances in studies on milk oligosaccharides of cows and other domestic farm animals. Biosci. Biotechnol. Biochem. 77, 455–466 10.1271/bbb.120810 [DOI] [PubMed] [Google Scholar]
- 37. O'Connell Motherway M., Kinsella M., Fitzgerald G. F., and van Sinderen D. (2013) Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microbial Biotechnol. 6, 67–79 10.1111/1751-7915.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knoch E., Dilokpimol A., and Geshi N. (2014) Arabinogalactan proteins: focus on carbohydrate active enzymes. Front. Plant Sci. 5, 198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Voragen A. G. J., Coenen G. J., Verhoef R. P., and Schols H. A. (2009) Pectin, a versatile polysaccharide present in plant cell walls. Struct. Chem. 20, 263–275 10.1007/s11224-009-9442-z [DOI] [Google Scholar]
- 40. Ichinose H., Kotake T., Tsumuraya Y., and Kaneko S. (2006) Characterization of an exo-β-1,3-D-galactanase from Streptomyces avermitilis NBRC14893 acting on arabinogalactan-proteins. Biosci. Biotechnol. Biochem. 70, 2745–2750 10.1271/bbb.60365 [DOI] [PubMed] [Google Scholar]
- 41. Ichinose H., Kotake T., Tsumuraya Y., and Kaneko S. (2008) Characterization of an endo-β-1,6-galactanase from Streptomyces avermitilis NBRC14893. Appl. Environ. Microbiol. 74, 2379–2383 10.1128/AEM.01733-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Human Microbiome Project Consortium (2012) A framework for human microbiome research. Nature 486, 215–221 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 44. Ursby T., Unge J., Appio R., Logan D. T., Fredslund F., Svensson C., Larsson K., Labrador A., and Thunnissen M. (2013) The macromolecular crystallography beamline I911–3 at the MAX IV laboratory. J. Synchrotron Radiat. 20, 648–653 10.1107/S0909049513011734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Sanctis D., Beteva A., Caserotto H., Dobias F., Gabadinho J., Giraud T., Gobbo A., Guijarro M., Lentini M., Lavault B., Mairs T., McSweeney S., Petitdemange S., Rey-Bakaikoa V., Surr J., Theveneau P., Leonard G. A., and Mueller-Dieckmann C. (2012) ID29: a high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. J. Synchrotron Radiat. 19, 455–461 10.1107/S0909049512009715 [DOI] [PubMed] [Google Scholar]
- 46. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krug M., Weiss M. S., Heinemann U., and Mueller U. (2012) XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Crystallogr. 45, 568–572 10.1107/S0021889812011715 [DOI] [Google Scholar]
- 48. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 10.1107/S0907444909012098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. 3rd, Snoeyink J., Adams P. D., et al. (2018) MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Katoh K., and Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., and Drummond A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beitz E. (2000) TEXshade: shading and labeling of multiple sequence alignments using LATEX 2ϵ. Bioinformatics 16, 135–139 10.1093/bioinformatics/16.2.135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.