Abstract
Myoviruses, bacteriophages with T4-like architecture, must contract their tails prior to DNA release. However, quantitative kinetic data on myovirus particle opening are lacking, although they are promising tools in bacteriophage-based antimicrobial strategies directed against Gram-negative hosts. For the first time, we show time-resolved DNA ejection from a bacteriophage with a contractile tail, the multi-O-antigen–specific Salmonella myovirus Det7. DNA release from Det7 was triggered by lipopolysaccharide (LPS) O-antigen receptors and notably slower than in noncontractile-tailed siphoviruses. Det7 showed two individual kinetic steps for tail contraction and particle opening. Our in vitro studies showed that highly specialized tailspike proteins (TSPs) are necessary to attach the particle to LPS. A P22-like TSP confers specificity for the Salmonella Typhimurium O-antigen. Moreover, crystal structure analysis at 1.63 Å resolution confirmed that Det7 recognized the Salmonella Anatum O-antigen via an ϵ15-like TSP, DettilonTSP. DNA ejection triggered by LPS from either host showed similar velocities, so particle opening is thus a process independent of O-antigen composition and the recognizing TSP. In Det7, at permissive temperatures TSPs mediate O-antigen cleavage and couple cell surface binding with DNA ejection, but no irreversible adsorption occurred at low temperatures. This finding was in contrast to short-tailed Salmonella podoviruses, illustrating that tailed phages use common particle-opening mechanisms but have specialized into different infection niches.
Keywords: bacteriophage, lipopolysaccharide (YLPS), structural biology, DNA viruses, glycobiology, fluorescence, Salmonella enterica, contractile tail, DNA ejection, O-antigen specificity, Salmonella myovirus, tailspike protein, molecular machine
Introduction
Tailed dsDNA bacteriophages constitute one of the largest phage groups described so far (1). These viral particles are assemblies of icosahedral capsids containing the genome, a portal complex and a tail machine that provides effective means for host cell surface recognition and subsequent DNA transmission into the cytosol, thereby overcoming all bacterial outer protective layers. In some phages, their tails may additionally contain glycan-specific tailspike proteins (TSPs),2 often endoglycosidases, to mediate initial bacterial surface contacts (Fig. 1). It has thus been proposed that tailed phages have evolved to deliver much larger genomes to their hosts than nontailed phages (2). A wealth of biochemical and microscopy in vitro studies has addressed the function of bacteriophage tails as sophisticated molecular machines that exist in three different architectures (Fig. 1) (3–9).
Figure 1.
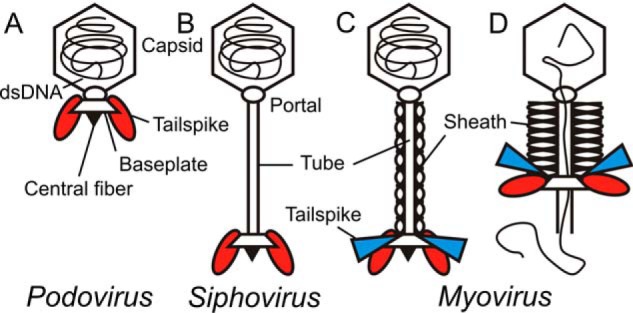
Schematic structures of the three morphology types found in tailed bacteriophages (2, 13, 19, 43). A, podovirus with short, noncontractile tail. B, siphovirus with long, noncontractile tail. C, mature myovirus with long, contractile tail, the subject of this work. D, myovirus after tail contraction during DNA release. Representation of viral particles is oversimplified (i.e. symbols for building blocks like capsids, tails, and baseplates correspond to multimeric protein assemblies). Other features like head-to-tail adaptors or long tail fibers have been omitted for clarity. Different TSP types may occur, represented by ellipsoids (red) or triangles (blue), respectively. However, phages without TSPs or a larger number of TSPs are also commonly found (43). Each individual TSP type is assembled to the baseplate in six copies, but only two copies are shown for clarity.
Bacteriophages of the class Myoviridae possess contractile tail systems (7, 10–13). For myovirus T4, the full genome transfer process has been reported to take place on a time scale between 30 s and several minutes in vivo (7, 10). Siphoviridae with long, noncontractile tails also transfer their genome within minutes (e.g. the Escherichia coli phages λ (5) or T5 (14) or the Bacillus subtilis phage SPP1 (15). The process is slower in Podoviridae with short, noncontractile tails like E. coli phage T7 or B. subtilis phage ϕ29, taking 10–30 min (16, 17).
In all tailed phages, genome internalization in vivo requires a cell membrane proton motive force, although its detailed functional relationships with DNA transfer are still debated (6, 7, 15). Moreover, protein assemblies and RNA polymerases of viral or host origin were found to interact with DNA in vivo during translocation, pulling the genome inside the cell (6, 18, 19). However, these data cannot resolve single molecular steps of genome internalization and do not link kinetics of DNA transfer to cell wall architecture and tail morphology (15, 20).
In vitro, EM has shown the initial structural rearrangements in the E. coli myovirus T4 baseplate tail leading to contraction (11, 21). Initial surface contacts by T4 long tail fibers to outer membrane proteins (Omps) or lipopolysaccharide (LPS) are followed by a dome-to-star transition of the baseplate (22). This exposes the short tail fibers for irreversible cell surface attachment, and, eventually, the bound phage contracts its sheath (11, 21). Similar mechanisms have been proposed for phages infecting Gram-positive hosts, emphasizing that these structural rearrangements are universally conserved in contractile injection systems irrespective of host cell wall composition (11, 23, 24). Following contraction, T4 phages do not yet release their DNA but need an additional signal from the membrane to start genome transfer (25). However, the nature of this putative less specific membrane trigger has so far not been described. EM analyses of T4 bound to E. coli cells captured later steps where the tail tube is inserted into the membrane, building a protein channel for translocation of the genome into the cytoplasm (7).
In vitro studies with noncontractile-tailed bacteriophages have shown that contact with isolated protein or LPS membrane receptors is sufficient to initiate conformational changes in the tail apparatus and in the portal complex leading to opening of the phage particle and release of its DNA contents (20, 26–28). The DNA ejection time courses obtained in these experiments suggest that particle-opening kinetics in vitro depend on tail morphology. DNA ejection kinetics of long-tailed Siphoviridae λ, SPP1, and T5 occurred within seconds, faster than genome translocation in vivo, and are driven by the osmotic pressure inside the capsid (29, 30). In contrast, Podoviridae P22 and T7 completed their DNA ejection only after 90 min, more slowly than in vivo (20, 26). For each of these noncontractile tail architectures, DNA release kinetics were characterized by a single rate-limiting step (14, 27). Given the structural conservation of bacteriophage tail components, this points to conserved conformational rearrangement steps within a given tail architecture.
In this work, we describe interactions of Salmonella myovirus Det7 with its purified LPS receptors in vitro. Det7 is a contractile-tailed Viunalikevirus with a 157-kbp dsDNA genome forming small clear plaques on Salmonella Typhimurium (31, 32). We have analyzed genome release from Det7 upon receptor contact in vitro. For the first time in a bacteriophage with a contractile tail machine, we show the kinetics of this process that has so far neither been quantified for T4 nor for any other myovirus. In contrast to the typical, single-step DNA ejection mechanism found in noncontractile-tailed phages, we describe an additional kinetic step for genome release in the myovirus Det7, associated with its contractile tail machine.
Results
Myovirus Det7 has an ϵ15-like TSP and recognizes serogroup E Salmonella
In O-antigen–specific phages, TSPs enzymatically modify the LPS O-antigen part as an essential step to start genome release (33–36). The Det7 genome encodes for four TSPs (gp206–209). gp207 (designated Det7TSP) has been characterized earlier and is highly homologous to the TSPs found in siphovirus 9NA and podovirus P22, infecting S. Typhimurium (27, 31). However, the function of the other three TSPs (gp206, -208, and -209) encoded by Det7 has not been described. The first 150 residues of gp207 were proposed to serve as adaptor domains, binding the TSP to the phage tail apparatus (31), a feature often found for N-terminal residues in phage TSPs (37). All four TSPs in Det7 share these 150 homologous residues at their N termini (Fig. S1). In gp206 and gp208, these parts are preceded by two additional N-terminal, nonhomologous extensions of 250 and 80 amino acids, respectively. Det7's TSPs hence have three different putative N-terminal tail adaptor domains.
The larger C-terminal TSP parts following the N-terminal tail adaptor domains are responsible for host recognition (33). We found that gp208 is homologous to the TSP of podovirus ϵ15, infecting Salmonella Anatum (serogroup E1) (ϵ15TSP, 33% identity over 798 residues; Fig. S1) and thus designated it as “DettilonTSP” (38). We therefore tested Det7's host specificity and found that it formed plaques on S. Anatum (Table 1). Additionally, it could infect S. Typhimurium or Salmonella Enteritidis. This is in contrast to ϵ15, P22, and 9NA, which only contain a single TSP type and accordingly showed a more restricted host range (Table 1) (27, 38).
Table 1.
Plaque-forming assay of O-antigen–specific bacteriophages Det7, P22, and 9NA
| Strain | Serogroup | Myovirus Det7a | Podovirus P22a | Siphovirus 9NAa |
|---|---|---|---|---|
| S. Typhimurium (DB7136) | B | +++ | +++ | +++ |
| S. Heidelberg O4 (13-00586) | ++ | − | + | |
| S. Typhimurium O4 (13-01225) | ++ | − | − | |
| S. Typhimurium O4,5 (06-01900) | ++ | ++ | − | |
| S. Enteritidis O9 | D1 | +++ | +++ | + |
| S. Anatum O3,10 (13-00188) | E | ++ | − | − |
| S. Typhimurium ΔWaaL (DB7155) | None | − | − | − |
| S. Typhimurium ΔWaaJ (DB7155) | − | − | − | |
| S. Typhimurium ΔWaaKL (DB7155) | − | − | − | |
| S. Typhimurium ΔLPS(DB7155) | − | − | − |
a Infectivity measured according to number of plaque-forming units. +++, 5 × 1011 to 5 × 1012; ++, 1 × 1010 to 4.9 × 1011; +, ≤1 × 1010; −, not infectious.
Det7 contains the S. Anatum-specific Dettilon tailspike endorhamnosidase
To further analyze how Det7 broadens its host range by employing different TSPs, we cloned and purified a C-terminal fragment of DettilonTSP (DettilonTSPΔN, residues 252–798). DettilonTSP was enzymatically active on purified S. Anatum LPS preparations, where it reduced the number of long O-antigen chains on LPS while shorter chains accumulated (Fig. 2A). DettilonTSP enzymatically produced oligosaccharides from S. Anatum O-polysaccharide. Oligosaccharides contained different numbers of O-antigen repeat units and were separated by size-exclusion chromatography (Fig. 2B). We obtained partially O-acetylated tri-, hexa-, and nonasaccharides, as shown by MALDI MS (Table S1). Their masses corresponded to the reported S. Anatum serogroup E1 O-antigen repeat composition (→3)-α-d-Galp-(1→6)-β-d-Manp-(1→4)-α-l-rhamnose-(1→) (39) (Table S1). From this product analysis, we conclude that DettilonTSP is an endorhamnosidase, hydrolyzing the α-1,3-glycosidic bond between rhamnose and galactose in the S. Anatum O-polysaccharide.
Figure 2.
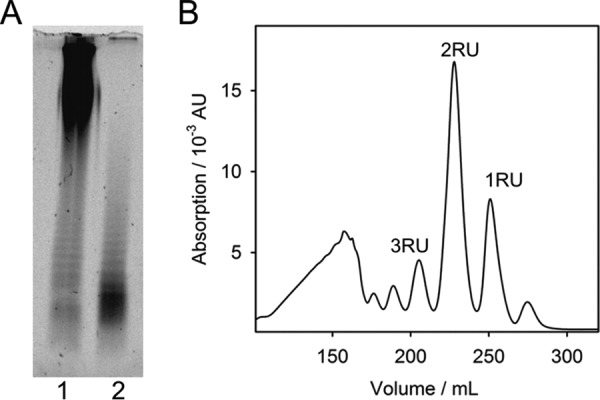
Enzymatic activity of DettilonTSP (gp208) of myovirus Det7 on the S. Anatum O-antigen. A, 150 ng of LPS before (lane 1) and after (lane 2) overnight incubation with DettilonTSP were analyzed on 15% polyacrylamide gels with LPS-specific fluorescence staining. B, size-exclusion chromatography of digestion products after incubation of O-polysaccharide with DettilonTSP. Labeled peaks contain oligosaccharides corresponding to 1–3 repeat units (RU) of the S. Anatum O-antigen as confirmed by MS (Table S1).
To confirm that the mature Det7 phage particle assembly contains different TSP types, we propagated Det7 either on its S. Anatum or S. Typhimurium hosts and tested its plaque-forming capacity on both hosts (Fig. 3A). Det7 retained its host range irrespective of the propagating strain. We conclude that both TSP types are simultaneously present in Det7; host specificity is thus not a transient feature of the phage acquired during growth, as also shown for other phages containing multiple TSPs (40).
Figure 3.
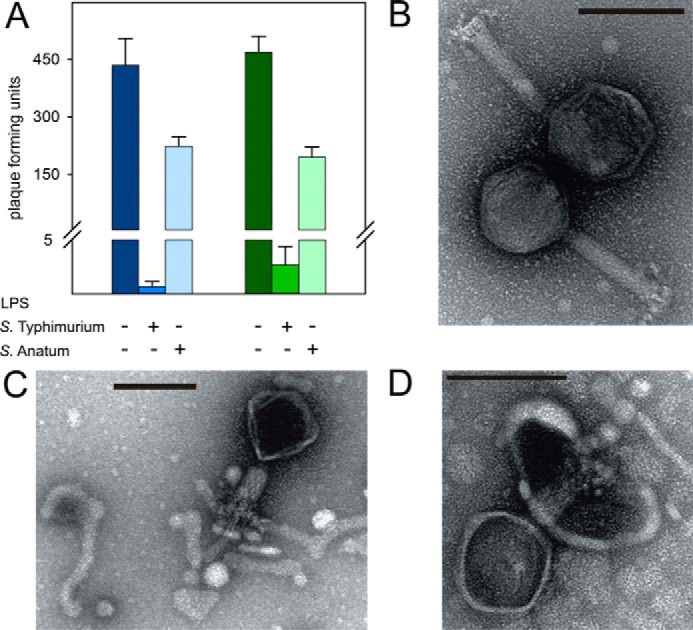
A, blocking myovirus Det7 with LPS. Det7 was incubated with 25 μg ml−1 purified LPS of S. Typhimurium (DB7155) or S. Anatum at 37 °C for 120 min as indicated. Infective particles were quantified as pfu on either S. Typhimurium (blue, left) or S. Anatum (green, right) host bacteria. Error bars, S.D. values from three independent experiments. B–D, transmission EM images of phage Det7 before (B) and after LPS incubation with 10 μg ml−1 LPS of S. Typhimurium or S. Anatum (C and D). 1010 pfu ml−1 Det7 particles were stained with 1% (w/v) uranyl acetate and viewed on carbon-covered grids. Bars, 100 nm.
Det7 is an O-antigen–specific myovirus
To show that LPS is the Det7 host receptor, we added LPS from S. Typhimurium or S. Anatum to Det7 prior to plating. Both host LPS types decreased the number of Det7 plaque-forming units (pfu) (Fig. 3A). In negative stain EM, Det7 particles incubated with LPS of either serotype had capsids appearing dark in the negative stain, pointing to DNA-emptied capsids (Fig. 3, B–D). These particles had contracted tails and were bound to tubular LPS filament structures. Salmonella rough mutants lacking O-antigen were not infected by phage Det7, P22, or 9NA (Table 1), confirming that myovirus Det7 infectivity depended on the presence of O-antigen. This is in contrast to other broadly specific Salmonella myoviruses that are able to penetrate outer membranes in the absence of O-antigen or even most of the LPS core saccharides (41). Myovirus Det7 hence belongs to the group of O-antigen–specific phages (36).
Crystal structure of Dettilon tailspike protein
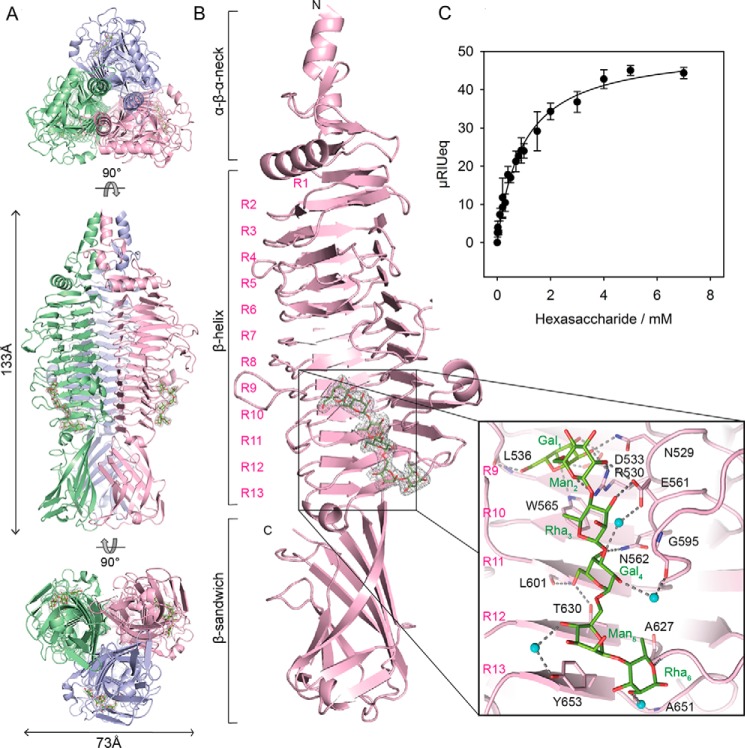
From the infectivity and inactivation assays, we hypothesized that myovirus Det7 has broadened its host range to S. Anatum because it is equipped with the ϵ15-like DettilonTSP. We crystallized DettilonTSP lacking the N-terminal head-binding domain (DettilonTSPΔN, residues 252–798) and obtained a structural model of the trimer at 1.63 Å resolution (Fig. 4A and Table S2). Each DettilonTSPΔN subunit is composed of an N-terminal α-β-α neck (aa 252–283) forming a helix bundle in the trimer, followed by a β-β-α transition (aa 284–317) into the 13 rungs containing β-helix (aa 318–667), which shows insertions of several large loops and two short helices (Fig. 4B). A β-sandwich domain (aa 668–798) is located at the C terminus. It contains 11 antiparallel β-strands forming two β-sheets with short insertions (Fig. S2). DettilonTSPΔN showed overall structural similarity to diverse polysaccharide-binding proteins that contain parallel β-helix folds (Fig. S3) (42).
Figure 4.
X-ray crystal structure of bacteriophage Det7 DettilonTSP (gp208), residues 252–798. A, trimeric assembly with chains highlighted in different colors shown from top to bottom as N-terminal, side, and C-terminal views, respectively. B, structure of a single chain in the asymmetric unit with rungs of the β-helix numbered (R1–R13). The 2Fo − Fc density (gray mesh) for an S. Anatum O-antigen hexasaccharide is contoured at 1σ. Inset, hexasaccharide with the structure (→3)-α-d-Galp-(1→6)-α-d-Manp-(1→4)-β-l-Rhap-(1→)2 (green sticks) and contacting side chains (pink sticks). Water molecules mediating hexasaccharide binding are shown as cyan spheres, and hydrogen bonds are shown as dashed black lines. C, hexasaccharide binding to surface-immobilized DettilonTSP at 20 °C. The isothermal curve was fitted to data from surface plasmon resonance equilibrium signals (cf. Fig. S5) and resulted in KD = 0.97 ± 0.09 mm for S. Anatum hexasaccharide. Error bars, S.D. values from three independent experiments.
DettilonTSPΔN in complex with hexasaccharide, at 2.1 Å resolution, shows a binding cleft of 22 Å in length and 13–15 Å in width. It is located in the lower region of the central β-helix involving rungs 9–13 and is sufficient to accommodate two O-antigen repeat units (Fig. 4B and Figs. S4 and S5). The nonreducing end repeat unit forms a partly water-mediated hydrogen bond network between galactose and mannose residues, the carbonyl oxygen of Leu-536 and the side chains of Asn-529, Arg-530, Asp-533, Trp-565, and Glu-561. Only one hydrogen-bond interaction to Glu-561 is found for the following rhamnose. The second repeat unit is fixed via galactose, hydrogen-bonding to the Glu-561, Asn-562, and Thr-630 side chains and the carbonyl oxygen of Leu-601. A single aromatic stacking interaction is found for the following mannose with Tyr-653. The reducing end rhamnose forms three hydrogen bonds with the backbone atoms of Gly-625, Ala-627, and Ala-651.
S. Anatum O-antigen hexasaccharide fragments bound to surface-immobilized DettilonTSP with a dissociation constant of 1 mm when monitored with surface plasmon resonance (Fig. 4C and Fig. S6). This overall low hexasaccharide affinity is in agreement with a rather loose and hydrogen bond–dominated interaction network in a preformed rigid binding site. Multispecific myoviruses typically contain multiple TSPs (43). In the Det7 baseplate assembly, multiple TSPs with three low-affinity binding sites each can act in concerted O-antigen binding. This provides a multivalent and thus high-affinity S. Anatum adsorption platform.
LPS from either S. Anatum or S. Typhimurium triggers DNA ejection from bacteriophage Det7
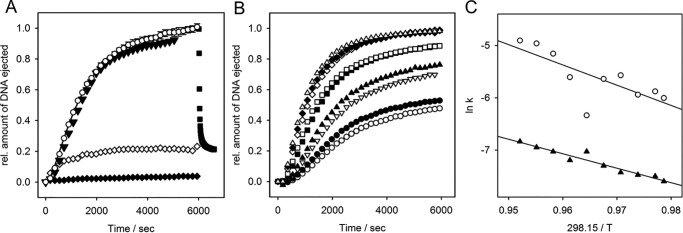
We incubated bacteriophage Det7 with LPS from S. Anatum and S. Typhimurium in the presence of the DNA-specific dye Yo-Pro and observed an increasing fluorescence signal that could be decreased by the addition of DNase (Fig. 5A and Fig. S7). This indicates that Det7 ejected its DNA upon LPS contact. Particle opening was specifically triggered by LPS only, whereas signal increase was not observed with polysaccharide alone nor with lipid A lacking the O-antigen part, nor with smooth LPS from an unrelated E. coli nonhost strain (Fig. S7). Full DNA release was observed at temperatures between 38 and 40 °C, whereas the amount of DNA liberated decreased at higher or lower temperatures (Fig. 5B and Fig. S8).
Figure 5.
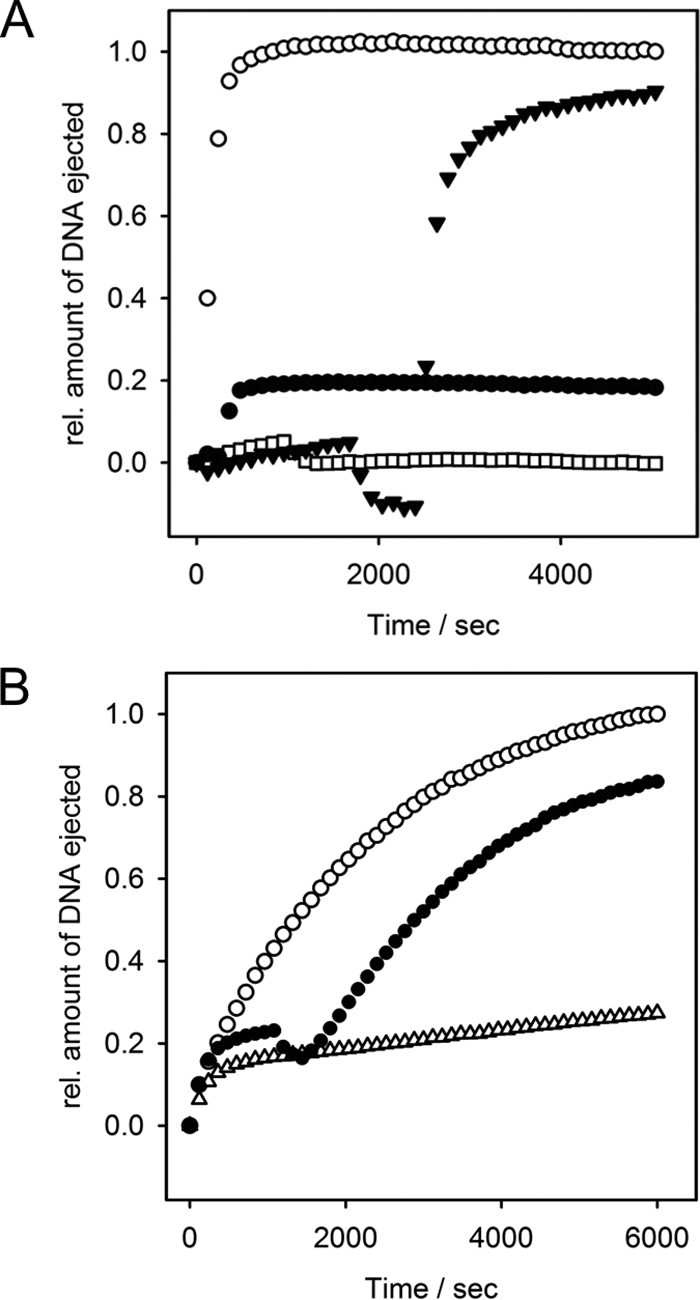
In vitro LPS-triggered DNA ejection from phage Det7. 8 × 109 pfu mixed with 67 μg ml−1 LPS in the presence of a fluorescent DNA-binding dye. A, fluorescence signal obtained at 37 °C with S. Typhimurium LPS (white circles, mean values of three independent experiments with error bars representing the S.D.), with 10 nm purified Det7TSP added after 2 min (white diamonds), with DNase added after 6000 s (black squares), with S. Anatum LPS (black triangles), with S. Typhimurium LPS lacking O-antigen (white triangles). B, fluorescence signals obtained in the presence of S. Typhimurium LPS at different temperatures: 31.5 °C (white circles), 32 °C (black circles), 33 °C (inverted white triangles), 34 °C (black triangles), 36 °C (black squares), 37 °C (white squares), 38 °C (white diamonds), 39 °C (black diamonds), and 40 °C (white triangles). C, Arrhenius plots of rate constants obtained from fits of kinetic traces to a set of two consecutive first-order processes yielded activation barriers k1 = 19.2 kcal mol−1 (white circles) and k2 = 26.8 kcal mol−1 (black triangles). For curves determining the maximum receptor-saturating LPS concentration, see Fig. S8. For validation of the fitting procedure, see the supporting Methods.
We varied the LPS concentration and found that at 37 °C, an absolute LPS amount of 200 μg was sufficient to quantitatively trigger opening of all phage particles present in the cuvette (2.4 × 109 pfu). Here, the DNA-specific signal reached after about 100 min was comparable with the signal obtained after the same number of phage particles were treated with proteinase K, confirming that all particles had quantitatively released their DNA into the solution upon LPS contact (Fig. S9).
We therefore conclude that LPS is sufficient to elicit DNA release from myovirus Det7. Using either S. Anatum or S. Typhimurium host cell LPS resulted in similar DNA ejection velocities and yields (i.e. kinetics were independent of O-antigen composition). Moreover, to trigger DNA ejection from Det7, long O-antigen chains on the LPS were necessary. When the O-antigen part of LPS preparations from S. Typhimurium or S. Anatum was cleaved off with the respective specific TSP (Det7TSP for S. Typhimurium or DettilonTSP for S. Anatum) 2 min after initiation of ejection, the yield of ejected phage DNA decreased significantly, indicating that TSPs had deprived the mixture of functional O-antigen receptors (Fig. 5A and Fig. S7).
DNA ejection from Det7 was monitored in the presence of different LPS amounts, with velocities not further increasing above 50 μg ml−1, defining the saturating LPS concentration for all measurements (Fig. S9). As a result, we consider receptor binding as a fast, pseudo-first-order step, as it has been previously shown for DNA ejection from phages with short or long noncontractile tails (14, 27, 28). In these podo- or siphoviruses, in vitro particle opening could then be described as a single, first-order process. In contrast, DNA release from myovirus Det7 showed an initial lag phase. Kinetic analysis used a two-step process, A → B → C, describing two conformational transitions with rates k1 and k2 during Det7 particle opening for time courses measured at different temperatures. Models with more kinetic steps or with a pre-equilibrium step were excluded using the Akaike information criterion (see supporting Methods).
The two conformational transitions found in the myovirus Det7 particle upon DNA release have Arrhenius activation energies of 19.2 and 26.8 kcal mol−1, respectively, a range typically observed during phage particle opening (Fig. 5, B and C and Table S3). DNA release kinetics from Det7 were observed independently from the type of host receptor used to trigger in vitro DNA ejection (i.e. either S. Typhimurium or S. Anatum LPS), further emphasizing that we monitored rearrangements in the Det7 tail apparatus rather than receptor adsorption.
O-antigen cleavage and particle opening are coupled processes in myovirus Det7 but not in podovirus P22
At temperatures below 31 °C, only about 20% of the phages ejected their DNA, at 25 °C DNA ejection ceased completely (Fig. S8). To analyze whether Det7 remained ejection-competent in the presence of its receptor at temperatures nonpermissive for particle opening, we heated LPS/phage mixtures after different times of pre-incubation at 25 °C (Fig. 6A). Temperature increase to 37 °C, after 2 min, yielded about 20% of ejected particles, whereas heating after 17 min failed to elicit DNA ejection. The same result was obtained with a 3-fold increased LPS concentration.
Figure 6.
In vitro DNA ejection of myovirus Det7 and podovirus P22 after pre-incubation with LPS receptor at nonpermissive temperature. Fluorescence was monitored in the presence of S. Typhimurium LPS at 25 °C. Mixtures were heated to 37 °C after different waiting times. A, 8 × 109 Det7 pfu mixed with 67 μg ml−1 LPS and heated after 2 min (green circles), 17 min (white squares), and 30 min plus the addition of new LPS receptor (67 μg ml−1) (cyan triangles). B, 7.2 × 109 P22 pfu mixed with 10 μg ml−1 LPS at 25 °C (white triangles) or with heating after 17 min (black circles). For comparison, ejection at 37 °C is shown for both phages (white circles).
However, when phage/LPS mixtures were pre-incubated at 25 °C and heated to 37 °C together with the addition of new LPS receptor, all Det7 particles ejected their DNA (Fig. 6A). This is in contrast to short, noncontractile-tailed podovirus P22 that also ejected DNA without the addition of new LPS. P22 still readily ejected its DNA after pre-incubation at 25 °C for more than 15 min with the same S. Typhimurium LPS as Det7 (Fig. 6B). Velocities of O-antigen cleavage were similar for Det7TSP and P22TSP (Fig. S10). We therefore conclude that bacteriophage Det7 with a contractile tail needs a permissive temperature and an intact O-antigen receptor for particle opening. Det7 cannot irreversibly adsorb to LPS at low temperatures, in contrast to O-antigen–specific podoviruses (28).
Discussion
Det7 is a multi-O-antigen–specific myovirus
Upon initial cell encounter bacteriophages are in contact with a variety of bacterial envelope structures (44). This has created diverse niches for bacteriophages in which they exploit chemically highly diverse surface matrices to initiate cell entry with different degrees of specialization in their tails (45, 46). Bacteriophage Det7 uses a set of different TSP enzymes to bind and enzymatically cleave LPS receptors on different hosts. As is typical for O-antigen–specific phages, also in Det7, a TSP-mediated LPS enzymatic cleavage and adsorption step is essential for infection (26, 36). As a consequence, Det7 cannot use rough bacterial strains as hosts and is unable to open its particle upon challenge with O-antigen–deficient LPS.
Details of the Det7 baseplate structure are so far unknown, but it is conceivable that myoviruses can arrange their different TSP adhesion tools very specifically in their tails to be functional in guiding particle orientation. This has been shown for example in bacteriophage G7C, which has evolved a complex baseplate hub to accommodate different TSPs (47). In Det7, each of the four different TSPs has a distinct N-terminal adapter sequence (Fig. S1). This suggests a unique position for each TSP in the Det7 baseplate to expand the phage's host range. In this way, all TSPs in Det7 could play similar roles in particle opening, each providing an individual O-antigen specificity. It is thus conceivable that the other two, so far uncharacterized, TSPs (gp206 and gp209) encoded in the Det7 genome further broaden the host range (32). This host-targeting strategy has also been exploited in biotechnology to guide specificities of phage-derived antimicrobials like pyocins by equipping them with an appropriate TSP (48).
Occurrence of surface-binding fibers or spikes with different specificities within the same phage particle has been described in all tail morphologies (40, 49, 50). Here, the extended baseplate architecture of myoviruses usually allows for accommodating the largest numbers of different TSPs (43, 47, 51). TSPs are usually enzymes and serve to destroy the extracellular matrix (i.e. capsules or biofilms) to uncover parts of the bacterial cell surface serving as phage receptors (52). When less specialized phages are equipped with TSPs, their action may be nonessential for infection, enabling these phages to attack hosts without O-antigen. For example, podovirus SP6 or myovirus φ92 possess multiple TSPs to cleave capsules or O-antigen but also can grow on rough strains (40, 43).
By contrast, bacteriophage Det7 needs cleavage of the O-antigen receptor for infection and is unable to interact with other cell wall features that become exposed in rough strains. This is in contrast, for example, to myovirus S16, which infects a broad Salmonella host range and, like T4, uses long tail fibers to recognize Omps (22, 41). Det7 is hence a multi-O-antigen–specific phage that has acquired different TSPs for different host cell O-antigen receptors and does not rely on Omp receptors for particle opening in vitro.
Opening kinetics of bacteriophage particles observed in vitro point to general, tail architecture–dependent mechanisms
Studying the function of the bacteriophage tail molecular machine in vitro with EM or biochemical methods cannot capture the full in vivo process of genome transfer. Conformational rearrangements within the tail that occur upon host binding address the whole phage particle assembly, ultimately leading to DNA release, but many single steps of this process are so far not understood. For example, in Det7, information is lacking on the molecular rearrangements in its portal complex, linking the capsid to the tail. We do not know whether mature Det7 particles have a closed portal as found in phage P22 or whether DNA already protrudes into the tail as in T5, where ejection of the tape measure protein regulates DNA egress (8, 53).
Analysis of DNA release as a consequence of conformational rearrangements in the phage particle, however, might elucidate some crucial steps during early phage infection at the Gram-negative outer membrane. In vitro, this bacteriophage particle-opening process has been studied in short-tailed (Podoviridae) and long noncontractile-tailed (Siphoviridae) systems (20, 26–28, 54–56). In general, DNA ejection in vitro was more rapid in siphoviruses than in podoviruses. However, the type of receptor used to trigger DNA release did not influence the in vitro ejection velocity when comparing phages with similar tail structures. For example, genome release occurred on similar time scales in siphoviruses using Omp receptors compared with a siphovirus using an LPS receptor.
Complete ejection from all Det7 particles in our setup was reached at LPS amounts approximately equivalent to 5 × 1010 bacterial cells. At the given phage particle number, this would correspond to a multiplicity of infection of only 0.16 (i.e. the infection of only one of eight cells). This illustrates that pure LPS molecules in solution, usually forming multilamellar aggregates, apparently only in part mimic an intact bacterial membrane (57). However, studying the myovirus Det7 molecular machine under these conditions in vitro showed that the phage particles opened and ejected their DNA upon contact with the pure, protein-free LPS. Importantly, this bulk experiment revealed a defined kinetic profile for Det7 DNA release distinct from those found for phages with other tail architectures, providing the first quantitative kinetic study of DNA release via a contractile tail system.
Det7 DNA ejection in vitro occurs more slowly than in Siphoviridae but on a time scale similar to that seen in the Podoviridae. However, whereas all noncontractile-tailed phages in the given experimental setups under manual mixing regimes showed single first-order processes, for the myovirus Det7, two kinetic steps became visible. Clearly, both steps are related to conformational rearrangements in the tail and not to receptor binding, because different LPS preparations with different O-antigen compositions elicited similar DNA ejection velocities. In contrast, the DNA ejection itself is assumed to be fast due to densely packaged and pressurized DNA in the capsid and cannot be resolved in the experiment (14).
In vitro experiments on particle opening showed that in all cases studied so far with podo- or siphoviruses, the numbers of kinetic steps resolved as well as the velocities of DNA release were found to be highly similar in phages of the same tail morphology. We therefore propose that the relatively slow two-step time course observed in Det7 is typical for in vitro opening of a contractile-tailed particle. In support of this hypothesis is early work from the 1950s analyzing phage T4 mixtures with Shigella sonnei “lipocarbohydrate” (58). Release of T4 capsid contents, measured as viscosity increase over time, appeared two-step-like and reached saturation at similar velocities as now found for myovirus Det7.
Connection of receptor adsorption with particle opening
We have analyzed Det7 at saturating LPS receptor concentrations (Fig. S9) and consider all kinetic steps during DNA release associated with tail conformational rearrangements, occurring after a fast and irreversible LPS adsorption step. For noncontractile tail systems, it has been argued that the main barrier to DNA release is conformational changes that open the portal (27, 56) (Table S3). Accordingly, lower barriers were found for siphovirus SPP1 or λ, where the portal channel is already filled with DNA (59, 60). In vitro DNA ejection of Det7 has two activation barriers. The larger one is similar to those found in SPP1 or λ and thus might be linked to a step liberating DNA to proceed through the Det7 tail. In myoviruses, DNA release must be preceded by tail sheath contraction, but this contraction alone does not yet liberate DNA from the capsid (11, 21, 23–25).
In fact, the kinetic profile of Det7 myovirus DNA ejection showed a second step with a significant barrier. We propose that this faster kinetic step is related to tail contraction, a step absent in sipho- and podoviruses. To substantiate this hypothesis, it will be necessary to define the opening state of the portal within the mature Det7 head-tail assembly and to experimentally assign the observed kinetic barriers to specific conformational changes leading to contraction in the Det7 tail.
The yield of ejected Det7 DNA decreased stepwise with decreasing temperatures. This points to a complex mixture of populations both of phage particles and LPS receptors. Mobility of DNA inside the capsid is strongly dependent on temperature and ionic strength, leading to different populations of DNA-ejecting particles, as described for phage λ (61). Moreover, a gel-liquid crystalline transition midpoint of 37 °C in S. Typhimurium LPS indicates a homogeneous LPS state only above this temperature (62). Accordingly, we observed a notable deviation of the rates determined at 36 and 37 °C for the fast kinetic step from linear Arrhenius behavior, whereas the slow step was not affected by the LPS phase transition (cf. Fig. 4C).
This observation might be in favor of a mechanism with a fast step that involves LPS interaction of the phage particle and a slower rearrangement step in the tail independent of LPS. LPS interaction could thus induce tail contraction, followed in a second step by DNA release. For podovirus P22, it has been proposed that after LPS adsorption, a mechanical signal may trigger particle opening and that the protruding tail needle gp26 might act as sensor contacting the rigid lipid A part (27). So far, we lack detailed structural descriptions of the baseplate to suggest a similar model for myovirus Det7 that might involve an LPS contact step initiating tail contraction.
Temperature shift experiments did not show irreversible Det7 adsorption onto its LPS receptor at low temperatures. In contrast, podovirus P22 could irreversibly stick to LPS at 25 °C and readily release its DNA upon heating. This means that in myovirus Det7, surface attachment is coupled to all following opening steps. Presumably, a rapid and consecutive process of TSP O-antigen binding, enzymatic cleavage, dissociation, and rebinding is necessary here, because diffusion free energy alone would not effectively position the Det7 particle near the bacterial surface. However, only in close proximity to the membrane, conformational rearrangements in the tail should start that ultimately lead to DNA ejection. In the same way, TSP enzymes in podovirus SP6 were suggested to supply the driving forces for its membrane orientation (63).
It has been proposed earlier that temperature-controlled phage infection is adapted to the bacterial life cycle (64). Myovirus Det7 with a lytic life style may be adapted to hosts with fast replication rates at 37 °C, whereas it might be advantageous to remain mobile and infection-competent at low temperatures and replication rates. In contrast, podoviruses like P22 or HK620 may have adapted to remain bound to suitable hosts even at low temperatures due to their temperate life style (28). Here, bacteria might regulate phage adsorption by varying the length of O-antigen chains at different temperatures, although this was not found for the Salmonella strains analyzed in this work (65).
In conclusion, the analysis of Salmonella phage Det7 genome release illustrates that we are only beginning to explore the diverse functional niches of bacteriophage infection mechanisms populated by generalist and specialist phages. Myovirus Det7 starts a concerted set of reorganization steps in the particle upon contact with its LPS O-antigen receptor, ultimately leading to DNA release. Understanding the kinetics of this process will be important for timing EM snapshots to analyze all tail rearrangements leading to particle opening in contractile tail systems. Det7 uses highly specialized TSPs to adsorb to smooth LPS-covered bacterial surfaces. This strategy can be seen as a unique infection control point, where a universal myovirus particle-opening mechanism is coupled to a specific trigger. Det7 exploits hosts with an O-antigen at permissive temperature only and might thus be part of a more complex community of bacteriophages that cooperatively act on their hosts but use different infection strategies.
Experimental procedures
Materials
Salmonella enterica strains were obtained from the National Reference Centre for Salmonellae and other Enterics (Robert Koch Institute, Wernigerode, Germany). Salmonella rough mutants (41) were kindly provided by Prof. Martin Loessner (Swiss Federal Institute of Technology Zurich, Switzerland). Lipopolysaccharides were prepared according to published procedures from cells grown at 37 °C (66, 67). O-antigen chain length distributions did not change for LPS grown at lower temperatures (Fig. S11). Polysaccharides and oligosaccharides were obtained by methods described previously (68). The Pro-Q® Emerald 300 Glycoprotein Gel and Blot Stain Kit and Yo-Pro®-1 Iodide (491/509) were purchased from Thermo Fisher Scientific Life Technologies GmbH (Darmstadt, Germany). Phage Det7 was propagated at 37 °C, its Salmonella host physiological growth temperature, and purified as described (26, 31). Phage concentrations are given in pfu. All TSPs used were N-terminally shortened (TSPΔN) and lacked the capsid adaptor domains. Standard buffer in all experiments was 50 mm Tris-HCl, pH 7.6, 4 mm MgCl2 if not stated otherwise.
DettilonTSP cloning, purification, and oligosaccharide binding
DNA fragments encoding amino acids 253–781 of DettilonTSP were amplified from guanidinium hydrochloride–treated phage lysate with primers 5′-GGCCCTCCATATGTTAAGAGGTGAGTTAAATAATGAAGGGGTA and 5′-CCGCTCGAGTTAGTTAAAATCAACAAAACCTTTTAATATAAACGATG and cloned into expression vector pET23a (Novagen, Madison, WI) using NdeI and XhoI restriction sites. Proteins were expressed in E. coli BL21(DE3) and purified using standard protocols (35). Binding of S. Anatum O-antigen hexasaccharide fragments was analyzed on an SR7500DC surface plasmon resonance spectrometer (Reichert, Buffalo NY). DettilonTSP was immobilized on a carboxymethyldextran surface (CMD200D, Xantec Bioanalytics, Düsseldorf, Germany) with standard amine-coupling procedures (69), and oligosaccharides were injected for 2.5 min at 25 μl min−1 at 20 °C in 50 mm sodium phosphate buffer, pH 7. A one-site independent binding isotherm was fitted to the equilibrium plateau signals (Fig. S5).
Crystallization, diffraction data collection, and structure determination
200 nl of 10 mg ml−1 DettilonTSP were mixed with an equal volume of reservoir solution containing 1 m (native crystals) or 0.85 m (crystals for hexasaccharide soaks) succinate, pH 7.0, 0.1 m HEPES, pH 7.0, 1% (w/v) PEG methyl ether 2000 and crystallized with sitting-drop vapor diffusion. For heavy-atom derivatives, protein solutions contained 3 mm mercury(II) chloride or 3 mm ammonium tetrachloroplatinate(II). Crystals (140 × 50 × 40 μm) appeared within 2–3 days at 20 °C and were flash-frozen in liquid nitrogen with the addition of 20% glycerol as cryoprotectant. Prior to freezing, native crystals were soaked for 30 min in precipitant solution containing 5 mm S. Anatum O-antigen hexasaccharide.
Diffraction data were recorded at BL14.1 at BESSY II (Helmholtz-Zentrum Berlin), processed, and scaled using XDSapp (70) to obtain native (1.63 Å), Hg (1.65 Å), Pt (1.97 Å), and hexasaccharide complex (2.1 Å) data sets (Table S2). Phases for ligand-free model building and refinement were obtained from two heavy-atom derivative data sets using Phenix (71) and from molecular replacement with Phaser (72) for the ligand complex using the native data set. Structures were manually built using Coot (73) and refined using Phenix with resulting Rwork/Rfree values of 15.8%/18.8% and 18.4%/21.4%, respectively (Table S2). The final coordinates were deposited in the Protein Data Bank with accession numbers 6F7D and 6F7K. Figures were generated with PyMOL (version 2.0, Schrödinger, LLC, New York).
Bacteriophage Det7 DNA ejection experiments
DNA ejection from Det7 was monitored as described (26). Briefly, 8 × 109 pfu ml−1 Det7 were mixed with LPS from S. Typhimurium or S. Anatum at different concentrations in the presence of 1.1 μm Yo-Pro®-1 Iodide, and fluorescence increase was monitored over time. Eventually, DNase I (10 μg ml−1) was added as a control for DNA accessibility at the end of an ejection time course. 100 data points were recorded in each measurement; for clarity, figures display fewer points. Kinetic data were fitted to Equation 1 with MatLab using the routine lsqnonlin. The routine nlparci computed 95% confidence intervals of the parameters (MatLab script available on request). See supporting Methods and Table S4 for kinetic model discrimination with the Akaike information criterion and all fitting results.
| (Eq. 1) |
Author contributions
N. K. B., Y. R., D. A., U. H., and S. B. conceptualization; N. K. B., Y. R., J. K., U. H., and S. B. supervision; N. K. B. and S. B. funding acquisition; N. K. B., Y. R., A. V., and M. S. S. investigation; N. K. B., Y. R., A. V., and S. B. visualization; N. K. B., A. V., D. A., U. H., and S. B. methodology; N. K. B., Y. R., A. V., D. A., U. H., and S. B. writing-original draft; N. K. B., Y. R., and S. B. project administration; N. K. B., Y. R., U. H., and S. B. writing-review and editing; Y. R., A. V., and D. A. data curation; Y. R., A. V., and D. A. formal analysis; A. V. and D. A. software; J. K., U. H., and S. B. resources.
Supplementary Material
Acknowledgments
We thank Mandy Schietke and Sibylle Rüstig for excellent technical assistance and Tobias Irmscher for oligosaccharide mass analysis.
This work was supported by Deutsche Forschungsgemeinschaft Grant BA 4046/1-2 (to S. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting Methods, Tables S1–S4, and Figs. S1–S11.
- TSP
- tailspike protein
- Omp
- outer membrane protein
- LPS
- lipopolysaccharide
- pfu
- plaque-forming protein
- aa
- amino acid(s).
References
- 1. Ackermann H. W., and Prangishvili D. (2012) Prokaryote viruses studied by electron microscopy. Arch. Virol. 157, 1843–1849 10.1007/s00705-012-1383-y [DOI] [PubMed] [Google Scholar]
- 2. Davidson A., Cardarelli L., Pell L., Radford D., and Maxwell K. (2012) Long noncontractile tail machines of bacteriophages. in Viral Molecular Machines (Rossmann M., and Rao V., eds) pp. 115–142, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 3. Lander G. C., Khayat R., Li R., Prevelige P. E., Potter C. S., Carragher B., and Johnson J. E. (2009) The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure 17, 789–799 10.1016/j.str.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lhuillier S., Gallopin M., Gilquin B., Brasilès S., Lancelot N., Letellier G., Gilles M., Dethan G., Orlova E. V., Couprie J., Tavares P., and Zinn-Justin S. (2009) Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. U.S.A. 106, 8507–8512 10.1073/pnas.0812407106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Valen D., Wu D., Chen Y. J., Tuson H., Wiggins P., and Phillips R. (2012) A single-molecule Hershey–Chase experiment. Curr. Biol. 22, 1339–1343 10.1016/j.cub.2012.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu B., Margolin W., Molineux I. J., and Liu J. (2013) The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339, 576–579 10.1126/science.1231887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu B., Margolin W., Molineux I. J., and Liu J. (2015) Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc. Natl. Acad. Sci. U.S.A. 112, E4919–E4928 10.1073/pnas.1501064112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arnaud C. A., Effantin G., Vivès C., Engilberge S., Bacia M., Boulanger P., Girard E., Schoehn G., and Breyton C. (2017) Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Nat. Commun. 8, 1953 10.1038/s41467-017-02049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C., Tu J., Liu J., and Molineux I. J. (2019) Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol. 4, 1049–1056 10.1038/s41564-019-0403-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boulanger P., and Letellier L. (1988) Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J. Biol. Chem. 263, 9767–9775 [PubMed] [Google Scholar]
- 11. Taylor N. M. I., Prokhorov N. S., Guerrero-Ferreira R. C., Shneider M. M., Browning C., Goldie K. N., Stahlberg H., and Leiman P. G. (2016) Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346–352 10.1038/nature17971 [DOI] [PubMed] [Google Scholar]
- 12. Nobrega F. L., Vlot M., de Jonge P. A., Dreesens L. L., Beaumont H. J. E., Lavigne R., Dutilh B. E., and Brouns S. J. J. (2018) Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 16, 760–773 10.1038/s41579-018-0070-8 [DOI] [PubMed] [Google Scholar]
- 13. Leiman P. G., and Shneider M. M. (2012) Contractile tail machines of bacteriophages. in Viral Molecular Machines (Rossmann M. G., and Rao V. B., eds) pp. 93–114, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 14. Chiaruttini N., de Frutos M., Augarde E., Boulanger P., Letellier L., and Viasnoff V. (2010) Is the in vitro ejection of bacteriophage DNA quasistatic? a bulk to single virus study. Biophys. J. 99, 447–455 10.1016/j.bpj.2010.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernandes S., Labarde A., Baptista C., Jakutytè L., Tavares P., and São-José C. (2016) A non-invasive method for studying viral DNA delivery to bacteria reveals key requirements for phage SPP1 DNA entry in Bacillus subtilis cells. Virology 495, 79–91 10.1016/j.virol.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 16. García L. R., and Molineux I. J. (1995) Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 177, 4066–4076 10.1128/jb.177.14.4066-4076.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. González-Huici V., Salas M., and Hermoso J. M. (2004) The push-pull mechanism of bacteriophage phi29 DNA injection. Mol. Microbiol. 52, 529–540 10.1111/j.1365-2958.2004.03993.x [DOI] [PubMed] [Google Scholar]
- 18. Chang J. T., Schmid M. F., Haase-Pettingell C., Weigele P. R., King J. A., and Chiu W. (2010) Visualizing the structural changes of bacteriophage ϵ15 and its Salmonella host during infection. J. Mol. Biol. 402, 731–740 10.1016/j.jmb.2010.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casjens S. R., and Molineux I. J. (2012) Short noncontractile tail machines: adsorption and DNA delivery by Podoviruses. in Viral Molecular Machines (Rossmann M. G., and Rao V. B., eds) pp. 143–179, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 20. González-Garcia V. A., Pulido-Cid M., García-Doval C., Bocanegra R., van Raaij M. J., Martín-Benito J., Cuervo A., and Carrascosa J. L. (2015) Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J. Biol. Chem. 290, 10038–10044 10.1074/jbc.M114.614222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yap M. L., Klose T., Arisaka F., Speir J. A., Veesler D., Fokine A., and Rossmann M. G. (2016) Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc. Natl. Acad. Sci. U.S.A. 113, 2654–2659 10.1073/pnas.1601654113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Washizaki A., Yonesaki T., and Otsuka Y. (2016) Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiologyopen 5, 1003–1015 10.1002/mbo3.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Habann M., Leiman P. G., Vandersteegen K., Van den Bossche A., Lavigne R., Shneider M. M., Bielmann R., Eugster M. R., Loessner M. J., and Klumpp J. (2014) Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 92, 84–99 10.1111/mmi.12539 [DOI] [PubMed] [Google Scholar]
- 24. Nováček J., Šiborová M., Benešík M., Pantuček R., Doškař J., and Plevka P. (2016) Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc. Natl. Acad. Sci. U.S.A. 113, 9351–9356 10.1073/pnas.1605883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldberg E., Grinius L., and Letellier L. (1994) Recognition, attachment, and injection. in Molecular Biology of Bacteriophage T4 (Karam J. D., ed) pp. 347–356, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 26. Andres D., Hanke C., Baxa U., Seul A., Barbirz S., and Seckler R. (2010) Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J. Biol. Chem. 285, 36768–36775 10.1074/jbc.M110.169003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andres D., Roske Y., Doering C., Heinemann U., Seckler R., and Barbirz S. (2012) Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Mol. Microbiol. 83, 1244–1253 10.1111/j.1365-2958.2012.08006.x [DOI] [PubMed] [Google Scholar]
- 28. Broeker N. K., Kiele F., Casjens S. R., Gilcrease E. B., Thalhammer A., Koetz J., and Barbirz S. (2018) In vitro studies of lipopolysaccharide-mediated DNA release of podovirus HK620. Viruses 10, E289 10.3390/v10060289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grayson P., Han L., Winther T., and Phillips R. (2007) Real-time observations of single bacteriophage λ DNA ejections in vitro. Proc. Natl. Acad. Sci. U.S.A. 104, 14652–14657 10.1073/pnas.0703274104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. São-José C., de Frutos M., Raspaud E., Santos M. A., and Tavares P. (2007) Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J. Mol. Biol. 374, 346–355 10.1016/j.jmb.2007.09.045 [DOI] [PubMed] [Google Scholar]
- 31. Walter M., Fiedler C., Grassl R., Biebl M., Rachel R., Hermo-Parrado X. L., Llamas-Saiz A. L., Seckler R., Miller S., and van Raaij M. J. (2008) Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J. Virol. 82, 2265–2273 10.1128/JVI.01641-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Casjens S. R., Jacobs-Sera D., Hatfull G. F., and Hendrix R. W. (2015) Genome sequence of Salmonella enterica phage Det7. Genome Announc. 3, e00279–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steinbacher S., Baxa U., Miller S., Weintraub A., Seckler R., and Huber R. (1996) Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. U.S.A. 93, 10584–10588 10.1073/pnas.93.20.10584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Müller J. J., Barbirz S., Heinle K., Freiberg A., Seckler R., and Heinemann U. (2008) An intersubunit active site between supercoiled parallel β helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6. Structure 16, 766–775 10.1016/j.str.2008.01.019 [DOI] [PubMed] [Google Scholar]
- 35. Barbirz S., Müller J. J., Uetrecht C., Clark A. J., Heinemann U., and Seckler R. (2008) Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol. Microbiol. 69, 303–316 10.1111/j.1365-2958.2008.06311.x [DOI] [PubMed] [Google Scholar]
- 36. Broeker N. K., and Barbirz S. (2017) Not a barrier but a key: how bacteriophages exploit host's O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 105, 353–357 10.1111/mmi.13729 [DOI] [PubMed] [Google Scholar]
- 37. Seul A., Muller J. J., Andres D., Stettner E., Heinemann U., and Seckler R. (2014) Bacteriophage P22 tailspike: structure of the complete protein and function of the interdomain linker. Acta Crystallogr. D Biol. Crystallogr. 70, 1336–1345 10.1107/S1399004714002685 [DOI] [PubMed] [Google Scholar]
- 38. Kropinski A. M., Kovalyova I. V., Billington S. J., Patrick A. N., Butts B. D., Guichard J. A., Pitcher T. J., Guthrie C. C., Sydlaske A. D., Barnhill L. M., Havens K. A., Day K. R., Falk D. R., and McConnell M. R. (2007) The genome of ϵ 15, a serotype-converting, group E1 Salmonella enterica-specific bacteriophage. Virology 369, 234–244 10.1016/j.virol.2007.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robbins P. W., and Uchida T. (1962) Studies on chemical basis of phage conversion of O-antigens in E-group Salmonellae. Biochemistry 1, 323–335 10.1021/bi00908a020 [DOI] [PubMed] [Google Scholar]
- 40. Gebhart D., Williams S. R., and Scholl D. (2017) Bacteriophage SP6 encodes a second tailspike protein that recognizes Salmonella enterica serogroups C-2 and C-3. Virology 507, 263–266 10.1016/j.virol.2017.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marti R., Zurfluh K., Hagens S., Pianezzi J., Klumpp J., and Loessner M. J. (2013) Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 87, 818–834 10.1111/mmi.12134 [DOI] [PubMed] [Google Scholar]
- 42. Kang Y., Gohlke U., Engström O., Hamark C., Scheidt T., Kunstmann S., Heinemann U., Widmalm G. R., Santer M., and Barbirz S. (2016) Bacteriophage tailspikes and bacterial O-antigens as a model system to study weak-affinity protein-polysaccharide interactions. J. Am. Chem. Soc. 138, 9109–9118 10.1021/jacs.6b00240 [DOI] [PubMed] [Google Scholar]
- 43. Schwarzer D., Buettner F. F. R., Browning C., Nazarov S., Rabsch W., Bethe A., Oberbeck A., Bowman V. D., Stummeyer K., Mühlenhoff M., Leiman P. G., and Gerardy-Schahn R. (2012) A multivalent adsorption apparatus explains the broad host range of phage φ92: a comprehensive genomic and structural analysis. J. Virol. 86, 10384–10398 10.1128/JVI.00801-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bertozzi Silva J., Storms Z., and Sauvageau D. (2016) Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363, fnw002 10.1093/femsle/fnw002 [DOI] [PubMed] [Google Scholar]
- 45. Bono L. M., Gensel C. L., Pfennig D. W., and Burch C. L. (2015) Evolutionary rescue and the coexistence of generalist and specialist competitors: an experimental test. Proc. Biol. Sci. 282, 20151932 10.1098/rspb.2015.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yosef I., Goren M. G., Globus R., Molshanski-Mor S., and Qimron U. (2017) Extending the host range of bacteriophage particles for DNA transduction. Mol. Cell 66, 721–728.e3 10.1016/j.molcel.2017.04.025 [DOI] [PubMed] [Google Scholar]
- 47. Prokhorov N. S., Riccio C., Zdorovenko E. L., Shneider M. M., Browning C., Knirel Y. A., Leiman P. G., and Letarov A. V. (2017) Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 105, 385–398 10.1111/mmi.13710 [DOI] [PubMed] [Google Scholar]
- 48. Buth S. A., Shneider M. M., Scholl D., and Leiman P. G. (2018) Structure and analysis of R1 and R2 pyocin receptor-binding fibers. Viruses 10, 427 10.3390/v10080427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scholl D., Rogers S., Adhya S., and Merril C. R. (2001) Bacteriophage K1–5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75, 2509–2515 10.1128/JVI.75.6.2509-2515.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Golomidova A. K., Kulikov E. E., Prokhorov N. S., Guerrero-Ferreira R. C., Knirel Y. A., Kostryukova E. S., Tarasyan K. K., and Letarov A. V. (2016) Branched lateral tail fiber organization in T5-like bacteriophages DT57C and DT571/2 is revealed by genetic and functional analysis. Viruses 8, 26 10.3390/v8010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pan Y. J., Lin T. L., Chen C. C., Tsai Y. T., Cheng Y. H., Chen Y. Y., Hsieh P. F., Lin Y. T., and Wang J. T. (2017) Klebsiella phage φ K64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 91, e02457–02416 10.1128/JVI.02457-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pires D. P., Oliveira H., Melo L. D. R., Sillankorva S., and Azeredo J. (2016) Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151 10.1007/s00253-015-7247-0 [DOI] [PubMed] [Google Scholar]
- 53. Parent K. N., Schrad J. R., and Cingolani G. (2018) Breaking symmetry in viral icosahedral capsids as seen through the lenses of X-ray crystallography and cryo-electron microscopy. Viruses 10, 67 10.3390/v10020067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu D., Van Valen D., Hu Q., and Phillips R. (2010) Ion-dependent dynamics of DNA ejections for bacteriophage λ. Biophys. J. 99, 1101–1109 10.1016/j.bpj.2010.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chiaruttini N., Letellier L., and Viasnoff V. (2013) A novel method to couple electrophysiological measurements and fluorescence imaging of suspended lipid membranes: the example of T5 bacteriophage DNA ejection. PLoS One 8, e84376 10.1371/journal.pone.0084376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raspaud E., Forth T., São-José C., Tavares P., and de Frutos M. (2007) A kinetic analysis of DNA ejection from tailed phages revealing the prerequisite activation energy. Biophys. J. 93, 3999–4005 10.1529/biophysj.107.111435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Richter W., Vogel V., Howe J., Steiniger F., Brauser A., Koch M. H., Roessle M., Gutsmann T., Garidel P., Mäntele W., and Brandenburg K. (2011) Morphology, size distribution, and aggregate structure of lipopolysaccharide and lipid A dispersions from enterobacterial origin. Innate Immun. 17, 427–438 10.1177/1753425910372434 [DOI] [PubMed] [Google Scholar]
- 58. Jesaitis M. A., and Goebel W. F. (1955) Lysis of phage T4 by the specific lipocarbohydrate of phase-II Shigella sonnei. J. Exp. Med. 102, 733–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chaban Y., Lurz R., Brasilès S., Cornilleau C., Karreman M., Zinn-Justin S., Tavares P., and Orlova E. V. (2015) Structural rearrangements in the phage head-to-tail interface during assembly and infection. Proc. Natl. Acad. Sci. U.S.A. 112, 7009–7014 10.1073/pnas.1504039112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tavares P., Zinn-Justin S., and Orlova E. V. (2012) Genome gating in tailed bacteriophage capsids. in Viral Molecular Machines (Rossmann M. G., and Rao V. B., eds) pp. 585–600, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 61. Evilevitch A. (2018) The mobility of packaged phage genome controls ejection dynamics. eLife 7, e37345 10.7554/eLife.37345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brandenburg K., and Seydel U. (1990) Investigation into the fluidity of lipopolysaccharide and free lipid A membrane systems by Fourier-transform infrared spectroscopy and differential scanning calorimetry. Eur. J. Biochem. 191, 229–236 10.1111/j.1432-1033.1990.tb19114.x [DOI] [PubMed] [Google Scholar]
- 63. Tu J. G., Park T., Morado D. R., Hughes K. T., Molineux I. J., and Liu J. (2017) Dual host specificity of phage SP6 is facilitated by tailspike rotation. Virology 507, 206–215 10.1016/j.virol.2017.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Freeman K. G., Behrens M. A., Streletzky K. A., Olsson U., and Evilevitch A. (2016) Portal stability controls dynamics of DNA ejection from phage. J. Phys. Chem. B 120, 6421–6429 10.1021/acs.jpcb.6b04172 [DOI] [PubMed] [Google Scholar]
- 65. Raetz C. R., and Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Westphal O., and Jann K. (1965) Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. in Methods in Carbohydrate Chemistry (Whistler R. L., and Wolfan M. L., eds) pp. 83–91, Academic Press, New York [Google Scholar]
- 67. Darveau R. P., and Hancock R. E. (1983) Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155, 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zaccheus M. V., Broeker N. K., Lundborg M., Uetrecht C., Barbirz S., and Widmalm G. (2012) Structural studies of the O-antigen polysaccharide from Escherichia coli TD2158 having O18 serogroup specificity and aspects of its interaction with the tailspike endoglycosidase of the infecting bacteriophage HK620. Carbohydr. Res. 357, 118–125 10.1016/j.carres.2012.05.022 [DOI] [PubMed] [Google Scholar]
- 69. Andres D., Gohlke U., Broeker N. K., Schulze S., Rabsch W., Heinemann U., Barbirz S., and Seckler R. (2013) An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O-antigen fragments to bind as nonsolution conformers. Glycobiology 23, 486–494 10.1093/glycob/cws224 [DOI] [PubMed] [Google Scholar]
- 70. Krug M., Weiss M., Heinemann U., and Mueller U. (2012) XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Cryst. 45, 568–572 10.1107/S0021889812011715 [DOI] [Google Scholar]
- 71. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.