Abstract
Bacteria can rapidly and reversibly respond to changing environments via complex transcriptional and post-transcriptional regulatory mechanisms. Many of these adaptations are specific, with the regulatory output tailored to the inducing signal (for instance, repairing damage to cell components or improving acquisition and use of growth-limiting nutrients). However, the general stress response, activated in bacterial cells entering stationary phase or subjected to nutrient depletion or cellular damage, is unique in that its common, broad output is induced in response to many different signals. In many different bacteria, the key regulator for the general stress response is a specialized sigma factor, the promoter specificity subunit of RNA polymerase. The availability or activity of the sigma factor is regulated by complex regulatory circuits, the majority of which are post-transcriptional. In Escherichia coli, multiple small regulatory RNAs, each made in response to a different signal, positively regulate translation of the general stress response sigma factor RpoS. Stability of RpoS is regulated by multiple anti-adaptor proteins that are also synthesized in response to different signals. In this review, the modes of signaling to and levels of regulation of the E. coli general stress response are discussed. They are also used as a basis for comparison with the general stress response in other bacteria with the aim of extracting key principles that are common among different species and highlighting important unanswered questions.
Keywords: Hfq protein, RNA, RNA polymerase, prokaryotic signal-transduction, proteolysis, Escherichia coli (E. coli), anti-adaptor, ClpXP, general stress response, RpoS
Introduction
All organisms depend upon their ability to respond appropriately to changes in their environments and/or temporary disappearance of nutrients. As important as the response to these stresses is the ability to reverse these stress adaptations when conditions improve. Bacteria have long been a wonderful example of how cells carry out such adaptations.
Many stress adaptations are specifically tailored to the inducing stress, and we can identify many of genes that should help deal with the stress (Fig. 1A). For instance, high temperature induces chaperones and proteases that help deal with proteins misfolded or aggregated in response to the heat shock; the SOS response to DNA damage includes induction of repair proteins and polymerases that can bypass DNA damage. Loss of nutrients similarly induces regulons that may redirect metabolism to minimize the necessity for the missing nutrient and may induce systems for improving the cell's ability to find and import the nutrient. That said, in most cases we only understand the contribution of a small subset of the members of any given stress regulon to the response.
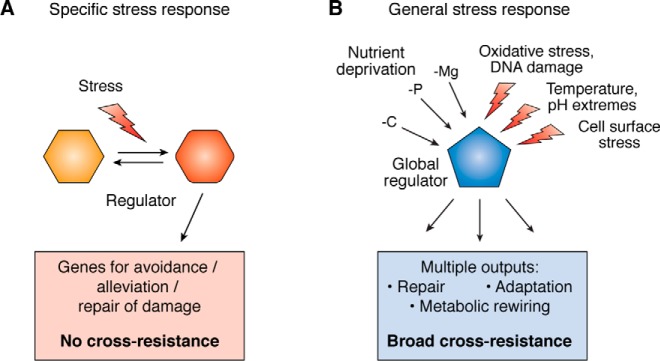
Figure 1.
Specific stress responses versus general stress responses. A, specific stress responses respond to environmental signals to change the state of the specific regulator; the set of induced and repressed genes (the regulon) includes those encoding proteins that help the cell avoid or repair damage or reduce the need for and increase import for a limiting nutrient in the case of a starvation response. B, in contrast, general stress responses are triggered by multiple different stresses, and the output is multipronged, leading to cross-resistance to stresses not used in the original induction. In all cases studied thus far, the global regulator that mediates the general stress response is a specialized sigma factor.
However, in addition to these responses many (and possibly all) bacteria also encode a robust “general stress response,” characterized by the ability of the cell to defend itself not only from the specific inducing stress but also from a variety of other seemingly unrelated stresses (Fig. 1B). We imagine that this response allows cells to anticipate and prepare for what are usually linked stresses and limiting growth conditions; this response presumably provides insight into what these bacterial cells are likely to encounter in their natural environments. Exactly when and why the cell chooses to invoke a general stress response rather than the more specific one is not well-understood, but frequently the general stress response is also associated with stationary phase growth, when nutrients are likely limiting.
Our lab has focused on the regulatory mechanisms that lead to induction of the Escherichia coli general stress response. Intriguingly, and possibly reflecting the nature of this response and when it must be used, much of the regulation is post-transcriptional. Complicating matters, a given stress may affect multiple levels of induction (1, 2). I review and discuss these mechanisms and regulatory circuits, comparing them with general stress responses in other bacteria. Interestingly, there are a number of conserved characteristics that may help in analyzing the critical roles of these systems in disparate bacteria.
In E. coli, the scope of resistance mediated by the general stress response is most clearly seen by the consequences of mutating rpoS, encoding the central transcription factor for the response. rpoS mutants are sensitive to starvation for carbon, treatment with hydrogen peroxide, survival at high temperature, survival at low pH, and after osmotic stress (3–5). Mutant cells change their ability to grow on limiting levels of different carbon sources, fail to accumulate glycogen, and change their ability to form biofilms (3, 6, 7). This list is far from comprehensive. In some cases, the downstream genes responsible for these phenotypes have been characterized. For instance, a catalase, capable of destroying hydrogen peroxide in stationary phase, is transcribed dependent on RpoS, as is Dps, an abundant DNA-binding protein that helps protect cells from oxidative damage (8, 9). A variety of global approaches have identified multiple genes dependent upon RpoS (10–14), although for many of these we do not know how they contribute to the general stress response. A detailed understanding of the full downstream output of the general stress response remains to be undertaken and is outside the scope of this review.
Common characteristics of general stress responses
The definition I will use here for a general stress response requires evidence that, upon induction by a given stress, cells become resistant to multiple stress treatments that are, to the best of our current knowledge, in different repair or adaptation pathways than the response to the original inducing stress. In most bacterial systems, this response is frequently also found associated with stationary phase, when bacteria stop growing exponentially, as they run out of nutrients and accumulate inhibitory by-products. Certainly, in the environment, bacteria will generally not be in the type of rich and uncrowded conditions we provide in the lab, and thus, many bacteria may be in a stationary phase-like state for much of the time. The assumption has been and continues to be that induction of this response reflects the likelihood that there is a frequently encountered growth/stress condition that requires the broad resistance mechanisms, and that any single inducing treatment can serve, under the right conditions, as the harbinger of the other stresses.
Intriguingly, in the bacterial general stress responses discussed here, the key regulator responsible for transcription of the downstream regulon genes is a specialized sigma factor, the component of RNA polymerase that combines with the core subunits of polymerase to endow promoter specificity on the polymerase holoenzyme. The primary or vegetative sigma factor, RpoD or σ70 in E. coli, is responsible for transcribing genes for the basic machinery of cell growth and replication. Under various stress or during developmental pathways, sigma factors specialized to transcribe genes for that condition accumulate and are used (reviewed in Refs. 15, 16). The sigma factors are, by nature of how they act, almost exclusively positive regulators of transcription and thus should increase transcription of their targets when they are available and active, although they also must compete with the vegetative sigma factor for limiting levels of the core RNA polymerase.
In principle, the ability of the general stress response sigma factor to act can be regulated at any level, and to some extent it is. The plethora of levels of regulation may well-reflect the necessity for multiple signals to feed into inducing a common factor. However, it is also striking that a great deal of the regulation is post-transcriptional, particularly at the level of negative regulation of sigma factor activity by proteins that bind and sequester specific sigma factors (anti-sigmas) and by proteolysis. Having a system under negative post-transcriptional regulation may have advantages in terms of keeping the response off when it is not needed and rapidly mounting a response when it is needed. Constant expression of the general stress sigma factors is generally detrimental for rapid cell growth, so that efficient negative regulation can be important. However, if the sigma factor is needed rapidly, under conditions when energy and nutrients are running out, it may be very useful for the cell to have the sigma factor ready for activation when it is needed, rather than starting the induction process when a stress is encountered with new transcription and translation. An additional important consequence of negative post-transcriptional regulation of a sigma factor, whether by degradation or by anti-sigma inhibition, is the potential for rapid recovery from the stress state by shutting off the stress transcriptional program.
Induction mechanisms for the general stress response in E. coli and related bacteria
In E. coli K12, the lab strain that has been most studied, RpoS (also called σS or σ38) is the sigma factor that mediates the general stress response, also called the stationary phase response. A variety of studies suggest that multiple signals and multiple levels of regulation contribute to whether RpoS accumulates and is active in mediating transcription (Fig. 2). As a result, a detailed look at the levels of regulation of RpoS provide a view of the very many modes of regulation in use in bacterial cells. A first comprehensive sense of the complexity of the regulation is reflected in work by Lange and Hengge-Aronis in 1994 (17), in which a set of parallel transcriptional and translational fusions revealed evidence of regulation at the level of transcription, translation, and protein stability, with somewhat different patterns depending on growth in minimal media, rich media, or after osmotic shock (18). Over the 25 years since that paper, experiments by many groups have uncovered some of the pathways for each of these levels of regulation, but we are still far from a full understanding of the ways in which cells induce and recover from the general stress response.
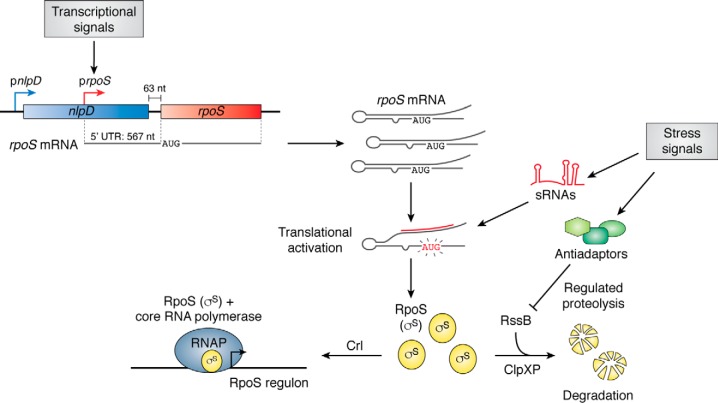
Figure 2.
Multiple levels of regulation affect availability of the RpoS (σS) general stress response sigma factor in E. coli. The major regulators of RpoS synthesis and function are outlined here. Transcription is primarily from the prpoS promoter, embedded within the upstream nlpD gene. The resulting transcript includes a long 5′ UTR, which folds back to occlude ribosome entry. sRNAs, made in response to specific signals (see Fig. 3 and text), open this structure, promoting translation. Other stress signals lead to synthesis of anti-adaptors (green shapes) that block the rapid degradation of RpoS. When levels of RpoS (yellow spheres, σS) rise as a consequence of these changes in regulation, RpoS combines with core RNA polymerase (blue), inducing transcription of the genes of the RpoS regulon; Crl helps to promote RpoS capture of core polymerase.
Regulation of RpoS synthesis
Transcriptional regulation
The gene for rpoS is downstream from nlpD, encoding an outer membrane lipoprotein involved in cell division via an effect on peptidoglycan metabolism, and the primary promoter for rpoS is embedded in the middle of nlpD (Fig. 2) (19). The promoter upstream of nlpD apparently contributes to basal level expression of rpoS in exponential phase (17, 19, 20). This organization is well-conserved in many gammaproteobacteria (and beyond) (21), although the basis for this relationship is not currently known.
Conclusions about the regulation of transcription initiation come from a variety of evidence, not all of them measuring only promoter activity. Measurements of levels of the rpoS mRNA, for instance, under different growth conditions or in cells mutant for transcriptional regulators, can be confounded if these conditions change mRNA stability and/or have effects on transcription elongation or termination. The same caveats are true for measurements of expression of rpoS transcriptional fusions that include significant extents of the transcribed message. For instance, the long untranslated leader for RpoS (Fig. 2), present in the most-studied transcriptional fusions, may be subject to regulated Rho-dependent termination (22). Conclusions from the literature, which sometimes disagree, are confounded by different groups using different strains and different growth conditions. The best evidence of direct and significant regulation of transcription initiation would presumably include identification of the regulator, establishment of its ability to bind to the rpoS promoter, coupled with identification of the binding site(s), and demonstration that mutation of the binding site eliminates the regulatory effect in vivo.
A short discussion of the data on the role of cAMP and CRP2 in regulation of RpoS may illustrate the difficulties in making any simple conclusions about rpoS transcriptional regulation. cAMP is synthesized when the cell is using secondary carbon sources (rather than glucose), and the global transcriptional regulator CRP, in complex with cAMP, positively regulates the genes necessary for using these less-favored carbon sources. A number of studies show higher levels of RpoS early in exponential phase in the absence of cAMP and CRP (3, 17, 23, 24), suggesting negative transcriptional regulation by CRP/cAMP, and based on these studies, EcoCyc currently shows the rpoS promoter as negatively regulated by CRP (25). However, other studies show positive effects of cAMP and CRP (26), and mutations in the suggested CRP-binding sites upstream and downstream of the major rpoS promoter significantly reduced RpoS expression (23), providing compelling evidence for an important positive role for CRP for RpoS transcription. Possibly, CRP regulates RpoS differently at different stages of growth, or the negative effects early in growth are indirect.
ArcA, a response regulator responsible for regulation of many genes as cells switch growth from aerobic to anaerobic conditions and back to aerobic conditions, was found to negatively regulate an rpoS transcriptional fusion in exponential phase, and ArcA directly binds to sites upstream and downstream of the transcriptional start site (27), reminiscent of what was seen with CRP. Based on the fusion results (which could possibly reflect other levels of regulation by ArcA), ArcA would help to keep basal levels of RpoS low during exponential phase. Although binding sites were identified for ArcA, mutations in the sites were not tested.
Other regulators, including Fis and the antitoxin MqsA, have also been implicated in transcriptional regulation of rpoS (24, 28). Fis is an abundant nucleoid-associated protein; in a recent study, mutations in fis increased expression of an rpoS transcriptional fusion, and possible sites for Fis binding were identified by sequence comparisons. This fusion contains the full leader, leaving open the possibility of a more indirect effect of Fis (24, 29). The physiological implications of this regulation have not been explored. MqsA, like many antitoxins, is also a transcriptional regulator and was shown to repress expression of RpoS by binding to a site significantly upstream of the rpoS transcription start. Mutations in mqsA led to modest up-regulation of RpoS (2-fold) in stationary phase (28).
Translational regulation
The existence of a long 5′ UTR for RpoS, conserved in enterobacteria, suggests the possibility of regulation at the level of transcription elongation and/or translation (Fig. 2). In fact, studies on translational regulation of RpoS in the mid-1990s provided some of the initial evidence for how small RNAs could regulate gene expression. We now know that regulated translation of RpoS, much of it mediated by small regulatory RNAs (sRNAs), is critical for proper expression of the general stress response.
A short history of Hfq, sRNAs, and regulation of RpoS
As noted above, it was clear from work published in 1994 by Lange and Hengge-Aronis (17) that RpoS was subject to regulation beyond the promoter, as reflected clearly in the up-regulation of translational fusions upon entry to stationary phase. In an independent set of experiments, published the same year, Winkler and co-workers (30) made mutants in the hfq gene of E. coli, embedded in a complex operon the Winkler lab was also investigating. Hfq had been previously described for its role in the replication of the RNA phage Qβ, was clearly an RNA-binding protein, and had been found associated with ribosomes (31), but biological roles in the cell had not previously been described. The reported phenotypes of the hfq mutant (30), increased osmosensitivity, UV sensitivity, and a failure to form the usual rounded cells in stationary phase, were recognized by Hengge and coworkers to resemble cells devoid of RpoS. As predicted from this observation, hfq mutants were in fact found to have lost the increase in expression of rpoS–lacZ translational fusions upon osmotic upshift and had significantly decreased levels of RpoS (32). In parallel studies in Salmonella, Brown and Elliott (33) also found decreased RpoS translation in an hfq mutant and decreased expression of RpoS-dependent genes.
In our lab, in studies meant to help us understand the role and regulation of RcsA, a component of the capsule regulatory cascade, Sledjeski et al. (34) had come across a small noncoding RNA (sRNA), DsrA, and had found that DsrA was necessary for the expression of RpoS at low temperature. Our models for thinking about how this sRNA might work were informed by the elegant studies of RNA-based regulation of plasmid copy number (35), in which pairing between RNAs can change the folding and thus behavior of a target RNA.
This work on regulating with RNAs and the work on Hfq came together in a number of ways. Howard Nash, our colleague at the National Institutes of Health and member of the “lambda lunch” prokaryotic seminar series, had suggested to us, early in our work with DsrA, that Hfq's association with the ribosome and RNA-binding properties might suggest it could be involved in DsrA function. This proved to be the case (36). Brown and Elliott (37) followed up on their observation that there was very little expression of an rpoS–lacZ translational fusion in the absence of Hfq by screening for mutations in the fusion that bypassed or suppressed the defect. Those mutants defined two regions in the rpoS 5′ leader, separated by more than 60 nt, that were capable of pairing with each other to form a hairpin, inhibiting ribosome binding; the Hfq-independent mutants disrupted the hairpin (Fig. 3) (37). The results strongly suggested that Hfq somehow overcame the inhibitory pairing and led us directly to a testable model for DsrA regulation of rpoS; DsrA was able to pair with the inhibitor region, thus opening up the hairpin and allowing ribosome entry (Fig. 3), as confirmed in studies published by us and others in 1998 (38, 39). Gisela Storz, our colleague at the National Institutes of Health, had come across another sRNA, OxyS (40). They found, in collaboration with the Hengge-Aronis lab, that OxyS bound to and depended upon the Hfq protein for function (41). These sets of observations suggested that there might be many previously unrecognized sRNA regulators and provided the impetus for global searches for other sRNAs (see Refs. 42, 43). This work also served as the foundation for our basic understanding of bacterial sRNAs that act by pairing (reviewed in Ref. 44). The regulation of RpoS translation by sRNAs has been the paradigm for many in vitro studies of sRNA function (45, 46) as well as for our general thinking about positive regulation by sRNAs. Here, I focus on the regulatory circuitry associated with sRNA-based stimulation of RpoS translation.
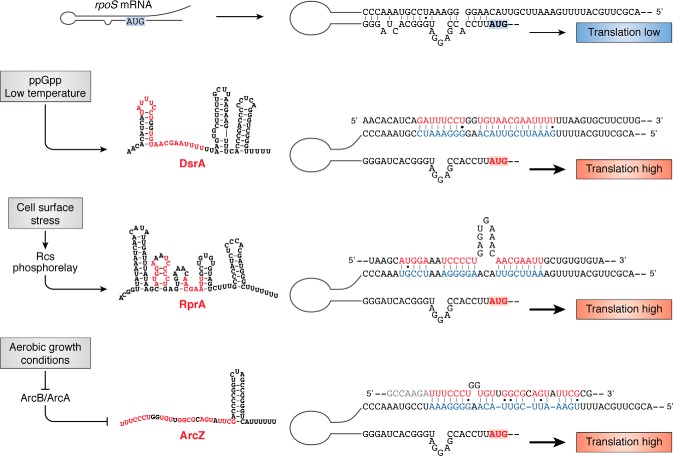
Figure 3.
Translational activation by sRNAs. In the absence of the activating sRNAs or the RNA chaperone Hfq (not shown), translation of RpoS is low, reflecting both occlusion of ribosome entry and Rho-dependent termination within the long leader. Each of the sRNAs shown has been found to pair with the upper region of the hairpin, as shown, to allow translation. The red portion of each sRNA has been predicted to pair with the rpoS 5′ UTR, although in all cases only a subset of the predicted pairs has been tested for function. ArcZ is rapidly processed to a short sRNA; only the processed form is shown here. Each sRNA is regulated at the level of transcription, by the signals and regulators shown to the left, and discussed in the text.
Current status of understanding: multiple sRNAs activate RpoS
We now know of three separate sRNAs, DsrA, ArcZ, and RprA, that activate RpoS translation; each uses Hfq to promote annealing to the rpoS 5′ UTR (Fig. 3). Hfq also protects the sRNAs from degradation prior to pairing (47). Because each sRNA is made in response to a different set of signals, these sRNAs can each provide a different signal transduction pathway for induction of the general stress response. Each of these sRNAs also regulates many other genes, likely leading to a nuanced general stress response, with different sRNAs fine-tuning the set of genes that are on and off.
Recent studies suggest a second level of activation by these sRNAs. In addition to opening up the ribosome-binding site for rpoS translation, pairing of sRNAs with the long 5′ UTR can also block access of the Rho termination protein, thus stimulating RpoS expression by anti-termination (22). For Rho to act to terminate transcription, it must first bind to a region of “naked” single-stranded RNA, not engaged by ribosomes, and then translocate along the RNA to the elongating polymerase, stimulating termination at somewhat ill-defined sites (reviewed in Ref. 48). Long 5′ UTRs can provide such a Rho-binding site. sRNAs, by pairing with the 5′ UTR, may interfere with Rho access and thus block termination (22).
DsrA
The DsrA sRNA is induced at low temperature (34, 49, 50) and recently was shown to be positively regulated by the ppGpp stress alarmone (2). ppGpp, a general signal of starvation in many bacteria, accumulates in response to amino acid starvation or in response to starvation for fatty acids, phosphate, and likely other insults, and it changes promoter utilization via interactions with RNA polymerase (51–53). DsrA is reasonably abundant in cells grown under many conditions. Deletion of this sRNA reduces the basal level of RpoS in exponential phase and abolishes the increase in RpoS normally seen at low temperature (34). Other physiologically important regulators of dsrA transcription may well exist. LeuO, a transcriptional regulator, represses dsrA expression, but because this was observed only when LeuO was overproduced, the physiological significance is not yet clear (49, 54).
In addition to activation of RpoS translation, DsrA also represses the synthesis of the transcriptional repressor H-NS (39, 55). H-NS is known as a general silencer of transcription of many genes in E. coli and related bacteria, including many horizontally acquired genes (reviewed in Ref. 56). Most of the studies of DsrA repression of H-NS involve overexpression of DsrA, leaving the critical physiological role unclear and worth further investigation, particularly under the conditions (low temperature, for instance) where DsrA is most abundant. Among the genes repressed by H-NS are those encoding two different proteins, called anti-adaptors, that under stress conditions contribute to stabilization of RpoS (57) (Fig. 2, discussed further below). Thus, induction of DsrA will contribute to inducing RpoS at two levels, by increasing RpoS translation and, via inhibition of H-NS expression, by induction of anti-adaptors that increase RpoS stability. The possible contributions of other DsrA targets in terms of the RpoS response remain to be investigated.
Overall, one might suggest that DsrA effectively induces RpoS directly and indirectly, possibly favoring expression of the subset of RpoS-dependent genes that are also H-NS repressed, expressing them both via RpoS and via relief of H-NS.
ArcZ
ArcZ sRNA is negatively regulated by the two-component ArcB/ArcA system, active under anaerobic conditions (58). ArcB is a membrane-localized histidine kinase; when it is active, it phosphorylates the ArcA response regulator. Phosphorylated ArcA binds DNA, activating or repressing a large regulon, and this serves as a critical transcriptional switch from aerobic to anaerobic growth conditions (59). A 55-nt processed form of the 120-nt primary ArcZ transcript activates RpoS translation in vivo (Fig. 3) and binds to the rpoS mRNA in vitro, dependent on Hfq (46, 58). ArcZ is fairly abundant under many growth conditions and contributes significantly to the levels of RpoS grown either in rich or in minimal media (58). It is striking that a selection for mutants that decrease stationary phase mutagenesis, dependent upon RpoS, identified multiple members of the electron transfer pathways upstream of ArcB and lower levels of expression of RpoS-dependent genes (60). The Arc regulon thus may have multiple effects in signaling to RpoS, with ArcZ contributing significantly under many growth conditions.
ArcZ has a large number of other targets (61, 62); only a few have been studied in any detail, so it is unknown whether they too have roles in the RpoS-dependent general stress response. A recent approach that captures in vivo sRNA–mRNA pairs on Hfq (RIL-Seq) identified hundreds of likely ArcZ targets, particularly during stationary phase (62). Given the large number of partners suggested by this global approach, it is not surprising that multiple roles for ArcZ have been identified. Overexpression of ArcZ in Salmonella led to changes in >16% of mRNAs, and cells were nonmotile (61). ArcZ was necessary, via an unknown mechanism, for expression of curli regulator CsgD, needed for development of biofilm (63). In E. coli, cells overexpressing ArcZ are also nonmotile, and deletion of ArcZ increased motility of otherwise WT cells. These effects are in part explained by ArcZ's ability to directly repress expression of the FlhDC master regulator for the flagellar genes needed for motility (64). Deletion of arcZ reduced biofilm formation dramatically in E. coli cells in which biofilm depended upon CsgD (7, 65), consistent with the observations in Salmonella. These effects of ArcZ, decreasing motility and promoting biofilm, should reinforce one aspect of the RpoS response. ArcZ also represses expression of mutS, a component of the mismatch repair pathway; in the absence of ArcZ, mutagenesis in stationary phase and during long-term starvation is decreased due to higher MutS levels and therefore more effective monitoring of mismatches (66). Increased mutagenesis in stationary phase has been interpreted as a type of bet-hedging, allowing starving/nutrient-poor bacteria to explore alternative pathways. Further work will be necessary to understand whether the large number of ArcZ targets are generally tied to the general stress response or have broader roles.
RprA
RprA is the third sRNA activator of rpoS expression (67). It pairs to rpoS in a similar region of the rpoS 5′ UTR hairpin and was identified by a screen for genes which, upon overexpression, induce RpoS (Fig. 3) (67). Synthesis of rprA is fully dependent on activation of the Rcs phosphorelay (68). This complex phosphorelay, first defined because it is necessary for regulation of capsular polysaccharide synthesis, senses a variety of disruptions to the cell surface, including treatment with antimicrobial peptides such as polymyxin and disruption of peptidoglycan biosynthesis by β-lactam antibiotics. Activating signals lead to phosphate transfer from a hybrid histidine kinase, RcsC, through a phosphorelay protein, RcsD, to the response regulator RcsB (reviewed in Ref. 69). Activated RcsB positively regulates the rprA promoter, promoters for capsule synthesis, as well as many others, and it negatively regulates motility.
Because the basal level expression of RprA is low in the absence of an inducing stress, it probably does not contribute significantly to the basal translation of RpoS, even on the entry to stationary phase. Sublethal treatment of cells with β-lactams, such as ampicillin, has been found to induce RpoS, likely by increasing Hfq-dependent translation (70). Given that low doses of β-lactam antibiotics are also known to induce the Rcs regulon (71), it is quite possible that RprA plays a significant role in the induction of RpoS by these antibiotics. Thus, RprA may only contribute to inducing RpoS under particular but important cell envelope stress conditions, when the levels of this sRNA will increase.
Interestingly, RprA also directly represses CsgD, the curli regulator, and this repression can be seen even at moderate levels of RprA (72). Thus, the type of signals that lead to RpoS synthesis may determine the timing of CsgD expression or whether CsgD is induced at all. ArcZ activates CsgD expression and RprA represses it. Induction of the Rcs phosphorelay will have a range of other effects in addition to induction of the RprA sRNA, including repressing motility and inducing synthesis of capsular polysaccharide, thus likely also affecting when/if the CsgD-dependent biofilm pathway is used (69).
Effects of single, double, and triple mutants for the three sRNAs were examined for expression of an rpoS–lacZ translational fusion when cells are grown in rich medium at 37 °C to stationary phase; deletion of all three sRNAs reduced expression to 28% of the WT level. Deletion of single sRNAs suggested the most significant contribution was by DsrA, followed by ArcZ and very little contribution of RprA (58). In the same strains in minimal media, ArcZ had a bigger effect than DsrA (58).
Another sRNA, OxyS, was implicated in negative regulation of RpoS (40). However, there is no evidence OxyS directly interacts with the rpoS mRNA, but rather it may, when overproduced, titrate Hfq, leading to lower levels of RpoS (73).
Translation effects on the general stress response regulator beyond sRNAs
Although it is clear that sRNAs are necessary for translation, there is also evidence for increased translation initiation of RpoS in stationary phase independent of sRNAs. For the most part, how these other levels of translation regulation occur remains to be fully understood.
Hirsch and Elliott (74) found that much of the stationary phase induction of RpoS in E. coli depended on a region just upstream of the AUG start, independent of the region necessary for sRNA translational induction. In our lab, mutations in subunits of pyruvate dehydrogenase, which slow growth and thus possibly are sensed as entry into starvation, were found to cause induction of RpoS, with much of the induction occurring at the level of RpoS translation, independent of sRNAs (75); the mechanism of this regulation is still under investigation.
The translational regulation pathways above have focused on effects of environmental signals or mutants on translation initiation. However, there is growing evidence that signals may also perturb the efficiency and/or accuracy of translation of the RpoS ORF, beyond initiation. In general, significant differences have been found between translation of RpoS and translation of the vegetative sigma factor RpoD, suggesting the existence of specific mechanisms that may affect the general stress response. Mutations in the quality control ssrA gene reduce RpoS translation, independent of sRNAs (76). SsrA encodes tmRNA, an RNA that mimics a tRNA in that it can use an empty ribosomal A site, created either by translation that gets to the end of the translated mRNA without a stop signal or when the ribosome encounters problems (rare codons and starvation for an amino acid, for instance); tmRNA enters the A site and adds a short C-terminal 11-amino acid tail to the translating protein. The protein sequence is sufficient to target the tagged protein for degradation (reviewed in Ref. 77). The loss of full-length RpoS protein in the ssrA mutant suggests that normally there are translational pause sites within RpoS that engage tmRNA and that releasing the stalled ribosome with tmRNA is important for subsequent ribosomes to proceed to the end of the protein. To what extent these pauses and the need for tmRNA varies with growth conditions is not known.
A second mutant shown to reduce translation within the rpoS ORF is miaA, encoding a tRNA-modifying enzyme specific to UXX codons, including four out of six codons for leucine. Among leucine codons, rpoS has a significantly enriched use of these MiaA-dependent codons than rpoD, suggesting an evolutionary selection for this increased MiaA dependence (78). Consistent with an important role of these leucine codons, recoding them from UUN to CUN made expression of an rpoS–lacZ fusion relatively MiaA-independent (79). Other genes affecting tRNA modifications also reduce RpoS levels (80)
If the rpoS ORF (and other stress proteins) are enriched for MiaA-dependent codons, are there conditions where translation of this protein would be restricted, due to limiting modifications? Although that has not yet been directly examined, at least one report highlights the advantages and disadvantages of RpoS, depending on growth conditions and its sensitivity to translation conditions (81). A mutation in the ribosomal protein S12 that improves translation accuracy (and thus increases stalling within the translating ORF) had reduced ability to increase RpoS levels when grown in poor-carbon sources. This led to more rapid growth of cells in the short term, but lack of induction of stress resistance genes dependent upon RpoS. Cells with decreased translational accuracy, from a mutation in rpsD, encoding ribosomal protein S4 (I199N), made more RpoS, rendering cells resistant to hydrogen peroxide as a result of making more RpoS-dependent genes katE or osmC (82). This increase in RpoS may be dependent on an observed increase in DsrA synthesis and thus translation initiation, but part of the effect may also reflect more readthrough of pauses in rpoS.
Finally, a recent study highlighted the unexpected finding that variations in sequences within the multiple rRNA genes in E. coli affect expression of RpoS (83). One particular rRNA copy, encoded by rrsH, shows the most increase in RpoS upon switching cells from modestly rich medium to minimal growth medium. Overexpression of a single rRNA operon, with or without the sequence variations found in rrsH, shows that RpoS-dependent genes and RpoS protein itself are up-regulated only when the rrsH variant is expressed, suggesting the possibility that a particular subset of ribosomes, containing the rrsH-encoded rRNA, are up-regulated during starvation and help to improve RpoS translation (or stability). It is not yet known what drives the preference for rrsH during starvation, whether this effect is important in other stress responses, is linked to effects of tmRNA or RNA modifications, or is another distinct level of regulation.
Regulated proteolysis of RpoS
A striking feature of RpoS is the robust regulation of its stability. A starting point was the recognition in the early 1990s that RpoS is unstable under exponential growth conditions, and it becomes more stable upon entry to stationary phase and/or upon certain stress conditions (17, 26). The initial components of the degradation machinery were identified soon after, but understanding how degradation is regulated took a decade more and still remains a topic of active investigation.
RpoS degradation depends upon the ATP-dependent ClpXP protease (84) and the adaptor protein RssB, also called SprE, a member of the response regulator family (Fig. 2) (85, 86). In bacteria, a small number of energy-dependent proteases are responsible for protein degradation within the cytoplasm; these proteases, such as ClpXP, include a proteolytic core (ClpP, for instance) gated by ATPase subunits (ClpX), organized in a complex comparable to the eukaryotic proteasome (87). Because E. coli and most other bacteria do not have tagging systems such as ubiquitin, selectivity for proteolysis depends upon degradation tags within proteins and/or the participation of adaptors that can deliver specific substrates to the protease (reviewed in Ref. 88).
RssB, like other response regulators, has a conserved aspartate that can be phosphorylated. The small molecule acetyl phosphate can carry out this phosphorylation in vitro. Rapid degradation of RpoS in a purified system requires ClpXP, ATP, RssB, and a source of RssB phosphorylation (89). In vivo, it was assumed for many years that changes in RssB phosphorylation might explain the shift from rapid degradation to slower degradation. However, the source of phosphorylation remained unclear, because there was clearly no cognate dedicated histidine kinase for RssB. A critical experiment by Silhavy and co-workers (90), published in 2004, demonstrated that RssB phosphorylation could not be the primary signal for regulation of RpoS stability. Cells in which the chromosomal copy of rssB was mutated at the site of phosphorylation, D58A, had a somewhat slower but still significant degradation of RpoS, and upon carbon or phosphate starvation, RpoS became stable (90). Therefore, regulation of degradation in response to starvation had to operate independently of phosphorylation.
Anti-adaptors mediate regulated proteolysis
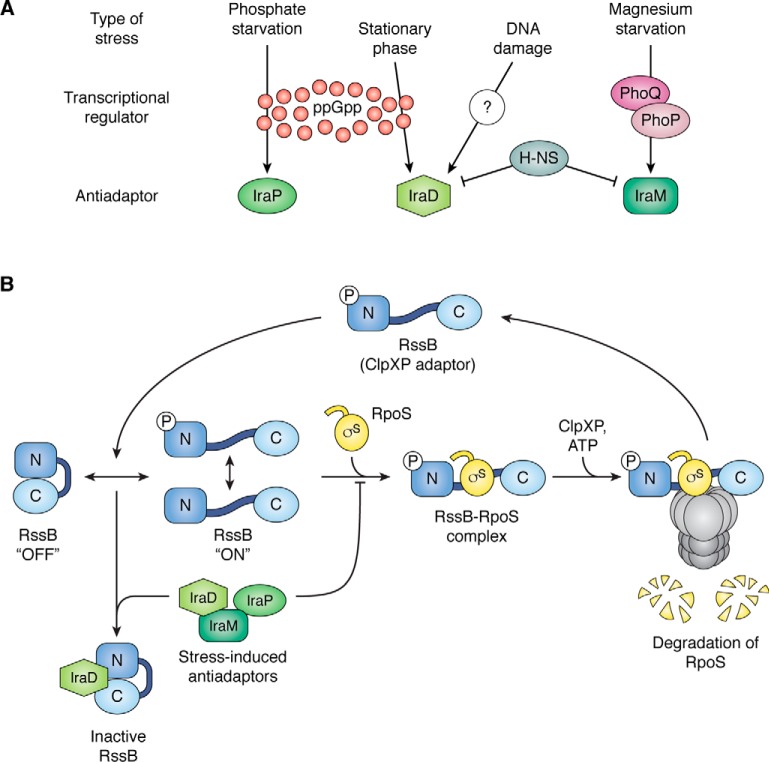
The next major step in understanding this process came from studies of Alex Bougdour in our lab, first published in 2006 (91). A screen of a multicopy library of E. coli genome fragments with an RpoS–lacZ translational fusion identified a previously uncharacterized 86-amino acid protein, IraP, capable of stabilizing RpoS in vivo when overproduced and in vitro in a purified system (91). IraP, named as an Inhibitor of RssB activity under Phosphate starvation, interacts with RssB and blocks the interaction of RpoS; IraP is not itself degraded (Fig. 4). This class of protease regulators, perturbing the function of the adaptors that deliver specific substrates to proteases, at least in some cases without themselves being degraded, were termed anti-adaptors (Fig. 4). Synthesis of IraP is, as implied by its name, high under phosphate starvation and is primarily responsible for the observed stabilization of RpoS when cells are deprived of phosphate. However, in E. coli, IraP does not have a role when cells are starved of glucose or magnesium (92, 93). Two other anti-adaptors have since been characterized, IraM (stabilization during Mg starvation) and IraD (stabilization during DNA damage) (Fig. 4). Although each of these proteins can stabilize RpoS when expressed, they interact in distinct ways with RssB and do not share similarity in sequence (94). Recently, a structure of IraD in complex with RssB has been determined and shows IraD binding to and presumably stabilizing a “closed” collapsed form of RssB, with both domains of RssB bound by IraD (Fig. 4) (95). Further understanding of how other anti-adaptors inhibit RssB and how RssB delivers RpoS to the protease await future structures.
Figure 4.
Anti-adaptor regulation of RpoS degradation. A, three differently characterized anti-adaptors are shown in green, with their known upstream transcriptional regulators. The regulator mediating the response to DNA damage is not known. B, pathway for adaptor-mediated degradation of RpoS and anti-adaptor regulation of this process is shown. RpoS (σS) is shown in yellow. Adaptor RssB is shown in blue, both in phosphorylated and unphosphorylated form. The ClpXP protease is shown in gray; the details of binding of RpoS and RssB to ClpXP are not fully understood.
Examination of the iraP promoter showed that the induction under phosphate starvation depended on ppGpp, an alarmone that increases under phosphate starvation; the iraP promoter is directly positively regulated by ppGpp (2, 93). IraM is positively regulated by the two-component PhoQ histidine kinase and PhoP response regulator system in E. coli, activated in response to limiting Mg2+ or antimicrobial peptides (92).
IraD was independently identified as a protein necessary for resistance to DNA damage; based on its identification as an anti-adaptor in our multicopy screen, the resistance to DNA damage was shown to be dependent upon RpoS, by blocking RpoS degradation (96). It was also found to be induced after treatment with hydrogen peroxide (97), although the induction pathway is still undefined. Like IraP, IraD is also subject to positive regulation by ppGpp, possibly indirectly (Fig. 4) (98). IraD translation is also regulated. IraD translation is coupled to translation of a short upstream ORF, which is in turn repressed by CsrA, a global translational regulatory protein (99). CsrA repression is relieved when sRNAs that can bind to it are produced. Therefore, any signals that lead to the expression of the titrating RNAs (primarily CsrB and CsrC) (reviewed in Ref. 100) will contribute to IraD-dependent RpoS stabilization, at least under the conditions when iraD is being transcribed. The extent of this effect on RpoS has not yet been examined and presumably would be important only under conditions in which iraD is being transcribed. Translational regulation of other anti-adaptors has not been investigated.
Carbon starvation was one of the first known stresses for induction, leading to resistance to hydrogen peroxide and heat shock, dependent upon RpoS, and it was this treatment that was first used to define the general stress response (3). RpoS is clearly stabilized after carbon starvation (17), but none of the known anti-adaptors are needed for this stabilization (92). Instead, it has been suggested that the lower levels of ATP after starvation lead to differential stabilization of RpoS (101). ClpXP uses ATP for degradation, via ATP-dependent unfolding and threading of proteins through the ClpX ATPase; the model presented here is that the RssB-dependent unfolding of RpoS is a relatively unfavored/difficult substrate that requires more ATP than some other targets.
Does phosphorylation of RssB play a role in the general stress response?
Given that mutations in the phosphorylation site of RssB slow degradation only modestly and do not block regulation by the anti-adaptors or stabilization of RpoS under most tested starvation conditions (90, 102), it is unclear whether there are in fact conditions under which phosphorylation plays a critical role in induction of or recovery from the general stress response. The situation is further complicated by the lack of understanding of the source of the phosphate in vivo. Two in vivo sources of RssB phosphorylation have been reported: the small molecule acetyl phosphate and phosphorylation by the ArcB histidine kinase (27, 103). Acetyl phosphate is synthesized from acetyl-CoA by Pta and degraded by AckA to acetate, and deletion of these two genes, which should render cells devoid of AcP, does show increased levels of RpoS (103). However, we found that either a pta deletion (no AcP) or an ackA deletion (high AcP) leads to a similar increase in RpoS to that seen in the double deletion (104). Therefore, this increase in RpoS is likely not due to AcP phosphorylation of RssB. Mutations in arcB, encoding the histidine kinase for the ArcB/ArcA phosphorelay, slow RpoS degradation in vivo, dependent upon the phosphorylation site in rssB, and ArcB was able to phosphorylate RssB in vitro (27). Levels of RpoS were higher in arcA and arcB mutants (27). This is likely due in part to increased synthesis of the sRNA ArcZ and therefore increased RpoS synthesis levels (58), although there is certainly evidence that ArcA and ArcB act at multiple levels to down-regulate RpoS (27). Possibly phosphorylation is via a variety of sources (cross-phosphorylation by multiple histidine kinases?), or it is only critical under a yet-to-be-defined stress condition.
Regulated activity of RpoS
In addition to all of these ways to modulate RpoS accumulation, once made, RpoS may not necessarily be active. It must act in concert with core RNA polymerase, and thus must compete with other sigma factors, including RpoD, the vegetative sigma factor, for RNA polymerase. Its intrinsic binding to core polymerase is significantly less than that for RpoD (105, 106), and levels of RpoS in stationary phase are still well below those for RpoD (107, 108). Several factors help to promote RpoS binding. For instance, Crl protein directly binds RpoS and increases its ability to compete for core (109, 110). When bound to core RNA polymerase, RpoS is both active for initiating transcription and resistant to RssB delivery for proteolysis (89). Thus, proteins that affect core association should also affect RpoS stability in vivo.
In the competition between RpoS and RpoD for core RNA polymerase, RpoS access to core polymerase is aided by 6S RNA and Rsd sequestration of RpoD. Rsd is the most straightforward, binding free RpoD sigma factor (111). Levels of Rsd increase modestly in stationary phase, under the control of a variety of transcription factors, some of which are known to act at other levels of induction of the general stress response (112). 6S RNA, however, binds RpoD/RNA polymerase holoenzyme, mimicking promoter elements and trapping the complex until the cell exits starvation conditions, contributing to survival in stationary phase by keeping RpoD from engaging in further rounds of competition for core or possibly by specifically blocking expression of some RpoD-dependent genes (reviewed in Ref. 113).
The activity of many sigma factors, including those for general stress responses in alphaproteobacteria and many Gram-positive bacteria (see below), is primarily regulated by the availability of anti-sigma factors. In E. coli, however, the cell takes the more drastic step of degrading the RpoS sigma factor. That said, RssB, in the absence of the degradation machinery, can act as an anti-sigma (114, 115), which is not surprising given its ability to bind RpoS (Fig. 5). It seems quite possible, based on this behavior as well as on observations with related proteins in other organisms (see Fig. 5 and below), that the RssB adaptor function has evolved from a more standard anti-sigma protein.
Figure 5.
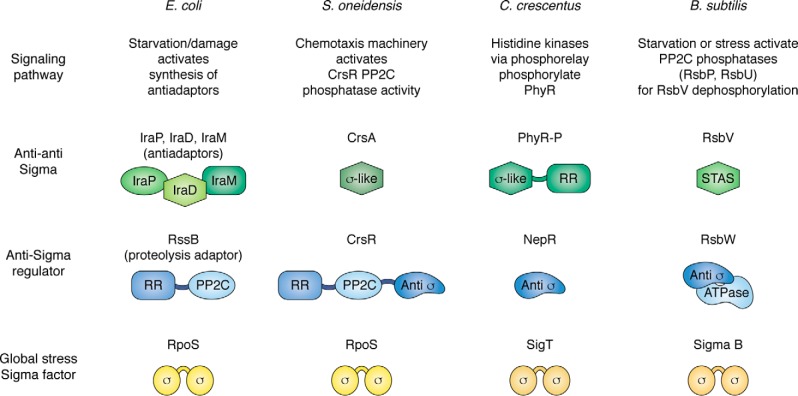
Anti-sigmas and anti-anti–sigmas in general stress responses. On this chart, sigma factors mediating general stress responses are shown in shades of yellow and brown. Alphaproteobacteria use an ECF family sigma factor, named SigT in C. crescentus, shown here, and are referred to more generally as σEcfG in other alphaproteobacteria, to mediate their general stress response. RpoS (E. coli and S. oneidensis) and sigma B (in B. subtilis and some other Gram-positive bacteria) are related to the vegetative sigma factors. Blue symbols show general organization of the proteins that operate as “anti-sigmas,” although RssB, as noted, targets RpoS for proteolysis rather than simply binding it, as for other anti-sigmas. RR domains are response regulators. The PP2C domain in RssB is inactive as a phosphatase (pale blue oval). Some of the anti-anti–sigmas have sigma-like domains, providing mimics for binding to the anti-sigmas. STAS stands for sulfate transporter and anti-sigma factor antagonist domain; CrsA is also a STAS protein. PP2C domains (ovals in anti-sigmas) are also found in the upstream signaling pathways of many of these organisms (for more detail see Ref. 117). Not shown here is the signaling pathway upstream of PhyR–P, in which multiple histidine kinases converge on response regulator MrrA, which in turns transfers phosphate via PhyK to PhyR (131).
Specialization within the general stress response
Although there is ample evidence that inducing the RpoS-dependent stress response by different stresses leads to cross-resistance to noninducing stresses, one can imagine that this apparently “general” stress response may in fact be further specialized under some growth conditions. For instance, when different levels of RpoS are expressed, not all RpoS-dependent promoters react similarly. Some promoters are highly sensitive to even low levels of RpoS, whereas others are relatively insensitive, possibly reflecting promoters important at different stages of the transition into stationary phase (11). In complex bacterial communities, it is clear that there are geographical differences in where genes are expressed, and secondary signals contribute to complex downstream behaviors like biofilm formation (65, 116).
The sRNAs that are responsible for induction of RpoS, as discussed above, exemplify how the cell may end up specializing the general stress response. Although each of these sRNAs has in common the rpoS target, they each have other targets as well, and thus a general stress response induced primarily by expression of DsrA, with concomitant repression of H-NS, may have somewhat different characteristics than one induced by expression of RprA. To what extent these additional targets contribute to moderating the nature of the general stress response has not really been studied.
Do anti-adaptors also participate in “specialization” of the general stress response the way that sRNAs would be expected to? Thus far, no targets other than RssB are known for these proteins, suggesting each will act to stabilize RpoS, without any additional effects on gene expression. However, we might predict some specialization dependent on different inducing signals. Thus, one can imagine that other ppGpp-dependent genes, affected under the same conditions that lead to IraP induction, might collaborate or fine-tune the general stress response. Similarly, other targets of PhoQ/PhoP, the two-component system that activates transcription of iraM in response to Mg2+ starvation, could also affect the general stress response.
One conclusion from considering specialization within the general stress response is that extrapolations from a full understanding of how and when RpoS is induced (not yet available) and a complete list of RpoS-dependent genes will still leave us with much to unravel about the physiological consequences of this response in different settings. For instance, sophisticated and carefully constructed system-wide analysis of growth and gene expression comparing specific conditions of RpoS induction might start to identify the core (always present) response, and how/when it can be modified.
General stress responses in other bacteria: Many variations on common themes
E. coli and its close relatives generally share an RpoS-based general stress response, induced by the machinery and sensing pathways outlined above, with interesting variations that highlight the flexibility of this complex set of inputs. More distantly related bacteria still have general stress responses, but the sigma factor at the core of these responses does not necessarily closely resemble RpoS. Nonetheless, sigma factor activity in all of these bacteria is frequently under complex control, allowing the integration of multiple signals for a common response. What is known about how these stress sigma factors are regulated is reviewed briefly in this section (Fig. 5); other recent reviews of this process in a variety of bacteria include Refs. 117–119. In general, more is known about the mechanisms for regulating activity of these general stress response sigma factors via partner switch mechanisms, in which the sigma factor is sequestered when not needed, than about either regulated degradation or regulated translation. Possibly these are not used in other organisms, but it seems likely that they are present but have yet to be reported.
The sRNAs responsible for activation of RpoS in E. coli are present and functional in close relatives (Salmonella, Klebsiella, and other enterobacteria), although their roles have not been studied much beyond Salmonella. In general, less is currently known about whether there are important regulatory RNAs involved in the induction of general stress responses beyond the enterobacteria.
Even within enterobacteria, modifications in the network of RpoS regulators provide evidence for how systems are evolving and adapting to different bacterial lifestyles. IraL, an anti-adaptor related to IraM, is found in uropathogenic E. coli. However, unlike the situation in E. coli K12, it is expressed during exponential phase, reflecting a need for the RpoS regulon under the conditions this organism encounters in the urinary tract (120). In Salmonella, the iraP anti-adaptor has acquired a second promoter, activated by PhoQ/PhoP, and thus IraP is critical for PhoP-dependent stabilization of RpoS in Salmonella (121). An IraM-like protein exists in Salmonella (originally named RssC), but its regulation has not yet been characterized (92). A compendium of Salmonella expression experiments suggests it is poorly expressed, but may still have a weak response to Mg2+ starvation (condition for activation of PhoPQ) and NaCl osmotic shock (122).
Pseudomonas aeruginosa
As one moves away from enterobacterial species, more variations appear. P. aeruginosa encodes an RpoS-like sigma factor, transcribed from a promoter well-upstream from the translation start site, as in E. coli; expression of the mRNA increases when cells enter stationary phase (123), and transcriptional regulation may play a more significant role than in E. coli (124). Cells becomes resistant to a variety of stresses in stationary phase. However, although cells mutant in the rpoS homolog have increased sensitivity to stress, much of the stationary phase resistance is retained, suggesting the existence of additional mechanisms (125). In tests of transcriptional and translational fusions, increases in RpoS in stationary phase appeared to be primarily attributable to increased transcription, but mutations in clpX increased RpoS levels in exponential phase, similar to what is seen in E. coli (124). There are no close homologs of the anti-adaptor proteins in P. aeruginosa. The structure of an RssB homolog has been described (PDB code 3EQ2). This protein differs from RssB in having all the sites for an active phosphatase in the C-terminal domain (126) and a linker that appears to be significantly more extended than that found in E. coli RssB (95). Intriguingly, this protein, PA2798 in P. aeruginosa PA01, is found next to and likely translationally coupled with a protein annotated as an anti-anti–sigma, PA2797. Although it is not known whether PA2798 acts on RpoS, if it does, the active phosphatase domain and anti-anti–sigma suggest that Pseudomonas may regulate this response not by proteolysis but by upstream kinase activity and partner switching, akin to that found in Shewanella and Gram-positive bacteria (see below). In Acinetobacter baumanii, homologous proteins to PA2797 (GigA) and 2798 (GigB) act in a signaling pathway for resistance to a variety of environmental stresses, antibiotic resistance, and pathogenesis and have been suggested to be mediators of the global stress response in this organism (127). No activating sRNAs have yet been described for P. aeruginosa rpoS, although an inhibitory sRNA has been found (128). Thus, if Pseudomonas RpoS is mediating a general stress response, it seems likely that many of the signaling pathways that induce or inhibit its synthesis or activity remain to be characterized.
Shewanella oneidensis
In S. oneidensis, a Gram-negative aquatic bacteria, the RpoS protein is regulated at the activity level by anti-sigma factor CrsR and anti-anti–sigma CrsA (129), in response to signals apparently mediated via the components of the chemotaxis pathway (Fig. 5). The N-terminal domain of CrsR is a response regulator, followed by an active PP2C phosphatase domain; the protein also includes a third “anti-sigma” domain, akin to those identified in regulators of the general stress response in Gram-positive species. The model developed from in vitro and in vivo tests is that, in the absence of stress, the anti-sigma domain of CrsR binds and sequesters RpoS. Stress leads to activation of the phosphatase domain and dephosphorylation of CrsA; CrsA can then bind the anti-sigma domain and thus lead to release of RpoS (Fig. 5) (129). The anti-sigma factor may also play a protective role, preparing cells for rapid RpoS activation. Although RpoS-dependent transcription increases significantly on entry to stationary phase, there is detectable RpoS protein in exponential phase cells. However, in the absence of the anti-sigma protein, CrsR, RpoS activity could not be detected at all in exponential phase, and increased expression of the reporter in stationary phase was delayed, suggesting a positive role for CsrR. Consistent with this, RpoS appeared to be unstable in crude extracts in the absence but not the presence of CsrR (130). Although the degradation pathway has not been determined, these results provide a model in which an anti-sigma factor plays two roles: (i) keeping RpoS inactive but (ii) also keeping it available for rapid activation when needed. The components of this system are found in a variety of other aquatic bacteria. Not yet addressed is how the phosphatase activity of CsrR is regulated by stress, whether yet other levels of regulation of RpoS exist, and the full role of RpoS in Shewanella.
Gram-negative alphaproteobacteria
The sigma factor at the center of the alphabacterial general stress response is an extracytoplasmic function (ECF) sigma factor, σEcfG, named SigT in Caulobacter crescentus. As for most ECF sigma factors, it is regulated by an anti-sigma factor (NepR), and a second anti-anti–sigma factor (PhyR), by partner switching, and all three of these proteins are genetically linked in many alphaproteobacterial species (reviewed in Ref. 118). NepR binds to and sequesters σEcfG. PhyR contains an N-terminal response regulator domain, susceptible to phosphorylation by histidine kinases in response to appropriate signals, and C-terminal domains that mimic the ECF sigma factor regions. When PhyR is phosphorylated, the sigma-like domains are freed to bind NepR, thus releasing σEcfG and activating the general stress response. Recent studies in C. crescentus investigating phosphorylation and dephosphorylation of PhyR provide a clear example of the integration of multiple signals to turn on a general stress response (131). Multiple histidine kinases, from different families and presumably each responding to a different signal, all converge on phosphorylation of a single-domain response regulator, MrrA. MrrA–P then transfers phosphate to phosphotransfer proteins, including one, PhyK, that then phosphorylates PhyR. Another phosphotransfer protein integrates other aspects of the Caulobacter lifestyle and may serve as a feedback control for SigT.
Gram-positive bacteria: Bacillus subtilis and others
In B. subtilis and many other Gram-positive bacteria, SigB is the sigma factor that mediates the general stress/stationary phase response. There are sufficient studies in B. subtilis to make it clear that, as in E. coli, there are a broad range of inducing signals and robust mechanisms to keep the basal level of SigB activity low when it is not needed (reviewed in Ref. 119). Two major pathways of SigB regulation, in response to different stresses, have been described, both converging, as in the cases discussed above, on the activities of anti-sigmas and anti-anti–sigmas regulated by phosphorylation cascades. The major anti-sigma factor is RsbW, a protein kinase; the alternative partner for the anti-sigma is anti-anti–sigma RsbV, a substrate for RsbW phosphorylation (Fig. 5). In growing cells, RsbV is phosphorylated and inactive, and RsbW sequesters SigB. PP2C family phosphatases RsbU and RsbP, active in response to different signals of stress, dephosphorylate RsbV, allowing it to bind RsbW and thus free SigB.
The pathways to activation of these phosphatases are complex and depend on the nature of the stress, with RsbP mediating energy stress, dependent on a hydrolase, RsbQ. Current models suggest that a small molecule sensed and likely cleaved by RsbQ leads to activation of RsbP, but that molecule has not yet been identified (132). Environmental stress signals (changes in pH, salt, heat, etc.) signal through the “Stressosome,” a large protein complex including the switch protein, kinase RsbT. When RsbT is retained in the stressosome, RsbU is inactive; only when RsbT is released from the stressosome is RsbU activated as a phosphatase, leading to SigB activation as for RsbP. RsbT is released after phosphorylating the various inhibitors in the stressosome and then activates RsbU. Different protein members of the stressosome sense different signals, although in most cases the details of these signal cascades are not understood.
Challenges for dissecting the general stress response in other species
As outlined here, it is clear that the general stress response exists broadly in bacterial species, with variations in the details of what growth conditions are sensed as stresses, how stress is sensed, and how it leads to changes in regulation. However, studies of regulation in most bacteria have focused on the anti-sigma factors discussed above. Maybe one important lesson from the past and ongoing studies on E. coli is that, even when a complex regulatory circuit has been unraveled, and we think it is understood, there may well be more. Finding ways to bypass the regulation we know about allows us to explore what is left, and as we find new growth conditions that depend on the general stress response, it may become obvious that there must be other levels of regulation. The sophisticated genetic tools and deep history of E. coli research have been critical for uncovering different layers of regulation; as these tools become available in other species, further levels of regulation may well be found. For instance, is there also important regulation of the general stress response at the level of translation for organisms other than E. coli and its close relatives? It seems likely. The transcript for rpoS in P. aeruginosa, for instance, contains the same type of long 5′ UTR as in E. coli, although translational regulation of P. aeruginosa RpoS has not been reported. The first experiments that led to the discovery of multiple levels of regulation in E. coli can now be done in many bacteria of interest. Is there a change in translation of the stress sigma factor upon entry of cells into stationary phase or starvation, as was observed for RpoS, using appropriate reporter fusions (17, 74)? For bacteria in which the Hfq chaperone is present and important for the function of regulatory RNAs (true in many Gram-negative bacteria), a simple test of whether these bacteria can properly mount a general stress response in the absence of Hfq would be a worthwhile first step to whether it is worth a search for sRNA regulators of RpoS.
General conclusions and issues to consider
The general stress response in E. coli and many other bacteria highlight the likelihood that under natural growth conditions these cells encounter a linked set of stresses/starvation conditions. Thus, inducing a generally resistant phenotype serves the cell well. However, it is also apparent that these general stress responses, mediated by specialized sigma factors, are not favored when nutrients and growth conditions are optimal. Changes in metabolism, the cost to the cell of unneeded products, and likely competition for core RNA polymerase are all negative effects. This combination of clear advantages and disadvantages reinforces the need for robust and rapidly adaptable regulatory mechanisms, both for induction of the stress response and for recovery upon return to rapid growth. Coupled with the need for many different stresses to converge on availability of a given sigma factor, this makes studies on regulation of general stress factors an entertaining playground for those interested in novel regulatory mechanisms.
Why sigma factors as mediators rather than some other type of transcription factor? Certainly there are other regulators that are subject to multiple levels of regulation, suggesting that the need for many regulatory inputs is unlikely the only requirement for sigma factor use. FlhDC and CsgD, hubs for regulation of motility and biofilm, respectively, are good examples; these are reviewed in Refs. 65, 133, 134. It still remains to be determined whether all of these general stress responses are evolutionarily related, possibly reflecting the use of a sigma factor in the original ancestor. It is striking that some types of domains are used in very different ways and in different stress responses. Kinase and phosphatase cascades that, in the end, affect the activity of anti-sigma factors are found in multiple different geometries in different organisms (Fig. 5), possibly suggesting evolutionary connections. More analysis of these systems will be necessary to understand whether this is a convergent or divergent evolution. One major characteristic of using a sigma factor switch is the change from a vegetative (or other developmental) program to the specific set of genes necessary for the general stress response. In this case, shutting off or significantly decreasing use of other sigmas may be as important a characteristic as turning on the genes of the new sigma factor regulon.
Not discussed here in any detail are the following: the nature of the downstream genes of the general stress regulons; how related these are in different organisms; and how and why these responses differ or overlap with the more specific responses to specific stresses. What is the proper condition under which these general stress responses are most important? We are only starting to have a sense of this and clearly there is a lot more to learn.
Acknowledgments
I thank the members of my laboratory and our collaborators in the Storz, Wickner, and Woodson labs, past and present, for all they have contributed over the years to our knowledge of RpoS and how it is regulated. I thank S. Wickner for providing the initial version of Fig. 4B, a modification of a figure used in Ref. 95, and I thank K. Ramamurthi and J. Chen for comments on the manuscript.
This work was supported by the Intramural Research Program of the NCI, Center for Cancer Research, National Institutes of Health. The author declares that she has no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
- CRP
- cAMP receptor protein
- sRNA
- small regulatory RNA
- ppGpp
- guanosine tetraphosphate
- nt
- nucleotide
- AcP
- acetyl phosphate
- ECF
- extracytoplasmic function
- PP2C
- protein phosphatase 2C.
References
- 1. Hamdallah I., Torok N., Bischof K. M., Majdalani N., Chadalavada S., Mdluli N., Creamer K. E., Clark M., Holdener C., Basting P. J., Gottesman S., and Slonczewski J. L. (2018) Experimental evolution of Escherichia coli K-12 at high pH and with RpoS induction. Appl. Environ. Microbiol. 84, e00520–18 10.1128/AEM.00520-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Girard M. E., Gopalkrishnan S., Grace E. D., Halliday J. A., Gourse R. L., and Herman C. (2018) DksA and ppGpp regulate the σS stress response by activating promoters for the small RNA DsrA and the anti-adaptor protein IraP. J. Bacteriol. 200, e00463–17 10.1128/JB.00463-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lange R., and Hengge-Aronis R. (1991) Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5, 49–59 10.1111/j.1365-2958.1991.tb01825.x [DOI] [PubMed] [Google Scholar]
- 4. Li C., and Clarke S. (1992) A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat shock resistance. Proc. Natl. Acad. Sci. U.S.A. 89, 9885–9889 10.1073/pnas.89.20.9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Small P., Blankenhorn D., Welty D., Zinser E., and Slonczewski J. L. (1994) Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176, 1729–1737 10.1128/jb.176.6.1729-1737.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lacour S., and Landini P. (2004) σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195 10.1128/JB.186.21.7186-7195.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parker A., Cureoglu S., De Lay N., Majdalani N., and Gottesman S. (2017) Alternative pathways for Escherichia coli biofilm formation revealed by sRNA overproduction. Mol. Microbiol. 105, 309–325 10.1111/mmi.13702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Almirón M., Link A. J., Furlong D., and Kolter R. (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6, 2646–2654 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- 9. Mulvey M. R., Sorby P. A., Triggs-Raine B. L., and Loewen P. C. (1988) Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene 73, 337–345 10.1016/0378-1119(88)90498-2 [DOI] [PubMed] [Google Scholar]
- 10. Peano C., Wolf J., Demol J., Rossi E., Petiti L., De Bellis G., Geiselmann J., Egli T., Lacour S., and Landini P. (2015) Characterization of the Escherichia coli σS core regulon by chromatin immunoprecipitation-sequencing (ChIP-seq) analysis. Sci. Rep. 5, 10469 10.1038/srep10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong G. T., Bonocora R. P., Schep A. N., Beeler S. M., Lee Fong A. J., Shull L. M., Batachari L. E., Dillon M., Evans C., Becker C. J., Bush E. C., Hardin J., Wade J. T., and Stoebel D. M. (2017) Genome-wide transcriptional response to varying RpoS levels in Escherichia coli K-12. J. Bacteriol. 199, e00755–16 10.1128/JB.00755-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patten C. L., Kirchhof M. G., Schertzberg M. R., Morton R. A., and Schellhorn H. E. (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Gen. Genet. 272, 580–591 10.1007/s00438-004-1089-2 [DOI] [PubMed] [Google Scholar]
- 13. Weber H., Polen T., Heuveling J., Wendisch V. F., and Hengge R. (2005) Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603 10.1128/JB.187.5.1591-1603.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong T., and Schellhorn H. E. (2009) Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol. Genet Genomics 281, 19–33 10.1007/s00438-008-0389-3 [DOI] [PubMed] [Google Scholar]
- 15. Helmann J. D. (2019) Where to begin? sigma factors and selection of transcription initiation in bacteria. Mol. Microbiol. 2019 10.1111/mmi.14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Felkístov A., Sharon B. D., Darst S. A., and Gross C. A. (2014) Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376 10.1146/annurev-micro-092412-155737 [DOI] [PubMed] [Google Scholar]
- 17. Lange R., and Hengge-Aronis R. (1994) The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8, 1600–1612 10.1101/gad.8.13.1600 [DOI] [PubMed] [Google Scholar]
- 18. Barth M., Marschall C., Muffler A., Fischer D., and Hengge-Aronis R. (1995) Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 177, 3455–3464 10.1128/jb.177.12.3455-3464.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lange R., and Hengge-Aronis R. (1994) The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13, 733–743 10.1111/j.1365-2958.1994.tb00466.x [DOI] [PubMed] [Google Scholar]
- 20. Takayanagi Y., Tanaka K., and Takahashi H. (1994) Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol. Gen. Genet. 243, 525–531 10.1007/BF00284200 [DOI] [PubMed] [Google Scholar]
- 21. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sedlyarova N., Shamovsky I., Bharati B. K., Epshtein V., Chen J., Gottesman S., Schroeder R., and Nudler E. (2016) sRNA-mediated control of transcription termination in E. coli. Cell 167, 111–121.e13 10.1016/j.cell.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo M., Wang H., Xie N., and Xie Z. (2015) Positive effect of carbon sources on natural transformation in Escherichia coli: role of low-level cyclic AMP (cAMP)-cAMP Receptor protein in the derepression of rpoS. J. Bacteriol. 197, 3317–3328 10.1128/JB.00291-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amores G. R., de Las Heras A., Sanches-Medeiros A., Elfick A., and Silva-Rocha R. (2017) Systematic identification of novel regulatory interactions controlling biofilm formation in the bacterium Escherichia coli. Sci. Rep. 7, 16768 10.1038/s41598-017-17114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keseler I. M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martinez C., Caspi R., Fulcher C., Gama-Castro S., Kothari A., Krummenacker M., Latendresse M., Muniz-Rascado L., Ong Q., Paley S., Peralta-Gil M., et al. (2017) EcoCyc: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 41, D543–D550 10.1093/nar/gkw1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCann M. P., Fraley C. D., and Matin A. (1993) The putative sigma factor KatF is regulated posttranscriptionally during carbon starvation. J. Bacteriol. 175, 2143–2149 10.1128/jb.175.7.2143-2149.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mika F., and Hengge R. (2005) A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev. 19, 2770–2781 10.1101/gad.353705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X., Kim Y., Hong S. H., Ma Q., Brown B. L., Pu M., Tarone A. M., Benedik M. J., Peti W., Page R., and Wood T. K. (2011) Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366 10.1038/nchembio.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amores G. (2017) Unveiling the Effects of Global Regulators in the Regulatory Network for Biofilm Formation in Escherichia coli. Ph.D. thesis, Universidade de Sao Paulo, Sao Paulo, Brazil [Google Scholar]
- 30. Tsui H. C., Leung H. C., and Winkler M. E. (1994) Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13, 35–49 10.1111/j.1365-2958.1994.tb00400.x [DOI] [PubMed] [Google Scholar]
- 31. Kajitani M., Kato A., Wada A., Inokuchi Y., and Ishihama A. (1994) Regulation of the Escherichia coli hfq gene encoding the host factor for phage Qβ. J. Bacteriol. 176, 531–534 10.1128/jb.176.2.531-534.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muffler A., Fischer D., and Hengge-Aronis R. (1996) The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10, 1143–1151 10.1101/gad.10.9.1143 [DOI] [PubMed] [Google Scholar]
- 33. Brown L., and Elliott T. (1996) Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178, 3763–3770 10.1128/jb.178.13.3763-3770.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sledjeski D. D., Gupta A., and Gottesman S. (1996) The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15, 3993–4000 10.1002/j.1460-2075.1996.tb00773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eguchi Y., Itoh T., and Tomizawa J. (1991) Antisense RNA. Annu. Rev. Biochem. 60, 631–652 10.1146/annurev.bi.60.070191.003215 [DOI] [PubMed] [Google Scholar]
- 36. Sledjeski D. D., Whitman C., and Zhang A. (2001) Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183, 1997–2005 10.1128/JB.183.6.1997-2005.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown L., and Elliott T. (1997) Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 179, 656–662 10.1128/jb.179.3.656-662.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Majdalani N., Cunning C., Sledjeski D., Elliott T., and Gottesman S. (1998) DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U.S.A. 95, 12462–12467 10.1073/pnas.95.21.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lease R. A., Cusick M. E., and Belfort M. (1998) Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. U.S.A. 95, 12456–12461 10.1073/pnas.95.21.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., and Storz G. (1997) A small stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90, 43–53 10.1016/S0092-8674(00)80312-8 [DOI] [PubMed] [Google Scholar]
- 41. Zhang A., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., and Storz G. (1998) The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17, 6061–6068 10.1093/emboj/17.20.6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wassarman K. M., Repoila F., Rosenow C., Storz G., and Gottesman S. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15, 1637–1651 10.1101/gad.901001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E. G., Margalit H., and Altuvia S. (2001) Novel small RNA-encoding genes in the intergenic region of Escherichia coli. Curr. Biol. 11, 941–950 10.1016/S0960-9822(01)00270-6 [DOI] [PubMed] [Google Scholar]
- 44. Updegrove T. B., Zhang A., and Storz G. (2016) Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138 10.1016/j.mib.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soper T. J., and Woodson S. A. (2008) The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14, 1907–1917 10.1261/rna.1110608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soper T., Mandin P., Majdalani N., Gottesman S., and Woodson S. A. (2010) Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U.S.A. 107, 9602–9607 10.1073/pnas.1004435107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Battesti A., Majdalani N., and Gottesman S. (2011) The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213 10.1146/annurev-micro-090110-102946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roberts J. W. (2019) Mechanisms of bacterial transcription termination. J. Mol. Biol. 2019, pii S0022–2836(0019)30185–30188 [DOI] [PubMed] [Google Scholar]
- 49. Repoila F., and Gottesman S. (2001) Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183, 4012–4023 10.1128/JB.183.13.4012-4023.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Repoila F., and Gottesman S. (2003) Temperature sensing by the dsrA promoter. J. Bacteriol. 185, 6609–6614 10.1128/JB.185.22.6609-6614.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sanchez-Vazquez P., Dewey C. N., Kitten N., Ross W., and Gourse R. L. (2019) Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 116, 8310–8319 10.1073/pnas.1819682116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Potrykus K., and Cashel M. (2008) (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- 53. Gourse R. L., Chen A. Y., Gopalkrishnan S., Sanchez-Vazquez P., Myers A., and Ross W. (2018) Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184 10.1146/annurev-micro-090817-062444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klauck E., Böhringer J., and Hengge-Aronis R. (1997) The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25, 559–569 10.1046/j.1365-2958.1997.4911852.x [DOI] [PubMed] [Google Scholar]
- 55. Sledjeski D., and Gottesman S. (1995) A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 2003–2007 10.1073/pnas.92.6.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Singh K., Milstein J. N., and Navarre W. W. (2016) Xenogeneic silencing and its impact on bacterial genomes. Annu. Rev. Microbiol. 70, 199–213 10.1146/annurev-micro-102215-095301 [DOI] [PubMed] [Google Scholar]
- 57. Battesti A., Tsegaye Y. M., Packer D. G., Majdalani N., and Gottesman S. (2012) H-NS regulation of IraD and IraM anti-adaptors for control of RpoS degradation. J. Bacteriol. 194, 2470–2478 10.1128/JB.00132-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mandin P., and Gottesman S. (2010) Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 29, 3094–3107 10.1038/emboj.2010.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cronan J. E., J, and LaPorte D. (2005) Tricarboxylic acid cycle and glyoxylate bypass. American Society for Microbiology, Washington, D. C. 10.1128/ecosalplus.3.5.2 [DOI] [PubMed] [Google Scholar]
- 60. Al Mamun A. A., Lombardo M.-J., Shee C., Lisewski A. M., Gonzalez C., Lin D., Nehring R. B., Saint-Ruf C., Gibson J. L., Frisch R. L., Lichtarge O., Hastings P. J., and Rosenberg S. M. (2012) Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338, 1344–1348 10.1126/science.1226683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Papenfort K., Said N., Welsink T., Lucchini S., Hinton J. C. D., and Vogel J. (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 74, 139–158 10.1111/j.1365-2958.2009.06857.x [DOI] [PubMed] [Google Scholar]
- 62. Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y. E., Reiss N., Bar A., Altuvia Y., Argaman L., and Margalit H. (2016) Global mapping of small RNA–target interactions in bacteria. Mol. Cell 63, 884–897 10.1016/j.molcel.2016.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Monteiro C., Papenfort K., Hentrich K., Ahmad I., Le Guyon S., Reimann R., Grantcharova N., and Römling U. (2012) Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 9, 489–502 10.4161/rna.19682 [DOI] [PubMed] [Google Scholar]
- 64. De Lay N., and Gottesman S. (2012) A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 86, 524–538 10.1111/j.1365-2958.2012.08209.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mika F., and Hengge R. (2014) Small RNAs in the control of RpoS, CsgD, and biofilm architecture in Escherichia coli. RNA Biol. 11, 494–507 10.4161/rna.28867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen J., and Gottesman S. (2017) Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev. 31, 1382–1395 10.1101/gad.302547.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Majdalani N., Chen S., Murrow J., St John K., and Gottesman S. (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39, 1382–1394 10.1111/j.1365-2958.2001.02329.x [DOI] [PubMed] [Google Scholar]
- 68. Majdalani N., Hernandez D., and Gottesman S. (2002) Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46, 813–826 10.1046/j.1365-2958.2002.03203.x [DOI] [PubMed] [Google Scholar]
- 69. Wall E., Majdalani N., and Gottesman S. (2018) The complex Rcs regulatory cascade. Annu. Rev. Microbiol. 72, 111–139 10.1146/annurev-micro-090817-062640 [DOI] [PubMed] [Google Scholar]
- 70. Gutierrez A., Laureti L., Crussard S., Abida H., Rodríguez-Rojas A., Blázquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J., and Matic I. (2013) β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 4, 1610 10.1038/ncomms2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laubacher M. E., and Ades S. E. (2008) The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190, 2065–2074 10.1128/JB.01740-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mika F., Busse S., Possling A., Berkholz J., Tschowri N., Sommerfeldt N., Pruteanu M., and Hengge R. (2012) Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol. Microbiol. 84, 51–65 10.1111/j.1365-2958.2012.08002.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moon K., and Gottesman S. (2011) Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 82, 1545–1562 10.1111/j.1365-2958.2011.07907.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hirsch M., and Elliott T. (2005) Stationary-phase regulation of RpoS translation in Escherichia coli. J. Bacteriol. 187, 7204–7213 10.1128/JB.187.21.7204-7213.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Battesti A., Majdalani N., and Gottesman S. (2015) Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc. Natl. Acad. Sci. U.S.A. 112, 5159–5164 10.1073/pnas.1504639112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ranquet C., and Gottesman S. (2007) Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J. Bacteriol. 189, 4872–4879 10.1128/JB.01838-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Keiler K. C. (2008) Biology of trans-translation. Annu. Rev. Microbiol. 62, 133–151 10.1146/annurev.micro.62.081307.162948 [DOI] [PubMed] [Google Scholar]
- 78. Thompson K. M., and Gottesman S. (2014) The MiaA tRNA modification enzyme is necessary for robust RpoS expression in Escherichia coli. J. Bacteriol. 196, 754–761 10.1128/JB.01013-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Aubee J. I., Olu M., and Thompson K. M. (2016) The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation. RNA 22, 729–742 10.1261/rna.053165.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Aubee J. I., Olu M., and Thompson K. (2017) TrmL and TusA are necessary for rpoS and MiaA is required for hfq expression in Escherichia coli. Biomolecules 7, E39 10.3390/biom7020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Paulander W., Maisnier-Patin S., and Andersson D. I. (2009) The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source, and RpoS (σS). Genetics 183, 539–546, 1SI–2SI 10.1534/genetics.109.106104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fan Y., Wu J., Ung M. H., De Lay N., Cheng C., and Ling J. (2015) Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 43, 1740–1748 10.1093/nar/gku1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kurylo C. M., Parks M. M., Juette M. F., Zinshteyn B., Altman R. B., Thibado J. K., Vincent C. T., and Blanchard S. C. (2018) Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 25, 236–248.e6 10.1016/j.celrep.2018.08.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schweder T., Lee K.-H., Lomovskaya O., and Matin A. (1996) Regulation of Escherichia coli starvation sigma factor (σS) by ClpXP protease. J. Bacteriol. 178, 470–476 10.1128/jb.178.2.470-476.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Muffler A., Fischer D., Altuvia S., Storz G., and Hengge-Aronis R. (1996) The response regulator RssB controls stability of the σS subunit of RNA-polymerase in Escherichia coli. EMBO J. 15, 1333–1339 10.1002/j.1460-2075.1996.tb00475.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pratt L. A., and Silhavy T. J. (1996) The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U.S.A. 93, 2488–2492 10.1073/pnas.93.6.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Olivares A. O., Baker T. A., and Sauer R. T. (2016) Mechanistic insights into bacterial AAA+ proteases and protein-remodeling machines. Nat. Rev. Microbiol. 14, 33–44 10.1038/nrmicro.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kuhlmann N. J., and Chien P. (2017) Selective adaptor dependent protein degradation in bacteria. Curr. Opin. Microbiol. 36, 118–127 10.1016/j.mib.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhou Y., Gottesman S., Hoskins J. R., Maurizi M. R., and Wickner S. (2001) The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev. 15, 627–637 10.1101/gad.864401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Peterson C. N., Ruiz N., and Silhavy T. J. (2004) RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J. Bacteriol. 186, 7403–7410 10.1128/JB.186.21.7403-7410.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bougdour A., Wickner S., and Gottesman S. (2006) Modulating RssB activity: IraP, a novel regulator of σS stability in Escherichia coli. Genes Dev. 20, 884–897 10.1101/gad.1400306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bougdour A., Cunning C., Baptiste P. J., Elliott T., and Gottesman S. (2008) Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68, 298–313 10.1111/j.1365-2958.2008.06146.x [DOI] [PubMed] [Google Scholar]
- 93. Bougdour A., and Gottesman S. (2007) ppGpp regulation of RpoS degradation via an anti-adaptor protein IraP. Proc. Natl. Acad. Sci. U.S.A. 104, 12896–12901 10.1073/pnas.0705561104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Battesti A., Hoskins J. R., Tong S., Milanesio P., Mann J. M., Kravats A., Tsegaye Y. M., Bougdour A., Wickner S., and Gottesman S. (2013) Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 27, 2722–2735 10.1101/gad.229617.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dorich V., Brugger C., Tripathi A., Hoskins J. R., Tong S., Suhanovsky M. M., Sastry A., Wickner S., Gottesman S., and Deaconescu A. M. (2019) Structural basis for inhibition of a response regulator of σS stability by a ClpXP anti-adaptor. Genes Dev. 33, 718–732 10.1101/gad.320168.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Merrikh H., Ferrazzoli A. E., Bougdour A., Olivier-Mason A., and Lovett S. T. (2009) A DNA damage response in Escherichia coli involving the alternative sigma factor RpoS. Proc. Natl. Acad. Sci. U.S.A. 106, 611–616 10.1073/pnas.0803665106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., and Storz G. (2001) DNA Microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 10.1128/JB.183.15.4562-4570.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Merrikh H., Ferrazzoli A. E., and Lovett S. T. (2009) Growth phase and (p)ppGpp control of IraD, a regulator of rpoS stability, in Escherichia coli. J. Bacteriol. 191, 7436–7446 10.1128/JB.00412-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Park H., McGibbon L. C., Potts A., Yakhnin H., Romeo T., and Babitzke P. (2017) Translational repression of the RpoS anti-adaptor IraD by CsrA is mediated via translational coupling to a short upstream open reading frame. MBio 8, e01355–17 10.1128/mBio.01355-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Romeo T., and Babitzke P. (2018) Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 6, RWR-0009–2017 10.1128/microbiolspec.RWR-0009-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Peterson C. N., Levchenko I., Rabinowitz J. D., Baker T. A., and Silhavy T. J. (2012) RpoS proteolysis is controlled directly by ATP levels in Escherichia coli. Genes Dev. 26, 548–553 10.1101/gad.183517.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mandel M. J., and Silhavy T. J. (2005) Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability. J. Bacteriol. 187, 434–442 10.1128/JB.187.2.434-442.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bouché S., Klauck E., Fischer D., Lucassen M., Jung K., and Hengge-Aronis R. (1998)) Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 27, 787–795 10.1046/j.1365-2958.1998.00725.x [DOI] [PubMed] [Google Scholar]
- 104. De Mets F., Van Melderen L., and Gottesman S. (2019) Regulation of acetate metabolism and coordination with the TCA cycle via a processed small RNA. Proc. Natl. Acad. Sci. U.S.A. 116, 1043–1052 10.1073/pnas.1815288116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Maeda H., Fujita N., and Ishihama A. (2000) Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28, 3497 10.1093/nar/28.18.3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Colland F., Fujita N., Ishihama A., and Kolb A. (2002) The interaction between σS, the stationary phase sigma factor, and the core enzyme of Escherichia coli RNA polymerase. Genes Cells 7, 233–247 [DOI] [PubMed] [Google Scholar]
- 107. Jishage M., and Ishihama A. (1995) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 177, 6832–6835 10.1128/jb.177.23.6832-6835.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jishage M., Iwata A., Ueda S., and Ishihama A. (1996) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178, 5447–5451 10.1128/jb.178.18.5447-5451.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bougdour A., Lelong C., and Geiselmann J. (2004) Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279, 19540–19550 10.1074/jbc.M314145200 [DOI] [PubMed] [Google Scholar]
- 110. Typas A., Barembruch C., Possling A., and Hengge R. (2007) Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. EMBO J. 26, 1569–1578 10.1038/sj.emboj.7601629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jishage M., and Ishihama A. (1998) A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 95, 4953–4958 10.1073/pnas.95.9.4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yoshida H., Shimada T., and Ishihama A. (2018) Coordinated hibernation of transcriptional and translational apparatus during growth transition of Escherichia coli in stationary phase. mSystems 3, e00057 10.1128/mSystems.00057-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wassarman K. M. (2018) 6S RNA, a global regulator of transcription. Microbiol. Spectr. 6, 10.1128/microbiolspec.RWR-0019-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhou Y., and Gottesman S. (1998) Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180, 1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Becker G., Klauck E., and Hengge-Aronis R. (2000) The response regulator RssB, a recognition factor for σS proteolysis in Escherichia coli, can act like an anti-σS factor. Mol. Microbiol. 35, 657–666 [DOI] [PubMed] [Google Scholar]
- 116. Klauck G., Serra D. O., Possling A., and Hengge R. (2018) Spatial organization of different sigma factor activities and c-di-GMP signalling within the three-dimensional landscape of a bacterial biofilm. Open Biol. 8, 180066 10.1098/rsob.180066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bouillet S., Arabet D., Jourlin-Castelli C., Méjean V., and Iobbi-Nivol C. (2018) Regulation of sigma factors by conserved partner switches controlled by divergent signalling systems. Environ. Microbiol. Rep. 10, 127–139 10.1111/1758-2229.12620 [DOI] [PubMed] [Google Scholar]
- 118. Francez-Charlot A., Kaczmarczyk A., Fischer H.-M., and Vorholt J. A. (2015) The general stress response in Alphaproteobacteria. Trends Microbiol. 23, 164–171 10.1016/j.tim.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 119. Hecker M., Pané-Farré J., and Völker U. (2007) SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236 10.1146/annurev.micro.61.080706.093445 [DOI] [PubMed] [Google Scholar]
- 120. Hryckowian A. J., Battesti A., Lemke J. J., Meyer Z. C., and Welch R. A. (2014) IraL is an RssB anti-adaptor that stabilizes RpoS during logarithmic phase growth in Escherichia coli and Shigella. MBio 5, e01043–14 10.1128/mBio.01043-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tu X., Latifi T., Bougdour A., Gottesman S., and Groisman E. A. (2006) The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 103, 13503–13508 10.1073/pnas.0606026103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kröger C., Colgan A., Srikumar S., Händler K., Sivasankaran S. K., Hammarlöf D., Canals R., Grissom J. E., Conway T., Hokamp K., and Hinton J. C. (2013) An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14, 683–695 10.1016/j.chom.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 123. Fujita M., Tanaka K., Takahashi H., and Amemura A. (1994) Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol. Microbiol. 13, 1071–1077 10.1111/j.1365-2958.1994.tb00498.x [DOI] [PubMed] [Google Scholar]
- 124. Bertani I., Sevo M., Kojic M., and Venturi V. (2003) Role of GacA, LasI, RhlI, Ppk, PsrA, Vfr and ClpXP in the regulation of the stationary-phase sigma factor in rpoS/RpoS in Pseudomonas. Arch. Microbiol. 180, 264–271 10.1007/s00203-003-0586-8 [DOI] [PubMed] [Google Scholar]
- 125. Jørgensen F., Bally M., Chapon-Herve V., Michel G., Lazdunski A., Williams P., and Stewart G. S. (1999) RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145, 835–844 10.1099/13500872-145-4-835 [DOI] [PubMed] [Google Scholar]
- 126. Galperin M. Y. (2006) Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 10.1128/JB.01887-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gebhardt M. J., and Shuman H. A. (2017) GigA and GigB are master regulators of antibiotic resistance, stress responses, and virulence in Acinetobacter baumannii. J. Bacteriol. 199, e00066–17 10.1128/JB.00066-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Thi Bach Nguyen H., Romero A. D., Amman F., Sorger-Domenigg T., Tata M., Sonnleitner E., and Bläsi U. (2018) Negative control of RpoS synthesis by the sRNA ReaL in Pseudomonas aeruginosa. Front. Microbiol. 9, 2488 10.3389/fmicb.2018.02488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bouillet S., Genest O., Jourlin-Castelli C., Fons M., Méjean V., and Iobbi-Nivol C. (2016) The general stress response σS is regulated by a partner switch in the Gram-negative bacterium Shewanella oneidensis. J. Biol. Chem. 291, 26151–26163 10.1074/jbc.M116.751933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bouillet S., Genest O., Méjean V., and Iobbi-Nivol C. (2017) Protection of the general stress response σS factor by the CrsR regulator allows a rapid and efficient adaptation of Shewanella oneidensis. J. Biol. Chem. 292, 14921–14928 10.1074/jbc.M117.781443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lori C., Kaczmarczyk A., de Jong I., and Jenal U. (2018) A single-domain response regulator functions as an integrating hub to coordinate general stress response and development in alphaproteobacteria. MBio 9 e00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Nadezhdin E. V., Brody M. S., and Price C. W. (2011) An α/β hydrolase and associated Per-ARNT-Sim domain comprise a bipartite sensing module coupled with diverse output domains. PLoS One 6, e25418 10.1371/journal.pone.0025418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Simm R., Ahmad I., Rhen M., Le Guyon S., and Römling U. (2014) Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 9, 1261–1282 10.2217/fmb.14.88 [DOI] [PubMed] [Google Scholar]
- 134. Mika F., and Hengge R. (2013) Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int. J. Mol. Biol. 14, 4560–4579 10.3390/ijms14034560 [DOI] [PMC free article] [PubMed] [Google Scholar]