Abstract
There is recent evidence to indicate the existence of an inverse association between the incidence of neurological disorders and cancer development. Concurrently, the transcriptional pathways responsible for the onset of glioblastoma multiforme (GBM) and Alzheimer's disease (AD) have been found to be mutually exclusive between the two pathologies. Despite advancements being made concerning the knowledge of the molecular mechanisms responsible for the development of GBM and AD, little is known about the identity of the microRNA (miRNAs or miRs) involved in the development and progression of these two pathologies and their possible inverse expression patterns. On these bases, the aim of the present study was to identify a set of miRNAs significantly de-regulated in both GBM and AD, and hence to determine whether the identified miRNAs exhibit an inverse association within the two pathologies. For this purpose, miRNA expression profiling datasets derived from the Gene Expression Omnibus (GEO) DataSets and relative to GBM and AD were used. Once the miRNAs significantly de-regulated in both pathologies were identified, DIANA-mirPath pathway prediction and STRING Gene Ontology enrichment analyses were performed to establish their functional roles in each of the pathologies. The results allowed the identification of a set of miRNAs found de-regulated in both GBM and AD, whose expression levels were inversely associated in the two pathologies. In particular, a strong negative association was observed between the expression levels of miRNAs in GBM compared to AD, suggesting that although the molecular pathways behind the development of these two pathologies are the same, they appear to be inversely regulated by miRNAs. Despite the identification of this set of miRNAs which may be used for diagnostic, prognostic and therapeutic purposes, further functional in vitro and in vivo evaluations are warranted in order to validate the diagnostic and therapeutic potential of the identified miRNAs, as well as their involvement in the development of GBM and AD.
Keywords: microRNA, glioblastoma, Alzheimer's disease, biomarker, diagnosis, prognosis, therapy
Introduction
Over the past century, human life expectancy has increased significantly, settling at approximately 76.3 years for males and 81.2 for females (1). Such an increment in lifespan has been associated with a huge surge in the most diffused life-threatening diseases, including cardiovascular, neurodegenerative and oncological pathologies (2).
Although aging represents a common risk factor, the physiological and molecular mechanisms behind the development of age-related disorders differ completely, particularly when comparing cancer with neurodegenerative disorders (3,4). Supporting this evidence, recent epidemiological data have demonstrated that there is an inverse association between cancer and neurodegeneration (5,6).
Despite this inverse association between the incidence of tumors and the rise of neurodegenerative disorders, some conditions, such as inflammation (7–9), the alteration of the intestinal microbiota (10–13), diet-related diseases (14–18) and risks related to the exposure to environmental pollutants (19–25) are involved in the development of both pathologies. Consistently, the association between the development of neurodegenerative diseases and the onset of tumors may be either direct or indirect. As a direct consequence, it has been shown that several benign and malignant types of cancer are associated with neurological, as well as neurodegenerative disorders (26–28).
In particular, it has been demonstrated that although glioblastoma multiforme (GBM) and Alzheimer's disease (AD) share the same molecular pathways, substantial differences exist in their modulation (29,30). In fact, while rapid cell proliferation and apoptotic cell arrest are typical features of GBM, cellular damage and subsequent cell death are common consequences in AD (31,32).
To date, the only available treatments for AD are only palliative, as they are capable of delaying memory and cognitive function impairment, without actually blocking neuronal loss. The main drugs used in the treatment of AD are cholinesterase inhibitors and glutamatergic N-methyl D-aspartate (NMDA) receptors antagonists, such as memantine (Namenda), respectively able to improve neuropsychiatric symptoms and neuronal cell-to-cell communication (33). Importantly, the benefits of such therapies are not long-lasting and are coupled with adverse impairing effects.
GBM is a widely diffused brain malignancy, as well as the most aggressive tumor of the central nervous system (34,35). Currently, GBM treatment options are limited, typically represented by surgical resection of the tumor mass (when the lesion does not involve vascular and nerve structures), followed by radiotherapy and chemotherapy (36). Despite advancements being made in anticancer treatments (37), the therapeutic approaches available for GBM are often ineffective, given the high rate of GBM relapse and drug resistance (36). Recently, in vitro studies have demonstrated that treatment with nitric oxide-releasing HIV protease inhibitors previously adopted for other tumors (38,39), is effective in reducing the proliferation of GBM cancer cells (40,41).
Notably, the diagnostic and therapeutic approaches that are normally used to recognize and to treat cognitive deficits and dementia symptoms, appear to be slightly effective in the early detection and treatment of brain precancerous lesions. Although they need to be further validated in a larger cohort of patients, transcranial magnetic stimulation (TMS) and transcranial Doppler ultrasonography, normally used for vascular cognitive impairments (42–47), AD (48), restless leg syndrome (49–52), and other neurological syndromes (53–56), appear to be promising approaches suitable for the diagnosis and cure of precancerous brain lesions.
A concern about GBM management is relative to the lack of effective biomarkers. In particular, a number of biomarkers have been proposed to solve this issue. Several studies have highlighted the possible application of extracellular protein biomarkers, such as extracellular matrix proteins, vascular endothelial growth factor (VEGF), angiogenesis-associated proteins, matrix metalloproteinases (MMPs; MMP-2, MMP-9) and astrocyte elevated gene-1 (AEG-1), macrophage migration inhibitory factor (MIF) and functionally-related genes (DD-T; CD74, CD44, CXCR2 and CXCR4) (57–59). Other proteins can be also used for the treatment or prognostic evaluation of tumor development (60,61).
Given the lack of effective diagnostic strategies, as well as treatments able to effectively cure both GBM and AD, there is an urgent need for the identification of novel diagnostic biomarkers and therapeutic targets for the effective treatment of such pathologies. Moreover, the understanding of the expression patterns of such biomarkers may prove to be useful in order to further demonstrate the existence of an inverse association between GBM and AD. In this context, several studies have demonstrated that the evaluation of microRNA (miRNA or miR) expression levels in patients compared with their healthy controls, may provide information on the development of several diseases, including cancer and neurodegenerative disorders (62–65). Indeed, miRNAs are involved in both physiological and pathological processes; therefore studying their alterations in GBM and AD may prove to be helpful in detecting early the onset of such pathologies.
On this ground, the aim of this study was to analyze miRNA expression profiling datasets of GBM and AD obtained from the Gene Expression Omnibus (GEO) DataSets portal in order to identify specific miRNAs de-regulated in both diseases. Once the presence of altered miRNAs shared between GBM and AD would be established, the second aim of this study was to determine whether their expression levels are inversely associated.
Materials and methods
Selection and analysis of GBM and AD miRNA profiling datasets
The selection of both GBM and AD miRNA expression profiling datasets was performed using the publicly available GEO DataSets database, as previously reported (66,67). Briefly, for the selection of GBM miRNA datasets the following search terms were used: ‘{[‘non coding rna profiling by array’(DataSet Type)] AND glioblastoma} AND ‘Homo sapiens’[porgn:__txid9606]’; while for the selection of AD datasets the search terms used were as follows: ‘{[‘non coding rna profiling by array’(DataSet Type)] AND Alzheimer} AND ‘Homo sapiens’[porgn:__txid9606]’. These search criteria allowed the identification of different miRNA expression datasets of which only those with >10 samples (total of normal and pathological samples) were selected for the further computational analyses. Given the low number of AD miRNA microarray platforms, datasets containing <10 samples were considered. Datasets including the expression data of GBM or AD within in vitro models were not considered for the analyses.
Following dataset selection, the data matrices were downloaded and differential analyses were performed between normal and pathological samples using the GEO2R tool available on GEO DataSets. Since different miRNA microarray platforms were adopted, the differentially expressed miRNAs of each dataset were annotated using the last version of miRBase (miRBase version 22) (68). The miRNA expression fold change (FC) was expressed as base-2 logarithm of FC (log2FC) to normalize the miRNA expression values obtained from different microarray platforms.
All the miRNAs with a value of P≤0.01 were considered for the merging analyses and the following identification of miRNAs involved in GBM and AD.
Identification of miRNAs potentially involved in GBM and AD
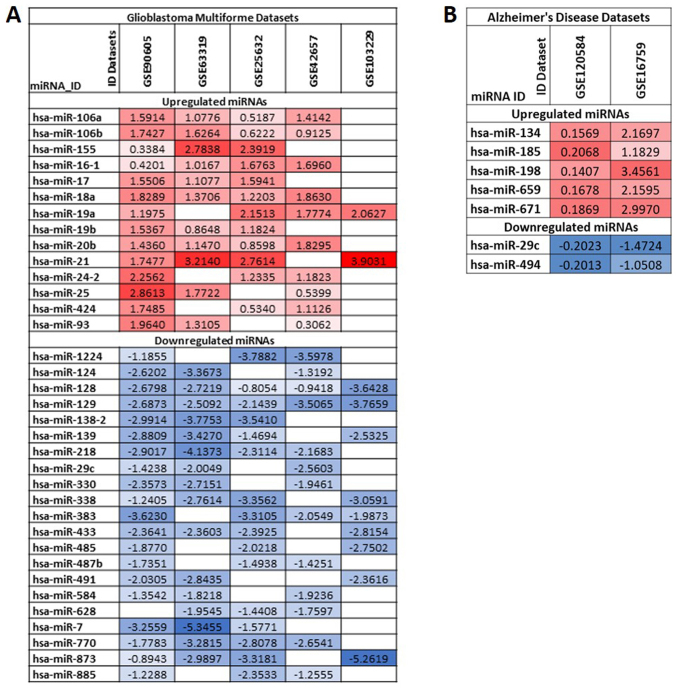
The lists of differentially expressed miRNAs obtained from the GBM and AD datasets were merged through a Venn diagram calculating tool, in order to obtain miRNAs shared with >1 dataset (http://bioinformatics.psb.ugent.be/webtools/Venn/). In particular, only the miRNAs contained at least in the 50% of GBM and AD datasets were considered. The log2FC levels of each miRNA are reported in a graphic table indicating the level of upregulation and downregulation, using red boxes and blue boxes, respectively with different gradient.
Schemes of de-regulated miRNAs were generated for the GBM datasets, AD datasets and for the GBM and AD datasets together, in order to establish the existence of an inverse association between the expression levels of GBM- and AD-related miRNAs.
Involvement of GBM- and AD-related miRNAs in the modulation of glioma- and AD-related pathways
To establish the involvement of the identified miRNAs in the modulation of glioma- and AD-related pathways and their relative target genes, a pathway prediction analysis was performed using the prediction tool DIANA-mirPath version 3 (69), as previously described (70). The DIANA-mirPath analysis was performed comparing the identified miRNAs with the panel of miRNAs involved in both glioma- (hsa05214) and AD-related (hsa05010) pathways.
Gene Ontology (GO) and roles of GBM and AD miRNA-target genes
The GO analysis and the functional roles of the miRNAs-targeted genes related to GBM and AD were evaluated using the enrichment software ‘STRING: Functional protein association networks’ (https://string-db.org/) (71). In particular, for the genes related to GBM and AD the ‘Biological process’, ‘Molecular function’ and ‘Cellular component’ were determined. Furthermore, the interaction network between genes was determined for both GBM and AD. The STRING analyses were performed for the 13 selected miRNAs resulting from the comparison of GBM and AD miRNA expression profiling datasets.
Statistical analyses
The GEO2R software already normalized the miRNA expression data derived from GEO DataSets. Only miRNAs with a value of P≤0.01 were considered for further analyses. Furthermore, the GEO2R software automatically calculated the GEO DataSets data P-values. The P-values obtained from the prediction pathway analysis were already calculated using DIANA-mirPath software (V.3.0).
Results
miRNA profiling dataset selection for GBM and AD
The research of miRNA expression profiling datasets performed with GEO2R using specific search terms allowed for the identification of 51 and 9 miRNA profiling datasets for GBM and AD, respectively. The datasets either relative to in vitro studies or built with less than 8 samples, including normal and pathological specimens, were excluded from the analysis. Following this filtering, five datasets for GBM and two datasets for AD, were selected for the study. The information of all selected datasets is reported in Table I.
Table I.
Information pertaining to the selected datasets for glioblastoma and Alzheimers disease.
| Series accession | n. normal | n. cancer | Samples | Platform | Author/(Ref.) | Total samples |
|---|---|---|---|---|---|---|
| Glioblastoma datasets | ||||||
| GSE90604 | 7 | 16 | Fresh frozen brain tissues and GBM tumor tissues | GPL21572 [miRNA-4] affymetrix multispecies miRNA-4 array | Gulluoglu et al, 2017 (No Ref.) | 25 |
| GSE25632 | 5 | 82 | Normal brain tissues and GBM tumor samples | GPL8179 Illumina Human v2 MicroRNA expression beadchip | (112,113) | 87 |
| GSE103229 | 5 | 5 | Normal brain tissues and GBM tumor samples | GPL18058 Exiqon miRCURY LNA microRNA array, 7th generation | Chun et al, 2018 (No Ref.) | 10 |
| GSE63319 | 4 | 11 | Normal brain tissues (epileptic) and GBM tumor samples | GPL16384 [miRNA-3] affymetrix multispecies miRNA-3 array | Sarkar et al, 2016 (No Ref.) | 18 |
| GSE42657 | 7 | 5 | Normal brain tissues and GBM tumor samples | GPL8179 Illumina Human v2 MicroRNA expression beadchip | (114) | 61 |
| Alzheimers disease datasets | ||||||
| GSE120584 | 288 | 1021 | Serum samples | GPL21263 3D-Gene Human miRNA V21_1.0.0 | (115) | 1601 |
| GSE16759 | 4 | 4 | Normal brain tissues and AD tissue samples | GPL8757 USC/XJZ Human 0.9 K miRNA-940-v1.0 | (116) | 8 |
GBM, glioblastoma multiforme; AD, Alzheimers disease.
In particular, for GBM, two datasets were developed by Affymetrix (Affymetrix miRNA Array), two developed by Illumina (Illumina Human MicroRNA expression beadchip) and one developed by Exiqon (Exiqon miRCURY LNA microRNA array, 7th generation). For the AD datasets, one was developed by 3D-Gene (3D-Gene Human miRNA V21_1.0.0) and the other one was a custom platform (USC/XJZ Human 0.9 K miRNA-940-v1.0) (Table I).
Identification of miRNAs involved in the development of GBM and AD
Merging the lists of differentially expressed miRNAs in GBM datasets allowed the identification of a set of miRNAs strictly involved in the development and progression of GBM. In particular, among all miRNAs, 35 were found to be de-regulated with concordant expression levels in at least 3 out of 5 GBM miRNA expression datasets (downregulated or upregulated in all datasets) (Fig. 1A). In particular, 14 miRNAs were upregulated and 21 downregulated. Of these miRNAs, the most upregulated miRNAs were the following: hsa-miR-21, hsa-miR-18a, hsa-miR-19a, hsa-miR-25, hsa-miR-16-1, hsa-miR-106a and hsa-miR-106b; and the most downregulated miRNAs were the following: hsa-miR-128, hsa-miR-129, hsa-miR-7, hsa-miR-873, hsa-miR-218, hsa-miR-139 and hsa-miR-770.
Figure 1.
(A) Differentially expressed miRNAs between GBM samples and normal brain tissues; (B) Differentially expressed miRNAs between samples of AD and normal controls. log2FC values relative to upregulated miRNAs were reported with a gradient of red boxes while a gradient of blue boxes were used for downregulated miRNAs. GBM, glioblastoma multiforme; AD, Alzheimer's disease.
Similarly, following the merging of the 2 AD miRNA lists, 7 miRNAs were uncovered. All of them were related to the development of AD. Among these, 5 were upregulated (hsa-miR-134, hsa-miR-185, hsa-miR-198, hsa-miR-659 and hsa-miR-671) and 2 downregulated (hsa-miR-29c and hsa-miR-494) (Fig. 1B).
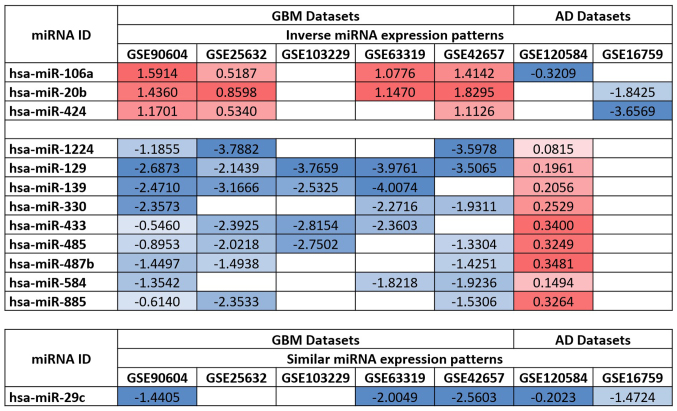
By comparing the de-regulated miRNAs in both the GBM and AD datasets, it was observed that the expression levels of 12 miRNAs were inversely associated between GBM and AD. Of these, 3 were upregulated in GBM and downregulated in AD (hsa-miR-106a, hsa-miR-20b and hsa-miR-424) and 9 were downregulated in GBM and upregulated in AD (hsa-miR-1224, hsa-miR-129, hsa-miR-139, hsa-miR-330, hsa-miR-433, hsa-miR-485, hsa-miR-487b, hsa-miR-584 and hsa-miR-885). Additionally, hsa-miR-29c was downregulated in both the GBM and AD datasets, suggesting its involvement in both pathologies (Fig. 2).
Figure 2.
Comparison of the expression levels of miRNAs in GBM and AD datasets. In total, 12 out of 13 miRNAs exhibited inverse expression patterns in GBM and AD, while hsa-miR-29c was found to be downregulated in both GBM and AD. GBM, glioblastoma multiforme; AD, Alzheimer's disease.
Pathway prediction analysis of selected miRNAs
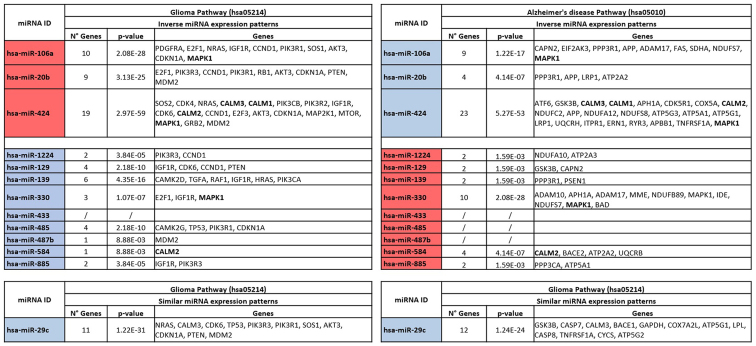
To elucidate the role of the de-regulated miRNAs shared between the GBM and AD datasets, DIANA-mirPath analysis was performed. The analysis revealed that all the 13 selected miRNAs, apart from hsa-miR-433, were involved in the regulation of GBM by modulating the expression levels of 34 different genes (Fig. 3). As regards AD, the pathway prediction analysis revealed that, with the exclusion of hsa-miR-433, hsa-miR-485 and hsa-miR-487b, all the remaining identified miRNAs were able to interact with the AD pathway and to target 51 different genes. Notably, the analysis revealed that the miRNAs, hsa-miR-106a, hsa-miR-424 and hsa-miR-330, were able to modulate the expression levels of MAPK1 in both GBM and AD. In particular, hsa-miR-106a and hsa-miR-424 were able to induce the downregulation and upregulation of MAPK1 in GBM and AD, respectively. This could be explained by the inverse association of miR-106a expression levels in GBM and AD. Similarly, hsa-miR-330 was able to induce the upregulation of MAPK1 in GBM, while in AD, this gene was downregulated. In the same manner, the miRNAs, hsa-miR-424, hsa-miR-885 and hsa-miR-29c, were involved in the regulation of several genes belonging to the family of calmodulins (CALM family). In detail, hsa-miR-424 and hsa-miR-885 exhibited an inverse association in modulating the expression of CALM genes, while hsa-miR-29c was downregulated in both the GBM and AD datasets, thus determining the upregulation of CALM3 in both pathologies (Fig. 3).
Figure 3.
Diana-mirPath pathway prediction analysis showing the genes modulated by the selected miRNAs in glioma (hsa05214) and Alzheimer's disease (hsa05010) pathways. In red are reported the miRNAs upregulated, while in blue those downregulated. The genes shared in both glioma and Alzheimers’ disease pathways are presented in bold.
In addition, the pathway prediction analysis identified the most targeted genes in GBM as IGF1R (targeted by 6 miRNAs), CCND1, CDKN1A, MDM2 (targeted by 5 miRNAs), AKT3, CDK6, E2F1, MAPK1, PIK3R1, PIK3R3 (targeted by 4 miRNAs). Notably, the mostly targeted gene families were PI3K (targeted by 11 miRNAs), CDK (targeted by 10 miRNAs) and E2F (targeted by 5 miRNAs) (data not shown). As regards the AD pathway, the 13 selected miRNAs were able to target mainly the MAPK1 (targeted by 4 miRNAs), APH1A, APP, GSK3B (targeted by 3 miRNAs), ADAM17, ATP2A2, CALM2 and CALM3 (targeted by 2 miRNAs). Of note, the most altered gene families were the ATP (9 miRNAs), NDUF (8 miRNAs), CALM (5 miRNAs) and ADAM (3 miRNAs) families, strictly involved in the development of AD (data not shown).
GO enrichment analysis of selected miRNAs and target genes
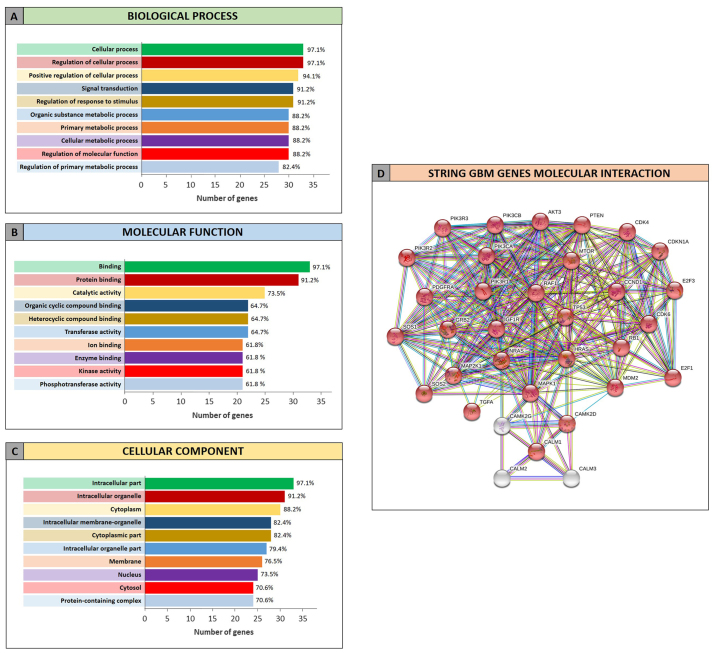
The GO analysis performed by STRING allowed to determine the functional roles of the genes related to GBM and AD and targeted by the selected miRNAs. The STRING analysis performed on GBM miRNA-targeted genes revealed that of the 34 genes, 33 were recognized as proteins. Among these 33 proteins, 30 were directly involved in the glioma pathway (Fig. 4D). The clustering of proteins according to the ‘Biological process’, ‘Molecular function’ and ‘Cellular component’ categories revealed that the majority of proteins were involved in the regulation of cellular processes and metabolic processes (Fig. 4A), in the binding of molecules and in catalytic activities (Fig. 4B). Moreover they belong to intracellular organelle and cytoplasm (Fig. 4C).
Figure 4.
Gene Ontology enrichment analysis by STRING of miRNAs-targeted genes in GBM. (A) Genes clustered according to molecular function; (B) Genes clustered according to biological process; (C) Genes clustered according to cellular component; (D) Interaction network between genes targeted by the selected miRNAs in GBM. In red the genes involved in glioma pathway (hsa05214). GBM, glioblastoma multiforme.
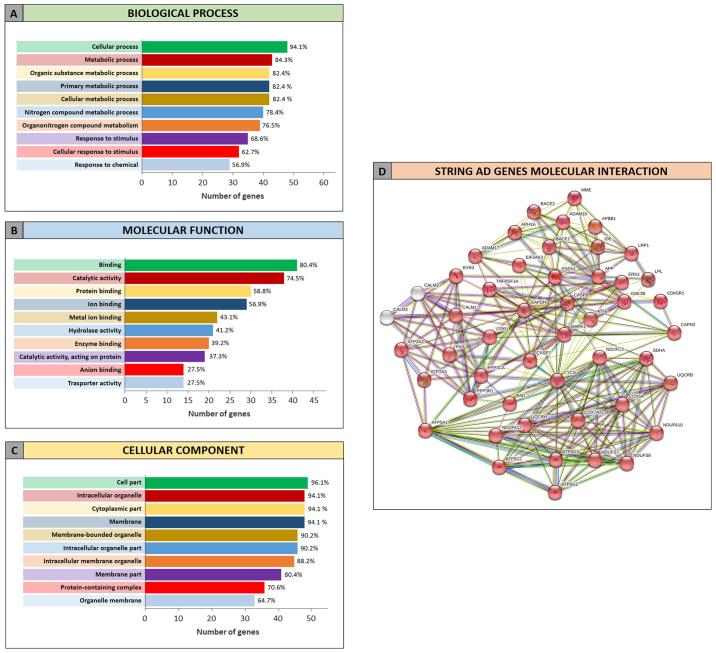
Similarly, the STRING analysis performed on the list of miRNA-targeted genes obtained for the AD highlighted that 47 of the 51 identified genes were involved in AD pathway (Fig. 5D). As described for GBM, regarding the ‘Biological process’ category, the identified genes were involved in the cellular and metabolic processes (metabolism of different compounds), but also in the response to stimuli (Fig. 5A). The AD proteins were also significantly involved in the binding of several molecules and less in the catalytic activity, in contrast to what it was observed for the GBM proteins clustered in the ‘Molecular function’ category (Fig. 5B). Finally, the analyzed proteins were part of the intracellular organelle, as well as the cell membrane, therefore playing a fundamental role in the regulation of cellular homeostasis (Fig. 5C).
Figure 5.
Gene Ontology enrichment analysis by STRING of miRNAs-targeted genes in AD. (A) Genes clustered according to molecular function; (B) Genes clustered according to biological process; (C) Genes clustered according to cellular component; (D) Interaction network between genes targeted by the selected miRNAs in AD. In red the genes involved in the AD pathway (hsa05010). AD, Alzheimer's disease.
Discussion
Despite tremendous advancements being made in the characterization of the clinicopathological features typical of tumors and neurodegenerative diseases, little is known about the association between the molecular mechanisms responsible for the development and progression of brain cancer and neurodegeneration (72–74). In detail, an indirect association in the incidence rates of GBM and AD has been widely reported, thus suggesting that this inverse association may be coupled with an inverse regulation of the same molecular mechanisms involved in the development of GBM and AD (29,30).
To shed light on the molecular mechanisms potentially responsible for the rise of these pathologies, and to demonstrate the existence of an inverse association between the molecular alterations in GBM and AD, the present study identified the miRNAs found altered in GBM and AD. In particular, in this study, we analyzed the potential involvement of these miRNAs in the onset of both diseases although oppositely expressed. For the first time, at least to the best of our knowledge, the existence of a strong inverse association was demonstrated in human samples between selected miRNA expression levels in GBM and AD through the comparison of altered miRNAs.
The choice of analyzing the expression levels of miRNAs in both pathologies depends on the numerous profiling data collected during the years for GBM and AD, and the increasing number of studies coupling the analysis of miRNAs in cancer and neurodegenerative disorders (75–78).
The independent analysis of GBM miRNA expression datasets revealed that 35 miRNAs were de-regulated in tumor samples compared to the normal controls. Almost all of these de-regulated miRNAs have been widely associated with the development and progression of GBM. In particular, several studies have already described the role of hsa-miR-21 and hsa-miR-155 overexpression in the promotion of the development of glioblastoma (79–81). Other studies have demonstrated the therapeutic potential of the overexpression of several miRNAs normally downregulated in glioblastoma, such as hsa-miR-7, hsa-miR-93 and hsa-miR-139 (82–84). As regards the most downregulated miRNAs, hsa-miR-128 and hsa-miR-129, found in the GBM dataset analysis, some studies have highlighted their possible use in the context of novel therapeutic strategies aimed at inhibiting the molecular pathways involved in GBM aggressive phenotypes (85–87).
Concerning the analysis performed on the 2 AD miRNA datasets, the results revealed significant data for the downregulated miRNA, hsa-miR-29c. Several studies have demonstrated that the downregulation of this miRNA may play a potential role either as biomarker or as therapeutic agent in AD in vitro models and in patients (88–91). Of note, the downregulation of hsa-miR-29c has also been observed in GBM where it was observed that its induced overexpression led to the suppression of glioma (92).
However, the most robust and interesting data derived from the comparison between miRNAs de-regulated in both the GBM AD datasets, was that all miRNAs identified, with the exception of hsa-miR-29c, exhibited inverse patterns of expression in GBM compared with AD. This result is in accordance with the hypothesis of the existence of an inverse regulation of molecular pathways in GBM and AD, as postulated by Liu et al (30) and Sánchez-Valle et al (29). In line with this theory, miRNAs were found inversely regulated in the two pathologies by our analyses. Noteworthy, the inverse regulation of miRNAs in GBM and AD is coupled with the inverse regulation of targeted genes by selected miRNAs. Such inverse miRNAs and genes regulatory patterns may explain the inverse comorbidity existing between neurological disorders and cancers (6).
The preliminary data obtained comparing miRNA expression levels in GBM and AD were further confirmed by the pathway prediction and gene ontology enrichment analyses. In particular, the DIANA-mirPath analysis revealed that the 13 selected miRNAs were in common between GBM and AD and they were able to modulate several genes within the glioma pathway (hsa05214). In particular, the miRNAs were involved in the regulation of genes, such as MAPK1, IGF1R, and genes belonging to the PIK3 and RAS families, known to be involved in the development of GBM and other tumors (93–97).
Conversely, the same miRNAs were shown to also target fundamental genes involved in AD, such as APP, responsible for the β-amyloid plaque formation (98); GSK3B, hyperactivated in Alzheimer's neurons (99); NDUF family, responsible for mitochondrial alterations in AD (100); LRP1 able to regulate the metabolism of amyloid-β peptides thus maintaining brain homeostasis (101) and other genes.
Therefore, it is clear how miRNA de-regulation and other epigenetic mechanisms may lead to the modulation of these genes and, in turn, to the acquisition of a more aggressive tumor phenotype or neurodegenerative disorder susceptibility (102,103).
In addition, STRING analysis revealed that the selected miRNAs were able to modulate genes involved in both GBM and AD and those genes performed, in general, the same function and processes within the two pathologies. Therefore, differential expression levels of miRNAs in GBM and AD may be responsible for the onset of GBM rather than the AD, and vice versa.
Overall, the identification of altered miRNAs in both GBM and AD, as well as the definition of the inverse patterns of expression, may pave the way for new studies to better elucidate the involvement of these miRNAs in GBM and AD. In particular, further studies are required to examine the therapeutic potential of such identified miRNAs at the early stages of disease, since several treatments for AD are administered only when the pathology is at an advanced stage (104–106). On this matter, the results of the present study and the potential applications of this research methodology also to other brain diseases, may improve the diagnostic and therapeutic strategies mainly based on the analysis of low-specific biomarkers and on the use of low-sensitive and low-efficacy instrumental procedures (107–111).
In conclusion, in the present study, at least to the best of our knowledge, for the first time a set of de-regulated miRNAs in both GBM and AD was identified, demonstrating the existence of an inverse association between the expression levels of miRNAs in GBM and AD. The findings of the present study may pave the way for other functional in vitro and in vivo studies to validate the diagnostic or prognostic significance of the identified miRNAs, as well as to depict their possible use as novel therapeutic approaches for GBM and AD.
Acknowledgements
The authors would like to thank the Italian League Against Cancer (LILT) for its support.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request. The analyzed datasets are publicly available on the GEO DataSet database.
Authors' contributions
LF, DAS and ML conceived and designed the study. SC, LF, MP and GG performed all the analyses. GL, CDA and MSB provided all the information useful to describe the features of Glioblastoma Multiforme, while MP was involved in the analysis of AD datasets and gave information about neurodegenerative disorders and Alzheimer's disease. MP, MCP and SV were involved in the pathway prediction analysis and in the definition of the functional roles of selected miRNAs. LF, SC, MCP, MP and SV were involved in the interpretation of all data. GG, MCP and LF were involved in the preparation of the figures and tables. SV, LF and ML were involved in the preparation of the original draft of the manuscript, while MCP, SC, SV, GB, CDA and MP reviewed and edited the article. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
All patient data were derived from publicly available datasets.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no influence in the reviewing process, or any involvement in terms of adjudicating on the final decision, for this article. The other authors declare that there are not competing interests.
References
- 1.Crimmins EM, Zhang YS. Aging populations, mortality, and life expectancy. Annu Rev Sociol. 2019;45:annurev–soc-073117-041351. doi: 10.1146/annurev-soc-073117-041351. [DOI] [Google Scholar]
- 2.Franceschi C, Garagnani P, Morsiani C, Conte M, Santoro A, Grignolio A, Monti D, Capri M, Salvioli S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front Med (Lausanne) 2018;5:61. doi: 10.3389/fmed.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganguli M. Cancer and dementia: It's complicated. Alzheimer Dis Assoc Disord. 2015;29:177–182. doi: 10.1097/WAD.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens MI, Lendon C, Roe CM. A common biological mechanism in cancer and Alzheimer's disease? Curr Alzheimer Res. 2009;6:196–204. doi: 10.2174/156720509788486608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder HM, Ahles T, Calderwood S, Carrillo MC, Chen H, Chang CH, Craft S, De Jager P, Driver JA, Fillit H, et al. Exploring the nexus of Alzheimer's disease and related dementias with cancer and cancer therapies: A convening of the Alzheimer's Association & Alzheimer's Drug Discovery Foundation. Alzheimers Dement. 2017;13:267–273. doi: 10.1016/j.jalz.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibáñez K, Boullosa C, Tabarés-Seisdedos R, Baudot A, Valencia A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014;10:e1004173. doi: 10.1371/journal.pgen.1004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trovato Salinaro A, Pennisi M, Di Paola R, Scuto M, Crupi R, Cambria MT, Ontario ML, Tomasello M, Uva M, Maiolino L, et al. Neuroinflammation and neurohormesis in the pathogenesis of Alzheimer's disease and Alzheimer-linked pathologies: Modulation by nutritional mushrooms. Immun Ageing. 2018;15:8. doi: 10.1186/s12979-017-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennisi M, Crupi R, Di Paola R, Ontario ML, Bella R, Calabrese EJ, Crea R, Cuzzocrea S, Calabrese V. Inflammasomes, hormesis, and antioxidants in neuroinflammation: Role of NRLP3 in Alzheimer disease. J Neurosci Res. 2017;95:1360–1372. doi: 10.1002/jnr.23986. [DOI] [PubMed] [Google Scholar]
- 9.Cornelius C, Trovato Salinaro A, Scuto M, Fronte V, Cambria MT, Pennisi M, Bella R, Milone P, Graziano A, Crupi R, et al. Cellular stress response, sirtuins and UCP proteins in Alzheimer disease: Role of vitagenes. Immun Ageing. 2013;10:41. doi: 10.1186/1742-4933-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vivarelli S, Falzone L, Basile MS, Nicolosi D, Genovese C, Libra M, Salmeri M. Benefits of using probiotics as adjuvants in anticancer therapy (Review) World J Acad Sci June. 2019;1:125–135. [Google Scholar]
- 11.Vivarelli S, Salemi R, Candido S, Falzone L, Santagati M, Stefani S, Torino F, Banna GL, Tonini G, Libra M. Gut microbiota and cancer: From pathogenesis to therapy. Cancers (Basel) 2019;11:E38. doi: 10.3390/cancers11010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T, Han Y, Wang J, Hou D, Deng H, Deng YL, Song Z. Comparative epidemiological Investigation of Alzheimer's disease and colorectal cancer: The possible role of gastrointestinal conditions in the pathogenesis of AD. Front Aging Neurosci. 2018;10:176. doi: 10.3389/fnagi.2018.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banna GL, Torino F, Marletta F, Santagati M, Salemi R, Cannarozzo E, Falzone L, Ferraù F, Libra M. Lactobacillus rhamnosus GG: An overview to explore the rationale of its use in cancer. Front Pharmacol. 2017;8:603. doi: 10.3389/fphar.2017.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanza G, Bella R, Cantone M, Pennisi G, Ferri R, Pennisi M. Cognitive impairment and celiac disease: Is transcranial magnetic stimulation a trait d'Union between gut and brain? Int J Mol Sci. 2018;19:E2243. doi: 10.3390/ijms19082243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotakopoulos G, Brotis AG, Kotlia P, Fountas K. Glioblastoma multiforme in a patient with celiac disease: Management of seizures after gross total tumor resection. World Neurosurg. 2018;118:209–211. doi: 10.1016/j.wneu.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Pennisi M, Bramanti A, Cantone M, Pennisi G, Bella R, Lanza G. Neurophysiology of the ‘Celiac Brain’: Disentangling gut-brain connections. Front Neurosci. 2017;11:498. doi: 10.3389/fnins.2017.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bella R, Lanza G, Cantone M, Giuffrida S, Puglisi V, Vinciguerra L, Pennisi M, Ricceri R, D'Agate CC, Malaguarnera G, et al. Effect of a gluten-free diet on cortical excitability in adults with celiac disease. PLoS One. 2015;10:e0129218. doi: 10.1371/journal.pone.0129218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pennisi G, Lanza G, Giuffrida S, Vinciguerra L, Puglisi V, Cantone M, Pennisi M, D'Agate CC, Naso P, Aprile G, et al. Excitability of the motor cortex in de novo patients with celiac disease. PLoS One. 2014;9:e102790. doi: 10.1371/journal.pone.0102790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rapisarda V, Ledda C, Matera S, Fago L, Arrabito G, Falzone L, Marconi A, Libra M, Loreto C. Absence of t(14;18) chromosome translocation in agricultural workers after short-term exposure to pesticides. Mol Med Rep. 2017;15:3379–3382. doi: 10.3892/mmr.2017.6385. [DOI] [PubMed] [Google Scholar]
- 20.Garozzo A, Falzone L, Rapisarda V, Marconi A, Cinà D, Fenga C, Spandidos DA, Libra M. The risk of HCV infection among health-care workers and its association with extrahepatic manifestations (Review) Mol Med Rep. 2017;15:3336–3339. doi: 10.3892/mmr.2017.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenga C, Gangemi S, Di Salvatore V, Falzone L, Libra M. Falzone L amd Libra M: Immunological effects of occupational exposure to lead (Review) Mol Med Rep. 2017;15:3355–3360. doi: 10.3892/mmr.2017.6381. [DOI] [PubMed] [Google Scholar]
- 22.Rapisarda V, Salemi R, Marconi A, Loreto C, Graziano AC, Cardile V, Basile MS, Candido S, Falzone L, Spandidos DA, et al. Fluoro-edenite induces fibulin-3 overexpression in non-malignant human mesothelial cells. Oncol Lett. 2016;12:3363–3367. doi: 10.3892/ol.2016.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falzone L, Marconi A, Loreto C, Franco S, Spandidos DA, Libra M. Occupational exposure to carcinogens: Benzene, pesticides and fibers (Review) Mol Med Rep. 2016;14:4467–4474. doi: 10.3892/mmr.2016.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pennisi M, Malaguarnera G, Puglisi V, Vinciguerra L, Vacante M, Malaguarnera M. Neurotoxicity of acrylamide in exposed workers. Int J Environ Res Public Health. 2013;10:3843–3854. doi: 10.3390/ijerph10093843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alessandria I, Pennisi M, Cataudella E, Frazzetto PM, Malaguarnera M, Rampello L, Rampello L. Neurotoxicity in cadmium-exposed workers. Acta Med Mediter. 2012;28:253–526. [Google Scholar]
- 26.Pennisi M, Lanza G, Cantone M, Schepis C, Ferri R, Barone R, Bella R. Unusual neurological presentation of nevoid basal cell carcinoma syndrome (Gorlin-Goltz syndrome) J Clin Neurol. 2017;13:439–441. doi: 10.3988/jcn.2017.13.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantone M, Lanza G, Pennisi M, Bella R, Schepis C, Siragusa M, Barone R, Ferri R. Prominent neurological involvement in Dercum disease. J Neurol. 2017;264:796–798. doi: 10.1007/s00415-017-8415-1. [DOI] [PubMed] [Google Scholar]
- 28.Uccello M, Vacante M, Giordano M, Malaguarnera M, Biondi A, Basile F, Malaguarnera G, Pennisi M, Motta M. Osteoblastoma of cervical spine causing an unusual neck pain. Eur Rev Med Pharmacol Sci. 2012;16(Suppl 4):17–20. [PubMed] [Google Scholar]
- 29.Sánchez-Valle J, Tejero H, Ibáñez K, Portero JL, Krallinger M, Al-Shahrour F, Tabarés-Seisdedos R, Baudot A, Valencia A. A molecular hypothesis to explain direct and inverse co-morbidities between Alzheimer's disease, glioblastoma and lung cancer. Sci Rep. 2017;7:4474. doi: 10.1038/s41598-017-04400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Ren D, Zhu X, Yin Z, Jin G, Zhao Z, Robinson D, Li X, Wong K, Cui K, et al. Transcriptional signaling pathways inversely regulated in Alzheimer's disease and glioblastoma multiform. Sci Rep. 2013;3:3467. doi: 10.1038/srep03467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrer S. Glioma and Alzheimer's disease. J Alzheimers Dis Rep. 2018;2:213–218. doi: 10.3233/ADR-180084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roe CM, Behrens MI, Xiong C, Miller JP, Morris JC. Alzheimer disease and cancer. Neurology. 2005;64:895–898. doi: 10.1212/01.WNL.0000152889.94785.51. [DOI] [PubMed] [Google Scholar]
- 33.Parsons CG, Danysz W, Dekundy A, Pulte I. Memantine and cholinesterase inhibitors: Complementary mechanisms in the treatment of Alzheimer's disease. Neurotox Res. 2013;24:358–369. doi: 10.1007/s12640-013-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee ShU. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18:3–9. doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis ME. Glioblastoma: Overview of Disease and Treatment. Clin J Oncol Nurs. 2016;20(Suppl 5):S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alphandéry E. Glioblastoma Treatments: An Account of Recent Industrial Developments. Front Pharmacol. 2018;9:879. doi: 10.3389/fphar.2018.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falzone L, Salomone S, Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front Pharmacol. 2018;9:1300. doi: 10.3389/fphar.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paskaš S, Krajnović T, Basile MS, Dunđerović D, Cavalli E, Mangano K, Mammana S, Al-Abed Y, Nicoletti F, Mijatović S, et al. Senescence as a main mechanism of Ritonavir and Ritonavir-NO action against melanoma. Mol Carcinog. 2019 Apr 17; doi: 10.1002/mc.23020. (Epub ahead of print). doi: 10.1002/mc.23020. [DOI] [PubMed] [Google Scholar]
- 39.Paskas S, Mazzon E, Basile MS, Cavalli E, Al-Abed Y, He M, Rakocevic S, Nicoletti F, Mijatovic S, Maksimovic-Ivanic D. Lopinavir-NO, a nitric oxide-releasing HIV protease inhibitor, suppresses the growth of melanoma cells in vitro and in vivo. Invest New Drugs. 2019 Feb 1; doi: 10.1007/s10637-019-00733-3. (Epub ahead of print). doi: 10.1007/s10637-019-00733-3. [DOI] [PubMed] [Google Scholar]
- 40.Basile MS, Mazzon E, Krajnovic T, Draca D, Cavalli E, Al-Abed Y, Bramanti P, Nicoletti F, Mijatovic S, Maksimovic-Ivanic D. Anticancer and differentiation properties of the nitric oxide derivative of lopinavir in human glioblastoma cells. Molecules. 2018;23:E2463. doi: 10.3390/molecules23102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarević M, Mazzon E, Momčilović M, Basile MS, Colletti G, Petralia MC, Bramanti P, Nicoletti F, Miljković Đ. The H2S donor GYY4137 stimulates reactive oxygen species generation in BV2 cells while suppressing the secretion of TNF and nitric oxide. Molecules. 2018;23:E2966. doi: 10.3390/molecules23112966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinciguerra L, Lanza G, Puglisi V, Pennisi M, Cantone M, Bramanti A, Pennisi G, Bella R. Transcranial Doppler ultrasound in vascular cognitive impairment-no dementia. PLoS One. 2019;14:e0216162. doi: 10.1371/journal.pone.0216162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puglisi V, Bramanti A, Lanza G, Cantone M, Vinciguerra L, Pennisi M, Bonanno L, Pennisi G, Bella R. Impaired cerebral haemodynamics in vascular depression: Insights from transcranial doppler ultrasonography. Front Psychiatry. 2018;9:316. doi: 10.3389/fpsyt.2018.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanza G, Bramanti P, Cantone M, Pennisi M, Pennisi G, Bella R. Vascular cognitive impairment through the looking glass of transcranial magnetic stimulation. Behav Neurol. 2017;2017:1421326. doi: 10.1155/2017/1421326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pennisi M, Lanza G, Cantone M, Ricceri R, Spampinato C, Pennisi G, Di Lazzaro V, Bella R. Correlation between motor cortex excitability changes and cognitive impairment in vascular depression: Pathophysiological insights from a longitudinal TMS study. Neural Plast. 2016;2016:8154969. doi: 10.1155/2016/8154969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Concerto C, Lanza G, Cantone M, Pennisi M, Giordano D, Spampinato C, Ricceri R, Pennisi G, Aguglia E, Bella R. Different patterns of cortical excitability in major depression and vascular depression: A transcranial magnetic stimulation study. BMC Psychiatry. 2013;13:300. doi: 10.1186/1471-244X-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bella R, Ferri R, Lanza G, Cantone M, Pennisi M, Puglisi V, Vinciguerra L, Spampinato C, Mazza T, Malaguarnera G, et al. TMS follow-up study in patients with vascular cognitive impairment-no dementia. Neurosci Lett. 2013;534:155–159. doi: 10.1016/j.neulet.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Pennisi G, Ferri R, Lanza G, Cantone M, Pennisi M, Puglisi V, Malaguarnera G, Bella R. Transcranial magnetic stimulation in Alzheimer's disease: A neurophysiological marker of cortical hyperexcitability. J Neural Transm (Vienna) 2011;118:587–598. doi: 10.1007/s00702-010-0554-9. [DOI] [PubMed] [Google Scholar]
- 49.Lanza G, Lanuzza B, Aricò D, Cantone M, Cosentino FII, Bella R, Pennisi G, Ferri R, Pennisi M. Impaired short-term plasticity in restless legs syndrome: A pilot rTMS study. Sleep Med. 2018;46:1–4. doi: 10.1016/j.sleep.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Lanza G, Cantone M, Aricò D, Lanuzza B, Cosentino FII, Paci D, Papotto M, Pennisi M, Bella R, Pennisi G, et al. Clinical and electrophysiological impact of repetitive low-frequency transcranial magnetic stimulation on the sensory-motor network in patients with restless legs syndrome. Ther Adv Neurol Disorder. 2018;11:1756286418759973. doi: 10.1177/1756286418759973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanza G, Cantone M, Lanuzza B, Pennisi M, Bella R, Pennisi G, Ferri R. Distinctive patterns of cortical excitability to transcranial magnetic stimulation in obstructive sleep apnea syndrome, restless legs syndrome, insomnia, and sleep deprivation. Sleep Med Rev. 2015;19:39–50. doi: 10.1016/j.smrv.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Lanza G, Lanuzza B, Aricò D, Cantone M, Cosentino FI, Pennisi M, Bella R, Pennisi G, Ferri R. Direct comparison of cortical excitability to transcranial magnetic stimulation in obstructive sleep apnea syndrome and restless legs syndrome. Sleep Med. 2015;16:138–142. doi: 10.1016/j.sleep.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Cantone M, Bramanti A, Lanza G, Pennisi M, Bramanti P, Pennisi G, Bella R. Cortical Plasticity in Depression. ASN Neuro. 2017;9:1759091417711512. doi: 10.1177/1759091417711512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pennisi M, Lanza G, Cantone M, Ricceri R, Ferri R, D'Agate CC, Pennisi G, Di Lazzaro V, Bella R. Cortical involvement in celiac disease before and after long-term gluten-free diet: A Transcranial Magnetic Stimulation study. PLoS One. 2017;12:e0177560. doi: 10.1371/journal.pone.0177560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanza G, Bella R, Giuffrida S, Cantone M, Pennisi G, Spampinato C, Giordano D, Malaguarnera G, Raggi A, Pennisi M. Preserved transcallosal inhibition to transcranial magnetic stimulation in nondemented elderly patients with leukoaraiosis. BioMed Res Int. 2013;2013:351680. doi: 10.1155/2013/351680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spampinato C, Aguglia E, Concerto C, Pennisi M, Lanza G, Bella R, Cantone M, Pennisi G, Kavasidis I, Giordano D. Transcranial magnetic stimulation in the assessment of motor cortex excitability and treatment of drug-resistant major depression. IEEE Trans Neural Syst Rehabil Eng. 2013;21:391–403. doi: 10.1109/TNSRE.2013.2256432. [DOI] [PubMed] [Google Scholar]
- 57.Presti M, Mazzon E, Basile MS, Petralia MC, Bramanti A, Colletti G, Bramanti P, Nicoletti F, Fagone P. Overexpression of macrophage migration inhibitory factor and functionally-related genes, D-DT, CD74, CD44, CXCR2 and CXCR4, in glioblastoma. Oncol Lett. 2018;16:2881–2886. doi: 10.3892/ol.2018.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mammana S, Fagone P, Cavalli E, Basile MS, Petralia MC, Nicoletti F, Bramanti P, Mazzon E. The role of macrophages in neuroinflammatory and neurodegenerative pathways of Alzheimer's disease, amyotrophic lateral sclerosis, and multiple sclerosis: Pathogenetic cellular effectors and potential therapeutic targets. Int J Mol Sci. 2018;19:E831. doi: 10.3390/ijms19030831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mangano K, Mazzon E, Basile MS, Di Marco R, Bramanti P, Mammana S, Petralia MC, Fagone P, Nicoletti F. Pathogenic role for macrophage migration inhibitory factor in glioblastoma and its targeting with specific inhibitors as novel tailored therapeutic approach. Oncotarget. 2018;9:17951–17970. doi: 10.18632/oncotarget.24885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicoletti F, Mazzon E, Fagone P, Mangano K, Mammana S, Cavalli E, Basile MS, Bramanti P, Scalabrino G, Lange A, et al. Prevention of clinical and histological signs of MOG-induced experimental allergic encephalomyelitis by prolonged treatment with recombinant human EGF. J Neuroimmunol. 2019;332:224–232. doi: 10.1016/j.jneuroim.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 61.Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134:505–512. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maniati MS, Maniati M, Yousefi T, Ahmadi-Ahangar A, Tehrani SS. New insights into the role of microRNAs and long noncoding RNAs in most common neurodegenerative diseases. J Cell Biochem. 2019;120:8908–8918. doi: 10.1002/jcb.28361. [DOI] [PubMed] [Google Scholar]
- 63.Battaglia R, Palini S, Vento ME, La Ferlita A, Lo Faro MJ, Caroppo E, Borzì P, Falzone L, Barbagallo D, Ragusa M, et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci Rep. 2019;9:84. doi: 10.1038/s41598-018-36452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falzone L, Lupo G, Rosa GRM, Crimi S, Anfuso CD, Salemi R, Rapisarda E, Libra M, Candido S. Identification of novel MicroRNAs and their diagnostic and prognostic significance in oral cancer. Cancers (Basel) 2019;11:E610. doi: 10.3390/cancers11050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hafsi S, Candido S, Maestro R, Falzone L, Soua Z, Bonavida B, Spandidos DA, Libra M. Correlation between the overexpression of Yin Yang 1 and the expression levels of miRNAs in Burkitt's lymphoma: A computational study. Oncol Lett. 2016;11:1021–1025. doi: 10.3892/ol.2015.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falzone L, Scola L, Zanghì A, Biondi A, Di Cataldo A, Libra M, Candido S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging (Albany NY) 2018;10:1000–1014. doi: 10.18632/aging.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falzone L, Candido S, Salemi R, Basile MS, Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M, Libra M. Computational identification of microRNAs associated to both epithelial to mesenchymal transition and NGAL/MMP-9 pathways in bladder cancer. Oncotarget. 2016;7:72758–72766. doi: 10.18632/oncotarget.11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falzone L, Romano GL, Salemi R, Bucolo C, Tomasello B, Lupo G, Anfuso CD, Spandidos DA, Libra M, Candido S. Prognostic significance of deregulated microRNAs in uveal melanomas. Mol Med Rep. 2019;19:2599–2610. doi: 10.3892/mmr.2019.9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nixon DW. The inverse relationship between cancer and Alzheimer's disease: A possible mechanism. Curr Alzheimer Res. 2017;14:883–893. doi: 10.2174/1567205014666170216152905. [DOI] [PubMed] [Google Scholar]
- 73.Shafi O. Inverse relationship between Alzheimer's disease and cancer, and other factors contributing to Alzheimer's disease: A systematic review. BMC Neurol. 2016;16:236. doi: 10.1186/s12883-016-0765-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park S, Yu SJ, Cho Y, Balch C, Lee J, Kim YH, Nam S. Network comparison of inflammation in colorectal cancer and Alzheimer's disease. BioMed Res Int. 2015;2015:205247. doi: 10.1155/2015/205247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Godlewski J, Lenart J, Salinska E. MicroRNA in brain pathology: Neurodegeneration the other side of the brain cancer. Noncoding RNA. 2019;5:E20. doi: 10.3390/ncrna5010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ham S, Kim TK, Ryu J, Kim YS, Tang YP, Im HI. Comprehensive MicroRNAome analysis of the relationship between Alzheimer disease and cancer in PSEN double-knockout mice. Int Neurourol J. 2018;22:237–245. doi: 10.5213/inj.1836274.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polo A, Crispo A, Cerino P, Falzone L, Candido S, Giudice A, De Petro G, Ciliberto G, Montella M, Budillon A, et al. Environment and bladder cancer: Molecular analysis by interaction networks. Oncotarget. 2017;8:65240–65252. doi: 10.18632/oncotarget.18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCubrey JA, Fitzgerald TL, Yang LV, Lertpiriyapong K, Steelman LS, Abrams SL, Montalto G, Cervello M, Neri LM, Cocco L, et al. Roles of GSK-3 and microRNAs on epithelial mesenchymal transition and cancer stem cells. Oncotarget. 2017;8:14221–14250. doi: 10.18632/oncotarget.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jesionek-Kupnicka D, Braun M, Trąbska-Kluch B, Czech J, Szybka M, Szymańska B, Kulczycka-Wojdala D, Bieńkowski M, Kordek R, Zawlik I. MiR-21, miR-34a, miR-125b, miR-181d and miR-648 levels inversely correlate with MGMT and TP53 expression in primary glioblastoma patients. Arch Med Sci. 2019;15:504–512. doi: 10.5114/aoms.2017.69374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.ParvizHamidi M, Haddad G, Ostadrahimi S, Ostadrahimi N, Sadeghi S, Fayaz S, Fard-Esfahani P. Circulating miR-26a and miR-21 as biomarkers for glioblastoma multiform. Biotechnol Appl Biochem. 2019;66:261–265. doi: 10.1002/bab.1707. [DOI] [PubMed] [Google Scholar]
- 81.Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res. 2019;38:133. doi: 10.1186/s13046-019-1132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin CY, Kong W, Jiang J, Xu H, Zhao W. miR-7-5p inhibits cell migration and invasion in glioblastoma through targeting SATB1. Oncol Lett. 2019;17:1819–1825. doi: 10.3892/ol.2018.9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi JY, Shin HJ, Bae IH. miR-93-5p suppresses cellular senescence by directly targeting Bcl-w and p21. Biochem Biophys Res Commun. 2018;505:1134–1140. doi: 10.1016/j.bbrc.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 84.Shi L, Yuan Y, Li HY. MicroRNA-139-3p suppresses growth and metastasis of glioblastoma via inhibition of NIN1/RPNI2 binding protein 1 homolog. Eur Rev Med Pharmacol Sci. 2019;23:4264–4274. doi: 10.26355/eurrev_201905_17931. [DOI] [PubMed] [Google Scholar]
- 85.Zeng A, Yin J, Li Y, Li R, Wang Z, Zhou X, Jin X, Shen F, Yan W, You Y. miR-129-5p targets Wnt5a to block PKC/ERK/NF-κB and JNK pathways in glioblastoma. Cell Death Dis. 2018;9:394. doi: 10.1038/s41419-018-0343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu H, Hu Y, Qiu W. Potential mechanisms of microRNA-129-5p in inhibiting cell processes including viability, proliferation, migration and invasiveness of glioblastoma cells U87 through targeting FNDC3B. Biomed Pharmacother. 2017;87:405–411. doi: 10.1016/j.biopha.2016.12.100. [DOI] [PubMed] [Google Scholar]
- 87.Shan ZN, Tian R, Zhang M, Gui ZH, Wu J, Ding M, Zhou XF, He J. miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3. Oncotarget. 2016;7:78813–78826. doi: 10.18632/oncotarget.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Y, Xu J, Xu J, Cheng J, Jiao D, Zhou C, Dai Y, Chen Q. Lower serum levels of miR-29c-3p and miR-19b-3p as biomarkers for Alzheimer's disease. Tohoku J Exp Med. 2017;242:129–136. doi: 10.1620/tjem.242.129. [DOI] [PubMed] [Google Scholar]
- 89.Yang G, Song Y, Zhou X, Deng Y, Liu T, Weng G, Yu D, Pan S. MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep. 2015;12:3081–3088. doi: 10.3892/mmr.2015.3728. [DOI] [PubMed] [Google Scholar]
- 90.Lei X, Lei L, Zhang Z, Zhang Z, Cheng Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer's disease. Int J Clin Exp Pathol. 2015;8:1565–1574. [PMC free article] [PubMed] [Google Scholar]
- 91.Zong Y, Yu P, Cheng H, Wang H, Wang X, Liang C, Zhu H, Qin Y, Qin C. miR-29c regulates NAV3 protein expression in a transgenic mouse model of Alzheimer's disease. Brain Res. 2015;1624:95–102. doi: 10.1016/j.brainres.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, Li Y, Sun J, Wang Q, Sun C, Yan Y, Yu L, Cheng D, An T, Shi C, et al. Tumor-suppressive effects of miR-29c on gliomas. Neuroreport. 2013;24:637–645. doi: 10.1097/WNR.0b013e3283630126. [DOI] [PubMed] [Google Scholar]
- 93.Song K, Yuan Y, Lin Y, Wang YX, Zhou J, Gai QJ, Zhang L, Mao M, Yao XX, Qin Y, et al. ERBB3, IGF1R, and TGFBR2 expression correlate with PDGFR expression in glioblastoma and participate in PDGFR inhibitor resistance of glioblastoma cells. Am J Cancer Res. 2018;8:792–809. [PMC free article] [PubMed] [Google Scholar]
- 94.Salemi R, Falzone L, Madonna G, Polesel J, Cinà D, Mallardo D, Ascierto PA, Libra M, Candido S. MMP-9 as a candidate marker of response to BRAF inhibitors in melanoma patients with BRAFV600E mutation detected in circulating-free DNA. Front Pharmacol. 2018;9:856. doi: 10.3389/fphar.2018.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leonardi GC, Falzone L, Salemi R, Zanghì A, Spandidos DA, Mccubrey JA, Candido S, Libra M. Cutaneous melanoma: From pathogenesis to therapy (Review) Int J Oncol. 2018;52:1071–1080. doi: 10.3892/ijo.2018.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guarneri C, Bevelacqua V, Polesel J, Falzone L, Cannavò PS, Spandidos DA, Malaponte G, Libra M. NF-κB inhibition is associated with OPN/MMP 9 downregulation in cutaneous melanoma. Oncol Rep. 2017;37:737–746. doi: 10.3892/or.2017.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weber GL, Parat MO, Binder ZA, Gallia GL, Riggins GJ. Abrogation of PIK3CA or PIK3R1 reduces proliferation, migration, and invasion in glioblastoma multiforme cells. Oncotarget. 2011;2:833–849. doi: 10.18632/oncotarget.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hardy J. The discovery of Alzheimer-causing mutations in the APP gene and the formulation of the ‘amyloid cascade hypothesis’. FEBS J. 2017;284:1040–1044. doi: 10.1111/febs.14004. [DOI] [PubMed] [Google Scholar]
- 99.Ochalek A, Mihalik B, Avci HX, Chandrasekaran A, Téglási A, Bock I, Giudice ML, Táncos Z, Molnár K, László L, et al. Neurons derived from sporadic Alzheimer's disease iPSCs reveal elevated TAU hyperphosphorylation, increased amyloid levels, and GSK3B activation. Alzheimers Res Ther. 2017;9:90. doi: 10.1186/s13195-017-0317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cadonic C, Sabbir MG, Albensi BC. Mechanisms of mitochondrial dysfunction in Alzheimer's disease. Mol Neurobiol. 2016;53:6078–6090. doi: 10.1007/s12035-015-9515-5. [DOI] [PubMed] [Google Scholar]
- 101.Shinohara M, Tachibana M, Kanekiyo T, Bu G. Role of LRP1 in the pathogenesis of Alzheimer's disease: Evidence from clinical and preclinical studies. J Lipid Res. 2017;58:1267–1281. doi: 10.1194/jlr.R075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Falzone L, Salemi R, Travali S, Scalisi A, McCubrey JA, Candido S, Libra M. MMP-9 overexpression is associated with intragenic hypermethylation of MMP9 gene in melanoma. Aging (Albany NY) 2016;8:933–944. doi: 10.18632/aging.100951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer's disease. Neurobiol Aging. 2011;32:1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Z, He C, Shi JS. Natural products for the treatment of neurodegenerative diseases. Curr Med Chem. 2019 May 27; doi: 10.2174/0929867326666190527120614. (Epub ahead of print). doi: 10.2174/0929867326666190527120614. [DOI] [PubMed] [Google Scholar]
- 105.Lanza G, Centonze SS, Destro G, Vella V, Bellomo M, Pennisi M, Bella R, Ciavardelli D. Comment on ‘Shiatsu as an adjuvant therapy for depression in patients with Alzheimer's disease: A Pilot Study’. J Evid Based Integr Med. 2019;24:2515690X18825105. doi: 10.1177/2515690X18825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lanza G, Centonze SS, Destro G, Vella V, Bellomo M, Pennisi M, Bella R, Ciavardelli D. Shiatsu as an adjuvant therapy for depression in patients with Alzheimer's disease: A pilot study. Complement Ther Med. 2018;38:74–78. doi: 10.1016/j.ctim.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 107.Calabrese V, Dattilo S, Petralia A, Parenti R, Pennisi M, Koverech G, Calabrese V, Graziano A, Monte I, Maiolino L, et al. Analytical approaches to the diagnosis and treatment of aging and aging-related disease: Redox status and proteomics. Free Radic Res. 2015;49:511–524. doi: 10.3109/10715762.2015.1020799. [DOI] [PubMed] [Google Scholar]
- 108.Giordano D, Kavasidis I, Spampinato C, Bella R, Pennisi G, Pennisi M. An integrated computer-controlled system for assisting researchers in cortical excitability studies by using transcranial magnetic stimulation. Comput Methods Programs Biomed. 2012;107:4–15. doi: 10.1016/j.cmpb.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 109.Pennisi G, Ferri R, Cantone M, Lanza G, Pennisi M, Vinciguerra L, Malaguarnera G, Bella R. A review of transcranial magnetic stimulation in vascular dementia. Dement Geriatr Cogn Disord. 2011;31:71–80. doi: 10.1159/000322798. [DOI] [PubMed] [Google Scholar]
- 110.Faro A, Giordano D, Kavasidis I, Pino C, Spampinato C, Cantone GM, Lanza G, Pennisi M. An Interactive Tool for Customizing Clinical Transacranial Magnetic Stimulation (TMS) Experiments. XII Mediterranean Conference on Medical and Biological Engineering and Computing. 2010:200–203. [Google Scholar]
- 111.Faro A, Giordano D, Pennisi M, Scarciofalo G, Spampinato C, Tramontana F. Transcranial Magnetic Stimulation (TMS) to Evaluate and Classify Mental Diseases Using Neural Networks. Conference on Artificial Intelligence in Medicine in Europe AIME Artificial Intelligence in Medicine. 2005:310–314. [Google Scholar]
- 112.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14:712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen L, Zhang W, Yan W, Han L, Zhang K, Shi Z, Zhang J, Wang Y, Li Y, Yu S, et al. The putative tumor suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in glioma. Carcinogenesis. 2012;33:2276–2282. doi: 10.1093/carcin/bgs261. [DOI] [PubMed] [Google Scholar]
- 114.Jones TA, Jeyapalan JN, Forshew T, Tatevossian RG, Lawson AR, Patel SN, Doctor GT, Mumin MA, Picker SR, Phipps KP, et al. Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-κB pathways. Acta Neuropathol Commun. 2015;3:86. doi: 10.1186/s40478-015-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shigemizu D, Akiyama S, Asanomi Y, Boroevich KA, Sharma A, Tsunoda T, Matsukuma K, Ichikawa M, Sudo H, Takizawa S, et al. Risk prediction models for dementia constructed by supervised principal component analysis using miRNA expression data. Commun Biol. 2019;2:77. doi: 10.1038/s42003-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS One. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request. The analyzed datasets are publicly available on the GEO DataSet database.