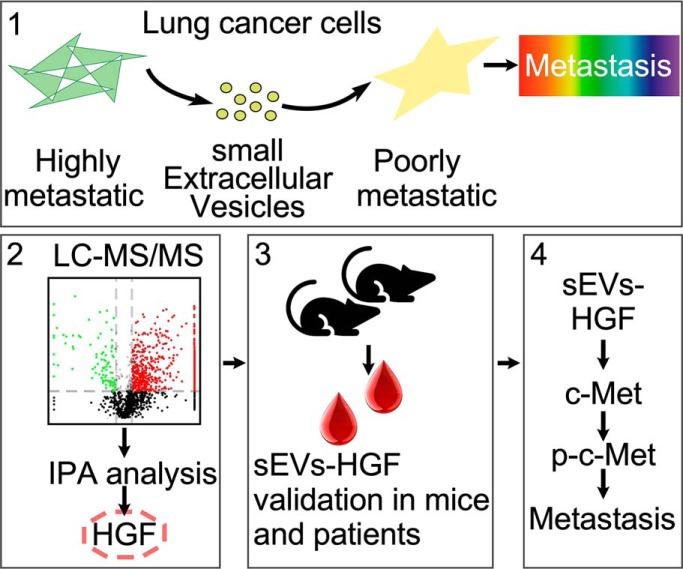
We found that highly metastatic lung cancer cell derived sEVs can regulate the phenotype of recipient cells. With the help of quantitative proteomics, cancer cell metastasis related sEVs proteins were revealed. Through in vitro and in vivo experiments, we found that sEVs-HGF can promote cancer metastasis through activating c-Met pathway. This finding provides insights into how cancer cells send carcinogenic information to remote cells and regulate their fate, which might in turn be targeted in cancer therapy.
Keywords: Exosomes, Lung cancer, Metastasis, Mass Spectrometry, Subcellular analysis, HGF/c-Met pathway, Proteomics
Graphical Abstract
Highlights
Quantitative proteomics analysis of cancer cell derived small extracellular vesicles (sEVs) reveals metastasis related proteins.
HGF/c-Met signaling pathway is mainly activated by cancer cell-secreted sEVs-HGF.
sEVs-HGF plays essential role in the metastasis of cancer cells.
Abstract
Cancer progression is frequently caused by metastasis and leads to significantly increased mortality. Cell derived extracellular vesicles, including exosomes, in the microenvironment play key roles in cellular signal transduction, whereas their biological function in cancer metastasis and progression needs in-depth investigation. Here, we initially demonstrate that the small extracellular vesicles (sEVs) derived from highly metastatic lung cancer cells exhibited great capacity to promote the progression of recipient cells. Quantitative proteomics was employed to comprehensively decipher the proteome of cell derived sEVs and more than 1400 sEVs proteins were identified. Comparison analysis indicates that sEVs-HGF is a potential metastasis related protein and our verification data from clinical lung cancer plasma samples and in vivo experiments further confirmed the association. We found that sEVs-HGF could induce epithelial-mesenchymal transition and the coordination between HGF and c-Met was confirmed through corresponding target knockdown and kinase inhibition. Our data collectively demonstrate that cancer cell derived sEVs contribute to recipient cell metastasis through promoting HGF/c-Met pathway, which are potential targets for the prevention and treatment of cancer metastasis.
Extracellular vesicles (EVs)1 are cell derived vesicles in the microenvironment that include exosomes, microvesicles, ectosomes, etc. EVs can be named by their physical characteristics, such as “small EVs” (sEVs) (<200 nm) and “medium/large EVs” (m/lEVs) (>200 nm) according to MISEV2018 (1). Exosomes are a kind of subtype of EVs and range from 30 to 150 nm in diameter belonging to sEVs (2, 3). Exosomes originating from multivesicular bodies are usually secreted into the extracellular matrix by fusion with cytomembrane and harbor various cargos, including proteins, lipids, and nucleic acids, etc. (4). According to recent studies, exosomes play essential roles in cellular signaling transduction as well as intercellular communication. Studies suggest that exosomes may induce alteration in tumor microenvironment and promote cancer metastasis and progression (5). Exosomal nucleic acids, including miRNA, mRNA, DNA fragments, have been explored for their contribution to cellular immunomodulation, chemotherapy resistance, as well as cancer progression (4–6). Exosomes' proteome has also been studied for their role in biomarker discovery and cancer research by quantitative proteomics (7–9). For instance, comparative proteome research reveals that exosomal proteins of saliva and serum can be used for the diagnostics of multiple cancers, including lung cancer (10, 11).
Lung cancer is the leading cause of cancer death worldwide with top-ranked morbidity and mortality. On diagnosis, its five-year survival rate is only 15.9%, which has not improved for decades (12). The most frequent invasive progression of lung cancer is metastasis, which is one of the major causes of death, including metastasis to the liver, bone, and leptomeninges (5, 13). Cancer metastasis is a complicated process that has frequently been linked to epithelial-mesenchymal transition (EMT) (14). The hallmarks of EMT include loss of epithelial cell adherence and cell polarity and the development of mesenchymal phenotype with increased ability to invade and metastasize (15). Recent studies show that tumor-derived exosomes may serve as a bridge for EMT-initiating signals and deliver amounts of EMT inducers (8, 16). Accordingly, recipient cells have physiological changes associated with increasing of N-cadherin and Vimentin and reducing of E-cadherin, the marks of EMT (17). However, the mechanism of how tumor-derived exosomal proteins induce lung cancer metastasis through EMT has not been thoroughly elucidated.
In the clinic, abnormal c-Met signaling is associated with the poor prognosis, lymph node metastasis, and drug resistance in lung cancer (18, 19). As a transmembrane receptor of hepatocyte growth factor (HGF), c-Met has been found overexpressed in lung cancer, which can only be activated by HGF to promote EMT (20). Silencing of c-Met has been shown to cause decreased viability of cancer cells. Hence, it has become a therapeutic target for cancer treatment (21). Highly metastatic melanoma-derived exosomes could increase the metastatic behavior of primary tumors by permanently “educating” bone marrow progenitors through c-Met (22).
Quantitative proteomics is a robust approach for large-scale proteome analysis and biomarker discovery in biomedical research. We previously demonstrated that mass spectrometry-based proteomics can decipher the proteome of saliva as well as oral epithelium cells (23, 24). In the present study, we aimed to reveal the mechanism of lung cancer cell metastasis mediated by sEVs through quantitative proteomics. sEVs were isolated from highly metastatic and poorly metastatic lung cancer cells and their protein profiling were quantitatively compared with cancer cell metastasis related candidates. We found that HGF was specifically enriched in sEVs of highly metastatic cancer cells, which was the main inducer to active c-Met signaling in recipient cells. In addition, verification of sEVs-HGF in the plasma of lung cancer patients demonstrated that cell-secreted sEVs-HGF could be a promising indicator for the progression and metastasis of lung cancer.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
To reveal the mechanism of how sEVs proteins promote the recipient cells' metastasis, the proteomes of highly metastatic lung cancer cells 95D and poorly metastatic lung cancer cells 95C derived sEVs were analyzed and compared through label free quantification. The expression levels of confidently identified proteins were compared using two-sided Student's t test with fold change > 2 and p value< 0.01. Metastasis related protein was selected for further verification in clinical samples as well as in vivo experiments. The potential mechanism was further tested in vitro. Student's t test was applied to evaluate the significant difference between groups and p < 0.05 was considered as significant.
Cell Culture and Lentivirus Package
Human lung carcinoma cell line A549, 95C, and 95D, were cultured in RPMI 1640. 95C and 95D cell lines were subcloned from a poorly differentiated human large-cell lung carcinoma cell line PLA-801, which was of different metastatic potential (25, 26). Human lung fibroblast cell line MRC-5 was cultured in MEM medium. All cell lines were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China) and cultured at 37 °C in a 5% CO2 incubator. All culture media were from Thermo Fisher Scientific (Waltham, MA) and supplemented with 10% sEV-free FBS (Gibco, Grand Island, NY, ultracentrifugation at 110,000 × g for 16 h), 100 U/ml penicillin, and 100 mg/ml streptomycin sulfate. All cultured cells were tested for mycoplasma contamination before use.
Lentivirus packaging was performed according to previously published work (27). Briefly, target sequences (TCGAGGTCTCATGGATCATAC#1 and CCCGTAATATCTTGTGCCAAA#2) against human HGF and a control scrambled sequence (GCGCGCTTTGTAGGATTCGTT) that has no significant homology with the human genome were inserted into the pLKO.1 vector, according to the manufacturer's protocol (Addgene, MA). Lentiviral particles were transferred to 95D cells for further assay.
sEV Isolation and Sucrose Gradient Ultracentrifugation
About 200 ml conditioned medium was harvested from cultured cells (5 × 107 cells for 48 h). All sample preparation was performed at 4 °C. sEVs were isolated by differential centrifugation at 300 × g for 10 min, 2000 × g for 10 min, 10,000 × g for 10 min, and 110,000 × g for 70 min. Equal volume of cold PBS in the last step was used to wash the pellet once. The pellets were re-suspended in 50 μl PBS with Proteinase Inhibitor Mixture (Roche Applied Science, Basel, Switzerland) and then stored at −80 °C for subsequent experiments. For validation of HGF and CD81, the pellets were resuspended in 1 ml PBS and loaded to sucrose solution with the gradient from 20% to 65% (low concentration to high concentration, 1 ml per layer was injected in that order), which was centrifuged at 180,000 × g overnight. Ten fractions were collected from the second layer to the bottom and diluted to 6 ml with cold PBS. They were then centrifuged at 100,000 × g for 70 min.
Transmission Electron Microscopy
sEVs were imaged by Philips CM120 transmission electron microscope (Eindhoven, Netherlands), which operated at 120 kV. 10 μl of purified sEVs were loaded onto an ultrathin carbon film 300 mesh copper grid, dried for fixation, and stained with 2% phosphotungstic acid (PTA).
Nanoparticle Tracking Analysis
NanoSight NS300 (Malvern, UK) was used for measuring the number and size of sEVs. Purified sEVs were resuspended in PBS and injected into the sample chamber. Each sample was measured three times.
Quantitative Real-time PCR and siRNA-mediated Knockdown
Total RNA was isolated by the TRIzol reagent (Invitrogen, Carlsbad, CA). RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) was used for reverse transcription. Detection was performed in the StepOnePlus Real-Time PCR System (Life Science, Carlsbad) with Power Up SYBR Green Master Mix (Thermo Fisher Scientific). Absolute mRNA copy numbers were normalized to reference gene GAPDH. Relative fold expression values were determined according to the ΔΔCt method. siRNA was transfected into 95D cells using RNAi-Mate (GenePharma, Shanghai, China) according to the manufacturer's protocol. The primers are presented in supplemental Table S1.
LC-MS/MS
sEVs isolated from 95C and 95D cells through ultracentrifugation were dissolved in RIPA buffer, and sEVs proteins were extracted and then digested overnight at 37 °C by trypsin (Promega, Madison, WI) using the FASP approach (28). Briefly, 30 μg sEVs proteins were used and three biological repeats of each treatment were prepared. Equal amounts of peptides (measured by Nanodrop, A280) were injected into Easy-nLC 1000 coupled with a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Peptides were eluted to analytical column (75 μm × 15 cm) packed with Jupiter Proteo resin (3 μm, C18, 300 Å, Phenomenex, Torrance, CA). The mobile phase consisted of buffer A (2% acetonitrile and 0.1% formic acid in water) and buffer B (0.1% formic acid in 95% acetonitrile). A flow rate of 250 nL/min and 60 min of the gradient from 12% B to 32% B was applied for the separation of peptides. MS Scan range was from 300 to 1600 m/z with the resolution of 70,000. For MS/MS, scan range was from 200 to 2000 m/z with the resolution of 17,500. The raw data were submitted for database search by MaxQuant (version 1.6.1.0) software against with the UniProt Human database (release 2017_11_28, 20244 entries). The maximum of missed cleavages was set as two, and trypsin was selected for protein digestion. Oxidation of modifications and acetylation of protein N termini were considered as variable modifications. The mass tolerance values for precursor and fragment ions were 6 ppm and 0.5 Da, respectively. Both maximum peptide and protein false discovery rates (FDR) were limited to 1%. Label-free quantification was performed by intensity-based absolute quantification (iBAQ), which was based on at least two unique peptides to quantify the different protein profiling in the exosomes derived from 95C cells and 95D cells. Quantile normalization was performed to ensure that each sample had the same distribution, the 2-fold change and p < 0.01 cut-off was set up for the screening of differentially expressed proteins.
Cell Migration Assay
The migration ability of cells was tested in a Transwell Permeable Support (Corning, NY) with polycarbonate membranes (8 μm pore size). The membrane was coated with Matrigel matrix (Corning) in advance. Cells were cocultured with isolated sEVs for different times in advance. Cells suspended in serum-free medium were seeded into the upper chamber and 10% fetal bovine serum was added to the lower compartment. Finally, cells that moved through the membrane were fixed with 100% methyl alcohol at −20 °C for 20 min and stained with 0.1% crystal violet solution. The image was captured by photomicroscope (Canon, Tokyo, Japan).
Western Blotting
Antibodies were all purchased from Abcam (Cambridge, MA) and listed as follows: TSG101, CD63, CD81, E-cadherin, N-cadherin, Vimentin, HGF, c-Met, p-c-Met, Granulin, Calnexin, GRP78, Elastin, and GAPDH. ECL (SuperSignal West Pico, Thermo) was used for visualization.
ELISA Assay
The proteins in patients' plasma and the supernatant of cells were measured with human HGF ELISA kit (eBioscience, Thermo scientific) following the manufacturer's instructions.
In Vivo Metastasis Assays in Mice Xenograft Model
NOD-SCID mice were purchased from Vital River Laboratory. 1 × 106 (volume 0.1 ml) 95C or 95D cells were intravenously injected into female NOD-SCID mice (6–7 weeks old). The cell-injected animals were housed under sterile conditions at Experimental Animal Facility of Shanghai Jiao Tong University (Shanghai, China). The percentage of cancer cells was monitored using human-CD47 antibody (R&D, Minneapolis, MN) by flow cytometry. Animal care and sacrifice were conducted according to methods approved by the Animal Care and Use Committee, the Center for Animal Experiments of Shanghai Jiao Tong University.
Coimmunoprecipitation Assay
Antibodies of HGF and Granulin were incubated with sEVs lysates from 95D cells or 95D cell lysates to precipitate target proteins using Protein A/G Magnetic Beads (Thermo Fisher Scientific) according to the manufacturer's instructions. Precipitated proteins were analyzed by LC-MS/MS.
Cell Proliferation and Cell Cycle Assays
1 × 103 cells were seeded into 96-well plates and sEVs were added for coculture. Cell proliferation was assessed at 1–6 days. Twenty microliters MTS assay (Promega) reagent was added into each well and incubated at 37 °C for 4 h. According to the manufacture's protocol, cell cycle was measured using APC-BrdU Flow Kit (BD Pharmingen, San Diego, CA). Cells staining with fluorochromes was acquired using flow cytometer and acquired data were analyzed by FlowJo software.
Plasma Samples Collection
Normal subjects and lung cancer patients at different stages were recruited for this study. Plasma samples were collected at the Shanghai Chest Hospital between 2016 and 2017, according to approved protocols (IRB#M15017) by Institutional Review Board (IRB) of Shanghai Jiao Tong University. All subjects provided written informed consent. Briefly, venous blood was collected to an evacuated blood collection tube with EDTA and left to stand for 30 mins. Samples were centrifuged for 30 mins at 4000 × g to remove all cell debris and platelets and then stored at −80 °C. All experimental protocols were approved by Bio-X Ethics Committee of Shanghai Jiao Tong University. Patient demographics and detailed clinical information are presented in supplemental Table S2.
Bioinformatics Analysis
Omicsbean (http://www.omicsbean.com/) was used for gene ontology (GO) analysis of identified proteins, including their cellular component, molecular function, and biological process. Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, http://www.ingenuity.com/) was used for analyzing “Canonical pathways,” “Upstream analysis,” “Diseases & Functions,” and “Networks.”
Statistical Analysis
The experimental results were presented as the mean ± S.D. The comparisons of quantitative data between two groups were assessed using the Student's t test, and p < 0.05 was considered as statistical significance. The Cox proportional hazards model was used to analyze overall survival (OS), and the Kaplan-Meier method was used for the construction of survival curves for OS.
RESULTS
sEVs Derived from Highly Metastatic Lung Cancer Cells Promote the Proliferation and Migration of Recipient Cells
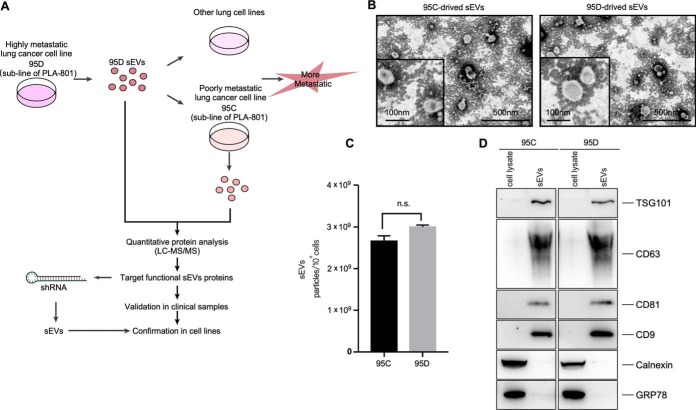
sEVs secretion is an essential way for tumors to induce systemic changes, whereas how they influence and regulate the behavior of recipient cells is still unclear (5). In the present work, the highly metastatic lung cancer cell line 95D and poorly metastatic lung cancer cell line 95C as well as normal lung cell line MRC-5 were selected as models for discovery (Fig. 1A) (25). We first isolated sEVs from 95C and 95D cell culture media through ultracentrifugation, which were then comprehensively characterized. The sEVs released by 95C and 95D cells were both around 100 nm (Fig. 1B). The secreted amount of sEVs by 95C and 95D cells was similar (Fig. 1C). Exosome representative markers, including TSG101, CD63, and CD81, were specifically enriched in these isolated sEVs (Fig. 1D), whereas negative markers were negligible, including GRP78 and Calnexin. These results demonstrated that sEVs were successfully isolated, most of which were exosomes.
Fig. 1.
Characterization of sEVs secreted by 95C cells and 95D cells. A, Workflow for dissecting sEVs functions through proteomics; B, Transmission electron microscopy of 95C-sEVs and 95D-sEVs; C, Nanoparticle tracking analysis of purified sEVs secreted by 95C cells and 95D cells; D, Western blot assay of maker proteins for 95C-sEVs and 95D-sEVs.
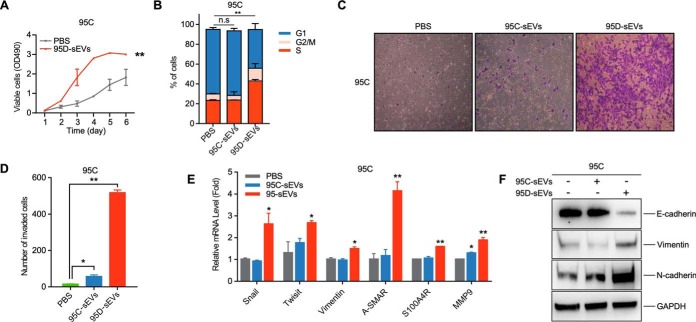
Tumor cells can release differentially expressed proteins into the microenvironment. Because sEVs can readily be absorbed by recipient cells (29), we wondered whether oncogenic phenotypes could be transferred from more aggressive cells to indolent poorly metastatic cells. We performed in vitro experiments by incubating 95C or MRC-5 cells with sEVs derived from 95D cells. When compared with PBS control, 95C-derived sEVs (95C-sEVs) slightly improved the proliferation of 95C, MRC-5, and 95D cells (supplemental Fig. S1A and S1D). Of interest, 95D derived sEVs (95D-sEVs) dramatically enhanced the proliferation of 95C, 95D, and MRC-5 cells (Fig. 2A and supplemental Fig. S1A and S1D). We then analyzed the cell cycle distribution of these treated cells by flow cytometry. As expected, the percentage of 95C cells and MRC-5 cells in S-phase significantly increased after the treatment with 95D-sEVs (Fig. 2B and supplemental Fig. S1B). For 95C-sEVs treated cells, however, there was no dramatic difference (Fig. 2B and supplemental Fig. S1B).
Fig. 2.
sEVs secreted from 95D cells regulate the proliferation and the metastasis of recipient cells. A, MTS assay on cell viability of 95C cells treated with 10 μg/mL 95D-sEVs, and blank PBS, respectively. B, Flow cytometry on cell cycle profiling of the 95C cells treated with 10 μg/mL 95C-sEVs, 10 μg/mL 95D-sEVs, and blank PBS for 48 h, respectively. The percentages of cells in G1, S and G2/M are show by stacked bar plot. C, D, Migration assays of 95C cells treated for 72 h with 10 μg/mL 95C-sEVs, 10 μg/mL 95D-sEVs, or blank PBS, respectively. The number of migrated cells shows in the histogram. E, F, Gene expression of key EMT markers in 95C treated with 10 μg/mL 95C-sEVs, 10 μg/mL 95D-sEVs or blank PBS were detected by qRT-PCR analysis and western blot. Each experiment was performed in triplicates and results are presented as mean ±S.D. (*p < 0.05; **p < 0.01; ***p < 0.001).
We also assessed the effects of sEVs on the migration ability of recipient cells. When compared with the PBS control, migration assay data showed that 3.60 times more 95C cells, 1.50 times more A549 cells, and 1.55 times more MRC-5 cells migrated across the membrane after the 95C-sEVs treatment. After incubation with 95D-sEVs, 32 times more 95C cells, 5.55 times more A549 cells, and 3.36 times more MRC-5 cells crossed the membrane (Fig. 2C and 2D, and supplemental Fig. S1C). There was minor increase when 95C and 95D cells were treated with 95C-sEVs and 95D-sEVs, respectively (Fig. 2C and supplemental Fig. S1E). We further assessed the expression of EMT markers in recipient cells because EMT plays essential roles in all stages of cancer progression (30). After 95D-sEVs treatment, the relative mRNA level of Snail, Twisit, Vimentin, A-SMAR, S100A4R, MMP9, and the protein expression of Vimentin and N-cadherin were notably increased in 95C cells; the expression of E-cadherin was dramatically decreased (Fig. 2E and 2F). These results indicated that sEVs from highly metastatic lung cancer cells strongly promoted the proliferation and migration of recipient cells and regulated the cell cycle and gene expression that related to migration.
Cancer Cell Metastasis-related sEVs Protein Identification by LC-MS/MS
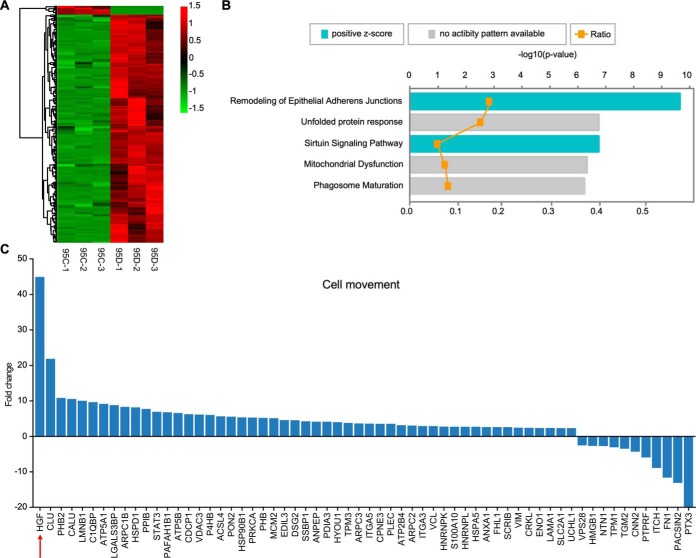
The apparent sEVs-mediated intercellular transfer of phenotype raises the question of sEVs proteins in promoting cancer cell metastasis. To address this question, quantitative proteomics was used to compare the sEVs proteome derived from highly metastatic and poorly metastatic lung cancer cells. The proteome data of three biological replicates had good reproducibility, as principal component analysis (PCA) showed that PC2 was 1.5%. Besides, the PC1 (97.3%) significantly separated 95D-sEVs from 95C-sEVs (supplemental Fig. S2A). In total, 1484 proteins were identified with at least 2 unique peptides and less than 1% FDR. Among them, 1411 proteins were derived from 95D-sEVs, and 989 proteins belonged to 95C-sEVs, with 61.7% overlap (supplemental Fig. S2B and supplemental Table S3–S5). A volcano plot illustrated their differential protein abundance (expressed as the mean ratio of 95D-sEVs/95C-sEVs, n = 3) (supplemental Fig. S2C). GO analysis indicated that the cellular component and biological function of these proteins were similar between 95C-sEVs and 95D-sEVs (supplemental Fig. S2D and S2E). Among them, 268 proteins (18.1% of the proteome) with significant changes (p < 0.01 and fold change >2 or <0.5, supplemental Table S6) were analyzed and their abundance was presented as a heatmap in Fig. 3A. Interestingly, 259 of 268 proteins were upregulated in 95D-sEVs, whereas only 9 of them were downregulated.
Fig. 3.
Quantitative proteomics analysis of sEVs from 95C cells and 95D cells. A, The Heatmap of 268 differentially expressed sEVs proteins. B, IPA analysis of “Canonical pathway” on datasets obtained from distinction protein. C, The fold change distribution of these 62 sEVs proteins that related to “cell movement” by IPA. The red arrow marks the most changed protein HGF.
IPA analysis was performed to analyze these 268 proteins and found vital sEVs proteins that related to cancer cell metastasis. The canonical pathway enrichment analysis highlighted that remodeling of epithelial adherent junctions was the most significant pathway (Fig. 3B). The cell movement and migration was the most potential downstream effect predicted by disease and functional enrichment analysis (p value = 5.98e−07 and activation z-score = 3.93). We detected sEV proteins enriched in cell movement, cell spreading of epithelial cells, migration of tumor cells, as well as invasion of lung cancer (supplemental Table S7). Moreover, 62 proteins were significantly enriched in cell movement.
We then focused on these 62 enriched proteins. Among them, HGF was the most notably upregulated one (fold change was 44.65 times with p value of 5.4e−06) (Fig. 3C and supplemental Fig. S2F). HGF can induce EMT through canonical c-Met pathways, resulting in cell migration and proliferation, which is the intercellular junction between polarized intestinal and kidney epithelium (31). Therefore, we focused on sEVs-HGF and its contribution to cancer cell metastasis. The expression level of HGF was measured both in vivo and in vitro.
Confirmation of sEVs-HGF in Mouse Model and Lung Cancer Sample Set
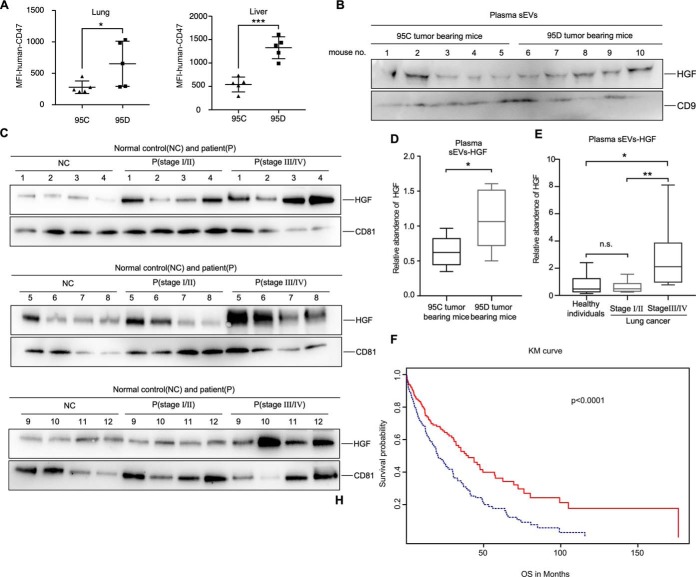
We evaluated the aggressiveness of 95C and 95D cells in vivo through NOD-SCID mice intravenously. CD47 is a transmembrane protein often expressed on tumor cells (32). Staining of 95C and 95D cells with human-CD47 antibodies for flow cytometric analysis allows determination of human lung cancer cell populations (supplemental Fig. S3A), whereas the cells from mice organs cannot be labeled with human-CD47 antibodies (supplemental Fig. S3A). In contrast to 95C cells from injected mice, the tumor in 95D cells in injected mice grew and spread rapidly into lung, liver, spleen, vertebral column, and shoulder (Fig. 4A and supplemental Fig. S3B–S3H). The number of metastatic tumors significantly increased in the mice injected with 95D cells when compared with those injected with 95C cells (supplemental Fig. S3B, S3C, S3E and S3H). Mean fluorescence intensity (MFI) of CD47 on tumor cells indicated that 95D cells were more aggressive than 95C cells in vivo (Fig. 4A and supplemental Fig. S3G). Plasma sEVs were isolated from 95C tumor bearing mice (n = 5) and 95D tumor bearing mice (n = 5) through ultracentrifugation. HGF was increased in the plasma sEVs of 95D tumor bearing mice (p = 0.0235) (Fig. 4B and 4D).
Fig. 4.
Validation of HGF in clinical sEVs samples. A, MFI of human-CD47 in lung and liver tissues of 95C tumor bearing mice and 95D tumor bearing mice. B, Western blot of HGF in the plasma sEVs from 95C tumor bearing mice and 95D tumor bearing mice. CD9 was used as the internal control. The corresponding quantification data are shown in (D). C, Western blot of HGF in the plasma sEVs from normasl control subjects and lung cancer patients at different stages. CD81 was used as the internal control. The corresponding quantification data are shown in (E). F, Patients were divided into two groups according to the expression values of HGF. Red line indicates patients with HGF RPKM < 1.12 and blue line indicates patients with HGF RPKM >1.12. Results are presented as mean ± S.D. (*p < 0.05; **p < 0.01; ***p < 0.001).
Although HGF is usually upregulated in many cancers (33), few studies focused on HGF delivered by sEVs. To check the presence of sEVs-HGF in human plasma, a clinical sample set (including plasma samples from 12 normal control subjects, 12 lung cancer patients at stageI/II, and 12 lung cancer patients at stage III/IV) was used for confirmation. There was no significant difference between early stage (stageI/II) and normal control (p = 0.5014) (Fig. 4C and 4E). Surprisingly, HGF was specifically enriched in the plasma sEVs of late stage cancer patients (stage III/IV) (Fig. 4C and 4E). There was a significant difference between late stage and early stage or normal control (stage III/IV versus normal control, p = 0.0111; stage III/IV versus stageI/II, p = 0.0051). Another clinical sample set (including plasma samples from 5 normal control subjects, 5 lung cancer patients at stageI/II, and 5 lung cancer patients at stage III/IV) was used for confirmation of plasma HGF by ELISA. There was no significant difference among normal control, early stage (stage I/II) and late stage cancer patients (stage III/IV) (supplemental Fig. 4I). The ROC curve for sEVs-HGF in Fig. 4C and 4E is shown in supplemental Fig. S3J.
We analyzed the expression of HGF in patients with NSCLC from TCGA (https://tcga-data.nci.nih.gov/) for their OS. The number of observed events were 320 cases, with the median OS of 14.9 months. The average expression of HGF in the 247 early stage patients was RPKM = 1.29 (Reads Per Kilobase per Million mapped reads), whereas it climbed to RPKM = 3.15 in the 73 late stage patients. Fig. 4F shows the Kaplan-Meier (Km) estimates for OS of all cases. With the HGF cutoff of RPKM = 1.12, the OS of lung cancer patients decreased dramatically (p < 0.0001) if HGF level was upregulated.
These results suggest that sEVs-HGF from plasma is consistent with the expression level of HGF in tissues, which is a promising indicator for late stage lung cancer or poor prognosis.
sEVs-HGF Might Contribute to the Metastasis of Lung Cancer Cells Through c-Met
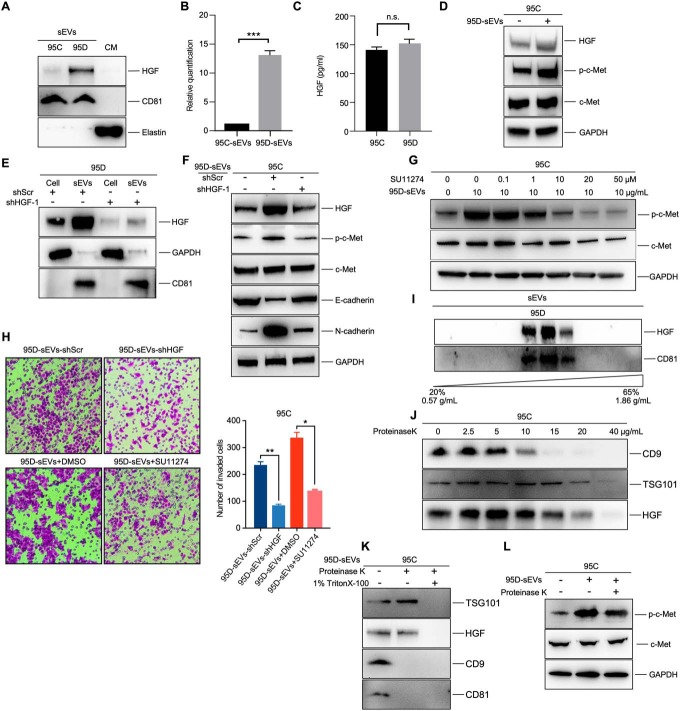
We then measured the HGF expression in 95C-sEVs and 95D-sEVs by Western blotting. The results showed that HGF was significantly upregulated in the 95D-sEVs and 13.5 times higher than that of 95C-sEVs, which was consistent with our LC-MS/MS results (Fig. 5A and 5B). Elastin is a kind of extracellular matrix protein that worked as the negative control to exclude sEVs-HGF with extracellular matrix. ELISA assay showed that there was no significant difference in the culture media between 95C and 95D (Fig. 5C). This was consistent with the intracellular expression of HGF between the 95C and 95D cells by Western blotting (supplemental Fig. S4A and S4B).
Fig. 5.
sEVs-HGF actives c-Met signaling in recipient cells. A, B, Western blot of HGF protein level in 95C-sEVs and 95D-sEVs. Elastin was used as the negative control for intercellular matrix. Histogram shows the corresponding quantification data. C, ELISA assay of HGF in the culture media from 95C and 95D cells. D, Western blot of c-Met and p-c-Met in recipient cells treated with 10 μg/mL 95D-sEVs or PBS for 3 h. E, Effect of HGF knockdown in 95D-sEVs and cells measured by western blot. F, Target protein expression of 95C cells treated with 10 μg/mL 95D-sEVs-shScr, 10 μg/mL 95D-sEVs-shHGF, or blank PBS for 3 h. G, The phosphorylation of c-Met in 95C cells treated with 95D-sEVs and inhibitor SU11274 were detected by western blot. H, Migration assays of 95C cells treated with 10 μg/mL 95D-sEVs-shScr, 10 μg/mL 95D-sEVs-shHGF, 10 μg/mL 95D-sEVs +DMSO, or 10 μg/mL 95D-sEVs + 20 μM SU11274. The number of migrated cells shows in the right histogram. I, Western blot of HGF protein level in the sEVs, which were purified by sucrose density gradient centrifugation. J, Western blot of HGF and indicated protein markers in 95D-sEVs treated with proteinase K for 30 min at 4 °C. K, Western blot of HGF and indicated protein markers in 95D-sEVs treated with proteinase K for 30 min at 4 °C with or without 1% Triton X-100. L, The phosphorylation of c-Met in 95C cells treated with 10 μg/mL 95D-sEVs or 10 μg/mL protease-treated 95D-sEVs were detected by western blot. Each experiment was performed in triplicates and results are presented as mean ± S.D. (*p < 0.05; **p < 0.01; ***p < 0.
HGF/c-Met pathway is involved in cell invasion, proliferation, and angiogenesis (13). Therefore, we assessed the effects of 95D-sEVs secreted HGF on the activation of downstream c-Met signaling. As expected, phosphorylation of c-Met (p-c-Met) was increased in 95C cells after the treatment of 95D-sEVs (Fig. 5D). To reveal the specific requirement for HGF in inducing cancer cell metastasis, we performed a knockdown of the HGF gene in 95D cells with lentivirus short hairpin RNA (shRNA), and the levels of HGF in 95D cells and 95D-sEVs were both decreased (Fig. 5E and supplemental Fig. S4C). Then 95C cells were cocultured with sEVs from 95D cells infected with shScramble (95D-sEVs-shScr) or shHGF (95D-sEVs-shHGF), respectively. The results showed that 95D-sEVs-shHGF had no significant induction of migration on recipient cells when compared with 95D-sEVs-shScr (Fig. 5F). We further verified the necessity of 95D-sEVs in activation of c-Met signaling through using the kinase inhibitor SU11274 of c-Met (34). Inhibition of c-Met led to a significant decrease in inducing the phosphorylation of c-Met by 95D-sEVs (Fig. 5G). Further, transwell assay revealed that either knockdown of HGF or inhibition of phosphorylation of c-Met could dramatically reduce 95D-sEVs' effect on cell invasion and migration (Fig. 5H). 95D cells were co-cultured with 95D-sEVs or 95D-sEVs-shHGF, respectively. The results showed that 95D-sEVs could increase the proliferation of 95D cells, which was HGF-dependent (supplemental Fig. S1D). However, transwell assay revealed that 95D-sEVs had no significant influence on the migration ability of itself (supplemental Fig. S1E).
We then sought to explore the potential mechanism behind the activation of c-Met by sEVs-HGF. HGF is a secretory protein, which binds to receptors and modulates intracellular signaling cascades (35). It is necessary to confirm whether this protein is packed inside or on the surface of the sEVs. Continuous sucrose gradient centrifugation verified that HGF was only presented in 40–50% fractions, which was consistent with the presence of exosomal marker CD81 (FIG. 5I). We used proteinase K to digest the proteins on the surface of the sEVs to exclude the HGF adhered to the sEVs' surface. Different concentration of proteinase K was added into sEVs to determine the critical concentration by measuring sEVs surface molecules CD9 and internal protein constituents TSG101 (36). When CD9 was almost degraded while TSG101 was intact, it indicated that the concentration of 15 μg/ml was the critical point to remove the adherent HGF completely (Fig. 5J). The sEVs' membrane was disrupted by the addition of detergent allowing the proteinase K to enter, which did not degrade HGF in the absence of detergent (Fig. 5K). We found that protease-treated 95D-sEVs could still activate c-Met (Fig. 5L), confirming that HGF, identified by LC-MS/MS, was coming from sEVs instead of extracellular matrix. Taken together, these results revealed that HGF was residing within 95D-sEVs, which was resistant to protease. sEVs-HGF is one of the major sources of HGF and plays a critical role in regulating the invasion of recipient cells through HGF/c-MET pathway.
We preliminarily explored the packaging mechanism of HGF into sEVs. Immunoprecipitation and LC-MS/MS were coupled to reveal proteins that may physically interact with HGF in the lysate of 95D-sEVs (supplemental Fig. S4D). Biological repeats and corresponding control samples were processed and analyzed. Fifteen proteins were revealed as candidate HGF-interaction proteins and 7 of them were identified in our sEVs proteome experiments, including Granulin, EEF2, PRDX1, RPL13, RPL18, HSPA8, and RPS3 (supplemental Table S9). The fold change of Granulin between 95D-sEVs and 95C-sEVs was 5.5 (p < 0.05), the highest one in these 7 proteins. Granulin is a secreted glycoprotein that mediates wound healing (37). Co-IP assays of 95D-sEVs validated that HGF and Granulin may have interaction (supplemental Fig. S4F). However, the interaction was not observed in 95D cells (supplemental Fig. S4G). To verify the specific role of Granulin in HGF enrichment, 95D cells were transfected with siRNA targeting Granulin, which significantly decreased the expression of HGF in 95D-sEVs (supplemental Fig. S4H). The nanoparticle tracking analysis showed that the knockdown of Granulin could reduce the release of 95D-sEVs (supplemental Fig. S4I). These results suggest that the packaging of HGF may be related to Granulin.
DISCUSSION
Growing evidence has tightly associated sEVs, including exosomes, with cancer metastasis and progression. For instance, exosomal miR-1247–3p secreted by high-metastatic Hepatocellular Carcinoma cells could activate β1-integrin-NF-κB signaling to promote secretion of pro-inflammatory cytokines, resulting in cancer progression (5). Recently, exosomal proteins are the subject of interests under intense investigation. Of note is that exosomal integrins are associated with cancer metastasis and different protein isoforms of integrins may be used for predicting organ-specific metastasis (38). Therefore, it is necessary to reveal the potential mechanism behind cancer cell metastasis that mediated by sEVs proteins.
In the present study, we found that sEVs released by highly metastatic cells could increase the percentage of S-phase in recipient cells to enhance cell proliferation. 95D-sEVs could reinforce cell invasion and migration by promoting EMT signaling. Quantitative proteomics was used to discover critical sEVs proteins in promoting proliferation and migration of recipient cells. IPA analysis revealed that these 268 altered proteins were significantly associated with remodeling of epithelial adherens junctions, which is consistent with the expectation that differential signaling pathway exists in 95D-sEVs and associates with epithelial morphology modulating. We found that these differential proteins also had significant association with different cancers, including breast cancer (p = 3.6e−11), ovarian cancer (p = 1.26e−08), and lung cancer (p = 3.8e−08) (supplemental Table S8). Notably, 62 proteins were involved in increasing “cell movement” (supplemental Table S7), among which HGF had the highest fold change.
We noted that HGF is an attractive molecule, not only because HGF was significantly upregulated in our results, but also because of its intimate link to cancer cell metastasis, which has been used as a biomarker for prognosis of liver cancer, breast cancer, and prostatic carcinoma (39, 40). For instance, paracrine HGF upregulated by cancer derived exosomal EGFR could facilitate the proliferation of metastatic cancer cells (13). Although cellular HGF has been reported (41), the role of secreted sEVs-HGF in the process of cancer cell metastasis requires in-depth investigation. Further, HGF is a regulator of EMT, which regulates the activation of c-Met by increasing the phosphorylation level of c-Met (13), subsequently active Ras-Raf-MEK-ERK1/2 kinase cascade (42), leading to the translocation of ERK1/2 into nuclei and expression of Snail (43), a transcriptional repressor involved in EMT. Snail represses the expression of E-cadherin and increases the expression of N-cadherin, followed by losing cell-cell adhesion, dissociation of epithelial structures, and increasing cell motility and tumor invasion (43, 44). Especially, we found that 95D-sEVs can transport HGF into the recipient cells, which might eventually initiate the activation of c-Met. We revealed that the HGF/c-Met signaling pathway was mainly activated by cancer cell-secreted sEVs-HGF. The mechanism of sEVs-HGF derived from 95D-sEVs affect the proliferation of 95D cells is still unclear (supplemental Fig. S1D), which require further investigation. The uptake of sEVs is accomplished via endocytosis, receptor-ligand interaction, or by direct fusion, which is dependent on the pH of the microenvironment (45). Herein we proved that it is the HGF inside the sEVs that can activate c-Met in recipient cells. Further research is needed to determine the role of sEVs in activating HGF/c-Met pathway, how 95D-sEVs are absorbed by the recipient cells, and how their sEVs-HGF is released and activates c-Met inside the recipient cells.
The HGF level in plasma sEVs was significantly increased in late/metastatic stages of NSCLC patients, which is consistent with our in vivo results. The ELISA data of soluble HGF in plasma indicated that there is no significant difference between normal group and lung cancer groups at different stages. Because our verification data are only based on a small sample set, this result, however, does not mean the plasma HGF is irrelevant with the progression of lung cancer. A large sample set is needed to verify the sensitivity and specificity of plasma sEVs-HGF and soluble HGF in differentiating late stage NSCLC from early stage NSCLC and normal control group. The finding preliminarily indicated that HGF from plasma sEVs was correlated to carcinoma deterioration, which is a promising indicator of initial cancer progression or a prognostic marker for cancer treatment.
The mechanisms for sorting proteins into sEVs are still poorly understood. The endosomal sorting complex required for transport (ESCRT) machinery is critical for budding and scission of intraluminal vesicles into multivesicular bodies (46). In particular, the ESCRT-I component TSG101 recruits cargo proteins through binding of the tetrapeptide motif P(S/T)AP (47). A recent report showed that the beta-galactoside binding lectin galectin-3 (Gal3) is recruited into exosomes through direct interaction with TSG101, and depends on a highly conserved P(S/T)AP motif in the N-terminal region of Gal3 (48). However, it is still unclear how sEVs-HGF is packed into sEVs. Our preliminary data show that Granulin might contribute to the packaging of HGF. Besides, whether HGF has other binding partners and which protein domain or motif is necessary for this interaction should be studied in future research.
In conclusion, our results indicate that cancer cell-derived sEVs-HGF contribute to metastasis of recipient cells by activating the c-Met signaling pathway and promoting EMT. More importantly, high expression of HGF in plasma sEVs shows a positive correlation with metastasis of lung cancer in patients. Our results reveal a new molecular mechanism underlying the crosstalk between cancer cells and adjacent normal cells, which will contribute to developing therapeutic strategies for efficient prevention and treatment of lung cancer metastasis.
Data availability
The mass spectrometry proteomics data and the search data by MaxQuant (version 1.6.1.0) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013419.
Supplementary Material
Footnotes
* This work was supported by grants from the National Key Research and Development Program (No. 2017YFC1200204), the National Natural Science Foundation of China (No. 21675110, No. 21475086, No. 21305087), the Committee of Shanghai Science and Technology (No. 14DZ0501200 and No. 15142200300), and the Key Scientific Project of Shanghai Jiao Tong University (No. YG2014QN21, YG2015MS48 and YG2017MS80). H.X. is supported by the Recruitment Program of Global Youth Experts of China and National High-tech R&D Program of China (863 Program, No. 2014AA020545).
 This article contains supplemental Figures and Tables. The authors declare no competing financial interests.
This article contains supplemental Figures and Tables. The authors declare no competing financial interests.
1 The abbreviations used are:
- EVs
- extracellulat vesicles
- sEVs
- small extracellular vesicles
- mEVs
- medium EVs
- EMT
- epithelial-mesenchymal progression.
REFERENCES
- 1. Thery C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., et al. (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thery C., Zitvogel L., and Amigorena S. (2002) Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 3. Thery C., Amigorena S., Raposo G., and Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3 22 [DOI] [PubMed] [Google Scholar]
- 4. Milane L., Singh A., Mattheolabakis G., Suresh M., and Amiji M. M. (2015) Exosome mediated communication within the tumor microenvironment. J. Control Release 219, 278–294 [DOI] [PubMed] [Google Scholar]
- 5. Fang T., Lv H., Lv G., Li T., Wang C., Han Q., et al. (2018) Tumor-derived exosomal miR-1247–3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 9, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fong M. Y., Zhou W., Liu L., Alontaga A. Y., Chandra M., Ashby J., Chow A., O'Connor S. T., Li S., Chin A. R., Somlo G., Palomares M., Li Z., Tremblay J. R., Tsuyada A., Sun G., Reid M. A., Wu X., Swiderski P., Ren X., Shi Y., Kong M., Zhong W., Chen Y., and Wang S. E. (2015) Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tauro B. J., Greening D. W., Mathias R. A., Mathivanan S., Ji H., and Simpson R. J. (2013) Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell Proteomics 12, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garnier D., Magnus N., Meehan B., Kislinger T., and Rak J. (2013) Qualitative changes in the proteome of extracellular vesicles accompanying cancer cell transition to mesenchymal state. Exp. Cell Res. 319, 2747–2757 [DOI] [PubMed] [Google Scholar]
- 9. Xu R., Greening D. W., Rai A., Ji H., and Simpson R. J. (2015) Highly purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11–25 [DOI] [PubMed] [Google Scholar]
- 10. Sun Y., Liu S., Qiao Z., Shang Z., Xia Z., Niu X., Qian L., Zhang Y., Fan L., Cao C. X., and Xiao H. (2017) Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Anal. Chim. Acta 982, 84–95 [DOI] [PubMed] [Google Scholar]
- 11. Sun Y., Huo C., Qiao Z., Shang Z., Uzzaman A., Liu S., Jiang X., Fan L. Y., Ji L., Guan X., Cao C. X., and Xiao H. (2018) Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J. Proteome Res. 17, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 12. Ettinger D. S., Akerley W., Borghaei H., Chang A. C., Cheney R. T., Chirieac L. R., D'Amico T. A., Demmy T. L., Govindan R., Grannis F. W. Jr, Grant S. C., Horn L., Jahan T. M., Komaki R., Kong F. M., Kris M. G., Krug L. M., Lackner R. P., Lennes I. T., Loo B. W. Jr, Martins R., Otterson G. A., Patel J. D., Pinder-Schenck M. C., Pisters K. M., Reckamp K., Riely G. J., Rohren E., Shapiro T. A., Swanson S. J., Tauer K., Wood D. E., Yang S. C., Gregory K., Hughes M., and National comprehensive cancer network. (2013) Non-small cell lung cancer, version 2.2013. J. Natl. Compr. Canc. Netw 11, 645–653; quiz 653 [DOI] [PubMed] [Google Scholar]
- 13. Zhang H., Deng T., Liu R., Bai M., Zhou L., Wang X., Li S., Wang X., Yang H., Li J., Ning T., Huang D., Li H., Zhang L., Ying G., and Ba Y. (2017) Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 8, 15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moustakas A. and de Herreros A. G. (2017) Epithelial-mesenchymal transition in cancer. Mol. Oncol. 11, 715–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nieto M. A. (2013) Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342, 1234850. [DOI] [PubMed] [Google Scholar]
- 16. Xiao D., Barry S., Kmetz D., Egger M., Pan J., Rai S. N., Qu J., McMasters K. M., and Hao H. (2016) Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 376, 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. You Y., Shan Y., Chen J., Yue H., You B., Shi S., Li X., and Cao X. (2015) Matrix metalloproteinase 13-containing exosomes promote nasopharyngeal carcinoma metastasis. Cancer Sci. 106, 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knowles L. M., Stabile L. P., Egloff A. M., Rothstein M. E., Thomas S. M., Gubish C. T., Lerner E. C., Seethala R. R., Suzuki S., Quesnelle K. M., Morgan S., Ferris R. L., Grandis J. R., and Siegfried J. M. (2009) HGF and c-Met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin. Cancer Res. 15, 3740–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma P. C., Kijima T., Maulik G., Fox E. A., Sattler M., Griffin J. D., Johnson B. E., and Salgia R. (2003) c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 63, 6272–6281 [PubMed] [Google Scholar]
- 20. Rothenberger N. J., and Stabile L. P. (2017) Hepatocyte Growth Factor/c-Met Signaling in Head and Neck Cancer and Implications for Treatment. Cancers 9, pii: E39. doi: 10.3390/cancers9040039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sadiq A. A. and Salgia R. (2013) MET as a possible target for non-small-cell lung cancer. J. Clin. Oncol. 31, 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C. M., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., Yuan J., Martins V. R., Skog J., Kaplan R. N., Brady M. S., Wolchok J. D., Chapman P. B., Kang Y., Bromberg J., and Lyden D. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiao H., Langerman A., Zhang Y., Khalid O., Hu S., Cao C. X., Lingen M. W., and Wong D. T. W. (2015) Quantitative proteomic analysis of microdissected oral epithelium for cancer biomarker discovery. Oral. Oncol. 51, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 24. Xiao H., Zhang L., Zhou H., Lee J. M., Garon E. B., and Wong D. T. (2012) Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell Proteomics 11, M111 012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He C., He P., Liu L. P., and Zhu Y. S. (2001) Analysis of expressions of components in the plasminogen activator system in high- and low-metastatic human lung cancer cells. J. Cancer Res. Clin. Oncol. 127, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu Y. L. (1989) [Spontaneous metastasis of clonal cell subpopulations of human lung giant cell carcinoma after subcutaneous inoculation in nude mice]. Zhonghua Zhong Liu Za Zhi 11, 1–7 [PubMed] [Google Scholar]
- 27. Ge M., Luo Z., Qiao Z., Zhou Y., Cheng X., Geng Q., Cai Y., Wan P., Xiong Y., Liu F., Wu K., Liu Y., and Wu J. (2017) HERP binds TBK1 to activate innate immunity and repress virus replication in response to endoplasmic reticulum stress. J. Immunol. 199, 3280–3292 [DOI] [PubMed] [Google Scholar]
- 28. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., and Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalluri R. (2016) The biology and function of exosomes in cancer. J. Clin. Invest. 126, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nieto M. A., Huang R. Y., Jackson R. A., and Thiery J. P. (2016) Emt: 2016. Cell 166, 21–45 [DOI] [PubMed] [Google Scholar]
- 31. Kalluri R. and Neilson E. G. (2003) Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 112, 1776–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vonderheide R. H. (2015) CD47 blockade as another immune checkpoint therapy for cancer. Nat. Med. 21, 1122–1123 [DOI] [PubMed] [Google Scholar]
- 33. Han S. U., Lee J. H., Kim W. H., Cho Y. K., and Kim M. W. (1999) Significant correlation between serum level of hepatocyte growth factor and progression of gastric carcinoma. World J. Surg. 23, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 34. Kentsis A., Reed C., Rice K. L., Sanda T., Rodig S. J., Tholouli E., Christie A., Valk P. J., Delwel R., Ngo V., Kutok J. L., Dahlberg S. E., Moreau L. A., Byers R. J., Christensen J. G., Vande Woude G., Licht J. D., Kung A. L., Staudt L. M., and Look A. T. (2012) Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat. Med. 18, 1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okazaki M., Yoshimura K., Suzuki Y., Uchida G., Kitano Y., Harii K., and Imokawa G. (2003) The mechanism of epidermal hyperpigmentation in cafe-au-lait macules of neurofibromatosis type 1 (von Recklinghausen's disease) may be associated with dermal fibroblast-derived stem cell factor and hepatocyte growth factor. Br. J. Dermatol. 148, 689–697 [DOI] [PubMed] [Google Scholar]
- 36. Shurtleff M. J., Temoche-Diaz M. M., and Schekman R. (2018) Extracellular vesicles and cancer: caveat lector. Ann. Rev. Cancer Biol. 2, 395–411 [Google Scholar]
- 37. Nielsen S. R., Quaranta V., Linford A., Emeagi P., Rainer C., Santos A., Ireland L., Sakai T., Sakai K., Kim Y. S., Engle D., Campbell F., Palmer D., Ko J. H., Tuveson D. A., Hirsch E., Mielgo A., and Schmid M. C. (2016) Macrophage-secreted granulin supports pancreatic cancer metastasis by inducing liver fibrosis. Nat. Cell Biol. 18, 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoshino A., Costa-Silva B., Shen T. L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., Singh S., Williams C., Soplop N., Uryu K., Pharmer L., King T., Bojmar L., Davies A. E., Ararso Y., Zhang T., Zhang H., Hernandez J., Weiss J. M., Dumont-Cole V. D., Kramer K., Wexler L. H., Narendran A., Schwartz G. K., Healey J.H., Sandstrom P., Labori K. J., Kure E. H., Grandgenett P. M., Hollingsworth M. A., de Sousa M., Kaur S., Jain M., Mallya K., Batra S. K., Jarnagin W. R., Brady M. S., Fodstad O., Muller V., Pantel K., Minn A. J., Bissell M. J., Garcia B. A., Kang Y., Rajasekhar V. K., Ghajar C. M., Matei I., Peinado H., Bromberg J., and Lyden D. (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Llovet J. M., and Beaugrand M. (2003) Hepatocellular carcinoma: present status and future prospects. J. Hepatol. 38, S136–S149 [DOI] [PubMed] [Google Scholar]
- 40. McLarty J., Bigelow R. L., Smith M., Elmajian D., Ankem M., and Cardelli J. A. (2009) Tea polyphenols decrease serum levels of prostate-specific antigen, hepatocyte growth factor, and vascular endothelial growth factor in prostate cancer patients and inhibit production of hepatocyte growth factor and vascular endothelial growth factor in vitro. Cancer Prev. Res. 2, 673–682 [DOI] [PubMed] [Google Scholar]
- 41. Jia X., Chen J., Megger D. A., Zhang X., Kozlowski M., Zhang L., Li J., Chu Q., Wu M., Li Y., Sitek B., and Yuan Z. (2017) Label-free proteomic analysis of exosomes derived from inducible hepatitis B virus-replicating HepAD38 cell line. Mol. Cell Proteomics 16, S144–S160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y. (2007) Mitogen-activated protein kinases in heart development and diseases. Circulation 116, 1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grotegut S., von Schweinitz D., Christofori G., and Lehembre F. (2006) Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 25, 3534–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng J. C., Auersperg N., and Leung P. C. (2012) EGF-induced EMT and invasiveness in serous borderline ovarian tumor cells: a possible step in the transition to low-grade serous carcinoma cells? PLoS ONE 7, e34071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steinbichler T. B., Dudas J., Riechelmann H., and Skvortsova I. I. (2017) The role of exosomes in cancer metastasis. Semin. Cancer Biol. 44, 170–181 [DOI] [PubMed] [Google Scholar]
- 46. Henne W. M., Stenmark H., and Emr S. D. (2013) Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sundquist W. I., Schubert H. L., Kelly B. N., Hill G. C., Holton J. M., and Hill C. P. (2004) Ubiquitin recognition by the human TSG101 protein. Mol. Cell 13, 783–789 [DOI] [PubMed] [Google Scholar]
- 48. Banfer S., Schneider D., Dewes J., Strauss M. T., Freibert S. A., Heimerl T., Maier U. G., Elsässer H. P., Jungmann R., and Jacob R. (2018) Molecular mechanism to recruit galectin-3 into multivesicular bodies for polarized exosomal secretion. Proc. Natl. Acad. Sci. U.S.A. 115, E4396–E4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data and the search data by MaxQuant (version 1.6.1.0) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013419.