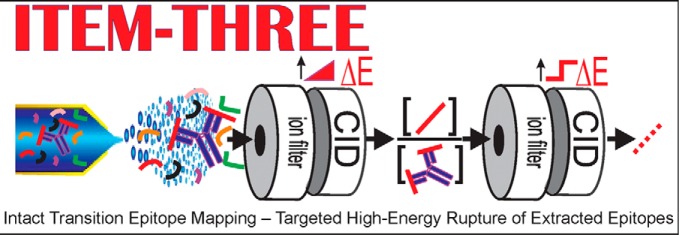
ITEM-THREE enables rapid epitope mapping. Sample consumption is minimized and in-solution handling reduced to mixing of antibody and antigen peptide solutions. After immune complex formation in solution, epitope mapping is performed in the gas phase using the mass spectrometer for sophisticated ion manipulation and filtering. Because amino acid sequence information is obtained from the epitope peptide, unknown antigens can be identified. Knowing the epitope broadens the application of antibodies to unspecified target proteins from any organism.
Keywords: Affinity proteomics, Antibodies*, Assay development, Immunology*, Ligand -Receptor Interactions*, Mass Spectrometry, Non-covalent interaction MS*, Protein complex analysis, Targeted mass spectrometry, Therapeutic antibodies*, BLAST, data base search, epitope mapping, peptide sequencing
Graphical Abstract
Highlights
Multiplex epitope mapping/antigenic determinant identification in the gas phase.
Intact transition and controlled dissociation of immune complexes by MS.
Simultaneous identification and amino acid sequence determination of epitopes.
Simplified in-solution sample handling because of ion manipulation and filtering by MS.
Abstract
Epitope mapping, which is the identification of antigenic determinants, is essential for the design of novel antibody-based therapeutics and diagnostic tools. ITEM-THREE is a mass spectrometry-based epitope mapping method that can identify epitopes on antigens upon generating an immune complex in electrospray-compatible solutions by adding an antibody of interest to a mixture of peptides from which at least one holds the antibody′s epitope. This mixture is nano-electrosprayed without purification. Identification of the epitope peptide is performed within a mass spectrometer that provides an ion mobility cell sandwiched in-between two collision cells and where this ion manipulation setup is flanked by a quadrupole mass analyzer on one side and a time-of-flight mass analyzer on the other side. In a stepwise fashion, immune-complex ions are separated from unbound peptide ions and dissociated to release epitope peptide ions. Immune complex-released peptide ions are separated from antibody ions and fragmented by collision induced dissociation. Epitope-containing peptide fragment ions are recorded, and mass lists are submitted to unsupervised data base search thereby retrieving both, the amino acid sequence of the epitope peptide and the originating antigen. ITEM-THREE was developed with antiTRIM21 and antiRA33 antibodies for which the epitopes were known, subjecting them to mixtures of synthetic peptides of which one contained the respective epitope. ITEM-THREE was then successfully tested with an enzymatic digest of His-tagged recombinant human β-actin and an antiHis-tag antibody, as well as with an enzymatic digest of recombinant human TNFα and an antiTNFα antibody whose epitope was previously unknown.
The identification of epitopes or antigenic determinants is essential for the design of novel antibody-based therapeutics and vaccines (1–4). With current personalized medicine concepts (4, 5), epitope mapping, i.e. accurate identification of antigenic determinants (epitopes) of protein antigens (6–8), is very useful in the design of novel antibody-based diagnostic tools, particularly for companion diagnostics (9, 10). Although structure-based methods, such as X-ray crystallography (11, 12) and NMR (13, 14) have been regarded as “gold standard” to map epitopes because they achieve atomic resolution, they are not always readily applicable because a given antigen-antibody pair may lie beyond the scope of either or both of these methods, e.g. when the immune complex is not crystallizable or is too large for NMR (15, 16). One great disadvantage of X-ray crystallography and NMR is that both require rather large sample amounts (17, 18).
By contrast, the relatively low amounts of samples required (19) and the rapidity (6) by which mass spectrometric epitope mapping is executed is of great advantage in this respect (20). Chemical cross-linking mass spectrometry (21, 22), hydrogen/deuterium exchange (HDX)1 mass spectrometry (23) and mass spectrometric methods that employ chemical modification on proteins, such as Fast Photochemical Oxidation of Proteins (FPOP) (24, 25) or chemical modification of surface exposed residues (26, 27) have been applied in epitope mapping experiments (28) and in determinations of protein - protein interaction sites in general (29), but their application may be limited when rather demanding chemistries are involved, or when performing such experiments becomes laborious, and/or requires sophisticated laboratory equipment (20, 30). Significant advances in epitope mapping protocols/methods have been reached with the two most commonly used mass spectrometric methods: epitope extraction and epitope excision (20, 31, 32). These techniques have matured either through automation of solution handling procedures (33) or by minimizing in-solution handling, i.e. avoiding immobilization procedures and other chemical reactions (6, 34).
Advanced mass spectrometer designs have led to increased flexibility by coupling various ion filtering devices with different mass analyzers, and have opened new opportunities for performing ion reactions, such as CID and SID (19, 35–39) in the gas phase and/or laser irradiation and UV irradiation of ions, respectively (36, 40, 41). The availability of mass spectrometers equipped with ion-mobility separation chambers provide an additional dimension for the separation of ions based on not only their m/z values but also on their shapes and sizes (42–44). This new generation of mass spectrometers led to the development of fast and easy to apply epitope mapping methods by which epitope peptides of an antibody of interest can be identified in a relatively simple and robust fashion (6, 20). Based on our gas phase epitope mapping strategy, termed ITEM-ONE (6), where epitopes of known antigens have been identified by precisely determining the mass of the extracted epitope peptide, we have now advanced to ITEM-THREE, where mass spectrometric amino acid sequencing of unknown epitope peptides is performed to identify an antigenic determinant on an antigen surface.
MATERIALS AND METHODS
Proteins and Peptides
Mouse antiRA33 antibody (monoclonal anti-hnRNP-A2/B1; clone DP3B3 lot: 044K4766) was obtained from Sigma-Aldrich (Steinheim, Germany). Rabbit antiTRIM21 antibody (polyclonal anti-52kDa Ro/SSA antibody; sc-20960 lot: F0503) raised against amino acids 141–280 of TRIM21 (52kDa Ro/SSA) of human origin was obtained from Santa Cruz Biotechnology, Inc. (Heidelberg, Germany). Mouse antiHis-tag antibody (monoclonal antibody MCA 1396; Batch no. 0309) was supplied by Bio-Rad, (Munich, Germany) and mouse antiTNFα antibody (monoclonal antibody; catalogue no. MA5-23720) was produced by ThermoFisher Scientific GmbH (Ulm, Germany). Recombinant human TNF alpha (rhTNFα) was a gift from Prof. Harald Illges, Hochschule Bonn-Rhein-Sieg University of Applied Sciences, Germany. Actin, cytoplasmic 1 recombinant protein was purchased from GenWay Biotech (Catalogue no. 10-288-23014F, San Diego, CA). RA33 peptide (MAARPHSIDGRVVEP-NH2), GPI peptide (ALKPYSPGGPR), Angiotensin II (DRVYIHPF), TRIM21A peptide (LQELEKDEREQLRILGE), TRIM21B peptide (LQPLEKDEREQLRILGE) and TRIM21C peptide (LQELEKDEPEQLRILGE) were synthesized by Peptides and Elephants GmbH (Potsdam, Germany). The synthetic FLAG peptide (DYKDDDDK; article no. 020015) was obtained from ThermoFisher Scientific GmbH and sequencing grade, modified trypsin was obtained from Promega Corporation (Madison, WI).
Preparation of the Synthetic Peptide Mixture Solution (Solution 1)
A mixture of equimolar concentrations of seven synthetic peptides (10 μm each; GPI peptide, FLAG peptide, Angiotensin II, TRIM21A peptide, TRIM21B peptide, TRIM21C peptide, and RA33 peptide) was prepared by dissolving the appropriate amounts of the individual lyophilized peptide powders in freshly prepared 200 mm ammonium acetate, pH 7.1 and mixing the appropriate volumes. The peptide mixture-containing solution was shock-frozen and kept at −20 °C until either mass spectrometric analysis or immune complex formation were performed.
Tryptic Digestion of Recombinant Human Beta Actin (Solution 1)
Recombinant human beta actin (rhβactin) was digested with trypsin (26, 34, 45) using a modified Filter Aided Sample Preparation (FASP) protocol. To 10 μl of 200 mm DTT, dissolved in 0.1 m Tris/HCl containing 8 m urea was added 20 μl of rhβactin solution (protein concentration 1 μg/μl). This mixture was incubated at 37 °C for 30 min. Then, this solution was transferred into an equilibrated 30K Amicon centrifugal filter (equilibration with 1% formic acid according to protocol (46)) and 170 μl of 8 m urea in 0.1 m Tris/HCl, pH 8.5, were added and centrifuged at 13,000 rpm for 15 min. After discarding the filtrate, 150 μl of 8 m urea in 0.1 m Tris/HCl, pH 8.5, were added to the retentate in the filter unit and centrifuged again at 13,000 rpm for 15 min. A further wash of the retentate was done by adding 100 μl of 8 m urea in 0.1 m Tris/HCl, pH 8.5, to the filter unit and centrifuging at 13,000 rpm for 12 min. The filtrates were discarded and the retentate was further washed for three times, first by adding 100 μl, then 75 μl, and lastly 50 μl of 50 mm ammonium bicarbonate solution, pH 8.6, and each time centrifugation was performed at 13,000 rpm for 10 min, 12,000 rpm for 8 min, and 12,000 rpm for 6 min, respectively. After the three washings, the filter unit containing the retentate (ca. 5 μl) was transferred into a new collection tube. A volume of 35 μl of 11.42 ng/μl of trypsin in 50 mm ammonium bicarbonate, pH 8.6, was added to the protein that was dissolved in the solution on the filter unit to obtain an enzyme/substrate ratio of 1:50 (w/w). The mixture was incubated at room temperature in a wet chamber for 16 h and then centrifuged at 8000 rpm for 5 min and at 12,000 rpm for 3 min. Next, a volume of 40 μl of 10 mm ammonium bicarbonate, pH 8.6, and a further amount of 4 μl of 0.1 μg/μl trypsin solution (composition see above) was added and incubated at 37 °C for 2 h. Finally, the mixture was centrifuged at 12,000 rpm for 8 min and the filtrate (ca. 80 μl), which contained the tryptic peptides, was collected for further analysis. The peptide concentration of the solution was determined using the Qubit® 2.0 Fluorometer (Carlsbad, CA), following described procedures (6, 47). Aliquots (10 μl, each), were shock-frozen and kept at −20 °C until either mass spectrometric analysis or immune complex formation were performed.
Tryptic Digestion of Recombinant Human TNF Alpha (Solution 1)
Tryptic digestion of recombinant human TNF alpha (26, 34, 45) (rhTNFα, 1 μg/μl) was performed by adding 15 μl of the rhTNFα dissolved in 200 mm ammonium acetate, pH 7.1, to 32 μl of trypsin solution (9.4 ng/μl in 4.8 mm Tris/HCl with 5 mm DTT) to yield an enzyme/substrate ratio of 1:50 (w/w). This mixture was incubated at 37 °C for 20 h. The resulting tryptic peptide solution was divided into nine aliquots, each of which contained a volume of 5 μl. Each aliquot was desalted by loading the entire 5 μl volume onto one C18 ZipTip Pipette Tip (Merck Millipore Ltd, Tullagreen, Carrigtwohill, Co. Cork, Ireland) that had been wetted with a mixture of deionized H2O/ACN (50:50, v/v). Equilibration and washing solutions consisted of 1% HCOOH in deionized H2O; two times 10 μl were used for each step. The affinity-bound peptides were eluted into 5 μl of 1% HCOOH in H2O : 1% HCOOH in ACN (50:50, v/v) (46, 48). Next, all nine desalted portions of the tryptic peptide-containing solutions were pooled to obtain a total volume of 45 μl. This solution was aliquoted into 10 μl volumes, shock-frozen and kept at −20 °C until either mass spectrometric analysis or immune complex formation were performed.
Preparation of Antibody Solutions (Solution 2)
As described previously (6), a volume of 30 μl of 0.8 μg/μl of antiRA33 antibody solution was loaded onto a 50K Amicon centrifugal filter (Merck Millipore Ltd.) and 470 μl of 200 mm ammonium acetate, pH 7.1, was added. The resulting solution was centrifuged at 13,000 rpm for 10 min for eight times. Each time the filtrates were discarded and 470 μl of 200 mm ammonium acetate, pH 7.1, were added to the retentates. After the last centrifugation the filter units were inverted and placed into new tubes and centrifuged at 4500 rpm for 5 min to collect the retentates of ca. 20 μl antibody solution in each case. Protein concentrations were determined using the Qubit® 2.0 Fluorometer following described procedures (6). Similarly, 100 μl of 0.2 μg/μl of antiTRIM21 antibody solution, 20 μl of 1.0 μg/μl antiHis-tag antibody solution, and 40 μl of 0.5 μg/μl of antiTNFα antibody solution, which were all obtained from suppliers in PBS buffer, were buffer exchanged into 200 mm ammonium acetate buffer, pH 7.1.
Preparation of Immune Complex-containing Solutions (Solution 3)
For immune complex formation, a final concentration of ca. 0.2 μg/μl of each antibody-containing solution (solution 2) was prepared by diluting the solutions obtained from the buffer exchange with 200 mm ammonium acetate, pH 7.1. Solutions 3 were binary mixtures of one Solution 1 with one Solution 2 to obtain the molar ratios of ∼2.2:1 (epitope peptide/antibody) in each of Solutions 3. Immune complex-containing mixtures (Solutions 3) were incubated at room temperature for at least 1 h.
NanoESI-IMS-MS/MS Instrument Settings and Spray Needle Preparation
NanoESI-IMS-MS/MS measurements were carried out in positive ion mode on a quadrupole ion-mobility separation time-of-flight mass spectrometer (Synapt G2-S, Waters MS-Technologies, Manchester, United Kingdom) as described (6). The m/z range 200–8000 of the time-of-flight analyzer was calibrated externally using a 1 mg/ml sodium iodide solution dissolved in an isopropanol/water mixture (50:50, v/v). Measurements were performed with the following instrumental settings: source temperature, 50 °C; capillary voltage, 1.60–1.90 kV; source offset, 80–100 V; sample cone voltage, 90–120 V; TRAP cell gas flow, 6.0 ml/min; cone gas flow, 100 liters/h. Gas controls were set to automatic as follows: TRAP cell gas flow, 2.0 ml/min; helium cell gas flow, 180 ml/min; IMS cell gas flow, 90 ml/min. IMS wave velocity and wave height were manually set to 650 m/s and 40 V, respectively. Start wave height and end wave height were also optimized with 30–35 V and 20–25 V, respectively, for each experiment to obtain adequate ion mobility separation. Pusher width and pusher cycle times were both set to automatic. Scan duration of 1.0 s and inter scan delay of 0.015 s were set for both IMS and MS measurements. Reflectron grid, flight tube and reflectron voltages were 1.46 kV, 10.00 kV and 3.78 kV, respectively, and detector sensitivity was set to normal. Pressure settings within the various parts of the mass spectrometer were as follows: TRAP cell, ∼2.2 × 10−2 mbar; Helium cell, ∼1.35 × 103 mbar; IMS cell, ∼3.5 × 100 mbar; TRANSFER cell, ∼2.6 × 10−2 mbar; ToF analyzer, ∼8.0 × 10−7 mbar. Spray needles were prepared in-house from borosilicate glass tubes of 1 mm outer and 0.5 mm inner diameters with a P-1000 Flaming/BrownTM Micropipette Puller System (Sutter Instruments, Novato, CA, USA) followed by gold coating, applying the Sputter Coater SCD 005 (BAL-TEC Inc., Balzers, Liechtenstein) (6, 47).
NanoESI-IMS-MS/MS Measurements for Epitope Mapping
To perform ITEM-THREE experiments, solutions 3 (ca. 3 μl, each) were loaded into spray needles with the aid of 20 μl microloader pipette tips (Eppendorf AG, Hamburg, Germany) and were electrosprayed without any purification. In the mass spectrometer, the quadrupole analyzer was first used to block transmission of lower molecular weight ions (filter off ions below m/z 5000) by manually setting the quadrupole appropriately (6). The epitope peptide-antibody complexes were able to transit the quadrupole intact. Next, dissociation of the epitope peptide-antibody complexes in the TRAP cell (first collision cell) was achieved by increasing the collision cell voltage difference (ΔCV) to between 50 and 80 V. The collision voltage differences in the TRAP cell were raised in a stepwise manner (5–20 V/step) and were so optimized for each experiment to ensure adequate dissociation of the antibody - peptide complex with minimal antibody fragmentation. The dissociated complex constituents then entered the ion mobility chamber where they were separated according to their m/z values, sizes and shapes. Finally, the collision cell voltage differences (ΔCV) in the TRANSFER cell (second collision cell) were also increased to 40–70 V to cause enough fragmentation of the complex released peptides (CoRPs). Again, the collision voltage differences in the TRANSFER cell were raised in a stepwise manner (5–20 V/step) to adjust optimized peptide ion fragmentation conditions for each experiment. Both ion mobility raw data (arrival time of ions) and mass spectral raw data were collected and stored using MassLynx software 4.1 (Waters MS-Technologies). The mass spectrometry data have been deposited in the PRIDE database (49).
Mascot Database Search with MS/MS Fragment Ions
After selecting the arrival time of the released epitope peptide ion, raw data from an MS/MS spectrum were de-convoluted and de-isotoped and then converted into peak lists using the MaxEnt3 algorithm on the MassLynx version 4.1 software. The peak list was then saved as a SEQUEST file (*.DTA) that was then uploaded onto the Mascot (Matrix Science Ltd., London, UK) search engine (50, 51), using the UniProt database (release 2018_06) that contained 557,713 sequences and an “amended UniProt” database that was generated to contain the amino acid sequences of the recombinant antigens in addition to all the amino acid sequence entries of the UniProt database (release 2018_06). The search parameters were set as follows: taxonomy, all entries; enzyme, none or trypsin (where tryptic digests were used), and up to 1 missed cleavage was allowed. Fixed modifications, carbamidomethylation of cysteine, and variable modifications, dicarbamidomethylation of lysine, were selected where necessary. Peptide mass tolerance and MS/MS ion mass tolerance were both set to 0.3 Da. Threshold ion scores were used in accepting the individual spectra. The ion scores above these thresholds indicated amino acid sequences with significant homology to the sequence entries from the data base.
NCBI BLAST of Identified Epitope Peptides
The amino acid sequence of the first hit from the Mascot search (epitope peptide) was submitted to an NCBI BLAST search using the following parameters: database, Uniprot (release 2018_06); organisms, all; max target sequence, 250; expect threshold, 2000; matrix PAM50 (52).
RESULTS
Method Development—
Immune Complex Formation and Epitope Identification for the antiRA33 Antibody
On electrospraying a peptide mixture that consists of seven synthetic peptides dissolved in aqueous ammonium acetate (solution 1) and operating the mass spectrometer in positive ion and ion mobility separation modes, the arrival times of the peptide ions at the end of the ion mobility separation chamber were recorded between 4 ms and 14 ms, respectively. The mass spectrum showed all ion signals from this arrival time regime as mainly singly and doubly charged ion signals within the mass range between m/z 300 and m/z 1600 (Fig. 1A–1B). In addition, fragment ions from the FLAG peptide and the GPI peptide were recorded as well (supplemental Table S1), despite low voltage differences (ca. 2 V to 4 V) in both, the TRAP cell that is the first collision cell located behind the quadrupole but in front of the ion mobility separation chamber, and the TRANSFER cell, which is the second collision cell, located behind the ion mobility separation chamber but in front of the ToF analyzer. As expected, in later arrival time regimes, i.e. above 16 ms, the mass spectrum of this peptide mixture did not show any ion signals (supplemental Fig. S1A–S1B).
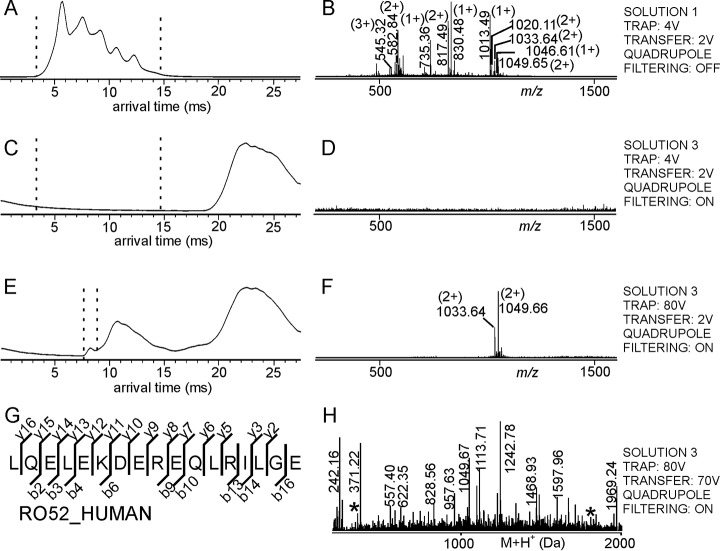
Fig. 1.
Mass spectrometric dissociation of the RA33 epitope peptide – antiRA33 antibody complex and amino acid sequence determination of the complex-released peptide by mass spectrometric fragmentation. Ion mobility arrival time plots of A, Solution 1, C, and E, Solution 3. Dashed lines mark the regions for mass spectra selections. Settings of the TRAP cell, the TRANSFER cell, and the quadrupole are indicated at the right. B, D, and F, nanoESI mass spectra (low m/z range) of ions from selected arrival time ranges. Selected m/z values are given and charge states are indicated in parentheses. For ion signal assignments see supplemental Table S1. G, Amino acid sequence of the complex-released peptide (single letter code) as determined by the matched mass spectrometric fragment ions (fragment ion types and numbers are indicated). The Uniprot protein ID of the peptide source protein (first hit) is shown. H, Pseudo mass spectrum (after charge deconvolution and de-isotoping) of fragment ions derived by selecting arrival time of the complex-released peptide with m/z 817.48. For ion signal assignments see supplemental Table S2.
Adding the antiRA33 antibody (solution 2) to the synthetic peptide mixture (solution 1) produced solution 3 in which formation of the immune complex occurred, consisting of RA33 epitope peptide and antiRA33 antibody. Electrospraying solution 3 and setting the quadrupole to block transmission of ions with m/z values below m/z 5000, only the intact immune complex ions as well as free antibody ions were able to traverse the quadrupole ion filter. The high molecular weight components of solution 3 produced multiply charged ions, ranging from 24+ to 30+ charge states, which all appeared above m/z 5000 (supplemental Fig. S1C–S1D, cf. supplemental Table S3). All these multiply charged ion signals were recorded with arrival times between 16 ms and 22 ms at the end of the ion mobility separation chamber (Fig. 1C). On looking at the arrival time regime where the unbound peptide ions (UBPs) were expected to reach the end of the ion mobility separation chamber (4 ms to 14 ms), there appeared no peptide ion signals within the mass range of m/z 300 to m/z 1600 (Fig. 1D).
However, when higher collision cell voltage differences (50 V) were applied in the TRAP cell, collision of the immune complex ions with argon gas atoms caused dissociation of singly and/or doubly charged peptide ions from the multiply charged immune complex ions. The complex-released peptide ions (CoRPs) were detected with arrival times of around 13 ms and the corresponding arrival time-matched mass spectrum showed doubly charged ions at m/z 817.48 and at m/z 825.48 for the RA33 epitope peptide and its oxygenated product, both with mass accuracies of 61 ppm (Fig. 1E–1F). Obviously, after intact transition into the gas phase, the respective multiply charged immune complex ions traversed the quadrupole mass filter while the quadrupole effectively blocked transmission of all UBPs (Fig. 1C–1D).
Next, the collision cell voltage difference in the TRANSFER cell was increased to 60 V to fragment (rupture) the CoRPs, which had been generated in the TRAP cell. Thus, at high collision cell voltage differences in both the TRAP cell (50 V) and the TRANSFER cell (60 V) fragment ions of the RA33 epitope peptide were recorded with the same arrival times (ca. 13 ms) as the precursor CoRP ions (Fig. 1H). Additionally, when high collision cell voltage differences in both collision cells are applied, multiply charged antibody fragment ions appeared at around m/z 2000 with arrival times similar to those of the intact immune complex ions and/or the ions of the free antibody (supplemental Fig. S1E–S1F).
After processing the raw data from the CoRP fragment ion mass spectra by de-isotoping and deconvolution of charge states, and after submitting the list of fragment ions to data base search using the Mascot search engine, the best hit from the search result reported the amino acid sequence of the RA33 peptide (Fig. 1G). In addition to b-type and y-type ions, some b-type and y-type ions that had lost ammonia were identified as well (supplemental Table S2). The best hit reported a score of 43 (threshold 34), indicating that the determined amino acid sequence had a significant homology to the sequence entry from the data base. In addition to the amino acid sequence the data base entry revealed the name of the originating protein, hence the antigen.
Although the CoRPs, i.e. epitope peptides, which had been generated in the TRAP cell could be identified with high mass accuracies by comparing their monoisotopic masses to the known masses of all the peptides in the peptide mixture (solution 1), fragmenting the CoRP ions by increasing the collision cell voltage difference in the TRANSFER cell enabled experimental determination of the respective partial amino acid sequence on data base search, thereby substantiating epitope identification.
Immune Complex Formation and Epitope Identification for the antiTRIM21 Antibody
To further test our ITEM-THREE workflow an antiTRIM21 antibody (solution 2) was added to the mixture of synthetic peptides (solution 1), thereby obtaining TRIM21 epitope peptide - antiTRIM21 antibody complexes in solution 3. As before, the arrival times of the peptide ions at the end of the ion mobility separation chamber were recorded between 4 ms and 14 ms, respectively. The mass spectrum showed all ion signals from this arrival time regime as mainly singly and doubly charged ion signals within the mass range between m/z 300 and m/z 1600 (Fig. 2A–2B). Again, and as expected, the mass spectrum of this peptide mixture did not show any ion signals in later arrival time regimes, i.e. above 16 ms (supplemental Fig. S2A–S2B).
Fig. 2.
Mass spectrometric dissociation of the TRIM21 epitope peptide – antiTRIM21 antibody complex and amino acid sequence determination of the complex-released peptides by mass spectrometric fragmentation. Ion mobility arrival time plots of A, Solution 1, C, and E, Solution 3. Dashed lines mark the regions for mass spectra selections. Settings of the TRAP cell, the TRANSFER cell, and the quadrupole are indicated at the right. B, D, and F, nanoESI mass spectra (low m/z range) of ions from selected arrival time ranges. Selected m/z values are given, and charge states are indicated in parentheses. For ion signal assignments see supplemental Table S1. G, Amino acid sequence of the complex-released peptide (single letter code) as determined by the matched mass spectrometric fragment ions (fragment ion types and numbers are indicated). The Uniprot protein ID of the peptide source protein (first hit) is shown. H, Pseudo mass spectrum (after charge deconvolution and de-isotoping) of fragment ions derived by selecting arrival time of the complex-released peptide with m/z 1049.66. The “*” marks ions that belong to the peptide with ion signal at m/z 1033.64. For ion signal assignments see supplemental Tables S4 and S5.
Electrospraying solution 3 and setting the quadrupole to block transmission of ions below m/z 5000, only the multiply charged ions of the intact immune complex and of the free antibody traversed the quadrupole and the ion mobility separation chamber with arrival times of between 19 ms and 28 ms (Fig. 2C). Like before, within the time regime at which the peptides' arrivals at the end of the ion mobility separation chamber were expected (4 ms to 14 ms), no peptide ion signals were seen in the mass range below m/z 1600 (Fig. 2D).
However, contrary to the case of antiRA33 antibody-RA33 epitope peptide complex, the mass spectrum that covered the arrival time range of 19 ms to 28 ms of the TRIM21 epitope peptide - antiTRIM21 antibody complex showed the presence of unresolved antiTRIM21 antibody together with TRIM21 epitope peptide-antiTRIM21 antibody complex ion signals at above m/z 5000. Their charge states ranged from 23+ to 28+ (supplemental Fig. S2C–S2D, cf. supplemental Table S6). Despite the unresolved multiply charged ion signals, this result repeatedly showed that the quadrupole was effectively filtering off UBPs.
On raising the TRAP cell voltage difference to 80 V and looking at the mass spectra that matched to the arrival times of CoRPs, in this case approx. 8 ms (Fig. 2E–2F), there appeared two doubly charged peptide ions, one at m/z 1049.66 and one at m/z 1033.64. By comparing the monoisotopic masses of these ions to the masses of the peptide ions that were contained in the peptide mixture (solution 1), the ion signal at m/z 1049.66 was assigned to the TRIM21A peptide with a mass accuracy of 85 ppm. Likewise, the ion signal at m/z 1033.64 was assigned to the TRIM21B peptide with a mass accuracy of 68 ppm (cf. supplemental Table S1). It is worth noting that the TRIM21C peptide, which was a constituent of the peptide mixture (solution 1) had not bound to the antiTRIM21 antibody and, hence, did not show up as a CoRP but remained a UBP.
When the TRANSFER cell voltage difference was increased to 70 V, both doubly charged precursor CoRP ions were simultaneously fragmented. The respective fragment ion spectrum that corresponded to the arrival time of the doubly charged precursor ions (ca. 8 ms) showed a mixture of the fragment ions. On performing data base search with the deconvoluted and de-isotoped fragment mass list, only the TRIM21A peptide sequence was reported as best hit with a score of 46 (threshold 37; Fig. 2G–2H, cf. supplemental Table S4), because only the wild type epitope peptide sequence was included in the Uniprot database. The TRIM21B peptide was identified (score 27; threshold 35) when repeating the data base search and applying the amended Uniprot database that contained the manually added amino acid sequence with the respective amino acid exchange (Fig. 2H, cf. supplemental Table S5). For the TRIM21A peptide both, b-type and y-type ions were assigned. Additionally, ion signals were observed for b-type and y-type fragment ions that had lost ammonia or water (supplemental Table S4). For the TRIM21B peptide barely b-type ions with loss of ammonia or water were seen (supplemental Table S5).
The mass spectra that were recorded at high collision cell energies in both, the TRAP cell and the TRANSFER cell, and selection of arrival times from 19 ms to 28 ms, where the multiply charged ion signals of unresolved TRIM21 epitope peptide - antiTRIM21 antibody immune complexes and free antibody had appeared, showed in addition to these ion signals the presence of multiply charged antibody fragment ions (supplemental Fig. S2E–S2F, cf. supplemental Table S6). To unequivocally assign antibody fragment ion signals we performed control experiments by individually electrospraying the antibody solutions (solutions 2), one after the other, under the same fragmentation conditions for comparisons (data not shown).
Application Examples—
Immune Complex Formation and Epitope Identification for the antiHis-tag Antibody
As a first application example, we performed our ITEM-THREE method to identify a His-tag epitope peptide from the 6-times histidine tagged recombinant human beta actin (rhβactin) using an antiHis-tag antibody. To generate solution 1 an in-solution tryptic digestion of the His-tagged rhβactin was performed. The arrival times of the tryptic peptide ions from solution 1, after traversing the quadrupole and the ion mobility separation chamber, ranged from 4 ms to 14 ms when measured at low collision cell voltage differences in both, the TRAP cell (4V) and the TRANSFER cell (2V). The corresponding mass spectrum showed ion signals below m/z 1600, which covered 55% of the amino acid sequence of the full length rhβactin (Fig. 3A–3B, cf. supplemental Fig. S5, supplemental Table S7). However, none of the peptide masses from the mass spectrum could be assigned to the C-terminal tryptic peptide, which contained the His-tag. In the mass range above m/z 5000, there were no ion signals present (supplemental Fig. S4A–S4B).
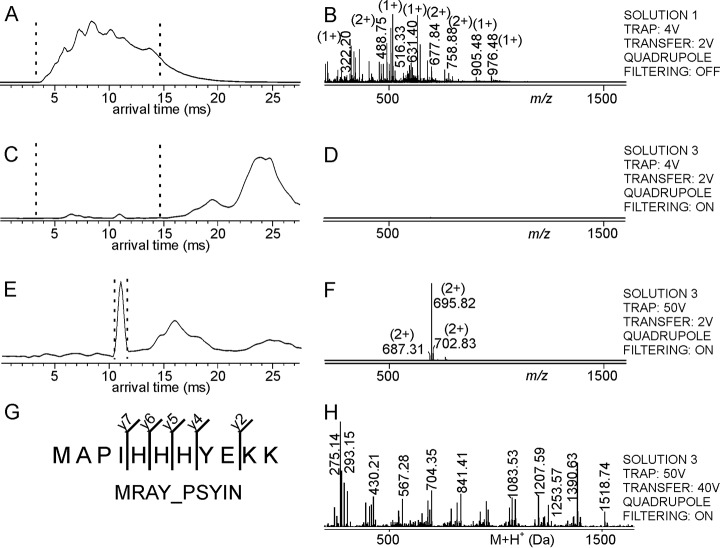
Fig. 3.
Mass spectrometric dissociation of the His-tag epitope peptide – antiHis-tag antibody complex and amino acid sequence determination of the complex-released peptide by mass spectrometric fragmentation. Ion mobility arrival time plots of A, Solution 1, C, and E, Solution 3. Dashed lines mark the regions for mass spectra selections. Settings of the TRAP cell, the TRANSFER cell, and the quadrupole are indicated at the right. B, D, and F, nanoESI mass spectra (low m/z range) of ions from selected arrival time ranges. Selected m/z values are given and charge states are indicated in parentheses. For ion signal assignments see supplemental Table S7. G, Amino acid sequence of the complex-released peptide (single letter code) as determined by the matched mass spectrometric fragment ions (fragment ion types and numbers are indicated). The Uniprot protein ID of the peptide source protein (first hit) is shown. H, Pseudo mass spectrum (after charge deconvolution and de-isotoping) of fragment ions derived by selecting arrival time of the complex-released peptide with m/z 695.82. For ion signal assignments see supplemental Table S8.
The antiHis-tag antibody (solution 2) was added to solution 1 and the mixture (solution 3) was electrosprayed without any further purification after an incubation period of 1 h at room temperature. When analyzing solution 3 at low collision cell voltage differences in both the TRAP cell and the TRANSFER cell and with setting the quadrupole to block transmission of UBPs, no ion signals appeared in the mass spectrum below m/z 1600 (Fig. 3C–3D). In the mass range above m/z 5000 multiply charged ion signals of unresolved antiHis-tag antibody and His-tag peptide-antiHis-tag antibody complex were recorded with charge states ranging from 22+ to 27+ (supplemental Fig. S4C–S4D, cf. supplemental Table S10).
After raising the collision cell voltage difference to 50 V in the TRAP cell, there appeared a doubly charged CoRP ion signal at m/z 695.82 with an arrival time of around 11 ms (Fig. 3E–3F). Surprisingly, the mass of this ion signal did not match to any of the predicted tryptic peptide masses from rhβactin. With an additional high collision cell voltage difference (40 V) in the TRANSFER cell, we were able to obtain the fragment ions of the corresponding CoRP, i.e. the peptide that was pulled out by the antiHis-tag antibody, hence the epitope containing peptide. After processing the ion signals from the fragment ion spectrum of the low m/z range and submitting the mass list to Uniprot data base search using the Mascot search engine, an assignment of the doubly charged CoRP was obtained for a peptide with three histidine residues in a row (Fig. 3G–3H), originating from Phospho-N-acetylmuramoyl-pentapeptide-transferase (MRAY_PSYIN) from Psychromonas ingrahamii (strain 37). The reported peptide sequence only provided 33% homology to the His-tag peptide from rhβactin that was used in the experiment. Although the search engine reported a false positive hit (score 18), the thereby suggested amino acid sequence indicated that the epitope peptide indeed contained a set of consecutively arranged histidine residues. After including the amino acid sequence of the His-tagged rhβactin into the amended Uniprot database, we obtained the C-terminal peptide from rhβactin as chemically modified six histidine residue encompassing peptide as best hit (score 22; threshold 23) for the fragmented epitope peptide, i.e. the precursor ion at m/z 695.82. Of note, the mass difference of 114.04 Da between the measured and the theoretical masses of the tryptic His-tag peptide KC(carb)FHHHHHH was assumed to be because of di-carbamidomethylation on the lysine K373 residue (supplemental Fig. S3, cf. supplemental Table S9) in addition to carbamidomethylation of the cysteine C374 residue. Chemical modification on the K373 residue stands in agreement with shielding this lysine residue from enzymatic cleavage; explaining the “missed cleavage” at this amino acid residue (cf. supplemental Table S7).
At high collision cell energies in both the TRAP cell and the TRANSFER cell, the mass spectrum of high m/z range (above m/z 5000) with arrival times between 15 ms and 23 ms showed multiply charged ion signals of intact antibody unresolved from immune complex ions together with antibody fragments ions (supplemental Fig. S4E–S4F).
Immune Complex Formation and Epitope Identification for the antiTNFα Antibody
Because the ITEM-THREE method successfully identified epitope peptides from either synthetic peptide mixtures or from peptide mixtures originating from digested antigen proteins when exposed to the respective antibody, we applied our procedure to the identification of the unknown epitope of an antiTNFα antibody. Like before, solution 1 was generated by tryptic digestion of the protein, in this case the trimeric recombinant human TNFα (rhTNFα). The rhTNFα tryptic peptides' arrival times ranged from 4 ms to 12 ms and produced ion signals below m/z 1600 that covered 100% of the entire amino acid sequence of the full length protein monomer (Fig. 4A–4B). After addition of the antiTNFα antibody (solution 2) and incubation for 1 h at room temperature, solution 3 was electrosprayed. Setting the quadrupole analyzer to block transmission of unbound peptide ions from solution 3, there appeared no peptide ions in the lower mass range (m/z below 1600) but in the higher mass range (m/z above 5000) there appeared unresolved ion signals of antiTNFα antibody and rhTNFα epitope peptide-antiTNFα antibody immune complexes with charge states ranging from 22+ to 27+ (Fig. 4C–4D and supplemental Fig. S7). Then, on raising the TRAP cell voltage difference to 50 V, two singly charged ion signals, one at m/z 857.60 and one at m/z 909.57 showed up with significant intensities in the corresponding mass spectrum (Fig. 4E–4F). Albeit these two CoRPs overlapped with respect to their arrival times of approx. 11 ms, they could be separately fragmented by applying different TRANSFER cell voltage differences. A TRANSFER cell voltage difference of 50 V was required to efficiently fragment the precursor CoRP ion at m/z 857.60 whereas a TRANSFER cell voltage difference of 55 V was needed to obtain high enough yields of fragment ions of the precursor CoRP ion at m/z 909.57. Subsequent submission of the two different mass lists from the processed fragment ion mass spectra to Uniprot data base search revealed two amino acid sequences, VNLLSAIK for the CoRP with m/z 857.60 (score 14; threshold 26, aa79–86) and IAVSYQTK for the CoRP with m/z 909.57 (score 35; threshold 26, aa87–94), respectively (Fig. 4G–4H and supplemental Fig. S6, cf.supplemental Tables S12–S13). The lower score for VNLLSAIK can be explained by the fact that at the TRANSFER cell collision voltage of 50 V that was applied to sufficiently fragment the precursor ion at m/z 857.60 some rather high intensity fragments of the IAVSYQTK precursor peptide were seen as well, now characterized as noise. Because unmatched ion signals generate a penalty in the Mascot algorithm, the ion score for the amino acid sequence of the target peptide is low. Interestingly, the two peptides are located adjacent to each other in the amino acid sequence of the rhTNFα (supplemental Fig. S8).
Fig. 4.
Mass spectrometric dissociation of the TNFα epitope peptide – anti TNFα antibody complex and amino acid sequence determination of the complex-released peptide by mass spectrometric fragmentation. Ion mobility arrival time plots of A, Solution 1, C, and E, Solution 3. Dashed lines mark the regions for mass spectra selections. Settings of the TRAP cell, the TRANSFER cell, and the quadrupole are indicated at the right. B, D, and F, nanoESI mass spectra (low m/z range) of ions from selected arrival time ranges. Selected m/z values are given, and charge states are indicated in parentheses. For ion signal assignments see supplemental Table S11. G, Amino acid sequence of the complex-released peptide (single letter code) as determined by the matched mass spectrometric fragment ions (fragment ion types and numbers are indicated). The Uniprot protein ID of the peptide source protein (first hit) is shown. H, Pseudo mass spectrum (after charge deconvolution and de-isotoping) of fragment ions derived by selecting arrival time of the complex-released peptide with m/z 909.57. For ion signal assignments see supplemental Table S12.
To examine whether both peptides together form the antiTNFα antibody epitope or, alternatively, to decide which of the two peptides contained the specific epitope, we compared their hydrophobicity values and solvent accessible surface areas (asa) using X-ray crystallographic data (1tnf.pdb). Our computational investigations revealed that the amino acid residues of the VNLLSAIK peptide (aa79–86) were highly hydrophobic and only surface accessible when assuming the presence of monomeric rhTNFα. By contrast, the residues of the IAVSYQTK peptide were highly surface accessible in both, trimeric and monomeric rhTNFα. The IAVSYQTK peptide (aa87–94) is rather hydrophilic as compared with the VNLLSAIK peptide (supplemental Fig. S9).
From this comparison it can be concluded that on the trimeric rhTNFα the IAVSYQTK peptide is highly surface exposed and, hence, is better accessible for antibody (paratope) recognition than the VNLLSAIK peptide. On the other hand it can be argued that the residues of VNLLSAIK are involved in the trimerization interface of TNFα and, consistent with the three dimensional structure of TNFα, only become surface exposed on destroying the rhTNFα trimer (Fig. 5). Thus, the appearance of the VNLLSAIK peptide in the ITEM-THREE experiment may be a result of nonspecific interaction of this peptide to the antibody that could be attributed to its “stickiness” because of its many hydrophobic residues.
Fig. 5.

3D structure images of the human TNFα protein trimer. A, The cartoon display shows ribbons with backbone atom coordinates. B, Display of vanderWaals surfaces. Monomer surfaces are shown in different green shades. The epitope peptides′ surfaces on each monomer (aa79 – aa86: IAVSYQTK) are displayed in red, the adjacent peptides′ surfaces (aa87 – aa94: VNNLSAIK) in purple (cf. supplemental Fig. S9).
However, when in control experiments either rituximab or a mouse monoclonal anti-actin antibody (solutions 2) was added to the rhTNFα tryptic peptide mixtures (solution 1) not a single peptide was fished out by either of the two negative control antibodies in ITEM-THREE (data not shown). Hence, we are tempted to speculate that the antiTNFα antibody may not be monoclonal. If we were to assume that the commercially available antiTNFα antibody was in fact containing two antibody clones, then, one might bind to the surface accessible peptide (IAVSYQTK) as the “expected” epitope and the other could bind to the “sticky” peptide (VNLLSAIK). In this case both peptides fully satisfied the properties of epitope peptides.
DISCUSSION
ITEM-THREE differs from MALDI MS-based epitope mapping methods (20, 53–55) in several aspects. Most currently available MALDI-MS approaches for epitope mapping require immobilization of the capturing antibody on a protein A (or protein G) substrate or on some other sort of a substrate (beads or columns) when chemically immobilized. Using a substrate is usually associated with the risk of nonspecific adsorption of the antigen/epitope peptide to its surface. Respective control experiments that test for nonspecific adsorption of antigens and/or epitope peptides onto the substrate material become mandatory, thereby doubling or tripling the amount of antigen/epitope peptides to be consumed. In ITEM-THREE one does not need to fix the capturing antibody on any substrate and therefore a respective negative control experiment with only substrate (bead or column surfaces, protein A or protein G, etc.) is obsolete. Hence, with ITEM-THREE sample consumption is reduced and the experimenter′s time is saved as well. Perhaps more importantly, ITEM-THREE allows one to check whether an antibody-epitope peptide complex has been formed in the first place. This is done by recording mass spectra with higher m/z range before dissociation of the complex. Using a MALDI MS-based method, the formation of the immune complex is not directly observable as on addition of the matrix (mostly because of the acidic solution in which the matrix is typically dissolved in), immune complexes are destroyed.
Of note, in MALDI-MS-based methods, mass spectrometry is applied “only” as a readout for the in-solution enzymatic/chemical processing steps, whereas in ITEM-THREE the mass spectrometer′s capabilities of ion manipulation and sorting become part of the experimental process. For one, we have observed that loosely (nonspecifically) attached peptides can be “shaken off” from the antibody surface during transition between the condensed and the gas phase by adjusting desorption/ionization conditions. Stronger bound epitope-containing peptides mostly survive this “cleansing” step and are dissociated later by applying CID conditions in a collision cell within the mass spectrometer. A comparable “clean-up” effect within the ionization regime may be difficult to achieve by applying a MALDI-based method, as dissociation of peptides is not controlled but forced by denaturation of the antibody/immune complex under acidic matrix preparation conditions.
Also worth mentioning is the fact that dissociation of the epitope-containing peptide from the antibody by CID generates the charged epitope peptide ions in an ionization process that is completely different than that in an ionization source of a mass spectrometer. In the ion source all analytes compete for obtaining charge carriers (protons in positive ion mode) and there is a selection taking place by which in general more basic peptides become better ionized than less basic peptides. In MALDI-MS ionization the “lucky survivor concept” has found acceptance (56, 57). This in the end may have an impact in the detection sensitivity because there is the risk that some peptides simply may not ionize well and, hence their ion signals are weak and, depending on the noise, may be overlooked. By contrast, as has been amply described (6, 47, 58, 59) CID results in an asymmetric charge distribution and the epitope-containing peptide, being the smaller partner of the to be dissociated complex, receives at least one proton. At least in our hands, ionization yields were not limiting detection of the epitope peptides.
Further, additional in-solution handling steps, such as trimming of the epitope by applying different enzymes (60) can be done equally well in both of the two methods, MALDI-based and ITEM-THREE, at least as long as the proteases that in ITEM-THREE are to be added to one and the same sample without purification are stable enough and not digest each other faster than the time needed for trimming of the epitope peptide. Another prerequisite for ITEM-THREE is that the applied proteases should work in ammonium acetate buffer at pH 7. Yet, also in MALDI-based epitope mapping methods, during “trimming” the experimental conditions have to be kept such that the immune complex is not destroyed, i.e. a pH value of about neutral.
The fact that ITEM-THREE reveals partial amino acid sequences of epitope peptides as well as the respective protein name of the antigen in one experiment, principally allows one to identify unknown antigens of an antibody of interest in a given protein extract. An example for the identification of an unknown epitope amino acid sequence is presented by the chemically modified His-tag containing C-terminal epitope peptide, KCFHHHHHH, of rhβactin that was found to be triply carbamidomethylated. Both, the epitope peptide amino acid sequence and the location of the modifications were clearly identified by mass spectrometric fragmentation, thereby proving the versatility of ITEM-THREE. The observed double-carbamidomethylation at the lysine residue of the epitope peptide is consistent with reports that have shown that lysine residues might be modified by iodoacetamide, depending on the lysine residues' microenvironment (61, 62). In addition to the highly abundant [M+2H]2+ ion at m/z 695.82 that was released from the immune complex, we also observed ion signals of two low-abundance doubly charged ion signals (m/z 687.31 and 702.83) with mass differences of −17.04 Da and +14.02 Da with respect to the epitope peptide′s mass (Fig. 3F). The −17.04 Da likely was because of a loss of ammonia (63), whereas +14.02 Da can be explained by methylation of the K373 residue (64). Although loss of ammonia is produced from the peptide at elevated collision energies, methylation of the peptide was observed already in the peptide mixture of the tryptic digest of rhβactin (supplemental Table S7). Obviously, epitope peptides that contain modified residues because of post-translational modifications or as a result of chemical conversions during sample preparation are still identified by ITEM-THREE, provided that the modification does not affect recognition of the epitope by the antibody′s paratope. This stands in agreement with findings of methionine oxidized His-tag containing peptides, where oxidation did not prevent recognition of the epitope by the antibody under investigation or with partially carbamylated epitope peptides, where the chemical modification of a lysine residue was tolerated by the antibody as well (34). Such results illustrate the importance of ITEM-THREE as a method that can study amino acid modification-related effects on antigen-antibody binding.
Once an epitope peptide′s sequence has been determined, one becomes able to search for amino acid sequence similarities on other proteins, thereby allowing to estimate cross-reactivities of the investigated antibody, or even to predict which unrelated proteins might bind to an antibody of interest in addition to its specific antigen. Such information becomes particularly relevant when one aims at using antibodies to identify proteins in species for which an antibody has not been produced or specified. When the amino acid sequence of the protein of interest is known, one can predict whether a given antibody might be a good binder, i.e. suitable for immune assays within a research project of a different species. Based on epitope peptide sequence similarities, the applicability of a precious antibody was securely broadened (8, 65). Practicality of this approach is illustrated by NCBI BLAST searches using the here identified epitope peptide sequences without taxonomy restrictions. The obtained lists of proteins that shared amino acid sequences that were like the query sequences from the identified epitope peptides, for instance, provided more than 250 entries, each. Within the top 50 listed proteins in the report from the BLAST search applying the RA33 epitope peptide, only the top 6 proteins contained amino acid sequence stretches that were 100% identical to the query sequence. The next 16 proteins in the list had 100% identity to shorter query sequence stretches (Table I). The following amino acid sequences in the list were from unrelated proteins that possessed partial amino acid sequences with still high homologies to the original epitope peptide sequence.
Table I. BLAST search results with identified epitope peptide amino acid sequences.
| Epitope peptide sequence (query)a,b | Protein IDs of 1st hit list entrya,c | No. of peptides identicald/similare to query | Query/urp hit lengthf | 1st urp's epitope-related amino acid sequenceg | Protein IDs of 1st urpg | List pos. of 1st urpg |
|---|---|---|---|---|---|---|
| MAARPHSIDGRVVEP | ROA2_HUMAN | 6/16 | 15/15 | LASRPHTLDGRNIDP | DAZP1_XENLA | 24 |
| LQELEKDEREQLRILGE | RO52_HUMAN | 1/26 | 17/17 | TFLEKTERLEQLRILEN | KDSB_RICTY | 3 |
| KCFHHHHHHh | rhβ-actin_HUMAN | 1/0 | 9/22 | QHHHQHHFHHHHHHHHHHHNHG | SUV42_DROME | 2 |
| IAVSYQTK | TNFA_HUMAN | 15/17 | 8/8 | GAVNYQTK | HPUB_NEIMC | 20 |
aUniprot database was searched.
bSequence parts which are identical to the query sequence are printed in bold.
cNumber of subjects (entries in hit list) was limited to 250.
dNumber of subjects (entries in hit list) identified with 100% sequence coverage and 100% sequence identity to query.
eNumber of subjects (entries in hit list) identified with 100% sequence coverage and less than 100% sequence identity to query.
furp: unrelated protein.
gFirst subject (entry in hit list) which belongs to an unrelated protein.
hAmended Uniprot database was searched.
The same type of results, which match with reports from cross-reactivity-studies with diagnostic heart muscle troponin T antibodies (65), can be obtained with any epitope peptide amino acid sequence (Table I). Because of amino acid sequence similarities it is likely that the antibody of interest was able to recognize binding motifs on unrelated proteins, as long as the targeted amino acid sequence was surface exposed and the partial peptide assumed a somewhat similar three dimensional structure compared with that of the original epitope peptide, even when the respective antibody was not raised against the unrelated protein.
ITEM-THREE is an electrospray mass spectrometry-based method that determines an utmost important antibody feature, its molecular recognition code. Except for mixing the antibody of interest with a peptide mixture that contains the epitope peptide, all experimental steps, such as epitope extraction and epitope peptide sequencing are performed in a single mass spectrometry experiment. Followed by an in-silico search, which starts with subjecting the experimentally determined mass lists to unsupervised data base search, the epitopes′ peptide amino acid sequences are defined, and the originating antigens thereby unequivocally determined while homologues are retrievable and cross-reactivities estimated using BLAST search tools.
DATA AVAILABILITY
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013823 (http://www.ebi.ac.uk/pride/archive/projects/PXD013823).
Supplementary Material
Acknowledgments
We thank Mr. Michael Kreutzer for providing his expertise on bioinformatics.
Footnotes
* We acknowledge the German Academic Exchange Service (DAAD) for providing scholarships for BD (No. 91566064). The WATERS Synapt G2S mass spectrometer has been bought through an EU grant [EFRE-UHROM 9] made available to MOG.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HDX
- hydrogen deuterium exchange
- Nano-ESI
- nano-electrospray ionization
- IMS
- ion mobility separation
- ToF
- time of flight
- CID
- collision induced dissociation
- ΔCV
- collision cell voltage difference
- UBPs
- unbound peptide ions
- CoRPs
- complex-released peptide ions
- BLAST
- basic local alignment search tool.
REFERENCES
- 1. Baraniak I., Kropff B., McLean G. R., Pichon S., Piras-Douce F., Milne R. S. B., Smith C., Mach M., Griffiths P. D., and Reeves M. B. (2018) Epitope-specific humoral responses to human cytomegalovirus glycoprotein-B vaccine with MF59: anti-AD2 levels correlate with protection from viremia. J. Infect. Dis. 217, 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu K., Acharya P., Kong R., Cheng C., Chuang G. Y., Liu K., Louder M. K., O'Dell S., Rawi R., Sastry M., Shen C. H., Zhang B., Zhou T., Asokan M., Bailer R. T., Chambers M., Chen X., Choi C. W., Dandey V. P., Doria-Rose N. A., Druz A., Eng E. T., Farney S. K., Foulds K. E., Geng H., Georgiev I. S., Gorman J., Hill K. R., Jafari A. J., Kwon Y. D., Lai Y. T., Lemmin T., McKee K., Ohr T. Y., Ou L., Peng D., Rowshan A. P., Sheng Z., Todd J. P., Tsybovsky Y., Viox E. G., Wang Y., Wei H., Yang Y., Zhou A. F., Chen R., Yang L., Scorpio D. G., McDermott A. B., Shapiro L., Carragher B., Potter C. S., Mascola J. R., and Kwong P. D. (2018) Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Volk A. L., Hu F. J., Berglund M. M., Nordling E., Stromberg P., Uhlen M., and Rockberg J. (2016) Stratification of responders towards eculizumab using a structural epitope mapping strategy. Sci. Rep. 6, 31365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pritchard A. L. (2018) Targeting neoantigens for personalised immunotherapy. BioDrugs 32, 99–109 [DOI] [PubMed] [Google Scholar]
- 5. Linnebacher M., Lorenz P., Koy C., Jahnke A., Born N., Steinbeck F., Wollbold J., Latzkow T., Thiesen H. J., and Glocker M. O. (2012) Clonality characterization of natural epitope-specific antibodies against the tumor-related antigen topoisomerase IIa by peptide chip and proteome analysis: a pilot study with colorectal carcinoma patient samples. Anal. Bioanal. Chem. 403, 227–238 [DOI] [PubMed] [Google Scholar]
- 6. Yefremova Y., Opuni K. F. M., Danquah B. D., Thiesen H. J., and Glocker M. O. (2017) Intact transition epitope mapping (ITEM). J. Am. Soc. Mass. Spectrom. 28, 1612–1622 [DOI] [PubMed] [Google Scholar]
- 7. Al-Majdoub M., Koy C., Lorenz P., Thiesen H. J., and Glocker M. O. (2013) Mass spectrometric and peptide chip characterization of an assembled epitope: analysis of a polyclonal antibody model serum directed against the SjOgren/systemic lupus erythematosus autoantigen TRIM21. J. Mass Spectrom. 48, 651–659 [DOI] [PubMed] [Google Scholar]
- 8. El-Kased R. F., Koy C., Deierling T., Lorenz P., Qian Z., Li Y., Thiesen H. J., and Glocker M. O. (2009) Mass spectrometric and peptide chip epitope mapping of rheumatoid arthritis autoantigen RA33. Eur. J. Mass Spectrom. 15, 747–759 [DOI] [PubMed] [Google Scholar]
- 9. Weidele K., Stojanovic N., Feliciello G., Markiewicz A., Scheitler S., Alberter B., Renner P., Haferkamp S., Klein C. A., and Polzer B. (2018) Microfluidic enrichment, isolation and characterization of disseminated melanoma cells from lymph node samples. Int. J. Cancer 145, 232–241 [DOI] [PubMed] [Google Scholar]
- 10. Mankoff D. A., Edmonds C. E., Farwell M. D., and Pryma D. A. (2016) Development of companion diagnostics. Semin. Nucl. Med. 46, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies D. R., Padlan E. A., and Sheriff S. (1990) Antibody-antigen complexes. Annu. Rev. Biochem. 59, 439–473 [DOI] [PubMed] [Google Scholar]
- 12. Toride King M., and Brooks C. L. (2018) Epitope mapping of antibody-antigen interactions with X-ray crystallography. Methods Mol. Biol. 1785, 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bardelli M., Livoti E., Simonelli L., Pedotti M., Moraes A., Valente A. P., and Varani L. (2015) Epitope mapping by solution NMR spectroscopy. J. Mol. Recognit. 28, 393–400 [DOI] [PubMed] [Google Scholar]
- 14. Simonelli L., Pedotti M., Bardelli M., Jurt S., Zerbe O., and Varani L. (2018) Mapping antibody epitopes by solution NMR spectroscopy: practical considerations. Methods Mol. Biol. 1785, 29–51 [DOI] [PubMed] [Google Scholar]
- 15. Chiliveri S. C., and Deshmukh M. V. (2016) Recent excitements in protein NMR: Large proteins and biologically relevant dynamics. J. Biosci. 41, 787–803 [DOI] [PubMed] [Google Scholar]
- 16. Pan J. X., Zhang S. P., Chou A., and Borchers C. H. (2016) Higher-order structural interrogation of antibodies using middle-down hydrogen/deuterium exchange mass spectrometry. Chem. Sci. 7, 1480–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gershoni J. M., Roitburd-Berman A., Siman-Tov D. D., Tarnovitski Freund N., and Weiss Y. (2007) Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs 21, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mackay J. P., Landsberg M. J., Whitten A. E., and Bond C. S. (2017) Whaddaya know: a guide to uncertainty and subjectivity in structural biology. Trends Biochem. Sci. 42, 155–167 [DOI] [PubMed] [Google Scholar]
- 19. Al-Majdoub M., Opuni K. F., Koy C., and Glocker M. O. (2013) Facile fabrication and instant application of miniaturized antibody-decorated affinity columns for higher-order structure and functional characterization of TRIM21 epitope peptides. Anal. Chem. 85, 10479–10487 [DOI] [PubMed] [Google Scholar]
- 20. Opuni K. F. M., Al-Majdoub M., Yefremova Y., El-Kased R. F., Koy C., and Glocker M. O. (2018) Mass spectrometric epitope mapping. Mass Spectrom. Rev. 37, 229–241 [DOI] [PubMed] [Google Scholar]
- 21. Pimenova T., Nazabal A., Roschitzki B., Seebacher J., Rinner O., and Zenobi R. (2008) Epitope mapping on bovine prion protein using chemical cross-linking and mass spectrometry. J. Mass Spectrom. 43, 185–195 [DOI] [PubMed] [Google Scholar]
- 22. Horne J. E., Walko M., Calabrese A. N., Levenstein M. A., Brockwell D. J., Kapur N., Wilson A. J., and Radford S. E. (2018) Rapid mapping of protein interactions using tag-transfer photocrosslinkers. Angewandte Chemie-International Edition 57, 16688–16692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Artigues A., Nadeau O. W., Rimmer M. A., Villar M. T., Du X., Fenton A. W., and Carlson G. M. (2016) Protein structural analysis via mass spectrometry-based proteomics. Adv. Exp. Med. Biol. 919, 397–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones L. M., Sperry J. B., Carroll J. A., and Gross M. L. (2011) Fast photochemical oxidation of proteins for epitope mapping. Anal. Chem. 83, 7657–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J., Wei H., Krystek S. R. Jr, Bond D., Brender T. M., Cohen D., Feiner J., Hamacher N., Harshman J., Huang R. Y., Julien S. H., Lin Z., Moore K., Mueller L., Noriega C., Sejwal P., Sheppard P., Stevens B., Chen G., Tymiak A. A., Gross M. L., and Schneeweis L. A. (2017) Mapping the energetic epitope of an antibody/interleukin-23 interaction with hydrogen/deuterium exchange, fast photochemical oxidation of proteins mass spectrometry, and alanine shave mutagenesis. Anal. Chem. 89, 2250–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glocker M. O., Nock S., Sprinzl M., and Przybylski M. (1998) Characterization of surface topology and binding area in complexes of the elongation factor proteins EF-Ts and EF-Tu center dot GDP from Thermus thermophilus: A study by protein chemical modification and mass spectrometry. Chemistry 4, 707–715 [Google Scholar]
- 27. Suckau D., Mak M., and Przybylski M. (1992) Protein surface topology-probing by selective chemical modification and mass spectrometric peptide mapping. Proc. Natl. Acad. Sci. U.S.A. 89, 5630–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dhungana S., Fessler M. B., and Tomer K. B. (2009) Epitope mapping by differential chemical modification of antigens. Methods Mol. Biol. 524, 119–134 [DOI] [PubMed] [Google Scholar]
- 29. Glocker M. O., Borchers C., Fiedler W., Suckau D., and Przybylski M. (1994) Molecular characterization of surface topology in protein tertiary structures by amino-acylation and mass spectrometric peptide mapping. Bioconjug. Chem. 5, 583–590 [DOI] [PubMed] [Google Scholar]
- 30. Yefremova Y., Danquah B. D., Opuni K. F., El-Kased R., Koy C., and Glocker M. O. (2017) Mass spectrometric characterization of protein structures and protein complexes in condensed and gas phase. Eur. J. Mass Spectrom. 23, 445–459 [DOI] [PubMed] [Google Scholar]
- 31. Suckau D., Kohl J., Karwath G., Schneider K., Casaretto M., Bitter-Suermann D., and Przybylski M. (1990) Molecular epitope identification by limited proteolysis of an immobilized antigen-antibody complex and mass spectrometric peptide mapping. Proc. Natl. Acad. Sci. U.S.A. 87, 9848–9852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macht M., Fiedler W., Kurzinger K., and Przybylski M. (1996) Mass spectrometric mapping of protein epitope structures of myocardial infarct markers myoglobin and troponin T. Biochemistry 35, 15633–15639 [DOI] [PubMed] [Google Scholar]
- 33. Baschung Y., Lupu L., Moise A., Glocker M., Rawer S., Lazarev A., and Przybylski M. (2018) Epitope ligand binding sites of blood group oligosaccharides in lectins revealed by pressure-assisted proteolytic excision affinity mass spectrometry. J. Am. Soc. Mass Spectrom. 29, 1881–1891 [DOI] [PubMed] [Google Scholar]
- 34. El-Kased R., Koy C., Lorenz P., Montgomery H., Tanaka K., Thiesen H., and Glocker M. (2011) A novel Mass spectrometric epitope mapping approach without immobilization of the antibody. J. Proteomics Bioinform. 4, 001–009 [Google Scholar]
- 35. Uetrecht C., Rose R. J., van Duijn E., Lorenzen K., and Heck A. J. (2010) Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 39, 1633–1655 [DOI] [PubMed] [Google Scholar]
- 36. Valentine S. J., Liu X., Plasencia M. D., Hilderbrand A. E., Kurulugama R. T., Koeniger S. L., and Clemmer D. E. (2005) Developing liquid chromatography ion mobility mass spectometry techniques. Expert Rev. Proteomics 2, 553–565 [DOI] [PubMed] [Google Scholar]
- 37. Wyttenbach T., Pierson N. A., Clemmer D. E., and Bowers M. T. (2014) Ion mobility analysis of molecular dynamics. Annu. Rev. Phys. Chem. 65, 175–196 [DOI] [PubMed] [Google Scholar]
- 38. VanAernum Z. L., Gilbert J. D., Belov M. E., Makarov A. A., Horning S. R., and Wysocki V. H. (2019) Surface-induced dissociation of noncovalent protein complexes in an extended mass range orbitrap mass spectrometer. Anal. Chem. 91, 3611–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Busch F., VanAernum Z. L., Ju Y., Yan J., Gilbert J. D., Quintyn R. S., Bern M., and Wysocki V. H. (2018) Localization of protein complex bound ligands by surface-induced dissociation high-resolution mass spectrometry. Anal. Chem. 90, 12796–12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X., Li H., Moore B., Wongkongkathep P., Ogorzalek Loo R. R., Loo J. A., and Julian R. R. (2014) Radical-directed dissociation of peptides and proteins by infrared multiphoton dissociation and sustained off-resonance irradiation collision-induced dissociation with Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 28, 2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Halim M. A., Girod M., MacAleese L., Lemoine J., Antoine R., and Dugourd P. (2016) Combined infrared multiphoton dissociation with ultraviolet photodissociation for ubiquitin characterization. J. Am. Soc. Mass Spectrom. 27, 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ben-Nissan G., and Sharon M. (2018) The application of ion-mobility mass spectrometry for structure/function investigation of protein complexes. Curr. Opin. Chem. Biol. 42, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clemmer D. E., and Jarrold M. F. (1997) Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 32, 577–592 [Google Scholar]
- 44. Hoaglund C. S., Valentine S. J., Sporleder C. R., Reilly J. P., and Clemmer D. E. (1998) Three-dimensional ion mobility/TOFMS analysis of electrosprayed biomolecules. Anal. Chem. 70, 2236–2242 [DOI] [PubMed] [Google Scholar]
- 45. Glocker M. O., Arbogast B., and Deinzer M. L. (1995) Characterization of disulfide linkages and disulfide bond scrambling in recombinant human macrophage colony stimulating factor by fast-atom bombardment mass spectrometry of enzymatic digests. J. Am. Soc. Mass Spectrom. 6, 638–643 [DOI] [PubMed] [Google Scholar]
- 46. Yang J., Rower C., Koy C., Russ M., Ruger C. P., Zimmermann R., von Fritschen U., Bredell M., Finke J. C., and Glocker M. O. (2015) Mass spectrometric characterization of limited proteolysis activity in human plasma samples under mild acidic conditions. Methods 89, 30–37 [DOI] [PubMed] [Google Scholar]
- 47. Yefremova Y., Melder F. T. I., Danquah B. D., Opuni K. F. M., Koy C., Ehrens A., Frommholz D., Illges H., Koelbel K., Sobott F., and Glocker M. O. (2017) Apparent activation energies of protein-protein complex dissociation in the gas-phase determined by electrospray mass spectrometry. Anal. Bioanal. Chem. 409, 6549–6558 [DOI] [PubMed] [Google Scholar]
- 48. Wolter M., Okai C. A., Smith D. S., Russ M., Rath W., Pecks U., Borchers C. H., and Glocker M. O. (2018) Maternal apolipoprotein B100 serum levels are diminished in pregnancies with intrauterine growth restriction and differentiate from controls. Proteomics Clin. Appl. 12, e1800017. [DOI] [PubMed] [Google Scholar]
- 49. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., and Vizcaino J. A. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pecks U., Seidenspinner F., Rower C., Reimer T., Rath W., and Glocker M. O. (2010) Multifactorial analysis of affinity-mass spectrometry data from serum protein samples: a strategy to distinguish patients with preeclampsia from matching control individuals. J. Am. Soc. Mass Spectrom. 21, 1699–1711 [DOI] [PubMed] [Google Scholar]
- 51. Kienbaum M., Koy C., Montgomery H. V., Drynda S., Lorenz P., Illges H., Tanaka K., Kekow J., Guthke R., Thiesen H. J., and Glocker M. O. (2009) MS characterization of apheresis samples from rheumatoid arthritis patients for the improvement of immunoadsorption therapy - a pilot study. Proteomics Clin. Appl. 3, 797–809 [DOI] [PubMed] [Google Scholar]
- 52. Madi A., Mikkat S., Ringel B., Ulbrich M., Thiesen H. J., and Glocker M. O. (2003) Mass spectrometric proteome analysis for profiling temperature-dependent changes of protein expression in wild-type Caenorhabditis elegans. Proteomics 3, 1526–1534 [DOI] [PubMed] [Google Scholar]
- 53. Al-Majdoub M., Opuni K. F., Yefremova Y., Koy C., Lorenz P., El-Kased R. F., Thiesen H. J., and Glocker M. O. (2014) A novel strategy for the rapid preparation and isolation of intact immune complexes from peptide mixtures. J. Mol. Recognit. 27, 566–574 [DOI] [PubMed] [Google Scholar]
- 54. Rinaldi F., Lupu L., Rusche H., Kukacka Z., Tengattini S., Bernardini R., Piubelli L., Bavaro T., Maeser S., Pollegioni L., Calleri E., Przybylski M., and Temporini C. (2019) Epitope and affinity determination of recombinant Mycobacterium tuberculosis Ag85B antigen towards anti-Ag85 antibodies using proteolytic affinity-mass spectrometry and biosensor analysis. Anal. Bioanal. Chem. 411, 439–448 [DOI] [PubMed] [Google Scholar]
- 55. Raska C. S., Parker C. E., Sunnarborg S. W., Pope R. M., Lee D. C., Glish G. L., and Borchers C. H. (2003) Rapid and sensitive identification of epitope-containing peptides by direct matrix-assisted laser desorption/ionization tandem mass spectrometry of peptides affinity-bound to antibody beads. J. Am. Soc. Mass Spectrom. 14, 1076–1085 [DOI] [PubMed] [Google Scholar]
- 56. Jaskolla T. W., and Karas M. (2011) Compelling evidence for Lucky Survivor and gas phase protonation: the unified MALDI analyte protonation mechanism. J. Am. Soc. Mass Spectrom. 22, 976–988 [DOI] [PubMed] [Google Scholar]
- 57. Karas M., Gluckmann M., and Schafer J. (2000) Ionization in matrix-assisted laser desorption/ionization: singly charged molecular ions are the lucky survivors. J. Mass Spectrom. 35, 1–12 [DOI] [PubMed] [Google Scholar]
- 58. Sciuto S. V., Liu J., and Konermann L. (2011) An electrostatic charge partitioning model for the dissociation of protein complexes in the gas phase. J. Am. Soc. Mass Spectrom. 22, 1679–1689 [DOI] [PubMed] [Google Scholar]
- 59. Zhou M., Dagan S., and Wysocki V. H. (2012) Protein subunits released by surface collisions of noncovalent complexes: nativelike compact structures revealed by ion mobility mass spectrometry. Angew Chem. Int. Ed. Engl. 51, 4336–4339 [DOI] [PubMed] [Google Scholar]
- 60. Parker C. E., Papac D. I., Trojak S. K., and Tomer K. B. (1996) Epitope mapping by mass spectrometry: determination of an epitope on HIV-1 IIIB p26 recognized by a monoclonal antibody. J. Immunol. 157, 198–206 [PubMed] [Google Scholar]
- 61. Svozil J., and Baerenfaller K. (2017) A cautionary tale on the inclusion of variable posttranslational modifications in database-dependent searches of mass spectrometry data. Proteomics Biol. 586, 433–452 [DOI] [PubMed] [Google Scholar]
- 62. Mukouyama E. B., Oguchi M., Kodera Y., Maeda T., and Suzuki H. (2004) Low pKa lysine residues at the active site of sarcosine oxidase from Corynebacterium sp. U-96. Biochem. Biophys. Res. Commun. 320, 846–851 [DOI] [PubMed] [Google Scholar]
- 63. Sun S., Yu C., Qiao Y., Lin Y., Dong G., Liu C., Zhang J., Zhang Z., Cai J., Zhang H., and Bu D. (2008) Deriving the probabilities of water loss and ammonia loss for amino acids from tandem mass spectra. J. Proteome Res. 7, 202–208 [DOI] [PubMed] [Google Scholar]
- 64. Lanouette S., Mongeon V., Figeys D., and Couture J. F. (2014) The functional diversity of protein lysine methylation. Mol. Syst. Biol. 10, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Macht M., Marquardt A., Deininger S. O., Damoc E., Kohlmann M., and Przybylski M. (2004) 'Affinity-proteomics': direct protein identification from biological material using mass spectrometric epitope mapping. Anal. Bioanal. Chem. 378, 1102–1111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013823 (http://www.ebi.ac.uk/pride/archive/projects/PXD013823).