Abstract
Melanoma is the deadliest form of skin cancer, and little is known about the impact of deregulated expression of long noncoding RNAs (lncRNAs) in the progression of this cancer. In this study, we explored RNA-Seq data to search for lncRNAs associated with melanoma progression. We found distinct lncRNA gene expression patterns across melanocytes, primary and metastatic melanoma cells. Also, we observed upregulation of the lncRNA ZEB1-AS1 (ZEB1 antisense RNA 1) in melanoma cell lines. Data analysis from The Cancer Genome Atlas (TCGA) confirmed higher ZEB1-AS1 expression in metastatic melanoma and its association with hotspot mutations in BRAF (B-Raf proto-oncogene, serine/threonine kinase) gene and RAS family genes. In addition, a positive correlation between ZEB1-AS1 and ZEB1 (zinc finger E-box binding homeobox 1) gene expression was verified in primary and metastatic melanomas. Using gene expression signatures indicative of invasive or proliferative phenotypes, we found an association between ZEB1-AS1 upregulation and a transcriptional profile for invasiveness. Enrichment analysis of correlated genes demonstrated cancer genes and pathways associated with ZEB1-AS1. We suggest that the lncRNA ZEB1-AS1 could function by activating ZEB1 gene expression, thereby influencing invasiveness and phenotype switching in melanoma, an epithelial-to-mesenchymal transition (EMT)-like process, which the ZEB1 gene has an essential role.
Subject terms: Melanoma, Gene expression, Transcriptomics, Long non-coding RNAs
Introduction
Melanoma is the most lethal form of skin cancer. Although it comprises only 5% of skin cancers, it corresponds to 80% of patient deaths1. While trends for most cancers have been declining worldwide, the incidence of melanoma has risen in the last decades2. Melanoma progression strongly impacts on patient lifespan, with the 5-year survival rate decreasing from 98% of patients with local melanoma to only 16% of patients with distant melanoma3,4. As for other cancers, metastasis is the leading cause of melanoma deaths, making detection of early disease, when curative resection is possible, considered the most effective treatment.
Currently, the tumour, lymph node, metastasis (TNM) staging system does not provide an accurate prognosis for melanoma3, reflecting the need to identify biomarkers5 with better diagnostic and staging and predictive values. Furthermore, the identification of molecules and mechanisms to confront melanoma development has improved rapidly in the last decades and could in the future provide a new source of treatment strategies. Recent genomic studies have identified a functionally diverse class of noncoding RNAs, defined as long noncoding RNAs (lncRNAs)6, which has been associated with gene regulatory functions. These molecules are transcripts longer than 200 nucleotides lacking protein-coding capacity. They are versatile molecules involved in gene expression regulation by diverse interactions with DNA, RNA and/or proteins7,8. Studies on lncRNAs expression patterns and function have demonstrated enough evidence of their key role in developmental processes9 and diseases10, including many cancers11. In melanoma, lncRNAs have recently been associated with important biological features, such as proliferation12–14, invasion15,16 and apoptosis17,18; however, details of their influence on melanoma development are still missing.
Previous findings of our laboratory demonstrated the influence of the lncRNA HOTAIR (HOX transcript antisense RNA) in stemness and epithelial-to-mesenchymal transition (EMT) in cancer19. The EMT process is a highly conserved critical step for embryogenesis and melanocyte lineage differentiation. Importantly, reactivation of EMT in many epithelial cancers is common and implicated in the loss of cell polarity and cell-to-cell adhesion, cytoskeleton reorganization and gaining of mesenchymal-like morphology, including the ability of migration20,21. A similar process has been observed in melanoma progression, in which melanocytic cells can reversibly change their status towards a more mesenchymal-like state22,23. This melanoma “phenotype switching” model is concordant with gene expression profiling studies, which demonstrated two alternating phenotypic profiles switching from a rapidly proliferative/low invasive condition to slower proliferative/highly migratory with high invasive potential24. Remarkably, proliferative melanoma samples express high levels of MITF (melanocyte inducing transcription factor), SOX10 (SRY-box 10) and PAX3 (paired box 3) genes22,25,26. In contrast, samples with invasive phenotype demonstrate low levels of MITF and high expression of ZEB1 and TGFB1 (transforming growth factor, beta 1) pathway genes27,28. Recently, it was confirmed that the melanoma proliferative phenotype is regulated by SOX10/MITF and that AP1/TEADs are regulators of the invasive phenotype29. Although several protein-coding genes involved in melanoma phenotype switching have been characterized, the role of noncoding genes in this process remains poorly understood.
Herein, we performed RNA sequencing (RNA-Seq) and downloaded public data to search for lncRNAs associated with melanoma progression. Differential gene expression analysis revealed lncRNA ZEB1-AS1 upregulation in melanoma in comparison to normal melanocytes. Importantly, significantly higher levels of ZEB1-AS1 were detected in metastatic melanoma when compared with primary melanoma. Using GSVA (Gene Set Variation Analysis), we found a significant correlation between ZEB1-AS1 expression and a melanoma invasive transcriptional signature. Our study provides insights into the ZEB1-AS1 relationship with melanoma progression and suggests its contribution to ZEB1 regulation, which in turn influences melanoma phenotype switching.
Results
Overview of RNA-Seq data from melanocytes and melanoma cell lines
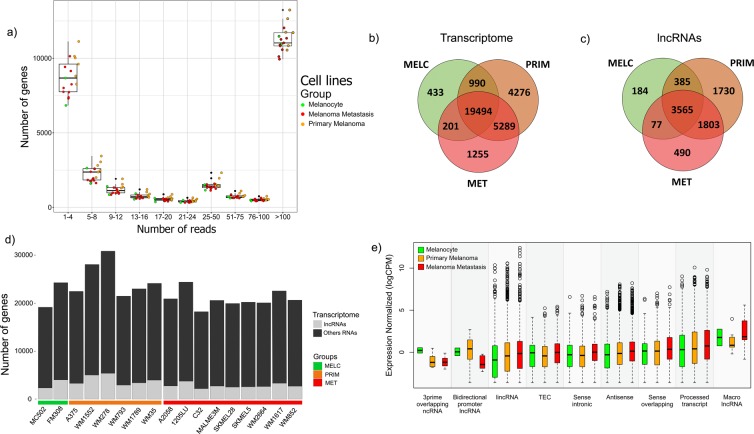
We performed RNA-Seq in melanocytic cell lines (melanocytes = 1, primary melanoma = 5, and metastatic melanoma = 3). Subsequently, we downloaded raw public data (melanocyte = 1, primary melanoma = 1, and metastatic melanoma = 6) from a previous study30 and assembled all data into a single dataset for subsequent genome mapping and analysis. We obtained read mapping efficiency of 64,39% on average (Supplementary Table S1). Figure 1a shows the number of genes in the human transcriptome per range of mapped reads from the dataset. To avoid non-biological overrepresentation of reads detected and to allow further transcriptome characterization, we only considered a gene being expressed when it presented at least five mapped reads, as previously applied by Tarazona et al.31. As expected, we detected the expression of a lower number of lncRNAs in comparison to protein-coding genes in the total transcriptome for each cell line (Fig. 1d). When exploring data for exclusive and common transcripts, the highest number of genes expressed exclusively in a group was found in primary melanoma cell lines (PRIM), which presented a total of 4276 genes, of which 1730 were lncRNA genes. Transcripts exclusively expressed in melanocyte cell lines (MELC) represented a total of 433 genes, of which 184 were lncRNA genes. Besides, metastatic melanoma cell lines (MET) exclusive transcripts represented a total of 1255 genes, of which 490 were lncRNAs genes (Fig. 1b,c). According to the GENCODE lncRNA biotypes classification, the two predominant categories in our dataset were antisense (n = 976) and long intergenic non-coding RNAs – lincRNAs (n = 972) genes. Although being the most frequently detected, the median expression level of lncRNAs in those two biotypes categories were not higher than that from lncRNAs in other categories (Fig. 1e).
Figure 1.
Overview of transcriptome data analysis. (a) Boxplot showing the number of all genes detected within regular range of mapped count reads in assembled RNA-Seq data. (b) Venn diagram showing transcriptome common and exclusive expressed genes (with mapped reads ≥5) in melanocytes (433 genes), primary melanoma (4276 genes) and metastatic melanoma (1255 genes) cell lines. (c) Venn diagram showing only shared and exclusive lncRNAs expressed (with mapped reads ≥5) in melanocytes (184 lncRNAs genes), primary melanoma (1730 lncRNAs genes) and in metastatic melanoma (490) cell lines. (d) Characterization of transcriptome composition showing lncRNAs and other genes expressed (with mapped reads ≥5) in melanocytes, primary and metastatic melanoma cell lines. (e) Boxplot showing lncRNA biotypes according to GENCODE classification in melanoma progression groups (only considering genes with mapped reads ≥5). Middle lines in boxplot represent median.
Differential gene expression analysis between melanocytes and primary and metastatic melanoma cell lines
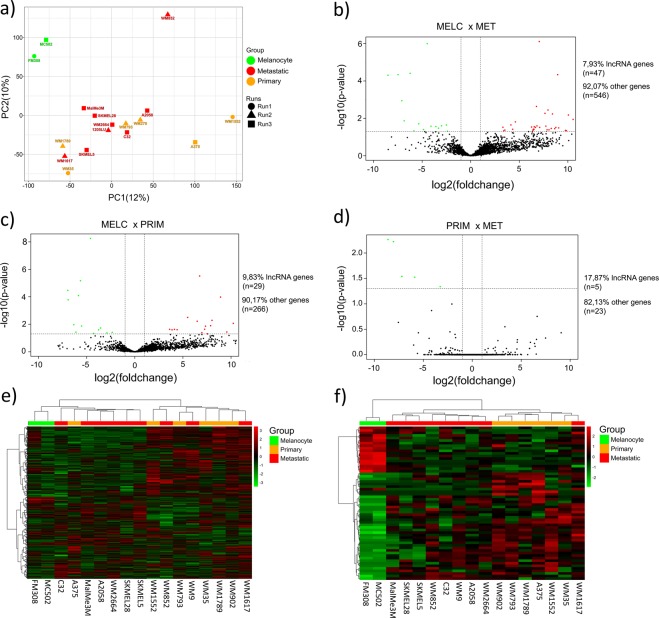
For data quality control and to check cell lines disparities, we performed Principal Component Analysis (PCA). We detected two major clusters distinct from each other, which consists of melanoma (including primary and metastatic) cell lines and melanocyte cell lines (Fig. 2a). We next evaluated differentially expressed genes to detect genes deregulated at different melanoma stages. The analysis comparing melanocytes and metastatic melanoma cell lines (MELC × MET) presented 593 differentially expressed genes, of which 47 were lncRNA genes, including 32 upregulated and 15 downregulated in metastatic melanoma (Fig. 2b). The comparison between melanocytes and primary tumour melanoma cell lines (MELC × PRIM) revealed 295 differentially expressed genes, of which 29 were lncRNA genes, including 16 upregulated and 13 downregulated in primary tumour melanoma cell lines (Fig. 2c). Analysis of primary tumour versus metastatic melanoma cell lines (PRIM × MET) presented 28 differentially expressed genes including five lncRNA genes, all of them downregulated in the metastatic stage (Fig. 2d). We further analyzed expression patterns of differentially expressed genes by unsupervised hierarchical clusterization. We found that melanocytes always clustered together in a distinct manner away from melanoma samples, based either on the total transcriptome (Fig. 2e) or only on the lncRNA gene set (Fig. 2f). However, better separation between the primary and metastatic melanoma cell lines was achieved when performing clusterization based on the lncRNA gene set only.
Figure 2.
PCA and deregulated expressed genes in melanocyte and melanoma. (a) PCA plot showing disparities of melanocytes and melanoma cell lines. Each colour represents a cell line group and shapes represent different runs from the two RNA-Seq data sources. (b) Volcano plot showing deregulated lncRNAs in MELC × MET, including upregulated (red dots) and downregulated (green dots) lncRNAs in metastatic melanoma (MET) in comparison with melanocyte (MELC). (c) Volcano plot showing deregulated lncRNAs in MELC × PRIM, including upregulated (red dots) and downregulated (green dots) lncRNAs in primary melanoma (PRIM) in comparison with melanocyte (MELC). (d) Volcano plot showing deregulated lncRNAs in PRIM × MET, with only downregulated lncRNAs (green dots) in metastatic melanoma (MET) in comparison with primary melanoma (PRIM). (e,f) Hierarchical clusterization using Euclidean distance of all differentially expressed genes and considering only differentially expressed lncRNA genes, respectively.
The lncRNA ZEB1-AS1 is upregulated in both primary and metastatic melanoma cell lines in comparison to melanocytes
To further investigate deregulated lncRNA genes, we then selected top upregulated lncRNAs genes according to their Fold Change (FC) values. Pursuing the characterization of lncRNAs potentially associated with both melanoma development and metastatic spread, we prioritised candidates that were deregulated in both comparisons (MELC × PRIM and MELC × MET). The lncRNA with the highest differential expression in both comparisons (MELC × PRIM and MELC × MET) was the recently characterized melanoma-specific lncRNA SAMMSON18. Similarly, we found the frequently cancer deregulated lncRNA H1932 amongst the top differentially expressed lncRNAs in our comparisons (see Tables 1 and 2). The third lncRNA most upregulated in primary melanoma cell lines in comparison to melanocytes was ZEB1-AS1, which is located in the opposite strand of gene ZEB1. Importantly, the lncRNA ZEB1-AS1 was also amongst the top five lncRNAs upregulated in metastatic melanoma cell lines when compared to melanocytes. The ZEB1-AS1 persistence as a deregulated lncRNA in different comparisons, as well as the fact that ZEB1 gene (that we also found upregulated in melanoma cell lines in comparison to melanocytes) is known to contribute to EMT and cancer progression, lead us to prioritize ZEB1-AS1 for further analysis. Interestingly, in the melanoma cell lines, our analysis revealed a robust positive correlation between ZEB1-AS1 and ZEB1 expression levels (Supplementary Fig. S1).
Table 1.
Top 10 lncRNAs upregulated in primary melanoma cell lines in comparison to melanocytes.
| GENE ID | GENE NAME | LOGFC | ADJ. P-VALUE | GENE TYPE |
|---|---|---|---|---|
| ENSG00000240405.5 | SAMMSON | 10,23 | 5,93E-05 | lincRNA |
| ENSG00000130600.16 | H19 | 9,56 | 4,76E-04 | processed transcript |
| ENSG00000237036.4 | ZEB1-AS1 | 8,91 | 1,48E-07 | antisense |
| ENSG00000268812.3 | RP1-102K2.8 | 8,02 | 2,81E-05 | antisense |
| ENSG00000271857.1 | RP1-244F24.1 | 7,88 | 1,02E-04 | antisense |
| ENSG00000220891.1 | LL22NC03-63E9.3 | 7,55 | 7,64E-04 | antisense |
| ENSG00000231528.2 | FAM225A | 7,34 | 1,18E-04 | lincRNA |
| ENSG00000257497.2 | RP11-585P4.5 | 7,25 | 2,31E-04 | antisense |
| ENSG00000272872.1 | LL22NC03-N14H11.1 | 6,97 | 5,74E-04 | sense intronic |
| ENSG00000206195.10 | DUXAP8 | 6,74 | 2,83E-09 | processed transcript |
Table 2.
Top 10 lncRNAs upregulated in metastatic melanoma cell lines in comparison to melanocytes.
| GENE ID | GENE NAME | LOGFC | ADJ. P-VALUE | GENE TYPE |
|---|---|---|---|---|
| ENSG00000240405.5 | SAMMSON | 10,06 | 6,60E-05 | lincRNA |
| ENSG00000272068.1 | RP11-284F21.9 | 10,54 | 1,42E-04 | lincRNA |
| ENSG00000260604.2 | RP1-140K8.5 | 9,86 | 1,36E-03 | lincRNA |
| ENSG00000244300.2 | GATA2-AS1 | 9,75 | 1,26E-03 | antisense |
| ENSG00000130600.16 | H19 | 9,06 | 7,44E-04 | processed transcript |
| ENSG00000237036.4 | ZEB1-AS1 | 8,95 | 8,68E-08 | antisense |
| ENSG00000259153.1 | RP6-65G23.3 | 8,76 | 6,79E-04 | lincRNA |
| ENSG00000282057.1 | RP4-621F18.2 | 8,65 | 8,85E-04 | lincRNA |
| ENSG00000272502.1 | RP11-713M15.2 | 8,42 | 5,63E-05 | antisense |
| ENSG00000204860.4 | FAM201A | 8,29 | 8,24E-04 | antisense |
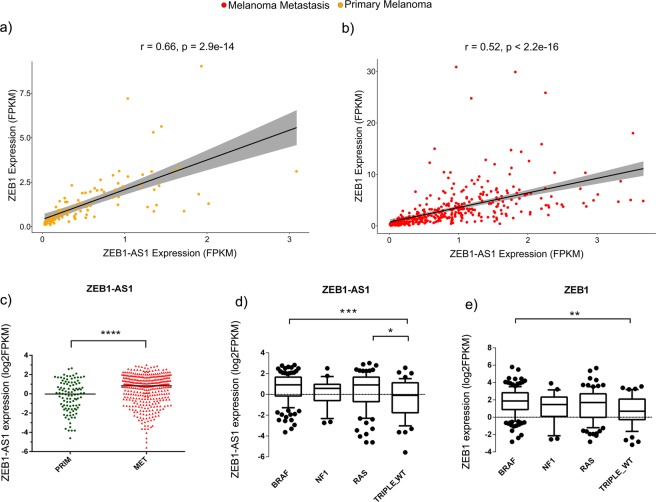
Expression levels of lncRNA ZEB1-AS1 and ZEB1 gene are positively correlated in melanoma tumours and associated with frequent melanoma mutations
According to normal tissue data available in the GTEx (Genotype-Tissue Expression) database33, ZEB1-AS1 is a ubiquitously expressed lncRNA, with a tissue distribution pattern similar to ZEB1 transcription factor (Supplementary Figs S2 and S3). We then evaluated the ZEB1-AS1 expression in cutaneous melanoma samples (Skin Cutaneous Melanoma - SKCM) from the TCGA data. ZEB-AS1 expression showed a strong positive correlation with ZEB1 mRNA in both primary melanomas (Fig. 3a, r = 0.66 and P-value < 2.9E-14) and metastatic melanoma (Fig. 3b, r = 0.52 and P-value < 2.2E-16) samples. Although melanoma presented lower levels of ZEB1-AS1 expression in comparison to other tumours (Supplementary Fig. S4), significantly higher levels of ZEB1-AS1 (P-value < 0.0001) were found in metastasis in comparison to primary melanomas (Fig. 3c). Also, we evaluated the molecular subtypes defined by the TCGA network analysis34. Melanoma samples carrying BRAF or RAS mutations presented significantly higher expression levels of ZEB1-AS1 (P-value < 0.001 and P-value < 0.05, respectively) when compared to the Triple Wild-Type group (Fig. 3d). Comparatively, ZEB1 gene demonstrated higher expression (P-value < 0.001) only in the BRAF group (Fig. 3e).
Figure 3.
Analysis of ZEB1 and ZEB1-AS1 gene expression in TCGA melanoma samples. (a,b) High positive correlation between ZEB1 and ZEB1-AS1 expression in primary melanoma and metastatic melanoma samples, respectively. Analysis from Pearson’s statistical test. Grey band represents the confidence interval. (c) Graph showing higher expression of ZEB1-AS1 in metastatic melanoma compared to primary melanoma samples from SKCM TCGA. T-test analysis was used. (d,e) Boxplot showing expression levels of ZEB1-AS1 or ZEB1, respectively, across different mutational groups of melanoma. Statistics based on ANOVA test. P-values statistical significance labels: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
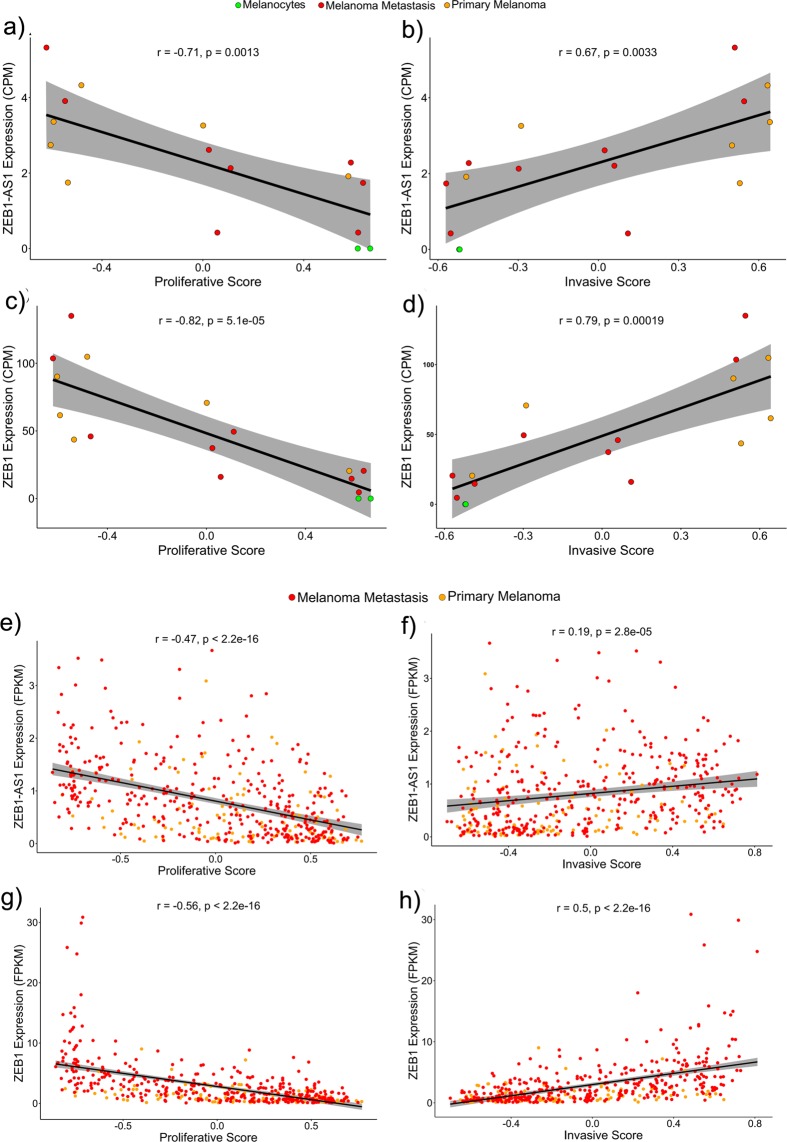
Association between lncRNA ZEB1-AS1 gene expression with melanoma invasive phenotype
It has been proposed that during tumour progression melanoma cells switch between a proliferative and an invasive state35. Specific gene expression signatures had been associated with each one of these phenotypic states22. Using GSVA and the proliferative or the invasive gene signatures we scored each melanoma cell line and each of the tumours sample from the TCGA database for both phenotypic states, based on their expression profile (RNA-Seq data). Then, we evaluated the correlation between ZEB1-AS1 or ZEB1 expression levels and the proliferative or invasive scores. Our results revealed that in melanoma cell lines, ZEB1-AS1 expression presents a robust positive correlation with the invasive score (Fig. 4b, r = 0.67, and P = 0.0033) and is inversely correlated with the proliferative score (Fig. 4a, r = −0.71, and P = 0.0013). Similar results were observed for ZEB1 gene expression, with even higher positive correlation with the invasive score (Fig. 4d, r = 0.79, and P = 0.00019) and higher negative correlation with the proliferative score (Fig. 4c, r = −0.82, and P = 5.1E-05). It is interesting to note that melanocytes, which have the lowest expression levels of ZEB1 and ZEB1-AS1, presented the highest proliferative score and the lowest invasive score values when compared to other melanoma cell lines. Likewise, analysis in the TCGA data set showed ZEB1-AS1 and ZEB1 expressions positively correlated with the invasive score and inversely correlated with the proliferative score in both primary and metastatic melanoma samples (Fig. 4e–h).
Figure 4.
ZEB1 and ZEB1-AS1 association with the invasive phenotype. (a,c) Correlation plots showing a negative correlation between ZEB1-AS1 or ZEB1 expression levels with melanocytic cell lines proliferative score, respectively. (b,d) Correlation plots showing a positive correlation of ZEB1-AS1 or ZEB1 expression levels with melanocytic cell lines invasive score, respectively. (e,g) Correlation plots showing a negative correlation between ZEB1-AS1 or ZEB1 expression levels with proliferative score in SKCM TCGA samples, respectively. (f,h) Correlation plots showing a positive correlation between ZEB1-AS1 or ZEB1 expression levels with an invasive score in SKCM TCGA samples, respectively. Grey band corresponds to the confidence interval.
Functional annotation of ZEB1-AS1 correlated genes in melanoma
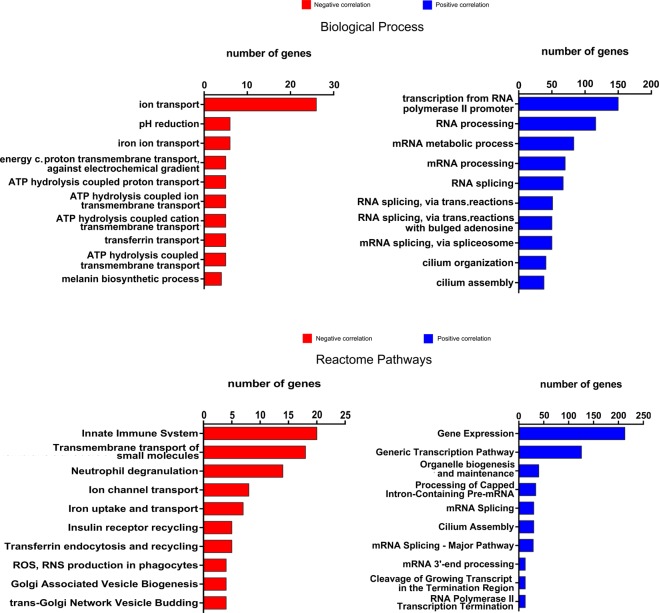
To gain insights on the biological pathways being regulated by ZEB1-AS1 in melanoma, we searched the RNA-Seq data from melanomas in the TCGA database for genes positively or negatively correlated with ZEB1-AS1 expression (correlated genes). This analysis revealed 2059 Positively Correlated genes (PC, Pearson r ≥ 0.3 and P-value < 0.01) and 116 Negatively Correlated genes (NC, Pearson r ≤ −0.3 and P-value < 0.01) with ZEB1-AS1 expression (see Supplementary Files S1 and S2). Comparatively, the same analysis for ZEB1 gene demonstrated 1617 PC genes and 242 NC genes (Supplementary Files S3 and S4). We performed pathway enrichment analysis according to Gene Ontology (see also Supplementary Fig. S5) or Reactome annotations for the genes correlated (positively or negatively) with ZEB1-AS1 expression. In the biological process, positively correlated genes in the top enriched pathways included transcriptional regulation, RNA processing, and cilium organization/assembly (Fig. 5). For the negative correlated genes, top-ranked biological processes were mostly associated with ion transport and intracellular pH regulation (Fig. 5).
Figure 5.
Gene Ontology (GO) and Reactome enrichment analysis showing top-10 ranked GO terms for ZEB1-AS1 correlated genes. Bar plots showing the number of significant genes for enriched Biological Process and Reactome pathways analysis, respectively. Red bars: ZEB1-AS1 negatively correlated genes (Pearson r = <−0.03). Blue bars: ZEB1-AS1 positively correlated genes (Pearson r > = 0.03).
Discussion
In recent years, gene expression and transcriptome analysis revealed lncRNAs with aberrant expression in tumour progression and many evidences support their key role in multiple cancers36,37. Also, it has been demonstrated that lncRNAs present lower expression levels and are more tissue-specific than protein-coding genes, even for cancer-related lncRNAs38–40. We revealed that lncRNAs gene expression produces a more consistent pattern of discrimination between melanocytes and both primary and metastatic melanoma cell lines. These results demonstrate the value of lncRNA profiling as a better way to distinguish the different stages (normal melanocytes, primary tumours, and metastasis) of melanoma development.
Among the top ten upregulated lncRNAs in either primary or metastatic melanoma cell lines in comparison to melanocytes, we found ZEB1-AS1. Previous studies reported the involvement of ZEB1-AS1 in different cancers, but no skin cancer associations until now41. ZEB1-AS1 upregulation was first reported in hepatocellular carcinoma and associated with poor prognosis and tumour growth and metastasis42. Interestingly, epigenetic studies demonstrated mechanistic variance according to different tumours. Liu and Lin (2016), suggested that ZEB1-AS1 directly binds and recruits the histone acetyltransferase P300 (E1A binding protein P300) to the ZEB1 promoter region, activating ZEB1 transcription and resulting in poor prognosis in osteosarcoma43. On the other hand, Su et al. (2017), showed that ZEB1-AS1 binds and recruits MLL1 (mixed lineage leukemia protein-1) to ZEB1 promoter region, inducing H3K4me3 modification and activating ZEB1 transcription44. Until now, lncRNA ZEB1-AS1 has not been associated with melanoma and invasiveness, although there is significant evidence of ZEB1 influence in EMT and melanoma development27,28. For the first time, we demonstrated lncRNA ZEB1-AS1 upregulation in melanoma cell lines and reported significantly higher expression of this lncRNA in metastatic melanomas in comparison to primary tumours using TCGA data. In addition, we confirmed prior expectations of ZEB1 and ZEB1-AS1 positive correlation expression in melanoma as previously reported in some cancers42,44. Moreover, our analysis also increased the findings associated with this positive correlation according to normal tissues or tumours analysed (Supplementary Files S7 and S8, respectively).
Next-generation sequencing studies have identified genetic mutations that provide insights into melanoma heterogeneity, which could have implications in determining patient prognosis or targeted therapy45–47. Previous characterization of TCGA samples according to mutational status was utilized to identify higher expression of ZEB1-AS1 in the BRAF and RAS mutant groups. Since both genes are found within the MAPK pathway, our findings could suggest a role of ZEB1-AS1 in the MAPK pathway regulatory network34. Indeed, a recent study has suggested that resistance to MAPK inhibitors in melanoma could be due to cell plasticity mediated by high levels of ZEB1 gene expression48.
As expected, by reported induction of ZEB1 by ZEB1-AS142, we tested well-established gene signatures associated with an invasive or proliferative phenotype of melanomas49. We showed a positive correlation between higher levels of ZEB1-AS1 and the invasive profile in melanoma cell lines and tumours. Conversely, there was a significant negative correlation with the proliferative profile score of melanocytic cells. This suggests the association of ZEB1 and ZEB1-AS1 with the phenotype switching in melanoma, towards the invasive phenotype. It was interesting to note that normal melanocytes demonstrated the highest scores for the proliferative profile and highest negative correlation with ZEB-AS1. The high proliferative score detected in cultured melanocytes is probably associated with the presence in those cells of a melanocytic cell lineage program coordinated by specific regulators such as MITF gene. This gene codified a transcription factor fundamental to differentiation and development of melanocytic cell lineage and associated with the proliferative profile of melanoma cells50. In this sense, the crucial role of MITF besides determining embryonic melanocyte fate includes regulation of downstream genes involved in cell cycle progression such as CDK251 (cyclin-dependent kinase 2) and cell proliferation inducers as DIAPH152(diaphanous related formin 1). In fact, it is suggested that MITF regulatory potential in normal endogenous levels stimulate proliferation while down or overexpression inhibits cell proliferation competence53.
Although the role of ZEB1, and other key transcription factors, in promoting invasion and malignant progression in many cancers54–56 is well established, associations of lncRNAs and invasiveness and tumour development still need more research. We observed a positive association between ZEB1-AS1 expression with the invasive phenotype in melanoma tumours. In addition, ZEB1-AS1 expression levels positively correlated with several genes involved in transcription regulation, indicative of a role in transcriptional reprogramming, a central process for phenotype switching. An example of a gene in this pathway is the transcription factor MZF1 (myeloid zinc finger 1) gene. This gene is known to contribute to malignancy and invasion in many tumours57. Indeed, it has been recently associated with PRAME (preferentially expressed antigen of melanoma) upregulation and colony formation, which is associated with metastatic potential in melanoma cell line studies58. Also, interestingly, genes involved in cilium formation were considered enriched among the genes positively correlated with ZEB1-AS1 in melanomas. Although the loss of primary cilia has been more frequently implicated in the development of melanoma59 and other cancers60, a recent study unveiled primary ciliogenesis as a key mechanism by which EMT programs induce stemness in a specific subset (claudin-low subtype) of breast tumours61. In this sense, it would be interesting to investigate whether ZEB1-AS1 has any role in the regulation of primary cilium formation and if this regulation may affect melanoma development and invasion in general or in a subset of melanoma tumours.
The association of ZEB1-AS1 expression with melanoma invasion profile is certainly dependent in part of ZEB1 function. However, our analysis pointed out also for a ZEB1 independent role of this lncRNA in the transcriptional regulation, since 82,1% of the positively and 25% of the negatively correlated genes with ZEB1-AS1 were not shared with ZEB1. Despite ZEB1 gene being known as a transcriptional repressor, our data analysis supports ZEB-AS1 as a positive modulator of gene expression62, like some antisense lncRNAs have been described to act like63.
In summary, we firstly identified lncRNA ZEB1-AS1 upregulation in melanoma when compared with melanocytes and metastatic tumours in comparison to primary melanomas. In addition, we showed an association between ZEB1-AS1 expression levels with BRAF/RAS status and with an invasive phenotype in melanoma and suggested this lncRNA directly or indirectly regulates potential genes and pathways. These findings support the notion of an important function of ZEB1-AS1, which may act with some independence of ZEB1, in melanoma progression and melanoma phenotype switching. To this date, lncRNAs interactions with diverse molecules have proved to be an essential aspect as modulators of cancer phenotypes64. Altogether, these evidences and our findings reinforce the need for further functional analysis, including the characterization of mechanistically associated molecules, of ZEB1-AS1 in melanoma progression.
Methods
Cell culture
Primary melanoma (WM35, WM1552, WM278, WM793 and WM1789) and metastatic melanoma (1205lu, WM1617 and WM852) cell lines were grown in conditions of 5% CO2 atmosphere and temperature of 37 °C. These melanoma cell lines were maintained in TU growth medium (80% of MCDB-153 medium, 20% of Leibovitz’s L-15 medium, 5 µg/mL of Insulin and 2 mM of CaCl2) supplemented with 2% Fetal Bovine Serum (Gibco) as previously described65, until harvesting at 80% of plate confluence. Dr. Meenhard Herlyn at the Wistar Institute (Philadelphia, PA) kindly provided all melanoma cell lines. Primary cultures of skin cells (melanocytes – FM308) were obtained from the foreskins of University Hospital (Hospital Universitário – HU-USP) patients, donated by Dr. Linda Maximiano. To this end, the project has undergone review and approval by the Ethics Committee of HU (HU no. CEP Case 943/09) coordinated by Dr. Silvya Stuchi Maria-Engler. Melanocytes were grown in medium 254CF (Thermo Fischer Scientific), supplemented with HMGS solution (Thermo Fischer Scientific) and 200 µM calcium chloride and maintained as previously described66,67. All cell lines utilized in this work were tested for mycoplasma infection.
RNA extraction
Total RNA was extracted with AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) following the manufacturer’s instructions. Density and purity of RNA were tested by the ratio 260/280 nm measurement in NanoDrop (Thermo Fischer Scientific). The RNA quality was tested by Agilent Bioanalyzer, which generates an RNA Integrity Number (RIN) between 1 (more degraded) and 10 (least degraded) and only samples with threshold RIN ≥7 were utilized in this study.
Library construction and RNA sequencing
RNA fragments sized 200–500 bp (base pairs) obtained by fragmentation were utilized to generate sequencing libraries using TruSeq Stranded Total RNA LT Sample Prep Kit (Illumina Inc). The RiboZero technology (Illumina Inc) was utilized to remove abundant rRNA and keep both poly(A) and non-poly(A) transcripts. Clustering was made in an automated system cBot (Illumina Inc), and samples were sequenced with TruSeq SBS kit v5, single-read 72 cycles in Genome Analyzer IIx equipment (Illumina Inc). All reagents were utilized following manufacturer’s usage protocols.
Public raw data and pre-processing
Raw sequencing data from melanocyte (MC502), primary melanoma (A375) and metastatic melanoma cell lines (A2058, C32, MALME3M, SKMEL28, SKMEL5, and WM2664) were downloaded from Gene Expression Omnibus (GEO), under access number GSE46818. Then, the data was converted to compatible fastq format file using the SRA Toolkit (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=toolkit_doc&f=fastq-dump), and quality control was tested using FastQC (Babraham Bioinformatics).
Transcriptome mapping, annotation, and quantification
All data were mapped and quantified by STAR v2.568. We considered a count feature as a count read per gene while mapping, as package default parameters from STAR, and quantifications were displayed as CPM (Counts Per Million) for cell lines. The lncRNA gene annotations were downloaded from GENCODE Release 25 (GRCh38). Therefore, different biotypes of lncRNAs were categorized according to those provided from the long noncoding genes annotation in the GTF file.
Data normalization and differential gene expression analysis
Public and in house-generated RNA-Seq data were normalized using the default method Trimmed Mean of M-values (TMM) to avoid bias effects. To detect differentially expressed lncRNAs, edgeR package version 3.16.5 from Bioconductor in R statistical platform was used. We considered differentially expressed genes in analysis with adjusted P-values < 0.05 and Fold Change (FC) ≤−2 or ≥2 for down and up-regulated genes, respectively.
TCGA data analysis and mutational classification of melanoma samples
TCGA gene expression data was downloaded directly from the TCGA portal and via UCSC repository (xenabrowser.net). TCGA samples were matched by barcode and divided into four main groups according to the presence or not of mutations established by genomic molecular classification of melanoma provided by TCGA team34. This classification relied on the division of SKCM according to the presence of melanoma hotspot mutations (BRAF, NF1, and RAS family group) or their absence (Triple Wild-Type).
Gene Set Variation Analysis and melanoma proliferative/invasive gene expression signatures
Gene Set Variation Analysis (GSVA)69 allowed us to define an enrichment score (ES) that represents the degree of enrichment of a gene set in each sample within a given dataset70. Based on the systematic classification of melanoma by phenotype-specific gene expression profile49, we retrieved melanoma gene expression signatures associated with either invasive (n = 45 genes, Supplementary File S5) or proliferative status (n = 52 genes, Supplementary File S6). Accordingly, we evaluated the correlation between either ZEB1 or ZEB1-AS1 expression with the proliferative or invasive ES presented by melanoma cell lines and TCGA melanoma tumours.
Correlated genes, Gene Ontology, and KEGG analysis
Pearson’s correlation coefficient was calculated between lncRNA ZEB1-AS1 and each other gene in melanoma samples from the TCGA. Genes significantly correlated (r ≤ −0.3 or r ≥ 0.3 and P-value < 0.01) were submitted to the functional annotation in Gene Ontology (GO) terms for biological processes (BP), cellular components (CC), and molecular function (MF). Then, top-ranked GO terms were displayed according to the highest number of genes assigned. Positive and negative ZEB1-AS1 correlated genes were also submitted to pathway enrichment analysis using the Reactome database. Both analyses were performed using the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt)71.
Statistical analysis
Statistical tests and plots were calculated and generated using R platform and PRISM V.6.01 (GraphPad Software, La Jolla California USA).
Supplementary information
Acknowledgements
This work was financed by The National Council for Scientific and Technological Development (CNPq), grant #141880/2016-3, #309187/2015-0; São Paulo Research Foundation (FAPESP), #2013/08135-2, #2018/04017-9, and by Research Support of the University Sao Paulo, CISBi-NAP/USP Grant #12.1.25441.01.2. We would like to thank Cibele Cardoso by technical support.
Author Contributions
A.D.D.S. designed and performed experiments, performed data and bioinformatics analysis, prepared figures and drafted the manuscript L.F.A., I.I.B., K.P., G.M. performed experiments J.R.P. and C.A.O.B.J. performed bioinformatics and statistical analysis. E.M.E. helped in the design of experiments and reviewed the manuscript. J.F.S. and W.A.S.J. designed experiments and drafted the manuscript. All authors reviewed the final manuscript.
Data Availability
The datasets generated during the current study are available in the SRA database repository, under SRA accession PRJNA530784 (http://www.ncbi.nlm.nih.gov/sra/PRJNA530784).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47363-6.
References
- 1.Miller AJ, Mihm MC. Melanoma. N. Engl. J. Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA. Cancer J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Sandru A, Voinea S, Panaitescu E, Blidaru A. Survival rates of patients with metastatic malignant melanoma. J. Med. Life. 2014;7:572–6. [PMC free article] [PubMed] [Google Scholar]
- 4.American Cancer Society. Cancer Facts & Figures (2014).
- 5.Dickson PV, Gershenwald JE. Staging and prognosis of cutaneous melanoma. Surg. Oncol. Clin. N. Am. 2011;20:1–17. doi: 10.1016/j.soc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2013;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 10.Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. J. Mol. Med. (Berl). 2014;92:337–46. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 11.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, et al. Long Non-Coding RNA BANCR Promotes Proliferation in Malignant Melanoma by Regulating MAPK Pathway Activation. PLoS One. 2014;9:e100893. doi: 10.1371/journal.pone.0100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goedert L, et al. RMEL3, a novel BRAF<sup>V600E</sup>-associated long noncoding RNA, is required for MAPK and PI3K signaling in melanoma. Oncotarget. 2016;7:36711–36718. doi: 10.18632/oncotarget.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med. Oncol. 2016;33:88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 15.Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res. Int. 2013;2013:251098. doi: 10.1155/2013/251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, et al. LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth. in vivo. Onco. Targets. Ther. 2016;9:4075–87. doi: 10.2147/OTT.S98203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khaitan D, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 18.Leucci E, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 19.Pádua Alves C, et al. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31:2827–32. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 20.Fenouille N, et al. The epithelial-mesenchymal transition (EMT) regulatory factor SLUG (SNAI2) is a downstream target of SPARC and AKT in promoting melanoma cell invasion. PLoS One. 2012;7:e40378. doi: 10.1371/journal.pone.0040378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larue L, Bellacosa A. Epithelial–mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 22.Hoek KS, et al. In vivo Switching of Human Melanoma Cells between Proliferative and Invasive States. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 23.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23:746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 24.Hoek KS, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 25.Shakhova O, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat. Cell Biol. 2012;14:882–890. doi: 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- 26.Scholl FA, et al. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61:823–6. [PubMed] [Google Scholar]
- 27.Caramel J, et al. A Switch in the Expression of Embryonic EMT-Inducers Drives the Development of Malignant Melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Wels C, Joshi S, Koefinger P, Bergler H, Schaider H. Transcriptional Activation of ZEB1 by Slug Leads to Cooperative Regulation of the Epithelial–Mesenchymal Transition-Like Phenotype in Melanoma. J. Invest. Dermatol. 2011;131:1877–1885. doi: 10.1038/jid.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verfaillie A, et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 2015;6:6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlikowski JS, et al. Wnt signaling potentiates nevogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:16009–14. doi: 10.1073/pnas.1303491110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–23. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol. Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akbani R, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li FZ, Dhillon AS, Anderson RL, McArthur G, Ferrao PT. Phenotype switching in melanoma: implications for progression and therapy. Front. Oncol. 2015;5:31. doi: 10.3389/fonc.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 37.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, et al. Exploring functions of long noncoding RNAs across multiple cancers through co-expression network. Sci. Rep. 2017;7:754. doi: 10.1038/s41598-017-00856-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016;48:1142–1150. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 41.Li J, et al. ZEB1-AS1: A crucial cancer-related long non-coding RNA. Cell Prolif. 2018;51:e12423. doi: 10.1111/cpr.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 43.Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am. J. Transl. Res. 2016;8:4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 44.Su W, et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol. Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Unamuno Bustos B, et al. Towards Personalized Medicine in Melanoma: Implementation of a Clinical Next-Generation Sequencing Panel. Sci. Rep. 2017;7:495. doi: 10.1038/s41598-017-00606-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richard G, et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol. Med. 2016;8:1143–1161. doi: 10.15252/emmm.201505971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widmer DS, et al. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 2012;25:343–353. doi: 10.1111/j.1755-148X.2012.00986.x. [DOI] [PubMed] [Google Scholar]
- 50.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 51.Du J, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 52.Carreira S, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–39. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kido K, et al. Simultaneous suppression of MITF and BRAF V600E enhanced inhibition of melanoma cell proliferation. Cancer Sci. 2009;100:1863–1869. doi: 10.1111/j.1349-7006.2009.01266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Tillo E, et al. ZEB1 Promotes Invasiveness of Colorectal Carcinoma Cells through the Opposing Regulation of uPA and PAI-1. Clin. Cancer Res. 2013;19:1071–1082. doi: 10.1158/1078-0432.CCR-12-2675. [DOI] [PubMed] [Google Scholar]
- 55.Larsen JE, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J. Clin. Invest. 2016;126:3219–3235. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–7. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eguchi T, Prince T, Wegiel B, Calderwood SK. Role and Regulation of Myeloid Zinc Finger Protein 1 in Cancer. J. Cell. Biochem. 2015;116:2146–2154. doi: 10.1002/jcb.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y-K, et al. Tumor antigen PRAME is up-regulated by MZF1 in cooperation with DNA hypomethylation in melanoma cells. Cancer Lett. 2017;403:144–151. doi: 10.1016/j.canlet.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Zingg D, et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell. 2018;34:69–84.e14. doi: 10.1016/j.ccell.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Cao M, Zhong Q. Cilia in autophagy and cancer. Cilia. 2015;5:4. doi: 10.1186/s13630-016-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guen VJ, et al. EMT programs promote basal mammary stem cell and tumor-initiating cell stemness by inducing primary ciliogenesis and Hedgehog signaling. Proc. Natl. Acad. Sci. 2017;114:E10532–E10539. doi: 10.1073/pnas.1711534114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koirala P, et al. LncRNA AK023948 is a positive regulator of AKT. Nat. Commun. 2017;8:14422. doi: 10.1038/ncomms14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zong X, et al. Natural antisense RNA promotes 3′ end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res. 2016;44:2898–2908. doi: 10.1093/nar/gkw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sousa JF, Espreafico EM. Suppression subtractive hybridization profiles of radial growth phase and metastatic melanoma cell lines reveal novel potential targets. BMC Cancer. 2008;8:19. doi: 10.1186/1471-2407-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sousa JF, et al. Novel primate-specific genes, RMEL 1, 2 and 3, with highly restricted expression in melanoma, assessed by new data mining tool. PLoS One. 2010;5:e13510. doi: 10.1371/journal.pone.0013510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Araujo LF, et al. Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Sci. Rep. 2018;8:14190. doi: 10.1038/s41598-018-31170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the SRA database repository, under SRA accession PRJNA530784 (http://www.ncbi.nlm.nih.gov/sra/PRJNA530784).