Abstract
Immature phenotypes of cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs) limit the utility of these cells in clinical application and basic research. During cardiac development, postnatal cardiomyocytes experience high oxygen tension along with a concomitant downregulation of hypoxia-inducible factor 1α (HIF-1α), leading to increased fatty acid oxidation (FAO). We hypothesized that targeting HIF-1α alone or in combination with other metabolic regulators could promote the metabolic maturation of hiPSC-CMs. We examined the effect of HIF-1α inhibition on the maturation of hiPSC-CMs and investigated a multipronged approach to promote hiPSC-CM maturation by combining HIF-1α inhibition with molecules that target key pathways involved in the energy metabolism. Cardiac spheres of highly-enriched hiPSC-CMs were treated with a HIF-1α inhibitor alone or in combination with an agonist of peroxisome proliferator activated receptor α (PPARα) and three postnatal factors (triiodothyronine hormone T3, insulin-like growth factor-1 and dexamethasone). HIF-1α inhibition significantly increased FAO and basal and maximal respiration of hiPSC-CMs. Combining HIF-1α inhibition with PPARα activation and the postnatal factors further increased FAO and improved mitochondrial maturation in hiPSC-CMs. Compared with mock-treated cultures, the cultures treated with the five factors had increased mitochondrial content and contained more cells with mitochondrial distribution throughout the cells, which are features of more mature cardiomyocytes. Consistent with these observations, a number of transcriptional regulators of mitochondrial metabolic processes were upregulated in hiPSC-CMs treated with the five factors. Furthermore, these cells had significantly increased Ca2+ transient kinetics and contraction and relaxation velocities, which are functional features for more mature cardiomyocytes. Therefore, targeting HIF-1α in combination with other metabolic regulators significantly improves the metabolic maturation of hiPSC-CMs.
Keywords: cardiomyocyte, calcium transients, fatty acid, metabolism, stem cell
1. Introduction
The ability to derive cardiomyocytes from human induced pluripotent stem cells (hiPSC-CMs) holds great potential for clinical application, disease modeling and drug discovery for cardiovascular diseases. However, these hiPSC-CMs structurally and functionally resemble cardiomyocytes at an early fetal stage and remain immature [1]. Thus, there are serious caveats to their use in modeling adult-onset diseases and other applications. Multiple approaches have been used to improve hiPSC-CM maturity [2–7], including three-dimensional (3D) tissue engineering, long-term culture, modulation of substrate stiffness, mechanical stress, and electric stimulation. However, metabolic regulation in hiPSC-CMs remains to be further elucidated.
The human adult heart requires a high metabolic demand compared to other organs in the body and the energy metabolism and bioenergetics are crucial factors for maintaining its normal function. A transition in energy metabolism is implicated in cardiomyocyte development and differentiation. Fetal cardiomyocytes reside in a hypoxic environment and rely mainly on glycolysis to generate ATP, whereas adult cardiomyocytes depend mostly on mitochondrial oxidative phosphorylation (fatty acid oxidation—FAO) for energy production [8]. Thus, targeting cardiac energy metabolism could be a promising strategy to promote cardiomyocyte maturation.
The cardiac energy metabolism is regulated by several molecular pathways during cardiac development. The hypoxia-inducible factor 1α (HIF-1α) is a key regulator of genes associated with glycolysis in fetal cardiomyocytes [8]. After the transition from fetal to postnatal life, HIF-1α signaling decreases while the peroxisome proliferator activated receptor α (PPARα), an activator of fatty acid oxidation, increases as cardiomyocytes mature [8, 9]. Previous research has shown that treatment of neonatal rat cardiomyocytes with fatty acids and the PPARα agonist, WY-14643 (WY), increased the expression of enzymes involved in FAO [10]. The activation of PPARα has also been demonstrated to be a key regulator of cardiac metabolism and maturation [11].
Additionally, there are rapid increases in the levels of hormones shortly after birth, including glucocorticoid [12] and thyroid hormone T3 (tri-iodo-L-thyronine) [13], which play a role in increasing cardiac mitochondrial function to meet the need for FAO. Growth factors such as the insulin-like growth factor 1 (IGF-1) are also present in the developing heart to support the growth and metabolism of myocardial cells [14].
Using factors that are present during postnatal heart development is an attractive approach to promote cardiomyocyte maturation. For example, glucocorticoid promoted structural maturation and increased mitochondrial respiration in primary mouse fetal cardiomyocytes [15], and T3 promoted the maturation of fetal sheep cardiomyocytes [16] and hiPSC-CMs [17]. T3, together with IGF-1 and dexamethasone together (TID), also improved bioenergetics and electrophysiology of hiPSC-CMs [18]. The roles of postnatal factors such as TID in combination with 3D tissue engineering and other metabolic regulators in hiPSC-CM maturation have not been explored.
In this study, we investigated whether inhibiting the HIF-1α pathway can enhance FAO in hiPSC-CMs and improve the maturation of hiPSC-CMs in 3D culture. Additionally, we assessed whether a simultaneous inhibition of HIF-1α and activation of PPARα with the postnatal factors TID have a synergistic effect on improving the metabolic maturation of hiPSC-CMs.
2. Materials and Methods
2.1. Human induced piuripotent stem cell culture and cardiac differentiation
Undifferentiated IMR-90 hiPSCs (WiCell, Madison, WI) were maintained on Matrigel (BD Biosciences, San Jose, CA)-coated 100 mm dishes in a feeder-free culture condition [19]. For cardiac differentiation, hiPSCs at ~80% confluence were dissociated using Versene/EDTA (Thermo Fischer Scientific, Waltham, MA), plated onto Matrigel-coated 12-well plates at a density of 4 × 105 cells/well and cultured for two days. The cells were then treated with 100 ng/mL recombinant human activin A (R&D Systems, Minneapolis, MN) on days 0 to 1 and 10 ng/mL recombinant human bone morphogenic protein-4 (BMP4; R&D Systems, Minneapolis, MN) on days 1 to 4 in RPMI medium with 2% B27 insulin-free (Thermo Fischer Scientific, Waltham, MA). At day 4 of cardiac differentiation, cells were dissociated using 0.25% trypsin/EDTA (Thermo Fischer Scientific, Waltham, MA) and seeded into AggreWell 400 plates (Stem Cell Technologies, Vancouver, Canada) at 1,500 cells/microwell in RPMI+B27 medium with insulin (Thermo Fischer Scientific, Waltham, MA) [20]. The medium was supplemented with 10 μM Rock inhibitor Y-27632 (Stemgent, Cambridge, MA) to facilitate cell survival [21, 22]. After 24 h, cardiac spheres were transferred to suspension culture without Rock inhibitor and maintained for additional 15 days in RPMI medium with 2% B27 with insulin. Medium replacement was performed on alternate days.
2.2. Maturation factors
The following compounds—FM19G11 (FM), WY-14643 (WY), tri-iodo-L-thyronine (T3), insulin-like growth factor-l (IGF-1) and dexamethasone —were obtained from Sigma-Aldrich (St. Louis, MO) and were reconstituted according to manufacturer’s instructions. FM has been previously characterized as a novel chemical entity that inhibits HIF-1α in various tumor cell lines as well as rodent and human stem cells [23].
2.3. Treatments of hiPSC-CMs with maturation factors
For initial compound screening (Supplementary Figure 1), cardiomyocyte differentiation cultures were dissociated using 0.25% trypsin/EDTA and replated onto 12-well plates at a density of 1.3 × 106 cells/well. Cells at differentiation day 28 were treated with the following compounds in the cardiomyocyte maturation medium on alternate days for 5 days: (1) vehicle control (DMSO, 1:1000 dilution, D2438, Sigma-Aldrich, St. Louis, MO); (2) HIF-1α inhibitor, 0.5 μM FM; (3) 1 μM FM; (4) PPARα agonist, 50 μM WY; (5) 100 μM WY. The cardiomyocyte maturation medium contained DMEM, 10% fetal bovine serum, 2 mM L-glutamine, 1% penicillin-streptomycin, 0.1 mM non-essential amino acids and 0.2 mM oleic acid (Sigma-Aldrich, St. Louis, MO).
For subsequent experiments described in Methods 2.4 to 2.12, cardiac spheres were treated for 7 days with maturation factors or DMSO and cardiac spheres at day 28 were used. We examined the effect of maturation treatments on cardiac spheres since 3D culture can improve structural maturation of cardiomyocytes [19]. The RPMI+B27 medium containing insulin was replaced with the cardiomyocyte maturation medium at day 21 of cardiac differentiation. Cardiac spheres were then treated with the following molecules in the cardiomyocyte maturation medium on alternate days for 7 days: (1) vehicle (DMSO, 1:1000 dilution) control; (2) 1 μM FM; (3) 100 μM WY; (4) TID (100 nM T3 + 100 ng/mL IGF-1 + 1μM dexamethasone); (5) 1 μM FM + 100 μM WY + TID.
2.4. Immunocytochemical analysis and high-content imaging analysis
Following the treatment with maturation factors or DMSO control for 7 days, cardiac spheres at differentiation day 28 were dissociated, re-plated onto Matrigel-coated 48-well plates and cultured for one additional day before fixation. Cells were then washed with PBS, fixed in 4% paraformaldehyde for 15 min and permeabilized in cold ethanol for 2 min at room temperature. Cells were washed twice with PBS and blocked in PBS containing 20% normal goat serum (Invitrogen, Carlsbad, CA) for 1-2 h prior to the incubation with primary antibodies (Supplementary Table 1) overnight at 4°C. Cells were washed twice and incubated with corresponding conjugated secondary antibodies (Supplementary Table 1) for 45 min at room temperature in the dark. Cells were then washed twice again and nuclei were stained with Hoechst 33342 (1 μg/mL, Thermo Fisher Scientific, Waltham, MA) for 10 minutes. Imaging was performed using Axio Vert.A1 inverted microscope (Zeiss, Oberkochen, Germany). For quantitative analysis of NKX-2-5 and MLC2V, images were acquired and analyzed by high-content imaging using ArrayScan as described in Supplemental Information.
2.5. Seahorse XF24 metabolic flux analysis
The oxygen consumption rate (OCR) was determined using the Seahorse XF24 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA). Following the treatment with maturation factors or DMSO control for 7 days, cardiac spheres at differentiation day 28 were dissociated into single cells using 0.25% trypsin/EDTA. Cells were subsequently plated onto a Matrigel (1:50) precoated Seahorse XF-24 cell culture plate (Agilent Technologies, Santa Clara, CA) at a density of 2.5 × 105 cells/well in maturation medium supplemented with 10 μM Rock inhibitor Y-27632. At this concentration, the Rock inhibitor had been shown to facilitate cell survival [21, 22]. After 24 h, the Rock inhibitor-containing maturation medium was replaced with maturation medium alone. Cells were then maintained in maturation medium for 3 additional days to allow them to recover before the Seahorse assay.
One hour before the assay, the cells were washed once with XF assay medium (unbuffered DMEM + 5 mM glucose + 2 mM L-glutamine + 0.5 mM sodium pyruvate) and incubated in 525 pL base medium at 37°C in a non-CO2 incubator. The degree of FAO was analyzed as previously described [21]. Briefly, cells were treated with 100 μM etomoxir (ETO, a specific CPT-1 inhibitor, Sigma-Aldrich, St. Louis, MO) during real-time recording of OCR. The levels of FAO were then calculated based on the difference in absolute OCR before and after the addition of ETO. The oxidation of non-fatty acid substrates was calculated based on absolute OCR after the addition of ETO. The results were normalized to the cell number obtained by trypan blue assay. Measurement of metabolic fluxes has accuracies comparable to those obtained with the radiometric FAO assays [24].
Mitochondrial function was analyzed using the XF Cell Mito Stress Kit (Agilent Technologies, Santa Clara, CA). Three mitochondrial inhibitors—oligomycin (2 μM), carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP, 1 μM) and rotenone (0.5 μM) + antimycin A (0.5 μM)—were diluted in base medium and sequentially added into each well during the measurements. OCR (pmol/min) and extracellular acidification rate (ECAR, the H+ production fate, mpH/min) were measured according to the manufacture’s instruction from Agilent Technologies. The results were normalized to 106 cells as determined by trypan blue assay.
2.6. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from hiPSC-CMs using Aurum total RNA mini kit (Bio-Rad Laboratories, Inc., Hercules, CA) according to manufacturer’s instructions. One μg RNA sample was used to generate cDNA using the Superscript VILO cDNA synthesis kit (Thermo Fischer Scientific, Waltham, MA), and reaction mixture was incubated using a C1000 touch thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA) as follows: 25°C for 10 min, 42°C for 2 h and 25°C for 5 min. The reaction mixture was further diluted to 300 μL in order to use appropriate amount of cDNA for the subsequent quantitative PCR reaction. Two μL cDNA was subjected to the quantitative PCR, which was performed in triplicates using a SYBR Green reaction master mix (Bio-Rad Laboratories, Inc., Hercules, CA). The mRNA levels were normalized to GAPDH mRNA levels. Primer sequences were obtained from the NCI/NIH qPrimer Depot (Supplementary Table 2).
2.7. MitoTracker Red flow cytometry and immunostaining
For flow cytometry of the mitochondrial content, we used the fluorescent dye MitoTracker Red (30 nM, Thermo Fisher Scientific, Waltham, MA) that emits 599 nm light when accumulated in mitochondria. Following the treatment with maturation factors or DMSO control for 7 days, cardiac spheres at day 28 were washed with PBS and labeled with 30 nM MitoTracker Red in maturation medium for 30 mins at 37°C in a 5% CO2 incubator. After washing with PBS, cells were dissociated with 0.25% trypsin/EDTA and fixed in 4% paraformaldehyde for 15 min. Cells were then washed twice with PBS and analyzed by BD FACS Canto II (BD Biosciences, San Jose, CA). Forward versus side scatter quadrants were defined and at least 10,000 live cells were acquired.
For immunocytochemistry analysis of the mitochondrial distribution, cardiac spheres at differentiation day 28 were dissociated using 0.25% trypsin/EDTA, replated onto Matrigel-coated microscope cover glasses (18 mm, Marienfeld, Germany). Cells were cultured for 3-4 additional days in maturation media to allow cells to completely attach onto the glass coverslips and recover beating before they were stained with MitoTracker Red (250 nM). The cells were then fixed, permeabilized with cold ethanol for 2 min at room temperature, blocked with 20% normal goat serum and co-stained with α-actinin (Supplementary Table 1) overnight at 4°C. Cells were washed twice and incubated with appropriate secondary antibodies (Supplementary Table 1) for 45 min at room temperature. Cells were washed twice and nuclei were counterstained with Hoechst 33342 (Thermo Fisher Scientific, Waltham, MA). Imaging was performed using Axio Vert.A1 inverted microscopy (Zeiss, Oberkochen, Germany).
2.8. Quantification of mitochondrial DNA content
Cardiac spheres at differentiation day 28 were dissociated with 0.25% trypsin/EDTA and total genomic DNA (gDNA) was isolated using QIAamp DNA Mini Kit (Qiagen, Venlo, Netherlands), according to the manufacturer’s instructions. Following determination of genomic DNA (gDNA) concentration using a UV-vis spectrophotometer (NanoDrop, Thermo Fisher Scientific, Waltham, MA), samples were diluted to yield equal amounts of gDNA. Real-time PCR amplification was performed in an ABI Prism 7500 Real-Time PCR System (Applied BioSystems, Foster City, CA) for nuclear genes, succinate dehydrogenase subunit A [25] (SDHA: Fw-TCTCCAGTGGCCAACAGTGTT; Rw-GCCCTCTTGTTCCCATCAAC) and lipoprotein lipase [17] (LPL: Fw-CGAGTCGTCTTTCTCCTGAT; Rw-TTCTGGATTCCAATGCTTCGA), and mitochondrial genes, NADH dehydrogenase subunit I [17] (ND1: Fw-CCCTAAAACCCGCCACATCT; Rw-GAGCGATGGTGAGAGCTAAGGT) and mitochondrial cytochrome oxidase II[25] (mt-CO2: Fw-CGATCCCTCCCTTAC; Rw-GAGAGGGGAGAGCAAT). The mitochondrial DNA (mtDNA) was normalized to nuclear DNA (nDNA).
2.9. Calcium imaging
Following the treatment with maturation factors or DMSO control for 7 days, cardiac spheres at day 28 were dissociated with 0.25% Trypsin/EDTA and replated onto Matrigel (1:30)-coated 25×25×1 mm glass coverslips. The replated cells were then cultured in maturation medium supplemented with treatment compounds or DMSO control for an additional 3-7 days to allow them to completely attach onto the glass coverslips and recover beating.
For calcium imaging, cells were incubated in 5 μM fluo-4 AM dye (Thermo Fisher Scientific, Waltham, MA) for 30 minutes at 37°C in maturation medium and washed with Tyrode’s solution (148 mM NaCI, 4 mM KCI, 0.5 mM MgCl2·6H20, 0.3 mM NaPH2O4·H2O, 5 mM HEPES, 1.8 mM CaCl2·H2O, and 10 mM D-glucose, pH adjusted to 7.4 with NaOH). Cells were subsequently transferred to a temperature-controlled microscope chamber coupled to an inverted laser confocal scanning microscope (Olympus FV1000, Olympus, Tokyo, Japan) equipped with FluoView software (Olympus, Tokyo, Japan). Cells were continuously perfused at 37°C with Tyrode’s solution [26]. The intracellular calcium fluorescence signals were acquired at 40× magnification when cells were stimulated at 1 Hz. The recordings were analyzed with ClampFit 10.7 software (Molecular Devices, San Jose, CA).
2.10. Contractility assay
Cardiac spheres at differentiation day 28 were plated into 6-well plates and were video-recorded using a Leica DM IRBE inverted microscope (Leica, Germany) equipped with a controlled CO2 and temperature chamber (5% CO2, 37°C). Cells were stimulated at 1 Hz and recorded at 10× magnification, a frame rate of 80 fps and a resolution of 1024 × 1024 pixels. Movie images were analyzed with MATLAB R2016b software (MathWorks, Natick, MA), using the open source optical flow software [27].
2.11. RNA-sequencing (RNA-Seq) analysis
Total RNA was extracted from both hiPSC-CMs and tissue samples of left ventricle (LV) free-wall from a 6.5 months old male using Aurum total RNA mini kit (Bio-Rad Laboratories, Inc., Hercules, CA) according to manufacturer’s instructions. The LV samples were discarded tissues obtained with Emory approved IRB from a pediatric patient during cardiac repair surgery. Library preparation and sequencing were performed at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology. RNA quality was assessed on a Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA) and all samples had RNA integrity number (RIN) ≥ 7. cDNA was derived from RNA, hybridized and probe intensities were generated on Iliumina HumanHT-12 v3 (Iliumina, Inc., San Diego, CA). RNA sequence reads were aligned to the human reference genome (GRCh38) [28] using HISAT2 (hierarchical indexing for spliced alignment of transcripts) [29], followed by using UCSC reference annotation and HTSeq [30] to estimate gene abundance. All the downstream analysis were performed in R v3.3.2 [31]. In total, there were 26,485 genes detected, though only 15,359 genes were retained with count per million (cpm) greater than 1 in at least 3 samples. The Trimmed Mean of M-value (TMM) normalization method was applied for the retained genes, followed by log2-tranformed of the normalized count data, which was used for the downstream analysis. Differentially expressed genes were detected using edgeR [32], and Gene Ontology (GO) analysis was completed using ToppGene Suite [33].
2.12. Statistical analysis
The results are presented as the mean ± standard error of the mean (SEM). Data were compared using one-way ANOVA or student’s t-test (GraphPad) and p values < 0.05 were considered to be significant.
3. Results
3.1. Treatment of hiPSC-CMs with a HIF-1α inhibitor increases expression of genes involved in FAO
To examine the role of HIF-1α in regulating FAO of hiPSC-CMs, we first evaluated the effect of HIF-1α inhibition on the expression of genes involved in FAO. hiPSC-CMs at differentiation day 28 were treated for 5 days with HIF-1α inhibitor FM19G11 at 0.5 and 1 μM, or PPARα agonist WY-14643 at 50 and 100 μM, or DMSO (solvent for the compounds as a control). PPARα agonist was used as a positive control since it was reported to improve metabolic maturation of hPSC-CMs [11, 34]. We analyzed the expression of 6 genes to assess the ability of the HIF-1α inhibition to modulate FAO including (1) ACADVL (Acyl-coA dehydrogenase, very long chain) which catalyzes the first step of the FAO pathway, (2) CPT1B (carnitine palmitoyltransferase 1B) which transports fatty acids from the cytoplasm into the mitochondria, (3) FABP2 (fatty acid binding protein 2) which is involved in intracellular metabolism and/or transport of fatty acids, (4) CD36 (fatty acid translocase) which is a regulator of fatty acid transport, (5) PPARA (peroxisome proliterator-activated receptor alpha) which is a master regulator of FAO, and (6) PGC1A (peroxisome prol iterator-activated receptor gamma, coactivator 1 alpha, PGC-1α) which is a transcriptional coactivator that regulates the genes involved in energy metabolism and mitochondrial capacity.
Among the genes examined, the expression of ACADVL, CD36 and PPARA was significantly increased after the treatment with HIF-1α inhibitor FM19G11 at 0.5 and 1 μM (Supplementary Fig. 1A), suggesting that HIF-1α inhibition may modulate FAO. As expected, the expression of ACADVL, FABP2, CD36, PPARA and PGC1A was significantly increased after the treatment with PPARα agonist WY-14643 at 100 μM (Supplementary Fig. 1B).
3.2. HIF-1α inhibition increases FAO, and combining HIF-1α inhibition with PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) further increases FAO and expression of FAO-associated genes in hiPSC-CMs
We next examined the effect of metabolic regulation on FAO of hiPSC-CMs in 3D cardiac spheres using HIF-1α inhibition and its combination with other factors including a PPARα activator and TID (a combination of thyroid hormone T3, IGF-1 and dexamethasone) [35]. 3D cardiac spheres were generated from day 4 differentiated cells induced by activin A and BMP4 (Fig. 1A). Spontaneous contraction was observed by differentiation days 9-10 and all cardiac spheres were contracting at differentiation day 21. These cardiac spheres were then treated with the following molecules for 1 week in a medium containing 0.2 mM oleic acid (a fatty acid that naturally occurs in human milk): (1) vehicle control (DMSO); (2) HIF-1α inhibitor: 1 μM FM (FM19G11); (3) PPARα agonist: 100 μM WY (WY-14643); (4) TID: 100 nM T3 + 100 ng/mL IGF-1 + 1 μM dexamethasone [35]; and (5) FM+WY+TID. Doses of these factors were selected based on increased expression of FAO genes (Supplementary Fig. 1) and the Birket et al publication [35].
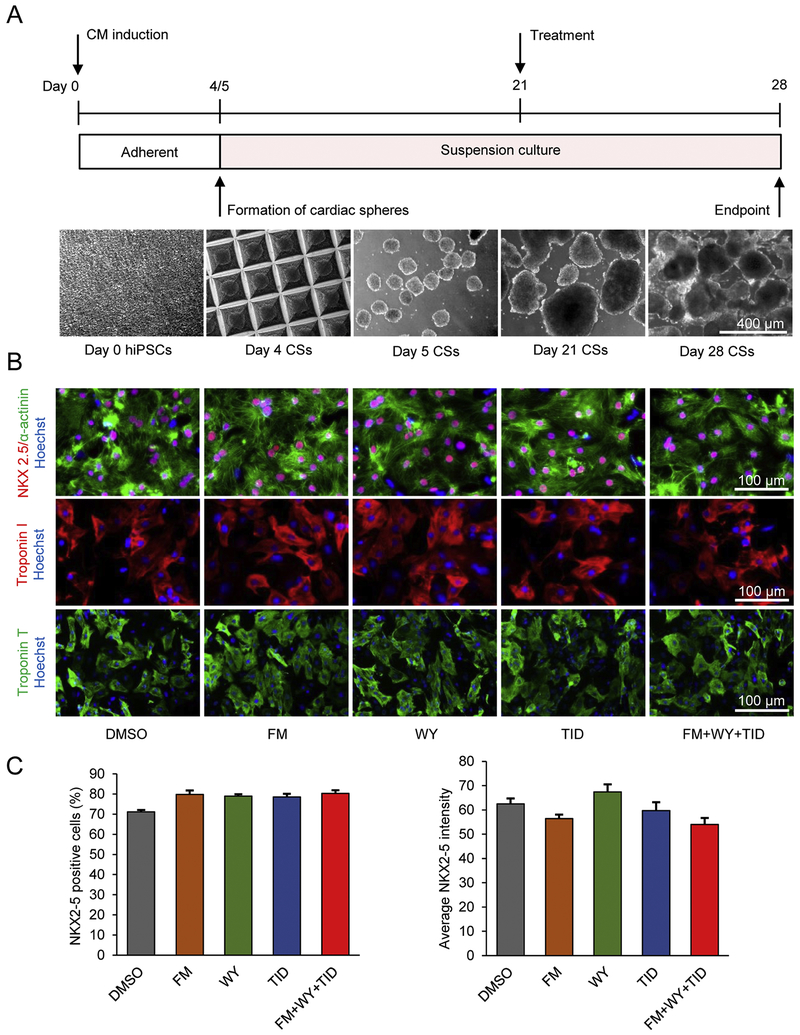
Figure 1.
Microscale tissue engineering generates enriched hiPSC-CMs and maturation treatments do not alter hiPSC-CM purity. (A) Schematic diagram of experimental design and cell morphology of hiPSCs and cardiac spheres (CSs). hiPSCs were treated with activin A (100 ng/mL) at day 0 and BMP4 (10 ng/mL) at day 1. Cells at differentiation day 4 were aggregated into cardiac spheres and the cardiac spheres at day 21 were treated with maturation factors or DMSO control for 1 week. (B) Immunostaining of differentiation cultures for cardiomyocyte markers. At day 28, cells were dissociated, replated and stained for NKX2-5 (red), α-actinin (green), cardiac troponin I (red) and cardiac troponin T (green). Cell nuclei were counterstained with Hoechst 33342 (blue). FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone. (C) Quantitative analysis of NKX2-5 by high-content imaging using ArrayScan. Data are presented as mean ± SEM (n=5).
All cardiac spheres were beating with no signs of cytotoxicity in all tested conditions after 1 week of the treatment. As expected, 3D culture generated enriched hiPSC-CMs as demonstrated by immunocytochemical analysis of CM-associated markers including NKX2-5, α-actinin, cardiac troponin T, and cardiac troponin I (Fig. 1B). Similar to the cells treated with DMSO control, the majority of the cells were positive for these cardiac markers in all treatment conditions (Fig. 1B) suggesting that the treatments did not change cardiomyocyte purity when compared to cells treated with DMSO control. As detected by high-content imaging, ~71-80% of the cells were positive for NKX2-5 (Fig. 1C and Supplementary Fig. 2) and the NKX2-5-positive cells were also positive for α-actinin (Fig. 1B). Compared with DMSO control, the FM+WY+TID treatment did not alter the proportion of cells that were positive for MLC2V (myosin regulatory light chain 2, ventricular/cardiac muscle isoform): ~45-48% MLC2V-positive cells were detected in the cultures (Supplementary Fig. 3).
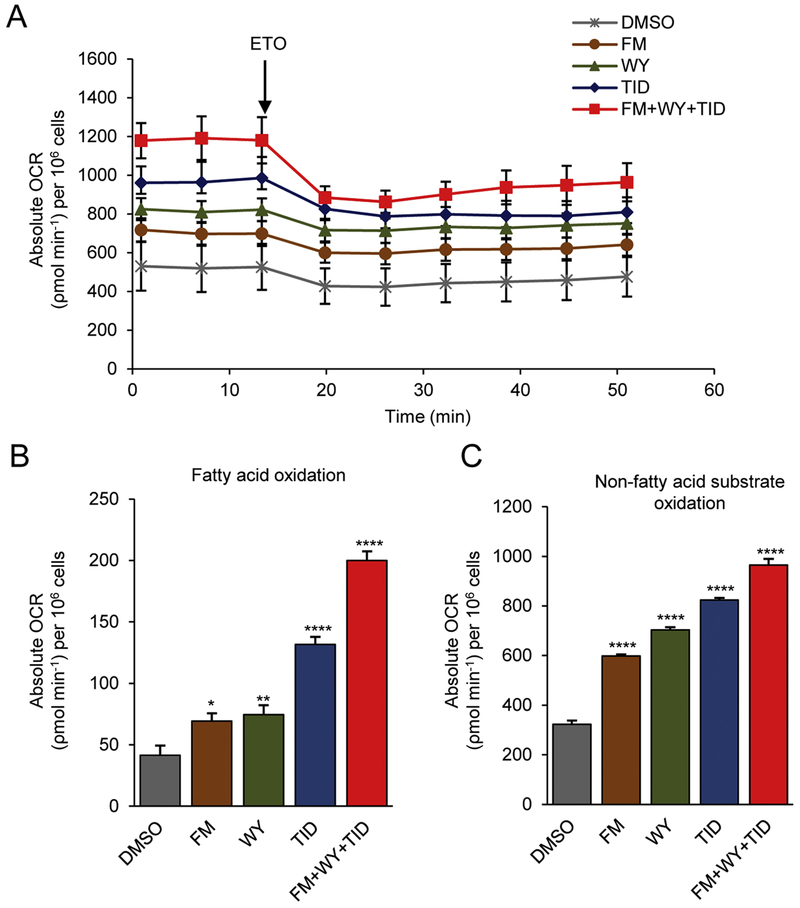
To assess the impact of these treatments on the metabolic status of hiPSC-CMs, we examined the level of FAO by monitoring OCR using the Seahorse XF24 Extracellular Flux Analyzer, which is an established assay for FAO measurement. During real-time recording of OCR, cells were treated with etomoxir (ETO), a specific inhibitor of carnitine palmitoyltransferase 1a (CPT-1a) which is involved in FAO by mediating the internalization of fatty acids into the mitochondrial matrix for oxidation. Following the treatment of ETO, a decrease in OCR was observed in cells under all maturation conditions (Fig. 2A). However, the largest decrease in OCR was observed when cells were cultured in maturation medium containing FM+WY+TID (Fig. 2A). When compared with the cells treated with DMSO control for the level of FAO calculated based on ETO-induced reduction of OCR, hiPSC-CM spheres treated with the HIF-1α inhibitor FM had increased levels of FAO (69.28 ± 6.34 vs. 41.53 ±7.88 p< 0.05) and those treated with FM+WY+TID had the highest levels of FAO (200.07 ± 6.01 vs. 41.53 ± 7.88, p<0.0001) (Fig. 2A, 2B). The level of FAO in TID-treated cells was higher than that in FM- or WY-treated cells but lower than that in FM+WY+TID-treated cells (Fig. 2A, 2B). In addition, treatment with HIF-1α inhibitor FM also increased the oxidation of non-fatty acid substrates when compared with control (598.25 ± 6.19 vs. 322.44 ± 15.69 p< 0.0001), and the treatment with FM+WY+TID resulted in the highest increase in oxidation of non-fatty acid substrates (964.93 ± 24.63 vs.322.44 ± 15.69, p<0.0001) (Fig. 2C).
Figure 2.
Treatments of 3D cardiac spheres with a HIF-1α inhibitor, a PPARα activator and the postnatal factors (T3, IGF-1 and dexamethasone) improve β-oxidation and other substrate oxidation. (A) Representative traces of real-time measurement of mitochondrial oxygen consumption rate (OCR) following addition of etomoxir (ETO, 100 μM). Quantification of amount of OCR derived from (B) fatty acid β-oxidation and (C) oxidation of non-fatty acid substrates in hiPSC-CMs treated with maturation factors or DMSO control. Data were normalized to cell counts and presented as mean ± SEM (n=8). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; by one-way ANOVA. FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
To further investigate the implication of FAO in hiPSC-CM maturation, we measured the expression of a subset of genes that are critically involved in multiple segments of the lipid flux pathway [36]. These included regulating fatty acid transport/activation (Supplementary Fig. 4A), lipid storage (Supplementary Fig. 4B), mitochondrial fatty acid oxidation (Supplementary Fig. 4C), transcriptional regulation (Supplementary Fig. 3D) and intracellular glucose uptake (Supplementary Fig. 4E). Among these genes, treatment with FM+WY+TID resulted in a 4.8-fold increase in the long chain fatty acid transporter CD36, 2.2-fold increase in the mitochondrial uncoupling protein UCP3, and 3.1-fold increase in the fatty acid synthase (FASN). The genes encoding the kruppel-like factor 15 (KLF15), the estrogen-related receptor alpha (ESRRA) and the mitochondrial trifunctional proteins HADHA and HADHB as well as the glucose transporter 4 (GLUT4) were also significantly upregulated after treatment with FM+WY+TID. In addition, we observed a 2.7-fold increase in pyruvate dehydrogenase kinase 4 (PDK4), which plays a key role in the regulation of glucose and fatty acid metabolism via phosphorylation and inhibition of pyruvate dehydrogenase and thereby inhibiting glucose oxidation [37].
The expression of some of FAO-related genes including CPT1B, DGAT2, NCOR1, RXRA, and ACADVL was not significantly altered by the treatment. In addition, FM treatment did not alter the expression of genes related to glucose metabolism, including GLUT1, GLUT4, PDK4, PDK1, PDK3 and PKM3 (Supplementary Figs. 4 and 5); however the effect of FM on the expression of other genes and posttranslational modification of proteins involved in glucose metabolism remains to be determined.
3.3. HIF-1α inhibition increases basal and maximal respiration and combining HIF-1α inhibition with PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) further improves mitochondrial maturation in hiPSC-CMs
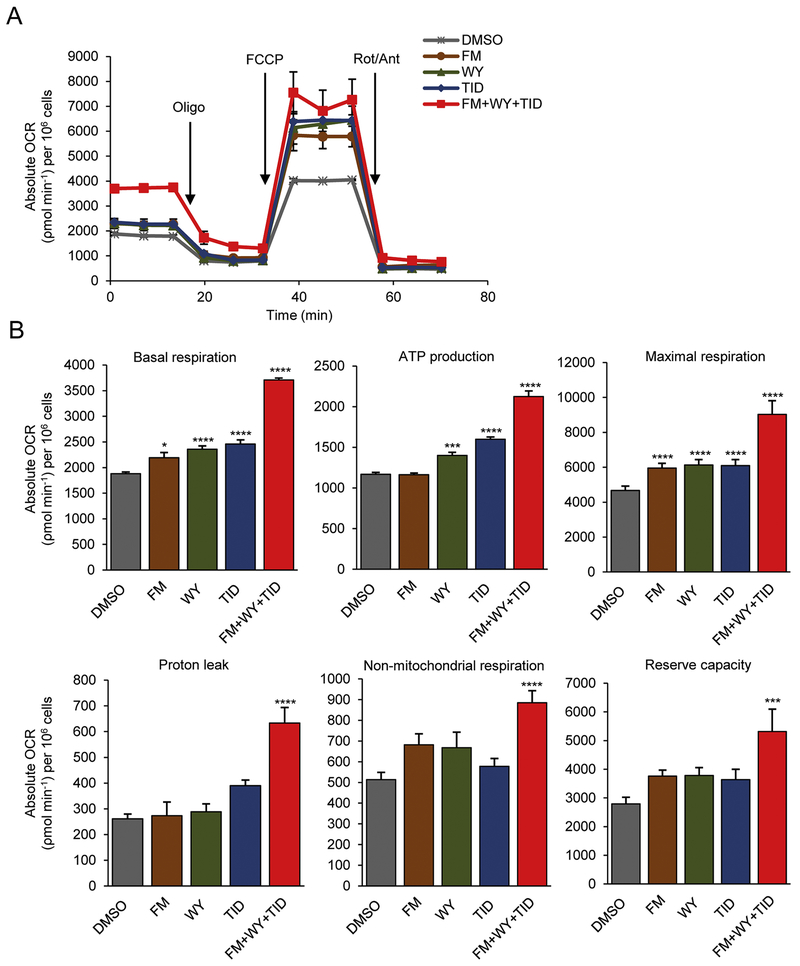
To characterize mitochondrial function, we measured the major aspects of mitochondrial coupling and respiratory control—basal respiration, maximal respiration, proton leak, non-mitochondrial respiration and reserve capacity using the Seahorse XF24 Extracellular Flux Analyzer [38]. As expected, changes in OCR and ECAR were detected following the sequential additions of the ATP synthase inhibitor oligomycin, a protonophoric uncoupler FCCP, and electron inhibitors rotenone and antimycin A (Fig. 3 and Supplementary Fig. 6). Fig. 3A shows representative traces of OCR in control and treated hiPSC-CMs. 3D hiPSC-CM spheres treated with the HIF-1α inhibitor FM had increased basal and maximal respiration and those treated with FM+WY+TID had the highest basal and maximal respiration and ATP production (Fig. 3A, 3B). The basal and maximal respiration were significantly increased following the treatments with FM, WY, TID, or FM+WY+TID (Fig. 3B, upper panel). However, only cells treated with FM+WY+TID displayed significantly increased OCR in all parameters of the respiratory chain, including proton leak, non-mitochondrial respiration and reserve capacity (Fig. 3B, lower panel). Basal respiration is correlated to the ability to generate contractile force and non-mitochondrial respiration is regulated by various enzymes that are involved in detoxification and oxidation [17]. Treatment with FM+WY+TID compared to control significantly increased both parameters, basal respiration (3709.26 ± 34.40 vs. 1879.13 ± 33.7, p<0.0001) and non-mitochondrial respiration (885.27 ± 58.01 vs. 513.36 ± 34.93, p<0.0001).
Figure 3.
Treatments of 3D cardiac spheres with a HIF-1α inhibitor, a PPARα activator and the postnatal factors (T3, IGF-1 and dexamethasone) improve mitochondrial function in hiPSC-CMs. (A) Representative traces showing the OCR of hiPSC-CMs following sequential addition of oligomycin (Oligo, 2 μM), FCCP (1 μM), and rotenone/antimycin A (Rot/Ant, 0.5 μM). (B) Quantification of basal respiration, maximal respiration, ATP production, proton leak, non-mitochondrial respiration and reserve capacity. All measurements were normalized to cell counts and presented as mean ± SEM (n=8). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; by one-way ANOVA. FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
3.4. Combining HIF-1α inhibition with PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) influences mitochondrial distribution and increases mitochondrial content in hiPSC-CMs
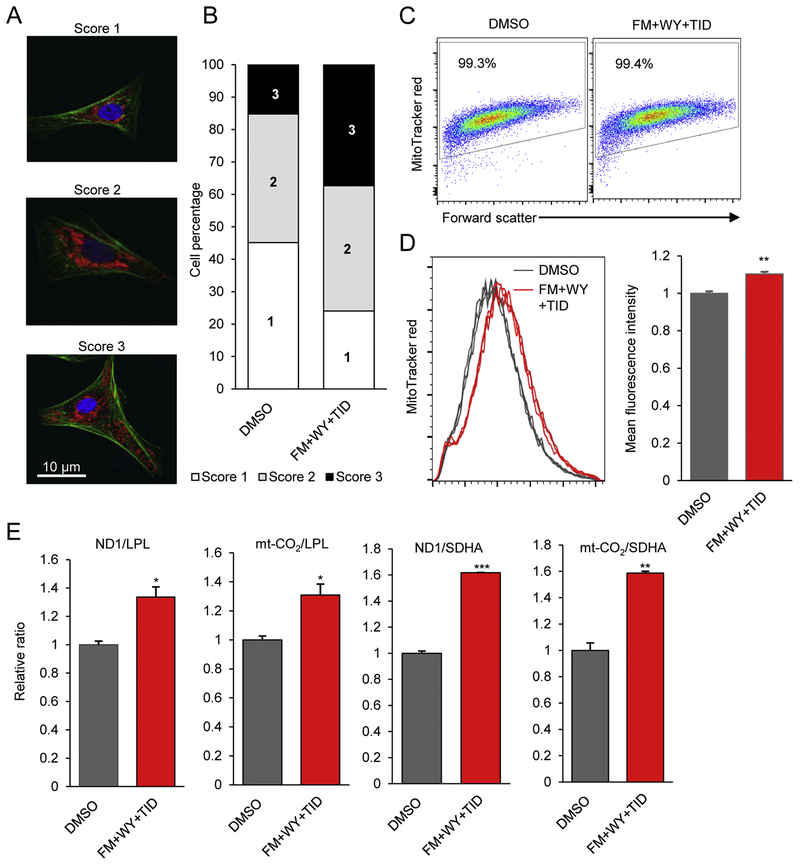
To examine the effect of FM+WY+TID on mitochondrial distribution in hiPSC-CMs, we evaluated the overall distribution of their mitochondria using the mitochondrial marker MitoTracker Red co-stained with a marker for myofibrillar Z-disks, α-actinin. The cells were categorized into 3 different levels: score 1 denotes cells with mitochondria that are localized to the perinucleus, score 2 cells have mitochondria that are distributed away from the nucleus but not throughout the cell, and the mitochondria of score 3 cells are highly dispersed throughout the entire cell area (Fig. 4A, 4B). Based on counting of 240 cells in each scoring group for each treatment, we observed a 2.5-fold increased number of score 3 cells in the FM+WY+TID-treated culture compared with the DMSO-treated culture (37% vs. 15%). On the other hand, the number of score 1 cells decreased in the FM+WY+TID-treated culture compared with the DMSO-treated culture (24% vs. 45%). These results suggest that FM+WY+TID improved mitochondrial distribution of hiPSC-CMs, which is a cardiomyocyte maturation indicator since cardiomyocyte maturation changes mitochondrial distribution.
Figure 4.
Treatment of 3D cardiac spheres with a HIF-1α inhibitor, a PPARα activator and the postnatal factors (T3, IGF-1 and dexamethasone) alters distribution and content of mitochondria in hiPSC-CMs. (A) Representative images showing the locations of mitochondria in hiPSC-CMs using MitoTracker dye (red), myofibrils with α-actinin (green), and nuclei with Hoechst (blue). (B)The overall mitochondrial distribution among FM+WY+TID-treated and control cells was categorized into 3 different levels: Score 1 cells with mitochondria clustered near the nucleus; Score 2 cells with mitochondria near the nucleus and at a low level in other areas; and Score 3 cells with mitochondria distributed throughout the cytoplasm. n=240 cells/group. (C) Representative dot plots of flow cytometric analysis of mitochondrial content of hiPSC-CMs using MitoTracker Red. (D) Representative histograms of MitoTracker Red fluorescence intensity and the relative fluorescence intensity of treated and control hiPSC-CMs. Data are represented as mean ± SEM (n=3). (E) Quantification of the ratio of mitochondria-encoded complex I ND1 or mt-CO2 to nuclear-encoded complex II LPL or SHDA DNA in treated compared to control cells. Data are presented as mean ± SEM (n=3). *P<0.05, ***P<0.001; by Student’s t test. FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
FM+WY+TID also strikingly increased mitochondrial content (mass) of hiPSC-CMs as detected by flow cytometry (Fig. 4C, 4D), although FM, WY or TID alone did not increase mitochondrial content (Supplementary Fig. 7). The mean fluorescence intensity of MitoTracker Red was significantly higher in FM+WY+TID-treated cells than in DMSO-treated cells. Mitochondrial content is an indication of cardiomyocyte maturation since mitochondria occupy ~20-40% of the cell volume in adult cardiomyocytes [1] compared with ~7% of cell volume in hiPSC-CMs [17]. Consistent with these observations, FM+WY+TID significantly increased mitochondrial DNA (mtDNA): nuclear DNA ratio (mitochondria-encoded complex I ND1 or mt-CO2 to nuclear-encoded complex II LPL or SHDA) as detected by quantitative real-time PCR (Fig. 4E).
3.5. HIF-1α inhibition together with PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) lead to faster calcium transient kinetics and contractility in hiPSC-CMs
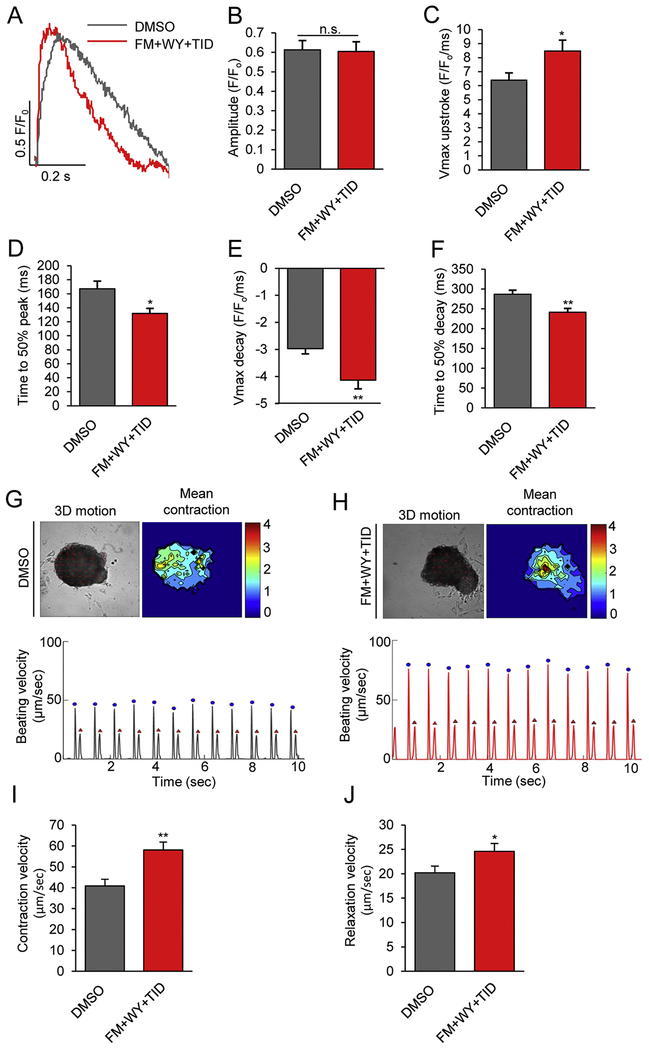
We further investigated the calcium-handling properties of hiPSC-CMs using line-scan confocal imaging after the cells were loaded with the intracellular calcium dye fluo-4AM. Cytosolic calcium transients were recorded at 1 Hz in hiPSC-CMs treated with DMSO control or FM+WY+TID (Fig. 5A–5F). After treatment with FM+WY+TID, the maximal upstroke velocity (Fig. 5C) was significantly faster (6.40 ± 0.51 vs. 8.47 ± 0.77 F/F0/ms, p<0.05) and the time to 50% peak [Ca2+], (Fig. 5D) was significantly shorter (167.21 ± 10.96 vs. 131.96 ± 7.18 ms, p<0.05), although the peak transient amplitude (Fig. 5B) remained unchanged after treatment (0.61 ± 0.05 vs. 0.60 ± 0.05 F/F0, p=0.85). In addition, hiPSC-CMs treated with FM+WY+TID had a faster Ca2+-transient decay rate (2.97 ± 0.20 vs. 4.14 ± 0.32 F/F0/ms, p<0.01) (Fig. 5E) and a shorter time to 50% decay (287 ± 9.91 vs. 241.51 ± 9.63 ms, p<0.01) (Fig. 5F). Therefore, FM+WY+TID significantly increased the kinetics of calcium transients, a functional characteristic of more mature cardiomyocytes [39, 40].
Figure 5.
Treatments of 3D cardiac spheres with a HIF-1α inhibitor, a PPARα activator and the postnatal factors (T3, IGF-1 and dexamethasone) improve calcium transient kinetics and contractility. Calcium transients were assessed by loading hiPSC-CMs with the intracellular calcium dye fluo-4 AM. (A) Representative transient traces from control and FM+WY+TID-treated hiPSC-CMs. (B) The calcium transient amplitude magnitudes, (C) maximal upstroke velocity, (D) time to 50% peak, (E) maximal decay velocity, (F) time to 50% decay, and (G) beating frequency were recorded at the stimulation frequency of 1 Hz. Results were mean ± SEM (n=30). *P<0.05, **P<0.01; by Student’s t test. Representative image and heat map (top) depicting time-averaged magnitude of all motion and tracing (bottom) of average beating speed vs. time of hiPSC-CMs treated with (G) DMSO or (H) FM+WY+TID. Contractions are denoted in blue by the first peak and relaxations are denoted in red by the second peak of the duplex. (I) Contraction velocity and (J) relaxation velocity of control and FM+WY+TID-treated hiPSC-CMs. Data are represented as mean ± SEM (n=14). *P<0.05, **P<0.01; by Student’s t test. FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
Next we measured the contractility using the MotionGUI program [27]. Both average maximum contraction (58.87 ± 3.84 vs. 40.87 ± 3.22 μm/sec, p<0.01) and relaxation (24.60 ± 1.60 vs. 20.22 ± 1.34 μm/sec, p<0.05) velocities were significantly higher in hiPSC-CMs treated with FM+WY+TID compared with DMSO (Fig. 5G–5J). Together these data suggest that FM+WY+TID promotes the functional maturation of hiPSC-CMs.
3.6. Gene expression profile in response to HIF-1α inhibition together with PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) in hiPSC-CMs
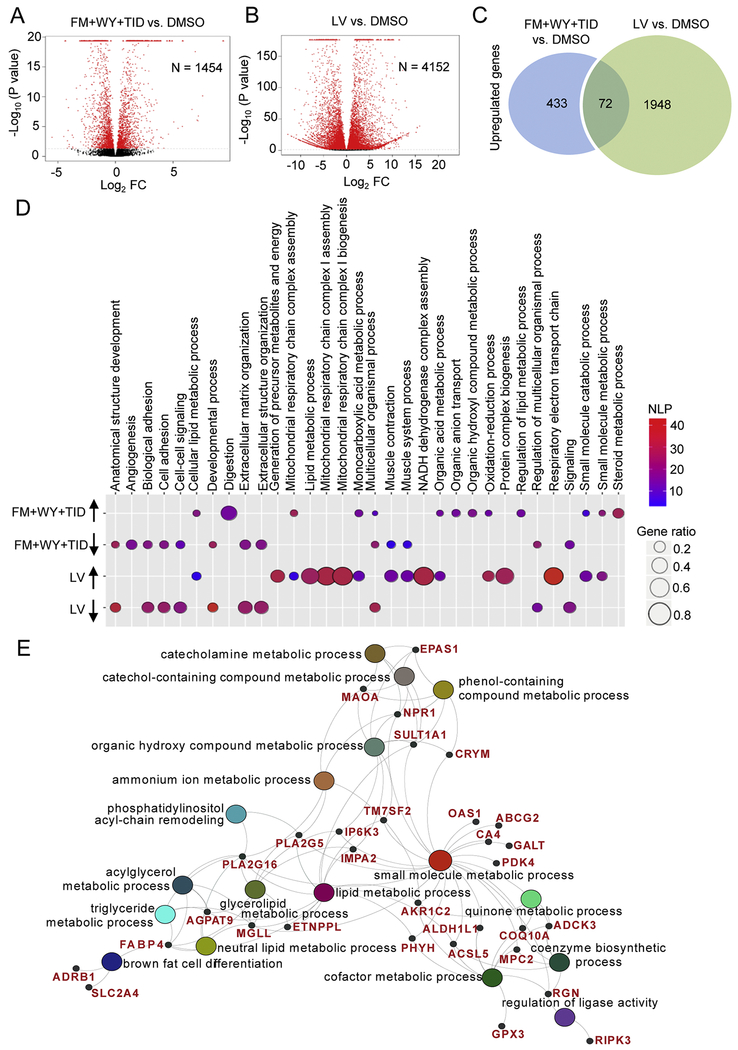
To examine the effect of HIF-1α inhibition together with WY and TID on gene expression of hiPSC-CMs, we performed RNA-Seq on hiPSC-CMs treated with FM+WY+TID or DMSO control as well as tissue samples from a pediatric left ventricle (LV, male, 6.5 months). We identified 1454 transcripts that were differentially expressed in FM+WY+TID-treated hiPSC-CMs compared with DMSO-treated hiPSC-CMs (Fig. 6A) and 4152 transcripts that were differentially expressed in LV compared with DMSO-treated hiPSC-CMs (Fig. 6B). Among the differentially expressed genes compared with DMSO-treated hiPSC-CMs, 505 genes were upregulated in FM+WY+TID-treated hiPSC-CMs, with 72 genes commonly upregulated in both FM+WY+TID-treated hiPSC-CMs and LV groups (Fig. 6C).
Figure 6.
Treatments of 3D cardiac spheres with a HIF-1α inhibitor, a PPARα activator and the postnatal factors (T3, IGF-1 and dexamethasone) alter gene expression. Volcano plots portray log2 fold change (FC) vs. −log10(P value) for differentially expressed genes in hiPSC-CMs treated with (A) FM+WY+TID and (B) tissues from human left ventricle (LV) compared with DMSO-treated hiPSC-CMs. The red dots denote significantly regulated genes (P<0.02); N indicates the number of differentially expressed genes. (C) Venn diagram showing the number of significantly up-regulated genes in FM+WY+TID-treated hiPSC-CMs vs. DMSO-treated hiPSC-CMs and LV vs. DMSO-treated hiPSC-CMs (P<0.002). (D) Bubble plot showing the enrichment analysis of differentially expressed genes in hiPSC-CMs treated with FM+WY+TID and LV compared with DMSO-treated hiPSC-CMs. Colors of displayed circles indicate the levels of significant enrichment of the gene ontology terms (GO terms) according to negative log10(P value) (NLP). The area of displayed circles corresponds to Gene_ratio which is the ratio of genes enriched within each GO term. (E) Genes within the identified GO terms that are commonly upregulated in hiPSC-CMs treated with FM+WY+TID and LV compared with DMSO-treated hiPSC-CMs. FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
GO-term analysis was performed using the DEGs in FM+WY+TID- vs. DMSO-treated hiPSC-CMs and LV vs. DMSO-treated hiPSC-CMs. Compared with DMSO-treated hiPSC-CMs, several biological processes were commonly enhanced in FM+WY+TID-treated hiPSC-CMs and LV. These included cellular lipid metabolic process, mitochondrial respiratory chain complex assembly, organic and monocarboxylic acid metabolic processes, oxidation-reduction process, small molecule catabolic and metabolic processes (Fig. 6D). GO-term analysis of upregulated DEGs focusing on GO categories under the metabolic process ontology indicated that the top commonly upregulated genes belong to metabolic processes that are functionally interconnected (Supplementary Fig. 8). As presented in a polyhierarchical graph, treatment of hiPSC-CMs with FM+WY+TID significantly upregulated GO categories: oxidation-reduction, small molecule and organic substance processes. The latter two categories are further subdivided into organic acid and lipid metabolic processes, leading to the upregulation of FA metabolic process (Supplementary Fig. 8).
We next further analyzed the commonly upregulated 72 genes in FM+WY+WY-treated hiPSC-CMs and LV when compared with DMSO-treated hiPSC-CMs. Consistent with the GO-term analysis, these commonly upregulated genes were involved in multiple biological processes that are functionally interconnected, including small molecule metabolic process, catechol-containing compound metabolic process and lipid metabolic process (Fig. 6E). Several of these upregulated genes have previously been shown to be highly expressed in the mature human heart [9]. These include genes that are involved in lipid and fatty acid metabolism: the carbonic anhydrase 4 (CA4), monoamine oxidase (MAOA), monoglyceride lipase (MGLL), glutathione peroxidase 3 (GPX3), and PDK4 (Fig. 6E; Supplementary Tables 4).
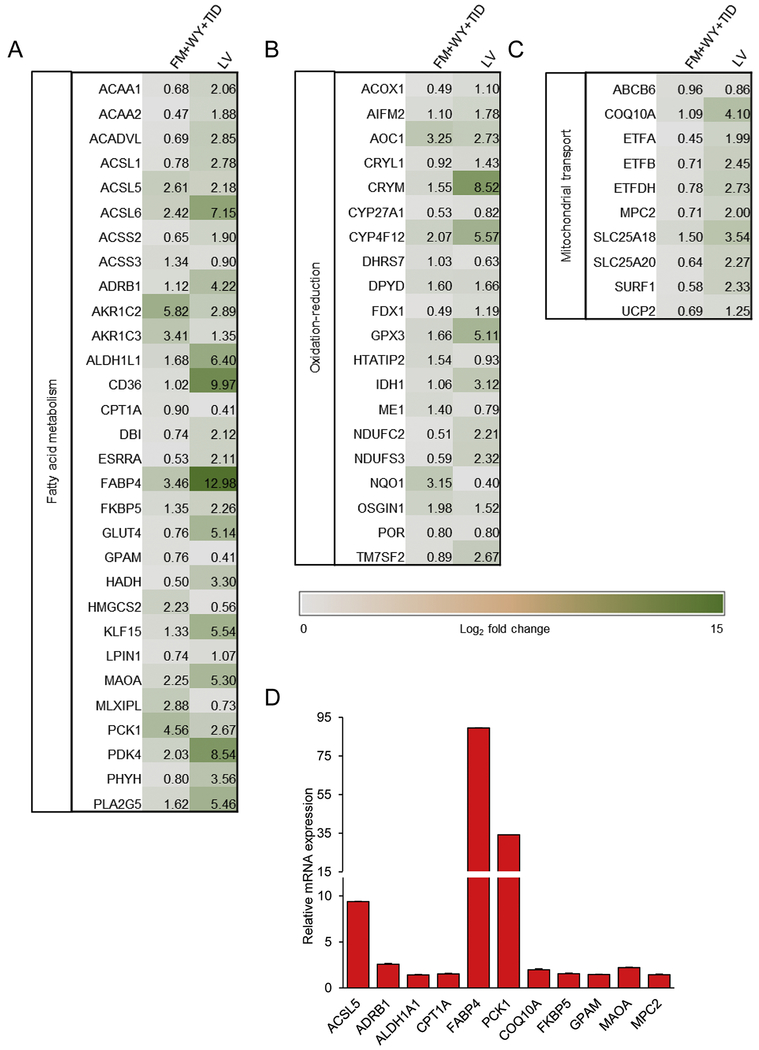
The majority of the commonly upregulated genes in both groups fell under fatty acid metabolism (Fig. 7A), oxidation-reduction (Fig. 7B) and mitochondrial transport (Fig. 7C). For example, both FM+WY+WY-treated hiPSC-CMs and LV had increased expression of oxidative-reduction-related genes including the NADH:ubiquinone oxidoreductase genes (NDUFC2 and NDUFS3) of the complex I in the electron transport chain of mitochondria and members of the cytochrome p450 gene family (CYP27A1 and CYP4F12) (Fig. 7B).
Figure 7.
Fold changes of significantly upregulated targets that are common in both FM+WY+TID-treated hiPSC-CMs vs. DMSO-treated hiPSC-CMs and LV vs. DMSO-treated hiPSC-CMs. Heat map showing upregulated genes that are categorized according to biological processes; (A) fatty acid oxidation, (B) oxidation-reduction, and (C) mitochondrial transport. Data are presented as Log2 fold change expression relative to mean for all conditions (n=3). (D) Quantitative measurement of gene expression in FM+WY+TID-treated hiPSC-CMs relative to those in DMSO-treated hiPSC-CMs was validated with qRT-PCR. Data are presented as mean ± SEM (n=3). FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
The mRNA expression of 11 genes associated with oxidative catabolic processes and mitochondrial transport were validated using qRT-PCR (Fig. 7D). Consistent with the RNA-Seq data, qRT-PCR detected a significant increase in the expression of all genes examined, including lipid metabolic genes: acyl-coA synthase long chain family member 5 (ACSL5), adrenoceptor beta 1 (ADRB1), aldehyde dehydrogenase 1 family member A1 (ALDH1A1), carnitine palmitoyltransferase 1A (CPT1A), fatty acid binding protein 4 (FABP4), phosphoenolpyruvate carboxykinase 1 (PCK1), FK506 binding protein 5 (FKBP5), glycerol-3-phosphate acyltransferase, mitochondrial (GPAM), and MAOA and mitochondrial genes: coenzyme Q10A (COQ10A) and mitochondrial pyruvate carrier 2 (MPC2).
Several members of the solute carriers (SLC) family of solute transporters were also upregulated in FM+WY+TID-treated hiPSC-CMs, including members of the SLC25 family of mitochondrial carriers, SLC25A5, SLC25A10, SLC25A18, and SLC25A20 (Fig. 7C and Supplementary Tables 3). FM+WY+TID treatment also increased the expression of mitochondrial uncoupling protein 2 (UCP2), which is a member of the family of mitochondrial anion carrier proteins (MACP) (Fig. 7C). Notably, SLCA18, SLC25A20, and UCP2 were commonly upregulated in both FM+WY+TID-treated hiPSC-CMs and LV compared with DMSO-treated hiPSC-CMs.
Other genes that were significantly up-regulated upon treatment with FM+WY+TID include several members of the voltage-gated potassium channel family (KCNE3, KCNJ16, KCNK5, and KCNIP1) as well as subunits of the sodium channels (SCNN1A and SCNN1G) (Supplementary Table 3). The expression of SCN5A and several other genes related to Ca2+ transients was not dramatically altered (P>0.05 or log2 fold changes within −0.5 to 0.5), including ATP1A, ATP2A2, CACNA1C, CaMK2D, FKBP12.6, PLN, RYR2 and SLC8A1, so did the expression of contractile/structure-related genes examined (except MYH7) (Supplementary Table 3). Additionally, FM+WY+TID increased the expression of ADRB1 (Supplementary Table 4), which is associated with cardiomyocyte maturation [41].
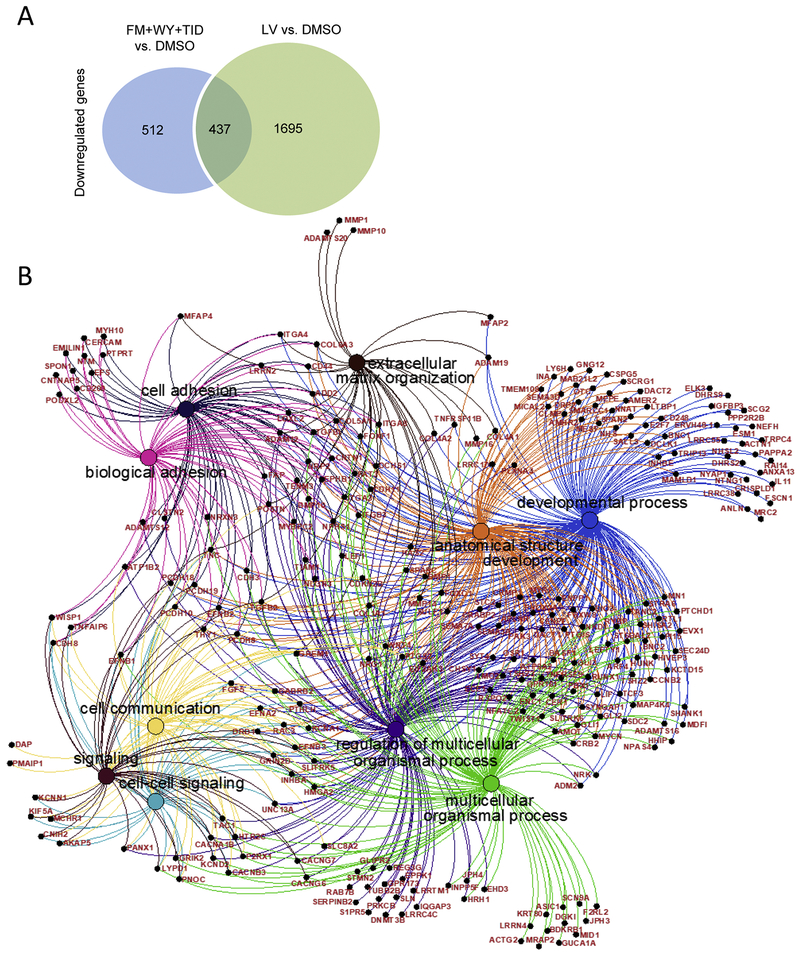
Our RNA-Seq analysis also identified that 949 genes were downregulated in FM+WY+TID-treated hiPSC-CMs and 2137 genes were downregulated in LV, with 437 genes downregulated in both FM+WY+TID-treated hiPSC-CMs and LV compared with DMSO-treated hiPSC-CMs (Fig. 8). Several biological processes were commonly downregulated in both groups, including signaling, regulation of multicellular organismal process, extracellular structure and matrix organizations, developmental process, cell-cell signaling and adhesion and anatomical structure development (Fig. 8). Further analysis of the 437 commonly decreased DEGs in FM+WY+WY-treated hiPSC-CMs and LV when compared to DMSO indicated that these DEGs were involved in multiple biological processes identified based on GO-term analysis (Fig. 8).
Figure 8.
Downregulated genes/biological processes. (A) Venn diagram of the numbers of genes downregulated genes in FM+WY+TID-treated hiPSC-CMs and LV as detected by RNA-Seq analysis. (B) Top 10 significant biological processes of the commonly downregulated genes with corresponding genes in both FM+WY+TID-treated hiPSC-CMs and LV compared with DMSO-treated hiPSC-CMs. FM, FM19G11; WY, WY-14643; TID, T3 + IGF-1 + dexamethasone.
4. Discussion
In this study, we demonstrated that targeting HIF-1α in combination with other regulators including PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) significantly improves the metabolic maturation of hiPSC-CMs. Specifically, we found that treatment of 3D hiPSC-CMs with a HIF-1α inhibitor significantly increased FAO and mitochondrial respiratory capacity and that combining HIF-1α inhibitor with the other 4 factors (WY, T3, IGF-1 and dexamethasone) was the most potent treatment to increase FAO and mitochondrial function in hiPSC-CMs. hiPSC-CMs treated with the combined molecules displayed increased mitochondrial content and enhanced calcium kinetics and contraction and relaxation velocities when compared to control cells. Furthermore, RNA-Seq revealed upregulation of genes involved in many cellular metabolic processes, including FAO in hiPSC-CMs treated with the combined molecules.
The increased FAO in hiPSC-CMs treated with HIF-1α inhibitor or HIF-1α inhibitor together with other factors (FM+WY+TID) is a hallmark of metabolic maturation in cardiomyocytes. A major difference between immature and mature cardiomyocytes is that fatty acids in mature cardiomyocytes are a primary source of energy generation whereas immature cardiomyocytes use glucose [42]. In mature cardiomyocytes, the majority of ATP utilized to drive cardiac contraction is generated by mitochondrial oxidative phosphorylation and the uptake of fatty acids into cardiomyocytes is facilitated by the action of a number of fatty acid transporters, such as CD36 and FATPs [43]. Our maturation treatment not only increased FAO but also increased the expression of many key genes involved in FAO. For example, the expression of CD36, the most important fatty acid translocase in cardiomyocytes [44], was increased significantly by HIF-1α inhibition together with other factors (FM+WY+TID). Fatty acid transporters translocate fatty acids from intracellular stores to the plasma membrane to facilitate fatty acid uptake in response to external trigger, such as insulin on adipocytes and an increase in muscular contraction of heart and skeletal muscle [45, 46]. CD36 is one of the main transporter in the heart that functions by this mechanism and is a multifunctional transmembrane glycoprotein that facilitates uptake of long-chain fatty acid. CD36 also regulates other cellular activities such as intracellular Ca2+ signaling [47] and AMPK activation [48]. Interestingly, HIF-1α inhibition also significantly increased the expression level of ESRRA, a known upstream activator of PPARGC1a pathway, which ultimately regulates mitochondrial biogenesis, fatty acid oxidation and contractile genes [11,49].
Treatment of hiPSC-CMs with 3 postnatal factors TID was reported to be able to synergistically improve the maturation of hPSC-CMs and permit the detection of disease phenotypes in hiPSC-CMs carrying disease-associated genetic mutations [18]. In our study, we observed that TID increased FAO to higher levels than FM or WY single factor treatment, although the combination treatment with FM+WY+TID was most potent to increase FAO. This observation highlights the need for multiple factors to achieve robust cardiomyocyte maturation. Further mechanistic study is needed to understand the contribution of multiple factors to cardiomyocyte maturation.
Mitochondrial bioenergetics and ATP production are key indications of more mature cardiomyocytes, because mitochondrial function is highly increased in adult and mature cardiomyocytes compared to neonatal and immature cardiomyocytes [1, 8]. A recent study published during the revision of our manuscript reported that inhibition of HIF-1α through a small molecule or siRNA knockdown increased metabolic maturation of hPSC-CMs as demonstrated by increased oxidative phosphorylation, mitochondrial content and ATP production [50]. In consistent with this finding, we found that the treatment of hiPSC-CMs with HIF-1α inhibitor and other factors (FM+WY+TID) also increased mitochondrial bioenergetics and ATP production. The enhanced mitochondrial oxidative metabolism in hiPSC-CMs treated with FM+WY+TID is consistent with an increased expression of oxidation-reduction-related genes detected by RNA-Seq including the NADH:ubiquinone oxidoreductase genes (NDUFC2 and NDUFS3) of the complex I in the electron transport chain of mitochondria and members of the cytochrome p450 gene family (CYP27A1 and CYP4F12). Changes in these mitochondrial carriers suggest a direct effect of FM+WY+TID on mitochondrial function in hiPSC-CMs.
Furthermore, the FM+WY+TID treatment led to increase in mitochondrial content and changes in mitochondrial distribution in hiPSC-CMs that are consistent with more mature cardiomyocyte phenotypes. Changes in mitochondrial content and distribution are critical components of cardiomyocyte maturation during heart development [51, 52]. In immature cardiomyocytes (such as hiPSC-CMs), the mitochondria are clustered around the nucleus or in the cell periphery [1, 53]. In contrast, the mitochondria in the adult heart are regularly distributed throughout the cell area [54]. Our data revealed that the combined treatment with FM+WY+TID enhanced mitochondrial distribution toward the phenotype observed in more mature cardiomyocytes. This structural change also correlates to an increase in mitochondrial oxidative capacity [17].
Immature cardiomyocytes are known to exhibit smaller calcium transient amplitudes as well as slower rise and decay kinetics in comparison to adult cardiomyocytes [55]. As cardiomyocytes mature, they have also been shown to display increased contractile kinetics to meet energy demand [5, 6, 56]. Our metabolic maturation treatment favorably altered the calcium-handling properties of hiPSC-CMs, including significantly faster rate of rise and decay of calcium transients when compared to control hiPSC-CMs. In addition, the treatment improved cardiomyocyte contractility as indicated by increased contraction and relaxation velocities. Consistent with these functional maturation characteristics, our RNA-Seq data implicated that the maturation treatment increased the expression of several members of the voltage-gated potassium channel family (KCNE3, KCNJ16, KCNK5, and KCNIP1), subunits of the epithelial sodium channels (SCNN1A and SCNN1G) as well as ADRB1, which are vital in regulating cadiomyocyte contraction. Ion channels are critical in shaping action potentials and maintaining a resting membrane potential and other electrophysiological properties. Whether changes in ion channel gene expression observed in our study can lead to electrophysiological maturation in hiPSC-CMs remains to be investigated. An electrophysiological analysis would help to determine if the HIF-1α inhibition and in combination with the other 4 metabolic factors 9 (FM+WY+TID) alter action potential profiles and ion currents in hiPSC-CMs. We note that the RNA-Seq data did not reveal increased expression of several Ca2+ transients and contractile/structure-related genes we examined. These observations highlight the need for further study on protein expression as well as post-translational modification which play an important role in the regulation of cardiomyocyte contraction and Ca2+ handling [57]. Since FM+WY+TID treatment increased cellular ATP levels which are needed for protein phosphorylation, the treatment could alter protein post-translational modification.
We also note that unbiased RNA-Seq provides a robust way to examine the expression of genes associated with FAO since the process involves numerous genes. The upregulated genes in FM+WY+TID-treated hiPSC-CMs showed significant increase in many metabolic processes which are also highly enriched in human pediatric heart samples. They include many interconnected metabolic processes that are upstream of lipid metabolism and FAO, agreeing with the shift to FAO for energy utilization in more mature cardiomyocytes. For example, the FM+WY+TID treatment increased FABP4 and CPT1A which are known to be involved in the maintenance of high levels of FAO [58]. In addition, the FM+WY+TID treatment increased several mitochondrial carriers in hiPSC-CMs. For example, SLC25A20 was upregulated in both FM+WY+TID-treated hiPSC-CMs and LV. Overall, treatment of hiPSC-CMs with FM+WY+TID altered genes (for example: FABP4, MAOA and MGLL) that have been previously identified as biomarkers of cardiomyocyte maturity [9, 59], confirming the maturation of hiPSC-CMs at the molecular level upon the treatment. In addition, genes involved in developmental processes, adhesion and signaling were commonly downregulated in both FM+WY+TID-treated hiPSC-CMs and LV. The overlap in both upregulated and downregulated genes in both groups confirmed an advanced degree of cardiomyocyte maturation in response to FM+WY+TID.
Since hiPSC-CMs are fetal-like cardiomyocytes and the maturation of these cells into adult-like cells may first reach to pediatric cardiomyocyte-like stage, we used pediatric LV for RNA-Seq as a control. A limitation of our RNA-Seq study is that the human heart samples we used were discarded tissues obtained from a pediatric patient during cardiac repair surgery instead of unaffected subjects. Nevertheless, our comprehensive transcriptomic data set should provide a great resource for the investigation of molecular cues for the developing human heart and the identification of novel strategies to promote the maturation of hiPSC-CMs. The RNA-Seq data reported in this manuscript are available on the GEO database with the accession number GSE 125862.
5. Conclusion
In conclusion, targeting HIF-1α together with regulation of multiple other pathways including PPARα activation and the postnatal factors (T3, IGF-1 and dexamethasone) can efficiently enhance the metabolic maturation of hiPSC-CMs during a one-week treatment period and within a culture duration of <1 month in a 3D culture format which is scalable.
Supplementary Material
Acknowledgements
The authors thank Drs. N. Anthony and A. Reedy from the Emory University Integrated Cellular Imaging Microscopy Core of the Emory+Children’s Pediatric Research Center, and A. Bryksin, S. Biliya, and D. Gulick from the core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology for the use of their shared equipment, services and expertise.
Funding
This work was supported in part by the Grant-in-Aid 16GRNT30090002 from the American Heart Association, and R01HL136345 from the NIH.
Non-standard abbreviations
- 2D
2-dimensional
- 3D
3-dimensional
- hiPSC
Human induced pluripotent stem cell
- hiPSC-CM
Human induced pluripotent stem cell-derived cardiomyocyte
- FAO
Fatty acid oxidation
- FM
FM19G11
- OCR
Oxygen consumption rate
- HIF-1α
hypoxia-inducible factor 1α
- PPARα
peroxisome proliferator activated receptor α
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- TID
T3 + IGF-1 + dexamethasone
- WY
WY-14643
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
None.
References
- [1].Yang X, Pabon L, Murry CE, Engineering Adolescence: Maturation of Human Pluripotent Stem Cell-derived Cardiomyocytes, Circ Res 114 (2014) 511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M, Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes, Nat Methods 10 (2013) 781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao ML, Levent E, Raad F, Zeidler S, Wingender E, Riegler J, Wang M, Gold JD, Kehat I, Wettwer E, Ravens U, Dierickx P, van Laake LW, Goumans MJ, Khadjeh S, Toischer K, Hasenfuss G, Couture LA, Unger A, Linke WA, Araki T, Neel B, Keller G, Gepstein L, Wu JC, Zimmermann WH, Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair, Circulation 135 (2017) 1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER, Hudson JE, Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest, Proc Natl Acad Sci U S A 114(2017)E8372–E8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ruan JL, Tulloch NL, Razumova MV, Saiget M, Muskheli V, Pabon L, Reinecke H, Regnier M, Murry CE, Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue, Circulation 134 (2016) 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Parikh SS, Blackwell DJ, Gomez-Hurtado N, Frisk M, Wang L, Kim K, Dahl CP, Fiane A, Tonnessen T, Kryshtal DO, Louch WE, Knollmann BC, Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes, Circ Res 121 (2017) 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Advanced maturation of human cardiac tissue grown from pluripotent stem cells, Nature 556 (2018) 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lopaschuk GD, Jaswal JS, Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation, J Cardiovasc Pharmacol 56 (2010) 130–40. [DOI] [PubMed] [Google Scholar]
- [9].Ellen Kreipke R, Wang Y, Miklas JW, Mathieu J, Ruohola-Baker H, Metabolic remodeling in early development and cardiomyocyte maturation, Semin Cell Dev Biol 52 (2016) 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Montessuit C, Palma T, Viglino C, Pellieux C, Lerch R, Effects of insulin-like growth factor-I on the maturation of metabolism in neonatal rat cardiomyocytes, Pflugers Arch 452 (2006) 380–6. [DOI] [PubMed] [Google Scholar]
- [11].Poon E, Keung W, Liang Y, Ramalingam R, Yan B, Zhang S, Chopra A, Moore J, Herren A, Lieu DK, Wong HS, Weng Z, Wong OT, Lam YW, Tomaselli GF, Chen C, Boheler KR, Li RA, Proteomic Analysis of Human Pluripotent Stem Cell-Derived, Fetal, and Adult Ventricular Cardiomyocytes Reveals Pathways Crucial for Cardiac Metabolism and Maturation, Circ Cardiovasc Genet 8 (2015) 427–36. [DOI] [PubMed] [Google Scholar]
- [12].Fowden AL, Li J, Forhead AJ, Glucocorticoids and the preparation for life after birth: are there long-term consequences of the life insurance?, Proc Nutr Soc 57 (1998) 113–22. [DOI] [PubMed] [Google Scholar]
- [13].Thorpe-Beeston JG, Nicolaides KH, Felton CV, Butler J, McGregor AM, Maturation of the secretion of thyroid hormone and thyroid-stimulating hormone in the fetus, N Engl J Med 324 (1991) 532–6. [DOI] [PubMed] [Google Scholar]
- [14].Troncoso R, Ibarra C, Vicencio JM, Jaimovich E, Lavandero S, New insights into IGF-1 signaling in the heart, Trends Endocrinol Metab 25 (2014) 128–37. [DOI] [PubMed] [Google Scholar]
- [15].Rog-Zielinska EA, Thomson A, Kenyon CJ, Brownstein DG, Moran CM, Szumska D, Michailidou Z, Richardson J, Owen E, Watt A, Morrison H, Forrester LM, Bhattacharya S, Holmes MC, Chapman KE, Glucocorticoid receptor is required for foetal heart maturation, Hum Mol Genet 22 (2013) 3269–82. [DOI] [PubMed] [Google Scholar]
- [16].Chattergoon NN, Giraud GD, Louey S, Stork P, Fowden AL, Thornburg KL, Thyroid hormone drives fetal cardiomyocyte maturation, FASEB J 26 (2012) 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, Sniadecki NJ, Ruohola-Baker H, Murry CE, Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells, J Mol Cell Cardiol 72 (2014) 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, Atsma DE, Passier R, Mummery CL, Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function, Cell Rep 13(2015) 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nguyen DC, Hookway TA, Wu Q, Jha R, Preininger MK, Chen X, Easley CA, Spearman P, Deshpande SR, Maher K, Wagner MB, McDevitt TC, Xu C, Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells, Stem Cell Reports 3 (2014) 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jha R, Wu Q, Singh M, Preininger MK, Han P, Ding G, Cho HC, Jo H, Maher KO, Wagner MB, Xu C, Simulated Microgravity and 3D Culture Enhance Induction, Viability, Proliferation and Differentiation of Cardiac Progenitors from Human Pluripotent Stem Cells, Sci Rep 6 (2016) 30956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y, A ROCK inhibitor permits survival of dissociated human embryonic stem cells, Nat Biotechnol 25 (2007) 681–6. [DOI] [PubMed] [Google Scholar]
- [22].Nguyen DC, Hookway TA, Wu Q, Jha R, Preininger MK, Chen X, Easley CA, Spearman P, Deshpande SR, Maher K, Wagner MB, McDevitt TC, Xu C, Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells, Stem Cell Reports 3 (2014) 260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Moreno-Manzano V, Rodriguez-Jimenez FJ, Acena-Bonilla JL, Fustero-Lardies S, Erceg S, Dopazo J, Montaner D, Stojkovic M, Sanchez-Puelles JM, FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status, J Biol Chem 285 (2010) 1333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ferrick DA, Neilson A, Beeson C, Advances in measuring cellular bioenergetics using extracellular flux, Drug Discov Today 13 (2008) 268–74. [DOI] [PubMed] [Google Scholar]
- [25].San Martin N, Cervera AM, Cordova C, Covarello D, McCreath KJ, Galvez BG, Mitochondria determine the differentiation potential of cardiac mesoangioblasts, Stem Cells 29 (2011) 1064–74. [DOI] [PubMed] [Google Scholar]
- [26].Preininger MK, Jha R, Maxwell JT, Wu Q, Singh M, Wang B, Dalal A, McEachin ZT, Rossoll W, Hales CM, Fischbach PS, Wagner MB, Xu C, A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses, Dis Model Mech 9 (2016) 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur A, Nguyen TN, Yoo JC, Judge LM, Spencer CI, Chukka AC, Russell CR, So PL, Conklin BR, Healy KE, Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales, Tissue Eng Part C Methods 21 (2015) 467–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, The human genome browser at UCSC, Genome Res 12 (2002) 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim D, Langmead B, Salzberg SL, HISAT: a fast spliced aligner with low memory requirements, Nat Methods 12 (2015) 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data, Bioinformatics 31 (2015) 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].R Core Team (2017) R A Language and Environment for Statistical Computing. - References - Scientific Research Publishing, 2018 http://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/ReferencesPapers.aspx?ReferenceID=2144573. [Google Scholar]
- [32].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26 (2010) 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization, Nucleic Acids Res 37 (2009) W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP, Chen HS, Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs, Nature 494 (2013) 105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, Atsma DE, Passier R, Mummery CL, Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function, Cell Rep 13 (2015) 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Haldar SM, Jeyaraj D, Anand P, Zhu H, Lu Y, Prosdocimo DA, Eapen B, Kawanami D, Okutsu M, Brotto L, Fujioka H, Kerner J, Rosca MG, McGuinness OP, Snow RJ, Russell AP, Gerber AN, Bai X, Yan Z, Nosek TM, Brotto M, Hoppel CL, Jain MK, Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation, Proc Natl Acad Sci U S A 109 (2012) 6739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harris RA, Bowker-Kinley MM, Huang B, Wu P, Regulation of the activity of the pyruvate dehydrogenase complex, Adv Enzyme Regul 42 (2002) 249–59. [DOI] [PubMed] [Google Scholar]
- [38].Burridge PW, Li YF, Matsa E, Wu H, Ong SG, Sharma A, Holmstrom A, Chang AC, Coronado MJ, Ebert AD, Knowles JW, Telli ML, Witteles RM, Blau HM, Bernstein D, Altman RB, Wu JC, Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity, Nat Med 22 (2016) 547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER, Hudson JE, Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest, Proc Natl Acad Sci U S A 114 (2017) E8372–e8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Racca AW, Klaiman JM, Pioner JM, Cheng Y, Beck AE, Moussavi-Harami F, Bamshad MJ, Regnier M, Contractile properties of developing human fetal cardiac muscle, J Physiol 594 (2016) 437–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jung G, Fajardo G, Ribeiro AJS, Kooiker KB, Coronado M, Zhao M, Hu DQ, Reddy S, Kodo K, Sriram K, Insel PA, Wu JC, Pruitt BL, Bernstein D, Time-dependent evolution of functional vs. remodeling signaling in induced pluripotent stem cell-derived cardiomyocytes and induced maturation with biomechanical stimulation, FASEB J 2016, pp. 1464–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bhute VJ, Bao X, Dunn KK, Knutson KR, McCurry EC, Jin G, Lee WH, Lewis S, Ikeda A, Palecek SP, Metabolomics Identifies Metabolic Markers of Maturation in Human Pluripotent Stem Cell-Derived Cardiomyocytes, Theranostics 7 (2017) 2078–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Steinbusch LK, Schwenk RW, Ouwens DM, Diamant M, Glatz JF, Luiken JJ, Subcellular trafficking of the substrate transporters GLUT4 and CD36 in cardiomyocytes, Cell Mol Life Sci 68 (2011) 2525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA, Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36, J Biol Chem 268 (1993) 17665–8. [PubMed] [Google Scholar]
- [45].Glatz JF, Luiken JJ, Bonen A, Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease, Physiol Rev 90 (2010) 367–417. [DOI] [PubMed] [Google Scholar]
- [46].Glatz JF, Angin Y, Steinbusch LK, Schwenk RW, Luiken JJ, CD36 as a target to prevent cardiac lipotoxicity and insulin resistance, Prostaglandins Leukot Essent Fatty Acids 88 (2013) 71–7. [DOI] [PubMed] [Google Scholar]
- [47].Pietka TA, Sulkin MS, Kuda O, Wang W, Zhou D, Yamada KA, Yang K, Su X, Gross RW, Nerbonne JM, Efimov IR, Abumrad NA, CD36 protein influences myocardial Ca2+ homeostasis and phospholipid metabolism: conduction anomalies in CD36-deficient mice during fasting, J Biol Chem 287 (2012) 38901–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Samovski D, Sun J, Pietka T, Gross RW, Eckel RH, Su X, Stahl PD, Abumrad NA, Regulation of AMPK activation by CD36 links fatty acid uptake to beta-oxidation, Diabetes 64 (2015) 353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dorn GW 2nd, Vega RB, Kelly DP, Mitochondrial biogenesis and dynamics in the developing and diseased heart, Genes Dev 29 (2015) 1981–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hu D, Linders A, Yamak A, Correia C, Kijlstra JD, Garakani A, Xiao L, Milan DJ, van der Meer P, Serra M, Alves PM, Domian IJ, Metabolic Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes by Inhibition of HIF1alpha and LDHA, Circ Res 123 (2018) 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hom JR, Quintanilla RA, Hoffman DL, de Mesy Bentley KL, Molkentin JD, Sheu SS, Porter GA Jr., The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation, Dev Cell 21 (2011) 469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Feric NT, Radisic M, Maturing human pluripotent stem cell-derived cardiomyocytes in human engineered cardiac tissues, Adv Drug Deliv Rev 96 (2016) 110–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gherghiceanu M, Barad L, Novak A, Reiter I, Itskovitz-Eldor J, Binah O, Popescu LM, Cardiomyocytes derived from human embryonic and induced pluripotent stem cells: comparative ultrastructure, J Cell Mol Med 15 (2011) 2539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Porter GA, Hom J, Hoffman D, Quintanilla R, de Mesy Bentley K, Sheu SS, Bioenergetics, mitochondria, and cardiac myocyte differentiation, Prog Pediatr Cardiol 31 (2011) 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Keung W, Boheler KR, Li RA, Developmental cues for the maturation of metabolic, electrophysiological and calcium handling properties of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Res Ther 2014, p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Correia C, Koshkin A, Duarte P, Hu D, Teixeira A, Domian I, Serra M, Alves PM, Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells, Scientific Reports 7 (2017) 8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Frey N, McKinsey TA, Olson EN, Decoding calcium signals involved in cardiac growth and function, Nat Med 6 (2000) 1221–7. [DOI] [PubMed] [Google Scholar]
- [58].Houten SM, Wanders RJ, A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation, J Inherit Metab Dis 33 (2010) 469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA, Kwon C, Transcriptional Landscape of Cardiomyocyte Maturation, Cell Rep 13 (2015) 1705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.