Abstract
The increasing antibiotic resistance evolved by Helicobacter pylori has alarmingly reduced the eradication rates of first-line therapies. To overcome the current circulating resistome, we selected a novel potential therapeutic target in order to identify new candidate drugs for treating H. pylori infection. We screened 1120 FDA-approved drugs for molecules that bind to the essential response regulator HsrA and potentially inhibit its biological function. Seven natural flavonoids were identified as HsrA binders. All of these compounds noticeably inhibited the in vitro DNA binding activity of HsrA, but only four of them, apigenin, chrysin, kaempferol and hesperetin, exhibited high bactericidal activities against H. pylori. Chrysin showed the most potent bactericidal activity and the most synergistic effect in combination with clarithromycin or metronidazole. Flavonoid binding to HsrA occurs preferably at its C-terminal effector domain, interacting with amino acid residues specifically involved in forming the helix-turn-helix DNA binding motif. Our results validate the use of HsrA as a novel and effective therapeutic target in H. pylori infection and provide molecular evidence of a novel antibacterial mechanism of some natural flavonoids against H. pylori. The results further support the valuable potential of natural flavonoids as candidate drugs for novel antibacterial strategies.
Subject terms: Screening, High-throughput screening
Introduction
Helicobacter pylori is considered the most prevalent human pathogen worldwide, more than half the world’s population is infected by this Gram-negative bacterium1. Unless treated, colonization of the human stomach usually persists lifelong causing gastric inflammation and contributing to the pathogenesis of peptic ulceration, gastric adenocarcinoma, and mucosa-associated lymphoid-tissue (MALT) lymphoma2,3. Nearly 90% of all gastric cancers, the third most common cause of cancer death worldwide, can be attributable to H. pylori infection1.
A standard triple therapy containing a proton-pump inhibitor (PPI) and two antibiotics, clarithromycin and either amoxicillin or metronidazole, has been traditionally considered the first-line regimen for treatment of H. pylori infection4,5. However, the PPI-based triple therapy has been described to be losing its efficacy for H. pylori, with eradication cure rates as low as 50% to 70%, due to high rates of antibiotic resistance, high rates of antibiotic-associated side effects and low compliance6,7. Because of increasing failure of the traditional therapy, the current guidelines recommend a quadruple therapy (PPI + amoxicillin + metronidazole + clarithromycin) as first-line strategy, while the PPI triple therapy is restricted to areas with known low clarithromycin resistance8. In February 2017, the WHO included H. pylori in its first ever list of antibiotic-resistant “priority pathogens”, a catalogue of 12 families of bacteria that pose at present the greatest threat to human health9. Nowadays, an effective novel therapy against this clinically relevant pathogen is mandatory to increase eradication rates and minimize both antimicrobial resistance and side effects on normal microbiota. To mitigate the challenges of antimicrobial resistance, the search for new bioactive compounds able to inhibit the growth of pathogens by acting specifically on new molecular targets can be a valuable route for drug discovery.
The H. pylori genome harbours the hp1043 (hsrA) gene, encoding for an OmpR-like “orphan” response regulator which has been shown to be essential for cell viability10,11. HsrA (for homeostatic stress regulator) is unique and highly conserved among members of the Epsilonproteobacteria12. The protein functions as a global homeostatic regulator synchronizing metabolic functions and virulence with availability of nutrients and cell division13, mediating also the response to oxidative stress14. HsrA binds to DNA acting as a transcriptional activator, modulating its own expression15, but also the expression of a plethora of genes involved in crucial cellular functions such as translation, transcription, energy metabolism, nitrogen metabolism and redox homeostasis14,16. NMR-spectroscopy and X-ray crystallography analyses proposed that HsrA exists as a symmetric dimer in vivo with two well-defined functional domains, an N-terminal regulatory domain and a C-terminal DNA-binding/effector domain17. Unlike most response regulators, where phosphorylation of the regulatory domain triggers conformational changes that promote dimerization of the protein and activate its DNA binding function, the two domains of HsrA seems to act independently. Thus, mutations in the N-terminal regulatory domain that inhibited dimerization did not affect DNA binding function17, while deletion of the C-terminal effector domain did not have effect on the dimerization ability of the protein15. Attempts to both deletion and overexpression of HsrA have resulted unsuccessful, supporting not only an essential function of the regulator but also a very tight post-transcriptional control of its expression that ensures appropriate levels of the protein into the cell12,15.
As an essential protein for microbial viability, easily produced under lab conditions, with a known crystal structure and no counterpart in humans, the response regulator HsrA become in a promising therapeutic target against H. pylori infection. In the present study, we screened the Prestwick Chemical Library®, a collection of 1120 FDA-approved, off-patent small molecules for compounds that specifically bind to HsrA and potentially inhibit its essential function. Four HsrA inhibitors, the natural flavonoids apigenin, chrysin, kaempferol and hesperetin exhibited strong bactericidal activities against both metronidazole- and clarithromycin-resistant H. pylori strains.
Materials and Methods
Chemicals
The Prestwick Chemical Library was purchased from Prestwick Chemical (France). All drugs were provided as 10 mM solutions in 100% DMSO in 96-well plates, which were stored at −20 °C until use. For some assays, compounds of interest were purchased from Sigma-Aldrich and properly stored according to the manufacturer indications. Stock solutions of each compound were prepared at 20 mM in 100% DMSO for EMSA and ITC, and at 10.24 g/L in 100% DMSO for MIC/MBC determinations. In all cases, stock solutions of drugs were stored at −20 °C.
Recombinant HsrA expression and purification
The complete sequence of gene hsrA (hp1043) was amplified from the genomic DNA of H. pylori strain 26695 (ATCC 700392) using the oligonucleotides Hpylori.hsrA_up (5′- GGAATTCCATATGCGCGTTCTACTGATTG-3′) and Hpylori.hsrA_dw (5′-CCCAAGCTTTTACTCTTCACACGCCGG-3′). The resulting PCR product was digested with NdeI and HindIII, and inserted between the same restriction sites of vector pET-28a (Novagen). The final vector was partially sequenced to ensure that no modifications in the nucleotide sequence occurred during PCR and cloning. N-terminal 6 × His-tagged HsrA was overexpressed in E. coli BL21(DE3) (EMD Biosciences). Recombinant E. coli carrying the pET-28a-hsrA vector was grown to late log phase (OD600 = 0.6–0.8) in Luria Bertani (LB) broth at 37 °C with vigorous shaking. At this time, hsrA expression was induced by addition of 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) and culture was further incubated by 6 h. Cells were harvested by centrifugation at 8,000 rpm for 10 min at 4 °C, washed twice with ice-cold PBS (pH 7.4) and stored at −20 °C.
For protein purification, cells were resuspended in ice-cold lysis buffer containing 50 mM Tris-HCl (pH 8), 500 mM NaCl, 10 mM imidazole, 1 mM PMSF (phenylmethylsulfonyl fluoride) and sonicated in an ice-water bath by ten 30 s bursts with 30 s cooling intervals. The resulting crude extracts were centrifuged twice at 15,000 rpm for 20 min at 4 °C to remove cell debris. Recombinant His-tagged HsrA was purified by immobilized metal-affinity chromatography (IMAC) using nickel charged Chelating Sepharose Fast Flow resin (GE Healthcare). The bound protein was eluted by using an imidazole gradient (10 mM to 1 M) and dialyzed against the store buffer (50 mM Tris-HCl [pH 8], 300 mM NaCl, 10% glycerol). Protein concentration was determined using the BCA™ Protein Assay kit (Thermo Fisher Scientific). The N-terminal His-tag was removed by overnight treatment of the 6 × His-tagged HsrA with thrombin (GE Healthcare), 10 units of enzyme for each mg of tagged protein. The cleaved HsrA protein was obtained in the flow throw of a second IMAC-Ni2+ purification step.
High-throughput screening
A target-based high-throughput screening (HTS) methodology was used to test the Prestwich Chemical Library for compounds that specifically bind to H. pylori HsrA regulator and potentially inhibit its biological function. Compound binding to HsrA was assessed by a fluorescence-based thermal shift assay18,19. The screening was performed in V-shape 96-well plates Thermo-Fast 96 (ABgene), adding a final volume of 100 μL of reaction mixture in each well of the plate. A solution of 10 µM of recombinant HsrA was freshly prepared in a buffer containing 50 mM Tris-HCl [pH 8], 150 mM NaCl, 10% glycerol and 5 mM DTT. The SYPRO® Orange ready-to-use fluorescent stain (Thermo Fisher Scientific) was added to this protein solution at a final concentration of 10X and thoroughly mixed. Next, 97.5 μL of this reaction mixture was dispensed into all the wells of a V-shape 96-well plate. While all wells of the columns 1 and 12 received 2.5 μL of DMSO (vehicle) instead of compounds and were used as reference controls, each of the eighty wells of columns 2 to 11 in the 96-well plate received 2.5 μL of a different compound from the chemical library (250 μM as final concentration each). After mixing, each well of the plate was overlaid with 50 μL of mineral oil to avoid evaporation. Unfolding curves were registered from 25 °C to 75 °C in 1 °C steps using a FluoDia T70 High Temperature Fluorescence Microplate Reader (Photon Technology International, UK). The system was allowed to equilibrate at each temperature for 1 min before each fluorescence acquisition. In practice, this represents an operational heating rate of 0.25 °C/min approximately. Because SYPRO Orange dye interacts with hydrophobic regions in the protein that become exposed upon denaturation, fluorescence emission of the dye serves as a read out of thermal denaturation of the protein20. The unfolding curves corresponding to each well were analysed using a homemade software that estimates the midpoint temperature of unfolding (Tm) of each well. Compounds that increased thermal stability of HsrA above the twofold standard deviation value of reference controls were identified as HsrA binders and selected as potential inhibitors.
Electrophoretic mobility shift assays
The DNA binding activity of the recombinant HsrA response regulator as well as its putative inhibition by HsrA binders were evaluated by electrophoretic mobility shift assays (EMSAs). A 300-bp promoter region of porGDAB operon was amplified by PCR and used as target sequence of HsrA in all EMSA experiments14. Recombinant HsrA protein (6 μM) was mixed with 120 ng of target promoter in a 20 μL reaction volume containing 10 mM bis-Tris (pH 7.5), 40 mM KCl, 100 mg/L BSA, 1 mM DTT, and 5% glycerol. During the inhibition assays, putative inhibitors were added to final concentrations of 2, 1, 0.5 and 0.1 mM to the mixtures of protein and DNA. Binding assays with DMSO instead of inhibitors were included as vehicle controls. To ensure the specificity of EMSAs, a 150-bp DNA fragment corresponding to the coding region of gene pkn22 (alr2502) from Anabaena sp. PCC 7120 was included as non-specific competitor DNA in all assays. Mixtures were incubated at room temperature for 20 min and subsequently separated on a 6% non-denaturing polyacrylamide gel in running buffer (25 mM Tris, 190 mM glycine) at 90 V. The gel was stained with SYBR Safe DNA gel stain (Invitrogen) and processed with a Gel Doc 2000 Image Analyzer (Bio-Rad).
Minimal inhibitory and bactericidal concentrations
The antibacterial activity of HsrA inhibitors was tested against three different strains of H. pylori: ATCC 700392, ATCC 43504 (metronidazole-resistant reference strain), and ATCC 700684 (clarithromycin-resistant reference strain). MIC determinations were carried out by the broth microdilution method as previously described18,21, with slight modifications. H. pylori strains were grown in Blood Agar Base N°2 (OXOID) supplemented with 8% defibrinated horse blood (OXOID) in a humidified microaerobic incubator (85% N2, 10% CO2, 5% O2) at 37 °C for 72 h. To prepare inoculums for MIC assays, bacteria were grown for 48 hours at 37 °C in brain heart infusion broth supplemented with 4% FBS (BHI + FBS) and next diluted in the same fresh medium to a final optical density at 660 nm of 0.01 (∼106 CFU/mL). From this inoculum, 195 µL were added to all wells of the first column of a sterile 96-well flat-bottom microtiter plate, while 100 µL were added to all wells from column 2 to 12. Next, 5 µL of a 10.24 g/L in 100% DMSO stock solution of each HsrA inhibitor were added to wells of the first column and two-fold serial dilutions were performed in order to test each inhibitor in a concentration range from 256 to 0.125 mg/L. DMSO, metronidazole and clarithromycin were included as controls in all assays. Plates were incubated under microaerobic conditions at 37 °C and examined visually after 72 h22,23. MIC values were defined as the lowest concentration of compound that completely inhibited the visible growth of bacteria after 72 h incubation. For MBC determinations, 10 μL aliquots of two compound dilutions around the MIC value were seeded on Blood Agar Base N°2 supplemented with 8% defibrinated horse blood and incubated for 72 h under microaerobic conditions at 37 °C. MBC was defined as the lowest concentration of compound that prevented the growth of ≥99.9% of H. pylori cells after subculture on the inhibitor-free medium. Each experiment was performed twice in triplicate to confirm the results.
Time-kill kinetics assays
Time-kill kinetics of selected flavonoids were carried out as previously described18 with slight modifications. A final concentration of 2 × MIC from each flavonoid was mixed with 200 µL of H. pylori strain ATCC 700684 at 1.0 × 105 CFU/mL in BHI + FBS. The experiments were performed twice in triplicate using sterile 96-well flat-bottom microtiter plates. Plates were incubated under microaerobic conditions at 37 °C with shaking. Mixtures of bacteria with the same amount of DMSO (vehicle) instead flavonoid were incubated under the same conditions and included as controls in all determinations. Aliquots of 20 µL taken at time intervals of 0, 2, 4, 8, and 24 h were serially diluted in BHI medium and plated in Blood Agar Base N°2 supplemented with 8% defibrinated horse blood nutrient agar for colony forming unit (CFU) determination. Plates were incubated at 37 °C for 72 h under microaerobic conditions. The amount of grown H. pylori colonies was counted and presented as log10 CFU/mL versus incubation time. The results were subjected to statistical analysis using the Mann-Whitney U test24. The criterion P < 0.1 was used to determine the statistical significance.
Checkerboard assay
The existence of synergism in the antimicrobial activity of selected flavonoids and the first-line antibiotics clarithromycin and metronidazole was determined against two H. pylori strains (clarithromycin-resistance strain ATCC 700684 and metronidazole-resistant strain ATCC 43504) using the checkerboard assay25,26.
Solutions at 4-fold the highest concentration to be tested were prepared for each compound. Then, antibiotic and flavonoid were two-fold serially diluted in BHI + FBS using two sterile 96-well flat-bottom microtiter plates; one of the compounds was diluted along the rows (horizontally) of a first plate, and the other compound was diluted along the columns (vertically) of the second plate. A matrix of both gradient was obtained by mixing 50 µL of the content of each well of plate 1 (except from first column) with 50 µL from the same well of plate 2 (except from last row), thereby diluting both compounds at two-fold the concentrations to be tested. Thus, the first column in plate 3 would have serial dilutions of just one of the compounds, and the last row would have serial dilutions of the other. Finally, 100 µL of freshly prepared inoculum (2×106 CFU/mL) of proper H. pylori strain were added to all wells. The plates were incubated for 72 h at 37 °C under microaerophilic conditions. Then, 30 µL of 0.1 mg/ml filter-sterilized resazurin (Sigma-Aldrich) was added as growth indicator25,27 to each well and plates were further incubated for 8 hours. Each tested panel was performed in duplicate. The interaction between the tested antimicrobial agents was determined by calculating the fractional inhibitory concentration (FIC) index. In the 96-well plate, the first column shows the MIC of one of the compounds (A), and the last row shows the MIC of the second compound (B). FIC index could be calculated as: FICA (MICA in the presence of B/MICA alone) + FICB (MICB in the presence of A/MICB alone). The FIC index allows objective identification of any interaction between two compounds against a particular bacterial strain, indicating whether there is synergy (FIC index ≤0.5), additive effect (FIC index >0.5 to ≤1), no interaction or neutral (FIC index >1 to ≤4), or antagonism (FIC index >4)26,28,29. The term “synergy” implies that the resulting effect of a combination is significantly greater than the sum of its individual parts, while “additive effect” occurs when substances added together will improve or increase their efficacies, albeit not to the extent of a synergistic interaction30.
Isothermal titration calorimetry assays
Direct interaction of the selected compounds with HsrA was assessed by isothermal titration calorimetry (ITC). Titrations were performed in a high-sensitivity AutoiTC200 microcalorimeter (MicroCal, Malvern Instruments). Protein samples and compound solutions were properly degassed and carefully loaded into the calorimetric cells to avoid bubble formation during stirring. Experiments were performed with freshly prepared buffer-exchanged protein solutions, at 25 °C in 50 mM Tris-HCl [pH 8], 150 mM NaCl, 10% glycerol, 1% DMSO. A solution of HsrA (20 µM) in the calorimetric cell was titrated with a solution of the corresponding compound (200 µM) located in the injecting syringe. A sequence of 19 injections of 2 µL volume was programmed with a time spacing of 150 s, a stirring speed of 750 rpm, and a reference power of 10 µcal/s in the sample cell31,32. The binding isotherm (ligand-normalized heats as a function of the molar ratio) was obtained through integration of the individual heat effect associated with each ligand injection observed in the thermogram (thermal power required to maintain the difference between the temperatures of the sample cell and the reference cell close to zero as a function of time). Dissociation constants and binding enthalpies were calculated by non-linear least squares regression data analysis in Origin 7.0 (OriginLab).
Molecular docking
The tridimensional structures of flavonoids (ligands) were downloaded from PubChem33 and were docked onto the structure of the H. pylori HsrA response regulator (2HQR, model 1, chain A) obtained from the PDB34, using AutoDock Vina35. The ligands were considered flexible, and a ligand-specific torsion tree was defined in each case, representing the rigid and rotatable pieces of the molecule. In each torsion tree rotatable bonds were allowed the torsional degrees of freedom corresponding to the specific bond, according to AutoDock torsion parameterizations. The structure of the protein was considered rigid and a grid enclosing the whole structure of the protein was defined, representing the search space for putative interaction sites. Separately, pre-calculated maps were defined for the ligands, encompassing independent maps for each atom type in the ligand, combining the desolvation and electrostatics potentials. The AutoGrid algorithm was then used to estimate the interaction energy of a ligand conformation (i.e. ligand pose) by summing up the contribution of atoms of a specific element at each point in the grid around the rigid receptor. After scanning the whole protein structure search space, the docking solutions passing a threshold of ΔG < −4.0 kcal/mol were ranked based on the estimated interaction energy. The top ranking pose for each ligand was selected as predicted model of interaction.
Results
High-throughput screening of Prestwick Chemical Library reveals specific flavonoid binders of H. pylori HsrA response regulator
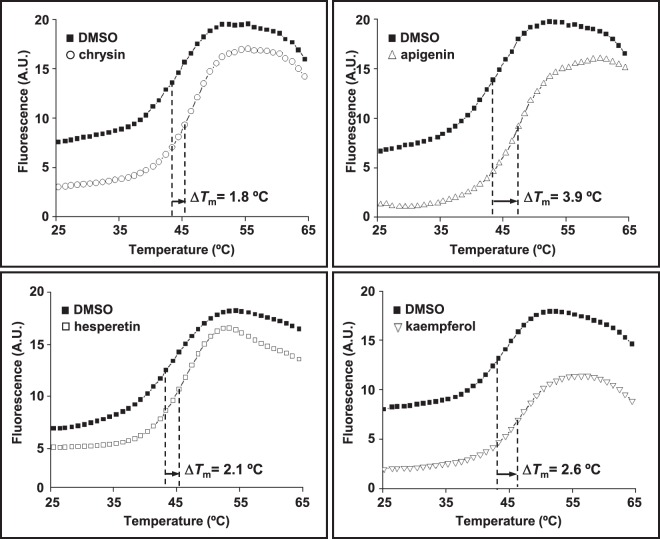
As the first step to identify new candidate drugs for treating H. pylori infection, we screened the Prestwich Chemical Library for small FDA-approved molecules that specifically bind to the essential response regulator HsrA. The HTS was carried out using a fluorescence-based thermal shift assay18,19. With this method, any compound of the chemical library that preferentially binds to the native state of HsrA increases the protein conformational stability and causes a shift of the protein unfolding curve to higher temperatures due to the increased melting temperature (Tm) of the protein-binder complex. To minimize the identification of false-positive binders, we defined as relevant compounds those which increased the thermal stability of HsrA above the twofold standard deviation of Tm value of reference controls. Following this selection criterion, the target-based HTS approach identified 14 compounds of the 1120 compound collection (1.25%) as HsrA binders. Notably, seven of these positive hits consisted in naturally occurring flavonoids, including apigenin, chrysin, hesperetin, kaempferol, luteolin, myricetin, and quercetin (Table 1). The increases in Tm values indicated that all of these seven natural flavonoids bound to HsrA and formed stable complexes. The thermal upshifts of the HsrA unfolding curve triggered by 250 μM of four different flavonoids (chrysin, apigenin, hesperetin and kaempferol) are shown in Fig. 1. Due to the high heterogeneity of the rest of HsrA binders (data not shown), we focused our further analyses to primarily discern the potentiality of flavonoid binders of HsrA as phytochemical therapies to H. pylori infections.
Table 1.
HsrA binders identified by a fluorescence-based HTS method using the thermal shift assay.
| Compound | ΔTm (°C)a | Class | Chemical structure | Pharmacological effects |
|---|---|---|---|---|
| chrysin | 1.8 | flavone | 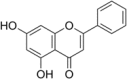 |
anti-inflammatory, antineoplastic, antioxidant, hepatoprotector |
| apigenin | 3.9 | flavone | 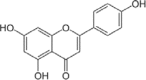 |
antiproliferative, anti-inflammatory |
| luteolin | 1.8 | flavone | 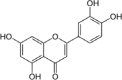 |
anti-inflammatory, antioxidant, antiproliferative |
| hesperetin | 2.1 | flavanone | 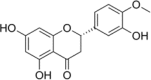 |
cholesterol lowering, hypolipidemic, antioxidant, anti-allergic, antineoplastic, anti-inflammatory |
| kaempferol | 2.6 | flavonol |  |
anti-inflammatory, diuretic, antioxidant |
| quercetin | 4.2 | flavonol |  |
antioxidant, anti-allergic, anti-inflammatory, antiproliferative |
| myricetin | 3.9 | flavonol |  |
antineoplastic, antioxidant, hypoglycemic |
aIncrease in the Tm value of protein-ligand complex with respect to the mean Tm value of controls (protein + DMSO). In all cases, ΔTm > twofold standard deviation of controls.
Figure 1.
Thermal upshifts of the HsrA unfolding curve triggered by 250 μM of the natural flavonoids chrysin, apigenin, hesperetin and kaempferol. The HTS was carried out using a fluorescence-based thermal shift assay. Any compound that preferentially binds to the native state of HsrA would increase the protein stability and cause an upshift of the protein unfolding curve due to the increased melting temperature (Tm) of the protein-binder complex. In all assays, DMSO (vehicle) instead of compounds were mixed with the protein and used as reference control of the protein unfolding curve.
Flavonoids inhibit DNA binding activity of HsrA in vitro
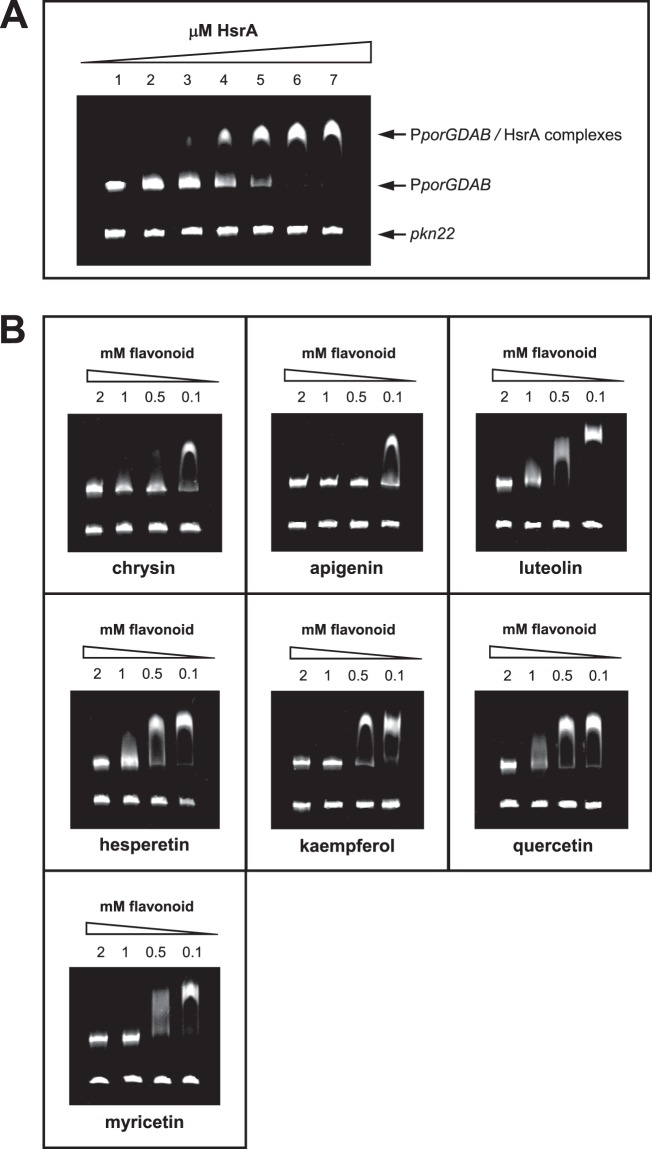
HsrA is an orphan response regulator that functions as a transcriptional activator of gene expression; hence, it interacts with DNA and binds to specific sequences located in target promoters. Biological activity of purified recombinant HsrA was evaluated before its use in the target-based HTS in order to ensure that the native conformation of the response regulator interacts with potential inhibitors of the chemical library. EMSA analyses of the recombinant response regulator in the presence of its target promoter PporGDAB14 demonstrated a high affinity of HsrA by its target DNA in a concentration-dependent manner (Fig. 2A). Under the experimental conditions used in EMSAs, 120 ng of target DNA were completely and specifically complexed to HsrA from 6 μM of the recombinant protein. Hence, this concentration was subsequently used for EMSA inhibition assays in the presence of flavonoid binders.
Figure 2.
Impact of several natural flavonoids on the in vitro affinity of HsrA for its target promoter. (A) EMSAs showing the ability of HsrA to specifically bind in vitro the promoter region of porGDAB operon. DNA fragments were mixed with increasing concentrations of recombinant HsrA protein (indicated in μM) and separated on a 6% PAGE. A 150-bp DNA fragment of Anabaena gene pkn22 was included as non-specific competitor DNA in all assays. (B) DNA fragments were mixed with 6 μM of recombinant HsrA protein in the presence of 2, 1, 0.5 and 0.1 mM of natural flavonoids.
To determine whether the flavonoid binders previously identified with our screening approach effectively inhibit the DNA binding activity of HsrA, we carried out EMSA analyses in the presence of different concentration of each flavonoids (from 2 mM to 100 μM). Since flavonoids were diluted in 100% DMSO, negative controls of EMSAs were carried out by mixing the same amounts of protein and DNA in the presence of DMSO instead of flavonoids. As shown in Fig. 2B, all flavonoids appreciably inhibited the in vitro binding activity of HsrA to its target promoter. However, the in vitro inhibitory effects of some flavonoid binders such as apigenin or chrysin on the DNA binding activity of HsrA appeared noticeable stronger than those exerted by other flavonoids such as quercetin.
Apigenin, chrysin, kaempferol and hesperetin exhibit potent bactericidal activity against H. pylori
To evaluate the antibacterial activity of the HsrA inhibitors, we determined both MIC and MBC values of each HsrA inhibitors against three different strains of H. pylori, including the metronidazole-resistant strain ATCC 43504 and the clarithromycin-resistant strain ATCC 700684. In all cases, the lowest concentration of compound that completely inhibited the visible growth of bacteria after 72 h incubation was also the same which prevented the growth of ≥99.9% of H. pylori cells after subculture on the inhibitor-free medium (Table 2). Thus, both MIC and MBC values corresponded to the same concentration of compound for all the HsrA inhibitors tested. Despite all the seven natural flavonoids specifically bound to HsrA and inhibited its biological function in vitro according to EMSA, only four of them, apigenin, chrysin, kaempferol and hesperetin, exhibited potent bactericidal activities (MBC ≤ 8 mg/L) against H. pylori. Hence, the bactericidal effect of the other three HsrA inhibitors, luteolin, quercetin, and myricetin were evidenced at concentrations quite higher than MIC breakpoints for antimicrobials traditionally used in anti-H. pylori therapies36,37, thereby these three flavonoids were considered poorly active against H. pylori. To further evaluate the bactericidal activities of apigenin, chrysin, kaempferol and hesperetin against H. pylori, time-kill kinetics were carried out at 2 × the MIC values (listed in Table 2) for each compound against the H. pylori strain ATCC 700684. As in shown Fig. 3, no live bacteria could be detected at 8 hours after exposition to 2 × MIC of each flavonoid, and this was found to be statistically significant according to the Mann-Whitney U test24 (P < 0.1). The bactericidal activity of chrysin at this concentration was significant (P < 0.1) even after only 4 hours of exposition to the microorganism.
Table 2.
Minimal inhibitory and bactericidal concentrations of HrsA inhibitors.
| Compound | MIC (MBC), mg/L | ||
|---|---|---|---|
| ATCC 700392 | ATCC 43504 (MTZ-R) | ATCC 700684 (CLR-R) | |
| chrysin | 8 (8) | 8 (8) | 4 (4) |
| apigenin | 8 (8) | 8 (8) | 8 (8) |
| luteolin | 64 (64) | 32 (32) | 64 (64) |
| hesperetin | 8 (8) | 8 (8) | 4 (4) |
| kaempferol | 8 (8) | 16 (16) | 16 (16) |
| quercetin | 64 (64) | 64 (64) | 64 (64) |
| myricetin | 128 (128) | 128 (128) | 128 (128) |
| metronidazole | 1 (2) | 64 (128) | 1 (2) |
| clarithromycin | ≤0.12 (≤0.12) | ≤0.12 (≤0.12) | 8 (16) |
MTZ-R, metronidazole resistant strain. CLR-R, clarithromycin resistant strain.
Figure 3.
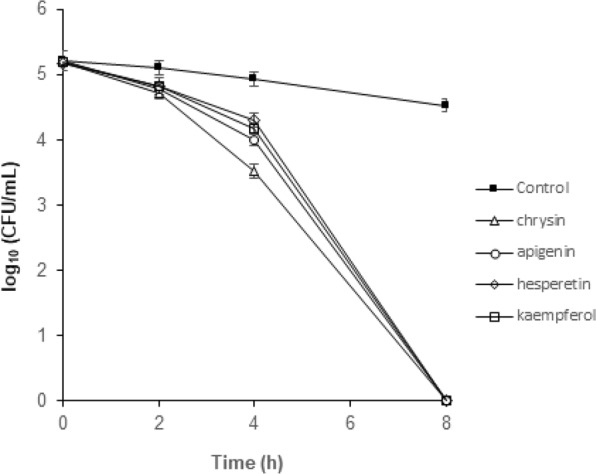
Time-kill kinetics of natural flavonoids chrysin, apigenin, hesperetin and kaempferol against H. pylori strain ATCC 700684. Bacterial counts were determined at time zero and after 2, 4, 8, and 24 hours of incubation with two times the MIC. Mixtures of bacteria with DMSO (vehicle) instead flavonoid were used as controls. Values are the averages of three independent determinations; vertical bars represent standard deviations. Please note that in some instances the error is smaller than the symbols used.
The in vitro interactions of the two flavonoids that exhibited the most potent bactericidal activities, chrysin and hesperetin (Table 2), with antibiotics commonly used as first-line H. pylori eradication therapies were analysed. As shown in Table 3, the combinations of flavonoids with antibiotics inhibited the growth of H. pylori at a lower concentration than when each single compound was assayed separately. In all cases, the combination of chrysin or hesperetin with clarithromycin or metronidazole showed a synergistic or additive antimicrobial effect against the two antibiotic-resistant strains used in the assays. Notably, chrysin reduced the MIC value of clarithromycin up to 8 times (FIC = 0.125) in the clarithromycin-resistant strain ATCC 700684, and up to 16 times the MIC value of metronidazole (FIC = 0.0625) in the metronidazole-resistant strain ATCC 43504 (Table 3).
Table 3.
Interaction of the natural flavonoids chrysin and hesperetin with antibiotics of first-line Helicobacter pylori eradication therapies in two H. pylori resistant strains.
| Strain | Combination tested | FICantibiotic | FICflavonoid | FICIa | Interactionb |
|---|---|---|---|---|---|
| ATCC 700684 (CLR-R) | CLR + chrysin | 0.125 | 0.25 | 0.375 | synergy |
| CLR + hesperetin | 0.5 | 0.5 | 1 | additive | |
| ATCC 43504 (MTZ-R) | MTZ + chrysin | 0.0625 | 0.0625 | 0.125 | synergy |
| MTZ + hesperetin | 0.25 | 0.25 | 0.5 | synergy |
aFractional inhibitory concentration (FIC) index could be calculated as: FICA (MICA in the presence of B/MICA alone) + FICB (MICB in the presence of A/MICB alone).
bAccording to the FICI value, the interaction between two compounds against a particular bacterial strain can be classified as: synergy (FICI ≤ 0.5), additive (FICI > 0.5 to ≤1), no interaction or neutral (FICI > 1 to ≤4), and antagonism (FICI > 4).
Bactericidal flavonoids interact with the C-terminal effector domain of HsrA
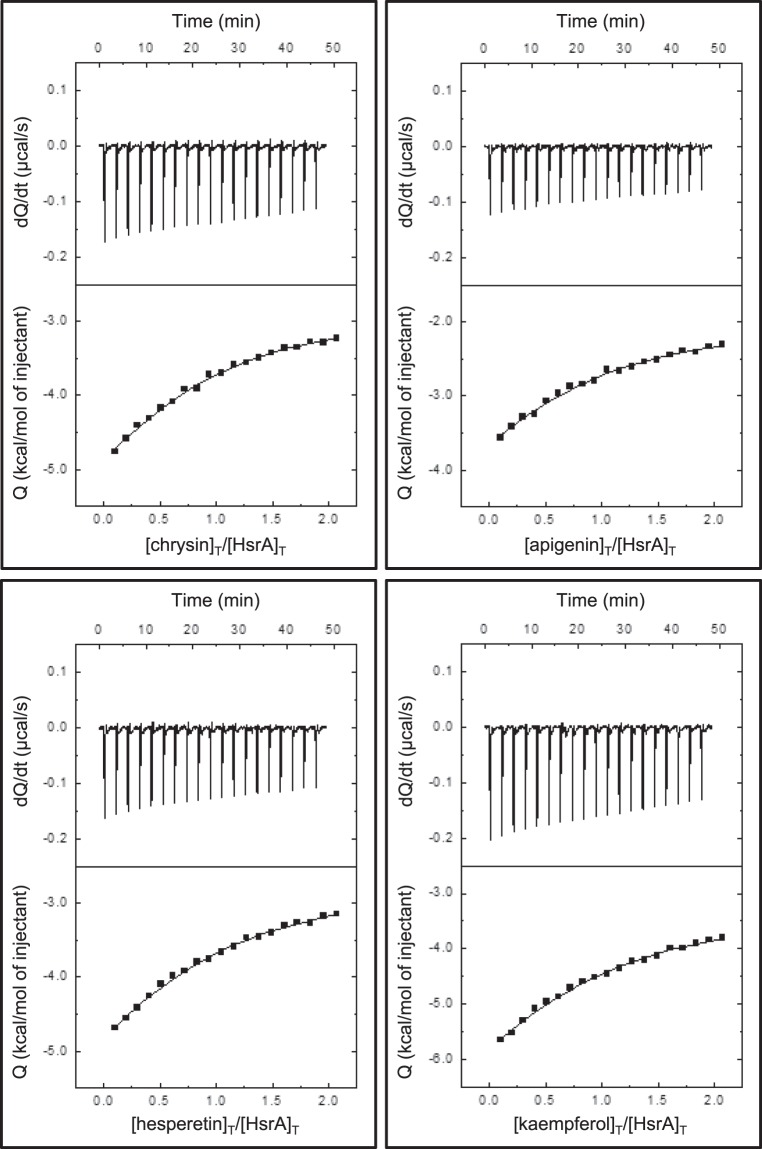
ITC and molecular docking analyses were carried out in order to analyze the binding affinity of bactericidal flavonoids (apigenin, chrysin, kaempferol and hesperetin) for the target protein as well as to understand the structural basis of this interaction. The ITC data indicated dissociation constants in the micromolar range in all cases, while complexes of a 1:1 stoichiometry were observed for all flavonoids (Table 4, Fig. 4). Thus, each HsrA monomer appeared to bind one molecule of flavonoid.
Table 4.
Analyses of the interaction between HsrA and its bactericidal inhibitors by isothermal titration calorimetry and molecular docking.
| Compound | ITCa | Molecular Dockingb |
|||
|---|---|---|---|---|---|
| n |
Kd (µM) |
ΔH (kcal/mol) |
ΔG (kcal/mol) |
Interacting residues | |
| chrysin | 0.77 | 17 | −4.1 | −6.5 | P181, K165, I184 |
| apigenin | 0.81 | 20 | −3.8 | −6.4 | P198, I135, K194, V144, G146, K145, L152 |
| hesperetin | 0.83 | 26 | −5.8 | −6.2 | P198, M195, I191, K194, P148, V144, K145, G146, L152 |
| kaempferol | 0.78 | 33 | −8.6 | −6.1 | Y137, K194, P148, G146 |
aAbsolute error in n is 0.06, relative error in Kd is 40%, absolute error in ΔH is 0.4 kcal/mol, absolute error in ΔG is 0.2 kcal/mol.
bAmino acid residues directly involved in forming the helix-turn-helix (HTH) DNA binding motif of HsrA are highlighted in bold fonts.
Figure 4.
Isothermal titration calorimetry analyses of the H. pylori HsrA response regulator interactions with the natural flavonoids chrysin, apigenin, hesperetin and kaempferol. In the figures, upper panels show the ITC thermograms while lower panels show the binding isotherms.
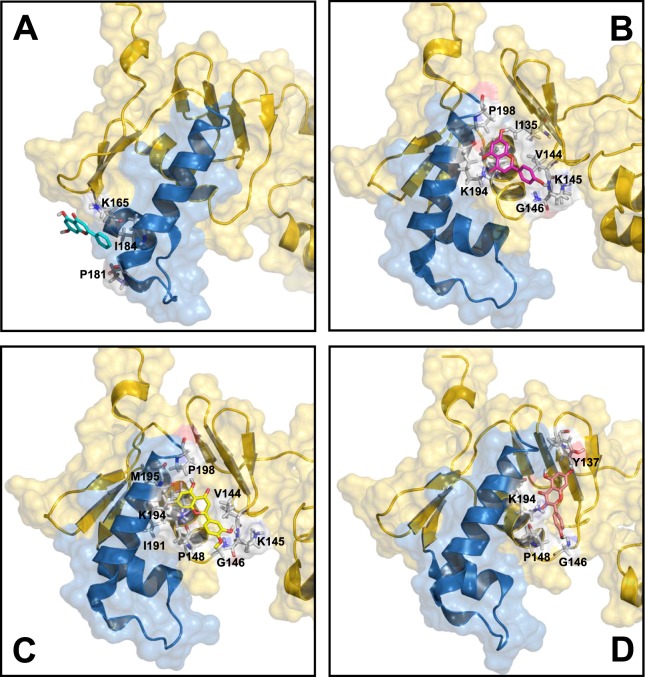
As shown in Fig. 5, flavonoid binding to the native conformation of HsrA occurs preferably at the C-terminal effector domain, as predicted by molecular docking analyses. Notably, the predicted binding site for chrysin differed from those occupied for the other three flavonoids. However, the best binding poses for all bactericidal flavonoids appeared to interact with amino acid residues specifically involved in forming the helix-turn-helix (HTH) DNA binding motif of the HsrA response regulator (Table 4)17.
Figure 5.
Local overviews of the best ranked docking poses of chrysin (A), apigenin (B), hesperetin (C), and kaempferol (D) interaction with HsrA. Ribbon model and transparent molecular surface showing the interacting residues of HsrA to each flavonoid. The helix-turn-helix (HTH) DNA binding motif of HsrA is highlighted in blue. Some interacting residues are indicated.
Discussion
The dramatic increase in bacterial resistance to antibiotics belonging to all pharmacological classes constitutes a major worldwide health concern and it must be a priority to the scientific community38. An important challenge in the current antibiotic resistance crisis is the identification of novel microbial targets, essential for in vivo growth or pathogenicity, whose inhibitors can overcome the currently circulating resistome of human pathogens. Among the plethora of genes indispensable for in vivo growth and virulence of bacterial pathogens, transcriptional regulators (TRs) stand out because of the potential multitargeting effect of anti-TR molecules on cellular virulence and physiology39.
In H. pylori, the essential response regulator HsrA appears as a promising target for drug development11–16. In the present study, we carried out an affinity-based HTS18,19 of the Prestwick Chemical Library for FDA-approved molecules that specifically bind to the HsrA response regulator and potentially inhibit its essential biological function. By using this screening methodology, seven natural flavonoids including apigenin, chrysin, hesperetin, kaempferol, luteolin, myricetin, and quercetin were identified as HsrA binders, since all of them significantly increased its thermal stability. All flavonoids identified as HsrA binders appreciably inhibited the DNA binding activity of HsrA in vitro according to EMSA; however, only four of them, apigenin, chrysin, kaempferol and hesperetin, exhibited potent bactericidal activities against three different strains of H. pylori, including strains resistant to metronidazole and clarithromycin. Notably, the flavonoids with two or more hydroxyl groups in ring B (luteolin, quercetin, myricetin) showed moderate to low bactericidal activities against H. pylori. Since more hydroxyl groups results in lower hydrophobicity40, the lowest bactericidal activity of myricetin despite its HsrA inhibitory effect could be the result of poor translocation across the cell membrane. Among all the flavonoids evaluated in this work, the flavone chrysin showed the most potent bactericidal activity and the most synergistic effect in combination with antibiotics of first-line H. pylori eradication therapies. Notably, chrysin strongly synergized with both clarithromycin and metronidazole, supporting its valuable potential as adjuvant in current combinatory therapies, especially in the treatment of refractory H. pylori infections or multidrug-resistance isolates41,42.
HsrA functions as a symmetric dimer in vivo with two well-defined functional domains in each monomer, an N-terminal regulatory domain (residues 1 to 119) and a C-terminal DNA-binding/effector domain (residues 120 to 223)17. In contrast to most response regulators, the two domains of HsrA seem to act independently, and mutations in the N-terminal regulatory domain did not appreciably affect DNA binding function17. Each monomer of HsrA contains eight α-helices and twelve β-strands, where two α-helices in the C-terminal effector domain (α7 and α8, comprising the residues 165 to 199) form the HTH DNA binding motif. In fact, single mutations at the amino acid residues involved in forming the HTH motif or its periphery noticeably inhibited or completely destroyed the DNA binding ability of the protein17. Molecular docking analyses suggested that the bactericidal flavonoids apigenin, chrysin, kaempferol and hesperetin bind to HsrA preferably in its C-terminal effector domain, interacting with several amino acids involved in both the structure of the HTH motif and the domain stabilization. According to these in silico predictions, the HsrA activity inhibition by flavonoids could be the result of direct blockage of key amino acid residues involved in protein-DNA interaction and/or conformational changes in the effector domain that destabilize the regulator binding to target promoters. In fact, one of the molecular mechanisms associated to the antibacterial action of flavonoids is the formation of complexes with proteins through different forces such as hydrogen bonding, hydrophobic interactions and covalent bonds, thereby inactivating microbial adhesins, enzymes, transport proteins, and other cell targets43.
Flavonoids comprise a wide class of compounds with variable phenolic structures, mostly found in plants44. Both medicinal plants and flavonoid rich extracts have been used for centuries to treat human pathologies45,46, including H. pylori infection and related diseases47–50. A large number of studies have demonstrated the strong antibacterial activity of natural, semisynthetic51 and synthetic flavonoids52; however, their mechanisms of action remain poorly characterized. In H. pylori, flavonoids have been found to inhibit enzymatic activities such as urease53, DNA gyrase48, dihydrofolate reductase48, N-acetyltransferase54, D-alanine:D-alanine ligase55, and β-hydroxyacyl-acyl carrier protein dehydratase56. Quercetin inhibited vacuolation and caspase-3 activation in HeLa cells induced by the vacuolating cytotoxin A, protecting from gastric inflammation and apoptosis associated with H. pylori infection57,58. Kaempferol decreased transcription of type IV secretion system (T4SS) components involved in CagA injection and secretion system subunit protein A (SecA) of type V secretion system (T5SS) involved in VacA secretion, thereby producing an anti-inflammatory effect by suppressing the translocation of CagA and VacA proteins59. Apigenin decreased atrophic gastritis and gastric cancer progression in Mongolian gerbils60, while hesperetin appears to damage the cell membrane triggering bacterial lysis51. Notably, the results of different studies conducted to decipher the mechanisms of action of flavonoids as antimicrobial agents suggest that a single flavonoid molecule could have multiple targets in the cell, triggering bacterial death or virulence decrease by inhibiting and/or affecting the activities of several biomolecules at the same time. Thus, apigenin, one of the HsrA inhibitors and potent anti-H. pylori bactericidal flavonoid identified in this work, also functions as inhibitor of the enzymes D-alanine:D-alanine ligase55 and β-hydroxyacyl-acyl carrier protein dehydratase56. Similarly, the lipophilic flavonoid hesperetin inhibited HsrA activity but also affected the integrity of H. pylori cell membrane51. In the last years, flavonoids have gained attention not only for its well-documented antimicrobial action, but also because some of these natural compounds manifest ability to reverse the antibiotic resistance in certain pathogens and enhance the action of some current antibiotic drugs61–63. Hence, combinatory therapies that take advantage of the clear evidences of synergism between flavonoids and several conventional antibiotics is now considered a promising strategy in the clinical therapy of infections with antibiotic-resistant microorganisms such as methicillin-resistance Staphylococcus aureus (MRSA)64.
Overall, the results of the present study validate the use of the response regulator HsrA as a novel and effective therapeutic target in H. pylori infection and further support the great potential of natural flavonoids as pharmaceutical candidates for novel antibacterial or antivirulence chemotherapies. In addition, our results provide molecular evidence of a novel antibacterial mechanism of some natural flavonoids against H. pylori infection and demonstrate that this mechanism of action efficiently overcome the strategies evolved by this pathogen to evade the effect of first-line eradication therapies such as metronidazole and clarithromycin.
Acknowledgements
Authors appreciate the help of Dr. José Antonio Aínsa (University of Zaragoza) in the interpretation of checkerboard assays. This work has been supported by Carlos III Health Institute [PI11/02578], the Spanish Ministry of Economy, Industry and Competitiveness [BFU2016-78232-P], the Government of Aragon [E45_17R], Campus Iberus [CI-2017/001-3] and FEDER funds from the European Union. Financial support by the Spanish Ministry of Economy, Industry and Competitiveness to A.G. through the “Juan de la Cierva” programmes (FJCI-2014-20704, IJCI-2016-27419) is gratefully acknowledged. S.S. is recipient of a predoctoral fellowship from the Government of Aragon, Spain.
Author Contributions
A.G. designed the study, coordinated the execution of experiments and wrote the manuscript. A.G., S.S., A.V.-C. and V.E.A. performed the experiments. M.F., J.S. and A.L. provided reagents and materials. A.G., S.S., A.V.-C., V.E.A., M.F., J.S. and A.L. analyzed data, discussed the results and edited the manuscript.
Data Availability
Materials, data and protocols are presented within the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hooi JKY, et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang JC, Lu CW, Lin CJ. Treatment of Helicobacter pylori infection: current status and future concepts. World J Gastroenterol. 2014;20:5283–5293. doi: 10.3748/wjg.v20.i18.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safavi M, Sabourian R, Foroumadi A. Treatment of Helicobacter pylori infection: Current and future insights. World J Clin Cases. 2016;4:5–19. doi: 10.12998/wjcc.v4.i1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vianna JS, Ramis IB, Ramos DF, Von Groll A, Silva PE. Drug resistance in Helicobacter pylori. Arq. Gastroenterol. 2016;53:215–223. doi: 10.1590/S0004-28032016000400002. [DOI] [PubMed] [Google Scholar]
- 7.Boyanova L, Evstatiev I, Yordanov D, Markovska R, Mitov I. Three unsuccessful treatments of Helicobacter pylori infection by a highly virulent strain with quadruple antibiotic resistance. Folia Microbiol. (Praha) 2016;61:307–310. doi: 10.1007/s12223-015-0439-2. [DOI] [PubMed] [Google Scholar]
- 8.Fallone CA, et al. The Toronto consensus for the treatment of Helicobacter pylori infection in adults. Gastroenterology. 2016;151:51–69 e14. doi: 10.1053/j.gastro.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Tacconelli E, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 10.Beier D, Frank R. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 2000;182:2068–2076. doi: 10.1128/JB.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDaniel TK, Dewalt KC, Salama NR, Falkow S. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter. 2001;6:15–23. doi: 10.1046/j.1523-5378.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- 12.Muller S, Pflock M, Schar J, Kennard S, Beier D. Regulation of expression of atypical orphan response regulators of Helicobacter pylori. Microbiol. Res. 2007;162:1–14. doi: 10.1016/j.micres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Olekhnovich IN, et al. Mutations to essential orphan response regulator HP1043 of Helicobacter pylori result in growth-stage regulatory defects. Infect. Immun. 2013;81:1439–1449. doi: 10.1128/IAI.01193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olekhnovich IN, Vitko S, Valliere M, Hoffman PS. Response to metronidazole and oxidative stress is mediated through homeostatic regulator HsrA (HP1043) in Helicobacter pylori. J. Bacteriol. 2014;196:729–739. doi: 10.1128/JB.01047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delany I, Spohn G, Rappuoli R, Scarlato V. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J. Bacteriol. 2002;184:4800–4810. doi: 10.1128/JB.184.17.4800-4810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelliciari S, et al. Insight into the essential role of the Helicobacter pylori HP1043 orphan response regulator: genome-wide identification and characterization of the DNA-binding sites. Sci. Rep. 2017;7:41063. doi: 10.1038/srep41063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong E, et al. Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J. Biol. Chem. 2007;282:20667–20675. doi: 10.1074/jbc.M609104200. [DOI] [PubMed] [Google Scholar]
- 18.Cremades N, et al. Discovery of specific flavodoxin inhibitors as potential therapeutic agents against Helicobacter pylori infection. ACS Chem Biol. 2009;4:928–938. doi: 10.1021/cb900166q. [DOI] [PubMed] [Google Scholar]
- 19.Velázquez-Campoy A, Sancho J, Abián O, Vega S. Biophysical screening for identifying pharmacological chaperones and inhibitors against conformational and infectious diseases. Curr Drug Targets. 2016;17:1492–1505. doi: 10.2174/1389450117666160201110449. [DOI] [PubMed] [Google Scholar]
- 20.Huynh, K. & Partch, C. L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci79, 28.29.21–28.29. 14, 10.1002/0471140864.ps2809s79 (2015). [DOI] [PMC free article] [PubMed]
- 21.Kennedy AJ, et al. Synthesis and antimicrobial evaluation of amixicile-based inhibitors of the pyruvate-ferredoxin oxidoreductases of anaerobic bacteria and Epsilonproteobacteria. Antimicrob. Agents Chemother. 2016;60:3980–3987. doi: 10.1128/AAC.00670-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piccolomini R, Di Bonaventura G, Catamo G, Carbone F, Neri M. Comparative evaluation of the E test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J. Clin. Microbiol. 1997;35:1842–1846. doi: 10.1128/jcm.35.7.1842-1846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sisto F, Scaltrito MM, Russello G, Bonomi A, Dubini F. Antimicrobial susceptibility testing of Helicobacter pylori determined by microdilution method using a new medium. Curr. Microbiol. 2009;58:559–563. doi: 10.1007/s00284-009-9368-0. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharyya, G. K. & Johnson, R. A. Statistical Concepts and Methods. 1 edn, (John Wiley & Sons, 1977).
- 25.Aguilar-Perez, C. et al. Synergy between circular bacteriocin AS-48 and ethambutol against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 62, 10.1128/AAC.00359-18 (2018). [DOI] [PMC free article] [PubMed]
- 26.Krzyżek Paweł, Franiczek Roman, Krzyżanowska Barbara, Łaczmański Łukasz, Migdał Paweł, Gościniak Grażyna. In Vitro Activity of 3-Bromopyruvate, an Anticancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Cancers. 2019;11(2):229. doi: 10.3390/cancers11020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HK, et al. Growth inhibitory, bactericidal, and morphostructural effects of dehydrocostus lactone from Magnolia sieboldii leaves on antibiotic-susceptible and -resistant strains of Helicobacter pylori. PLoS One. 2014;9:e95530. doi: 10.1371/journal.pone.0095530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abu-Qatouseh Luay, Abu-Sini Mohammad, Mayyas Amal, Al-Hiari Yusuf, Darwish Rula, Aburjai Talal. Synthesis of New Nitrofluoroquinolone Derivatives with Novel Anti-Microbial Properties against Metronidazole Resistant H. pylori. Molecules. 2017;22(1):71. doi: 10.3390/molecules22010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nostro A, et al. Effects of combining extracts (from propolis or Zingiber officinale) with clarithromycin on Helicobacter pylori. Phytother. Res. 2006;20:187–190. doi: 10.1002/ptr.1830. [DOI] [PubMed] [Google Scholar]
- 30.Cheesman MJ, Ilanko A, Blonk B, Cock IE. Developing new antimicrobial therapies: are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn Rev. 2017;11:57–72. doi: 10.4103/phrev.phrev_21_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velázquez-Campoy, A., Ohtaka, H., Nezami, A., Muzammil, S. & Freire, E. Isothermal titration calorimetry. Curr Protoc Cell Biol Chapter 17, Unit 17 18, 10.1002/0471143030.cb1708s23 (2004). [DOI] [PubMed]
- 32.Velázquez-Campoy A, Leavitt SA, Freire E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 2015;1278:183–204. doi: 10.1007/978-1-4939-2425-7_11. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 35.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogata SK, Gales AC, Kawakami E. Antimicrobial susceptibility testing for Helicobacter pylori isolates from Brazilian children and adolescents: comparing agar dilution, E-test, and disk diffusion. Braz J Microbiol. 2014;45:1439–1448. doi: 10.1590/S1517-83822014000400039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo VG, Fallone CA, De Souza E, Lavallee J, Barkun AN. In-vitro susceptibility of Helicobacter pylori to ampicillin, clarithromycin, metronidazole and omeprazole. J. Antimicrob. Chemother. 1997;40:881–883. doi: 10.1093/jac/40.6.881. [DOI] [PubMed] [Google Scholar]
- 38.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González A, Fillat MF, Lanas A. Transcriptional regulators: valuable targets for novel antibacterial strategies. Future Med Chem. 2018;10:541–560. doi: 10.4155/fmc-2017-0181. [DOI] [PubMed] [Google Scholar]
- 40.Wang T, Li Q, Bi K. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J Pharm Sci. 2018;13:12–23. doi: 10.1016/j.ajps.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayala G, Escobedo-Hinojosa WI, de la Cruz-Herrera CF, Romero I. Exploring alternative treatments for Helicobacter pylori infection. World J Gastroenterol. 2014;20:1450–1469. doi: 10.3748/wjg.v20.i6.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezaeimanesh N, Farzi N, Pirmanesh S, Emami S, Yadegar A. Management of multi-drug resistant Helicobacter pylori infection by supplementary, complementary and alternative medicine; a review. Gastroenterol Hepatol Bed Bench. 2017;10:S8–S14. [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. Scientific World Journal. 2013;2013:162750. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci. 2016;5:e47. doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cushnie TP, Lamb AJ. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cushnie TP, Lamb AJ. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents. 2011;38:99–107. doi: 10.1016/j.ijantimicag.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 47.Wang YC. Medicinal plant activity on Helicobacter pylori related diseases. World J Gastroenterol. 2014;20:10368–10382. doi: 10.3748/wjg.v20.i30.10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asha MK, et al. In vitro anti-Helicobacter pylori activity of a flavonoid rich extract of Glycyrrhiza glabra and its probable mechanisms of action. J. Ethnopharmacol. 2013;145:581–586. doi: 10.1016/j.jep.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 49.Zhang S, et al. Flavonoid glycosides of Polygonum capitatum protect against inflammation associated with Helicobacter pylori infection. PLoS One. 2015;10:e0126584. doi: 10.1371/journal.pone.0126584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ustun O, Ozcelik B, Akyon Y, Abbasoglu U, Yesilada E. Flavonoids with anti-Helicobacter pylori activity from Cistus laurifolius leaves. J. Ethnopharmacol. 2006;108:457–461. doi: 10.1016/j.jep.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Moon SH, et al. Antimicrobial effect of 7-O-butylnaringenin, a novel flavonoid, and various natural flavonoids against Helicobacter pylori strains. Int J Environ Res Public Health. 2013;10:5459–5469. doi: 10.3390/ijerph10115459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babii C, et al. A novel synthetic flavonoid with potent antibacterial properties: In vitro activity and proposed mode of action. PLoS One. 2018;13:e0194898. doi: 10.1371/journal.pone.0194898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu XD, et al. Biological evaluation and molecular docking of baicalin and scutellarin as Helicobacter pylori urease inhibitors. J. Ethnopharmacol. 2015;162:69–78. doi: 10.1016/j.jep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 54.Chung JG, et al. Inhibitory actions of luteolin on the growth and arylamine N-acetyltransferase activity in strains of Helicobacter pylori from ulcer patients. Toxicol In Vitro. 2001;15:191–198. doi: 10.1016/S0887-2333(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 55.Wu D, et al. D-Alanine:D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents. 2008;32:421–426. doi: 10.1016/j.ijantimicag.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, et al. Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay. Protein Sci. 2008;17:1971–1978. doi: 10.1110/ps.036186.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin JE, Kim JM, Bae EA, Hyun YJ, Kim DH. In vitro inhibitory effect of flavonoids on growth, infection and vacuolation of Helicobacter pylori. Planta Med. 2005;71:197–201. doi: 10.1055/s-2005-837816. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Shu, Huang Jian, Xie Xiaoqin, He Yun, Mo Fei, Luo Zhaoxun. Quercetin from Polygonum capitatum Protects against Gastric Inflammation and Apoptosis Associated with Helicobacter pylori Infection by Affecting the Levels of p38MAPK, BCL-2 and BAX. Molecules. 2017;22(5):744. doi: 10.3390/molecules22050744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeon MJ, et al. Anti-inflammatory effects of Kaempferol on Helicobacter pylori-induced inflammation. Biosci. Biotechnol. Biochem. 2019;83:166–173. doi: 10.1080/09168451.2018.1528140. [DOI] [PubMed] [Google Scholar]
- 60.Kuo CH, et al. Apigenin has anti-atrophic gastritis and anti-gastric cancer progression effects in Helicobacter pylori-infected Mongolian gerbils. J. Ethnopharmacol. 2014;151:1031–1039. doi: 10.1016/j.jep.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 61.Górniak I, Bartoszewski R, Króliczewski J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem Rev. 2019;18:241–272. doi: 10.1007/s11101-018-9591-z. [DOI] [Google Scholar]
- 62.Miklasińska-Majdanik Maria, Kępa Małgorzata, Wojtyczka Robert, Idzik Danuta, Wąsik Tomasz. Phenolic Compounds Diminish Antibiotic Resistance of Staphylococcus Aureus Clinical Strains. International Journal of Environmental Research and Public Health. 2018;15(10):2321. doi: 10.3390/ijerph15102321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanhueza L, et al. Synergistic interactions between phenolic compounds identified in grape pomace extract with antibiotics of different classes against Staphylococcus aureus and Escherichia coli. PLoS One. 2017;12:e0172273. doi: 10.1371/journal.pone.0172273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abreu AC, et al. Looking to nature for a new concept in antimicrobial treatments: isoflavonoids from Cytisus striatus as antibiotic adjuvants against MRSA. Sci Rep. 2017;7:3777. doi: 10.1038/s41598-017-03716-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Materials, data and protocols are presented within the manuscript.