Abstract
Background
The spectrum of classical and non-classical HLA genes related to the risk of multiple sclerosis (MS) and neuromyelitis optica spectrum disorder (NMOSD) in the Japanese population has not been studied in detail. We conducted a case-control analysis of classical and non-classical HLA genes.
Methods
We used next-generation sequencing (NGS)-based HLA genotyping methods for mapping risk for 45 MS patients, 31 NMOSD patients, and 429 healthy controls. We evaluated the association of the HLA variants with the risk of MS and NMOSD using logistic regression analysis and Fisher’s exact test.
Results
We confirmed that HLA-DRB1*15:01 showed the strongest association with MS (P = 2.1 × 10−5; odds ratio [OR] = 3.44, 95% confidence interval [95% CI] = 1.95–6.07). Stepwise conditional analysis identified HLA-DRB1*04:05, HLA-B*39:01, and HLA-B*15:01 as being associated with independent MS susceptibility (PConditional < 8.3 × 10−4). With respect to amino acid polymorphisms in HLA genes, we found that phenylalanine at HLA-DQβ1 position 9 had the strongest effect on MS susceptibility (P = 3.7 × 10−8, OR = 3.48, 95% CI = 2.23–5.43). MS risk at HLA-DQβ1 Phe9 was independent of HLA-DRB1*15:01 (PConditional = 1.5 × 10−5, OR = 2.91, 95% CI = 1.79–4.72), while HLA-DRB1*15:01 was just significant when conditioned on HLA-DQβ1 Phe9 (PConditional = 0.037). Regarding a case-control analysis for NMOSD, HLA-DQA1*05:03 had a significant association with NMOSD (P = 1.5 × 10−4, OR = 6.96, 95% CI = 2.55–19.0).
Conclusions
We identified HLA variants associated with the risk of MS and NMOSD. Our study contributes to the understanding of the genetic architecture of MS and NMOSD in the Japanese population.
Electronic supplementary material
The online version of this article (10.1186/s12974-019-1551-z) contains supplementary material, which is available to authorized users.
Keywords: Multiple sclerosis, Neuromyelitis optica spectrum disorder, HLA, Next-generation sequencing
Background
The major histocompatibility complex (MHC) region at 6p21.3 is associated with a wide range of complex human diseases and quantitative traits [1–3]. In autoimmune diseases, such as rheumatoid arthritis, psoriasis, and type 1 diabetes, fine-mapping studies have identified strong genetic risk variants in the MHC region [4–6].
Multiple sclerosis (MS) is an immune-mediated demyelinating disease of the central nervous system, which affects approximately 2.3 million people worldwide [7]. Genome-wide association studies (GWAS) have reported that the MHC region is the most significant locus associated with MS [8–12]. In Europeans, previous studies have reported a relationship between classical HLA genes and MS, and found that HLA-DRB1*15:01 had the strongest effect on MS susceptibility [13]. In 2013, Patsopoulos, et al. reported that both class II HLA genes and class I HLA genes independently confer susceptibility to MS [14].
These researchers used an HLA imputation method, which computationally imputed HLA variants from single nucleotide polymorphisms (SNP) genotype data obtained from GWAS. They analyzed MS associations with eight classical HLA genes and found HLA-A*02:01 had an independent protective effect on MS. In 2018, high-resolution HLA analysis of MS by using next-generation sequencing (NGS) was conducted in Europeans [15]. The researchers found that specific class I HLA polymorphisms, such as HLA-A*02:01, HLA-C*03:04, and HLA-B*40:01 were protective for MS, independently from HLA-DRB1*15:01. In a Japanese population, Yoshimura et al. reported in 2012 that HLA-DRB1*04:05 and HLA-DPB1*03:01 were significantly associated with MS susceptibility [16]. The authors also reported that MS patients with HLA-DRB1*04:05 have a younger onset of disease. In 2016, Nakamura et al. conducted HLA analysis and found that HLA-DRB1*04:05 and HLA-DRB1*15:01 were risk factors for MS in Japanese individuals [17]. They also found that living at higher latitude and HLA-DRB1*04:05 independently affect the severity of MS.
Neuromyelitis optica spectrum disorder (NMOSD) is an immune-mediated disorder of the central nervous system that mainly affects the optic nerve and spinal cord [18]. The MHC region also contributes to the genetic architecture of NMOSD like MS [18, 19]. In Europeans, HLA-DRB1*03:01 has been reported to be significantly associated with NMOSD from a whole-genome sequencing study [19]. In the Japanese population, HLA-DRB1*16:02 and HLA-DPB1*05:01 have been reported to be positively associated with NMOSD susceptibility, and HLA-DRB1*09:01 was negatively associated with NMOSD susceptibility [20]. These Japanese studies used sequence-specific oligonucleotide hybridization [21], and only two classical HLA genes (HLA-DRB1, HLA-DPB1) were genotyped [16, 20, 21]. Thus, the classical and non-classical HLA genes affecting MS and NMOSD risk in the Japanese population have not been studied in detail. In this research, we explored the MHC region comprehensively in the Japanese population. We utilized NGS-based HLA genotyping for fine-mapping of risk [22–24] for 45 MS patients, 31 NMOSD patients, and 429 controls.
Methods
Sample collection
We enrolled 45 MS patients and 31 NMOSD patients. MS and NMOSD cases were collected from three medical institutes in Japan (Osaka General Medical Center, Kindai University Hospital, and Osaka University Hospital) and assessed in a retrospective manner. All subjects were of Japanese origin and provided written informed consent that was approved by the ethics committee of each hospital. All participants lived in the Kinki region which is located in the central part of Honshu Island, Japan. The Kinki region is located at latitude 35° north and longitude 135° east. MS was diagnosed by the McDonald 2010 criteria for the diagnosis of MS [25]. NMOSD was diagnosed using the international consensus diagnostic tests for NMOSD [26]. We collected clinical data of patients including sex, age, disease onset, disease duration, MS subtype, Kurtzke’s Expanded Disability Status Scale (EDSS), and anti-AQP4 antibody (Table 1). Case genomic DNA was obtained from peripheral blood. We extracted DNA using QIAGEN’s QIAampBlood Midi (Mini) Kit. This is one of the integrated DNA extraction manuals recommended by the HLA typing Quality Workshop of the Japanese Society for Histocompatibility and Immunogenetics. We used HLA genotyping data of 429 healthy individuals which were used in a previous HLA study from the Japan Biological Informatics Consortium (JBIC), as described elsewhere [6, 27]. Control genomic DNA was obtained from Epstein-Barr virus-transformed B-lymphoblast cell lines of unrelated individuals. Control genomic DNA was extracted using phenol-chloroform extraction method. All samples were checked to ensure the quality was sufficient for DNA sequencing. (OD260nm/OD280nm = 1.8 to 2.0). We received DNA samples from JBIC.
Table 1.
Clinical features of multiple sclerosis and neuromyelitis optica spectrum disorder patients
| MS (n = 45) | NMOSD (n = 31) | |
|---|---|---|
| Sex (male/female) | 10/35 | 7/24 |
| Age (years) | 43.6 (20–65) | 52.7 (17–82) |
| Age at onset (years) | 34.3 (5–58) | 47.4 (15–81) |
| Disease duration (years) | 9.3 (1–38) | 5.3 (0–18) |
| EDSS | 3.1 (0–8) | 4.6 (0–9) |
| IgG index | 0.73 (n = 34) | 0.57 (n = 13) |
| Oligo clonal band | 21/38 | – |
| Anti-AQP4 antibody | – | 29/31 |
This study was approved by the ethical committee of Osaka University Graduate School of Medicine.
NGS-based HLA genotyping of classical and non-classical HLA genes
We conducted HLA genotyping (4-digit) of the 16 HLA genes in the subjects, 9 of which were classical HLA genes (class I: HLA-A, HLA-B, and HLA-C, class II: HLA-DRA, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1), seven were non-classical HLA genes (HLA-E, HLA-F, HLA-G, HLA-DOA, HLA-DOB, HLA-DMA, and HLA-DMB). HLA gene sequencing was carried out using sequence capture technology based upon hybridization between DNA of an adapter-ligated library (KAPA Hyper Prep Kit, Roche, CA, USA) and a biotinylated DNA probe (SepCap EZ choice kit, Roche, CA, USA), designed based upon target sequences of the genes in the MHC region [24]. Paired-end sequence reads were obtained using a MiSeq DNA sequencer (Illumina, the USA). Typing of 4-digit HLA alleles was conducted using Omixon Target Software (Omixon, Hungary) using the IPD-IMGT/HLA Database.
In order to supplement the HLA allele information, which was specific to the Japanese population, and was not correctly implemented in the Omixon, we updated the HLA typing results based on those obtained from a sequence-based typing method. We downloaded the GRCh37 reference genome from Ensembl, a genome browser for vertebrate genomes. GRCh37 reference genome was created by the Genome Reference Consortium (GRC) in 2009. The sequence reads were aligned to the GRCh37 of the MHC region using BWA (version 0.7.15) [28]. We obtained HLA allele sequences from the IPD and IMGT/HLA databases [29]. We used the GATK Unified Genotyper and Haplotypecaller for variant calling [30]. Details of the HLA genotyping protocol is described in our previous study [31, 32].
The statistical framework for analysis
We evaluated the association of the HLA variants with the risk of MS and NMOSD using a logistic regression model, which assumed additive effects of allele dosages on a log-odds scale using the R statistical software (version 3.4.1) when the number of cases was more than five. Otherwise, we used Fisher’s exact test. We analyzed HLA variants that were more than 0.1% both in case and control samples. We excluded HLA amino acid polymorphisms that were monomorphic in cases or controls. We assessed the significance level of the association study by applying a Bonferroni correction according to the number of the assessed variants (adjusted p < 0.05). We conducted conditional association analysis of the HLA variants by including the top-associated HLA variants as covariates. An omnibus p value was calculated for each HLA amino acid position which had more than two amino acid residues based on a log-likelihood ratio test. We assessed the significance of the improvement in the fit by calculating the deviance for the positions with n residues, which followed a χ2 distribution with n-1 degrees of freedom.
HLA amino acid 3D-structure models
We modeled the 3D-structure of HLA molecules and amino acids using the UCSF Chimera software, which is widely used for the visualization of molecular structures of proteins and three-dimensional ribbon models [33]. We downloaded the protein structure of HLA-DQ molecules from the Protein Data Bank in Europe entires 1jk8.
Results
NGS-based genotyping of the HLA genes in the Japanese MS and NMOSD patients and controls
For the 45 MS patients, 31 NMOSD patients, and 429 healthy controls, we conducted NGS-based genotyping of 16 HLA genes with 4-digit-level allele resolution (Additional file 1: Table S1, Table S2). Among the 16 sequenced HLA genes, 9 were classical HLA genes (class I: HLA-A, HLA-B, and HLA-C, class II: HLA-DRA, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1) and 7 were non-classical HLA genes (HLA-E, HLA-F, HLA-G, HLA-DOA, HLA-DOB, HLA-DMA, and HLA-DMB). We used a target capture technique and sequencing with relatively long read lengths to get high accuracy, as described in our previous study [27].
Associations of HLA alleles with MS susceptibility
We evaluated the associations of the 4-digit HLA alleles with MS risk and found the most significant association to be with HLA-DRB1*15:01 (P = 2.1 × 10−5, odds ratio [OR] = 3.44, 95% confidence interval [95% CI] = 1.95–6.07; Table 2). HLA-DQB1*06:02, which was in linkage disequilibrium with HLA-DRB1*15:01 (r2 = 0.94 in the controls), was also significantly associated with MS (P = 3.0 × 10−5, OR = 3.45, 95% CI = 1.93–6.17; Table 2). We found that HLA-B*15:01 was significantly associated with MS (P = 2.2 × 10−4, OR = 2.95, 95% CI = 1.66–5.24; Table 2). Previous studies in Japanese and European individuals reported the strongest association of HLA-DRB1*15:01 with MS [13, 17], and our study replicated these results.
Table 2.
HLA association analysis of multiple sclerosis in the Japanese population
| Variant | Frequency | Nominal analysis | |||
|---|---|---|---|---|---|
| HLA gene | MS (n = 45) | Control (n = 429) | OR (95% CI) | P | Fisher |
| HLA-B*15:01 | 0.200 | 0.078 | 2.95 (1.66–5.24) | 2.2 × 10−4 | |
| HLA-B*39:01 | 0.122 | 0.043 | 3.09 (1.52–6.29) | 0.0019 | |
| HLA-B*52:01 | 0.044 | 0.127 | 0.32 (0.08–0.88) | 0.017 | * |
| HLA-C*07:02 | 0.233 | 0.145 | 1.80 (1.07–3.04) | 0.028 | |
| HLA-C*12:02 | 0.044 | 0.127 | 0.32 (0.08–0.88) | 0.017 | * |
| HLA-DMB*01:07 | 0.044 | 0.008 | 5.64 (1.19–22.7) | 0.015 | * |
| HLA-DOA*01:01 | 0.978 | 0.997 | 0.15 (0.03–0.94) | 0.042 | |
| HLA-DPB1*05:01 | 0.522 | 0.356 | 1.98 (1.28–3.07) | 0.0021 | |
| HLA-DQA1*01:02 | 0.267 | 0.156 | 1.96 (1.19–3.25) | 0.0084 | |
| HLA-DQA1*03:03 | 0.222 | 0.141 | 1.74 (1.02–2.96) | 0.042 | |
| HLA-DQB1*03:01 | 0.033 | 0.104 | 0.30 (0.06–0.93) | 0.037 | * |
| HLA-DQB1*04:01 | 0.222 | 0.100 | 2.56 (1.49–4.42) | 7.0 × 10−4 | |
| HLA-DQB1*06:02 | 0.200 | 0.068 | 3.45 (1.93–6.17) | 3.0 × 10−5 | |
| HLA-DRA*01:01 | 0.733 | 0.579 | 2.00 (1.23–3.25) | 0.0053 | |
| HLA-DRA*01:02 | 0.267 | 0.421 | 0.50 (0.31–0.81) | 0.0053 | |
| HLA-DRB1*04:05 | 0.233 | 0.120 | 2.23 (1.31–3.79) | 0.0030 | |
| HLA-DRB1*15:01 | 0.211 | 0.072 | 3.44 (1.95–6.07) | 2.1 × 10−5 | |
| HLA-DRB1*15:02 | 0.033 | 0.120 | 0.25 (0.05–0.79) | 0.012 | * |
*Fisher’s exact test was used for association analysis, otherwise logistic regression was used
We then conducted stepwise conditional analysis with respect to the top-associated HLA alleles. Conditional analysis of HLA-DRB1*15:01 revealed an independent association with HLA-DRB1*04:05 (P = 2.2 × 10−4, OR = 2.83, 95% CI = 1.63–4.90; Additional file 1: Table S1). Subsequent conditional analysis regarding HLA-DRB1*15:01 and HLA-DRB1*04:05 revealed an independent association with HLA-B*39:01 (P = 8.3 × 10−4, OR = 3.51, 95% CI = 1.68–7.34; Additional file 1: Table S1). Conditional analysis of HLA-DRB1*15:01, HLA-DRB1*04:05, and HLA-B*39:01 revealed an independent association with HLA-B*15:01 (P = 2.3 × 10−4, OR = 3.10, 95% CI = 1.70–5.65; Additional file 1: Table S1). After conditioning HLA-DRB1*15:01, HLA-DRB1*04:05, HLA-B*39:01, and HLA-B*15:01, no significant association locus was observed (Additional file 1: Table S1).
We then conducted a multivariate regression analysis incorporating the four associated HLA alleles (HLA-DRB1*15:01, HLA-DRB1*04:05, HLA-B*39:01, and HLA-B*15:01) and found that all the HLA alleles were independently associated with MS (HLA-DRB1*15:01 for P = 5.7 × 10−6 and OR = 4.06, HLA-DRB1*04:05 for P = 1.2 × 10−4 and OR = 3.02, and HLA-B*39:01 for P = 1.9 × 10−4 and OR = 4.13, and HLA-B*15:01 for P = 2.3 × 10−4 and OR = 3.10; Table 3). Multivariate analysis confirmed that the HLA-B alleles were associated with MS susceptibility independently of the HLA-DRB1 alleles. As reported in a previous European study [14], we identified independent contributions of the class I HLA alleles to MS risk in Japanese subjects.
Table 3.
HLA conditional analysis of multiple sclerosis in the Japanese population
| Variant | Frequency | Conditional association | ||
|---|---|---|---|---|
| DRB1*15:01, DRB1*04:05, B*39:01, and B*15:01 | ||||
| HLA gene | MS (n = 45) | Control (n = 429) | OR (95% CI) | P |
| HLA-DRB1*15:01 | 0.211 | 0.072 | 4.06 (2.22–7.45) | 5.7 × 10−6 |
| HLA-DRB1*04:05 | 0.233 | 0.120 | 3.02 (1.72–5.31) | 1.2 × 10−4 |
| HLA-B*39:01 | 0.122 | 0.043 | 4.13 (1.96–8.71) | 1.9 × 10− 4 |
| HLA-B*15:01 | 0.200 | 0.078 | 3.10 (1.70–5.65) | 2.3 × 10−4 |
HLA alleles with MS susceptibility and MS clinical phenotype
We identified four HLA alleles associated with MS susceptibility. We then focused on whether these HLA alleles (HLA-DRB1*15:01, HLA-DQB1*06:02, HLA-B*15:01, and HLA-B*39:01) influence the clinical course of MS. We checked the clinical feature of HLA alleles associated with MS in our research. We focused on the onset of age and EDSS score, that is widely used for disease severity and measured in all participants in this study. The results showed that no HLA alleles have a significant association with EDSS score (P > 0.09; Table 4A) and the onset of age (P > 0.14; Table 4B).
Table 4.
HLA association analysis with the clinical course of MS
| Number of patients | EDSS | P (Student’s t) | |
| A | |||
| HLA-DRB1*15:01 positive | 18 | 3.08 | 0.990 |
| HLA-DRB1*15:01 negative | 27 | 3.09 | |
| HLA-DQB1*06:02 positive | 17 | 3.41 | 0.472 |
| HLA-DQB1*06:02 negative | 28 | 2.89 | |
| HLA-B*15:01 positive | 15 | 4.20 | 0.098 |
| HLA-B*15:01 negative | 30 | 2.53 | |
| HLA-B*39:01 positive | 11 | 3.50 | 0.504 |
| HLA-B*39:01 negative | 34 | 2.96 | |
| Number of patients | Age of onset | P (Student’s t) | |
| B | |||
| HLA-DRB1*15:01 positive | 18 | 33.3 | 0.618 |
| HLA-DRB1*15:01 negative | 27 | 35.0 | |
| HLA-DQB1*06:02 positive | 17 | 33.0 | 0.545 |
| HLA-DQB1*06:02 negative | 28 | 35.1 | |
| HLA-B*15:01 positive | 15 | 35.1 | 0.752 |
| HLA-B*15:01 negative | 30 | 33.9 | |
| HLA-B*39:01 positive | 11 | 30.0 | 0.142 |
| HLA-B*39:01 negative | 34 | 35.7 | |
*Student’s t test was used for association analysis
Association of HLA amino acid polymorphisms with MS susceptibility
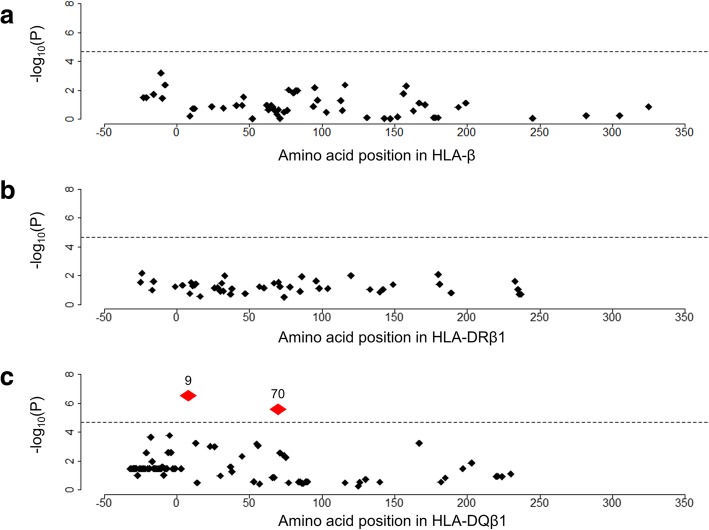
We then analyzed associations of the HLA amino acid polymorphisms with respect to MS susceptibility. We selected HLA-DRB1, HLA-DQB1, and HLA-B, which were associated with MS in our HLA allele-based study, for analysis (Additional file 1: Table S3–1). We found that the leading risk variant was the presence of phenylalanine at the HLA-DQβ1 position 9 (HLA-DQβ1 Phe9; P = 3.7 × 10− 8, OR = 3.48, 95%CI = 2.23–5.43; Additional file 1: Table S3–1, Fig. 1). We also found significant associations with the presence of arginine at HLA-DQβ1 position 70 (P = 2.7 × 10−6, OR = 0.34, 95% CI = 0.22–0.54; Additional file 1: Table S3–1), leucine at HLA-DQβ1 position − 5 (P = 8.9 × 10−6; OR = 3.54, 95% CI = 2.03–6.20; Additional file 1: Table S3–1), and valine at HLA-DQβ1 position − 18 (P = 2.1 × 10−4, OR = 2.80, 95% CI = 1.63–4.84; Additional file 1: Table S3–1). We assessed the significance of each amino acid position at HLA-DQβ1 using stepwise conditional analysis. After conditioning HLA-DQβ1 Phe9, no amino acid at HLA-DQβ1 was nominally associated with MS (Additional file 1: Table S3–2). We then conducted an omnibus test for each HLA amino acid position that had polymorphisms based on a log-likelihood ratio test [31, 32]. The HLA-DQβ1 position 9 showed the strongest association with MS (P = 3.6 × 10−7; Fig. 2), although its association was less significant than that of the binary test of HLA-DQβ1 Phe9.
Fig. 1.
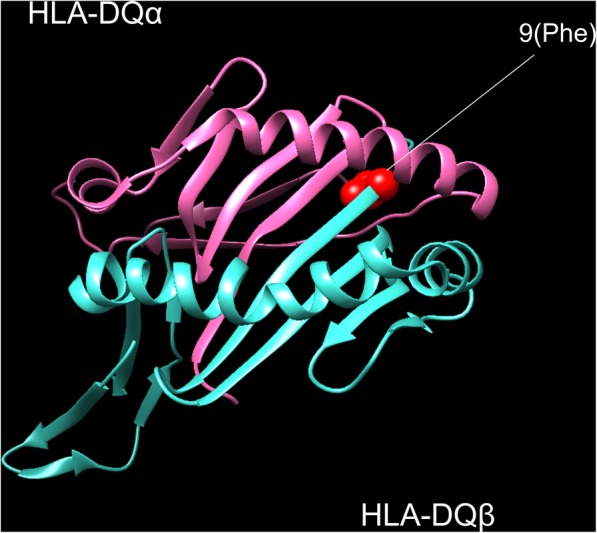
Multiple sclerosis risk-associated amino acid positions of the HLA genes in three-dimensional structure models. HLA amino acid positions with significant MS risk in HLA-DQ molecules. The protein structure is based on Protein Data Bank (PDB) entries 1jk8 and prepared using UCSF Chimera (version 1.7). Residues at the amino acid positions with significant MS risk is highlighted in red
Fig. 2.
Multiple sclerosis risk-associated amino acid positions of the HLA genes. Diamonds represent the –log10(P) of the amino acid positions of the tested HLA genes. Labeled red diamonds represent the –log10(P) values of the amino acid positions significantly associated with MS. The horizontal lines represent the association P value of HLA-DRB1*15:01. a No amino acid polymorphisms of HLA-B was significantly associated with MS. b No amino acid polymorphisms of HLA-DRB1 is significantly associated with MS. c The HLA-DQβ1 position 9 and 70 were associated with MS more significantly than HLA-DRB1*15:01
HLA-DQβ1 Phe9 independently confers MS risk with HLA-DRB1*15:01
Our study identified a significant MS risk associated with HLA-DRB1*15:01 and phenylalanine 9 at HLA-DQβ1, but it was not clear which of the variants caused these effects. We therefore conducted an analysis to investigate these effects. When conditioning HLA-DRB1*15:01, HLA-DQβ1 Phe9 was significantly associated with MS susceptibility (P = 1.5 × 10−5, OR = 2.91, 95% CI = 1.79–4.72; Additional file 1: Table S3–2). Conversely, when conditioning HLA-DQβ1 Phe9, HLA-DRB1*15:01 was just nominally significant (P = 0.037, OR = 1.94, 95% CI = 1.04–3.63). While further validation study should be warranted, our results suggested that HLA-DQβ1 Phe9 is a causal driver of HLA variants for MS risk in the Japanese population.
Associations of the HLA alleles with NMOSD susceptibility
We assessed the risk of NMOSD with 4-digit HLA alleles and found the most significant association to be at HLA-DQA1*05:03 (P = 1.5 × 10−4, OR = 6.96, 95% CI = 2.55–19.0; Table 5). We conducted conditional analysis controlling for the top HLA alleles associated with NMOSD. After conditioning for HLA-DQA1*05:03, no significant association was observed after Bonferroni correction, based on the number of alleles tested (Pcorr > 0.05; Additional file 1: Table S2). Our results indicated that HLA-DQA1*05:03 is a newly identified HLA allele associated with NMOSD in the Japanese population.
Table 5.
HLA association analysis of neuromyelitis optica spectrum disorder in the Japanese population
| Variant | Frequency | Nominal analysis | |||
|---|---|---|---|---|---|
| HLA gene | NMOSD (n = 31) | Control (n = 429) | OR (95% CI) | P | Fisher |
| HLA-A*26:03 | 0.065 | 0.020 | 3.40 (0.81–10.9) | 0.047 | * |
| HLA-B*07:02 | 0.129 | 0.059 | 2.34 (1.06–5.19) | 0.036 | |
| HLA-DPB1*05:01 | 0.516 | 0.356 | 1.93 (1.15–3.24) | 0.012 | |
| HLA-DQA1*01:01 | 0.129 | 0.058 | 2.39 (1.08–5.30) | 0.031 | |
| HLA-DQA1*03:02 | 0.048 | 0.147 | 0.30 (0.06–0.93) | 0.035 | * |
| HLA-DQA1*05:03 | 0.097 | 0.015 | 6.96 (2.55–19.0) | 1.5 × 10−4 | |
| HLA-DQB1*03:01 | 0.210 | 0.104 | 2.29 (1.20–4.39) | 0.012 | |
| HLA-DQB1*03:03 | 0.048 | 0.149 | 0.29 (0.06–0.91) | 0.024 | * |
| HLA-DRB1*01:01 | 0.129 | 0.058 | 2.39 (1.08–5.30) | 0.031 | |
| HLA-DRB1*09:01 | 0.016 | 0.135 | 0.10 (0.003–0.62) | 0.0027 | * |
| HLA-DRB1*14:06 | 0.081 | 0.012 | 7.40 (1.92–24.8) | 0.0021 | * |
*Fisher’s exact test was used for association analysis, otherwise, logistic regression was used
Associations of the HLA amino acid polymorphisms with NMOSD susceptibility
We analyzed the association of the HLA amino acid polymorphisms with NMOSD susceptibility. We selected HLA-DQA1, which was associated with NMOSD in our HLA allele-based association study. We found no amino acid variants with significant risk after Bonferroni correction, based on the number of amino acids tested (Pcorr > 0.05; Additional file 1: Table S4).
Discussion
In this study, we performed HLA analysis of MS/NMOSD in Japanese individuals. We replicated results indicating that HLA-DRB1*15:01 is the allele most strongly associated with MS susceptibility, a finding which was comparable with those of previous Japanese and European populations [13, 17]. HLA-B*39:01 and HLA-B*15:01 also appeared to be associated with MS. Previous European studies have reported an association of class I HLA alleles with MS susceptibility [14]. These results suggest a relationship between class I HLA alleles and MS susceptibility, as well as class II HLA alleles, in both Europeans and Japanese.
We also conducted an association study involving HLA amino acid polymorphisms and found that HLA-DQβ1 amino acid polymorphisms led to a significantly increased risk of MS. Previous studies have reported the importance of HLA-DRB1 with respect to MS [14, 34], but the role of HLA-DQB1 has not been studied in detail. In our study, even conditioning HLA-DRB1*15:01, HLA amino acid polymorphisms at HLA-DQβ1 were associated with increased risk of MS. This result indicates that HLA-DQβ1 polymorphisms could independently confer risk of MS. The association of HLA-DQB1 with MS has not been reported well. MHC class II molecules are heterodimers of an alpha and a beta chain, and have a peptide-binding site for presenting antigenic peptides to T cells [35]. In this study, we found that two amino acid positions associated with MS (HLA-DQβ1 position 9 and 70). Previous studies showed that HLA-DQβ1 position 9 is in the peptide-binding groove of HLA-DQ molecules and HLA-DQβ1 position 70 residues in the putative T cell receptor (TCR) [36]. Within the central nervous system, antigen-presenting cells such as microglia and astrocytes work as defenders against infections and inflammation [37]. Changes in antigen-presenting function of peptide binding amino acid changes could influence the susceptibility to MS. Animal model studies have found that susceptibility to myelin oligodendrocyte-binding protein or proteolipid protein were determined by HLA-DQB1*06:02 [38, 39]. These findings also suggest the possibility that HLA-DQB1 plays an important role in susceptibility to MS.
HLA-DQA1*05:03 was identified as being associated with NMOSD in our study. HLA-DQA1 has been reported to be associated with immune-mediated diseases such as celiac disease and type 1 diabetes [40, 41]. In Europeans, a recent GWAS identified rs28383224, which is located in HLA-DQA1, as having the strongest association with NMOSD susceptibility [19]. Although the role of the HLA-DQA1 risk SNP has not been investigated in detail, the importance of HLA-DQA1 in NMOSD pathophysiology was suggested.
A previous study in Japan reported that HLA-DRB1*16:02 and HLA-DPB1*05:01 presented a significantly elevated risk of NMOSD, while HLA-DRB1*09:01 reduced the risk. [22] In our analysis, HLA-DRB1*09:01 and HLA-DPB1*05:01 were nominally significant (HLA-DRB1*09:01; P = 0.0027, OR = 0.10, 95% CI = 0.003–0.62, HLA-DPB1*05:01; P = 0.012, OR = 1.93, 95% CI = 1.15–3.24; Additional file 1 Table S2), but after performing Bonferroni correction, they were not significantly associated with NMOSD. This result may be because our study sample size was smaller than that of the previous study in Japan. HLA-DRB1*16:02 was not significant in our study (P = 0.39, OR = 2.32, 95% CI = 0.05–19.6). These findings may indicate that the HLA alleles associated with NMOSD are localized in Japan. Further studies are needed to identify the details of the variability of HLA alleles in the Japanese population.
Conclusions
In summary, through NGS-based HLA typing of the MS and NMOSD patients and controls of Japanese ancestry, we analyzed the variants of the 16 HLA genes. We newly identified the risk HLA alleles associated with MS and NMOSD risk. We also identified that the amino acid polymorphisms at HLA-DQβ1 are more strongly associated with MS susceptibility than HLA alleles. Our study contributes to the understanding of the genetic background of MS and NMOSD in Japanese.
Additional file
Table S1 Results of the association analysis of HLA alleles (multiple sclerosis). Table S2 Results of the association analysis of HLA alleles (neuromyelitis optica spectrum disorder). Table S3–1 Results of the HLA amino acid association analysis (multiple sclerosis). Table S3–2 Results of the conditional HLA DQβ1 association analysis (multiple sclerosis). Table S4 Results of the HLA amino acid association analysis (neuromyelitis optica spectrum disorder). (PDF 542 kb)
Acknowledgements
Not applicable.
Abbreviations
- GWAS
Genome-wide association studies
- MHC
Major histocompatibility complex
- MS
Multiple sclerosis
- NGS
Next-generation sequencing
- NMOSD
Neuromyelitis optica spectrum disorder
- SNP
Single-nucleotide polymorphisms
Authors’ contributions
YO supervised the study. KO, TO, YO, and HM wrote the manuscript. KO, TO, KH, AH, JH, KS, SS, YA, and YO conducted the data analysis. TO, AH, MK, KM, SK, and HM collected the samples. KH and II conducted the experiments. All authors read and approved the final manuscript.
Funding
This research was supported by Bioinformatics Initiative of Osaka University Graduate School of Medicine, and Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University. Y.O. was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (15H05911 and 19H01021), AMED (19gm6010001h0004, 19ek0410041h0003, and 19ek0109413h0001). J.H. is an employee of Teijin Pharma Limited.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the ethical committee of Osaka University Graduate School of Medicine. All participants were of Japanese origin and provided written informed consent that was approved by the ethical committee of each hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kotaro Ogawa, Email: kogawa@sg.med.osaka-u.ac.jp.
Tatsusada Okuno, Email: okuno@neurol.med.osaka-u.ac.jp.
Kazuyoshi Hosomichi, Email: khosomic@med.kanazawa-u.ac.jp.
Akiko Hosokawa, Email: namba-hosokawa@neurol.med.osaka-u.ac.jp.
Jun Hirata, Email: jhirata@sg.med.osaka-u.ac.jp.
Ken Suzuki, Email: ksuzuki@sg.med.osaka-u.ac.jp.
Saori Sakaue, Email: ssakaue@sg.med.osaka-u.ac.jp.
Makoto Kinoshita, Email: mkinoshita@neurol.med.osaka-u.ac.jp.
Yoshihiro Asano, Email: asano@cardiology.med.osaka-u.ac.jp.
Katsuichi Miyamoto, Email: miyamoto@med.kindai.ac.jp.
Ituro Inoue, Email: itinoue@nig.ac.jp.
Susumu Kusunoki, Email: kusunoki@med.kindai.ac.jp.
Yukinori Okada, Phone: +81-6-6879-3971, Email: yokada@sg.med.osaka-u.ac.jp.
Hideki Mochizuki, Email: hmochizuki@neurol.med.osaka-u.ac.jp.
References
- 1.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dendrou CA, Petersen J, Rossjohn J, et al. HLA variation and disease. Nat Rev Immunol. 2018;18(5):325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 3.Kanai M, Akiyama M, Takahashi A, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50(3):390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 4.Silman AJ, Pearson JE. Epidemiology and genetics of rheumatoid arthritis. Arthritis Res. 2002;4(Suppl 3):S265–S272. doi: 10.1186/ar578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble JA. Immunogenetics of type 1 diabetes: a comprehensive review. J Autoimmun. 2015;64:101–112. doi: 10.1016/j.jaut.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Hirata J, Hirota T, Ozeki T, et al. Variants at HLA-A, HLA-C, and HLA-DQB1 confer risk of psoriasis vulgaris in Japanese. J Invest Dermatol. 2018;138(3):542–548. doi: 10.1016/j.jid.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Browne P, Chandraratna D, Angood C, et al. Atlas of multiple sclerosis 2013: a growing global problem with widespread inequity. Neurology. 2014;83(11):1022–1024. doi: 10.1212/WNL.0000000000000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Multiple Sclerosis Genetics C. Hafler DA, Compston A, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 9.Australia, New Zealand Multiple Sclerosis Genetics C Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 10.International Multiple Sclerosis Genetics C, Wellcome Trust Case Control C. Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Multiple Sclerosis Genetics C. Beecham AH, Patsopoulos NA, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa K, Okada Y. Statistical genetics and its application to neuroimmunology. Clin Exp Neuroimmunol. 2018;9(1):7–12. [Google Scholar]
- 13.Ramagopalan SV, Knight JC, Ebers GC. Multiple sclerosis and the major histocompatibility complex. Curr Opin Neurol. 2009;22(3):219–225. doi: 10.1097/WCO.0b013e32832b5417. [DOI] [PubMed] [Google Scholar]
- 14.Patsopoulos NA, Barcellos LF, Hintzen RQ, et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013;9(11):e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mack SJ, Udell J, Cohen F, et al. High resolution HLA analysis reveals independent class I haplotypes and amino-acid motifs protective for multiple sclerosis. Genes Immun. 2018;20:308–326. doi: 10.1038/s41435-017-0006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshimura S, Isobe N, Yonekawa T, et al. Genetic and infectious profiles of Japanese multiple sclerosis patients. PLoS One. 2012;7(11):e48592. doi: 10.1371/journal.pone.0048592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura Y, Matsushita T, Sato S, et al. Latitude and HLA-DRB1*04:05 independently influence disease severity in Japanese multiple sclerosis: a cross-sectional study. J Neuroinflammation. 2016;13(1):239. doi: 10.1186/s12974-016-0695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arellano B, Hussain R, Miller-Little WA, et al. A single amino acid substitution prevents recognition of a dominant human aquaporin-4 determinant in the context of HLA-DRB1*03:01 by a murine TCR. PLoS One. 2016;11(4):e0152720. doi: 10.1371/journal.pone.0152720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estrada K, Whelan CW, Zhao F, et al. A whole-genome sequence study identifies genetic risk factors for neuromyelitis optica. Nat Commun. 2018;9(1):1929. doi: 10.1038/s41467-018-04332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimura S, Isobe N, Matsushita T, et al. Distinct genetic and infectious profiles in Japanese neuromyelitis optica patients according to anti-aquaporin 4 antibody status. J Neurol Neurosurg Psychiatry. 2013;84(1):29–34. doi: 10.1136/jnnp-2012-302925. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita T, Matsuoka T, Isobe N, et al. Association of the HLA-DPB1*0501 allele with anti-aquaporin-4 antibody positivity in Japanese patients with idiopathic central nervous system demyelinating disorders. Tissue Antigens. 2009;73(2):171–176. doi: 10.1111/j.1399-0039.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- 22.Hosomichi K, Jinam TA, Mitsunaga S, et al. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics. 2013;14:355. doi: 10.1186/1471-2164-14-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosomichi K, Mitsunaga S, Nagasaki H, et al. A bead-based normalization for uniform sequencing depth (BeNUS) protocol for multi-samples sequencing exemplified by HLA-B. BMC Genomics. 2014;15:645. doi: 10.1186/1471-2164-15-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosomichi K, Shiina T, Tajima A, et al. The impact of next-generation sequencing technologies on HLA research. J Hum Genet. 2015;60(11):665–673. doi: 10.1038/jhg.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wingerchuk DM, Banwell B, Bennett JL, et al. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology. 2015;85(2):177–189. doi: 10.1212/WNL.0000000000001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirata J, Hosomichi K, Sakaue S, et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat Genet. 2019;51(3):470–480. doi: 10.1038/s41588-018-0336-0. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson J, Soormally AR, Hayhurst JD, et al. The IPD-IMGT/HLA database—new developments in reporting HLA variation. Hum Immunol. 2016;77(3):233–237. doi: 10.1016/j.humimm.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Pirooznia M, Kramer M, Parla J, et al. Validation and assessment of variant calling pipelines for next-generation sequencing. Hum Genomics. 2014;8:14. doi: 10.1186/1479-7364-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada Y, Momozawa Y, Ashikawa K, et al. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat Genet. 2015;47(7):798–802. doi: 10.1038/ng.3310. [DOI] [PubMed] [Google Scholar]
- 32.Okada Y, Suzuki A, Ikari K, et al. Contribution of a non-classical HLA gene, HLA-DOA, to the risk of rheumatoid arthritis. Am J Hum Genet. 2016;99(2):366–374. doi: 10.1016/j.ajhg.2016.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Brum DG, Barreira AA, dos Santos AC, et al. HLA-DRB association in neuromyelitis optica is different from that observed in multiple sclerosis. Mult Scler. 2010;16(1):21–29. doi: 10.1177/1352458509350741. [DOI] [PubMed] [Google Scholar]
- 35.Call MJ. Small molecule modulators of MHC class II antigen presentation: mechanistic insights and implications for therapeutic application. Mol Immunol. 2011;48(15–16):1735–1743. doi: 10.1016/j.molimm.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Jones EY, Fugger L, Strominger JL, et al. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6(4):271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 37.Chastain EM, Duncan DS, Rodgers JM, et al. The role of antigen presenting cells in multiple sclerosis. Biochim Biophys Acta. 2011;1812(2):265–274. doi: 10.1016/j.bbadis.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaushansky N, Altmann DM, Ascough S, et al. HLA-DQB1*0602 determines disease susceptibility in a new “humanized” multiple sclerosis model in HLA-DR15 (DRB1*1501;DQB1*0602) transgenic mice. J Immunol. 2009;183(5):3531–3541. doi: 10.4049/jimmunol.0900784. [DOI] [PubMed] [Google Scholar]
- 39.Kaushansky N, Altmann DM, David CS, et al. DQB1*0602 rather than DRB1*1501 confers susceptibility to multiple sclerosis-like disease induced by proteolipid protein (PLP) J Neuroinflammation. 2012;9:29. doi: 10.1186/1742-2094-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci. 2012;19:88. doi: 10.1186/1423-0127-19-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Amir MI, El-Feky MA, Laine AP, et al. Risk genes and autoantibodies in Egyptian children with type 1 diabetes - low frequency of autoantibodies in carriers of the HLA-DRB1*04:05-DQA1*03-DQB1*02 risk haplotype. Diabetes Metab Res Rev. 2015;31(3):287–294. doi: 10.1002/dmrr.2609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Results of the association analysis of HLA alleles (multiple sclerosis). Table S2 Results of the association analysis of HLA alleles (neuromyelitis optica spectrum disorder). Table S3–1 Results of the HLA amino acid association analysis (multiple sclerosis). Table S3–2 Results of the conditional HLA DQβ1 association analysis (multiple sclerosis). Table S4 Results of the HLA amino acid association analysis (neuromyelitis optica spectrum disorder). (PDF 542 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.