Abstract
Background
Two transmission patterns of Sarcoptes scabiei in host mammal communities have been reported based on microsatellite-level genetic studies in the last two decades. While one involves restrictions among different host taxa, the other is associated with predator–prey interactions between different host taxa. In contrast to these observations, the present study reports a possible irregular case of transmission of S. scabiei between herbivorous Japanese serow and omnivorous Caniformia mammals in Japan, though under very weak predator–prey relationships.
Methods
DNA from 93 Sarcoptes mites isolated from omnivorous Caniformia (such as the domestic dog, raccoon dog, raccoon and Japanese marten), omnivorous Cetartiodactyla (wild boar) and herbivorous Cetartiodactyla (Japanese serow) in Japan were analyzed by amplifying nine microsatellite markers. Principal components analyses (PCA), Bayesian clustering analyses using STRUCTURE software, and phylogenetic analyses by constructing a NeighborNet network were applied to determine the genetic relationships among mites associated with host populations.
Results
In all the analyses, the genetic differentiation of Sarcoptes mites from wild boars and Japanese serows was observed. Conversely, considerably close genetic relationships were detected between Caniformia-derived and Japanese serow-derived mites. Because the predator–prey interactions between the omnivorous Caniformia and herbivorous Japanese serow are quite limited and epidemiological history shows at least a 10-year lag between the emergence of sarcoptic mange in Japanese serow and that in Caniformia, the transmission of S. scabiei from Caniformia to Japanese serow is highly suspected.
Conclusions
The close genetic relationships among mites beyond Host–taxon relationships and without obvious predator–prey interactions in Caniformia and Japanese serow deviate from previously reported S. scabiei transmission patterns. This type of cryptic relationship of S. scabiei populations may exist in local mammalian communities worldwide and become a risk factor for the conservation of the remnant and fragmented populations of wild mammals.
Electronic supplementary material
The online version of this article (10.1186/s13071-019-3630-5) contains supplementary material, which is available to authorized users.
Keywords: Host–parasite relationship, Host specificity, Sarcoptic mange, Scabies, Genetic structure
Background
Sarcoptes scabiei is a notorious pathogenic mite species (Acari: Astigmata: Sarcoptidae) that causes a highly contagious dermatitis known as sarcoptic mange in mammalian hosts [1]. Among the mammalian species, and even within the species [2], the pathogenicity of the disease spans largely from devastating impacts on naive host individuals [1, 3] to relatively low pathogenic infections in less-vulnerable host individuals [4]. The impact of sarcoptic mange on mammalian host populations changes among populations and in the trade-offs among biological factors such as population density, host mortality rate and host reproductive rate [5–7]. However, this has often been viewed as a possible threat for the conservation of remnant and isolated mammal populations [1]. Although emerging (or re-emerging) outbreaks of sarcoptic mange in mammalian populations have been reported worldwide [1, 8], neither the origin of the epidemic lineages of S. scabiei nor the patterns of inter-species transmission of mites from the reservoir host species to the novel (or unobserved) host species have been sufficiently understood [9]. This is because the existence of a classical problem in the identification of mite strains, at least until the late 1990s, due to the nature of S. scabiei populations having different patterns of Host-specificities with indistinguishable morphology [8, 10]. To resolve this problem, a method for genetic analysis using microsatellite (simple sequence repeat; SSR) markers was developed by Walton et al. [11–13] and, since then, this technique has been utilized as an efficient way to elucidate the hidden transmission web of Sarcoptes mites in the multi-host system [9, 14].
Recent genetic studies using the SSR markers revealed two patterns of inter-species transmission of Sarcoptes mites: one is the transmission restricted by the “host taxon” [15–17] and the other is the transmission through “prey–predator” interaction [18–20]. For Host–taxon transmission restriction, Rasero et al. [15] found that Sarcoptes mite populations from ten European wild mammal species were genetically separated by three patterns of host taxon, named as carnivores, omnivores and herbivores (in the definition of Rasero et al. [15]). For predator–prey transmission, close genetic associations between mite populations derived from predators and their preferred prey were detected, e.g. between lion (Panthera leo) and wildebeest (Connochaetes taurinus), and between cheetah (Asinonyx jubatus) and Thomson’s gazelle (Gazelle thompsonii) [18]. The difference between these two patterns of transmission of Sarcoptes mites suggests the existence of diversity in their transmission web, associated with the interaction within the local mammalian communities co-existing in each region [9]. In Japan, the realization of the epidemic of sarcoptic mange in wild mammal hosts was started in early 1980s with the report of an outbreak in raccoon dogs (Nyctereutes procyonoides) [21, 22]. Since then, during the past 40 years, the disease has been drastically expanded in the omnivorous Carnivora species [e.g. red fox (Vulpes vulpes), masked palm civet (Paguma larvata)] and wild boars (Sus scrofa leucomystax) [21–23]. In addition, cases of infestation of Sarcoptes mites in Japanese serow (Capricornis crispus), an indigenous ruminant species designated as a Special National Monument in Japan, have been reported since the mid-1990s [21, 24, 25]. Because of the nearly simultaneous (within 10–20 years) and multiple beginnings of epidemics in wild mammals, it has been suspected that possible inter-species transmission occurred in these mammalian host communities, with the addition of transmission from domestic/companion animals (e.g. dog, swine and cattle) [21, 22]. However, the genetic relationships of Sarcoptes mites with mammalian hosts have not been elucidated at the level of SSR resolution, apart from limited reports on the relationship between raccoon dog populations and domestic/companion dogs (Canis familiaris) [26].
In the present study, we aim to illuminate the neglected relationships of Sarcoptes mite populations derived from five wild mammalian species and domestic/companion dogs in Japan by SSR analysis. This enables us to explicate whether the transmission within these multi-host species follows the host taxon and predator–prey patterns. The target hosts were medium-sized omnivorous Carnivora, more specifically Caniformia [domestic dog, raccoon dog, raccoon (Procyon lotor) and Japanese marten (Martes melampus melampus)], an omnivorous Cetartiodactyla (wild boar) and a herbivorous Cetartiodactyla (Japanese serow).
To our knowledge, the predator–prey relationships between these omnivorous Caniformia and Cetartiodactyla have not been previously reported, except for a few limited cases of predation of Japanese serow by feral dogs [27] and possible occasional scavenging on carcasses of omnivorous species [28–31]. Against these inadequate predator–prey relationships and the expected Host–taxon barriers for the inter-species transmission, we report here the observation of “cryptic,” close genetic relationships among the Sarcoptes mites derived from Japanese serows and those from omnivorous species in Caniformia.
Methods
Collection of S. scabiei from skin crusts of hosts
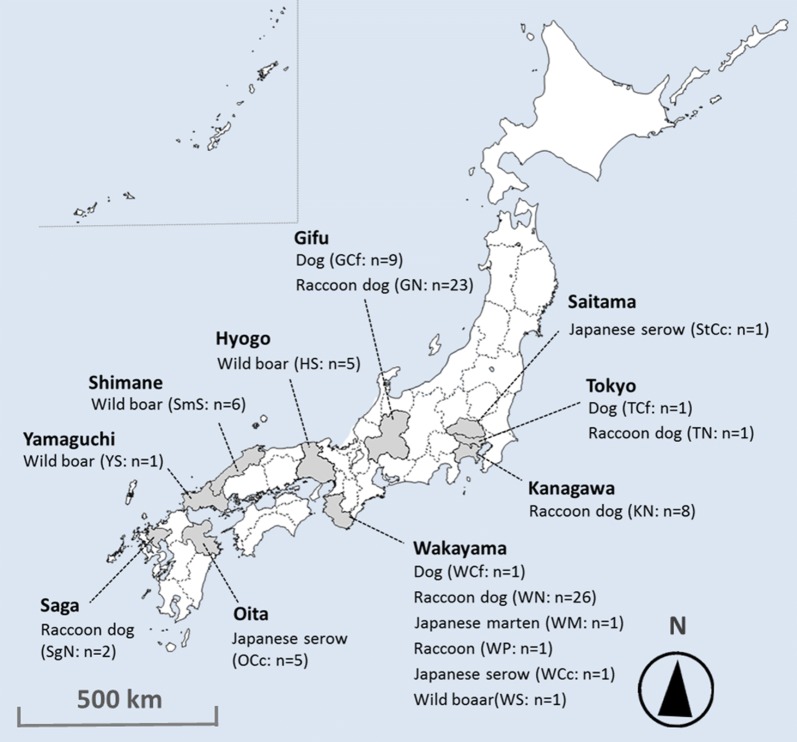
From 1996 to 2016, we collected skin crusts from 93 mangy animals from 10 different prefectures in Japan: Tokyo, Kanagawa, Saitama, Gifu, Wakayama, Hyogo, Shimane and Yamaguchi on Honshu Island, and Oita and Saga on Kyushu Island (Fig. 1).
Fig. 1.
The location of the 10 sampling areas and 93 sampled animals in the present study
The animals belonged to 17 populations from 6 species that consisted of raccoon dogs (n = 60), domestic/companion dogs (n = 11), raccoons (n = 1), Japanese martens (n = 1), Japanese serows (n = 7) and wild boars (n = 13) (Table 1). All skin crusts were stored at – 30 °C or in 70% ethanol until the mite collection. We collected one Sarcoptes mite per individual animal by the postponed (post-frozen) isolation method for frozen skins proposed by Alasaad et al. [32]. All mites were confirmed to be S. scabiei from the morphological criteria defined by Fain [10].
Table 1.
Host species, higher-level taxon and the number of mite samples in each area
| Prefecture | Host species | Host taxon | Sample size | Sampling year | Population name |
|---|---|---|---|---|---|
| Tokyo | Domestic dog | Carnivora (Caniformia) | 1 | 2012 | TCf |
| Raccoon dog | Carnivora (Caniformia) | 1 | 2012 | TN | |
| Kanagawa | Raccoon dog | Carnivora (Caniformia) | 8 | 2012–2014 | KN |
| Saitama | Japanese serow | Cetartiodactyla (Ruminantia) | 1 | 1996 | StCc |
| Gifu | Domestic dog | Carnivora (Caniformia) | 9 | 2014–2015 | GCf |
| Raccoon dog | Carnivora (Caniformia) | 23 | 2007–2015 | GN | |
| Wakayama | Domestic dog | Carnivora (Caniformia) | 1 | 2010 | WCf |
| Japanese marten | Carnivora (Caniformia) | 1 | 2010 | WM | |
| Raccoon dog | Carnivora (Caniformia) | 26 | 2009–2014 | WN | |
| Raccoon | Carnivora (Caniformia) | 1 | 2010 | WP | |
| Japanese serow | Cetartiodactyla (Ruminantia) | 1 | 2010 | WCc | |
| Wild boar | Cetartiodactyla (Suina) | 1 | 2011 | WS | |
| Hyogo | Wild boar | Cetartiodactyla (Suina) | 5 | 2011 | HS |
| Shimane | Wild boar | Cetartiodactyla (Suina) | 6 | 2011 | SmS |
| Yamaguchi | Wild boar | Cetartiodactyla (Suina) | 1 | 2011 | YS |
| Oita | Japanese serow | Cetartiodactyla (Ruminantia) | 5 | 1996, 2016a | OCc |
| Saga | Raccoon dog | Carnivora (Caniformia) | 2 | 2009 | SgN |
aOnly the OCc 5 was collected in 2016
DNA extraction and fluorescence-based PCR analysis
After the crushing individual mites, the DNA of each Sarcoptes mite was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Tokyo, Japan). Molecular analyses were conducted following the method of Alasaad et al. [33] with slight modifications. The DNA from each mite was extracted and the 9 specific SSRs (Sarms 33, 34, 36–38, 40, 41, 44 and 45; Walton et al. [12]) from Sarcoptes mites were amplified by multiplex polymerase chain reaction (PCR). Each 5 µl PCR mixture contained 1 µl of the isolated mite DNA together with the PCR mixture containing all primer pairs (each primer ranged from 0.04 to 0.1 µM), 200 µM dNTPs, 0.5 µl of 10× PCR buffer (200 mM KCl, 100 mM Tris-HCl, pH 8.0), 0.5 mM MgCl2 and 0.05 µl (0.5 U/reaction) Ex Taq (TaKaRa Shuzo, Kyoto, Japan). Fluorescent PCR amplicons were analyzed in an ABI PRISM 3130 Genetic Analyzer using GeneScan 500 Liz® Size Standard (Applied Biosystems, Foster City, CA, USA). Allele calling was performed using the GeneMapper® Software v.4.1 (Applied Biosystems, Foster City, CA, USA).
Molecular analysis
Observed (HO) and expected (HE) heterozygosity, linkage disequilibrium (LD) and Hardy–Weinberg equilibrium (HWE) tests were calculated using GENEPOP v.4.0. [34]. In this software, the exact P-values of LD tests were given by Chi-square tests and those of HWE were calculated by a Markov chain algorithm [34].
While some of the analyzed mite populations significantly deviated from HWE, LD was also detected between some SSR loci. Therefore, we used two methods that do not require assumptions, like HWE and linkage equilibrium (LE) of loci, to analyze the underlying population genetic model. First, to describe differentiation between mite populations, principal components analysis (PCA) was carried out using the package adegenet [35] of R v.3.2.2 [36]. Multivariate ordination in the PCA does not require data regarding the HWE and LD [37]. In this analysis, genetic relationships among mites associated with 17 host populations (Table 1) were analyzed. Secondly, to analyze the genetic relationships between mites, we calculated genetic distances (proportion of shared alleles; DSA [38]) among mites using Populations v.1.2.32 [39]. These genetic distances were then visualized using the NeighborNet network provided by SplitsTree4 [40].
However, HWE in populations and LE between SSR loci are often violated in natural populations and are viewed as unrealistic [37]. In such circumstances, Bayesian clustering analysis by STRUCTURE software v.2.3.4 [41] has often also been employed for populations deviated by HWE and/or for loci in LD (e.g. Rasero et al. [15]). Thus, we applied the STRUCTURE analysis to elucidate the genetic structure of 93 Sarcoptes mites and to support the former 2 analytical methods. We performed 100,000 MCMC (Marcov Chain Monte Carlo) simulations followed by 50,000 steps of burn-in for each K value (K = 1–17), and these calculations were repeated independently 10 times. Subsequently, the most likely clustering number of K was determined by the peak of ΔK calculated by the method of Evanno et al. [42]. The website STRUCTURE HARVESTER (taylor0.biology.ucla.edu/structureHarvester) [43] was used for this calculation. The results of 10 replicate runs for each value of K were averaged using the Greedy algorithm of CLUMPP v.1.1.2 [44], and clustering results for each value of K were displayed graphically using distruct v.1.1 [45].
Results
One hundred and eight alleles from the 9 SSR loci of all 93 mites were detected (Table 1); they contained 41 alleles from 11 domestic/companion dog-derived mites (TCf, GCf and WCf), 66 from 60 raccoon dog-derived mites (TN, KN, GN, WN and SgN), 10 from a marten-derived mite (WM), 12 from a raccoon-derived mite (WP), 25 from 6 serow-derived mites (StCc, WCc and OCc), and 61 from 13 boar-derived mites (WS, HS, SmS and YS). The number of alleles for each locus ranged from 7 (Sarms 36) to 18 (Sarms 33). HO and HE ranged from 0.152 (Sarms 44) to 0.337 (Sarms 37) and from 0.501 (Sarms 38) to 0.881 (Sarms 33), respectively.
A significant deviation from HWE was observed for some loci in Sarcoptes mite populations associated with KN, GN, GCf, WN, HS, OCc and SmS (P < 0.05, see Additional file 1: Table S1), and LD was confirmed between Sarms 33 and Sarms 34 (χ2 = 26.14, df = 12, P = 0.010), Sarms 34 and Sarms 40 (χ2 = 31.11, df = 8, P < 0.001), Sarms 34 and Sarms 45 (χ2 = 30.08, df = 12, P = 0.003), Sarms 40 and Sarms 41 (χ2 = 19.81, df = 8, P = 0.011) and Sarms 40 and Sarms 45 (χ2 = 15.61, df = 9, P = 0.048), when tested in the whole population.
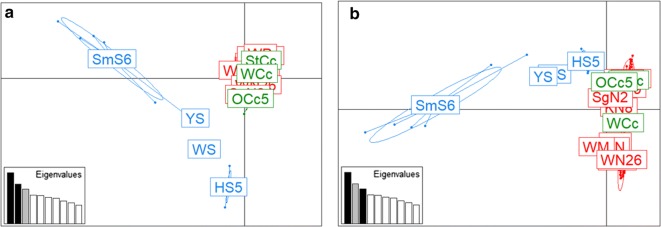
The results of the PCA are shown in Fig. 2. The projected inertia of components 1, 2 and 3 were 9.96, 7.68 and 6.76%, respectively. In components 1 and 2 (cumulative projected inertia: 17.65%), Caniformia- (TCf, TN, KN, GCf, GN, WCf, WM, WN, WP and SgN) and serow-derived mites (StCc, WCc and OCc) formed a cluster and were separated from boar-derived mites (WS, HS, SmS and YS). In components 1 and 3 (cumulative projected inertia: 16.73%), Caniformia- and serow-derived mites were slightly more scattered than components 1 and 2, but formed a cluster, and were separated from boar-derived mites (Fig. 2).
Fig. 2.
Results of the principle components analysis (PCA) of mite populations associated with 17 host populations. PCA showing the genetic structure of 93 mites from 17 host populations with component 1 (explaining 9.96% of the variance) versus component 2 (7.68%) (a) and component 3 (6.76%) (b). The eigenvalues of the two axes are displayed in each graph. Caniformia-, serow- and boar-derived mites are represented as red, green and blue, respectively. Each Host-associated mite population is indicated in the centre of component mites. Abbreviations for populations are provided in Table 1
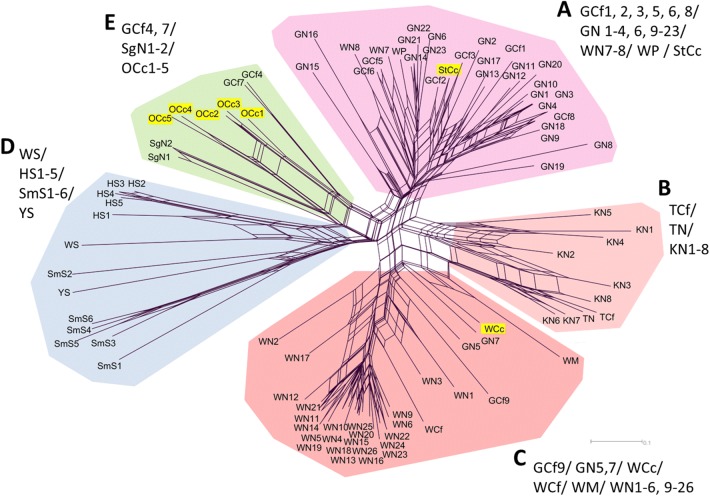
The NeighborNet network composed by DSA showed the 5 genetic clusters (Clusters A–E) in 93 mites (Fig. 3). Basically, mites from the same taxonomic or close host species within the same area were included in each cluster; Cluster A comprised Caniformia-derived mites from Gifu (GCf and GN), Cluster B comprised Caniformia-derived mites from Tokyo (TCf and TN) and Kanagawa (KN), Cluster C comprised Caniformia-derived mites from Wakayama (WCf and WN), Cluster D comprised boar-derived mites (WS, HS, SmS and YS) and Cluster E comprised serow-derived mites from Oita (OCc). However, some host species of other areas and/or different host taxa were also classified in each cluster. Raccoon- and raccoon dog-derived mites (WP1, WN7 and WN8) from Wakayama, and a Japanese serow-derived mite from Saitama (StCc) were grouped in Cluster A. A domestic/companion dog-derived mite from Gifu (GCf9) was grouped in Cluster C. Raccoon dog-derived mites from Saga (SgN) and domestic/companion dog-derived mites from Gifu (GCf4 and GCf7) were grouped in Cluster E. Raccoon dog-derived mites from Gifu (GN5, GN7), a Japanese serow-derived mite and a Japanese marten-derived mite from Wakayama (WCc, WM) showed just intermediate genotypes between Clusters B and C (we included them in Cluster C for the convenience in the discussion). All Japanese serow-derived mites were confirmed to be close to the Caniformia-derived mites.
Fig. 3.
NeighborNet network constructed by DSA between each pair of 93 mites. Main clusters (A, B, C, D and E) are separated by different colors. The genetic differentiation between clusters B and C is not clear, and WM, WCc, GN5, GN7 were experientially assorted into Cluster C. Japanese serow-derived mites (StCc, WCc, OCc1-5) are shown with the yellow background. Abbreviations for populations are provided in Table 1
The STRUCTURE analysis revealed the highest value of ΔK at K = 9 (Additional file 2: Figure S1). At K = 9, a similar result to that obtained using the NeighborNet network was seen, wherein the wild boar-derived mite population and Caniformia- with Japanese serow-derived mite population were assigned to different clusters. Subsequently, a second run of STRUCTURE analysis was performed for the 80 mites from the Caniformia- with Japanese serow-derived population, and the ΔK value showed the uppermost hierarchical cluster to be K = 4. The genetic structure in these 80 mites in K = 4 was also consistent with that observed using the NeighborNet network (Fig. 3 and Additional file 2: Figure S1).
Discussion
As previously described, Rasero et al. [15] comprehensively presented the genetic differentiation of Sarcoptes mites in mammals in western Europe. Nonetheless, we have to re-attempt the interpretation of the results of their study here, before going into the profound discussion of our results. In our opinion, the results presented by Rasero et al. [15] lack accuracy because of the ambiguous definitions of terms related to “host taxon” (i.e. “carnivore”, “omnivore” and “herbivore”). Although Rasero et al. [15] did not define “host taxon,” the words “carnivore,” “omnivore,” and “herbivore” are usually used in the context of the foraging ecology of animals. In their study, the red fox, stone marten (M. foina) and pine marten (M. martes) were defined as “carnivores.” These animals are widely recognized as omnivores based on their food habits [46, 47]. Therefore, if these animals were classified by their food habits, their sorting would be incorrect. Thus, more accurately, the differentiation of Sarcoptes mite populations observed in their study did not conform to the foraging ecology of mammals, but to the taxonomy at the suborder level: (i) Caniformia in Carnivora as a substitute for “carnivore”; (ii) Suina in Cetartiodactyla as a substitute for “omnivore”; and (iii) Ruminantia in Cetartiodactyla as a substitute for “herbivore”.
As far as the present study is concerned, the taxonomy of our target species was as follows: domestic dog, raccoon dog, raccoon and Japanese marten belong to the suborder Caniformia in Carnivora; wild boar belongs to the suborder Suina in Cetartiodactyla; and Japanese serow belongs to the suborder Ruminantia in Cetartiodactyla. Deficiency of observed heterozygosity and deviation from HWE in some of the SSR loci in a few mite populations were also confirmed in the previous reports of other Sarcoptes mite populations [15, 16, 18, 20]. This phenomenon is due to a lack of random mating of mite populations between host animals, because Sarcoptes mites do not have free-living stages and individual hosts function as their ephemeral habitats in patchy environments [33]. Bearing in mind the presence of deviations in HWE, though a PCA method which will not be influenced by those genetic biases existed, we obtained the following consistent results in the multiple analyses: (i) the close genetic relationship between the mite populations derived from hosts in the same taxon, represented in omnivorous Caniformia and in wild boar populations; (ii) the separation of genetic clusters among mites derived from omnivorous Caniformia and from wild boar populations; and (iii) the close genetic relationship between mites derived from Caniformia species and from Japanese serow against the difference of host taxon. The results (i) and (ii) correspond to the previously reported genetic relationships of Sarcoptes mite populations in the European mammalian fauna and are consistent with the “Host–taxon phenomenon” formulated by Rasero et al. [15]. Meanwhile, (iii) shows the deviation from the Host–taxon regulation in the transmission web. Those serow-derived mites collected in the middle of 1990s (StCc1 in 1996 and OCc1-4 between 1996 and 1998) and in the 2010s (WCc1 in 2011 and Occ5 in 2016) from different locations highly suggested the long-time maintenance of gene flow between Caniformia- and serow-derived mite communities at several distant locations.
In the previous studies, the prey-predator pattern was described as “Curse of prey” [18] and tended to focus on the one-directional infections of Sarcoptes mites from the herbivorous preys to their carnivorous predators. However, this one-directional concept is not supported in case of “very weak” prey-predator relationships, the epidemiological history and the ecology of Japanese serow. Rather, infection from Caniformia mammals to serows is highly suspected. Upon reviewing the recent epidemiological history of sarcoptic mange in Japan, it was seen that its outbreaks in Japanese serows started roughly ten years later than the epidemic in raccoon dogs [21]. From a questionnaire survey for the epidemiology of sarcoptic mange in wild mammals, observations of mangy cases in hunting as well as in wildlife rescue practices began in the 1970s and 1980s in raccoon dogs and other Caniformia species [21, 22]. In the mid-1990s, recordings in hunting and rescue cases in raccoon dogs had drastically increased [21]. During this time, cases of sarcoptic mange in the serows increased [21, 25]. It might be possible that the epidemic in serows was associated with the increase of mangy cases in raccoon dogs (and possibly other Caniformia species).
From the view point of Japanese serow ecology, which is a territorial and monogamous species, usually living solitarily [48], the transmission of sarcoptic mange between serows has been considered to be less frequent than that in mammals which exhibit strong sociality (i.e. making herds or family groups) such as the raccoon dog, domestic dog and wild boar [2, 49]. However, multi-locational outbreaks in serows were almost simultaneously recognized since the 1990s in several geographically distinct populations on the Honshu and Kyushu Islands of Japan [21]. This concurrent emergence of mange among distinct serow populations was possibly due to transmission from other Caniformia harboring S. scabiei. Based on the data obtained from a questionnaire survey conducted by the Wildlife Management Office [21], most of the epidemic areas of mange infection in Japanese serows overlapped with those of mange infection in raccoon dogs.
Given that the inter-species transmission of Sarcoptes mites arises through both direct or indirect contact [8], there are two likely explanations for the transmission from Caniformia populations to the serow populations: (i) the direct transmission upon touching the carcasses of mangy animals; and (ii) indirect transmission through the sharing of habitat intensively used by both mammal populations. For the direct contact, the probable reason is the exploring or foraging behavior for dead animals sometimes observed in ruminant species such as cattle [50] and deer [51]. There is no observation, but likelihood that serows behave similar to dead or debilitated mangy Caniformia animals. For the indirect contact, the common use of the same location (or microhabitats) by these species has often been recorded by sensor cameras [52]. Although the survival of Sarcoptes mites off the host is only a few days in the natural environment [8], infection through contaminated environments (e.g. animal trails, vegetation, soil, etc.) is not impossible [8, 53]. Further study on the Sarcoptes mite’s ecology and contact between host species, besides the transmission experiments, may furnish a clear answer for the observed gene flow between herbivore ruminants and Caniformia species.
Lastly, our data reveal that some Sarcoptes mites from Caniformia species and Japanese serows do not belong to dominant genotypes in the Caniformia-serow derived communities in their locations (Fig. 3 and Additional file 2: Figure S1). If we assume that the results of the present study represent the gene pool of Sarcoptes mites in Caniformia mammals and Japanese serows in Japan, it may imply the migration of Sarcoptes mites, which is perhaps associated with (i) natural events of host migration and dispersion, or (ii) artificial translocation of Sarcoptes mites. Note that GCf4 and GCf7 from the domestic/companion dogs in Gifu belonged to the mite group from raccoon dogs and serows in Kyushu Island (OCc1-5 from Oita and SgN from Saga). Kyushu and Gifu are geographically distinct (> 550 km) and separated by sea. Thus, it is unlikely that this genetic relationship was formed by a natural event. Rather, this link could probably be owed to the artificial translocation of mangy animals, such as domestic/companion dogs. However, there are, of course, possible alternative hypotheses for the observed close genetic relationships between distant Sarcoptes mite populations, such as (i) an effect of a small sample size in mite populations between the two distant mite populations (i.e. limited observation and un-detection of similar genotypes), and (ii) a chance effect of mutation and convergence of genetic characteristics. Future research is needed for understanding of this type of genetic relationships and also the possible artificial impact for the expanding distribution of sarcoptic mange.
Conclusions
The results of our study show that in Japan, Sarcoptes mite populations follow a “Host–taxon” regulation between wild boars and Caniformia with Japanese serow communities but deviate between the Caniformia and Japanese serow. To our knowledge, this result provides the first genetic evidence for a possible hidden transmission of sarcoptic mange between host species belonging to different taxa without unambiguous predator–prey systems. In the diverted transmission patterns of S. scabiei among multi-host systems worldwide, there may be cryptic relationships of S. scabiei populations in local mammalian communities. Although the effect of sarcoptic mange in wildlife species is still poorly understood [54], the understanding of the cryptic transmission of sarcoptic mange in local mammalian communities is necessary for the conservation of remnant and fragmented populations of wild mammals. In fact, sarcoptic mange is considered as one of the causes for the population decline in a remnant Japanese serow population on the Kyushu island of Japan [25]. For the preservation of vulnerable populations of Japanese serows and their related Capricornis species, sarcoptic mange infection from the Caniformia species, in addition to the possibility of artificial translocation of mangy animals (i.e. domestic/companion dogs), should be deliberated in wildlife management programs.
Additional files
Additional file 1: Table S1. Results of Hardy–Weinberg equilibrium tests for each microsatellite locus for each Host-associated Sarcoptes mite population.
Additional file 2: Figure S1. Population structure as inferred by STRUCTURE analysis of Sarcoptes mites in the present study. a The graph for K = 9 with the 93 mites. b The graph for K = 4 with 80 mites (Caniformia- and Japanese serow-derived mites, excluding wild boar-derived mites from 93 mites). Each mite genotype is represented by a single vertical bar plot. Each color represents one cluster, and the proportion of each color in each bar plot shows the likelihood of assignment in the inferred clusters. Mite populations associated with each host animals are separated by thick black lines. 1, TCf; 2, TN; 3, KN1-8; 4, GCf1-9; 5, GN1-23; 6, WCf; 7, WM; 8, WN1-26; 9, WP; 10, SgN1-2; 11, StCc; 12, WCc; 13, OCc1-5; 14, WS; 15, HS1-5; 16, SmS1-6; 17, YS. For host population abbreviations see Table 1.
Acknowledgements
We thank H. Kitagawa (Gifu University), S. Baba (Baba Dermatology Clinic), M. Kaneda (Biodiversity Network Japan), K. Ito (Yuinoshima Animal Hospital), S. Yamazaki (Mie Prefectural Government), K. Suzuki (Hikiiwa Park Center), R. Yasuda (Misato-Cho Town Office), H. Sato (Yamaguchi University), M. Yasuda (Forestry and Forest Products Research Center), A. Nakazono (Kyushu Natural Environment Research co., ltd) and T. Sakata (Kumamoto Wildlife Society) as well as many cooperative veterinary hospitals in Gifu Prefecture for their generous cooperation with mite collection. We also thank H. Tsuruga, M. Kondo (Hokkaido Research Institute for Environmental Sciences), T. Morimoto, J. Moribe, M. Sugiyama, M. Tsuruta (Gifu University) and H. Kawabata (National Institute of Infectious Diseases) for their invaluable technical advice and helpful comments.
Abbreviations
- HO
observed heterozygosity
- HE
expected heterozygosity
- HWE
Hardy–Weinberg equilibrium
- LD
linkage disequilibrium
- LE
linkage equilibrium
- MCMC
Marcov Chain Monte Carlo
- PCA
principal components analysis
- PCR
polymerase chain reaction
- SSR
simple sequence repeat
Authors’ contributions
RM designed the study. RM, TY, TO, TT, MB, MY, NK, TKd, TKt, MS and MA performed the sampling of mangy animals and their crafted skins in which S. scabiei were included. RM, TY and NS conducted the SSR genotyping and genetic analyses. RM wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Grant from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, for the Joint Research Program of the Research Center for Zoonosis Control, Hokkaido University.
Availability of data and materials
Data supporting the conclusions of this article are included within the article and its additional files. Raw data are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
All the companion dogs were sampled during the diagnosis or treatment after the informed consent of their owners.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ryota Matsuyama, Email: rmatasuyama@hiroshima-u.ac.jp.
Toshihiro Yabusaki, Email: m8023025@gmail.com.
Natsuko Senjyu, Email: haibeefriend@yahoo.co.jp.
Tsukasa Okano, Email: okano-tsukasa@pref.gifu.lg.jp.
Minoru Baba, Email: baba001@kmnh.jp.
Tomoka Tsuji-Matsukane, Email: tomoka.matsukane@gmail.com.
Mayumi Yokoyama, Email: yokoyama@wmi-hyogo.jp.
Nobuhide Kido, Email: kido@hama-midorinokyokai.or.jp.
Teruki Kadosaka, Email: kadosaka@aichi-med-u.ac.jp.
Takuya Kato, Email: tkato@nvlu.ac.jp.
Masatsugu Suzuki, Email: mszk@gifu-u.ac.jp.
Makoto Asano, Email: asanojr@gifu-u.ac.jp.
References
- 1.Pence DB, Ueckermann E. Sarcoptic mange in wildlife. Rev Sci Tech. 2002;21:385–398. doi: 10.20506/rst.21.2.1335. [DOI] [PubMed] [Google Scholar]
- 2.Kołodziej-Sobocińska M, Zalewski A, Kowalczyk R. Sarcoptic mange vulnerability in carnivores of the Białowieża Primeval Forest, Poland: underlying determinant factors. Ecol Res. 2014;29:237–244. doi: 10.1007/s11284-013-1118-x. [DOI] [Google Scholar]
- 3.Lindström ER, Andrén H, Angelstam P, Cederlund G, Hörnfeldt B, Jäderberg L, et al. Disease reveals the predator: sarcoptic mange, red fox predation, and prey populations. Ecology. 1994;75:1042–1049. doi: 10.2307/1939428. [DOI] [Google Scholar]
- 4.Oleaga Á, Casais R, Balseiro A, Espí A, Llaneza L, Hartasánchez A, et al. New techniques for an old disease: sarcoptic mange in the Iberian wolf. Vet Parasitol. 2011;181:255–266. doi: 10.1016/j.vetpar.2011.04.036. [DOI] [PubMed] [Google Scholar]
- 5.Soulsbury CD, Iossa G, Baker PJ, Cole N, Funk SM, Harris S. The impact of sarcoptic mange Sarcoptes scabiei on the British fox Vulpes vulpes population. Mamm Rev. 2007;37:278–296. [Google Scholar]
- 6.Uraguchi K, Ueno M, Iijima H, Saitoh T. Demographic analyses of a fox population suffering from sarcoptic mange. J Wildl Manag. 2014;78:1356–1371. doi: 10.1002/jwmg.794. [DOI] [Google Scholar]
- 7.Sugiura N, Doi K, Kato T, Morita T, Hayama S-I. Epizootic of sarcoptic mange in raccoon dogs (Nyctereutes procyonoides) in relation to population density. J Vet Med Sci. 2018;80:544–548. doi: 10.1292/jvms.17-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arlian L. Biology, host relations, and epidemiology of Sarcoptes scabiei. Annu Rev Entomol. 1989;34:139–161. doi: 10.1146/annurev.en.34.010189.001035. [DOI] [PubMed] [Google Scholar]
- 9.Alasaad S, Rossi L, Heukelbach J, Pérez JM, Hamarsheh O, Otiende M, et al. The neglected navigating web of the incomprehensibly emerging and re-emerging Sarcoptes mite. Infect Genet Evol. 2013;17:253–259. doi: 10.1016/j.meegid.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Fain A. Etude de la variabilité de Sarcoptes scabiei avec une révision des Sarcoptidae. Acta Zool Pathol Antverp. 1968;47:1–196. [Google Scholar]
- 11.Walton SF, Choy JL, Bonson A, Valle A, McBroom J, Taplin D, et al. Genetically distinct dog-derived and human-derived Sarcoptes scabiei in scabies-endemic communities in northern Australia. Am J Trop Med Hyg. 1999;61:542–547. doi: 10.4269/ajtmh.1999.61.542. [DOI] [PubMed] [Google Scholar]
- 12.Walton SF, Dougall A, Pizzutto S, Holt D, Taplin D, Arlian LG, et al. Genetic epidemiology of Sarcoptes scabiei (Acari: Sarcoptidae) in northern Australia. Int J Parasitol. 2004;34:839–849. doi: 10.1016/j.ijpara.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Walton SF, Currie BJ, Kemp DJ. A DNA fingerprinting system for the ectoparasite Sarcoptes scabiei. Mol Biochem Parasitol. 1997;85:187–196. doi: 10.1016/S0166-6851(96)02825-3. [DOI] [PubMed] [Google Scholar]
- 14.Alasaad S, Sarasa M, Heukelbach J, Mijele D, Soriguer RC, Zhu X-Q, Rossi L. Advances in studies of disease-navigating webs: Sarcoptes scabiei as a case study. Parasites Vectors. 2014;7:16. doi: 10.1186/1756-3305-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasero R, Rossi L, Soglia D, Maione S, Sacchi P, Rambozzi L, et al. Host taxon-derived Sarcoptes mite in European wild animals revealed by microsatellite markers. Biol Conserv. 2010;143:1269–1277. doi: 10.1016/j.biocon.2010.03.001. [DOI] [Google Scholar]
- 16.Alasaad S, Oleaga Á, Casais R, Rossi L, Min A, Soriguer RC, Gortázar C. Temporal stability in the genetic structure of Sarcoptes scabiei under the Host–taxon law: empirical evidences from wildlife-derived Sarcoptes mite in Asturias, Spain. Parasites Vectors. 2011;4:151. doi: 10.1186/1756-3305-4-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentería-Solís Z, Min A, Alasaad S, Müller K, Michler FU, Schmäschke R, et al. Genetic epidemiology and pathology of raccoon-derived Sarcoptes mites from urban areas of Germany. Med Vet Entomol. 2014;28:98–103. doi: 10.1111/mve.12079. [DOI] [PubMed] [Google Scholar]
- 18.Gakuya F, Rossi L, Ombui J, Maingi N, Muchemi G, Ogara W, et al. The curse of the prey: Sarcoptes mite molecular analysis reveals potential prey-to-predator parasitic infestation in wild animals from Masai Mara, Kenya. Parasites Vectors. 2011;4:193. doi: 10.1186/1756-3305-4-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gakuya F, Ombui J, Maingi N, Muchemi G, Ogara W, Soriguer RC, et al. Sarcoptic mange and cheetah conservation in Masai Mara (Kenya): epidemiological study in a wildlife/livestock system. Parasitology. 2012;139:1587–1595. doi: 10.1017/S0031182012000935. [DOI] [PubMed] [Google Scholar]
- 20.Oleaga A, Alasaad S, Rossi L, Casais R, Vicente J, Maione S, et al. Genetic epidemiology of Sarcoptes scabiei in the Iberian wolf in Asturias, Spain. Vet Parasitol. 2013;196:453–459. doi: 10.1016/j.vetpar.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Wildlife Management Office Inc . An urgent report of the epidemiological survey on sarcoptic mange of Japanese mammals. Tokyo: Ministry of the Environment; 1998. [Google Scholar]
- 22.Shibata A. Studies on mange of wildlife in Japan. Musashino: Nippon Veterinary Life Science University; 2004. [Google Scholar]
- 23.Kido N, Itabashi M, Takahashi M, Futami M. Epidemiology of sarcoptic mange in free-ranging raccoon dogs (Nyctereutes procyonoides) in Yokohama, Japan. Vet Parasitol. 2013;191:102–107. doi: 10.1016/j.vetpar.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi M, Nogami S, Misumi H, Matsumoto M, Takahama M, Uchikawa K. Mixed infestation of sarcoptic and chorioptic mange mites in Japanese serow, Capricornis crispus Temminck, 1845 in Japan, with a description of Chorioptes japonensis sp. nov. (Acari : Psoroptidia) Med Entomol Zool. 2001;52:297–306. doi: 10.7601/mez.52.297. [DOI] [Google Scholar]
- 25.Oita Prefectural Board of Education, Kumamoto Prefectural Board of Education, Miyazaki Prefectural Board of Education. Reports on the investigation of Japanese serow inhabiting in the Kyushu mountainous region. 2013. https://www.pref.miyazaki.lg.jp/ky-bunka/kanko/bunka/page00032.html. Accessed 20 June 2019.
- 26.Matsuyama R, Yabusaki T, Kuninaga N, Morimoto T, Okano T, Suzuki M, et al. Coexistence of two different genotypes of Sarcoptes scabiei derived from companion dogs and wild raccoon dogs in Gifu, Japan: the genetic evidence for transmission between domestic and wild canids. Vet Parasitol. 2015;212:356–360. doi: 10.1016/j.vetpar.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Namae M, Aoi T. Analysis of some topics related to dead Japanese serows (the surroundings and environment of the discovery spots, season, and the age structure of death of the serows) with a report of losses in Morioka. Bull Iwate Univ For. 2007;38:23–37. [Google Scholar]
- 28.Kauhala K. Reproductive strategies of the raccoon dog and the red fox in Finland. Acta Theriol. 1996;41:51–58. doi: 10.4098/AT.arch.96-4. [DOI] [Google Scholar]
- 29.Arai S, Takayuki A, Yoshiko K, Yoshida K. Food habit of the Japanese marten (Martes melampus) at Kuju Highland in Kyusyu, Japan. Honyurui-Kagaku (Mammalian Science). 2003;43:19–28. [Google Scholar]
- 30.Bartoszewicz M, Okarma H, Zalewski A, Szczesna J. Ecology of the raccoon (Procyon lotor) from western Poland. Ann Zool Fennici. 2008;45:291–298. doi: 10.5735/086.045.0409. [DOI] [Google Scholar]
- 31.Koba Y, Sakaguchi M, Muraoka R, Obitsu T, Tanida H. Food habitat of wild boars in Kami-Kamafari island in Hiroshima. Honyurui-Kagaku (Mammal Science). 2009;49:207–215. [Google Scholar]
- 32.Alasaad S, Soglia D, Maione S, Sartore S, Soriguer RC, Pérez JM, et al. Effectiveness of the postponed isolation (post-frozen isolation) method for PCR-quality Sarcoptes mite gDNA. Exp Appl Acarol. 2009;47:173–178. doi: 10.1007/s10493-008-9196-0. [DOI] [PubMed] [Google Scholar]
- 33.Alasaad S, Soglia D, Sarasa M, Soriguer RC, Pérez JM, Granados JE, et al. Skin-scale genetic structure of Sarcoptes scabiei populations from individual hosts: empirical evidence from Iberian ibex-derived mites. Parasitol Res. 2008;104:101–105. doi: 10.1007/s00436-008-1165-3. [DOI] [PubMed] [Google Scholar]
- 34.Raymond M, Rousset F. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. doi: 10.1093/oxfordjournals.jhered.a111573. [DOI] [Google Scholar]
- 35.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 36.R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 37.Jombart T, Devillard S, Dufour A-B, Pontier D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity. 2008;101:92–103. doi: 10.1038/hdy.2008.34. [DOI] [PubMed] [Google Scholar]
- 38.Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 39.Langella O. POPULATIONS 1.2.28. Population genetic software (individuals or populations distances, phylogenetic trees). 2002. http://bioinformatics.org/~tryphon/populations/. Accessed 20 June 2019.
- 40.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 41.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 43.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 44.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 45.Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2003;4:137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- 46.Grupe G, Krüger HH. Feeding ecology of the stone and pine marten revealed by element analysis of their skeletons. Sci Total Environ. 1990;90:227–240. doi: 10.1016/0048-9697(90)90195-Z. [DOI] [PubMed] [Google Scholar]
- 47.Macdonald D, Reynolds J. Red fox. In: Sillero-Zubiri C, Hoffmann M, Mcdonald D, editors. Status survey and conservation action plan, Canids: foxes, wolves, jackals dogs. Cambridge: International Union for Conservation of Nature and Natural Resources; 2004. pp. 129–136. [Google Scholar]
- 48.Kishimoto R, Kawamichi T. Territoriality and monogamous pairs in a solitary ungulate, the Japanese serow, Capricornis crispus. Anim Behav. 1996;52:673–682. doi: 10.1006/anbe.1996.0212. [DOI] [Google Scholar]
- 49.Chen CC, Pei KJC, Lai YC, Mortenson JA. Participatory epidmeiology to assess sarcoptic mange in serow of Taiwan. J Wildl Dis. 2012;48:869–875. doi: 10.7589/2011-07-189. [DOI] [PubMed] [Google Scholar]
- 50.Barron MC, Pech RP, Whitford J, Yockney IJ, De Lisle GW, Nugent G. Longevity of Mycobacterium bovis in brushtail possum (Trichosurus vulpecula) carcasses, and contact rates between possums and carcasses. N Z Vet J. 2011;59:209–217. doi: 10.1080/00480169.2011.595905. [DOI] [PubMed] [Google Scholar]
- 51.Meckel LA, McDaneld CP, Wescott DJ. White-tailed deer as a taphonomic agent: photographic evidence of white-tailed deer gnawing on human bone. J Forensic Sci. 2018;63:292–294. doi: 10.1111/1556-4029.13514. [DOI] [PubMed] [Google Scholar]
- 52.Tsujino R, Yumoto T. Habitat preferences of medium/large mammals in human disturbed forests in Central Japan. Ecol Res. 2014;29:701–710. doi: 10.1007/s11284-014-1159-9. [DOI] [Google Scholar]
- 53.Arlian LG, Morgan MS. A review of Sarcoptes scabiei: past, present and future. Parasites Vectors. 2017;10:297. doi: 10.1186/s13071-017-2234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Astorga F, Carver S, Almberg ES, Sousa GR, Wingfield K, Niedringhaus KD, et al. International meeting on sarcoptic mange in wildlife, June 2018, Blacksburg, Virginia, USA. Parasites Vectors. 2018;11:449. doi: 10.1186/s13071-018-3015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Results of Hardy–Weinberg equilibrium tests for each microsatellite locus for each Host-associated Sarcoptes mite population.
Additional file 2: Figure S1. Population structure as inferred by STRUCTURE analysis of Sarcoptes mites in the present study. a The graph for K = 9 with the 93 mites. b The graph for K = 4 with 80 mites (Caniformia- and Japanese serow-derived mites, excluding wild boar-derived mites from 93 mites). Each mite genotype is represented by a single vertical bar plot. Each color represents one cluster, and the proportion of each color in each bar plot shows the likelihood of assignment in the inferred clusters. Mite populations associated with each host animals are separated by thick black lines. 1, TCf; 2, TN; 3, KN1-8; 4, GCf1-9; 5, GN1-23; 6, WCf; 7, WM; 8, WN1-26; 9, WP; 10, SgN1-2; 11, StCc; 12, WCc; 13, OCc1-5; 14, WS; 15, HS1-5; 16, SmS1-6; 17, YS. For host population abbreviations see Table 1.
Data Availability Statement
Data supporting the conclusions of this article are included within the article and its additional files. Raw data are available from the corresponding author upon reasonable request.