Abstract
Background
The peroxiredoxins (PRDXs) gene family has been demonstrated to participate in carcinogenesis and development of numerous cancers and the prognostic values in several cancers have been evaluated already. Purpose of our research is to explore the expression profiles and prognostic values of PRDXs in breast cancer (BrCa).
Methods
The transcriptional levels of PDRX family members in primary BrCa tissues and their association with intrinsic subclasses were analyzed using UALCAN database. Then, the genetic alterations of PDRXs were examined by cBioPortal database. Moreover, the prognostic values of PRDXs in BrCa patients were investigated via the Kaplan-Meier plotter.
Results
The transcriptional levels of most PRDXs family members in BrCa tissues were significantly elevated compared with normal breast tissues. Meanwhile, dysregulated PRDXs expression was associated with intrinsic subclasses of BrCa. Besides, copy number alterations (CNA) of PRDXs positively regulated their mRNA expressions. Furthermore, high mRNA expression of PRDX4/6 was significantly associated with poor overall survival (OS) in BrCa patients, while high mRNA expression of PRDX3 was notably related to favorable OS. Simultaneously, high mRNA expression of PRDX1/2/4/5/6 was significantly associated with shorter relapse-free survival (RFS) in BrCa patients, while high mRNA expression of PRDX3 was notably related to favorable RFS. In addition, the prognostic value of PRDXs in the different clinicopathological features based on intrinsic subclasses and chemotherapeutic treatment of BrCa patients was further assessed in the KM plotter database.
Conclusion
Our findings systematically elucidate the expression profiles and distinct prognostic values of PRDXs in BrCa, which might provide novel therapeutic targets and potential prognostic biomarkers for BrCa patients.
Keywords: PRDX, Bioinformatic analysis, Gene expression, Prognostic, Breast cancer
Background
Breast cancer (BrCa) has the highest morbidity among all female’s cancers worldwide, which may cause 41,760 cancer-related deaths in the United States (US) in 2019 according to the prediction by the American Cancer Society (ACS) [1]. Being a multifaceted disease, BrCa can be classified into various subclasses based on the expression status of estrogen receptor (ER), progesterone receptor (PR), epidermal growth factor receptor 2 (HER2) and antigen ki-67 (Ki-67), which suggest different therapeutic guidance and prognostic implications for BrCa patients [2]. Although the constant amelioration of comprehensive therapies for BrCa has significantly decreased the mortality of BrCa in recent years, it is still necessary to further explore the potential mechanism of oncogenesis and progression of BrCa.
Peroxiredoxins (PRDXs), a family of antioxidant enzymes in eukaryotes, containing six isoforms (PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6), which catalyze the reduction reaction of peroxide and maintain the balance of intracellular hydrogen peroxide (H2O2) levels [3]. As momentous regulators in diverse signaling pathways, PRDXs are of great significance to the signal transduction and cells metabolism [4]. Expression of PRDXs will be significantly upregulated when cells are under oxidative stress conditions. Several researches have suggested that overexpression of PRDXs may play dichotomous role in oncogenesis of tumors, where they could either stimulate the progression of cancers or suppress the development of cancers [5]. An increasing number of studies have observed the preliminary functions and ambiguous prognostic values of PRDXs in cancerous diseases [6–9]. However, the expression profiles and prognostic values of PRDXs in BrCa samples are still elusive up to now. Our research aims to explore the differential expression and potential roles of PRDXs in BrCa.
The Cancer Genome Atlas (TCGA) program, which launched by the US National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), attempts to sequence the entire genome of more than 10,000 tumor samples and to distinguish the genetic changes specific for each cancer [10–12]. Along with the successful implementation of the TCGA project, massive genomic information is accumulating exponentially. Over the past few years, many interactive and user-friendly online platforms based on the TCGA database greatly elevate the efficiency of TCGA database analysis and increasing amounts of tumor biomarkers have been identified on the strength of these websites [13, 14].
Therefore, in the current research, we first compared the transcriptional levels of PRDXs in BrCa and adjacent breast tissues using UALCAN database. In addition, the cBioPortal database was used to analyze the genetic alterations of PDRXs and the correlation with transcriptional levels. Moreover, the Kaplan-Meier plotter database was used to assess the prognostic effects of PRDXs mRNA expression in patients with BrCa. Overall, our research preliminarily but systematically characterizes the expression profiles of PRDXs in BrCa and reveals that the detection of the PRDXs expression status of BrCa patients may be valuable and potential biomarkers for prognostic assessment.
Materials and methods
Gene expression analysis via UALCAN
UALCAN (http://ualcan.path.uab.edu/) is an online open-access platform based on level 3 RNA-seq and clinical information from TCGA database [15]. It can be used to analyze relative transcriptional levels of potential genes of interest between cancerous and paired normal tissues and association of the transcriptional levels with clinicopathologic features. In the current study, UALCAN was applied to analyze the transcriptional levels of PDRXs family members in primary BrCa tissues and their association with intrinsic subclasses. All the BrCa cases available on UALCAN were included in our research.
Data-mining analysis based on cBioPortal
cBioPortal (www.cbioportal.org/) is a user-friendly, interactive website resource and provides visualization, analysis, and download of large-scale cancer genomics datasets [16, 17]. In the current study, we analyzed the genetic alterations of PDRXs family members, which contained mutations and putative copy-number alterations (CNA) from GISTIC. Furthermore, we download the data of putative copy-number alterations and mRNA expression z-Scores to evaluate the association between various CNAs and transcriptional levels of PRDXs. Tumor samples with RNA-seq and CAN data on cBioPortal were included in our research which contains total 1076 BrCa samples.
Survival analysis by Kaplan-Meier plotter
Kaplan-Meier Plotter (KM Plotter, http://kmplot.com/analysis/) is an online database containing gene expression profiles and survival information of cancer patients [18]. The prognostic values of PRDXs (PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6) at mRNA level in BrCa was analyzed using all BrCa samples available on KM Plotter. The patients’ cohorts were split at the median expression of each PRDXs mRNA level. The subgroup analysis of the prognostic value of PRDXs in BrCa patients was further performed according to intrinsic subclasses and different regimens of chemotherapy. All cohorts were compared with Kaplan-Meier survival plots. Hazard ratio (HR), 95% confidence interval (95% CI), and log-rank P value were calculated and displayed online.
Statistical analysis
All statistical analyses were performed on the bioinformatics database online or using SPSS 25.0 software (Chicago, IL). The differential mRNA expression of PRDXs in BrCa tissues was analyzed by Student’s t-test. Kaplan-Meier survival plots were generated with survival curves compared by log-rank test. For all analyses, Differences were considered statistically significant if P values were less than 0.05.
Results
Transcriptional levels of PRDXs in BrCa samples
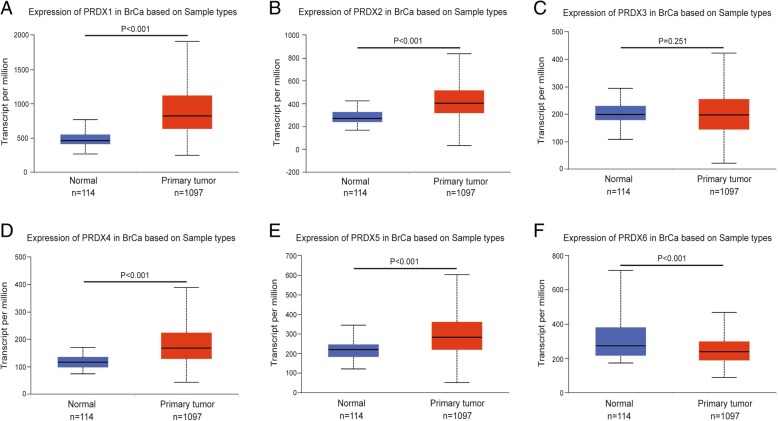
In order to evaluate the exact expression profiles of PRDXs members in BrCa patients, the differential transcriptional levels of PRDX family members between BrCa and paired normal breast tissue was evaluated by UALCAN database. As shown in Fig. 1, the transcriptional level of PRDX1 (Fig. 1a, P < 0.001), PRDX2 (Fig. 1b, P < 0.001), PRDX4 (Fig. 1d, P < 0.001), and PRDX5 (Fig. 1e, P < 0.001) was significantly upregulated in BrCa tissues compared with paracancerous tissues. However, the transcriptional level of PRDX3 (Fig. 1c, P = 0.251) showed a non-significant difference in BrCa tissues compared with paracancerous tissues. Besides, the transcriptional level of PRDX6 was significantly downregulated in BrCa tissues compared with paracancerous tissues (Fig. 1f, P < 0.001).
Fig. 1.
Transcriptional levels of PRDXs in paracancerous and BrCa tissues. Comparison of PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, PRDX6 mRNA expression in paracancerous (n = 114) and BrCa (n = 1097) tissues in TVGA dataset based on data mining via UALCAN. a, b, d, e) The transcriptional level of PRDX1, PRDX2, PRDX4, and PRDX5 was significantly upregulated in BrCa tissues compared with paracancerous tissues. c The transcriptional level of PRDX3 showed a non-significant difference in BrCa tissues compared with paracancerous tissues. f The transcriptional level of PRDX6 was significantly downregulated in BrCa tissues compared with paracancerous tissues
Transcriptional levels of PRDXs in different BrCa subclasses
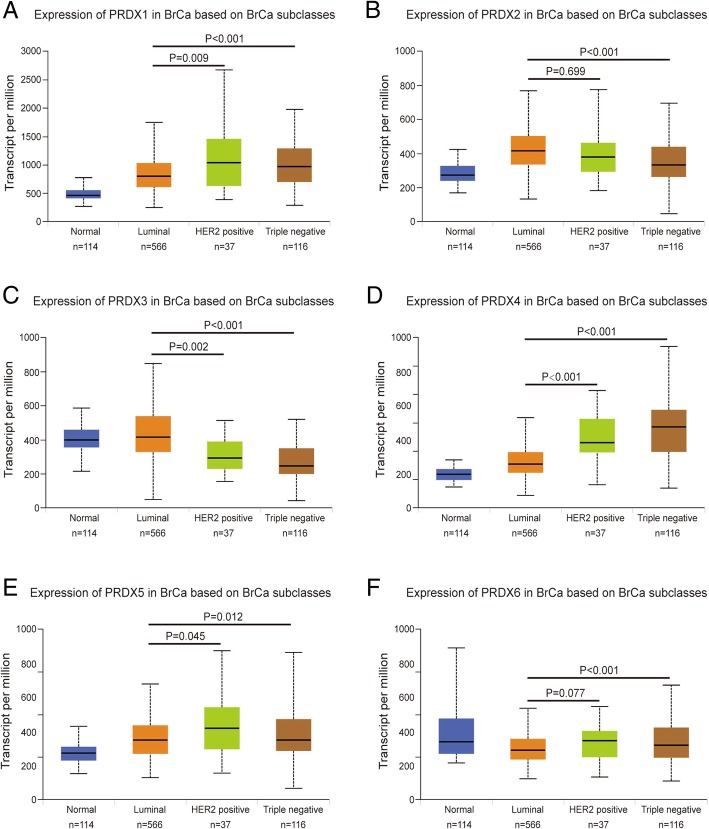
Classification of intrinsic subclasses is helpful in the prediction of therapeutic response and prognosis of BrCa [19]. So, we next compared the differential transcriptional levels of PRDX family members according to different intrinsic subclasses of BrCa. As shown in Fig. 2, mRNA expressions of PRDXs family members were significantly correlated with intrinsic subclasses of BrCa. Patients who were with HER2-positive and triple-negative subclasses BrCa tended to express higher PRDXs (exclude PRDX2, PRDX3) mRNA, while express lower PRDX2 and PRDX3 mRNA. The highest mRNA expressions of PRDX1/5/6 were found in HER2-positive tissues (Fig. 2a, e, f), and the highest mRNA expressions of PRDX4 were found in triple-negative tissues (Fig. 2d). Besides, the lowest mRNA expressions of PRDX2/3 were found in triple-negative tissues (Fig. 2b, c). Taken together, these findings above revealed that transcriptional levels of PRDXs family members were significantly correlated with intrinsic subclasses in BrCa patients.
Fig. 2.
Transcriptional levels of PRDXs in various BrCa subclasses. The transcriptional level of PRDXs in BrCa patients with different subclasses, PRDXs mRNA was significantly downregulated (PRDX3) or upregulated (other PRDXs) in HER2-positive and triple-negative BrCa tissues compared with luminal BrCa tissues. a PRDX1. b PRDX2. c PRDX3. d PRDX4. e PRDX5. f PRDX6
Genetic alterations of PRDXs in BrCa samples
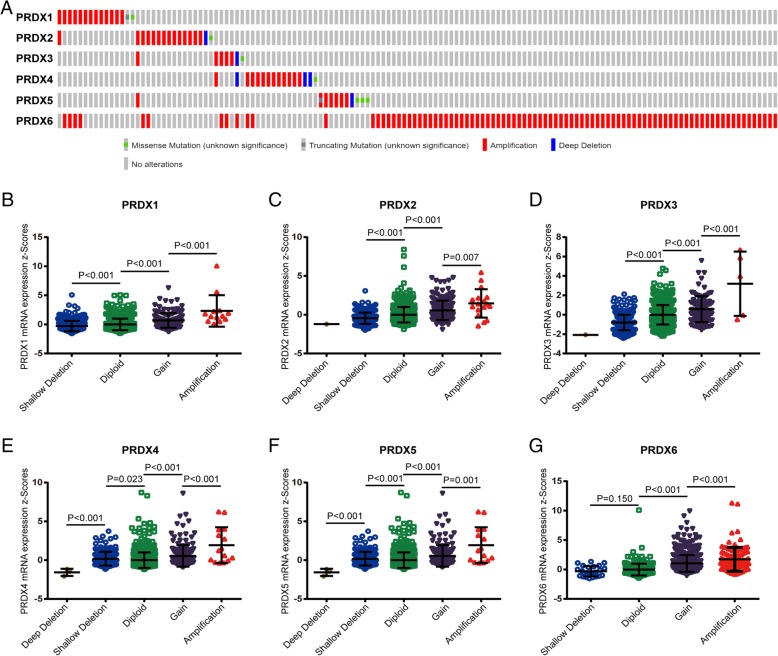
DNA copy number alterations (CNA) are most common genetic alterations which participates in oncogenesis of cancers via regulating cancer-related gene expression [20–22]. In the fact that most PRDX family members was dysregulated in BrCa tissues, we speculated that DNA CNA may regulate the transcriptional levels of PRDXs. Next, we analyzed genetic alteration in PRDXs and correlations with their mRNA expressions based on cBioPortal website. As shown in Fig. 3a and Table 1, low amplification rate of PRDXs was found in BrCa patients. However, although copy gain (gain and amplification) of PRDXs was not frequent, it was still associated with notably upregulated PRDXs mRNA levels compared with the copy-neutral (diploid) and copy-loss (shallow deletion and deep deletion) cases (Fig. 3b-g). To conclude, the results suggested that PRDX mRNA expressions were regulated by their DNA copy number alterations.
Fig. 3.
Correlation between the genetic alterations of PRDXs and mRNA levels in BrCa tissues. a Oncoprint in cBioPortal database exhibited the proportion and distribution of specimens with genetic alterations in PRDXs. The Figure was cropped on the right to exclude specimens without any alterations. b-g Copy gain (gain and amplification) of PRDXs was associated with notably upregulated PRDXs mRNA levels compared with the copy-neutral (diploid) and copy-loss (shallow deletion and deep deletion) cases. b PRDX1. c PRDX2. d PRDX3. e PRDX4. f PRDX5. g PRDX6
Table 1.
Frequency and proportion of genetic alterations of PRDXs in BrCa
| PRDXs | Deep deletion | Shallow deletion | Dipliod | Gain | Amplification |
|---|---|---|---|---|---|
| PRDX1 | 0 (0.00%) | 297 (27.60%) | 623 (57.90%) | 143 (13.29%) | 13 (1.21%) |
| PRDX2 | 1 (0.09%) | 226 (21.00%) | 646 (60.04%) | 186 (17.29%) | 17 (1.58%) |
| PRDX3 | 1 (0.09%) | 321 (29.83%) | 651 (60.50%) | 98 (9.11%) | 5 (0.46%) |
| PRDX4 | 3 (0.28%) | 184 (17.10%) | 706 (65.61%) | 168 (15.61%) | 15 (1.39%) |
| PRDX5 | 1 (0.09%) | 242 (22.49) | 657 (61.06%) | 167 (15.52%) | 9 (0.84%) |
| PRDX6 | 0 (0.00%) | 24 (2.23%) | 262 (24.35%) | 689 (64.03%) | 101 (9.39%) |
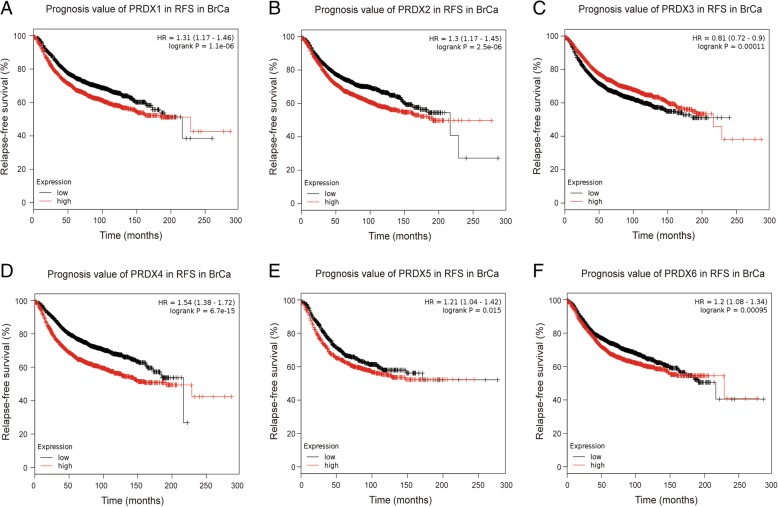
Prognostic values of PRDXs mRNA expression in all BrCa samples
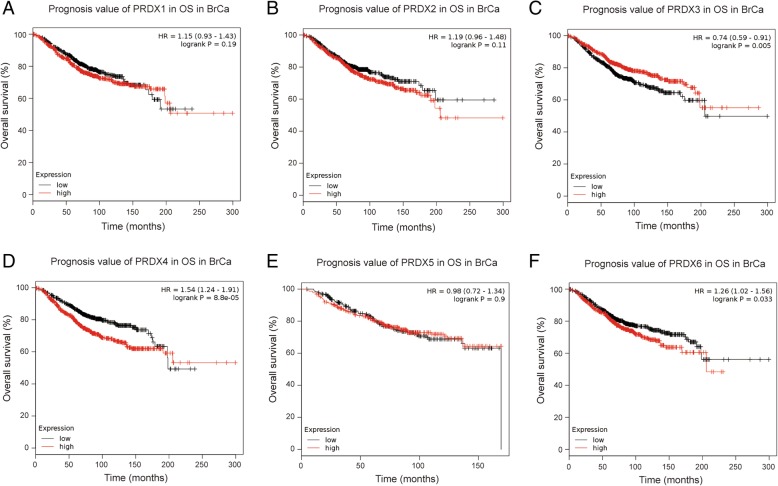
Further, we employed KM plotter to evaluate the prognostic values of PRDX family members. As shown in Fig. 4 and Fig. 5, high mRNA expression of PRDX4 (HR = 1.54, 95% CI: 1.24–1.91, P < 0.001), and PRDX6 (HR = 1.26, 95% CI: 1.02–1.56, P = 0.033) were significantly associated with poor overall survival (OS) of BrCa patients, while high mRNA expression of PRDX3 was notably related to favorable OS of BrCa patients (HR = 0.74, 95% CI: 0.59–0.91, P = 0.005). However, other PRDXs mRNA expression showed a null association with prognosis of BrCa patients.
Fig. 4.
Prognostic value of PRDXs mRNA in all BrCa patients (OS). OS curves were plotted to evaluate the prognostic value of PRDXs mRNA expression. a PRDX1 (208680_at). b PRDX2 (39729_at). c PRDX3 (201619_at). d PRDX4 (201923_at). e PRDX5 (1560587_s_at). f PRDX6 (200845_s_at)
Fig. 5.
Prognostic value of PRDXs mRNA in BrCa patients (RFS). RFS curves were plotted to evaluate the prognostic value of PRDXs mRNA expression. a PRDX1 (208680_at). b PRDX2 (39729_at). c PRDX3 (201619_at). d PRDX4 (201923_at). e PRDX5 (1560587_s_at). f PRDX6 (200845_s_at)
We next analyzed the associations between PRDXs mRNA expression and RFS of BrCa patients and found that high mRNA expression of PRDX1 (HR = 1.31, 95% CI: 1.17–1.46, P < 0.001), PRDX2 (HR = 1.30, 95% CI: 1.17–1.45, P < 0.001), PRDX4 (HR = 1.54, 95% CI: 1.38–1.72, P < 0.001), PRDX5 (HR = 1.21, 95% CI: 1.04–1.42, P = 0.015) and PRDX6 (HR = 1.20, 95% CI: 1.08–1.34, P < 0.001) were significantly associated with shorter relapse-free survival (RFS) of BrCa patients, while high mRNA expression of PRDX3 was notably related to favorable RFS of BrCa patients (HR = 0.81, 95% CI: 0.72–0.90, P < 0.001). Overall, the findings above implied that mRNA expressions of PRDX3/4/6 were remarkably correlated with BrCa patients’ both OS and RFS, which might be identified as promising biomarkers to predict the survival of BrCa patients.
Prognostic values of PRDXs mRNA in different BrCa subclasses
To further analyze the association of PRDXs mRNA expression with various BrCa subclasses, we detected the survival effects of PRDX family members in 4 subclasses of BrCa patients, including basal-like, luminal A, luminal B, and HER2 positive BrCa. As shown in Table 2, for PRDX1 (HR = 1.52, 95% CI: 1.06–2.17, P = 0.022), PRDX4 (HR = 1.58, 95% CI: 1.10–2.27, P = 0.012), and PRDX6 (HR = 1.49, 95% CI: 1.04–2.13, P = 0.027), high mRNA expression was associated with unfavorable OS in luminal A BrCa patients, respectively. Besides, high mRNA expression of PRDX4 (HR = 1.55, 95% CI: 1.06–2.27, P = 0.023) predicted poor OS in luminal B BrCa patients as well.
Table 2.
Association between prognostic value of PRDXs mRNA expression and different subclasses in BrCa
| Subclasses | Cases | HR | 95% CI | P value | Cases | HR | 95% CI | P value |
|---|---|---|---|---|---|---|---|---|
| OS | RFS | |||||||
| PRDX1 | ||||||||
| Basal-like | 241 | 0.66 | 0.40–1.09 | 0.104 | 618 | 1.02 | 0.80–1.32 | 0.850 |
| Luminal A | 611 | 1.52 | 1.06–2.17 | 0.022 | 1933 | 1.23 | 1.06–1.49 | 0.009 |
| Luminal B | 433 | 0.98 | 0.68–1.43 | 0.956 | 1149 | 1.20 | 0.99–1.46 | 0.060 |
| HER2 positive | 117 | 1.27 | 0.66–2.42 | 0.470 | 251 | 1.31 | 0.89–1.92 | 0.170 |
| PRDX2 | ||||||||
| Basal-like | 241 | 1.14 | 0.70–1.85 | 0.613 | 618 | 1.25 | 0.97–1.61 | 0.078 |
| Luminal A | 611 | 1.42 | 0.99–2.02 | 0.056 | 1933 | 1.23 | 1.04–1.46 | 0.016 |
| Luminal B | 433 | 0.99 | 0.67–1.44 | 0.960 | 1149 | 1.67 | 1.37–2.03 | < 0.001 |
| HER2 positive | 117 | 0.93 | 0.49–1.77 | 0.821 | 251 | 1.24 | 0.85–1.83 | 0.264 |
| PRDX3 | ||||||||
| Basal-like | 241 | 0.77 | 0.47–1.26 | 0.299 | 618 | 0.77 | 0.60–1.00 | 0.046 |
| Luminal A | 611 | 0.74 | 0.52–1.05 | 0.090 | 1933 | 0.87 | 0.73–1.03 | 0.112 |
| Luminal B | 433 | 1.02 | 0.70–1.48 | 0.919 | 1149 | 1.17 | 0.96–1.41 | 0.115 |
| HER2 positive | 117 | 0.59 | 0.30–1.15 | 0.118 | 251 | 0.73 | 0.49–1.07 | 0.102 |
| PRDX4 | ||||||||
| Basal-like | 241 | 0.67 | 0.41–1.11 | 0.119 | 618 | 1.04 | 0.81–1.33 | 0.784 |
| Luminal A | 611 | 1.58 | 1.10–2.27 | 0.012 | 1933 | 1.56 | 1.31–1.85 | < 0.001 |
| Luminal B | 433 | 1.55 | 1.06–2.27 | 0.023 | 1149 | 1.56 | 1.28–1.89 | < 0.001 |
| HER2 positive | 117 | 0.41 | 0.20–0.83 | 0.010 | 251 | 0.83 | 0.57–1.23 | 0.358 |
| PRDX5 | ||||||||
| Basal-like | 153 | 1.09 | 0.58–2.06 | 0.790 | 360 | 1.71 | 1.23–2.37 | 0.001 |
| Luminal A | 271 | 1.09 | 0.66–1.81 | 0.725 | 841 | 1.19 | 0.93–1.53 | 0.164 |
| Luminal B | 129 | 0.58 | 0.29–1.15 | 0.117 | 407 | 0.95 | 0.70–1.29 | 0.752 |
| HER2 positive | 73 | 1.59 | 0.71–3.54 | 0.253 | 156 | 1.34 | 0.85–2.11 | 0.213 |
| PRDX6 | ||||||||
| Basal-like | 241 | 1.08 | 0.66–1.76 | 0.761 | 618 | 1.01 | 0.78–1.29 | 0.963 |
| Luminal A | 611 | 1.49 | 1.04–2.13 | 0.027 | 1933 | 1.29 | 1.08–1.53 | 0.004 |
| Luminal B | 433 | 0.95 | 0.65–1.37 | 0.771 | 1149 | 1.27 | 1.05–1.54 | 0.015 |
| HER2 positive | 117 | 0.87 | 0.46–1.66 | 0.675 | 251 | 0.93 | 0.63–1.36 | 0.708 |
Moreover, we also analyzed the associations between PRDXs mRNA expression and RFS of various subclasses BrCa patients and the results indicated that high expressions of PRDX1 (luminal A: HR = 1.23, 95% CI: 1.06–1.49, P = 0.009), PRDX2 (luminal A: HR = 1.23, 95% CI: 1.04–1.46, P = 0.016; luminal B: HR = 1.67, 95% CI: 1.37–2.03, P < 0.001), PRDX4 (luminal A: HR = 1.56, 95% CI: 1.31–1.85, P < 0.001; luminal B: HR = 1.56, 95% CI: 1.28–1.89, P < 0.001), PRDX6 (luminal A: HR = 1.29, 95% CI: 1.08–1.53, P = 0.004; luminal B: HR = 1.27, 95% CI: 1.05–1.54, P = 0.015) were correlated with worse RFS in luminal BrCa patients. In addition, low expression of PRDX3 (HR = 0.77, 95% CI: 0.60–1.00, P = 0.046) and high expression of PRDX5 (HR = 1.71, 95% CI: 1.23–2.37, P = 0.001) predicted unfavorable RFS in basal-like BrCa patients. Thus, these results suggested the roles of PRDXs as potential prognostic predictors in BrCa patients with different subclasses.
Prognostic values of PRDXs mRNA in BrCa patients with diverse regimens of chemotherapy
Next, we also checked the prognostic effects of PRDX family members in BrCa patients with different chemotherapies, including adjuvant chemotherapy, neoadjuvant chemotherapy and non-chemotherapy. As shown in Table 3, high expression of PRDX1, PRDX2, PRDX3, and PRDX4 were significantly correlated with poor OS in BrCa patients with adjuvant chemotherapy. In addition, in BrCa patients who didn’t receive any chemotherapies, high expression PRDX1 (HR = 1.22, 95% CI: 1.03–1.44, P = 0.019), PRDX4 (HR = 1.37, 95% CI: 1.16–1.62, P < 0.001), PRDX6 (HR = 1.19, 95% CI: 1.00–1.40, P = 0.044) and low expression of PRDX3 (HR = 0.84, 95% CI: 0.71–1.00, P = 0.043) were associated with worse RFS. However, contrary to the prognostic effect of PRDX5 in total BrCa patients, high expression of PRDX5 (HR = 0.50, 95% CI: 0.30–0.81, P = 0.005) predicted better RFS in BrCa patients with adjuvant chemotherapy. Thus, these results suggested the roles of PRDXs as potential prognostic predictors in BrCa patients with different regimens of chemotherapy.
Table 3.
Association between prognostic value of PRDXs mRNA expression and various chemotherapies in BrCa
| Chemotherapies | Cases | HR | 95% CI | P value | Cases | HR | 95% CI | P value |
|---|---|---|---|---|---|---|---|---|
| OS | RFS | |||||||
| PRDX1 | ||||||||
| Adjuvant chemotherapy | 163 | 1.85 | 1.01–3.40 | 0.044 | 594 | 1.05 | 0.78–1.42 | 0.734 |
| Neoadjuvant chemotherapy | 156 | 0.74 | 0.34–1.60 | 0.448 | 223 | 1.19 | 0.69–2.03 | 0.539 |
| Non-chemotherapy | 549 | 1.13 | 0.79–1.60 | 0.502 | 1873 | 1.22 | 1.03–1.44 | 0.019 |
| PRDX2 | ||||||||
| Adjuvant chemotherapy | 163 | 2.02 | 1.09–3.73 | 0.023 | 594 | 1.33 | 0.98–1.80 | 0.064 |
| Neoadjuvant chemotherapy | 156 | 1.41 | 0.66–3.02 | 0.375 | 223 | 1.24 | 0.72–2.16 | 0.438 |
| Non-chemotherapy | 549 | 1.14 | 0.80–1.62 | 0.483 | 1873 | 1.07 | 0.90–1.26 | 0.444 |
| PRDX3 | ||||||||
| Adjuvant chemotherapy | 163 | 2.03 | 1.10–3.73 | 0.021 | 594 | 0.80 | 0.59–1.08 | 0.148 |
| Neoadjuvant chemotherapy | 156 | 1.05 | 0.49–2.23 | 0.906 | 223 | 0.95 | 0.55–1.65 | 0.851 |
| Non-chemotherapy | 549 | 0.79 | 0.56–1.12 | 0.185 | 1873 | 0.84 | 0.71–1.00 | 0.043 |
| PRDX4 | ||||||||
| Adjuvant chemotherapy | 163 | 1.97 | 1.07–3.65 | 0.027 | 594 | 1.02 | 0.75–1.38 | 0.906 |
| Neoadjuvant chemotherapy | 156 | 0.68 | 0.31–1.48 | 0.325 | 223 | 1.19 | 0.69–2.07 | 0.529 |
| Non-chemotherapy | 549 | 1.28 | 0.90–1.82 | 0.177 | 1873 | 1.37 | 1.16–1.62 | < 0.001 |
| PRDX5 | ||||||||
| Adjuvant chemotherapy | 0 | 1.09 | 0.58–2.06 | 0.790 | 255 | 0.50 | 0.30–0.81 | 0.005 |
| Neoadjuvant chemotherapy | 107 | 0.67 | 0.24–1.89 | 0.446 | 111 | 1.22 | 0.58–2.57 | 0.595 |
| Non-chemotherapy | 0 | 0.58 | 0.29–1.15 | 0.117 | 243 | 0.66 | 0.38–1.14 | 0.131 |
| PRDX6 | ||||||||
| Adjuvant chemotherapy | 163 | 0.86 | 0.48–1.56 | 0.625 | 594 | 1.10 | 0.81–1.49 | 0.532 |
| Neoadjuvant chemotherapy | 156 | 0.72 | 0.33–1.55 | 0.395 | 223 | 1.09 | 0.63–1.88 | 0.769 |
| Non-chemotherapy | 549 | 1.33 | 0.94–1.88 | 0.110 | 1873 | 1.19 | 1.00–1.40 | 0.044 |
Discussion
Reactive oxygen species (ROS), including the superoxide radical, the hydroxyl radical, H2O2 and etc., are the most important type of free radicals which produces secondary toxic metabolic products, such as peroxynitrites and nitrogen oxides, posing a lethal threat to cells by damaging DNA [23, 24]. Peroxiredoxins, one of the most significant antioxidant enzyme systems that include SOD, CAT, and GPx, were significnatly upregulated under oxidative stress conditions and mainly participate in the defense against oxidative [25, 26]. Several studies have observed that imbalances between the generation of ROS and PRDXs in tumor cells could lead to oxidative stress and the induction of cell apoptosis [27].
It has been demonstrated that PRDXs expression was significantly dysregulated during carcinogenesis of cancers and played dichotomous roles in oncogenesis. Overexpression of PRDX1 in BrCa has been observed to be positively associated with tumor grade and acted as dominant role in management of exogeneous oxidative stress [28, 29]. PRDX2, has been reported to specifically regulate the oxidative and metabolic stress response of metastatic breast cancer cells in lungs. Besides, overexpressed PRDX2 participates in chemo-resistant in BrCa cells [30, 31]. The function of PRDX3 and PRDX4 in BrCa is largely ambiguous, Liu et al. reveals that downregulation of PRDX3 potentiates PP2-induced apoptosis in MCF-7 cells, which suggests the tumor suppressor role of PRDX3 [32]. PRDX4 has been demonstrated to mediate osteoclast activation by human BrCa cells and enhance the aggressive phenotype [33]. Last but not least, there is no available research about the exact function of PRDX5 and PRDX6 in BrCa, but overexpression of PRDX5 and prognostic values have been observed in numerous cancers, including ovarian cancer and endometrial cancer [34, 35]. As well, the tumor promoter role of PRDX6 in cancers has also been suggested in colorectal cancer, lung cancer and so on [36, 37]. Although several studies that investigate the role of PRDXs in BrCa have been published, little is known about individual PRDXs expression and their effects on survival of BrCa patients.
In the present study, we first investigated the differential transcriptional levels of PRDX family members between BrCa and adjacent tissues and the results showed that the transcriptional levels of PRDX1, PRDX2, PRDX4, and PRDX5 were significantly upregulated in BrCa tissues. Besides, the transcriptional level of PRDX6 was significantly downregulated in BrCa tissues. However, the transcriptional level of PRDX3 showed a non-significant difference in BrCa tissues compared with adjacent tissues. we also compared the differential transcriptional levels of PRDX family members according to different intrinsic subclasses of BrCa and found mRNA expressions of PRDXs family members were significantly correlated with intrinsic subclasses of BrCa.
DNA copy number alterations (CNA) are most common genetic alterations which affect carcinogenesis and development of cancers by regulating cancer-related gene expression [20–22]. When we used cBioPortal to inspect genetic alteration in PRDXs and correlations with their mRNA expressions, the results showed that copy gain (gain and amplification) of most PRDXs was not frequent in BrCa, but it was still associated with notable upregulated PRDXs mRNA levels. Amplification is a positive factor to upregulate gene expression [38, 39]. However, to our largely surprise, the copy-gain frequency of PRDX6 accounts for a large proportion in total BrCa samples, but a significant decrease expression of PRDX6 was exhibited in BrCa tissues. We speculated that amplification of PRDX6 gene may upregulate PRDX6 expression in BrCa tissues compared with paired breast tissues. However, limited amounts of normal tissues expression data were included in TCGA dataset, thus, the opposite phenomenon that PRDX6 amplification decreased transcriptional level was exhibited.
The Kaplan-Meier plotter is an online database which is available to assess the prognostic effect of genes expression on survival in designate cancers. The primary purpose of the tool is a meta-analysis-based biomarker assessment and a lot of prognostic biomarkers have been identified based on this platform [40–43]. It has been reported that overexpression PRDX6 participates in cisplatin resistance in ovarian cancer and predicts poor OS and PFS [8, 44]. Several researches also observed the promising prognostic values of PRDXs in lung cancer and endometrial cancer [7, 35, 45]. However, the prognostic values of PRDXs in BrCa patients are largely unknown. Here, we found that high mRNA expression of PRDX4/6 were significantly associated with poor OS of BrCa patients and high mRNA expression of PRDX1/2/4/5/6 were significantly associated with shorter RFS of BrCa patients, while high mRNA expression of PRDX3 was notably related to favorable OS and DFS, which suggests the tumor suppressor role of PRDX3 in BrCa. Besides, the prognostic values of PRDXs mRNA in different BrCa subclasses and in BrCa patients with diverse regimens of chemotherapy were also assessed and results suggested the potential roles of PRDXs in predicting prognosis of BrCa patients with various subclasses and different regimens of chemotherapy.
Although this study systematically demonstrates the prognostic value of PRDXs in breast cancer, this research has several limitations as well. The major limitation is that online database only provides the expression of PRDXs mRNA level, which may not fully represent the expression of PRDXs at the protein level. In further study, western blotting, immumohistochemical staining and other protein detection techniques will be applied to determinate the protein level of PRDXs in breast cancer. Furthermore, the possible mechanisms that PRDXs is involved in the tumorigenesis and progression of breast cancer need to be further studied. Besides, although the large sample analyses based on meta-analysis have some advantages, but some essential information form one single center may be missing, such as some therapeutic information.
Conclusion
In summary, we systemically analyzed the expression profiles and prognostic values of PRDXs in BrCa. Our results revealed that PRDX1/2/4/5/6 might be the potential therapeutic targets for BrCa therapy, whereas PRDX3/4/6 were promising prognostic biomarkers for predicting OS and RFS of BrCa patients. Overall, our research provided a systematic insight into the heterogeneous and complex roles of PRDXs in the carcinogenesis of BrCa.
Acknowledgements
Not applicable.
Abbreviations
- ACS
The American Cancer Society
- BrCa
Breast cancer
- CAN
Copy-number alterations
- CI
Confidence interval
- ER
Estrogen receptor
- HER2
Epidermal growth factor receptor 2
- HR
Hazard ratio
- Ki-67
Antigen ki-67
- NCI
US National Cancer Institute
- NHGRI
The National Human Genome Research Institute
- OS
Overall survival
- PR
Progesterone receptor
- PRDX
Peroxiredoxin
- RFS
Relapse-free survival
- TCGA
The Cancer Genome Atlas
Authors’ contributions
Designed the experiments: CL, HW, and JM; Acquisition of data: JM and LH; Analysis and interpretation of data: JM, LH, XL, GS, and RX; Draft of the manuscript: JM; Critical revision of the manuscript for intellectual content: CL, HW, and JM; Funding Acquisition: CL and HW. All authors read and approved the final manuscript.
Funding
This work was founded by the National Natural Science Foundation of China (81602065), the Natural Science Foundation of Jiangsu Province of China (BE2017626), and the Foundation of Wuxi Health Commission (QNRC003).
Availability of data and materials
All data are included in the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jie Mei, Email: meijie199621@163.com.
Leiyu Hao, Email: leiyuhao316@163.com.
Xiaorui Liu, Email: liuxr7797@163.com.
Guangshun Sun, Email: s131823@163.com.
Rui Xu, Email: aqsw1465@126.com.
Huiyu Wang, Email: wanghuiyu2011@126.com.
Chaoying Liu, Email: chyliu666@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. doi: 10.3121/cmr.2008.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappaB activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- 4.Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park MH, Jo M, Kim YR, Lee CK, Hong JT. Roles of peroxiredoxins in cancer, neurodegenerative diseases and inflammatory diseases. Pharmacol Ther. 2016;163:1–23. doi: 10.1016/j.pharmthera.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Liu X, Chen L, Cai L, Li N, Zhu P, Chen J, Song X, Li G. Differential expression of peroxiredoxin 3 in laryngeal squamous cell carcinoma. Oncotarget. 2017;8:3471–3480. doi: 10.18632/oncotarget.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Huang C, Yang X, Zhang Q, Chen F. Prognostic roles of mRNA expression of peroxiredoxins in lung cancer. Onco Targets Ther. 2018;11:8381–8388. doi: 10.2147/OTT.S181314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Saisai, Hu Xiaoli, Ye Miaomiao, Zhu Xueqiong. The prognostic values of the peroxiredoxins family in ovarian cancer. Bioscience Reports. 2018;38(5):BSR20180667. doi: 10.1042/BSR20180667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding C, Fan X, Wu G. Peroxiredoxin 1 - an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21:193–202. doi: 10.1111/jcmm.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heng HH. Cancer genome sequencing: the challenges ahead. Bioessays. 2007;29:783–794. doi: 10.1002/bies.20610. [DOI] [PubMed] [Google Scholar]
- 11.Hanauer DA, Rhodes DR, Sinha-Kumar C, Chinnaiyan AM. Bioinformatics approaches in the study of cancer. Curr Mol Med. 2007;7:133–141. doi: 10.2174/156652407779940431. [DOI] [PubMed] [Google Scholar]
- 12.Li SC, Tachiki LM, Kabeer MH, Dethlefs BA, Anthony MJ, Loudon WG. Cancer genomic research at the crossroads: realizing the changing genetic landscape as intratumoral spatial and temporal heterogeneity becomes a confounding factor. Cancer Cell Int. 2014;14:115. doi: 10.1186/s12935-014-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ning G, Huang YL, Zhen LM, Xu WX, Jiao Q, Yang FJ, Wu LN, Zheng YY, Song J, Wang YS, Xie C, Peng L. Transcriptional expressions of Chromobox 1/2/3/6/8 as independent indicators for survivals in hepatocellular carcinoma patients. Aging (Albany NY) 2018;10:3450–3473. doi: 10.18632/aging.101658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Li G, Luo Q, Xie J, Gan C. Integrated TCGA analysis implicates lncRNA CTB-193M12.5 as a prognostic factor in lung adenocarcinoma. Cancer Cell Int. 18(2018):27. [DOI] [PMC free article] [PubMed]
- 15.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, Gyorffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 19.Goto W, Kashiwagi S, Takada K, Asano Y, Takahashi K, Fujita H, Takashima T, Tomita S, Hirakawa K, Ohira M. Significance of intrinsic breast cancer subtypes on the long-term prognosis after neoadjuvant chemotherapy. J Transl Med. 2018;16:307. doi: 10.1186/s12967-018-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi W, Ochoa A, McConkey DJ, Aine M, Hoglund M, Kim WY, Real FX, Kiltie AE, Milsom I, Dyrskjot L, Lerner SP. Genetic alterations in the molecular subtypes of bladder Cancer: illustration in the Cancer genome atlas dataset. Eur Urol. 2017;72:354–365. doi: 10.1016/j.eururo.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bigagli E, De Filippo C, Castagnini C, Toti S, Acquadro F, Giudici F, Fazi M, Dolara P, Messerini L, Tonelli F, Luceri C. DNA copy number alterations, gene expression changes and disease-free survival in patients with colorectal cancer: a 10 year follow-up. Cell Oncol (Dordr) 2016;39:545–558. doi: 10.1007/s13402-016-0299-z. [DOI] [PubMed] [Google Scholar]
- 22.Thomas LE, Winston J, Rad E, Mort M, Dodd KM, Tee AR, McDyer F, Moore S, Cooper DN, Upadhyaya M. Evaluation of copy number variation and gene expression in neurofibromatosis type-1-associated malignant peripheral nerve sheath tumours. Hum Genomics. 2015;9:3. doi: 10.1186/s40246-015-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pisoschi AM, Pop A. The role of antioxidants in the chemistry of oxidative stress: a review. Eur J Med Chem. 2015;97:55–74. doi: 10.1016/j.ejmech.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 24.Nicolussi A, D'Inzeo S, Capalbo C, Giannini G, Coppa A. The role of peroxiredoxins in cancer. Mol Clin Oncol. 2017;6:139–153. doi: 10.3892/mco.2017.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha MK, Kim HK, Kim IH. Thioredoxin-linked "thiol peroxidase" from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635–28641. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Wan XY, Wang HL, Yan ZY, Hou YD, Jin DY. Bacterial scavengase p20 is structurally and functionally related to peroxiredoxins. Biochem Biophys Res Commun. 1997;233:848–852. doi: 10.1006/bbrc.1997.6564. [DOI] [PubMed] [Google Scholar]
- 27.Song IS, Kim HK, Jeong SH, Lee SR, Kim N, Rhee BD, Ko KS, Han J. Mitochondrial peroxiredoxin III is a potential target for cancer therapy. Int J Mol Sci. 2011;12:7163–7185. doi: 10.3390/ijms12107163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cha MK, Suh KH, Kim IH. Overexpression of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J Exp Clin Cancer Res. 2009;28:93. doi: 10.1186/1756-9966-28-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bajor M, Zych AO, Graczyk-Jarzynka A, Muchowicz A, Firczuk M, Trzeciak L, Gaj P, Domagala A, Siernicka M, Zagozdzon A, Siedlecki P, Kniotek M, O'Leary PC, Golab J, Zagozdzon R. Targeting peroxiredoxin 1 impairs growth of breast cancer cells and potently sensitises these cells to prooxidant agents. Br J Cancer. 2018;119:873–884. doi: 10.1038/s41416-018-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stresing V, Baltziskueta E, Rubio N, Blanco J, Arriba MC, Valls J, Janier M, Clezardin P, Sanz-Pamplona R, Nieva C, Marro M, Petrov D, Sierra A. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32:724–735. doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
- 31.Wang T, Diaz AJ, Yen Y. The role of peroxiredoxin II in chemoresistance of breast cancer cells. Breast Cancer (Dove Med Press) 2014;6:73–80. doi: 10.2147/BCTT.S61281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Feng R, Du L. The role of enoyl-CoA hydratase short chain 1 and peroxiredoxin 3 in PP2-induced apoptosis in human breast cancer MCF-7 cells. FEBS Lett. 2010;584:3185–3192. doi: 10.1016/j.febslet.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Tiedemann K, Sadvakassova G, Mikolajewicz N, Juhas M, Sabirova Z, Tabaries S, Gettemans J, Siegel PM, Komarova SV. Exosomal release of L-Plastin by breast Cancer cells facilitates metastatic bone Osteolysis. Transl Oncol. 2019;12:462–474. doi: 10.1016/j.tranon.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sienko J, Gaj P, Czajkowski K, Nowis D. Peroxiredoxin-5 is a negative survival predictor in ovarian cancer. Ginekol Pol. 2019;90:1–6. doi: 10.5603/GP.2019.0001. [DOI] [PubMed] [Google Scholar]
- 35.Byun JM, Kim SS, Kim KT, Kang MS, Jeong DH, Lee DS, Jung EJ, Kim YN, Han J, Song IS, Lee KB, Sung MS. Overexpression of peroxiredoxin-3 and -5 is a potential biomarker for prognosis in endometrial cancer. Oncol Lett. 2018;15:5111–5118. doi: 10.3892/ol.2018.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang WS, Huang CY, Hsieh MC, Kuo YH, Tung SY, Shen CH, Hsieh YY, Teng CC, Lee KC, Lee KF, Kuo HC. Expression of PRDX6 correlates with migration and invasiveness of colorectal Cancer cells. Cell Physiol Biochem. 2018;51:2616–2630. doi: 10.1159/000495934. [DOI] [PubMed] [Google Scholar]
- 37.Yun HM, Park KR, Park MH, Kim DH, Jo MR, Kim JY, Kim EC, Yoon DY, Han SB, Hong JT. PRDX6 promotes tumor development via the JAK2/STAT3 pathway in a urethane-induced lung tumor model. Free Radic Biol Med. 2015;80:136–144. doi: 10.1016/j.freeradbiomed.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Kwak Y, Kim SI, Park CK, Paek SH, Lee ST, Park SH. C-MET overexpression and amplification in gliomas. Int J Clin Exp Pathol. 2015;8:14932–14938. [PMC free article] [PubMed] [Google Scholar]
- 39.Ohshima K, Hatakeyama K, Nagashima T, Watanabe Y, Kanto K, Doi Y, Ide T, Shimoda Y, Tanabe T, Ohnami S, Ohnami S, Serizawa M, Maruyama K, Akiyama Y, Urakami K, Kusuhara M, Mochizuki T, Yamaguchi K. Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1,454 solid tumors. Sci Rep. 2017;7:641. doi: 10.1038/s41598-017-00219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manicum Theasha, Ni Fubiao, Ye Yiming, Fan Xuhui, Chen Bi-Cheng. Prognostic values of E2F mRNA expression in human gastric cancer. Bioscience Reports. 2018;38(6):BSR20181264. doi: 10.1042/BSR20181264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gayed DT, Wodeyar J, Wang ZX, Wei X, Yao YY, Chen XX, Du Z, Chen JC. Prognostic values of inhibitory kappaB kinases mRNA expression in human gastric cancer. Biosci Rep. 2019;39:BSR20180617. [DOI] [PMC free article] [PubMed]
- 42.Zhao M, Li S, Zhou L, Shen Q, Zhu H, Zhu X. Prognostic values of excision repair cross-complementing genes mRNA expression in ovarian cancer patients. Life Sci. 2018;194:34–39. doi: 10.1016/j.lfs.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 43.Mei J, Yan T, Huang Y, Xia T, Chang F, Shen S, Hao L, Chen Y, Wang Z, Jiang X, Xu B, Zhu Y. A DAAM1 3′-UTR SNP mutation regulates breast cancer metastasis through affecting miR-208a-5p-DAAM1-RhoA axis. Cancer Cell Int. 2019;19:55. [DOI] [PMC free article] [PubMed]
- 44.Pak JH, Choi WH, Lee HM, Joo WD, Kim JH, Kim YT, Kim YM, Nam JH. Peroxiredoxin 6 overexpression attenuates cisplatin-induced apoptosis in human ovarian cancer cells. Cancer Investig. 2011;29:21–28. doi: 10.3109/07357907.2010.535056. [DOI] [PubMed] [Google Scholar]
- 45.Kim JH, Bogner PN, Baek SH, Ramnath N, Liang P, Kim HR, Andrews C, Park YM. Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clin Cancer Res. 2008;14:2326–2333. doi: 10.1158/1078-0432.CCR-07-4457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in the article.