Abstract
Genetics has long been considered to associate with many exercise-related traits and sport performance phenotypes. A genetic basis for elite international marathon running performance exists due to the heritability of endurance-related traits. This has prompted a generation of genomic study to identify marathon success. The aim of this study was to systematically review the evidence of genes, and their polymorphisms, that may play a role in marathon running performance. A search strategy was implemented on systematic databases following PRISMA guidelines. Studies were case-control, cohort or genome-wide association designs and provided data on the genotypes associated with elite marathon athlete status and/or marathon running performance. The search identified 241 studies, from which, 14 studies were deemed suitable for inclusion. A total of 160 different polymorphisms in 27 genes were identified in 10,442 participants, of which 2,984 were marathon distance runners. The review identified a possible 16 single nucleotide polymorphisms (SNPs) in 14 genes associated with marathon running performance. While multiple genes and their polymorphisms have been associated with marathon running performance, predicting future marathon success based on genomic data is premature due to the lack of replicated studies. There is limited replication of genotype-phenotype associations and there is possible publication bias, thus, further studies are required to strengthen our understanding of the genes involved in marathon running. Future research utilising genome-wide technologies in large cohorts is required to elucidate the multiple genetic factors that govern complex endurance-related traits and the impact of epigenetics should be considered.
Key points.
Elite marathon running has a polygenic nature of complex endurance-related traits.
There is limited replication of genotype-phenotype associations to currently determine the genes involved in marathon running.
Genome-wide technologies in large cohorts are required to elucidate the multiple genetic factors that govern the complex endurance-related traits of marathon running and the impact of epigenetics should be considered.
Key words: Genetics, exercise, performance, endurance, genotype
Introduction
Marathon running is predominantly determined by maximal oxygen uptake (V̇O2 max), lactate threshold, running economy and oxygen uptake kinetics (Joyner and Coyle, 2008). It has been acknowledged that to reach elite-level performance a synergy of physiological and psychological traits, combined with an optimal environment are required (Tucker et al., 2013). Physiological parameters of heritability such as V̇O2 max for endurance performance indicate significant genetic components. For example, Bouchard et al. (1999) demonstrated that the heritability of V̇O2max falls in the range of 40-50%, indicating such parameters present a considerable genetic influence. In addition, elite marathon runners of Kenyan or Ethiopian descent account for 90 of the top 100 marathon running performances of all time and hold the six previous world records (International Association of Athletics Federations, 2018). Based on this geographical concentration of unparalleled success, assertions of a genetic endowment for marathon running performance have been further amplified (Tuker et al., 2013; Vancini et al., 2014). Therefore, a major focus of sport genomics has been to identify specific genes and their polymorphisms associated with elite-level performance to facilitate in the identification of future athletic talent (Roth, 2012; Popovski et al., 2016). Specifically, one gene that has been extensively studied due to its associated role in endurance performance is angiotensin I-converting enzyme (ACE) (Gayagay et al., 1998; Montgomery et al., 1998). The ACE gene functional polymorphism is based on either the presence (insertion [I]) or absence (deletion [d]) of an intron sequence. The insertion (I) allele has been positively associated with elite endurance running performance in Caucasians due to lower circulating and tissue ACE activity (Myserson et al., 1999). No significant differences in the ACE I/deletion (D) genotype were found between Kenyan elite marathon runners and their respective general population (Scott et al., 2005). This ethnic disparity led to the contention that the East African running phenomenon is not a genetically mediated one (Wilber & Pitsiladis, 2012). On the contrary, the existing research to date, may be interpreted as evidence for the polygenic nature of complex endurance-related traits, due to the basis of researching the broader context of endurance performance, but also highlighting the limitations of a single candidate gene approach (Ahmetov et al., 2016). Furthermore, the use of case-control studies has given rise to inconsistent findings (Drozdovska et al., 2013; Tural et al., 2014), which may be attributed to small sample sizes resulting in insufficient statistical power to demonstrate significant effects of numerous genes each with small contributions (Wang et al., 2013). Where previous studies have been conducted on general endurance performance rather than specific to elite marathon running, ensuring sufficient statistical power is a particular challenge in elite-level performance since elite athletes are by definition, limited to a small number of superior individuals. Therefore, it has been suggested that the impact of these issues can be reduced by pooling single studies into a meta-analysis (Lopez-Leon et al., 2016). To date, meta-analyses have confirmed the associations of ACE and peroxisome proliferator-activated receptor alpha (PPARα) genes with endurance performance (Ma et al., 2013; Lopez-Leon et al., 2016). However, it has been suggested that at least 93 genetic markers are associated with elite endurance athletic status (Ahmetov et al., 2016). Yet, to date, no review has identified the genetic markers associated specifically to elite marathon running per se.
Therefore, the objectives of the current systematic review were to critically examine the evidence concerning genes that may play a role in the performance of elite-level international marathon athletes, and to provide insight into the predictive utility of genetic testing in identifying future marathon success. This investigation of multiple genes, and their polymorphisms, is advantageous as it allows a more comprehensive picture of genotype-phenotype relationships to emerge, thus improving the future application of genomics as a practical tool in marathon talent identification.
Methods
Search strategy
A published literature search that investigated the association between genes and marathon performance was conducted up to May 2018 and obtained according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al., 2009). A predetermined search strategy was conducted on systematic databases for publications indexed in PubMed, SPORTDiscus and Google Scholar using combinations of the following search terms: “genes” OR “genetics” OR “genomics” AND “marathon” OR “endurance” OR “elite performance” in the titles of papers. Additional publications were also considered by cross-referencing. A hand search of the reference lists of articles included in the final analysis that were identified via the database search was conducted, as were the first 20 “related articles” of those included database search articles on PubMed.
Study selection
All publications retrieved were screened by title and any duplicates or those irrelevant to the research question were removed. Abstracts of the remaining studies were then similarly screened and 24 studies were selected for full-text assessment against the predetermined inclusion and exclusion criteria outlined below.
Inclusion and exclusion criteria
The present review included case-control, cohort and genome-wide association studies (GWAS). To be included, studies were required to provide data on the genotypes associated with elite international marathon athlete status and/or marathon running performance published in peer-review journals. Those studies identified as ‘elite’ were based on participants having been international competitors and/or national representatives in the marathon distance as determined by entry standards from the International Association of Athletics Federations (IAAF, 2018). There were no restrictions applied regarding the age, gender or ethnicity of the participants. Studies were excluded if they were: (i) review articles, congress abstracts, editorials or other non-original articles; (ii) reported in a language other than English. Overall, 14 studies were included for qualitative synthesis. The study selection process and reasons for exclusion are illustrated in Figure 1.
Figure 1.
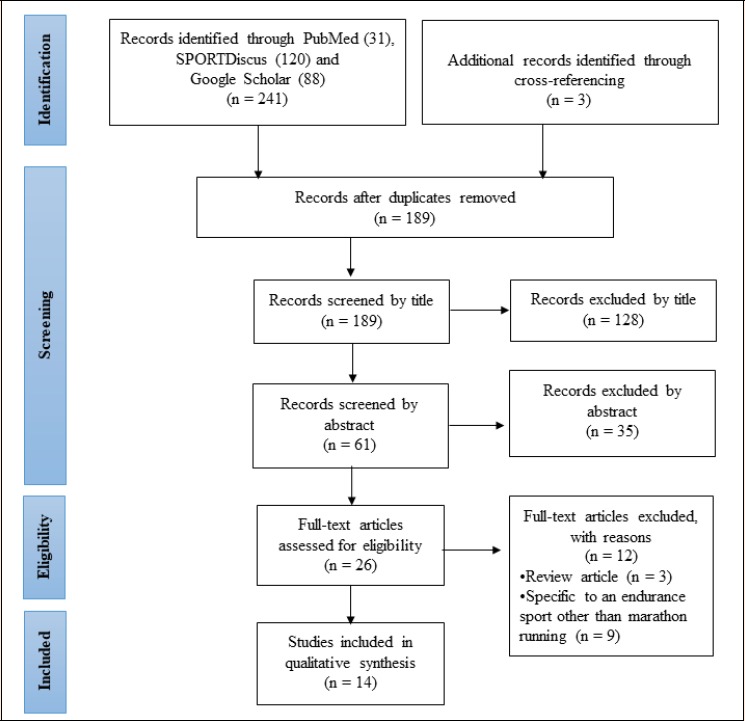
Flow diagram of the phases of study selection during the search process based on PRISMA.
Studies were assessed for inclusion by two independent reviewers, RK and DF, with disagreements resolved by discussion, and arbitration (HJM) if necessary. The reviewers, RK and DF were not blinded to authors, institutions or journals of publication. If a decision on whether to include or exclude a paper could not be made from the title and abstract, full text was obtained and checked.
Data extraction and quality assessment
For all selected studies, the following data were extracted: (i) first author name; (ii) publication date; (iii) participant characteristics; (iv) study design; (v) genetic markers measured; (vi) allelic frequencies; and (vii) bias. These outcomes were extracted for the narrative review.
Risk of bias assessment
The risk of bias of individual studies was assessed using the Cochrane Collaboration’s risk of bias tool (RoB 2.0; Higgins et al., 2016). Studies were given an overall risk of bias grade of either “high”, “some” or “low” calculated from the following five domains: a) sequence generation, b) allocation concealment, c) blinding, d) missing outcome data, e) selective reporting of results. If details for a particular domain were insufficient, the risk of bias was assessed as “unclear”. Assessments were performed independently by two authors (RK and DF) with disagreements resolved by discussion, and arbitration (HJM) if necessary. Components were assessed independently, as per PRISMA (Moher et al., 2009) and Cochrane collaboration (Higgins et al., 2016) recommendations.
Data synthesis
Qualitative analysis was carried out on the studies selected focusing on the association between genetic markers, elite marathon athlete status and running performance. A narrative review was provided in text and tables to summarise study characteristics. If deemed appropriate, a meta-analysis was planned for single nucleotide polymorphisms (SNPs) that had been investigated in at least three studies. However, a meta-analysis was not performed due to the heterogeneity of the study characteristics and SNPs measured with very few replication studies. Where data was not available in the full-text, original authors were contacted.
Results
Summary of studies retrieved
The search strategy identified 241 studies, of which, following cross-referencing and exclusions, 14 full-text articles were identified and included in the qualitative synthesis (see Figure 1).
Study characteristics
The characteristics of the 14 studies are summarised in Table 1. A total of 160 different polymorphisms in 27 genes were studied in 10,442 participants, of which 2,984 were marathon distance runners, 6,109 were non-athlete controls or 310 power athletes. Of the 160 polymorphisms, 159 were collectively investigated in two studies (Tsianos et al., 2010; He et al., 2015), where a total of 139 polymorphisms were studied in one paper alone by He et al., (2015). Where, the study investigated 84 polymorphisms in two genes, of which there were no reported associations (He et al., 2015). Only the ACE SNP I/D rs4646994 was investigated in three studies (Amir et al., 2007; Ash et al., 2011; Tobina et al., 2010). Thirteen of the studies were case-control or cohort designs with only one applying the new GWAS approach (Ahmetov et al., 2015), although GWAS was not applied to the comparison of the athlete groups with the control groups. While 11 studies discovered at least one endurance-related allele, Ash et al. (2011), Sawczuk et al. (2013) and Papadimitriou et al. (2018) found no genetic association.
Table 1.
Characteristics of studies included (bold indicates gene of interest).
| References | Participants | Study design | Genes and SNPs measured | Outcome(s) |
|---|---|---|---|---|
| Ahmetov et al., 2015 | Russian endurance athletes (n= 219; 2 marathon runners), power athletes (n=230), Russian controls (n=192) and European controls (n=1367) | GWAS |
NFIA-AS2 rs1572312 C/A TSHR rs7144481 T/C RBFOX1 rs7191721 G/A |
C alleles of NFIA-AS2 rs1572312 and TSHR rs7144481 associated with elite endurance athlete status including marathon runners. |
| Amir et al., 2007 | Israeli elite marathon runners (n=79), elite power athletes (n=42) and sedentary controls (n=247) | Case-control | ACE I/D rs4646994 | D allele associated with elite marathon athlete status. |
| Ash et al., 2011 | Ethiopian elite endurance runners (n=76), demographically matched controls (n=410), controls from general Ethiopian population (n=317), power athletes (n=38) | Case-control |
ACE I/D rs4646994; A22982G rs4363 |
No association with elite Ethiopian runners. |
| Döring et al., 2010 | Caucasian male elite endurance athletes (n=316; 39 runners) and Caucasian male sedentary controls (n=304) | Case-control |
HIF1A Pro582Ser; rs11549465; C/T rs17099207 G/A; rs1951795 C/A; rs11158358 C/G; rs2301113 A/C; rs11549467 G/A |
Pro582 C allele of rs11549465 and A allele of rs17099207 associated with elite endurance runners. |
| He et al., 2015 | Chinese elite endurance runners (n=235) and Chinese controls (n=504) | Case-control | PPARAGCIα (41 SNPs) PPARAGCIβ (43 SNPs) PPRCI1 (4 SNPs); TFAM (3 SNPSs); TFB1M (7 SNPs); TFB2M (3 SNPs); NRF1 (14 SNPs); GABPA (2 SNPs); GABPA (5 SNPs); ERRα (4 SNPs); SIRT1 (7 SNPs) |
No significant association between proliferator-activated receptor γ (PGC)-related genes and elite endurance running status after adjusting for multiple comparisons. |
| Martinez et al., 2009 | Hispanic marathon runners (n=784; 393 3rd percentile and 388 lowest 3rd percentile finishers) | Case-control | AQP1 rs1049305 C/G | C allele associated with elite performance in Hispanic marathon runners. |
| Myerson et al., 1999 | Elite runners (n=91; 79 Caucasian) and British controls (n=1906) | Case-control | ACE I/D rs1049305 C/G | I allele positively associated with elite endurance running performance. |
| Papadimitriou et al., 2018 | 1,5k, 3k, 5k, and 42k m running times of 698 male and female Caucasian endurance athletes | Cohort | ACTN3 R577X ACE I/D |
No association between ACTN3 or ACE I/D genotype and running performance at any distance. |
| Posthumus et al., 2011 | Caucasian male triathlon (incl. 42.2km run) finishers (n=313) | Cohort |
COL5A1 BstUI RFLP rs12722 T/C |
T allele associated with faster time to complete running component (42.2km) of triathlon. |
| Sawczuk et al., 2013 | Polish elite endurance athletes (n=123; 12 marathon runners) and sedentary controls (n=228) | Case-control | ADRA2A rs553668 C/T | No association with elite endurance athlete status including marathon runners. |
| Stebbings et al., 2018 | Male marathon runners (n=141) and recreationally active men (n=137) | Cohort | TTN rs10497520 | TTN gene is associated with shorter skeletal muscle fascicle length and conveys an advantage for marathon running performance in trained men. |
| Tobina et al., 2010 | Japanese male elite endurance runners (n=37) and non-athlete controls (n=335) | Case-control | ACE I/D rs4646994 | Frequency of the ACE I/D genotype was lower in elite endurance runners than controls. The D allele was associated with faster marathon -running speed. |
| Tsianos et al., 2010 | Greek Mount Olympus marathon runners (n=438) | Cohort |
ACTN3 rs1815739 AMPD1 rs17602729 BDKRB2 rs1799722 ADRB2 rs1042713 PPARGC1α rs8192678 PPARα rs4253778; rs6902123 rs1053049; rs2267668 APOE rs7412; rs429358 |
BDKRB2 rs1799722, ADRB2 rs1042713 and AMPD1 rs17602729 associated with endurance running performance. |
| Wolfarth et al., 2008 | Caucasian male elite endurance athletes (n=316; 39 runners) and sedentary male controls (n=299) | Case-control |
NOS3 Glu298Asp rsl799983 G/T (CA)n repeats; 27-bp repeats 4B/4A |
164 bp allele of (CA)n repeats associated with elite endurance runners. |
NFIA-AS2, nuclear factor I A- antisense RNA 2; TSHR, thyrotropin receptor precursor; RBFOX1, RNA binding protein fox-1 homolog; ACE, angiotensin-converting enzyme; HIF1A, hypoxia-inducible factor 1-alpha, PPARGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PPARGC1β, peroxisome proliferator-activated receptor gamma coactivator 1-beta; PPRC1, peroxisome proliferator-activated receptor gamma coactivator-related protein 1; TFAM, mitochondrial transcription factor A; TFB1M, mitochondrial transcription factor B1; TFB2M, mitochondrial transcription factor B2; NRF1, nuclear respiratory factor 1; GABPA, GA-binding protein transcription factor alpha; GABPB1, GA-binding protein transcription factor beta 1; ERRα, estrogen-related receptor alpha; SIRT1, sirtuin-1; AQP1, aquaporin 1; COL5A1, collagen type V alpha-1; ADRA2A, adrenergic receptor alpha 2A; ACTN3, alpha-actinin-3; AMPD1, adenosine monophosphate deaminase 1; BDKRB2, bradykinin receptor B2; ADRB2, adrenergic receptor beta 2; PPARα, peroxisome proliferator activated receptor alpha; PPARD, peroxisome proliferator activated receptor delta; APOE, apolipoprotein E; NOS3, nitric oxide synthase 3.
Due to the heterogeneity of the study characteristics and SNPs measured with very few replication studies, it was deemed inappropriate to amalgamate the results for a meta-analysis. Therefore, the results in the current review were only analysed qualitatively.
Risk of bias within studies
The risk of bias of each study is presented in Table 2. Overall, the majority of studies were considered to have a “low” risk of bias in random sequence generation, allocation concealment and blinding. However, “high” risk of bias arose from the domains of missing outcome data in both Döring et al. (2010) and Martinez et al. (2009) for whom additionally, an “unclear” risk of bias arose from selective reporting of results. Two studies were also deemed “unclear” for missing outcome data (Ash et al., 2011; Posthumus et al., 2011) and two “unclear” (Ahmetov et al., 2015; Myerson et al., 1999) for selective reporting of results.
Table 2.
Risk of bias assessment as analysed by the Cochrane risk of bias tool (Higgins et al., 2016).
| References | Domain A | Domain B | Domain C | Domain D | Domain E | Total |
|---|---|---|---|---|---|---|
| Ahmetov et al., 2015 | Low | Low | Low | Low | Unclear | Low |
| Amir et al., 2007 | Low | Low | Low | Low | Low | Low |
| Ash et al., 2011 | Low | Low | Low | Unclear | Low | Low |
| Döring et al., 2010 | Low | Low | Low | High | Low | Low |
| He et al., 2015 | Low | Low | Low | Low | Low | Low |
| Martinez et al., 2009 | Low | Low | Low | High | High | Unclear |
| Myerson et al., 1999 | Low | Low | Low | Low | Unclear | Low |
| Papadimitriou et al., 2018 | Unclear | Low | Low | Low | Low | Low |
| Posthumus et al., 2011 | Low | Low | Low | Unclear | Low | Low |
| Sawczuk et al., 2013 | Low | Low | Low | Low | Low | Low |
| Stebbings et al., 2017 | Low | Low | Low | Low | Low | Low |
| Tobina et al., 2010 | Low | Low | Low | Low | Low | Low |
| Tsianos et al., 2010 | Low | Low | Low | Low | Low | Low |
| Wolfarth et al., 2008 | Low | Low | Low | Low | Low | Low |
Definition of categories: Domain: (A) sequence generation; (B) allocation concealment; (C) blinding; (D) missing outcome data; (E) selective reporting of results. Total risk of bias grade calculated by assessing the five domains (A–E).
Results of individual studies
Of the ten studies which compared the distribution of SNPs between elite endurance athletes (marathon runners n= 1,832) with control groups (n= 6,109), eight found a significant difference between the two groups (Table 3). Notably, the results for the ACE gene were heterogeneous. For example, while Tobina et al. (2010) found a strong association between ACE I/D rs4646994 and elite endurance runners (p = 0.001; Table 1), Ash et al. (2011) found no significant association (p = 0.16; Table 1). Of the 16 endurance-related alleles identified, nuclear factor I A-antisense RNA 2 (NFIA-AS2) SNP rs1572312 C displayed the highest frequency among elite endurance athletes (95.5%), however, this study was limited to two elite marathon runners (Ahmetov et al., 2015).
Table 3.
Allelic frequencies of the SNPs identified by comparison between elite endurance runners and controls.
| References | SNP | Frequency of allele in elite endurance runners, % | Frequency of allele in controls, % |
P-value |
|---|---|---|---|---|
| Ahmetov et al., 2015 | NFIA-AS2 rs1572312 C TSHR rs7144481 C RBFOX1 rs7191721 G |
95.5 25.7 58.6 |
88.8 17.4 56.3 |
0.007* 0.032* 0 .60 |
| Amir et al., 2007 | ACE D rs4646994 ACE DD rs4646994 |
77.0 62.0 |
66.0 43.0 |
0.01* 0.004* |
| Ash et al., 2011 | ACE I/D rs4646994 ACE A22982G rs4363 |
44.7 44.7 |
45.6 45.8 |
0.16 0.97 |
| Döring et al., 2010 | HIF1A Pro582 rs11549465 C HIF1A rs17099207 A HIF1A rs1951795 C HIF1A rs11158358 C/G HIF1A rs2301113 A/C HIF1A rs11549467 G/A |
84.0 9.2 67.7 25.3 33.5 3.2 |
75.0 4.9 62.2 27.0 32.9 1.6 |
0.017* 0.045* 0.038* 0.07 0.06 0.21 |
| He et al., 2015 | PPARAGC1A rs4452416 T PPARAGC1B rs2161257 G NRF1 rs7794909 T SIRT1 rs11596401 C |
17.2 39.6 9.6 87.2 |
13.1 33.6 5.8 83.3 |
0.035* 0.026* 0.007* 0.050* |
| Martinez et al., 2009 | AQP1 rs1049305 C | 57.0 | 49.0 | 0.03* |
| Myerson et al., 1999 | ACE I rs4646994 | 52.0 | 49.0 | 0.01* |
| Sawczuk et al., 2013 | ADRA2A rs553668 C | 85.8 | 82.9 | 0.40 |
| Tobina et al., 2010 | ACE I/D rs4646994 | 44.0 | 44.0 | 0.001* |
| Wolfarth et al., 2008 | NOS3 Glu298Asp rs1799983 G/T NOS3 (CA)n repeats 164 bp NOS3 27-bp repeats 4B/4A |
41.0 24.1 25.0 |
47.0 16.7 25.0 |
0.27 0.01* 0.83 |
*Significant difference between elite endurance, including marathon runners, and controls at p < 0.05 level.
Another four studies evaluated the association of SNPs with marathon running performance as opposed to athlete status. Posthumus et al. (2011) demonstrated a significant association between the collagen type V alpha-1 (COL5A1) gene BstUI RFLP T/C (rs12722) polymorphism and time to complete the running component of a triathlon where participants with a TT genotype completed the 42.2km run significantly faster than those with a CC genotype (p = 0.019).
Tsianos et al. (2010) found a significant association between beta-2 adrenergic receptor (ADRB2) SNP rs1042713 and adenosine monophosphate deaminase 1 (AMPD1) SNP rs17602729 and the fastest reported marathon completion times among male athletes (p = 0.01 and p = 0.04, respectively). In addition, among the whole male cohort, and all the more so when limited to habitual male runners, bradykinin B2 receptor (BDKRB2) SNP rs1799722 significantly deviated from Hardy-Weinberg equilibrium with an excess of the TT genotype (18.8%) which may indicate an associated genotype to endurance performance.
Additionally, Stebbings et al. (2018) found a significant difference in personal running time between titin (TTN) CC homozygote and CT heterozygote carriers in trained marathon runners (p = 0.02). However, Papadimitriou et al. (2018) did not find any significant association between alpha-actinin-3 (ACTN3) R577X or ACE I/D polymorphisms and endurance running times in a sample of 698 Caucasian athletes, so that an influence on running performance based on these two genes cannot be assumed.
Discussion
In the present review, data from 3,889 elite endurance athletes; 2,984 of which were specified as marathon runners, and 6,109 control individuals were qualitatively synthesised to evaluate the contribution and role of identified polymorphisms and genes to differentiate elite marathon performance from non-elite, power athletes and controls. Advancing previous systematic reviews which had focused solely on the candidate genes ACE and PPARα (Ma et al., 2013; Lopez-Leon et al., 2016), the results of the current review demonstrated that the following 16 SNPs in 14 different genes may be associated with elite marathon running performance:
NFIA-AS2 rs1572312 C, thyrotropin receptor precursor (TSHR) rs7144481 C, ACE rs4646994 I/D, hypoxia-inducible factor 1-alpha (HIF1A) Pro582 rs11549465 C, HIF1A rs17099207 A, HIF1A rs1951795 C, PPARAGC1α rs4452416 T, PPARAGC1β rs2161257 T, nuclear respiratory factor 1 (NRF1) rs7794909 C, sirtuin-1 (SIRT1) rs11596401 C, aquaporin (AQP1) rs1049305 C, nitric oxide synthase 3 (NOS3) (CA)n repeats 164 bp, COL5A1 BstUI RFLP rs12722 T, ADRB2 rs1042713, AMPD1 rs17602729 and BDKRB2 rs1799722.
These findings provide support for the role of multiple endurance-related genes and are in line with previous literature that has reported that at least 93 genetic markers are associated with elite endurance athlete status in general (Ahmetov et al., 2016).
Key primary functions of identified genes
Although a detailed mechanistic explanation regarding the roles of each of the identified genes and their polymorphisms in elite marathon running performance is beyond the scope of this review, the following points can be noted.
Of the fourteen genes, the PPARAGC1α, PPARAGC1β, NRF1, SIRT1, HIF1A, AQP1, AMPD1, BDKRB2, NFIA-AS2 and TSHR genes code for transcription factors and coactivators primarily involved in metabolic pathways (i.e. adenosine triphosphate (ATP) generation, glucose and lipid metabolism, mitochondrial biogenesis, thermogenesis, angiogenesis and muscle fibre type composition).
The COL5A1 gene encodes type V collagen in ligaments and is involved in range of motion (Brown et al., 2011) and COL5A1 rs12722 may influence the energetic cost of running (Posthumus et al., 2011).
The ACE, NOS3 and ADRB2 genes code for enzymes involved in cardiovascular function such as blood pressure and vasodilation (Wolfarth et al., 2008; Ahmetov et al., 2009; Drozdovska et al., 2013).
In particular, the PPARAGC1α and PPARAGC1β genes have been suggested to play a key role in exercise-induced mitochondrial biogenesis through the co-activation of NRF1 (Joseph et al., 2006). In turn, NRF1 induces the transcription of mitochondrial transcription factor A (TFAM) resulting in mitochondrial DNA (mtDNA) replication (Yan et al., 2010). By increasing mitochondrial content, marathon running performance may be improved due to increased beta-oxidation resulting in the conservation of glycogen stores and reduction in lactate accumulation (Cantó et al., 2009).
Interestingly, it has been proposed that the activity of PPARAGC1α during endurance exercise is regulated by another of the identified genes SIRT1 (Philp et al., 2013). The AMP-activated protein kinase (AMPK)-induced activation of SIRT1 results in the deacetylation of PPARAGC1α, which in turn increases PPARAGC1α activity and upregulates mitochondrial biogenesis in skeletal muscle (Cantó et al., 2009). Therefore, SIRT1 may also be beneficial in marathon running performance due to its ability to deacetylate PPARAGC1α in a nicotinamide adenine dinucleotide (NAD+) dependent manner (Nemeto et al., 2005).
In addition, PPARAGC1α has been shown to stimulate the formation of type I slow-twitch fibres (Lin et al., 2002). This increased proportion of oxidative slow-twitch fibres is likely to improve marathon running performance as it is related to a higher oxidative capacity, mitochondrial content and fatigue resistance (Hawley and Spargo, 2007). Accordingly, Ahmetov et al. (2009) demonstrated that the PPARAGC1α rs8192678 polymorphism was positively associated with the percentage of slow-twitch fibres, V̇O2 max and elite endurance athlete status.
Conversely, in the present review, no association was found between PPARAGC1α rs8192678 and elite marathon running performance (p = 0.64; Tsianos et al., 2010). However, other allelic variants in the gene such as rs4452416 T were associated with elite endurance running status (p = 0.035; He et al., 2015). One possible explanation for the differences between the results of the current review and previous studies (Ahmetov et al., 2009; Tural et al., 2014) is the ethnicity of the population investigated. It has been demonstrated that a significant association between polymorphic variants and elite sport performance observed in one population is not always replicated in other populations (Ginevičienė et al., 2011). Therefore, the possibility cannot be excluded that the lack of association between PPARAGC1α rs819268 polymorphism and marathon running performance found by Tsianos et al. (2010) was distinctive to athletes of Greek ancestry.
Although studies set the per-allele effect typically at 80% power to detect at α= 0.05 associations (Tsianos et al., 2010), a small effect size could still remain and inter-individual differences in endurance performance may be influenced by genetic variations that may not be identified or consistently replicated at a genome-wide level (Papadimitriou et al., 2018). In genetic association studies, there is still uncertainty due to lack of statistical power, unknown connection with other variants and environmental interactions. Therefore, it cannot exclude the possibility that the associations are due to type 1 error or an unknown connection to another polymorphic functional variant (Döring et al., 2010).
The AQP1 rs1049035 C/G polymorphism (Martinez et al., 2009) encodes a water channel protein, which has been suggested to play a key role in the regulation of body temperature and the secretion and absorption of body fluid (Sui et al., 2001). During osmotic stress (i.e. endurance exercise), AQP1 facilitates the transport of water from the blood into the muscle and promotes water reabsorption (Sui et al., 2001). In support of this, carriers of the AQP1 rs1049305 C allele had a significantly greater adjusted body fluid loss (3.7 ± 0.9 kg) than non-carriers (1.5 ± 1.1 kg) in response to a 10km distance run (p < 0.05; Rivera et al., 2011). Therefore, the possession of the AQP1 rs1049305 C allele may enhance marathon running performance due to the maintenance of adequate body fluid balance, thereby improving blood flow and enhancing oxygen delivery to the working muscles and thermoregulation (Brooks et al., 2005).
The HIF1A gene functions as a master regulator of oxygen homeostasis and has been suggested to play a role in a number of cellular functions including angiogenesis, erythropoiesis and glucose metabolism (Ahmetov et al., 2009). The ability of HIF1A protein to inhibit ubiquitination and proteasomal degradation under hypoxic conditions results in an exponential increase in HIF1A levels as cellular oxygen concentration decreases, which in turn stimulates erythropoiesis and consequently improves the efficiency of oxygen delivery to the working muscles (Simenza, 2002). Furthermore, the HIF1A polymorphism Pro582 rs11549465 C has been associated with both a higher initial value and trainability of V̇O2 max (Prior et al., 2003) which is an important factor governing the capacity of marathon running performance (Joyner and Coyle, 2008).
With regard to the role of genes involved in cardiovascular functions, ACE has been the most extensively studied candidate gene (Ahmetov et al., 2009). It plays a key role in the regulation of blood volume and pressure by promoting the synthesis of vasoconstrictor angiotensin II (ANG II) and the degradation of vasodilator kinins (Woods et al., 2000). The I allele has been proposed to improve endurance performance due to lower ACE activity and increased bradykinin levels resulting in enhanced substrate delivery to the working muscles (Woods et al., 2000). In support of this, the present review found a positive association between the ACE polymorphism I rs4646994 and endurance running performance (Myerson et al., 1999). In contrast, two of the studies (Amir et al., 2007; Tobina et al., 2010) found that the D allele was favourable for elite marathon running performance. It is speculated that the D allele may result in improved cardiac function due to its association with increased exercise-induced left ventricular hypertrophy (Hernández et al., 2003).
In addition, the higher ACE activity associated with the D allele in comparison to the I allele, may hinder glucose uptake in skeletal muscle due to the increased degradation of bradykinin, a trigger for glucose transporter type 4 (GLUT4) translocation, predominantly expressed in the muscle (Kishi et al., 1998). This may be advantageous to marathon running performance as this regulates glucose homeostasis, allowing glucose uptake, thereby preventing hypoglycaemia and central fatigue (Newsholme et al., 1992). Moreover, Ash et al. (2011) found no association between ACE gene variation and East African elite marathon runners. These discrepancies highlight the limitations of the interpretation of association studies and may be attributable to sample sizes, ethnic disparities and the inclusion of athletes from a heterogeneous range of endurance sporting disciplines (Barley et al., 1994; Rankinen et al., 2000).
Risk of bias and reasons for divergent results
Despite the wide variety of genetic markers included in the present review, it should be emphasised that only one of the 160 SNPs, ACE I/D rs4646994, was investigated in three (Amir et al., 2007; Ash et al., 2011; Tobina et al., 2010). The lack of replicated studies raises the possibility that some of the results reported might be false-positive and highlights the infancy of the field of sport genomics. It is also important to highlight the impact of epigenetics, in which the environment influences the expression of genes. Of relevance, Raleigh (2012) highlights the implications of epigenetic factors on the regulation of the ACE gene beyond that of the I/D polymorphisms. As such, in addition to genotyping, understanding the impact of epigenetic regulation on genes such as ACE and the modification to endurance performance is of paramount importance.
As a consequence of limited replication of genotype-phenotype associations, the significant associations in the small candidate gene studies observed in 8 out of the 10 studies that compared elite athletes to control groups, illustrates a possible publication bias. Therefore, robust replication of studies in large cohorts of athletes is required before findings can be applied to practice in sport.
Furthermore, much of the existing research assessing the role of genes in sport performance has focused on endurance or power/strength as a whole (Ahmetov et al., 2016), which was evident in the present review with only four of the studies (Amir et al., 2007; Martinez et al., 2009; Tsianos et al., 2010; Ash et al., 2011) solely including elite marathon runners. The inclusion of different sporting disciplines in the majority of the studies identified a potential limitation as it dilutes the phenotypic characteristics and selection of elite athletes (Williams et al., 2014). Therefore, further empirical attention within a given sport, in this case marathon running, is required. The application of genomic data to practice in sport is currently considered premature, thus the second objective of this review, to provide some insight into the use of genetic testing in marathon talent identification was not achievable.
Although the overall risk of bias within the included studies was deemed low, some areas of concern included selective reporting bias and incomplete data. In particular, studies tended to only report the allelic frequencies of the most significant SNPs identified (Ahmetov et al., 2015). Therefore, it is recommended that future studies report all allelic frequencies measured rather than solely focusing on the most common genetic variants. It is envisaged that by moving away from the candidate gene approach a greater understanding of the polygenic nature of performance-related physical and mental traits will be established.
The lack of significant association in many studies despite the large number of SNPs included indicates how difficult it is to identify genes that influence the status of being a world-class elite athlete (He et al., 2015). This is likely due to the presence of small scale studies, the lack of replication of genotype-phenotype associations and the likelihood of publication bias. Future replication studies, however, are needed to determine if functional SNPs contribute to the wide variability in athletic performance and adaptation to endurance training (He et al., 2015).
To fully understand the mechanisms underlying the supposed association between polymorphisms and endurance performance, these functional studies are required. A GWAS approach may be warranted since this has been suggested to be advantageous in the detection of multiple, novel genetic variants due to its whole-genome analysis and hypothesis-free approach (Pitsiladis et al., 2013). Where GWAS has already shown success in elucidating the genetic basis for other complex traits such as obesity and Type II diabetes mellitus (Frayling et al., 2007; McCarthy and Zeggini, 2009). However, it cannot be ignored, that candidate genes have at times yielded reproducible outcomes, and thus GWAS may be used as an approach to identifying genetic variants to be followed by candidate gene studies. Additionally, the impact of epigenetics and therefore the role of environmental factors are also of importance to understanding the phenotype of marathon running and cannot be excluded from establishing the role and impact genes play in the success of sport performance.
Implications
In addition to the above-discussed natural influence of genetic differences on running performance, a potential risk and ethical issue has emerged by the manipulation of such genes to enhance physical performances through gene doping. Commonly defined as the transfer of genes or genetically modified cells into an individual as a potential method to illicit athletic performance enhancement. Gene doping is considered an arising and upcoming threat to clean sport and a prohibition of gene doping methods was added to the World Anti-Doping Agency (WADA) prohibited list since 2003, but although there are no known cases of gene doping use among high performance athletes as yet, the application of gene manipulation in sports cannot be ruled out (Haisma and De Hon, 2006).
Van der Gronde (2013) reviewed and assessed the usability of known human performance-related genes for the potential as a gene doping substances. Ultimately, peroxisome proliferator-activated receptor-delta (PPARr) and cytosolic phosphoenolpyruvate carboxykinase (PEPCK-C) were identified as high potential genes for abuse. However, to date, genetic science especially in terms of doping is in its infancy and therefore prone to unknown risks and linked to a high degree of potentially life-threatening dangers (Miah, 2004). Furthermore, a deliberate intervention into human genetics to enhance physical capacities goes against the notion and ethical aspects of sport as fair competition, just as conventional doping such as steroid use does (Breivik, 2005).
Conclusion
In conclusion, to the author’s knowledge this is the first systematic review investigating the role of multiple genetic markers in elite marathon running performance. Overall, a possible 16 different SNPs in 14 genes have been identified, which can be used as preliminary evidence for developing focused investigation of the polygenic nature of complex endurance-related traits.
It is still unknown which genes are specifically associated to elite marathon performance due to the small effect sizes, and that the variants of interest are not likely to be obligatory. Also limited replication of genotype-phenotype associations and possible publication bias exists, thus further studies are required to strengthen our understanding of the genes involved in elite marathon running. As a result, future research adopting the GWAS approach is warranted for the detection of multiple, novel genetic variants. Therefore, as the science of sport genomics matures through the utilisation of genome-wide technologies over the coming years there is potential for the use of genetic testing as a valid tool in the identification of young athletes. This early identification of potential physical and mental traits is of paramount interest to optimise training and marathon running performance but is also a potential threat to the integrity of sport, with the potential of gene doping.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The study comply with the current laws of the country in which were performed. The authors report no conflict of interest.
Biographies

Hannah J. MOIR
Employment
Applied & Human Sciences, School of Life Sciences, Pharmacy & Chemistry, Kingston University, UK.
Degree
PhD
Research interests
Allied Health Science, Ultra-endurance running, Sports Medicine and Immunochemistry.
E-mail: H.Moir@kingston.ac.uk

Rachael KEMP
Employment
School of Sport, Health and Exercise Sciences, Bangor University, UK.
Degree
MSc
Research interests
Exercise Immunology, Exercise Physiology
E-mail: rck18ttf@bangor.ac.uk

Dirk FOLKERTS
Employment
Department of Sport Psychology, Faculty of Sport and Exercise Sciences, University of Muenster, Germany.
Degree
MSc
Research interests
Sports Medicine, Sport Psychology, Doping in Sport
E-mail: dirk.folkerts@uni-muenster.de

Owen SPENDIFF
Employment
Applied & Human Sciences, School of Life Sciences, Pharmacy & Chemistry, Kingston University, UK.
Degree
PhD
Research interests
Exercise Science, Ergogenic aids such as Dark Chocolate and Keto diets, Nitrate and Flavanols in Exercise
E-mail: O.spendiff@kingston.ac.uk

Cristiana PAVLIDIS
Employment
Department of Pharmacy, University of Patras, Pátra, Greece.
Degree
PhD
Research interests
Clinical Nutrition and Dietetics, Nutrigenomics
E-mail: cristianapavlidis@hotmail.com

Elizabeth OPARA
Employment
Applied & Human Sciences, School of Life Sciences, Pharmacy & Chemistry, Kingston University, UK.
Degree
PhD
Research interests
Clinical Nutrition, Bioactive properties of plant derived foods and medicinal plants
E-mail: E.Opara@kingston.ac.uk
References
- Ahmetov I.I., Williams A.G., Popov D.V., Lyubaeva E.V., Hakimullina A.M., Fedotovskaya O.N., Mozhayskaya I.A., Vinogradova O.L., Astratenkova I.V., Montgomery H.E., Rogozkin V.A. (2009) The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Human Genetics 126 (6), 751. [DOI] [PubMed] [Google Scholar]
- Ahmetov I.I., Kulemin N.A., Popov D.V., Naumov V.A., Akimov E.B., Bravy Y.R., Egorova E.S., Galeeva A.A., Generozov E.V., Kostryukova E.S., Larin AK. (2015) Genome-wide association study identifies three novel genetic markers associated with elite endurance performance. Biology of Sport 32 (1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmetov I.I., Egorova E.S., Gabdrakhmanova L.J., Fedotovskaya O.N. (2016). Genes and Athletic Performance: An Update. Medicine and Sport Science 61, 41-54. [DOI] [PubMed] [Google Scholar]
- Amir O., Amir R., Yamin C., Attias E., Eynon N., Sagiv M., Sagiv M., Meckel Y. (2007) The ACE deletion allele is associated with Israeli elite endurance athletes. Experimental Physiology 92(5), 881-886. [DOI] [PubMed] [Google Scholar]
- Ash G.I., Scott R.A., Deason M., Dawson T.A., Wolde B., Bekele Z., Teka S., Pitsiladis Y.P. (2011) No association between ACE gene variation and endurance athlete status in Ethiopians. Medicine and Science in Sports and Exercise 43(4), 590-597. [DOI] [PubMed] [Google Scholar]
- Barley J., Blackwood A., Carter N.D., Crews D.E., Cruickshank J.K., Jeffery S., Ogunlesi A.O., Sagnella G.A. (1994) Angiotensin converting enzyme insertion/deletion polymorphism: association with ethnic origin. Journal of Hypertension 12(8), 955-957. [PubMed] [Google Scholar]
- Bouchard C., An P., Rice T., Skinner J.S., Wilmore J.H., Gagnon J., Pérusse L., Leon A.S., Rao D.C. (1999) Familial aggregation of VO2max response to exercise training: results from the HERITAGE Family Study. Journal of Applied Physiology 87(3), 1003-1008. [DOI] [PubMed] [Google Scholar]
- Breivik G. (2005). Sport, gene doping and ethics. Tamburrini C, Tansjö T, editors. Genetic Technology and Sport: Ethical questions. London: Routledge, Taylor and Francis, 165-177. [Google Scholar]
- Brooks G., Fahey T., Baldwin K. (2006) Exercise Physiology. Human Bioenergetics and its Applications. 4th edition. New York: McGraw Hill. [Google Scholar]
- Brown J.C., Miller C.J., Posthumus M., Schwellnus M.P., Collins M. (2011). The COL5A1 gene, ultra-marathon running performance, and range of motion. International Journal of Sports Physiology and Performance 6(4), 485-96. [DOI] [PubMed] [Google Scholar]
- Cantó C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458(7241), 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring F., Onur S., Fischer A., Boulay M.R., Pérusse L., Rankinen T., Rauramaa R., Wolfarth B., Bouchard C. (2010). A common haplotype and the Pro582Ser polymorphism of the hypoxia-inducible factor-1α (HIF1A) gene in elite endurance athletes. Journal of Applied Physiology, 108(6), 1497-500. [DOI] [PubMed] [Google Scholar]
- Drozdovska S.B., Dosenko V.E., Ahmetov I.I., Ilyin V.N. (2013). The association of gene polymorphisms with athlete status in Ukrainians. Biology of Sport 30(3), 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., Shields B. (2007) A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayagay G., Yu B., Hambly B., Boston T., Hahn A., Celermajer D.S., Trent R.J. (1998) Elite endurance athletes and the ACE I allele–the role of genes in athletic performance. Human Genetics 103(1), 48-50. [DOI] [PubMed] [Google Scholar]
- Ginevičienė V., Pranculis A., Jakaitienė A., Milašius K., Kučinskas V. (2011) Genetic variation of the human ACE and ACTN3 genes and their association with functional muscle properties in Lithuanian elite athletes. Medicina 47 (5), 40. [PubMed] [Google Scholar]
- Haisma H.J., De Hon O. (2006) Gene doping. International Journal of Sport Medicine, 27(04), 257-266. [DOI] [PubMed] [Google Scholar]
- Hawley J.A., Spargo F.J. (2007) Metabolic adaptations to marathon training and racing. Sports Medicine 37(4-5), 328-331. [DOI] [PubMed] [Google Scholar]
- He Z.H., Hu Y., Li Y.C., Gong L.J., Cieszczyk P., Maciejewska-Karlowska A., Leonska-Duniec A.G., Muniesa C.A., Marín-Peiro M., Santiago C.A., Garatachea N. (2015) PGC-related gene variants and elite endurance athletic status in a Chinese cohort: A functional study. Scandinavian Journal of Medicine and Science in Sports 25(2), 184-195. [DOI] [PubMed] [Google Scholar]
- Hernández D., De La Rosa A., Barragán A., Barrios Y., Salido E., Torres A., Martín B., Laynez I., Duque A., De Vera A., Lorenzo V. (2003) The ACE/DD genotype is associated with the extent of exercise-induced left ventricular growth in endurance athletes. Journal of the American College of Cardiology 42(3), 527-532. [DOI] [PubMed] [Google Scholar]
- Higgins J.P., Sterne J.A., Savović J., Page M.J., Hróbjartsson A. (2016) A revised tool for assessing risk of bias in randomized trials In: Chandler J, McKenzie J, Boutron I, Welch V. (editors). Cochrane Database of Systematic Reviews 10. [Google Scholar]
- Joseph A.M., Pilegaard H., Litvintsev A., Leick L., Hood D.A. (2006) Control of gene expression and mitochondrial biogenesis in the muscular adaptation to endurance exercise. Essays Biochemistry 42, 13-29. [DOI] [PubMed] [Google Scholar]
- Joyner M.J., Coyle E.F. (2008) Endurance exercise performance: the physiology of champions. Journal of Physiology 586(1), 35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi K., Muromoto N., Nakaya Y., Miyata I., Hagi A., Hayashi H., Ebina Y. (1998) Bradykinin directly triggers GLUT4 translocation via an insulin-independent pathway. Diabetes 47(4), 550-558. [DOI] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Lowell B.B. (2002) Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418 (6899), 797. [DOI] [PubMed] [Google Scholar]
- Lopez-Leon S., Tuvblad C., Forero D.A. (2016) Sports genetics: the PPARA gene and athletes’ high ability in endurance sports. A systematic review and meta-analysis. Biology of Sport 33 (1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Yang Y., Li X., Zhou F., Gao C., Li M., Gao L. (2013) The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PloS One 8 (1), e54685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J.L., Carrion A., Florian M.E., Martin J.A., Lopez-Taylor J.R., Fahey T.D., Rivera M.A. (2009) Aquaporin-1 Gene Dna Variation Predicts Performance in Hispanic Marathon Runners. Medicina Sportiva 13(4), 251-255. [Google Scholar]
- McCarthy M.I., Zeggini E. (2009) Genome-wide association studies in type 2 diabetes. Current Diabetes Reports 9(2), 164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miah A. (2004). Genetically Modified Athletes. London: Routledge. [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G. (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Medicine 151(4), 264-269. [DOI] [PubMed] [Google Scholar]
- Montgomery H. E., Marshall R., Hemingway H., Myerson S., Clark-son P., Dollery C., Hayward M., Holliman D.E., Jubb M., Thomas EL. (1998) Human gene for physical performance. Nature 393 (6682), 221. [DOI] [PubMed] [Google Scholar]
- Myerson S., Hemingway H., Budget R., Martin J., Humphries S., Montgomery H. (1999) Human angiotensin I-converting enzyme gene and endurance performance. Journal of Applied Physiology 87(4), 1313-1316. [DOI] [PubMed] [Google Scholar]
- Nemoto S., Fergusson M.M., Finkel T. (2005) SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. Journal of Biological Chemistry 280(16), 16456-16460. [DOI] [PubMed] [Google Scholar]
- Newsholme E.A., Blomstrand E., Ekblom B. (1992) Physical and mental fatigue: metabolic mechanisms and importance of plasma amino acids. British Medical Bulletin 48(3), 477-495. [DOI] [PubMed] [Google Scholar]
- Papadimitriou I. D., Lockey S. J., Voisin S., Herbert A. J., Garton F., Houweling P.J., Cieszczyk P., Maciejewska-Skrendo A., Sawczuk M., Massidda M., Calò C. M. (2018) No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. BMC Genomics 19 (1), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp A., Schenk S. (2013) Unraveling the complexities of SIRT1-mediated mitochondrial regulation in skeletal muscle. Exercise and Sport Science Reviews 41 (3), 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsiladis Y., Wang G., Wolfarth B., Scott R., Fuku N., Mikami E., He Z., Fiuza-Luces C., Eynon N., Lucia A. (2013) Genomics of elite sporting performance: what little we know and necessary advances. British Journal of Sports Medicine 47(9), 550-555. [DOI] [PubMed] [Google Scholar]
- Popovski Z. T., Nestorovski T., Wick M., Tufekchievski A., Aceski A., Gjorgjievski S. (2016) Molecular-genetic predictions in selection of sport talents and ethical aspect of their application. Research in Physical Education, Sport and Health 5(1), 57-63. [Google Scholar]
- Posthumus M., Schwellnus M.P., Collins M. (2011) The COL5A1 gene: a novel marker of endurance running performance. Medicine and Science in Sport and Exercise 43(4), 584-589. [DOI] [PubMed] [Google Scholar]
- Prior S. J., Hagberg J. M., Phares D. A., Brown M. D., Fairfull L., Ferrell R. E., Roth S. M. (2003) Sequence variation in hypoxia-inducible factor 1α (HIF1A): association with maximal oxygen consumption. Physiological Genomics 15(1), 20-26. [DOI] [PubMed] [Google Scholar]
- Raleigh S. (2012) Epigenetic regulation of the ACE gene might be more relevant to endurance physiology than the I/D polymorphism. Journal of Applied Physiology 112(6), 1082-1083. [DOI] [PubMed] [Google Scholar]
- Rankinen T., Wolfarth B., Simoneau J.A., Maier-Lenz D., Rauramaa R., Rivera M.A., Boulay M.R., Chagnon Y.C., Pérusse L., Keul J., Bouchard C. (2000) No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. Journal of Applied Physiology 88(5), 1571-1575. [DOI] [PubMed] [Google Scholar]
- Rivera M.A., Martínez J.L., Carrion A., Fahey T.D. (2011) AQP-1 association with body fluid loss in 10-km runners. International Journal of Sports Medicine 32(03), 229-33. [DOI] [PubMed] [Google Scholar]
- Roth S.M. (2012) Critical overview of applications of genetic testing in sport talent identification. Recent Patents on DNA and Gene Sequences 6(3), 247-255. [DOI] [PubMed] [Google Scholar]
- Sawczuk M., Maciejewska-Karlowska A., Cieszczyk P. (2013) A single nucleotide polymorphism rs553668 in the ADRA2A gene and the status of Polish elite endurance athletes. Trends in Sport Science 20(1), 30-35. [Google Scholar]
- Scott R.A., Moran C., Wilson R.H., Onywera V., Boit M.K., Goodwin W.H., Gohlke P., Payne J., Montgomery H., Pitsiladis Y.P. (2005) No association between Angiotensin Converting Enzyme (ACE) gene variation and endurance athlete status in Kenyans. Comparative Biochemistry and Physiology Part A: Molecular and Integrative Physiology 141(2), 169-175. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. (2002) Signal transduction to hypoxia-inducible factor 1. Biochemical Pharmacology 64(5-6), 993-998. [DOI] [PubMed] [Google Scholar]
- Stebbings G.K., Williams A.G., Herbert A.J., Lockey S.J., Heffernan S.M., Erskine R.M., Morse C.I., Day S.H. (2018) TTN genotype is associated with fascicle length and marathon running performance. Scandinavian Journal of Medicine and Science in Sports 28(2), 400-406. [DOI] [PubMed] [Google Scholar]
- Sui H., Han B. G., Lee J. K., Walian P., Jap B. K. (2001) Structural basis of water-specific transport through the AQP1 water channel. Nature 414 (6866), 872. [DOI] [PubMed] [Google Scholar]
- Tobina T., Michishita R., Yamasawa F., Zhang B., Sasaki H., Tanaka H., Saku K., Kiyonaga A. (2010) Association between the angiotensin I-converting enzyme gene insertion/deletion polymorphism and endurance running speed in Japanese runners. The Journal of Physiological Sciences 60(5), 325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsianos G.I., Evangelou E., Boot A., Carola Zillikens M., van Meurs J.B., Uitterlinden A.G., Ioannidis J.P. (2010) Associations of polymorphisms of eight muscle-or metabolism-related genes with performance in Mount Olympus marathon runners. Journal of Applied Physiology 108(3), 567-574. [DOI] [PubMed] [Google Scholar]
- Tucker R., Santos-Concejero J., Collins M. (2013) The genetic basis for elite running performance. British Journal of Sports Medicine 47(9), 545-549. [DOI] [PubMed] [Google Scholar]
- Tural E., Kara N., Agaoglu S. A., Elbistan M., Tasmektepligil M. Y., Imamoglu O. (2014) PPAR-α and PPARGC1A gene variants have strong effects on aerobic performance of Turkish elite endurance athletes. Molecular Biology Reports 41(9), 5799-5804. [DOI] [PubMed] [Google Scholar]
- Van der Gronde T., de Hon O., Haisma H. J., Pieters T. (2013) Gene doping: an overview and current implications for athletes. British Journal of Sports Medicine 47(11), 670-678. [DOI] [PubMed] [Google Scholar]
- Vancini R. L., Pesquero J. B., Fachina R. J., dos Santos Andrade M., Borin J. P., Montagner P. C., de Lira C. A. B. (2014) Genetic aspects of athletic performance: the African runners phenomenon. Open Access Journal of Sports Medicine 5, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Padmanabhan S., Wolfarth B., Fuku N., Lucia A., Ahmetov I.I., Cieszczyk P., Collins M., Eynon N., Klissouras V., Williams A. (2013) Genomics of elite sporting performance: what little we know and necessary advances. Advances in Genetics 84, 123-149. [DOI] [PubMed] [Google Scholar]
- Wilber R.L., Pitsiladis Y.P. (2012) Kenyan and Ethiopian distance runners: what makes them so good? The International Journal of Sports Physiology and Performance 2, 92-102. [DOI] [PubMed] [Google Scholar]
- Williams A. G., Day S. H., Lockey S. J., Heffernan S. M., Erskine R. M. (2014) Genomics as a practical tool in sport-have we reached the starting line? Cellular and Molecular Exercise Physiology 3(1), e6. [Google Scholar]
- Wolfarth B., Rankinen T., Mühlbauer S., Ducke M., Rauramaa R., Boulay M.R., Pérusse L., Bouchard C. (2008) Endothelial nitric oxide synthase gene polymorphism and elite endurance athlete status: the Genathlete study. Scandinavian Journal of Medicine and Science in Sports 18(4), 485-490. [DOI] [PubMed] [Google Scholar]
- Woods D. R., Humphries S. E., Montgomery H. E. (2000) The ACE I/D polymorphism and human physical performance. Trends in Endocrinology and Metabolism 11(10), 416-420. [DOI] [PubMed] [Google Scholar]
- Yan Z., Okutsu M., Akhtar Y.N., Lira V.A. (2010) Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. Journal of Applied Physiology 110(1), 264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Association of Athletics Federations. Senior Outdoor Marathon Men. 2018.. (2018) Available from URL: https://www.iaaf.org/records/toplists/road-running/marathon/outdoor/men/senior Accessed 11 June 2018


