Abstract
The outbreak of diseases ordinarily results from the disruption of the balance and harmony between hosts and pathogens. Devoid of adaptive immunity, shrimp rely largely on the innate immune system to protect themselves from pathogenic infection. Two nuclear factor-κB (NF-κB) pathways, the Toll and immune deficiency (IMD) pathways, are generally regarded as the major regulators of the immune response in shrimp, which have been extensively studied over the years. Bacterial infection can be recognized by Toll and IMD pathways, which activate two NF-κB transcription factors, Dorsal and Relish, respectively, to eventually lead to boosting the expression of various antimicrobial peptides (AMPs). In response to white-spot-syndrome-virus (WSSV) infection, these two pathways appear to be subverted and hijacked to favor viral survival. In this review, the recent progress in elucidating microbial recognition, signal transduction, and effector regulation within both shrimp Toll and IMD pathways will be discussed. We will also highlight and discuss the similarities and differences between shrimps and their Drosophila or mammalian counterparts. Understanding the interplay between pathogens and shrimp NF-κB pathways may provide new opportunities for disease-prevention strategies in the future.
Keywords: NF-κB pathways, shrimp, bacteria, white spot syndrome virus (WSSV), Toll-like receptor, IMD
Introduction
Shrimp farming is an important economic activity in China and many Southeast Asian countries, such as the Philippines, Indonesia, Malaysia, Thailand, and Vietnam, and it provides an important contribution to the diversity of income strategies for a large proportion of people living in these countries. According to the statistics of the Food and Agriculture Organization (FAO) of the United Nations, seawater and low-salinity brackish-water shrimp-culture production in the world expanded from 1,325 metric tons in 1950 to 4,875,793 metric tons in 2015. The main species of shrimp aquaculture include Litopenaeus vannamei (L. vannamei), Fenneropenaeus chinensis (F. chinensis), Penaeus monodon (P. monodon), Marsupenaeus japonicus (M. japonicus), Procambarus clarkii (P. clarkii), and Macrobrachium rosenbergii (M. rosenbergii) (1). Among them, L. vannamei accounted for nearly 80% of global shrimp production in 2015. Even though the production of shrimp has increased considerably in recent years, emerging diseases have been the main threat to restrict the sustainable development of the industry worldwide. A wide range of pathogens, including viruses, bacteria, fungi, and parasites, can infect shrimp, among which the white spot syndrome virus (WSSV) and Vibrio parahaemolyticus (VPAHPND) have led to the most serious economic losses in the shrimp-cultivation industry worldwide. WSSV is the causative agent of white spot syndrome (WSS), which causes 100% mortality within 7–10 days (2). VPAHPND contains two 69-kb plasmid-carrying binary Pir-like toxin genes, A and B, which are the causative agents of acute hepatopancreatic necrosis disease (AHPND) (3, 4). Outbreak of diseases often occurs when the homeostasis between pathogens and host resistance is disrupted. Therefore, better understanding the shrimp immune system will provide more effective strategies for disease prevention and control under specific culture ecosystems.
Innate immunity is characterized by the activation of pattern-recognition receptors or proteins (PRRs or PRPs) capable of sensing and binding with pathogen-associated molecular patterns (PAMPs) that are only presented in the pathogen but not in the host (5). The innate immune system of invertebrates is generally divided into humoral defenses and cellular defenses. Humoral defense include the production of soluble effector molecules, such as antimicrobial peptides (AMPs), while cellular defenses, such as phagocytosis and encapsulation, are mediated by circulating hemocytes (6–8). Like other invertebrates, shrimp mainly rely on the innate immune response to control and clear invading pathogens following infection. Upon PRRs-PAMPs interaction, the cellular and humoral immune responses are rapidly activated, which adopt a coordinated strategy to eliminate foreign pathogens. The nuclear factor-κB (NF-κB) family of transcription factors comprise key regulators of humoral immune responses that are indispensable for the host-defense system (9). In shrimp, there are two NF-κB transcription factors, namely Dorsal and Relish (10–14). Dorsal is the critical transcription factor in the Toll signaling pathway, while Relish plays a key role in the IMD signaling pathway (10–14). Since the first shrimp Toll receptor, LvToll or LvToll1, was reported in 2007 (15, 16) and the first shrimp IMD homolog, LvIMD, was identified in 2009 (17), a series of significant findings have been made over the last 20 years. Increased attention has been focused on the identification of pathway components and deciphering the molecular mechanisms underlying the Toll and IMD pathways related to infection. Herein, we present an overview of our current knowledge of the two NF-κB pathways in immune responses to bacterial and WSSV infection, with the hope of providing novel insights into the immune system of shrimp.
The Toll Pathway
Tolls and Toll-like receptors (TLRs, usually defined from the Toll homologs of vertebrates) have been recognized as major PRRs in innate immunity and they play an indispensable role in recognition of microbes during host defense (18, 19). In Drosophila, a total of nine Toll receptors have been identified (20). Drosophila Toll-1, or simply Toll, was the first Toll identified (21), and its mediated cellular-signaling cascades, comprising the MyD88/Tube/Pelle/Cactus/Dorsal axis, are widely considered to form the canonical Toll pathway. The Drosophila Toll pathway can be induced by Gram-positive bacteria, fungi, and some viruses (20). The activation of Toll leads to the initiation of cellular-signaling transduction and ultimately results in the systemic production of specific antimicrobial peptides (AMPs), such as the antifungal peptide Drosomycin, which is widely considered as a conventional readout of activation of the Drosophila Toll pathway (22, 23). After the identification of the Drosophila Toll pathway, many findings have been obtained in vertebrates, especially mammals, and some invertebrates, including shrimp. In recent years, many components of the shrimp canonical Toll pathway and its concomitant regulators—including Spätzle, Toll, MyD88, Tube, Pelle, Pellino, TRAF6, Dorsal, Cactus, Tollip, SARM, Flightless-I, and β-arrestin—have been identified and functionally characterized. These characterizations have provided some surprising insights into the shrimp Toll pathway in the context of receptor sensing, signaling transduction, and host-pathogen interactions. The identified components of the Toll-related pathway in penaeid shrimp are listed in Table 1.
Table 1.
Components of canonical Toll signaling pathway identified in shrimps.
| Components | Species | Gene names | Accession numbers | In response to pathogenic infections | References |
|---|---|---|---|---|---|
| Toll | L. vannamei | LvToll1 | DQ923424.1 | Gill: M. lysodeikticus, V. harveyi, V. anguillarum, and WSSV, up | (15, 16, 24, 25) |
| LvToll2 | JN180637 | Gill: WSSV, up | (26) | ||
| LvToll3 | JN180638 | Gill: WSSV, up | (26) | ||
| LvToll4 | Unsubmitted | Gill, Hemocytes: WSSV, up | (27) | ||
| LvToll5 | Unsubmitted | ND | (27) | ||
| LvToll6 | Unsubmitted | ND | (27) | ||
| LvToll7 | Unsubmitted | ND | (27) | ||
| LvToll8 | Unsubmitted | ND | (27) | ||
| LvToll9 | Unsubmitted | ND | (27) | ||
| P. monodon | PmToll | GU014556.1 | ND | (28, 29) | |
| PmToll9 | KY438975.1 | ND | (30) | ||
| F. chinensis | FcToll | EF407561 | Lymphoid organs: V. anguillarum, up; WSSV, down | (31) | |
| M. japonicus | MjToll1 | AB333779.1 | ND | (32) | |
| MjToll2 | AB385869.1 | ND | Direct Submission | ||
| M. rosenbergii | MrToll | JF895474 | Hemocytes: Aeromonas caviae, up | (33) | |
| MrToll | KX610955.1 | Gill: WSSV, up | (34) | ||
| MrToll1 | KJ188410.1 | ND | Direct Submission | ||
| MrToll2 | KJ188411.1 | ND | Direct Submission | ||
| MrToll3 | KJ188412.1 | ND | Direct Submission | ||
| P. clarkii | PcToll | KP259728.1 | Hemocytes, Hepatopancreas, Gill, Intestine: S. aureus and Vibrio, up | (19) | |
| PcToll1 | KP259728 | (19) | |||
| PcToll2 | KX505307 | Hemocytes: Vibro 0-12 hpi up, 12-48 hpi down | (35) | ||
| PcToll3 | KU680805.1 | Hemocytes: Vibrio and WSSV, up | (36) | ||
| PcToll4 | KU680806.1 | Intestine: WSSV, up | (37) | ||
| PcToll5 | KU680807.1 | ND | Direct Submission | ||
| Spätzle | L. vannamei | LvSpz1 | JN180646 | Gill: V. alginolyticus and WSSV, up | (26) |
| LvSpz2 | JN180647 | Gill: V. alginolyticus, down | (26) | ||
| LvSpz3 | JN180648 | Gill: V. alginolyticus and WSSV, up | (26) | ||
| LvSpz4 | KX060799 | Gill: S. aureus, V. alginolyticus, up | (38) | ||
| P. monodon | PmSpätzle 1 | KY053796 | hemocytes: S. aureus, V. harveyi and WSSV, up | (39, 40) | |
| PmSpätzle2 | KY053798 | ND | (39) | ||
| PmSpätzle3 | KY053797 | ND | (39) | ||
| F. chinensis | Fc-Spz | EU523114.1 | Hemocytes, Heart, Hepatopancreas, Gill, Stomach, Intestine: V. anguillarum and WSSV, up | (41) | |
| M. japonicus | MjSpz | KX424932 | Gill: WSSV, up | (42) | |
| M. rosenbergii | MrSpz | Unsubmitted | Hemocytes: A. caviae, up | (43) | |
| MyD88 | L. vannamei | LvMyD88 | JX073568.1 | Hemocytes: S. aureus, V. parahaemolyticus and WSSV, up | (44) |
| LvMyD88-1 | JX073567.1 | ND | (44) | ||
| P. monodon | PmMyD88 | KJ577578.1 | Hemocytes, Lymphoid organ, Gill, Stomach, Hepatopancreas, Midgut, Hindgut: WSSV, up | (45) | |
| F. chinensis | FcMyD88 | JX501341.1 | Hemocytes: M. lysodeikticus, V. anguillarium, up; WSSV, up | (46) | |
| Tube | L. vannamei | LvTube | JN180645.1 | Gill, Hepatopancreas: V. alginolyticus and WSSV, up, Intestine: V. alginolyticus and WSSV, down | (47) |
| LvTube-1 | KC346865 | Hemocytes: S. aureus, V. parahaemolyticus and WSSV, up | (48) | ||
| P. monodon | PmTube | KR136276.1 | Hemocytes: WSSV, up | (49) | |
| Pelle | L. vannamei | LvPelle | KC346864 | Hemocytes: S. aureus, V. parahaemolyticus and WSSV, up | (48) |
| TRAF6 | L. vannamei | LvTRAF6 | HM581680.1 | Intestine: V. alginolyticus and WSSV, down; Gill, Hepatopancreas: V. alginolyticus and WSSV, up | (50) |
| P. monodon | PmTRAF6 | KJ577579.1 | Hemocytes, Lymphoid organ, Stomach, Hepatopancreas: WSSV, up; Gill, Midgut, Hindgut: WSSV, down | (45) | |
| F. chinensis | FcTRAF6 | JQ693681.1 | ND | (51) | |
| IKKβ | L. vannamei | LvIKKβ | AEK86518 | Gill, Hepatpancreas, Intestine: V. alginolyticus and WSSV, up; Hemocytes: V. alginolyticus and WSSV, down | (52) |
| Dorsal | L. vannamei | LvDorsal | FJ998202.1 | Gill: WSSV, up | (11, 53) |
| F. chinensis | FcDorsal | EU815056.1 | Hemocytes, Lymphoid: M. lysodeikticus, V.anguillarium, up; Hemocytes: WSSV: 1 hpi up, 2–5 hpi down; Lymphoid: WSSV: 2 hpi up, 3–14 hpi down | (13) | |
| M. japonicus | MjDorsal | KU160503.1 | Gill, Hemocytes, Intestine: S. aureus, up | (54) | |
| M. rosenbergii | MrDorsal | KX219631.1 | Gill: WSSV, up | (10) | |
| Cactus | L. vannamei | LvCactus | JX014314.1 | Hemocytes: S. aureus, V. parahaemolyticus, up; WSSV, down | (55) |
| F. chinensis | FcCactus | JQ693681 | Hemocytes: M. lysodeikticus, V. anguillarium, up | (51) | |
| Tollip | L. vannamei | LvTollip | JN185616.1 | Hepatopancreas, Gill, Intestine: V. alginolyticus and WSSV, up, Hemocytes: V. alginolyticus, down; WSSV, up | (56) |
| Pellino | L. vannamei | LvPellino | KC346863.1 | Hemocytes: S. aureus, V. parahaemolyticus and WSSV, up | (57) |
| SARM | L. vannamei | LvSARM | JN185615 | Hemocyte, Gill, Intestine: V. alginolyticus and WSSV, up; Hepatopancreas: V. alginolyticus, down; WSSV, up | (58) |
| Flightless-I | L. vannamei | LvFli-I | KC800820 | Hemocytes: S. aureus, V. parahaemolyticus, up; WSSV, down | (59) |
| β-arrestin | M. japonicus | Mjβarr1 | KU160500 | ND | (54) |
| M. japonicus | Mjβarr2 | KU160501 | ND | (54) | |
| ATF | P. clarkii | PcATF4 | KX505308 | Hemocytes: Vibro, up | (35) |
up, differentially up-regulated; down, differentially down-regulated; ND, Not determined.
Toll and Spätzle
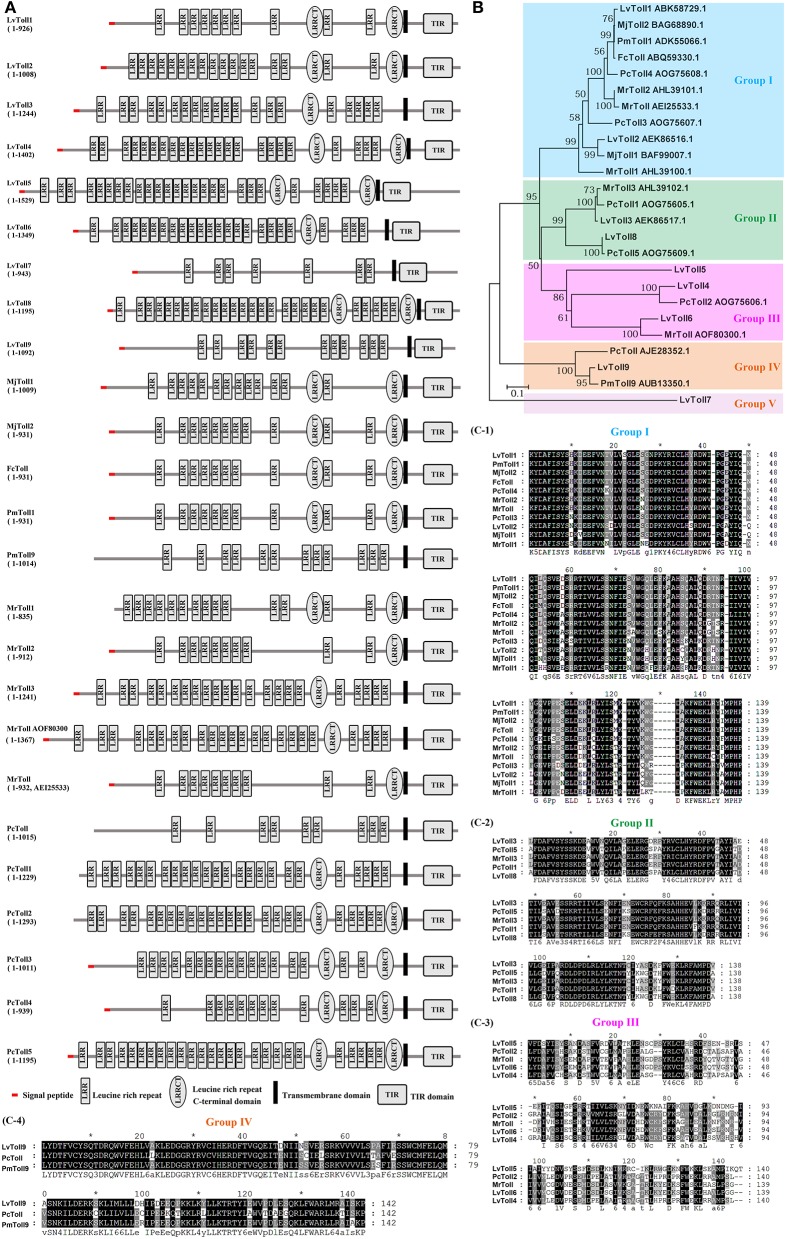
Tolls and TLRs are characterized by an extracellular domain containing various numbers of leucine-rich repeats (LRRs) and a cytoplasmic-signaling domain, Toll/IL-1R (TIR), that can interact with cytoplasmic-adaptor molecules, thereby activating downstream-signaling events (60). To date, 25 genes encoding distinct Toll homologs have been identified in shrimp from six different species including LvToll1-9 from L. vannamei (15, 16, 24–26, 61), FcToll from F. chinensis (31), PmToll1, and PmToll9 from P. monodon (28–30), MjToll1-2 from M. japonicas (32), PcToll and PcToll1-5 from P. clarkii (19, 35–37), and MrToll1-3 and two MrTolls from M. rosenbergii (33, 34) (Figure 1A). A phylogenetic tree based on TIRs of the 25 Tolls has been constructed, from which we observe that Tolls from shrimp can be divided into five groups (Figure 1B). Group I contain the majority of Tolls identified, with up to a total of 11 Tolls from all 6 shrimp species. Groups II, III, IV, and V contain five, five, three, and one Toll(s) from partial shrimp species, respectively (Figure 1B). Alignment of TIR domains in each corresponding group indicates that the TIR sequences in each group are highly conserved (Figure 1C). In addition to the shared LRRs in the ectodomain, Tolls usually possess one or two Leucine-rich-repeat C-terminal domains (LRRCTs) and/or a signal peptide in the N-terminal (Group I–III). In comparison with other Tolls, PmToll9, and PcToll clustered in Group IV have no LRRCT or signaling peptide (19, 30). The conserved TIR domains in each group suggest that they might focalize the same kind of adaptor molecules, suggesting that they could activate the same signaling pathway. However, in consideration that LRR-LRRCT ectodomains of Tolls function as ligand-recognition sites (62), it is likely that Tolls—even in the same group that harbor conserved cytoplasmic TIR domains but exhibit a variety of structures in the ectodomain—could recognize some specific ligands and respond to different pathogens. This may be in line with the observation that distinct Tolls in different species respond to diverse bacterial and viral infections but converge on the induction of the same kind of effectors, such as AMPs.
Figure 1.
Tolls identified from shrimp. (A) Architectural representation of characteristic domains of 25 Tolls from six different shrimp species. (B) Phylogenetic-tree analyses of TIR domains of the Tolls. Genbank accession numbers of some Tolls are shown after their scientific names. (C) Multiple sequence alignments of shrimp Toll proteins from Group I (C-1), Group II (C-2), Group III (C-3), and Group IV (C-4).
So far, the Tolls from L. vannamei are still the most studied Tolls in shrimp. LvToll1, LvToll2, and LvToll3 in gill are all up-regulated during WSSV infection, while only LvToll1 expression increases during Vibrio infection (16, 24–26). LvToll2 could significantly induce the activation of NF-κB pathway controlling AMPs in the background of insect cells, whereas these AMPs appear to be not induced by LvToll1 and LvToll3 (15, 26, 63). Further, knockdown of LvToll1 by RNA interference (RNAi) does not influence the susceptibility of shrimp to WSSV infection, but silencing of LvToll1 significantly increases the mortality of shrimp and reduces bacterial clearance after challenge with Vibrio harveyi (16, 64). In a recent study, another six Tolls of LvToll4–9 have been cloned, and the authors found that eight Tolls, except for Toll2, exhibit different levels of resistance to WSSV. Furthermore, LvToll4 has been shown to be a key factor (possibly as a receptor) for sensing WSSV and therefore activate the downstream Dorsal, resulting in the inducing of specific AMP production (27). As for other species, FcToll in lymphoid is significantly induced after 5-h-post Vibrio challenge but its expression markedly is reduced immediately after WSSV exposure (31). MrToll, PcToll1, and PcToll2 are up-regulated after bacterial infection, while PcToll4 can regulate the expression of AMPs to defend against WSSV (19, 33, 35, 37). Moreover, PcToll3 in hemocytes responds to both bacterial and WSSV infection (36). Although the RNAi phenotypes of most Tolls are unknown, their expression is responsive to bacterial or viral infection; it is therefore tempting to speculate that they are involved in host defense.
The subcellular localization of TLRs could somewhat influence PAMP accessibility (65). Some mammalian TLRs localized on cell surfaces, including TLR1, TLR2, TLR4, TLR5, TLR6, and TLR11, could recognize mainly microbial-membrane components, such as lipids, proteins, and lipoproteins, while there are also intracellularly expressed TLRs including TLR3, TLR7, TLR8, and TLR9, located in the endosome and mainly recognize microbial nucleic acids (65–67). Through ectopic expression in Drosophila S2 cells, LvToll1, and LvToll3 have been shown to localize at both the membrane and cytoplasm, while LvToll2 is ubiquitously distributed within the cytoplasm (15, 26). Over-expression of PmToll9 with GFP tag in Hela cells showed that PmToll9 is mainly located in the cytoplasm (30). Furthermore, more direct detection of cellular localization of shrimp Tolls should be conducted in vivo using immunofluorescent staining or immunohistochemical staining by using Toll-specific antibodies. Whether the shrimp Tolls localization is implicated with their potential roles in immune-signaling pathways remains unknown.
Different from mammalian TLRs, Toll in Drosophila cannot recognize PAMPs directly but need the cytokine-like molecule, Spätzle (Spz) as a ligand (45, 64, 68). Spätzle is a member of the cysteine-knot protein superfamily, which was cleaved from an inactive pro-Spätzle. Pro-Spätzle contains the pro-domain that prevents Spätzle from binding to the Toll receptor, via the serine protease, Easter, during dorsoventral patterning or Spätzle-processing enzymatic activity upon infection. The activated Drosophila Spätzle contains 106 residues in the C-terminal domain (C-106), which is sufficient to active the Toll pathway (41, 69). The interaction between Spätzle C-106 dimer and the extracellular domain of Toll rearranges cytoplasmic TIR domains conformation, thereby generating a docking site for recruiting TIR-domain-containing adaptors—such as MyD88, which in turn activate the Toll pathway (70). To be noted, Toll7 in Drosophila has been considered as a specific PRR for sensing vesicular-stomatitis viruses, because that it can bind to the plasma membrane of this virus (71). Surprisingly, several Tolls from shrimp are reported to detect some PAMPs directly. For example, in vitro, Toll1 and Toll3 from L. vannamei can combine with CpG ODN 2395 (72). Additionally, three Tolls from M. japonicas can directly interact with both PGN and LPS (73). Interestingly, two Tolls from M. japonicas are homologous to the above Toll1 and Toll3 from L. vananmei (73). These data indicate that in shrimp, one type of Toll could sense multiple types of PAMPs, which are more similar to TLRs in mammals.
The first Spätzle-like protein from shrimp was identified in F. chinensis in 2009 and was named as FcSpz (41). The activated form of FcSpz requires a seven cysteine residues on C-terminal which is essential for intra-molecular and inter-molecular disulfide bonds to form Spätzle homodimers (41). FcSpz respond to Vibro alginolyticus and WSSV infection, and the injection of C-terminal active FcSpz domain (114 residues) in vivo could activate the promoter of shrimp AMP Crustin2 (41). In addition to FcSpz, seven more Spätzles have been cloned and identified in different shrimp, including LvSpz1-4, PmSpz1-3, MjSpz, and MrSpz (26, 38, 39, 42, 43). All of these Spätzle genes can respond to bacterial and/or WSSV infections. Interestingly, multiple Spätzles—such as LvSpz1, LvSpz2, LvSpz3, and FcSpz3—lack the seventh Cys residue that is important to disulfide-linked homodimer formation, and that favors its binding to Toll receptors, which might explain their slight effects on induction of AMP expression (26, 41). Instead, the Spätzle domain of LvSpz4 containing the seventh Cys residue can strongly activate the NF-κB pathway that regulates AMPs such as Penaeidin 4 (PEN4), Drosomycin (Drs), Attacin (Atta), and Metchnikowin (Mtk) in Drosophila S2 cells. Moreover, the activating transcription factor 4 (ATF4) and X-Box-binding protein 1 (XBP1), components of the unfolded protein response (UPR), are capable of inducing the expression of LvSpz4, which suggests that LvSpz4 could be a regulator to link the Toll-NF-κB pathway and the UPR (38). Although a series of Tolls and Spzs have been identified, there are few studies that have investigated the interactions among these proteins. Detecting the interplay between Spz and Toll will be helpful to illustrate which pairs of Spzs/Tolls are function to mediate the shrimp Toll pathway inactivation, and induce immune-related genes expression.
Cascades of the Toll Pathway
The cascades of the Toll pathway have been well studied in both flies (e.g., Drosophila) and mammals (e.g., Human). In Drosophila, the Toll pathway can respond to Gram-positive bacteria, fungi, and some viruses (20). The activation of Toll triggers intracytoplasmic TIR domains dimerization, therefore recruiting the adaptor MyD88 via its own TIR domain (74). The second adaptor protein, Tube, binds with MyD88, and the protein kinase Pelle; these two interaction is formed via pairwise interactions of death domains (Figure 2) (75, 76). Pelle is able to phosphorylate itself, and the autophosphotylation results in the inhibitor of κB, Cactus, phosphorylation and destruction as well as the phosphorylation of the homolog of tumor necrosis factor receptor-associated factor 6 (TRAF6), dTRAF2. Then, depending on the context, the transcription factor, Dif, or Dorsal, then translocate from cytoplasm to nucleus (20, 77).
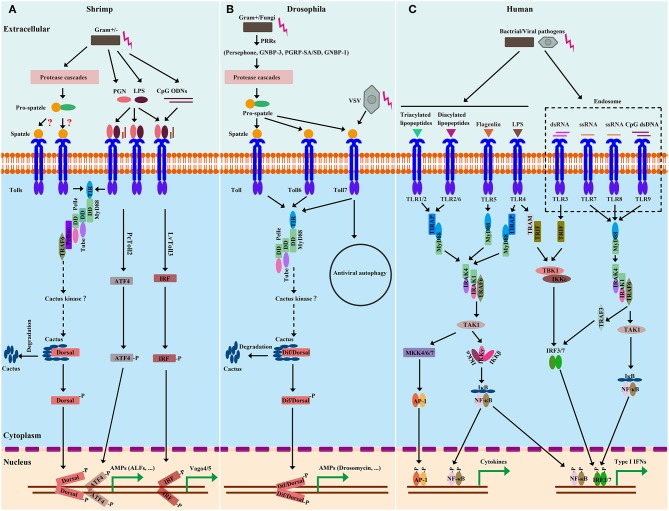
Figure 2.
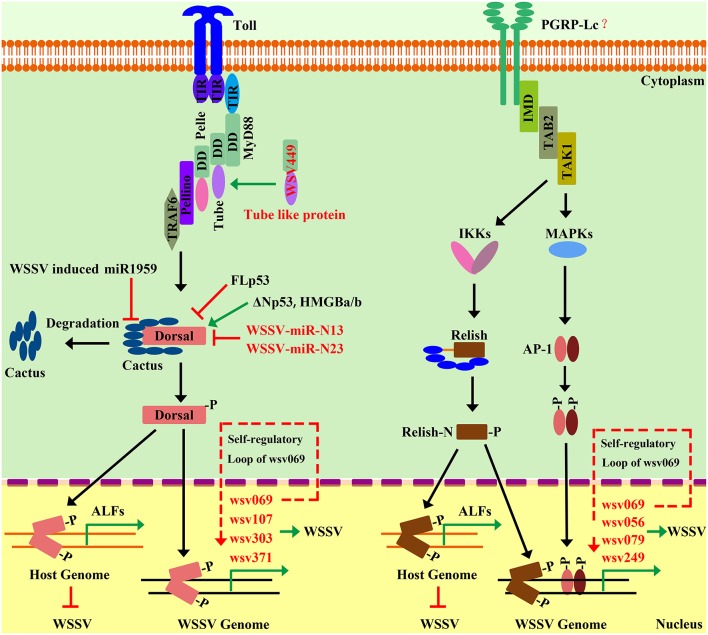
Comparison of Toll and TLR pathways from shrimp (A), Drosophila (B), and Human (C). Homologies between signaling components are depicted by similar shapes and colors. In shrimp, there are two extracellular-signaling routes leading to Toll pathway activation. Considering that many Spätzle (Spz) genes from shrimp have been identified and are able to induce Toll-Dorsal-controlled AMPs, the extracellular cleavage of Spz mediated by protease cascades could be similar to those of Drosophila. In the immune responses to microbial recognition, the protease cascades lead to the activation of Spz-processing enzymes (SPEs) to cleave full length Spz. Upon proteolytical processing, the Spz prodomain is cleaved, exposing the C-terminal Spz parts that are critical for binding of Toll. Spz binding to the Toll receptor initiates intracellular signaling. In contrast, shrimp Tolls are able to sense and directly bind to some conserved motifs, such as PGN, LPS, and ODN, from microbes, which are similar to those of Human. Upon binding to these motifs, shrimp Tolls are activated and initiate intracellular signaling. In the intracellular-signaling event, signaling cascades of shrimp Toll pathways broadly resemble those of Drosophila. After Toll activation, the adaptor MyD88 builds a signaling complex with Tube and Pelle. The MyD88-Tube-Pelle complex in turn recruits other regulators, such as Pellino and TRAF6, which leads to the phosphorylation and degradation of Cactus and thereby releases Dorsal (and/or Dif in Drosophila) to translocate to the nucleus and activate transcription. In Human, there are MyD88-dependent and MyD88-independent signal-transduction events. The intracellular signaling of Human can lead to active NF-κB, AP-1 and IFN regulatory factor 3/7 (IRF3/7) for their nuclear translocation and subsequent transcriptional activation of target genes. Of note, Toll3 from L. vannamei and Toll2 from P. clarkii have been shown to activate IRF and ATF4, respectively, leading to transcriptional synthesis of some antiviral effectors, such as Vago4/5 and ALFs.
In the TLR signaling pathway in Human, after binding with their ligands, all TLRs except for TLR3 initiate the MyD88-dependent pathway through recruiting MyD88 via the TIR domain (78). Similar to that of Drosophila, in Human, the signaling complex is composed of MyD88, the Tube ortholog, IRAK4, and the Pelle ortholog, IRAK1 (79, 80). The phosphorylated IRAK-1 gets associated with TRAF6 (81), resulting in the disassociating of IRAK1/TRAF6 complex from the above receptor complex, instead interacting with another complex consisting of transforming growth factor (TGF)-β activated kinase 1 (TAK1) and TAK1 binding protein (TAB) 1, TAB2 and TAB3 (57, 82). Pellino is a highly conserved E3-class ubiquitin ligase. It binds to the phosphorylated IRAK1, resulting in K63 polyubiquitination of IRAK1. The polyubiquitinated IRAK1 interacts with the ubiquitin-binding domain of NF-κB essential modifier (NEMO, also named IKKγ) (57, 83). This interaction leads to the TAK1-TAB1-TAB2 complex and IKKγ-IKKα-IKKβ complex into close proximity, which subsequently results in TAK1-mediated phosphorylation and activation of the IKKs (84). The activated IKKs phosphorylates the NF-κB cytoplasmic inhibitory protein, IκB, leading to polyubiquitylation and degradation of IκB and release of NF-κB from the IκB/NF-κB complex (57, 85). NF-κB subsequently undergoes nuclear translocation, where it induces the expression of a wide range of immune-modulatory genes, including pro-inflammatory cytokines. In the TIR-domain-containing adaptor protein-inducing IFN-β (TRIF)-dependent pathway, after recognizing dsRNA, TLR3 recruits TRIF, TRAF6, and TRAF3, leading to the activation of IKK-related kinases, including TANK-binding kinase I (TBK1) and IKKε, thereby resulting in the activation of IRF3/7 signaling pathways that eventually induce the transcription of type-I interferon (IFN; Figure 2) (77, 86). Of note, TLR4, located on the cell surface, can activate both of the distinct intracellular signaling pathways via the adaptor molecules, MyD88 and TRIF, that finally results in NF-κB and MAPK activation to trigger the expression of pro-inflammatory cytokines and/or lead to IRF3/7 activation to induce Type-I IFNs production (Figure 2) (87, 88).
Beginning with Toll, most components of the Drosophila Toll pathway have shrimp homologs (Table 1). Four MyD88s in three species (LvMyD88, LvMyD88-1, PmMyD88, and FcMyD88) (44, 46), three Tubes in two species (LvTube, LvTube-1, and PmTube) (48, 49), and LvPelle (47, 48) have been identified in shrimp. Each of these proteins contains a protein-interaction motif named death domain, which is originally described in apoptotic pathways (89). LvPellino has been the sole Pellino homolog found in shrimp until recently, and is able to interact with L. vannamei Pelle (LvPelle) and positively regulate the activity of LvDorsal (57). Three TRAF6s (LvTRAF6, PmTRAF6, and FcTRAF6) have been characterized in three species with a RING-type zinc-finger domain in the N-terminal, followed by two TRAF-type zinc-finger domains, a coiled region and a MATH domain in the C-terminal (45, 50, 51). So far, no Dif homolog has been found in shrimp, while four Dorsal proteins (LvDorsal, FcDorsal, MjDorsal, and MrDorsal) in shrimp are similar in sequence to the mammalian NF-κB (p65) and Drosophila Dorsal (10, 11, 13, 54). However, Drosophila Dorsal mainly responds to Gram-positive bacteria but does respond to a few Gram-negative bacteria (20, 64), whereas shrimp Dorsal—such as LvDorsal and FcDorsal—appear to respond to both Gram-positive and Gram-negative bacteria, as well as viruses such as WSSV (11, 13). In both Drosophila and shrimp, each Dorsal contains a conserved Rel-homology domain, which is a bearing sites for DNA binding, dimerization, and interaction with an inhibitor. The Cactus or its ortholog, IκB, function as an inhibitor of Dorsal (NF-κB), contain an N-terminal regulatory region responsible for ubiquitin recognition and proteasomal degradation, and have neighboring ankyrin repeats that are capable of binding with the Rel-homology region and a destabilizing C-terminal PEST domain that are required for inhibition of DNA binding (90). All three domains are found and conserved in LvCactus and FcCactus, suggesting that shrimp Cactus could comply with a similar function and regulatory fashion to those of Drosophila Cactus (51, 55). In addition to these classical pathway components, several members involved in regulating the Toll pathway directly or indirectly have also been identified including activating transcription factor 4 (PcATF4) (35), Tollip (LvTollip) (56), Flightless-I (LvFli-I) (59), β-arrestins (Mjβarr1-2) (54), and sterile-alpha and armadillo motif-containing protein (LvSARM) (58) (Figure 3 and Table 1).
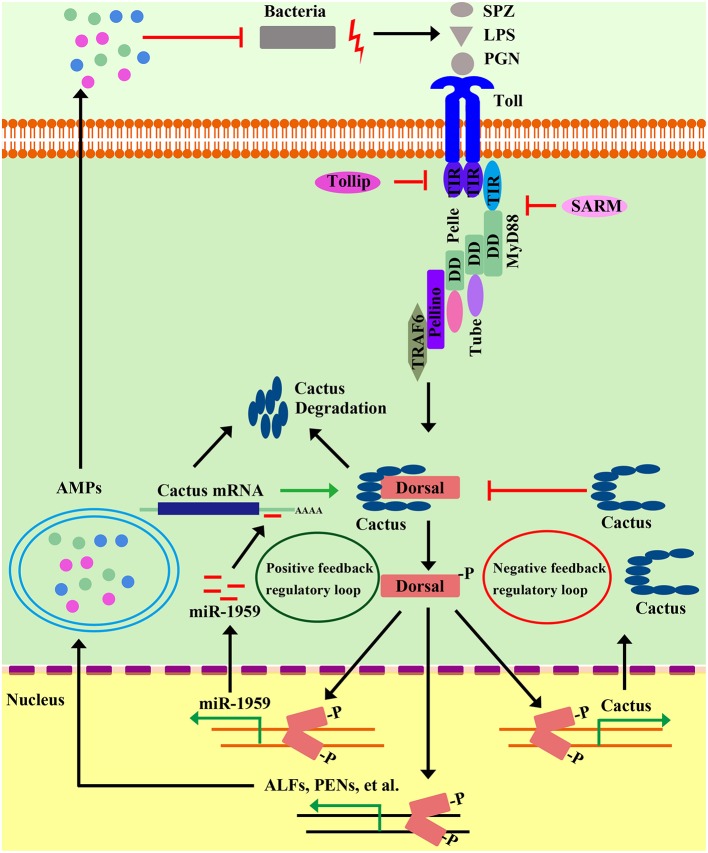
Figure 3.
Positive and negative regulatory events in shrimp Toll pathways. Positive regulation of the shrimp Toll pathway can amplify signaling cascades to rapidly limit and clear invading pathogens. In microbial recognition, intracellular signaling leads to activation of Dorsal, which in turn induces the transcription of a microRNA gene, miR1959. MiR1959 can target the mRNA of Cactus, an inhibitor of Dorsal, resulting in reduced Cactus protein levels, which leads to the up-regulation of Dorsal activity to boost some effectors, such as AMP expression against invading microbes. On the other hand, the activation of the Toll pathway must be controlled and inhibited to ensure a properly timed and adjusted response. Along with the clearing of invading pathogens, the gradually increased Cactus leads to the inhibition of Dorsal activity by their direct interaction, which establishes a negative-feedback loop to abate NF-κB pathway signaling to avoid the sustained activation of this pathway. In addition, there multiple negative regulators have been identified—including Tollip, SARM, FliI, and β-Arrestin—that modulate the Toll pathway in different ways.
Apart from Dorsal, the transcription factor of classical Toll pathway, in P. clarkii, PcATF4 is involved in PcToll2 signaling to promote AMP expression, which suggests that PcATF4 might be another crucial transcription factor involved in crustacean Toll signaling to prevent shrimp from Gram-negative bacterial invasion (35). Likewise, there is another case in which the Spätzle, LvSpz4, has been reported to be regulated by ATF4, one of the major components of the UPR pathway (38). Although there is no ATF4 homolog that has been found in Drosophila yet, in human, ATF4 has also been shown to participate in TLR4 pathway to defend against Gram-negative bacterial invasion by promoting inflammatory cytokines secretion, through working together with another transcription factor, c-Jun (36, 91). Similar to mammal TLR3 inducing IRF3/7 activation, LvToll3 is also involved in the induction of IRF-Vago-JAK/STAT pathway related genes, suggesting that LvToll3-mediated intracellular signaling may follow a cascade akin to those of vertebrates (Figure 2) (61, 92). These observations demonstrate that the canonical Toll pathway might crosstalk with other signaling pathways in a coordinated manner to launch a specific immune response.
Negative Regulators of the Toll Pathway
The Toll pathway is an important part of the innate immune system, but hypernomic activation of the Toll pathway can cause immune disorder and seriously affect the health, even the survival of an organism. So, the fine-tuned and subtle regulation—including negative regulators of the Toll pathway—has evolved to keep the balance of immunity (Figure 3). The IκB is the evolutionarily conserved inhibitor of canonical NF-κB pathway. As mentioned in above section, degradation of the NF-κB inhibitor, IκB, allows NF-κB free from the cytoplasmic NF-κB/IκB complex, and the freed NF-κB migrates from cytoplasm to nucleus to induce various target genes (93). Cactus is the homolog of IκB in invertebrates and mediates both negative- and positive-feedback regulatory loops of NF-κB via different pathways in shrimp. The induction of LvCactus is activated by LvDorsal but on the other hand is inhibited by LvCactus itself (55). LvCactus interacts with LvDorsal, which is confirmed by immunoprecipitation assays and fluorescent microscopy (55). Four of the five putative NF-κB-binding motifs in the promoter of LvCactus have been shown to be targeted by LvDorsal (55). Hence, we hypothesize that the Toll pathway is activated in some cases by bacterial infection, following by rapid up-regulation of LvDorsal activity that induces massive effectors that function against invaders. Additionally, LvDorsal activation also contributes to the increased expression of LvCactus, which feeds back to inhibit LvDorsal activity via their direct interactions. These interactions create a negative-feedback loop to abate NF-κB-pathway signaling in order to avoid the sustained activation of this pathway (Figure 3) (55). There is also a positive-feedback regulatory loop in the Toll pathway with the participation of Dorsal, Cactus, and a host microRNA, miR-1959 (94). Dorsal can directly bind the NF-κB-binding motif in the promoter region of miR-1959 and activate its transcription and, in turn, miR-1959 targets the 3′-untranslated region of Cactus, reducing the protein level of Cactus, further leading to enhanced activation of Dorsal (Figure 3) (94).
Additionally, Sun et al. showed that in the kuruma shrimp, M. japonicus, β-arrestin can negatively regulate Toll signaling in two different ways. β-arrestin can prevent Dorsal translocation via β-arrestin-Cactus-Dorsal heterotrimeric complex with Cactus as the bridge. β-arrestin and Dorsal do not come into contact with each other; instead, the ankyrin-repeat domain and C-terminal PEST domain of Cactus separately bind the arrestin-N domain of β-arrestin and RHD domain of Dorsal, respectively (54). After formation of the oligomeric β-arrestin-Cactus-Dorsal complex, Cactus phosphorylation, and degradation is prevented, which inhibits Dorsal translocation into the nucleus as well as the activation of Toll signaling pathway. On the other hand, β-arrestin can inhibit Dorsal phosphorylation and transcriptional activity. Extracellular signal-regulated protein kinase (ERK) can function as a kinase with a capacity for Dorsal phosphorylation. Meanwhile, β-arrestin can interact with non-phosphorylated ERK through its arrestin-C domain to inhibit ERK phosphorylation, which affects Dorsal phosphorylation and thus inhibits its transcriptional activity and nuclear localization (54).
In addition to these regulators mentioned above that target the transcription factor Dorsal or its inhibitor Cactus, Toll-interacting protein (Tollip) from L. vannamei functions as a negative modulator in the Toll pathway through interacting with up-stream Toll receptors (56). Tollip associates directly with some of the Tolls or TLRs through TIR-domain-mediated interactions and therefore inhibits Toll/TLR-mediated NF-κB activation by suppressing adaptor proteins, such as IRAK1, phosphorylation, and kinase activity (56). So far, no Tollip-like homolog has been found in the Drosophila genome. However, forcible expression of LvTollip in Drosophila S2 cells significantly inhibits the promoter activities of the Toll pathway controlling the antifungal-peptide gene, Drosomycin (Drs). In Human HEK 293T cells, LvTollip has been demonstrated to significantly suppress the inductions of both NF-κB and IFN-β (56). These findings might suggest that from shrimp to humans, the Tollips are functionally conserved in the TLR-NF-κB signaling pathway.
Mammalian MyD88 is known as a universal adaptor protein in the downstream signaling of many different kinds of TLRs, with the exception of TLR3, which instead recruits TRIF (56, 95). Hence, MyD88 is regarded as a perfect target for immunity regulation. Flightless-I (FliI) was originally characterized as a gene mutation that causes defects in the flight muscles in Drosophila melanogaster (96). The FliI protein belongs to the gelsolin superfamily of actin-remodeling proteins that usually contain six C-terminal gelsolin-like domains (GEL), and harbors multiple unique LRR domains in the N-terminal responsible for protein-protein or protein-lipid interactions (97). FliI is widely identified as a negative regulator to modulate NF-κB activity through interfering with MyD88-Toll receptor interactions (98). LvFliI, identified from L. vannamei, is up-regulated in vivo in response to the challenges of LPS, Poly (I: C), CpG-ODN 2006, V. parahaemolyticus, Staphyloccocus aureus, and WSSV, and it is shown to suppress the expression of the NF-κB-pathway-dependent AMPs, including LvPEN2, LvCrustin, LvALF1 (anti-LPS factor), and LvLyz1 (Lysozyme) in vivo (59). Also, over-expression of LvFliI in Drosophila S2 cells can negatively regulate the promoter activities of Drosophila and shrimp AMPs, such as Drs, Mtk, ALF2, PEN453, and PEN536, after LPS challenge in Drosophila S2 cells (56, 59). However, no significant differences have been observed in the mortality rates after V. alginolyticus, S. aureus, or WSSV infections in LvFliI-silenced shrimp compared to those of wild-type shrimp (59).
In mammals, SARM is the only negative regulator of the five TLR adaptor proteins—MyD88, TIRAP, TRIF, TRAM, and SARM. By associating with TRIF, SARM functions as an inhibitor of TRIF-dependent signaling (99). No TRIF-dependent signaling has yet been found in invertebrates. In contrast, SARM, such as amphioxus SARM, is able to inhibit the MyD88-dependent signaling pathway by interacting with MyD88 and TRAF6, indicating a negative regulatory role in the Toll pathway (56, 99). One shrimp SARM homolog has been cloned and identified from L. vannamei, which is also shown to interact with LvTRAF6, a positive regulatory member of the Toll pathway (58). Knockdown of endogenous LvSARM results in NF-κB activation and enhances the expression levels of NF-κB targeted AMPs such as PENs and ALFs, indicating its negative regulatory role in the shrimp Toll pathway. Unexpectedly, LvSRAM-silenced shrimp are more susceptible to infection by V. alginolyticus than that of control shrimp injected with GFP dsRNA, which might be explained LvSARM having additional roles beyond innate immunity, such as maintaining normal growth and development (58).
The IMD Signaling Pathway in Shrimp
The IMD pathway was originally defined in Drosophila by the identification of a mutation named immune deficiency (IMD) (7568155). The mutated IMD impairs the expression of several AMPs especially Diptericin (Dpt). For this reason, Dpt is often used as a readout of IMD pathway activation, but only marginally affects the Toll-pathway-targeted induction of Drosomycin (Drs) (100, 101). It is generally considered that IMD-deficient flies succumb to Gram-negative bacteria, which is different from that of Toll mutant flies that are more susceptible to fungi and Gram-positive bacteria (21, 100, 102). IMD encodes a death-domain–containing protein similar to the receptor-interacting protein (RIP) of the tumor necrosis factor receptor (TNF-R) pathway in mammals (103). In the TNFR pathway, RIP is essential for both NF-κB and mitogen-activated kinase (MAPK) activation. Drosophila IMD mediates a signaling cascade that broadly resembles the mammalian TNFR pathway (104) (Figure 4).
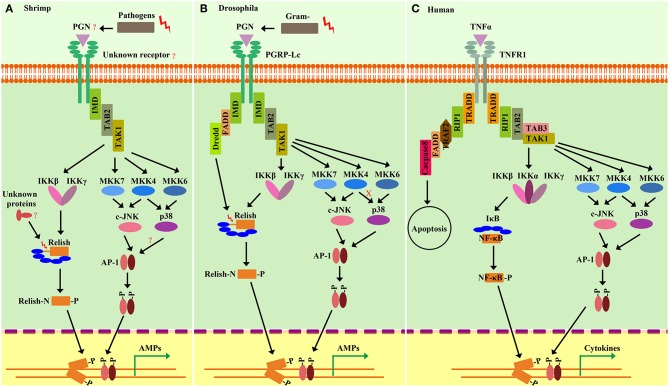
Figure 4.
Comparison of the shrimp IMD pathways (A) and Drosophila IMD pathways (B) and Human TNFR signaling (C). Homologies between signaling components are depicted by similar shapes and colors. Shrimp and Drosophila IMD pathway-signaling events resemble the Human tumor necrosis factor receptor (TNFR) signaling pathway. In shrimp, some pivotal components of the IMD pathway, such as the receptors, DREDD, and FADD, are still unknown. Similar to Drosophila, pathogen challenges can initiate intracellular signaling events involving IMD, TAB2, and TAK1, which in turn induce the activation of MAPK-AP-1 and IKK-Relish branches. The activation of Relish is required to be phosphorylated by the IKK complex and is cleaved by unknown factors in shrimp. In the MAPK-AP-1 branch of shrimp IMD pathways, the signaling cascade is more similar to that of Human than that of Drosophila, as manifested by the finding that shrimp p38 can be phosphorylated by MKK4, but Drosophila p38 cannot. In shrimp, activation of both of the two branches of IMD pathways can induce the expression of AMPs that confer protection from Gram-negative and Gram-positive bacterial infection.
The Canonical Components of the IMD Pathway
Specifically (Figure 4), the Drosophila IMD pathway is trigged by meso-diaminopimelic acid (DAP)-type peptidoglycan (PGN), which comprises the cell wall of most Gram-negative bacteria, as well as some Gram-positive bacteria (100). PGRP-LC and PGRP-LE are the two receptors, which are involved in the IMD pathway with different subcellular location. PGRP-LC is on the plasma membrane while the intracellular PGRP-LE binds specifically to DAP-type PGNs (100, 102). After binding to PGN, these receptors likely dimerize or multimerize and then lead to recruitment of a signaling complex consisting of IMD (101, 105), Fas-Associated protein with a death domain (FADD) (105, 106) and the caspase-8 homolog death-related ced-3/Nedd2-like protein (DREDD) (100, 102, 105, 106). Inhibitor of apoptosis 2 (IAP2), a ubiquitination-machinery component, function as an E3-ubiquitin ligase could activate DREDD via ubiquitination. Activated DREDD cleaves IMD, exposing a binding site for IAP2, leading to K63-ubiquitinate IMD (106). The K63 ubiquitin chain of IMD functions as a scaffold for interaction with TAK1/TAB2 complex. The TAK1/TAB2 complex is responsible for activating both MAPK/AP-1 and IKK/Relish branches of the IMD pathway (105–107). Relish activation is achieved by both IKK-mediated phosphorylation and cleavage by DREDD, after which the N-terminal Rel domain of Relish undergoes nuclear translocation and initiates the transcription of target genes (102, 108).
The studies of IMD pathway in shrimp have just begun in the last 10 years. In 2009, Wang et al. identified an IMD homolog (LvIMD) from L. vannamei, which was the first IMD homolog identified in shrimp (17, 64). LvIMD encodes a death domain-containing protein, which is moderately homologous to Drosophila IMD and mammalian RIP (17). Studies show that the IMD homologs from different shrimp species are distinctly different in the context of their tissue distributions and their responsiveness to infection. The FcIMD from F. chinensis is very abundant in the gills and stomach, whereas the PcIMD of P. clarkia exhibits high expression levels in the heart, hepatopancreas, and stomach. In contrast, LvIMD of L. vannamei exhibits expression abundantly in the nervous system, gill, intestine, and pyloric caecum (17, 109, 110). Additionally, FcIMD in gills is up-regulated by WSSV but not Vibrio challenge, while PcIMD in gills can respond to Vibrio but not WSSV (109, 110). LvIMD mRNA can be induced by multiple immune stimuli including LPS, V. alginolyticus, S. aureus, Saccharomyces cerevisiae (yeast), and WSSV, both in hepatopancreas and hemocytes but not in gills (17). Since the LvIMD was identified in L. vannamei, an increasing number of components of the canonical IMD pathway in shrimp have been identified, including TAK1, TAB1, TAB2, Relish, mitogen-activated protein kinase kinase 3 (MKK3), MKK4, MKK6, MKK7, c-Jun N-terminal kinase (JNK), c-Jun, c-Fos, p38, ATF2, NF-κB repressing factor (NKRF), Akirin, Bap60, and 14-3-3 (12, 17, 73, 105, 109–127). The canonical components of the IMD signaling pathway identified in penaeid shrimp are listed in Table 2.
Table 2.
Components of canonical IMD signaling pathway identified in shrimps.
| Components | Species | Gene names | Accession numbers | In response to pathogenic infections | References |
|---|---|---|---|---|---|
| IMD | L. vannamei | LvIMD | ACL37048.1 | Muscle: V. anguillarum and M. lysodeikticus, up; Hepatopancreas: V. alginolyticus, up; Hemocyte: V. alginolyticus, S. aureus and WSSV, up | (17, 24) |
| F. chinensis | FcIMD | JX867731.1 | Cephalothorax: V. anguillarum and M. lysodeikticus, up; Hemocytes: V. anguillarum, down; Hemocytes and gills: WSSV, up | (109, 110) | |
| P. clarkii | PcIMD | Unsubmitted | Hemocytes and gills: V. anguillarum, up | (109) | |
| TAK1 | L. vannamei | LvTAK1 | KU522004.1 | Gills: V. parahaemolyticus, S. saureus, and WSSV, up | (112) |
| TAB1 | L. vannamei | LvTAB1 | KY683840 | Gills: V. parahaemolyticus and S. saureus, up | (113) |
| TAB2 | L. vannamei | LvTAB2 | KP780842.1 | Gills: V. parahaemolyticus, S. saureus and WSSV, up; Hemocytes: V. parahaemolyticus, WSSV, up | (105) |
| IKKβ | L. vannamei | LvIKKβ | AEK86518 | Gill: V. alginolyticus and WSSV, up; Hemocyte: V. alginolyticus and WSSV, down; Intestine: V. alginolyticus and WSSV, up | (52) |
| IKKε | L. vannamei | LvIKKε1 | AEK86519 | Gill: V. alginolyticus and WSSV, up; Hemocyte: V. alginolyticus and WSSV, down; Hepatopancreas: V. alginolyticus and WSSV, up; Intestine: V. alginolyticus and WSSV, up | (52) |
| L. vannamei | LvIKKε2 | AEK86520 | Gill: V. alginolyticus and WSSV, up; Hemocyte: V. alginolyticus and WSSV, down; Hepatopancreas: V. alginolyticus and WSSV, up; Intestine: V. alginolyticus and WSSV, up | (52) | |
| Relish | L. vannamei | LvRelish | EF432734 | Gills: WSSV, up | (12, 12, 53) |
| P. monodon | PmRelish | KM204120 | Hemocytes, lymphoid organ, gill, hepatopancreas and heart: V. harveyi, WSSV, YHV, up | (114) | |
| F. chinensis | FcRelish | EU815055.1 | Gills: V. anguillarum, down | (110). | |
| M. rosenbergii | MrRelish | KR827675.1 | Hepatopancreas: V. anguillarum, up | (111) | |
| NKRF | L. vannamei | LvNKRF | KY864366 | Gills: WSSV, up | (117) |
| Akirin | L. vannamei | LvAkirin | KC415269.1 | Hepatopancreas: V. parahaemolyticus, up | (115) |
| M. japonicus | MjAkirin | AB503217.1 | Hemocytes: V. anguillarum, up | (116) | |
| Bap60 | M. japonicus | MjBap60 | KT892952.1 | Hemocytes: V. anguillarum, up | (116) |
| 14-3-3 | L. vannamei | Lv14-3-3EL | JF81119 | Gill and muscle: WSSV, up; lymphoid organ, WSSV, down | (118) |
| L. vannamei | Lv14-3-3ES | JF81120 | Muscle: WSSV, up Lymphoid organ: WSSV, down | (118) | |
| M. japonicus | Mj14-3-3 | KT892951.1 | Hemocytes: V. anguillarum, down | (116) | |
| JNK | L. vannamei | LvJNK | JN035903.1 | Gills: WSSV, up | (120) |
| c-Jun | L. vannamei | Lvc-Jun | KM401573.1 | Gills: WSSV and V. parahaemolyticus, up | (119, 121) |
| P. monodon | Pmc-Jun | KX216509 | Gill and hepatopancreas: V. harveyi and S. agalactiae, up. | (73) | |
| c-Fos | L. vannamei | Lvc-Fos | KP676567 | Gills: WSSV and V. parahaemolyticus, up | (119) |
| MKK3 | F. chinensis | FcMKK3 | KF994775 | Hemocytes and gills: WSSV, V. anguillarum and S. aureus, up | (122) |
| MKK4 | L. vannamei | LvMKK4 | KY693644 | Intestine and hepatopancreas: V. parahaemolyticus, S. aureus and WSSV, up. | (126) |
| F. chinensis | FcMKK4 | KJ023198 | Hemocytes and gills: WSSV, V. anguillarum and S. aureus, up | (122) | |
| MKK6 | L. vannamei | LvMKK6 | KR535627 | Gills: V. parahaemolyticus, S. aureus and WSSV, up | (123) |
| MKK7 | L. vannamei | LvMKK7 | KT719405 | Hepatopancreas: V. parahaemolyticus, S. aureus and WSSV, up | (125) |
| P38 | L. vannamei | LvP38 | JN035902.2 | Gills and hemocytes: V. alginolyticus, S. aureus and WSSV, up | (124, 127) |
| F. chinensis | FcP38 | KF991368 | Hemocytes and gills: WSSV, V. anguillarum and S. aureus, up | (122) | |
| ATF-2 | F. chinensis | FcATF-2 | KF991367 | Hemocytes and gills: WSSV, V. anguillarum and S. aureus, up | (122) |
| TRIM | L. vannamei | LvTRIM9 | Unsubmitted | Intestine: WSSV, up | (128) |
| β-TrCP | L. vannamei | Lvβ-TrCP | XM_027360659 | ND | (128) |
up, differentially up-regulated; down, differentially down-regulated; ND, Not determined.
Cascades of the IMD Pathway
As mentioned above, many members homologous to most of the components of the Drosophila canonical IMD pathway have been cloned and identified in shrimp, and the regulatory network of the shrimp IMD pathway has increasingly become further elucidated (Figure 4). The Drosophila IMD pathway uses the plasma-membrane-located PGRP-LC or the intracellular PGRP-LE to sense microbial DAP-type PGNs, but, until recently, no member of the PGRP family has been found in shrimp (100). We also were unable to find any PGRP homolog by homology searching against various transcriptome data from multiple shrimp species. Nevertheless, it is tempting to speculate that there could be a novel receptor mediating the sensing of microbial infection in shrimp, and that the signaling cascade of the shrimp IMD pathway generally resembles that of the Drosophila IMD pathway (Figure 4).
In particular, a signal-transduction complex consisting of TAK1, TAB1, and TAB2 has been observed in shrimp L. vannamei, where TAB2 could be an adaptor to link upstream IMD (105, 112, 113). Similar to the Drosophila IMD pathway, shrimp TAK1 is responsible for activating both the MAPK/AP-1 and IKK/Relish branches of the IMD pathway (102). There are two IKK-related kinases, IKKβ and IKKε, identified in shrimp, but whether they can receive the phosphorylation signaling from TAK1 is still unknown. Moreover, over-expression of shrimp TAK1 has been demonstrated to strongly regulate the promoter activities of the Drosophila IMD pathway controlling AMPs, such as Dpt in S2 cells, suggesting a conserved role of shrimp TAK1 in the IKK-Relish branch (112). Additionally, shrimp TAK1, functioning as a mitogen-activated protein kinase kinase kinase (MAPKKK), is able to activate and phosphorylate several MAPKKs, including MKK3, MKK4, MKK6, and MKK7, in vitro (unpublished data). Furthermore, MKK7 is recognized as the upstream-kinase target to JNK, while MKK6 is the upstream kinase responsible for p38 (123, 125). Of note, MKK4 from shrimp L. vannamei can activate and phosphorylate p38 (126). As to this point of the phosphorylation on p38, the cascade of the IMD pathway in shrimp is more similar to mammals than that of Drosophila. In shrimp, p38 can be activated and phosphorylated by both MKK4 and MKK6 (123, 126). Additionally, shrimp TAB1 is able to combine with p38, and, thus, it could be an important regulatory subunit for p38 (113). However, in Drosophila, there is no TAB1 homolog, and it has been definitively shown that MKK4 does not active p38 (129). So far, the activation of p38 in shrimp includes at least three routes: the MKK4-p38, MKK7-p38, and TAB1-p38 pathways. Such multiple pathways might supply a fine-tuning control of p38 activity to pathogenic invasion. It also suggests an important role of p38 in shrimp immunity. For example, when one or two routes are blocked under some specific condition, p38 can still work in the immune response.
c-Fos and c-Jun, belonging to the activator protein-1 (AP-1) family, are the transcription factors of the IMD-MAPK branch (130). Shrimp c-Jun has been observed to be mainly located in the nucleus of insect cells under non-stimulated signaling conditions. So, it is inferred that under stressed, immune conditions, JNK could translocate from the cytoplasm to the nucleus where it could phosphorylate and activate c-Jun, which in turn could induce the expression of JNK-pathway-target genes. This hypothesis is supported by the observation that JNK-phosphorylation levels are evidently reduced by JNK-inhibitor (SP600125) treatment in vivo (120, 121). More direct evidence should be demonstrated to further explore whether JNK can interact with and phosphorylate c-Jun in vitro (119). Next, c-Fos, another member of the AP-1 family, has been shown to form a heterodimer with c-Jun, which is also able to form a homodimer with itself, suggesting that c-Fos might also be downstream substrate of JNK (119). The ATF2, a member of the ATF/cAMP response-element-binding family of transcription factors, contains a common feature of the bZIP element, which is able to form homodimers or heterodimers with other proteins that contain bZIP elements, such as the AP-1 (131). A shrimp ATF2 homolog, FcATF2, has been reported in F. chinensis. Silencing of Fcp38 results in a reduction in the transcription of FcMKK3 and FcATF2, indicating that shrimp MKK3, p38, and ATF2 might function in the same signaling route (122). However, whether FcMKK3 can directly regulate the phosphorylation of Fcp38, and whether Fcp38 has the ability to activate FcATF2, need to be determined in further investigations.
Relish, another member of the NF-κB family, is the transcription factor of the IKK-Relish branch, both in Drosophila and shrimp. Similar to Drosophila Relish, shrimp Relish proteins are master regulators for the synthesis of a wide range of AMPs. Multiple Relish homologs have been identified from various shrimp species. The full length of LvRelish from L. vannamei consists of an N-terminal Rel homology domain (RHD), a nucleus-localization signal (NLS), and an IκB-like domain containing six ankyrin repeats (ANKs) and a death domain (DD) in the C-terminal. A truncated isoform of Relish, sLvRelish, has also been found in L. vannamei and it shares the RHD region with LvRelish but does not have several domains, including NLS, ANKs and DD (12). LvRelish can bind to a κB-response element from Drosophila and regulate the transcription of several AMPs such PEN2, PEN4, and Atta (12, 53). The PmRelish from P. monodon has been shown to regulate the synthesis of AMPs such as PEN5, PEN3, ALFPm3, and ALFPm6 in response to V. harveyi or yellow head virus (YHV) infection (132, 133). In M. rosenbergii, Shi et al. show that over-expression of MrRelish in S2 cells induce the expression of both Drosophila and shrimp AMPs, such as Drosophila Mtk, Atta, Drs, and Cecropin A (CecA) and shrimp PEN4 (111). Furthermore, RNAi of MrRelish leads to reduced expression of Crustin (Cru) 2, Cru5, Cru8, Lysozyme (Lyz) 1, and Lyz2, but not ALF1 and ALF3, in vivo (111). Wang et al. used the combined methods of RNA interference (RNAi) and suppression-subtractive hybridization (SSH) to screen F. chinensis Relish-regulated genes, and a large amount of genes were identified and could be involved in multiple biological processes, such as immunity, development, metabolism, and genetic-information processing (134). This study has provided a novel view to understand the function of Relish beyond its conventional role in regulating AMPs in innate immunity.
Regulators of the IMD Pathway
Recent reports have identified several regulators of the IMD-Relish pathway in shrimp, such as NF-κB repressing factor (NKRF), Akirin, and TRIpartite Motif 9 (TRIM9) (115, 116, 128). In mammals, NF-κB repressing factor (NKRF) is well-recognized as a suppression factor for NF-κB, which specifically counteracts the basal activity of several NF-κB-dependent promoters by binding directly to specific negative-regulatory DNA elements (NRE) (135). In contrast, L. vannamei NKRF shows no inhibitory effects but instead exhibits enhancing effects on activities of Dorsal and Relish, as observed by the fact that NKRF can directly interact with both of the NF-κB members to regulate the promoter activities of PEN4, a previously identified target gene of Toll and IMD pathways (117). Shrimp Akirin homologs are recently discovered nuclear factors that play important roles in innate immune system. Two Akirins have been identified in L. vannamei and M. japonicas, respectively (115, 116). By RNAi methods, both LvAkirin and MjAkirin have been shown to positively regulate the expression of several IMD-Relish-target AMPs in vivo (115, 117). MjAkirin is able to regulate IMD-Relish-target AMPs, which could be attributed to its direct interaction with Relish. Interestingly, MjAkirin could function as an important regulator for Bap60 and 14-3-3 to positively and negatively regulate the activity of the IMD-Relish pathway, respectively (116). As a bridge protein, MjAkirin links the transcription factors, Relish and Bap60, the latter of which is a component of the Brahma (SWI/SNF) ATP-dependent chromatin-remodeling complex and positively regulates AMP expression (115, 116). On the other side, the heterotrimeric complex, comprised of Akirin, Relish, and 14-3-3, has been shown to down-regulate AMP expression by an unknown mechanism (116, 118). Recently, an E3-ubiquitin ligase, TRIM9, has been identified in shrimp L. vannamei. LvTRIM9 can directly interact with beta-transducin repeat-containing protein (β-TrCP), an inhibitor of the NF-κB pathway, and down-regulate the expression levels of LvRelish and AMPs, which suggests that WSSV may hijack host LvTRIM9 for its propagation through inhibition of the NF-κB pathway and AMP production via the interaction of LvTRIM9 with Lvβ-TrCP (128). These observations suggest that the IKK-Relish branch may be under multiple layers of control and may crosstalk with many other pathways because 14-3-3 is conserved protein that is implicated with a wide variety of signal-transduction pathways.
NF-κB (Toll and IMD) Pathways Regulate AMPs Expression in Response to Bacterial Infection
Dorsal and Relish are the downstream NF-κB-family transcription factors of Toll and IMD pathways, respectively. In Drosophila, the Toll pathway responds to Gram-positive bacteria and fungi, while the IMD pathway responds to the Gram-negative pathway (20). However, in shrimp, it is very interesting to find that there is no specific response to Gram-positive or Gram-negative bacteria by Toll and IMD pathways, as demonstrated by the findings that many pivotal components from the shrimp Toll pathway—such as Toll, MyD88, and Dorsal—and from the IMD pathway—such as IMD, TAK1, and Relish—are activated in response to both Gram-negative bacteria and Gram-positive bacteria (12, 16, 17, 24, 25, 28, 30, 31, 35, 36, 39, 105, 110–113). Similar to the Drosophila NF-κB pathway, the activation of shrimp NF-κB pathways leads to the boosted expression of various AMPs, which are widely considered to be the major antimicrobial effectors in humoral immunity. AMPs are an group of molecules with molecular weights that are usually <10 kDa, which are effective on bacteria (Gram-positive and Gram-negative), fungi (yeasts and filamentous), and parasites, as well as in some cases on enveloped viruses (136). AMPs are found in evolutionarily diverse organisms ranging from prokaryotes, invertebrates, vertebrates, and to plants (137–139). To date, several classes of AMPs or effectors have been identified in shrimp, composed of Penaeidin (PEN), Crustin (Cru), anti-LPS-factor (ALF), C-type lectin (CTL), Lysozyme (Lyz), and thioester-containing protein (TEP). In Drosophila, AMPs are mainly regulated by NF-κB pathways, and the transcription of drosomycin (Drs) and diptericin (Dpt) have been identified as the hallmarks for the activation of the Toll pathway and the IMD pathway, respectively (22, 23, 136). However, the regulatory mechanism of AMPs is still not clear in shrimp. Herein, we will summarize these AMPs and other antimicrobial proteins that have been reported to be associated with the two NF-κB pathways, directly or indirectly (Table 3).
Table 3.
AMPs or effectors related to NF-κB pathways identified in shrimps.
| AMPs | Species | Types | Function identified | Signaling pathways | References |
|---|---|---|---|---|---|
| LvPEN2 | L. vannamei | Penaeidin | Anti-Vibrio | AP-1 (c-Fos, c-Jun) | (27, 119, 140) |
| LvPEN3 | L. vannamei | Penaeidin | Anti-Vibrio | Toll, AP-1 | (24, 27, 119, 140) |
| LvPEN4 | L. vannamei | Penaeidin | Anti-Vibrio | IMD; Toll2; NF-κB; AP-1 | (11, 12, 17, 27, 119, 140, 141) |
| LvCru1 | L. vannamei | Crustin | V. alginolyticus: down | Toll4; TAK1; IAP | (27, 112, 142, 143) |
| LvCru2 | L. vannamei | Crustin | ND | TAK1 | (112) |
| LvCru3 | L. vannamei | Crustin | ND | Toll4; TAK1; IAP | (27, 112, 143) |
| LvCrustinA | L. vannamei | Crustin | Anti-Vibrio; Anti-WSSV | NF-κB; AP-1 | (141) |
| LvCrustinP | L. vannamei | Crustin | ND | Toll; IMD; AP-1; NF-κB | (24, 144) |
| LvALF1 | L. vannamei | Anti-LPS-factor | Interact with VP19, VP26, VP28, wsv134, and wsv321 | IMD; Toll4; Dorsal | (24, 27) |
| LvALF2 | L. vannamei | Anti-LPS-factor | Anti-Vibrio; Anti-fungi; Anti-WSSV | Toll4; Dorsal | (27, 145) |
| LvALF3 | L. vannamei | Anti-LPS-factor | Anti-Vibrio; anti-WSSV | Toll4; Dorsal | (27) |
| LvCTL3 | L. vannamei | C-type lectin | Anti-Vibrio; anti-WSSV | Dorsal | (146) |
| LvCTL4 | L. vannamei | C-type lectin | Anti-Vibrio | NF-κB | (147) |
| LvLYZ1 | L. vannamei | Lysozyme | Anti-Vibrio; Anti-WSSV: Interact with VP26, VP28, wsv134, and wsv321 | Toll4; Dorsal; TAK1 | (27, 112) |
| LvLYZ2 | L. vannamei | Lysozyme | Anti-Vibrio; Anti-WSSV | Toll4; Dorsal; TAK1 | (27, 112) |
| LvLYZ3 | L. vannamei | Lysozyme | Anti-Vibrio; Anti-WSSV | Dorsal | (27) |
| LvLYZ4 | L. vannamei | Lysozyme | ND | Toll4; Dorsal; TAK1 | (27, 112) |
| LvTEP1 | L. vannamei | TEP | G+/G-: up; WSSV: up | NF-κB; AP-1 | (148) |
| FcPEN3 | F. chinensis | Penaeidin | ND | Relish | (149, 150) |
| FcPEN5 | F. chinensis | Penaeidin | Anti-bacteria | NF-κB | (13, 14, 151) |
| FcCru1 | F. chinensis | Crustin | ND | IMD; Relish | (109, 149, 152) |
| FcCru2 | F. chinensis | Crustin | ND | Spz | (41, 152) |
| FcCru3 | F. chinensis | Crustin | ND | IMD | (109, 152) |
| FcALF | F. chinensis | Anti-LPS-factor | ND | Relish | (149) |
| FcALF6 | F. chinensis | Anti-LPS-factor | ND | IMD | (109, 153) |
| FcALF8 | F. chinensis | Anti-LPS-factor | ND | IMD | (109) |
| FcLys2 | F. chinensis | Lysozyme | ND | IMD | (109) |
| PmPEN3 | P. monodon | Penaeidin | Anti-bacteria | Spz | (39, 154) |
| PmPEN411 | P. monodon | Penaeidin | ND | AP-1, Dorsal | (155) |
| PmPEN536 | P. monodon | Penaeidin | ND | AP-1, Dorsal | (119, 155) |
| PmPEN309 | P. monodon | Penaeidin | ND | Toll2 | (26) |
| Crus-likePm | P. monodon | Crustin | V. harveyi: up | NF-κB; STAT5; AP-1 | (26, 156) |
| crustinPm1 | P. monodon | Anti-LPS-factor | ND | Spz | (39) |
| crustinPm5 | P. monodon | Anti-LPS-factor | ND | NF-κB | (26, 157) |
| crustinPm7 | P. monodon | Anti-LPS-factor | ND | Spz | (39) |
| ALFPm2 | P. monodon | Anti-LPS-factor | V. harveyi: up | NF-κB; AP-1 | (158, 159) |
| ALFPm3 | P. monodon | Anti-LPS-factor | V. harveyi: up; WSSV: up | Spz | (39, 153, 158, 159) |
| MjLys1 | M. japonicas | Lysozyme | V. anguillarum: up; | Toll; IMD; Toll3 | (109, 160, 161) |
| Mj-Lys2 | M. japonicus | Lysozyme | ND | IMD | (109) |
| PcCru1 | P. clarkia | Crustin | V. anguillarum: up | Toll; IMD; Toll3 | (19, 36, 109) |
| PcCru2 | P. clarkia | Crustin | V. anguillarum: up | Toll; IMD | (19, 109) |
| PcALF1 | P. clarkii | Anti-LPS-factor | anti-bacteria | IMD; Toll4; Toll2; Toll3 | (35–37, 109, 162) |
| PcALF2 | P. clarkia | Anti-LPS-factor | V. anguillarum: up | IMD; Toll; Toll4; Toll2 | (19, 35, 37, 109) |
| PcALF4 | P. clarkia | Anti-LPS-factor | ND | Toll4 | (37) |
| PcALF7 | P. clarkia | Anti-LPS-factor | ND | Toll4 | (37) |
| PcALF10 | P. clarkia | Anti-LPS-factor | ND | Toll4 | (37) |
| PcLys1 | P. clarkii | Lysozyme | V. anguillarum: up; | Toll; IMD; Toll3 | (19, 36, 109) |
| MrCru2 | M. rosenbergii | Crustin | Anti-WSSV | Relish | (111) |
| MrCru3 | M. rosenbergii | Crustin | Anti-WSSV | Toll | (34) |
| MrCru5 | M. rosenbergii | Crustin | Anti-WSSV | Relish | (111) |
| MrCru7 | M. rosenbergii | Crustin | Anti-WSSV | Toll | (34) |
| MrCru8 | M. rosenbergii | Crustin | Anti-WSSV | Relish | (111) |
| MrALF2 | M. rosenbergii | Anti-LPS-factor | ND | Toll | (34) |
| MrALF3 | M. rosenbergii | Anti-LPS-factor | ND | Toll | (34) |
| MrALF4 | M. rosenbergii | Anti-LPS-factor | ND | Toll | (34) |
| MrALF5 | M. rosenbergii | Anti-LPS-factor | ND | Toll | (34) |
| MrLys1 | M. rosenbergii | Lysozyme | ND | Relish | (111) |
| MrLys2 | M. rosenbergii | Lysozyme | ND | Relish | (111) |
G+, Gram-positive bacteria; G−: Gram-negative bacteria; up, differentially up-regulated; down, differentially down-regulated; ND, Not determined.
PENs are a special class of AMPs that have only been identified in Penaeid shrimp. Delphine et al. isolated three PENs from the hemolymph of shrimp P. vannamei for the first time in 1997 (163). These peptides are defined as the name of Penaeidins after the genus Penaeus, which therefore cannot be associated to groups hitherto described (163). This family is highly cationic, consisting of a highly conserved leader peptide followed by an N-terminal proline-rich domain (PRD) and a C-terminal cysteine-rich domain (CRD). PENs have been shown to possess strong antimicrobial activities against Gram-positive and Gram-negative bacteria, as well as fungi (164). Until recently, PENs had only been discovered in several shrimp, including L. vannamei, F. chinensis, P. monodon, and M. japonicus. Unlike LvPEN2 and LvPEN3, the LvPEN4 upstream-regulatory region contains many putative transcription-factor-binding sites, including STATx, AP-1, Dorsal, and GATA (140). Luciferase-reporter assays have confirmed that LvToll2, LvToll4, LvMyD88, LvDorsal, LvIMD, and LvRelish, which belong to either IMD or Toll pathways, are able to induce the promoter activity of LvPEN4 in insect cells (11, 12, 17, 26, 27). Chromatin immunoprecipitation (CHIP) assays have also demonstrated that LvDorsal can bind with the promoter region of LvPEN4 after LPS challenge in vivo, which correlates well with the finding that over-expression of LvCactus represses the promoter activity of LvPEN4 in vitro (55). Moreover, LvPEN2, LvPEN3, and LvPEN4 have been shown to be up-regulated by IKKβ, TAK1, MKK4, MKK6, and AP-1 (c-Fos and c-Jun) in vivo or in vitro, which are key components of the IMD pathway (52, 112, 119, 123, 126). However, Hou et al. observed that knockdown of IMD only induced subtle effects on the expression of PEN3 in vivo after V. anguillarum and M. lysodeikticus infections (24). The discrepancy of these results may be due to differences between the evaluated methods on expression of PEN3 and/or different bacterial challenges used in the studies. In F. chinensis, knockdown of FcRelish results in down-regulation of FcPEN3 after bacterial infection in vivo (149). FcPEN5, with strong activities against Gram-positive and Gram-negative bacteria as well as fungi, has been shown to be greatly suppressed and delayed in Relish-silenced shrimp after Vibrio anguillarium and Micrococcus lysodeikticus challenges (13, 14, 151). Additionally, Ho et al. cloned the following two types of genomic fragments flanking the 5′ end of the Penaeidin gene in P. monodon: Type536 and Type411 sequences, both of which contain several transcription-factor-binding motifs, such as TATA box, GATA, dorsal, and AP-1 (155). Similar to PEN2-4 from L. vanamei, many members from either IMD or Toll pathways can induce the promoter activities of Type536 and Type411 in insect cells (119, 155). In summary, PENs have been found in several penaeid shrimp and have broad anti-microbial properties to Gram-positive and Gram-negative bacteria, as well as fungi (151, 164), but they do not exhibit any antiviral properties. Most PENs can be regulated by both the Toll pathway and two branches of the IMD pathway.
Crustins, functioning as protease inhibition or as an effector molecule, is a type of whey acidic protein (WAP) domain-containing and cationic cysteine-rich AMPs (141). Generally, the characterized WAP domain is composed of 50 amino-acid residues with eight cysteine residues at defined positions, which form four intracellular disulfide bonds to create a tightly packed structure (141, 144). Crustins are usually classified into five types (Type I–V) based on the differences in the domain organizations between the signal sequences and the WAP domain (165). Most Crustins found in shrimp belong to Type-II Crustins that contain an N-terminal signaling peptide, followed by a long glycine-rich domain, cysteine-rich domain, and a WAP domain at the C-terminal. There are also some Crustins in shrimp belonging to Type III, which are also named single-WAP-domain proteins (SWDs) (26, 144). Instead of the glycine-rich and the cysteine-rich regions, SWDs have a proline-arginine-rich domain between the signal sequence and the WAP domain (144). In shrimp, most Crustins are mainly expressed in the hemocytes or gills, but rarely in hepatopancreas (141, 144, 152, 166, 167). Surprisingly, PmCru5 from P. monodon could not be detectable in hemocytes but was highly expressed in the epipodite and eyestalks (157). The transcription of Crustins in shrimp might be regulated by both Toll and IMD pathways and other unknown signaling pathways. Analyzing 5′-upstream sequence of Crustins reveal that there are a putative TATA box and several putative binding sites for NF-κB, AP-1, and STAT5 (24, 26, 34, 36, 109, 141, 144, 149). Interestingly, in addition to the above potential binding sites, the promoter of PmCru5 also contains a complete heat-shock-regulatory element, indicating that heat shock may also induce the expression of PmCru5 (144, 157). Studies from Wang et al. have showed that silencing of LvIAP2, homologous to Drsosophila IAP2 of the IMD pathway, leads to a decrease in the expression of LvCrustin1 and LvCrustin3 in hemocytes (143). Zhang et al. have reported that the expression of a Crustin from L. vannamei is up-regulated when Flightless-I is silenced (168). Recently, Li et al. have reported a new Crustin from L. vannamei, named LvCrustinA, which is abundantly expressed in immune-related tissues, such as the gill, hemocyte, and epithelium (141). Dual-reporter assay in S2 cells shows that LvCrustinA can be induced by LvDorsal, LvRelish, and Lvc-Jun, suggesting that LvCrustinA could be regulated by both IMD and Toll pathways (141). Lan et al. have shown that the IMD pathway participated in inducing of three kinds of AMP genes, namely, Crustins, ALFs, and Lysozymes, in F. chinensis and P. clarkii. Specifically, Cru1, Cru2, ALF1, ALF2, and Lyz1 in P. clarkii, and Cru1, Cru3, ALF6, ALF8, and Lyz2 in F. chinensis are thought to be induced via the IMD pathway after V. anguillarum challenge (109). Feng et al. have reported that two Crustins, Cru3 and Cru7, from M. rosenbergii are down-regulated in gills of MrToll-knockdown shrimp in response to WSSV infection (34). However, a Crustin from L. vannamei has been shown to not be regulated by Toll or IMD genes in vivo (24). Most of the Crustins identified from shrimp have been reported to have antiviral or antibacterial roles (169–174), but their expressions implicated in signaling pathways are still unclear. For example, MjCru from M. japonicus has been shown to possess antibacterial activity against Gram-positive and Gram-negative bacteria through destroying the surface of bacterial cells (141, 172). Recently, two new Crustin isoforms, MjCRS8 and MjCRS9, from M. japonicus have been shown to only be expressed in gills, but they do not respond to V. parahaemolyticus or WSSV by immersion tests, which suggests that some Crustins could have additional roles beyond immunity (175).
ALFs are a group of AMPs that were firstly isolated from the hemocytes of the horseshoe crabs, Limulus polyphemus (144, 176). L. polyphemus ALF binds LPS, inhibited the LPS-mediated activation of the Limulus coagulation system, and could strongly inhibited the growth of Gram-negative bacteria (177). In shrimp, ALFs were first identified from the hemocytes of P. monodon (158). Antimicrobial in vitro assays performed with recombinant PmALFs have shown a strong activity against Gram-positive and Gram-negative bacteria and filamentous fungi (178). Subsequently, in vivo experiments, LvALF1-knockdown shrimps exhibited more sensitive to both bacterial and fungal infection than the control group (145). Recently, a new ALF isoform (LvALF AV-R) has been shown to have higher expression levels in hepatopancreas of VP_PirA/B-like toxin-resistant shrimp. The recombinant LvALF AV-R has been found to bind with bacterial proteins, but not Vp_PirAB-like toxin, which suggests that LvALF AV-R might be involved in the resistance mechanism in a non-direct manner (179). So far, several ALFs from several shrimp species—such as L. vannamei and F. chinensis—have been reported to be regulated by the IMD pathway, as indicated by the decreased expression of ALFs in IMD- or Relish-knockdown shrimp (24, 109, 149). Huang et al. have shown that 5 of 11 ALFs (PcALF1, PcALF2, PcALF4, PcALF7, and PcALF10) from P. charkii are regulated by PcToll4 after WSSV challenge in the intestine, but the expressions of other ALFs do not change significantly when PcToll4 is silenced (37). Interestingly, Lan et al. have reported that PcToll2 from P. charkii can positively regulate the expression of PcALF1 and PcALF2 through activating the transcription factor PcATF4, but not PcDorsal or PcSTAT, to defend against Gram-negative bacteria (35). Additionally, as mentioned earlier, Lan et al. also reported that several ALFs from F. chinensis and P. clarkia can be regulated by the IMD pathway to defend against bacterial infection (109). Feng et al. have determined that the MrToll from M. rosenbergii can regulate the expression of four ALF genes (MrALF2, MrALF3, MrALF4, and MrALF5 genes) in the gills after WSSV infection (34). ALFs have also been discovered in hydrothermal-vent shrimp Rimicaris sp. and exhibit activities against a wide range of bacteria (180). In a recent study, the analysis of the tissue distribution, regulation, and biological functions of ALF genes in shrimp suggest that functional diversification of ALFs may rely on multiple selection pressures (159). In addition to the Toll and IMD pathways, other signaling pathways, such as the JAK-STAT pathway, have been shown to participate in regulating the expression of ALFs and Crustins (181).
In crustaceans, non-self-recognition molecules, lectins, play a major role in immune responses mainly by inducing phagocytosis against bacterial pathogens through opsonization (182). C-type lectins (CTLs) are one of the lectin families and is widely existed in Metazoa. C-type lectin domain (CTLD) is the characteristic domain of CTLs, including two disulfide bridges composed of four conserved cysteine residues. Several kinds of CTLs have been identified in shrimp in recent years and have been well reviewed in a previous paper (160). Some CTLs could function in an AMP-like manner, such as FcLec1 and LvCTL3, which could agglutinate both Gram-positive and Gram-negative bacteria (160, 183). There are also reports that have shown that CTLs are regulated by the NF-κB pathway in L. vannamei. A NF-κB-binding site has been found in the LvCTL3 promoter and over-expression of LvDorsal can significantly induce the expression of LvCTL3, which came from the first report on the signaling pathway involved in shrimp CTL expression (146). Subsequently, LvCTL4 has also been found to be regulated by both of the two NF-κB proteins, LvDorsal and LvRelish (147). Considering that the expression of most lectins can been rapidly altered in response to diverse pathogens (160), the transcription of lectins induced by host-signaling pathways could be a generally important immune mechanism in shrimp.
Lysozyme (Lyz/ Lys) is known to be an important immune effector, especially for aquatic animals, in resisting bacterial pathogens by lysing bacterial cell walls. Lysozyme has already been identified in several shrimp species and the transcription levels of Lysozymes vary strikingly after bacterial and viral challenges. Only a few studies have reported that shrimp IMD pathway genes, such as IMD and IAP2, have the ability to regulate Lysozymes, including LvLys, FcLys2, Mj-Lys1, Mj-Lys2, and PcLys1 (109, 143, 160, 184). Additionally, PcLysi1 from P. clarkii possesses antimicrobial activity and has been shown to be regulated by Toll and Toll3 (184). Although many reports have indicated that shrimp Lysozymes have a broad spectrum of antimicrobial properties against multiple bacteria and viruses (19, 185–194), the information on their transcriptional regulation is still very limited.
In invertebrates, TEPs have been studied deeply in Anopheles gambiae (195–198). AgTEP1 has been reported to promote the uptake of bacteria and fungi (199, 200). The first TEP member that was found in crustaceans is Pacofastacus leniusculus TEP, which exclusively is expressed in cuticular tissues, such as the gill and intestine (201). In recent years another TEP has been reported in L. vannamei. Bacterial treatments degrade the full-length LvTEP1 into a processed fragment, which can bind to both Gram-negative and Gram-positive bacteria (148). Knockdown of LvTEP1 in vivo increases the susceptibility to both Gram-positive and Gram-negative bacteria, as well as WSSV. Additionally, the expression of LvTEP1 is dependent on two NF-κB factors, LvRelish and LvDorsal, via the sole NF-κB-binding motif and the AP-1 factors, Lvc-Jun and Lvc-Fos, via the AP-1 motifs in the LvTEP1 promoter (148). Therefore, the authors of this particular study proposed that TEP1 could be induced by both the Toll and IMD pathways (148).
Two NF-κB Pathways in Response to WSSV Infection
The function of shrimp NF-κB pathways is still elusive in response to viral infection, especially WSSV. WSSV is a large (80–120 × 250–380 nm), non-occluded, rod-shaped, enveloped, and double-stranded DNA virus with a genome of ~300 kbp. WSSV is highly pathogenic and virulent, especially in penaeid shrimp, and it has caused serious yearly economic damage to the shrimp industry worldwide. Shrimp with acute infection by WSSV generally begin to die after 24 h with cumulative mortality of 100% within 3–10 days. WSSV is the most studied viral pathogen in shrimp, which provides a promising possibility to elucidate the interplay between NF-κB pathways and this virus.
As mentioned above, the activation of the two shrimp NF-κB pathways (Toll and IMD pathways) leads to the expression of AMPs and other effectors, which confer resistance to a wide range of bacterial pathogens. It is noteworthy that some of these NF-κB pathway-controlled proteins, such C-type lectins and ALFs, possess direct antiviral activities. For instance, the Toll-Dorsal pathway controlling of several AMPs—such as PENs, ALFs, and LYZs—have been shown to interact with different structural proteins of WSSV (27). In addition, knockdown of LvCTL3, a Toll-Dorsal pathway-controlled C-type lectin, has been shown to render shrimp more susceptible to WSSV (146). There are many reported C-type lectins in shrimp with antiviral activity against WSSV—such as LvCTL1, FcLec3, MjLecA, MjLecB, and MjLecC—through their direct interactions with WSSV-envelope proteins; however, whether their expressions are regulated by NF-κB pathways requires further investigation (160, 161, 202, 203). Another case is LvTEP1, which is well identified to be regulated by several transcription factors, including LvDorsal, LvRelish, Lvc-Jun, and Lvc-Fos, which are downstream transcription factors of Toll and IMD pathways. Higher mortality has been observed in LvTEP1-silenced shrimp after WSSV infection, indicating its important antiviral role (63, 148). Additionally, shrimp ALFs have been extensively studied for their roles during WSSV infection. An increasing number of reports have shown that the NF-κB-pathway-targeted ALFs play important role to defend against WSSV. PcToll4 from P. clarkia is able to regulate the expressions of five PcALFs in the intestine to oppose WSSV (37). Moreover, five ALFs from M. rosenbergii have been shown to be regulated by MrToll to confer protection from WSSV (34). Mechanistically, shrimp ALFs exhibit their antiviral activity via interacting with WSSV structural proteins, therefore interfering with viral invasion (27). In accordance with this potential mechanism, PlALF has been shown to play important roles in protect shrimp from WSSV infection via interfering with viral replication in vitro and in vivo in crayfish P. leniusculus (204), while in red-claw crayfish C. quadricarinatus, CqALF can disrupt WSSV-envelope integrity that leads to a decrease of WSSV infectivity (205). PmALF3 from P. monodon has been shown to exhibit the ability to interact with several WSSV structural proteins, such as wsv131 (WSSV186), wsv134 (WSSV189), and wsv339 (WSSV395) (206). Therefore, we propose that the NF-κB pathway regulating AMPs to resist WSSV could be a conserved action in different shrimp species (Figure 5). Additionally, WSSV has been shown to have evolved some strategies to inhibit host NF-κB signaling. This strategy of WSSVs is executed via encoded microRNAs. Ren et al. have identified two viral microRNAs (WSSV-miR-N13 and WSSV-miR-N23), which can target Dorsal to suppress the Spz-Toll-Dorsal-ALF antiviral-signaling pathway in shrimp M. japonicus in vivo (42, 207).
Figure 5.
Interplay between shrimp NF-κB pathways and WSSV. The activation of the NF-κB pathway can be hijacked by WSSV to favor its gene expression and genome replication. WSSV infection activates the host Toll pathway, which leads to the activation of Dorsal that translocates into the nucleus to induce viral-gene expression and promote viral-genome replication. A similar situation is observed for the IMD pathway. The activation of wsv069 mediated by the NF-κB pathways can in turn induce its own expression, which creates a self-regulatory loop. Such a positive-feedback loop amplifies the signaling extent to further activate other viral genes. Until now, after WSSV infection, the NF-κB pathways have been shown to be subject to numerous regulatory controls by both host factors—such as p53, miR1959, and HMGB—and viral factor, such as WSSV449, WSSV-miR-N13, and WSSV-miR-N23. However, the activation of the shrimp canonical NF-κB pathway can also lead to AMP expression, such as ALFs, which have strong antiviral activity against WSSV. Therefore, WSSV could have evolved some currently unknown strategies to attenuate the antiviral role of the host NF-κB pathway to instead engage its activation to favor viral pathogenesis.
There are more and more studies showing that the activation of the NF-κB pathway could be hijacked by WSSVs to favor its own gene expression and genome replication (Table 4). The Toll-Dorsal pathway has been shown to be regulated by WSSV at multiple levels, especially via the Dorsal/Cactus complex (Figure 5). Until now, it has still been unclear as to how the host senses WSSV infection and activates the NF-κB pathway. WSSV encodes a WSSV449 protein that is homologous to the host, Tube, which is an adaptor of the MyD88-Tube-Pelle heterotrimeric complex in the Toll pathway. Similar to the host, Dorsal, WSSV449 is able to activate promoters of Toll-pathway-controlled AMPs, as well as the three viral genes, wsv069, wsv303, and wsv371, in insect cells. Therefore, the authors suspected that WSSV449 could activate the Toll-Dorsal pathway for regulating viral gene expression (47). At the layer of Cactus, WSSV infection can induce host miR-1959 in order to reduce the NF-κB inhibitor LvCactus level, thereby leading to the activation of LvDorsal, which in turn translocates into the nucleus to promote viral-gene expression and promote viral-genome replication (94). Additionally, several host factors—such as two p53 isoforms (LvΔNp53 and LvFLp53), two HMGB isoforms (LvHMGBa and LvHMGBb), and LvSTAT from L. vannamei—have been shown to regulate viral-gene expression through interacting with LvDorsal (121, 208, 209). In accordance with this, several studies have reported that the promoters of some WSSV genes contain binding sites for NF-κB (12, 42, 63, 117, 209). LvDorsal has been demonstrated to regulate the promoter activities of many viral genes, including wsv051, wsv056, wsv069, wsv078, wsv079, wsv080, wsv083, wsv091, wsv094, wsv098, wsv100, wsv101, wsv103, wsv108, wsv178, wsv249, wsv303, wsv358, wsv371, wsv403, and wsv465 (42, 109, 208). A similar situation exists in the IMD pathway (Figure 5). The two branches of the IMD pathway can be activated by WSSV infection in an unknown manner, and the activated transcription factors, LvRelish, Lvc-Jun and Lvc-Fos, have been shown to regulate a bulk of viral genes, such as wsv056, wsv069, wsv079, and wsv249 (52, 119, 123, 210, 211). Specifically, wsv069 was the first identified immediate-early gene in WSSV, is the most studied viral gene regulated by many host factors, and thus may be a promising case to investigate its regulation via signaling pathways. The promoter-regulatory region of wsv069 contains several binding motifs, including one NF-κB, one STAT, and two AP-1s (210). Until now, host transcription factors—Dorsal, Relish, STAT, YY1, c-Jun, and c-Fos—have all been shown to be involved in the regulation of wsv069 (119, 210, 212, 213); this suggests that after WSSV infection, the expression of wsv069 could be induced by the activation of multiple host-signaling pathways, such as the Toll-Dorsal pathway, the two branches of the IMD pathway, and the JAK-STAT pathway. Of note, the activation of wsv069 by host factors can in turn induce viral genes such as itself, which thus establishes a positive-feedback loop (210). Such positive-feedback loops could amplify the signaling extent to further activate other viral early and late genes. Based on these data, we hypothesize that WSSV may hijack host NF-κB pathways to achieve successfully infection. Some in vivo evidence supports this hypothesis, as shown by the fact that knockdown of many pivotal components of NF-κB pathways—such as Dorsal, Relish, and AP-1—have lower viral loads and render shrimp more resistant to viral infection (11, 53, 73, 114, 119).
Table 4.
WSSV proteins/genes related to host (shrimps) Toll and IMD pathways—proteins/genes.
| WSSV genes/proteins | Molecular interaction with host Toll and IMD pathways genes/proteins | Species | References |
|---|---|---|---|
| wsv390/WSSV449 | Tube-like protein | L. vannamei | (47) |
| WSSV-miR-N13, WSSV-miR-N23 | LvDorsal | M. japonicus | (42) |
| wsv069 (IE1) promoter | LvRelish | L. vannamei | (12) |
| wsv069 (IE1) promoter | LvHMGB, LvDorsal | L. vannamei | (208) |
| wsv069 (IE1) promoter | LvSpz4 mediated NF-κB activation | L. vannamei | (38) |
| wsv069 (IE1), wsv303, and wsv371 promoters | LvMyD88 | L. vannamei | (44) |
| wsv069 (IE1), wsv303, and wsv371 promoters | LvPellino | L. vannamei | (57) |
| wsv069 (IE1), wsv303, and wsv371 promoters | LvTube, wsv390 | L. vannamei | (47) |
| wsv069 (IE1) and wsv303 promoters | LvRelish, LvDorsal | L. vannamei | (53) |
| wsv069 and wsv249 promoters | Lvc-Jun, Lvc-Fos | L. vannamei | (119) |
| wsv051, wsv056, wsv069, wsv078, wsv079, wsv080, wsv083, wsv091, wsv094, wsv098, wsv100, wsv101, wsv103, wsv108, wsv178, wsv249, wsv358, wsv403, and wsv465 promoters | LvDorsal | L. vannamei | (209) |
| wsv051, wsv059, wsv069, wsv083, wsv090, wsv107, wsv244, wsv303, wsv371, and wsv445 promoters | LvIKKβ, LvIKKε | L. vannamei | (52) |
| wsv056, wsv069, wsv078, wsv079, wsv080, wsv083, wsv091, wsv094, wsv098, wsv099, wsv101, wsv103, wsv108, wsv178, wsv187, wsv249, wsv358, wsv403, and wsv465 promoters | LvMKK6 | L. vannamei | (123) |
Viruses have a very limited set of genes and, therefore, must use the host cellular resources to achieve their life cycles. WSSVs have evolved to use the activation of host NF-κB pathways—more specifically via the transcription factors Dorsal, Relish, c-Jun, and/or c-Jun—to promote its self-gene expression and genome replication. However, the activation of host NF-κB pathways leads to boost the synthesis of a specific set of AMPs or effectors with antiviral activities against WSSV. This is not inconceivable, but it seems apparent that WSSVs have evolved some unknown strategies to attenuate the antiviral role of host NF-κB pathways, but instead engage its activation to favor viral pathogenesis. This could be explained by the observation that many NF-κB-controlled AMPs are induced in the stage of early infection but are quickly inhibited later. However, the function of NF-κB pathways during other viral infection is still poorly understood. PmRelish from P. monodon seems to play an antiviral role against Yellow head virus (YHV) (114). More studies regarding the roles of host-signaling pathways in WSSV and other viral infections should be performed.
Conclusions and Future Studies
The innate immune system is of great importance for shrimp to defend against infection. Recently, studies on two NF-κB pathways in shrimp mainly contain the follow themes: the identification and characterization of components of NF-κB pathways; dissecting signal transduction components of NF-κB pathways; and determining the function of NF-κB pathways in response to bacterial and viral (WSSV) infection. However, there are also many key components that have not been uncovered, especially in the IMD pathway, such as the receptors for DREDD and FADD, and how the action of signal transduction in this pathway is mediated is still unclear. Recently, the genome sequence of L. vannamei, with the annotation of 25,596 protein-coding genes, has been reported (214). Decoding the L. vannamei genome will not only promote the discovery of some conserved components of signaling pathways, but will also provide the opportunity to understand various biological processes of shrimp at the genomic level. In the Drosophila genome, a total of nine Tolls have been found, but only the Toll1, or simply Toll, has been definitively identified as an upstream receptor of Dorsal or Dif. Until now, there have been five groups of Tolls identified in shrimp, but which of these Tolls can induce Dorsal or other transcription factors, such as ATF and IRF activation, is still unclear. Additionally, different from how Drosophila Toll recognizes infection via binding to Spätzle, shrimp Tolls can directly sense foreign pathogenic motifs similar to that of mammals. For example, three Tolls from M. japonicas can directly bind to both PGN and LPS (73) and two Tolls from L. vannamei can interact with CpG ODN 2395 (72) in vitro. Therefore, understanding how shrimp Toll receptors recognize invading pathogens will provide novel insights into the sensing of Tolls in invertebrates.
Additionally, most of these studies have only reported the phonotype of functions, which are insufficient to present the undergoing mechanisms of these genes mediated immune response. In mammals, crosslinking of different signaling pathways, such as NF-κB related pathways and IRF-related pathways, make the innate immune system a complex network for coordinating appropriate immune responses. Studies on the interplay between Toll and IMD pathways, as well as the interplay among other pathways, should be conducted in shrimp in future studies. Furthermore, it is generally accepted that most AMPs in shrimp could be regulated by both Toll and IMD pathways, but research in this field is still in its infancy and additional experiments are required to explore the individual roles of the two pathways in regulating AMPs. As mentioned above, the strategies by which WSSVs attenuate the antiviral role of host NF-κB pathways to instead engage its activation in favor of viral pathogenesis is still poorly characterized. Further studies on the relationship between host and viral proteins need to be investigated in detail, which will help us to better understand viral pathogenesis and to develop effective strategies for viral-disease control.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Funding. This research was supported by the following: the National Key Research and Development Program of China (2018YFD0900605, 2018YFD0900502) and the National Natural Science Foundation of China (31772883); Science and Technology Program of Guangzhou City of China (201804020013); Guangdong Natural Science Funds for Distinguished Young Scholars (2016A030306041); Tip-top Scientific and Technical Innovative Youth Talents of Guangdong Special Support Program (No. 2016TQ03N504); and Fundamental Research Funds for the Central Universities (17lgpy62). The funders had no role in the study design, data collection/analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li F, Xiang J. Recent advances in researches on the innate immunity of shrimp in China. Dev Comp Immunol. (2013) 39:11–26. 10.1016/j.dci.2012.03.016 [DOI] [PubMed] [Google Scholar]
- 2.Lo CF, Hsu HC, Tsai MF, Ho CH, Peng SE, Kou GH, et al. Specific genomic DNA fragment analysis of different geographical clinical samples of shrimp white spot syndrome virus. Dis Aquat Organ. (1999) 35:175–85. 10.3354/dao035175 [DOI] [Google Scholar]
- 3.Han JE, Tang KF, Tran LH, Lightner DV. Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis Aquat Organ. (2015) 113:33. 10.3354/dao02830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tassanakajon A, Rimphanitchayakit V, Visetnan S, Amparyup P, Somboonwiwat K, Charoensapsri W, et al. Shrimp humoral responses against pathogens: antimicrobial peptides and melanization. Dev Comp Immunol. (2018) 80:81–93. 10.1016/j.dci.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 5.Philpott DJ, Girardin SE. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol Immunol. (2004) 41:1099–108. 10.1016/j.molimm.2004.06.012 [DOI] [PubMed] [Google Scholar]
- 6.Zheng LP, Yu M, Zou XY, Hou L. Function of gram-negtive binding proteins and peptidoglycan-recognition proteins in invertebrate innate immune response. Chin J Microecol. (2010) 22:171–8. [Google Scholar]
- 7.Tang H, Kambris Z, Lemaitre B, Hashimoto C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev Cell. (2008) 15:617–26. 10.1016/j.devcel.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maningas MBB, Kondo H, Hirono I. Molecular mechanisms of the shrimp clotting system. Fish Shellfish Immunol. (2013) 34:968–72. 10.1016/j.fsi.2012.09.018 [DOI] [PubMed] [Google Scholar]
- 9.Mitchell S, Vargas J, Hoffmann A. Signaling via the NFκB system. Wiley Interdisc Rev Syst Biol Med. (2016) 8:227–41. 10.1002/wsbm.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Wang W, Ren Q. Dorsal transcription factor is involved in regulating expression of crustin genes during white spot syndrome virus infection. Dev Comp Immunol. (2016) 63:18–26. 10.1016/j.dci.2016.05.006 [DOI] [PubMed] [Google Scholar]
- 11.Huang XD, Yin ZX, Jia XT, Liang JP, Ai HS, Yang LS, et al. Identification and functional study of a shrimp Dorsal homologue. Dev Comp Immunol. (2010) 34:107–13. 10.1016/j.dci.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 12.Huang XD, Yin ZX, Liao JX, Wang PH, Yang LS, Ai HS, et al. Identification and functional study of a shrimp Relish homologue. Fish Shellfish Immunol. (2009) 27:230–8. 10.1016/j.fsi.2009.05.003 [DOI] [PubMed] [Google Scholar]
- 13.Li F, Wang D, Li S, Yan H, Zhang J, Wang B, et al. A Dorsal homolog (FcDorsal) in the Chinese shrimp Fenneropenaeus chinensis is responsive to both bacteria and WSSV challenge. Dev Comp Immunol. (2010) 34:874–83. 10.1016/j.dci.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 14.Li F, Yan H, Wang D, Priya TA, Li S, Wang B, et al. Identification of a novel relish homolog in Chinese shrimp Fenneropenaeus chinensis and its function in regulating the transcription of antimicrobial peptides. Dev Comp Immunol. (2009) 33:1093–101. 10.1016/j.dci.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 15.Yang LS, Yin ZX, Liao JX, Huang XD, Guo CJ, Weng SP, et al. A Toll receptor in shrimp. Mol Immunol. (2007) 44:1999–2008. 10.1016/j.molimm.2006.09.021 [DOI] [PubMed] [Google Scholar]
- 16.Han-Ching Wang K, Tseng CW, Lin HY, Chen IT, Chen YH, Chen YM, et al. RNAi knock-down of the Litopenaeus vannamei Toll gene (LvToll) significantly increases mortality and reduces bacterial clearance after challenge with Vibrio harveyi. Dev Comp Immunol. (2010) 34:49–58. 10.1016/j.dci.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 17.Wang PH, Gu ZH, Huang XD, Liu BD, Deng XX, Ai HS, et al. An immune deficiency homolog from the white shrimp, Litopenaeus vannamei, activates antimicrobial peptide genes. Mol Immunol. (2009) 46:1897–904. 10.1016/j.molimm.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 18.Sasai M, Yamamoto M. Pathogen recognition receptors: ligands and signaling pathways by toll-like receptors. Int Rev Immunol. (2013) 32:116–33. 10.3109/08830185.2013.774391 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Chen Y-H, Dai Y-J, Tan J-M, Huang Y, Lan J-F, et al. A novel vertebrates Toll-like receptor counterpart regulating the anti-microbial peptides expression in the freshwater crayfish, Procambarus clarkii. Fish Shellfish Immuno. (2015) 43:219–29. 10.1016/j.fsi.2014.12.038 [DOI] [PubMed] [Google Scholar]
- 20.Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J. immunol. (2011) 186:649–56. 10.4049/jimmunol.1002302 [DOI] [PubMed] [Google Scholar]
- 21.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. (1991) 351:355–6. 10.1038/351355b0 [DOI] [PubMed] [Google Scholar]
- 22.Ooi JY, Yagi Y, Hu X, Ip YT. The Drosophila Toll-9 activates a constitutive antimicrobial defense. Embo Rep. (2002) 3:82–7. 10.1093/embo-reports/kvf004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang ZT, Zhu SY. Drosomycin, an essential component of antifungal defence in Drosophila. Insect Mol Biol. (2009) 18:549. 10.1111/j.1365-2583.2009.00907.x [DOI] [PubMed] [Google Scholar]
- 24.Hou F, He S, Liu Y, Zhu X, Sun C, Liu X. RNAi knock-down of shrimp Litopenaeus vannamei Toll gene and immune deficiency gene reveals their difference in regulating antimicrobial peptides transcription. Dev Comp Immunol. (2014) 44:255–60. 10.1016/j.dci.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 25.Yao D, Su H, Zhu J, Zhao X, Aweya JJ, Wang F, et al. SNPs in the Toll1 receptor of Litopenaeus vannamei are associated with immune response. Fish Shellfish Immunol. (2018) 72:410–7. 10.1016/j.fsi.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 26.Wang PH, Liang JP, Gu ZH, Wan DH, Weng SP, Yu XQ, et al. Molecular cloning, characterization and expression analysis of two novel Tolls (LvToll2 and LvToll3) and three putative Spatzle-like Toll ligands (LvSpz1-3) from Litopenaeus vannamei. Dev Compar Immunol. (2012) 36:359–71. 10.1016/j.dci.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 27.Li H, Yin B, Wang S, Fu Q, Xiao B, Lu K, et al. RNAi screening identifies a new Toll from shrimp Litopenaeus vannamei that restricts WSSV infection through activating Dorsal to induce antimicrobial peptides. PLoS Pathog. (2018) 14:e1007109. 10.1371/journal.ppat.1007109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arts JA, Cornelissen FH, Cijsouw T, Hermsen T, Savelkoul HF, Stet RJ. Molecular cloning and expression of a Toll receptor in the giant tiger shrimp, Penaeus monodon. Fish Shellfish Immunol. (2007) 23:504–13. 10.1016/j.fsi.2006.08.018 [DOI] [PubMed] [Google Scholar]
- 29.Assavalapsakul W, Panyim S. Molecular cloning and tissue distribution of the Toll receptor in the black tiger shrimp, Penaeus monodon. Genet Mol Res. (2012) 11:484–93. 10.4238/2012.March.6.1 [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Xu D, Jiang S, Huang J, Zhou F, Yang Q, et al. Toll-receptor 9 gene in the black tiger shrimp (Penaeus monodon) induced the activation of the TLR-NF-κB signaling pathway. Gene. (2018) 639:27–33. 10.1016/j.gene.2017.09.060 [DOI] [PubMed] [Google Scholar]
- 31.Yang C, Zhang J, Li F, Ma H, Zhang Q, Jose Priya TA, et al. A Toll receptor from Chinese shrimp Fenneropenaeus chinensis is responsive to Vibrio anguillarum infection. Fish Shellfish Immunol. (2008) 24:564–74. 10.1016/j.fsi.2007.12.012 [DOI] [PubMed] [Google Scholar]
- 32.Mekata T, Kono T, Yoshida T, Sakai M, Itami T. Identification of cDNA encoding Toll receptor, MjToll gene from kuruma shrimp, Marsupenaeus japonicus. Fish Shellfish Immunol. (2008) 24:122–33. 10.1016/j.fsi.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 33.Srisuk C, Longyant S, Senapin S, Sithigorngul P, Chaivisuthangkura P. Molecular cloning and characterization of a Toll receptor gene from Macrobrachium rosenbergii. Fish Shellfish Immunol. (2014) 36:552–62. 10.1016/j.fsi.2013.12.025 [DOI] [PubMed] [Google Scholar]
- 34.Feng J, Zhao L, Jin M, Li T, Wu L, Chen Y, et al. Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii). Fish Shellfish Immunol. (2016) 57:148–59. 10.1016/j.fsi.2016.08.017 [DOI] [PubMed] [Google Scholar]
- 35.Lan JF, Zhao LJ, Wei S, Wang Y, Lin L, Li XC. PcToll2 positively regulates the expression of antimicrobial peptides by promoting PcATF4 translocation into the nucleus. Fish Shellfish Immunol. (2016) 58:59–66. 10.1016/j.fsi.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 36.Lan JF, Wei S, Wang YQ, Dai YJ, Tu JG, Zhao LJ, et al. PcToll3 was involved in anti-Vibrio response by regulating the expression of antimicrobial peptides in red swamp crayfish, Procambarus clarkii. Fish Shellfish Immunol. (2016) 57:17–24. 10.1016/j.fsi.2016.08.021 [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, Li T, Jin M, Yin S, Hui KM, Ren Q. Newly identified PcToll4 regulates antimicrobial peptide expression in intestine of red swamp crayfish Procambarus clarkii. Gene. (2017) 610:140–7. 10.1016/j.gene.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 38.Yuan K, Yuan FH, Weng SP, He JG, Chen YH. Identification and functional characterization of a novel Spätzle gene in Litopenaeus vannamei. Dev Comp Immunol. (2017) 68:46–57. 10.1016/j.dci.2016.11.016 [DOI] [PubMed] [Google Scholar]
- 39.Boonrawd S, Mani R, Ponprateep S, Supungul P, Masrinoul P, Tassanakajon A, et al. Characterization of PmSptzle 1 from the black tiger shrimp Peneaus monodon. Fish Shellfish Immunol. (2017) 65:88–95. 10.1016/j.fsi.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 40.Arayamethakorn S, Supungul P, Tassanakajon A, Krusong K. Characterization of molecular properties and regulatory pathways of CrustinPm1 and CrustinPm7 from the black tiger shrimp Penaeus monodon. Dev Comp Immunol. (2017) 67:18–29. 10.1016/j.dci.2016.10.015 [DOI] [PubMed] [Google Scholar]
- 41.Shi XZ, Zhang RR, Jia YP, Zhao XF, Yu XQ, Wang JX. Identification and molecular characterization of a Spatzle-like protein from Chinese shrimp (Fenneropenaeus chinensis). Fish Shellfish Immunol. (2009) 27:610–7. 10.1016/j.fsi.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 42.Ren Q, Huang X, Cui Y, Sun J, Wang W, Zhang X. Two white spot syndrome virus microRNAs target the dorsal gene to promote virus infection in Marsupenaeus japonicus Shrimp. J Virol. (2017) 91:e02261-16. 10.1128/JVI.02261-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaniksampanna A, Longyant S, Charoensapsri W, Sithigorngul P, Chaivisuthangkura P. Molecular isolation and characterization of a spatzle gene from Macrobrachium rosenbergii. Fish Shellfish Immunol. (2019) 84:441–50. 10.1016/j.fsi.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 44.Shuang Z, Li CZ, Hui Y, Wei Q, Chen YG, Wang PH, et al. Identification and function of myeloid differentiation factor 88 (MyD88) in Litopenaeus vannamei. PLoS ONE. (2012) 7:e47038 10.1371/journal.pone.0047038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deepika A, Sreedharan K, Paria A, Makesh M, Rajendran KV. Toll-pathway in tiger shrimp (Penaeus monodon) responds to white spot syndrome virus infection: evidence through molecular characterisation and expression profiles of MyD88, TRAF6 and TLR genes. Fish Shellfish Immunol. (2014) 41:441–54. 10.1016/j.fsi.2014.09.026 [DOI] [PubMed] [Google Scholar]
- 46.Wen R, Li F, Sun Z, Li S, Xiang J. Shrimp MyD88 responsive to bacteria and white spot syndrome virus. Fish Shellfish Immunol. (2013) 34:574–81. 10.1016/j.fsi.2012.11.034 [DOI] [PubMed] [Google Scholar]
- 47.Wang PH, Gu ZH, Wan DH, Zhang MY, Weng SP, Yu XQ, et al. The shrimp NF-κB pathway is activated by white spot syndrome virus (WSSV) 449 to facilitate the expression of WSSV069 (ie1), WSSV303 and WSSV371. PLoS ONE. (2011) 6:e24773. 10.1371/journal.pone.0024773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C1 CY, Weng S, Li S, Zuo H, Yu X, Li H, He J, Xu X. Presence of tube isoforms in Litopenaeus vannamei suggests various regulatory patterns of signal transduction in invertebrate NF-κB pathway. Dev Comp Immunol. (2014) 42:174–85. 10.1016/j.dci.2013.08.012 [DOI] [PubMed] [Google Scholar]
- 49.Sreedharan K, Deepika A, Paria A, Babu PPS, Makesh M, Rajendran KV. Ontogeny and expression analysis of tube (interleukin-1 receptor-associated kinase-4 homolog) from Penaeus monodon in response to white spot syndrome virus infection and on exposure to ligands. Agri Gene. (2017) 3:21–31. 10.1016/j.aggene.2016.10.002 [DOI] [Google Scholar]
- 50.Wang PH, Wan DH, Gu ZH, Deng XX, Weng SP, Yu XQ, et al. Litopenaeus vannamei tumor necrosis factor receptor-associated factor 6 (TRAF6) responds to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection and activates antimicrobial peptide genes. Dev Comp Immunol. (2011) 35:105–14. 10.1016/j.dci.2010.08.013 [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Li F, Li S, Chi Y, Wen R, Feng N, et al. An IκB homologue (FcCactus) in Chinese shrimp Fenneropenaeus chinensis. Dev Comp Immunol. (2013) 39:352–62. 10.1016/j.dci.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 52.Wang PH, Gu ZH, Wan DH, Liu BD, Huang XD, Weng SP, et al. The shrimp IKK-NF-κB signaling pathway regulates antimicrobial peptide expression and may be subverted by white spot syndrome virus to facilitate viral gene expression. Cell Mol Immunol. (2013) 10:423–36. 10.1038/cmi.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu W, Zhang S, Chen YG, Wang PH, Xu XP, Li CZ, et al. Litopenaeus vannamei NF-κB is required for WSSV replication. Dev Comp Immunol. (2014) 45:156–62. 10.1016/j.dci.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 54.Sun JJ, Lan JF, Shi XZ, Yang MC, Niu GJ, Ding D, et al. β-Arrestins negatively regulate the toll pathway in shrimp by preventing dorsal translocation and inhibiting dorsal transcriptional activity. J Biol Chem. (2016) 291:7488–504. 10.1074/jbc.M115.698134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C, Chen YX, Zhang S, Lu L, Chen YH, Chai J, et al. Identification, characterization, and function analysis of the Cactus gene from Litopenaeus vannamei. PLoS ONE. (2012) 7:e49711. 10.1371/journal.pone.0049711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang PH, Gu ZH, Wan DH, Zhu WB, Qiu W, Chen YG, et al. Litopenaeus vannamei Toll-interacting protein (LvTollip) is a potential negative regulator of the shrimp Toll pathway involved in the regulation of the shrimp antimicrobial peptide gene penaeidin-4 (PEN4). Dev Comp Immunol. (2013) 40:266–77. 10.1016/j.dci.2013.02.011 [DOI] [PubMed] [Google Scholar]
- 57.Li C, Chai J, Li H, Zuo H, Wang S, Qiu W, et al. Pellino protein from pacific white shrimp Litopenaeus vannamei positively regulates NF-κB activation. Dev Comp Immunol. (2014) 44:341–50. 10.1016/j.dci.2014.01.012 [DOI] [PubMed] [Google Scholar]
- 58.Wang PH, Gu ZH, Wan DH, Zhu WB, Qiu W, Weng SP, et al. Litopenaeus vannamei sterile-alpha and armadillo motif containing protein (LvSARM) is involved in regulation of Penaeidins and antilipopolysaccharide factors. PLoS ONE. (2013) 8:e52088. 10.1371/journal.pone.0052088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S, Qiu W, Chen YG, Yuan FH, Li CZ, Yan H, et al. Flightless-I (FliI) is a potential negative regulator of the Toll pathway in Litopenaeus vannamei. Fish Shellfish Immunol. (2015) 42:413–25. 10.1016/j.fsi.2014.10.023 [DOI] [PubMed] [Google Scholar]
- 60.Imler JL, Hoffmann JA. Toll receptors in innate immunity. Trends Cell Biol. (2001) 11:304–11. 10.1016/S0962-8924(01)02004-9 [DOI] [PubMed] [Google Scholar]
- 61.Guanzon DAV, Maningas MBB. Functional elucidation of LvToll 3 receptor from P. vannamei through RNA interference and its potential role in the shrimp antiviral response. Dev Comp Immunol. (2018) 84:172–80. 10.1016/j.dci.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 62.Jiménez-Dalmaroni MJ, Gerswhin ME, Adamopoulos IE. The critical role of toll-like receptors — From microbial recognition to autoimmunity: a comprehensive review. Autoimmun Rev. (2015) 15:1–8. 10.1016/j.autrev.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang P-H, Huang T, Zhang X, He J-G. Antiviral defense in shrimp: from innate immunity to viral infection. Antiviral Res. (2014) 108:129–41. 10.1016/j.antiviral.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 64.Li F, Xiang J. Signaling pathways regulating innate immune responses in shrimp. Fish Shellfish Immunol. (2013) 34:973–80. 10.1016/j.fsi.2012.08.023 [DOI] [PubMed] [Google Scholar]
- 65.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 66.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 67.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. (2009) 420:1–16. 10.1042/BJ20090272 [DOI] [PubMed] [Google Scholar]
- 68.Mizuguchi K, Parker JS, Blundell TL, Gay NJ. Getting knotted: a model for the structure and activation of Spätzle. Trends Biochem Sci. (1998) 23:239–42. 10.1016/S0968-0004(98)01216-X [DOI] [PubMed] [Google Scholar]
- 69.Delotto Y, Delotto R. Proteolytic processing of the Drosophila Spätzle protein by easter generates a dimeric NGF-like molecule with ventralising activity. Mech Dev. (1998) 72:141. 10.1016/S0925-4773(98)00024-0 [DOI] [PubMed] [Google Scholar]
- 70.Groppe J, Rumpel K, Economides AN, Stahl N, Sebald W, Affolter M. Biochemical and biophysical characterization of refolded Drosophila DPP, a homolog of bone morphogenetic proteins 2 and 4. J Biol Chem. (1998) 273:29052. 10.1074/jbc.273.44.29052 [DOI] [PubMed] [Google Scholar]
- 71.Nakamoto M, Moy R, Jie X, Bambina S, Yasunaga A, Shelly S, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. (2012) 36:658–67. 10.1016/j.immuni.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun R, Wang M, Wang L, Yue F, Yi Q, Huang M, et al. The immune responses triggered by CpG ODNs in shrimp Litopenaeus vannamei are associated with LvTolls. Dev Comp Immunol. (2014) 43:15. 10.1016/j.dci.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 73.Sun JJ, Xu S, He ZH, Shi XZ, Zhao XF, Wang JX. Activation of toll pathway is different between Kuruma Shrimp and Drosophila. Front Immunol. (2017) 8:1151. 10.3389/fimmu.2017.01151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tauszigdelamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. (2002) 3:91–7. 10.1038/ni747 [DOI] [PubMed] [Google Scholar]
- 75.Ji S, Sun M, Zheng X, Li L, Sun L, Chen D, et al. Cell-surface localization of Pellino antagonizes Toll-mediated innate immune signalling by controlling MyD88 turnover in Drosophila. Nat Commun. (2014) 5:3458. 10.1038/ncomms4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun H, Towb P, Chiem DN, Foster BA, Wasserman SA. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. Embo J. (2004) 23:100–10. 10.1038/sj.emboj.7600033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lindsay SA, Wasserman SA. Conventional and non-conventional Drosophila Toll signaling. Dev Comp Immunol. (2014) 42:16–24. 10.1016/j.dci.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin MS, Lee JO. Structures of the Toll-like receptor family and its ligand complexes. Immunity. (2008) 29:182–91. 10.1016/j.immuni.2008.07.007 [DOI] [PubMed] [Google Scholar]
- 79.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci USA. (2002) 99:5567–72. 10.1073/pnas.082100399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gottipati S, Rao NL, Fung-Leung WP. IRAK1: A critical signaling mediator of innate immunity. Cell Signal. (2008) 20:269–76. 10.1016/j.cellsig.2007.08.009 [DOI] [PubMed] [Google Scholar]
- 81.Muroi M, Tanamoto KI. IRAK-1-mediated negative regulation of Toll-like receptor signaling through proteasome-dependent downregulation of TRAF6. Biochim Biophys Acta. (2012) 1823:255–63. 10.1016/j.bbamcr.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 82.Qian Y, Commane M, Ninomiya-Tsuji J, Matsumoto K, Li X. IRAK-mediated translocation of TRAF6 and TAB2 in the interleukin-1-induced activation of NFkappa B. J Biol Chem. (2001) 276:41661–7. 10.1074/jbc.M102262200 [DOI] [PubMed] [Google Scholar]
- 83.Goh ET, Arthur JS, Cheung PC, Akira S, Toth R, Cohen P. Identification of the protein kinases that activate the E3 ubiquitin ligase Pellino 1 in the innate immune system. Biochem J. (2012) 441:339–46. 10.1042/BJ20111415 [DOI] [PubMed] [Google Scholar]
- 84.Tseng PH, Chen IT. O019 TLR4-mediated IKK and MAPK activation is regulated by differential post-translational modification of TAK1. Cytokine. (2012) 59:505–6. 10.1016/j.cyto.2012.06.050 [DOI] [Google Scholar]
- 85.Didonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. (1997) 388:548–54. 10.1038/41493 [DOI] [PubMed] [Google Scholar]
- 86.Narayanan KB, Park HH. Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis. (2015) 20:196–209. 10.1007/s10495-014-1073-1 [DOI] [PubMed] [Google Scholar]
- 87.Manfrere KC, Torrealba MP, Miyashiro DR, Oliveira LM, De GC, Lima JF, et al. Toll-like receptor agonists partially restore the production of pro-inflammatory cytokines and type I interferon in Sézary syndrome. Oncotarget. (2016) 7:74592–601. 10.18632/oncotarget.12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng Z, Taylor B, Ourthiague DR, Hoffmann A. Distinct single-cell signaling characteristics are conferred by the MyD88 and TRIF pathways during TLR4 activation. Sci Signal. (2015) 8:ra69. 10.1126/scisignal.aaa5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maxwell T. An apoptosis signaling pathway induced by the death domain of FADD selectively kills normal but not cancerous prostate epithelial cells. Cell Death Differ. (2001) 8:696–705. 10.1038/sj.cdd.4400866 [DOI] [PubMed] [Google Scholar]
- 90.Ernst MK, Dunn LL, Rice NR. The PEST-like sequence of I kappa B alpha is responsible for inhibition of DNA binding but not for cytoplasmic retention of c-Rel or RelA homodimers. Mol Cell Biol. (1995) 15:872–82. 10.1128/MCB.15.2.872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang C, Bai N, Chang A, Zhang Z, Yin J, Shen W, et al. ATF4 is directly recruited by TLR4 signaling and positively regulates TLR4-trigged cytokine production in human monocytes. Cell Mol Immunol. (2013) 10:84–94. 10.1038/cmi.2012.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li C, Li H, Chen Y, Chen Y, Wang S, Weng SP, et al. Activation of Vago by interferon regulatory factor (IRF) suggests an interferon system-like antiviral mechanism in shrimp. Sci Rep. (2015) 5:15078. 10.1038/srep15078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haas M, Page S, Page M, Neumann FJ, Marx N, Adam M, et al. Effect of proteasome inhibitors on monocytic IκB-α and -β depletion, NF-κB activation, and cytokine production. J Leukocyte Biol. (1998) 63:395–404. 10.1002/jlb.63.3.395 [DOI] [PubMed] [Google Scholar]
- 94.Zuo H, Yuan J, Chen Y, Li S, Su Z, Wei E, et al. A MicroRNA-mediated positive feedback regulatory loop of the NF-κB pathway in Litopenaeus vannamei. J Immunol. (2016) 196:3842. 10.4049/jimmunol.1502358 [DOI] [PubMed] [Google Scholar]
- 95.O'Neill LA, Fitzgerald KA, Bowie AG. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol. (2003) 24:286. 10.1016/S1471-4906(03)00115-7 [DOI] [PubMed] [Google Scholar]
- 96.Campbell HD, Schimansky T, Claudianos C, Ozsarac N, Kasprzak AB, Cotsell JN, et al. The Drosophila melanogaster flightless-I gene involved in gastrulation and muscle degeneration encodes gelsolin-like and leucine-rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc Natl Acad Sci USA. (1993) 90:11386–90. 10.1073/pnas.90.23.11386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. (2004) 61:2614–23. 10.1007/s00018-004-4225-6 [DOI] [PubMed] [Google Scholar]
- 98.Wang T, Chuang TH, Ronni T, Gu S, Du YC, Cai H, et al. Flightless I homolog negatively modulates the TLR pathway. J Immunol. (2006) 176:1355. 10.4049/jimmunol.176.3.1355 [DOI] [PubMed] [Google Scholar]
- 99.Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. (2006) 7:1074–81. 10.1038/ni1382 [DOI] [PubMed] [Google Scholar]
- 100.Myllymäki H, Valanne S, Rämet M. The Drosophila IMD signaling pathway. J Immunol. (2014) 192:3455–62. 10.4049/jimmunol.1303309 [DOI] [PubMed] [Google Scholar]
- 101.Lemaitre B, Kromermetzger E, Michaut L, Nicolas E, Meister M, Georgel P, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci USA. (1995) 92:9465–9. 10.1073/pnas.92.21.9465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. (2014) 42:25–35. 10.1016/j.dci.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila Immune Deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. (2001) 1:503–14. 10.1016/S1534-5807(01)00059-4 [DOI] [PubMed] [Google Scholar]
- 104.Kaneko T, Silverman N. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell Microbiol. (2010) 7:461–9. 10.1111/j.1462-5822.2005.00504.x [DOI] [PubMed] [Google Scholar]
- 105.Wang S, Li H, Qian Z, Song X, Zhang Z, Zuo H, et al. Identification and functional characterization of the TAB2 gene from Litopenaeus vannamei. Fish Shellfish Immunol. (2015) 46:206–16. 10.1016/j.fsi.2015.06.024 [DOI] [PubMed] [Google Scholar]
- 106.Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. (2002) 12:996–1000. 10.1016/S0960-9822(02)00873-4 [DOI] [PubMed] [Google Scholar]
- 107.Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, et al. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. (2006) 18:964. 10.1016/j.cellsig.2005.08.020 [DOI] [PubMed] [Google Scholar]
- 108.Dushay MS, Asling B, Hultmark D. Origins of immunity: relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci USA. (1996) 93:10343–7. 10.1073/pnas.93.19.10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lan JF, Zhou J, Zhang XW, Wang ZH, Zhao XF, Ren Q, et al. Characterization of an immune deficiency homolog (IMD) in shrimp (Fenneropenaeus chinensis) and crayfish (Procambarus clarkii). Dev Comp Immunol. (2013) 41:608–17. 10.1016/j.dci.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 110.Feng N, Wang D, Wen R, Li F. Functional analysis on immune deficiency (IMD) homolog gene in Chinese shrimp Fenneropenaeus chinensis. Mol Biol Rep. (2014) 41:1437–44. 10.1007/s11033-013-2988-2 [DOI] [PubMed] [Google Scholar]
- 111.Shi YR, Jin M, Ma FT, Huang Y, Huang X, Feng JL, et al. Involvement of Relish gene from Macrobrachium rosenbergii in the expression of anti-microbial peptides. Dev Comp Immunol. (2015) 52:236–44. 10.1016/j.dci.2015.05.008 [DOI] [PubMed] [Google Scholar]
- 112.Wang S, Li H, Lu K, Qian Z, Weng S, He J, et al. Identification and characterization of transforming growth factor beta-activated kinase 1 from Litopenaeus vannamei involved in anti-bacterial host defense. Fish Shellfish Immunol. (2016) 52:278–88. 10.1016/j.fsi.2016.03.149 [DOI] [PubMed] [Google Scholar]
- 113.Wang S, Li M, Yin B, Li H, Xiao B, Lu K, et al. Shrimp TAB1 interacts with TAK1 and p38 and activates the host innate immune response to bacterial infection. Mol Immunol. (2017) 88:10–9. 10.1016/j.molimm.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 114.Visetnan S, Supungul P, Hirono I, Tassanakajon A, Rimphanitchayakit V. Activation of PmRelish from Penaeus monodon by yellow head virus. Fish Shellfish Immunol. (2015) 42:335–44. 10.1016/j.fsi.2014.11.015 [DOI] [PubMed] [Google Scholar]
- 115.Hou F, Wang X, Qian Z, Liu Q, Liu Y, He S, et al. Identification and functional studies of Akirin, a potential positive nuclear factor of NF-kappaB signaling pathways in the Pacific white shrimp, Litopenaeus vannamei. Dev Comp Immunol. (2013) 41:703–14. 10.1016/j.dci.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 116.Liu N, Wang XW, Sun JJ, Wang L, Zhang HW, Zhao XF, et al. Akirin interacts with Bap60 and 14-3-3 proteins to regulate the expression of antimicrobial peptides in the kuruma shrimp (Marsupenaeus japonicus). Dev Comp Immunol. (2016) 55:80–9. 10.1016/j.dci.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 117.Qiu W, He JH, Zuo H, Niu S, Li C, Zhang S, et al. Identification, characterization, and function analysis of the NF-κB repressing factor (NKRF) gene from Litopenaeus vannamei. Dev Comp Immunol. (2017) 76:83–92. 10.1016/j.dci.2017.05.020 [DOI] [PubMed] [Google Scholar]
- 118.Wanna W, Thipwong J, Mahakaew W, Phongdara A. Identification and expression analysis of two splice variants of the 14-3-3 epsilon from Litopenaeus Vannamei during WSSV infections. Mol Biol Rep. (2012) 39:5487–93. 10.1007/s11033-011-1351-8 [DOI] [PubMed] [Google Scholar]
- 119.Li C, Li H, Wang S, Song X, Zhang Z, Qian Z, et al. The c-Fos and c-Jun from Litopenaeus vannamei play opposite roles in Vibrio parahaemolyticus and white spot syndrome virus infection. Dev Comp Immunol. (2015) 52:26–36. 10.1016/j.dci.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 120.Shi H, Yan X, Ruan L, Xu X. A novel JNK from Litopenaeus vannamei involved in white spot syndrome virus infection. Dev Comp Immunol. (2012) 37:421–8. 10.1016/j.dci.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 121.Yao D, Ruan L, Xu X, Shi H. Identification of a c-Jun homolog from Litopenaeus vannamei as a downstream substrate of JNK in response to WSSV infection. Dev Comp Immunol. (2015) 49:282–9. 10.1016/j.dci.2014.12.012 [DOI] [PubMed] [Google Scholar]
- 122.He Y, Yao W, Liu P, Li J, Wang Q. Expression profiles of the p38 MAPK signaling pathway from Chinese shrimp Fenneropenaeus chinensis in response to viral and bacterial infections. Gene. (2018) 642:381–8. 10.1016/j.gene.2017.11.050 [DOI] [PubMed] [Google Scholar]
- 123.Li H, Wang S, Qian Z, Wu Z, Lu K, Weng S, et al. MKK6 from pacific white shrimp Litopenaeus vannamei is responsive to bacterial and WSSV infection. Mol Immunol. (2016) 70:72–83. 10.1016/j.molimm.2015.12.011 [DOI] [PubMed] [Google Scholar]
- 124.Shi H, Ruan L, Yan X, Yao D, Xu X. The role of Litopenaeus vannamei p38 in white spot syndrome virus infection. Dev Comp Immunol. (2014) 44:180–5. 10.1016/j.dci.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 125.Wang S, Qian Z, Li H, Lu K, Xu X, Weng S, et al. Identification and characterization of MKK7 as an upstream activator of JNK in Litopenaeus vannamei. Fish Shellfish Immunol. (2016) 48:285–94. 10.1016/j.fsi.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 126.Wang S, Yin B, Li H, Xiao B, Lu K, Feng C, et al. MKK4 from Litopenaeus vannamei is a regulator of p38 MAPK kinase and involved in anti-bacterial response. Dev Comp Immunol. (2018) 78:61–70. 10.1016/j.dci.2017.09.015 [DOI] [PubMed] [Google Scholar]
- 127.Yan H, Zhang S, Li CZ, Chen YH, Chen YG, Weng SP, et al. Molecular characterization and function of a p38 MAPK gene from Litopenaeus vannamei. Fish Shellfish Immunol. (2013) 34:1421–31. 10.1016/j.fsi.2013.02.030 [DOI] [PubMed] [Google Scholar]
- 128.Sun M, Li S, Yu K, Xiang J, Li F. An E3 ubiquitin ligase TRIM9 is involved in WSSV infection via interaction with β-TrCP. Dev Comp Immunol. (2019) 97:57–63. 10.1016/j.dci.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 129.Geuking P, Narasimamurthy R, Lemaitre B, Basler K, Leulier F. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PLoS ONE. (2009) 4:60–71. 10.1371/journal.pone.0007709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. (1997) 9:240–6. 10.1016/S0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- 131.Morooka H, Bonventre JV, Pombo CM, Kyriakis JM, Force T. Ischemia and reperfusion enhance ATF-2 and c-Jun binding to cAMP response elements and to an AP-1 binding site from the c-jun promoter. J Biol Chem. (1995) 270:30084–92. 10.1074/jbc.270.50.30084 [DOI] [PubMed] [Google Scholar]
- 132.Suwattana V, Premruethai S, Sureerat T, Ikuo H, Anchalee T, Vichien R. YHV-responsive gene expression under the influence of PmRelish regulation. Fish Shellfish Immunol. (2015) 47:572–81. 10.1016/j.fsi.2015.09.053 [DOI] [PubMed] [Google Scholar]
- 133.Kamsaeng P, Tassanakajon A, Somboonwiwat K. Regulation of antilipopolysaccharide factors, ALFPm3 and ALFPm6, in Penaeus monodon. Sci Rep. (2017) 7:12694. 10.1038/s41598-017-12137-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang D, Li S, Li F. Screening of genes regulated by relish in Chinese shrimp Fenneropenaeus chinensis. Dev Comp Immunol. (2013) 41:209–16. 10.1016/j.dci.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 135.Froese N, Schwarzer M, Niedick I, Frischmann U, Köster M, Kröger A, et al. Innate Immune responses in NF-κB-repressing factor-deficient mice. Mol Cell Biol. (2006) 26:293–302. 10.1128/MCB.26.1.293-302.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Imler J, Bulet P. Antimicrobial Peptides in Drosophila: Structures, Activities and Gene Regulation. Basel: Karger Publishers; (2005). 10.1159/000086648 [DOI] [PubMed] [Google Scholar]
- 137.Tossi A, Sandri L. Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr Pharm Des. (2002) 8:743–61. 10.2174/1381612023395475 [DOI] [PubMed] [Google Scholar]
- 138.Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. (2004) 198:169–84. 10.1111/j.0105-2896.2004.0124.x [DOI] [PubMed] [Google Scholar]
- 139.Bulet P, Charlet M, Hetru C. Antimicrobial Peptides in Insect Immunity. Totowa, NJ: Humana Press; (2003). [Google Scholar]
- 140.O'Leary NA, Gross PS. Genomic structure and transcriptional regulation of the penaeidin gene family from Litopenaeus vannamei. Gene. (2006) 371:75–83. 10.1016/j.gene.2005.11.028 [DOI] [PubMed] [Google Scholar]
- 141.Li M, Ma C, Li H, Peng J, Zeng D, Chen X, et al. Molecular cloning, expression, promoter analysis and functional characterization of a new Crustin from Litopenaeus vannamei. Fish Shellfish Immunol. (2018) 73:42–9. 10.1016/j.fsi.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 142.Vargas-Albores F, Yepiz-Plascencia G, Jimenez-Vega F, Avila-Villa A. Structural and functional differences of Litopenaeus vannamei crustins. Comp Biochem Physiol B Biochem Mol Biol. (2004) 138:415–22. 10.1016/j.cbpc.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 143.Wang PH, Wan DH, Gu ZH, Qiu W, Chen YG, Weng SP, et al. Analysis of Expression, cellular localization, and function of three inhibitors of apoptosis (IAPs) from Litopenaeus vannamei during WSSV infection and in regulation of antimicrobial peptide genes (AMPs). PLos ONE. (2013) 8:e72592. 10.1371/journal.pone.0072592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tassanakajon A, Amparyup P, Somboonwiwat K, Supungul P. Cationic antimicrobial peptides in penaeid shrimp. Mar Biotechnol. (2011) 13:639–57. 10.1007/s10126-011-9381-8 [DOI] [PubMed] [Google Scholar]
- 145.de la Vega E, O'Leary NA, Shockey JE, Robalino J, Payne C, Browdy CL, et al. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): a broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection. Mol Immunol. (2008) 45:1916–25. 10.1016/j.molimm.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 146.Li M, Li C, Ma C, Li H, Zuo H, Weng S, et al. Identification of a C-type lectin with antiviral and antibacterial activity from pacific white shrimp Litopenaeus vannamei. Dev Comp Immunol. (2014) 46:231–40. 10.1016/j.dci.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 147.Li H, Chen Y, Li M, Wang S, Zuo H, Xu X, et al. A C-type lectin (LvCTL4) from Litopenaeus vannamei is a downstream molecule of the NF-κB signaling pathway and participates in antibacterial immune response. Fish Shellfish Immunol. (2015) 43:257–63. 10.1016/j.fsi.2014.12.024 [DOI] [PubMed] [Google Scholar]
- 148.Li C, Li H, Xiao B, Chen Y, Wang S, Lu K, et al. Identification and functional analysis of a TEP gene from a crustacean reveals its transcriptional regulation mediated by NF-κB and JNK pathways and its broad protective roles against multiple pathogens. Dev Comp Immunol. (2017) 70:45–58. 10.1016/j.dci.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 149.Wang D, Fuhua LI, Shihao LI, Wen R, Xiang J. Expression profiles of antimicrobial peptides (AMPs) and their regulation by Relish. Chin J Oceanol Limnol. (2012) 30:611–9. 10.1007/s00343-012-1195-7 [DOI] [Google Scholar]
- 150.Kang CJ, Wang JX, Zhao XF, Yang XM, Shao HL, Xiang JH. Molecular cloning and expression analysis of Ch-penaeidin, an antimicrobial peptide from Chinese shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. (2004) 16:513–25. 10.1016/j.fsi.2003.09.003 [DOI] [PubMed] [Google Scholar]
- 151.Kang CJ, Xue JF, Liu N, Zhao XF, Wang JX. Characterization and expression of a new subfamily member of penaeidin antimicrobial peptides (penaeidin 5) from Fenneropenaeus chinensis. Mol Immunol. (2007) 44:1535–43. 10.1016/j.molimm.2006.08.025 [DOI] [PubMed] [Google Scholar]
- 152.Chen S, Du XJ, Xu WT, Zhang HW, Zhao XF, Wang JX. Molecular cloning and characterization of three crustins from the Chinese white shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. (2010) 28:517–24. 10.1016/j.fsi.2009.12.001 [DOI] [PubMed] [Google Scholar]
- 153.Li S, Guo S, Li F, Xiang J. Functional diversity of anti-lipopolysaccharide factor isoforms in shrimp and their characters related to antiviral activity. Mar Drugs. (2015) 13:2602–16. 10.3390/md13052602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tassanakajon A, Klinbunga S, Paunglarp N, Rimphanitchayakit V, Udomkit A, Jitrapakdee S, et al. Penaeus monodon gene discovery project: the generation of an EST collection and establishment of a database. Gene. (2006) 384:104–12. 10.1016/j.gene.2006.07.012 [DOI] [PubMed] [Google Scholar]
- 155.Ho SH, Song YL. Cloning of penaeidin gene promoter in tiger shrimp (Penaeus monodon). Fish Shellfish Immunol. (2009) 27:73–7. 10.1016/j.fsi.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 156.Luo T, Li F, Lei K, Xu X. Genomic organization, promoter characterization and expression profiles of an antiviral gene PmAV from the shrimp Penaeus monodon. Mol Immunol. (2007) 44:1516–23. 10.1016/j.molimm.2006.09.015 [DOI] [PubMed] [Google Scholar]
- 157.Vatanavicharn T, Supungul P, Puanglarp N, Yingvilasprasert W, Tassanakajon A. Genomic structure, expression pattern and functional characterization of crustinPm5, a unique isoform of crustin from Penaeus monodon. Comp Biochem Physiol B Biochem Mol Biol. (2009) 153:244–52. 10.1016/j.cbpb.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 158.Tharntada S, Somboonwiwat K, Rimphanitchayakit V, Tassanakajon A. Anti-lipopolysaccharide factors from the black tiger shrimp, Penaeus monodon, are encoded by two genomic loci. Fish Shellfish Immunol. (2008) 24:46–54. 10.1016/j.fsi.2007.07.010 [DOI] [PubMed] [Google Scholar]
- 159.Matos GM, Schmitt P, Barreto C, Farias ND, Toledo-Silva G, Guzman F, et al. Massive gene expansion and sequence diversification is associated with diverse tissue distribution, regulation and antimicrobial properties of anti-lipopolysaccharide factors in shrimp. Mar Drugs. (2018) 16:E381 10.3390/md16100381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang XW, Wang JX. Diversity and multiple functions of lectins in shrimp immunity. Dev Comp Immunol. (2013) 39:27–38. 10.1016/j.dci.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 161.Song KK, Li DF, Zhang MC, Yang HJ, Ruan LW, Xu X. Cloning and characterization of three novel WSSV recognizing lectins from shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. (2010) 28:596–603. 10.1016/j.fsi.2009.12.015 [DOI] [PubMed] [Google Scholar]
- 162.Sun C, Xu WT, Zhang HW, Dong LP, Zhang T, Zhao XF, et al. An anti-lipopolysaccharide factor from red swamp crayfish, Procambarus clarkii, exhibited antimicrobial activities in vitro and in vivo. Fish Shellfish Immunol. (2011) 30:295–303. 10.1016/j.fsi.2010.10.022 [DOI] [PubMed] [Google Scholar]
- 163.Destoumieux D, Bulet P, Loew D, Van DA, Rodriguez J, Bachère E. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp Penaeus vannamei (Decapoda). J Biol Chem. (1997) 272:28398. 10.1074/jbc.272.45.28398 [DOI] [PubMed] [Google Scholar]
- 164.Cuthbertson BJ, Deterding LJ, Williams JG, Tomer KB, Etienne K, Blackshear PJ, et al. Diversity in penaeidin antimicrobial peptide form and function. Dev Comp Immunol. (2008) 32:167–81. 10.1016/j.dci.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fjell CD, Hiss JA, Hancock RE, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. (2012) 11:37–51. 10.1038/nrd3591 [DOI] [PubMed] [Google Scholar]
- 166.Zhang J, Li F, Wang Z, Xiang J. Cloning and recombinant expression of a crustin-like gene from Chinese shrimp, Fenneropenaeus chinensis. J Biotechnol. (2007) 127:605. 10.1016/j.jbiotec.2006.08.013 [DOI] [PubMed] [Google Scholar]
- 167.Amparyup P, Kondo H, Hirono I, Aoki T, Tassanakajon A. Molecular cloning, genomic organization and recombinant expression of a crustin-like antimicrobial peptide from black tiger shrimp Penaeus monodon. Mol Immunol. (2008) 45:1085–93. 10.1016/j.molimm.2007.07.031 [DOI] [PubMed] [Google Scholar]
- 168.Massry SG, Coburn JW, Kleeman CR. Evidence for suppression of parathyroid gland activity by hypermagnesemia. J Clin Invest. (1970) 49:1619–29. 10.1172/JCI106379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu N, Lan JF, Sun JJ, Jia WM, Zhao XF, Wang JX. A novel crustin from Marsupenaeus japonicus promotes hemocyte phagocytosis. Dev Comp Immunol. (2015) 49:313–22. 10.1016/j.dci.2014.11.021 [DOI] [PubMed] [Google Scholar]
- 170.Sun B, Wang Z, Zhu F. The crustin-like peptide plays opposite role in shrimp immune response to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol. (2017) 66:487. 10.1016/j.fsi.2017.05.055 [DOI] [PubMed] [Google Scholar]
- 171.Donpudsa S, Visetnan S, Supungul P, Tang S, Tassanakajon A, Rimphanitchayakit V. Type I and type II crustins from Penaeus monodon, genetic variation and antimicrobial activity of the most abundant crustinPm4. Dev Comp Immunol. (2014) 47:95–103. 10.1016/j.dci.2014.06.015 [DOI] [PubMed] [Google Scholar]
- 172.Hipolito SG, Shitara A, Kondo H, Hirono I. Role of Marsupenaeus japonicus crustin-like peptide against Vibrio penaeicida and white spot syndrome virus infection. Dev Comp Immunol. (2014) 46:461–9. 10.1016/j.dci.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 173.Chang YT, Lin CY, Tsai CY, Siva VS, Chu CY, Tsai HJ, et al. The new face of the old molecules: crustin Pm4 and transglutaminase type I serving as rnps down-regulate astakine-mediated hematopoiesis. PLoS ONE. (2017) 12:e0182405. 10.1371/journal.pone.0182405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sruthy KS, Nair A, Jayesh P, Antony SP, Singh I, Philip R. Molecular cloning, recombinant expression and functional characterization of an antimicrobial peptide, Crustin from the Indian white shrimp, Fenneropenaeus indicus. Fish Shellfish Immunol. (2017) 71:83–94. 10.1016/j.fsi.2017.09.071 [DOI] [PubMed] [Google Scholar]
- 175.Tandel GM, Kondo H, Hirono I. Gills specific type 2 crustin isoforms: Its molecular cloning and characterization from kuruma shrimp Marsupenaeus japonicus. Dev Comp Immunol. (2018) 85:25–30. 10.1016/j.dci.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 176.Muta T, Miyata T, Tokunaga F, Nakamura T, Iwanaga S. Primary structure of anti-lipopolysaccharide factor from American horseshoe crab, Limulus polyphemus. J Biochem. (1987) 58:1321 10.1093/oxfordjournals.jbchem.a121999 [DOI] [PubMed] [Google Scholar]
- 177.Schmitt P, Rosa RD, Destoumieuxgarzón D. An intimate link between antimicrobial peptide sequence diversity and binding to essential components of bacterial membranes. Biochim Biophys Acta. (2016) 1858:958–70. 10.1016/j.bbamem.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 178.Somboonwiwat K, Marcos M A, Klinbunga S, Aumelas A, Romestand B, Gueguen Y, et al. Recombinant expression and anti-microbial activity of anti-lipopolysaccharide factor (ALF) from the black tiger shrimp Penaeus monodon. Dev Comp Immunol. (2005) 29:841–51. 10.1016/j.dci.2005.02.004 [DOI] [PubMed] [Google Scholar]
- 179.Tinwongger S, Thawonsuwan J, Kondo H, Hirono I. Identification of an anti-lipopolysaccharide factor AV-R isoform (LvALF AV-R) related to Vp_PirAB-like toxin resistance in Litopenaeus vannamei. Fish Shellfish Immunol. (2019) 84:178–88. 10.1016/j.fsi.2018.10.005 [DOI] [PubMed] [Google Scholar]
- 180.Gu HJ, Sun QL, Jiang S, Zhang J, Sun L. First characterization of an anti-lipopolysaccharide factor (ALF) from hydrothermal vent shrimp: Insights into the immune function of deep-sea crustacean ALF. Dev Comp Immunol. (2018) 84:382–95. 10.1016/j.dci.2018.03.016 [DOI] [PubMed] [Google Scholar]
- 181.Sun JJ, Lan JF, Zhao XF, Vasta GR, Wang JX. Binding of a C-type lectin's coiled-coil domain to the Domeless receptor directly activates the JAK/STAT pathway in the shrimp immune response to bacterial infection. PLoS Pathog. (2017) 13:e1006626. 10.1371/journal.ppat.1006626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mrf M, Barracco MA. Lectins, as non-self-recognition factors, in crustaceans. Aquaculture. (2000) 191:23–44. 10.1016/S0044-8486(00)00417-8 [DOI] [Google Scholar]
- 183.Sun YD, Fu LD, Jia YP, Du XJ, Wang Q, Wang YH, et al. A hepatopancreas-specific C-type lectin from the Chinese shrimp Fenneropenaeus chinensis exhibits antimicrobial activity. Mol Immunol. (2008) 45:348–61. 10.1016/j.molimm.2007.06.355 [DOI] [PubMed] [Google Scholar]
- 184.Zhang HW, Sun C, Sun SS, Zhao XF, Wang JX. Functional analysis of two invertebrate-type lysozymes from red swamp crayfish, Procambarus clarkii. Fish Shellfish Immunol. (2010) 29:1066–72. 10.1016/j.fsi.2010.08.023 [DOI] [PubMed] [Google Scholar]
- 185.Sotelo-Mundo RR, Islas-Osuna MA, De-La-Re-Vega E, Hernández-López J, Vargas-Albores F, Yepiz-Plascencia G. cDNA cloning of the lysozyme of the white shrimp Penaeus vannamei. Fish Shellfish Immunol. (2003) 15:325–31. 10.1016/S1050-4648(02)00176-6 [DOI] [PubMed] [Google Scholar]
- 186.Hikima S, Ji H, Rojtinnakorn J, Hirono I, Aoki T. Characterization and function of kuruma shrimp lysozyme possessing lytic activity against Vibrio species. Gene. (2003) 316:187–95. 10.1016/S0378-1119(03)00761-3 [DOI] [PubMed] [Google Scholar]
- 187.de-la-Re-Vega E, Garcia-Galaz A, Diaz-Cinco ME, Sotelo-Mundo RR. White shrimp (Litopenaeus vannamei) recombinant lysozyme has antibacterial activity against Gram negative bacteria: Vibrio alginolyticus, Vibrio parahemolyticus and Vibrio cholerae. Fish Shellfish Immunol. (2006) 20:405–8. 10.1016/j.fsi.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 188.Burge EJ, Madigan DJ, Burnett LE, Burnett KG. Lysozyme gene expression by hemocytes of Pacific white shrimp, Litopenaeus vannamei, after injection with Vibrio. Fish Shellfish Immunol. (2007) 22:327–39. 10.1016/j.fsi.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 189.Bu X, Du X, Zhou W, Zhao X, Wang J. Molecular cloning, recombinant expression and characterization of lysozyme from Chinese shrimp Fenneropenaeus chinensis. Chin J Biotechnol. (2008) 24:723–32. 10.1016/S1872-2075(08)60037-0 [DOI] [PubMed] [Google Scholar]
- 190.Kaizu A, Fagutao FF, Kondo H, Aoki T, Hirono I. Functional analysis of C-type lysozyme in penaeid shrimp. J Biol Chem. (2011) 286:44344–9. 10.1074/jbc.M111.292672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Karthik V, Kamalakannan V, Thomas A, Sudheer NS, Singh IS, Narayanan RB. Functional characterization of a c-type Lysozyme from Indian Shrimp Fenneropenaeus indicus. Probiot Antimicrob Proteins. (2014) 6:114–21. 10.1007/s12602-013-9146-y [DOI] [PubMed] [Google Scholar]
- 192.Liu HT, Wang J, Mao Y, Liu M, Niu SF, Qiao Y, et al. Identification and expression analysis of a new invertebrate lysozyme in Kuruma shrimp (Marsupenaeus japonicus). Fish Shellfish Immunol. (2016) 49:336–43. 10.1016/j.fsi.2015.12.034 [DOI] [PubMed] [Google Scholar]
- 193.Koiwai K, Alenton RR, Kondo H, Hirono I. Extracellular trap formation in kuruma shrimp (Marsupenaeus japonicus) hemocytes is coupled with c-type lysozyme. Fish Shellfish Immunol. (2016) 52:206–9. 10.1016/j.fsi.2016.03.039 [DOI] [PubMed] [Google Scholar]
- 194.Mai WJ, Wang WN. Protection of blue shrimp (Litopenaeus stylirostris) against the White Spot Syndrome Virus (WSSV) when injected with shrimp lysozyme. Fish Shellfish Immunol. (2010) 28:727–33. 10.1016/j.fsi.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 195.Povelones M, Upton LM, Sala KA, Christophides GK. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. (2011) 7:e1002023. 10.1371/journal.ppat.1002023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Eldering M, Morlais I, van Gemert GJ, Van dVM, Graumans W, Siebelinkstoter R, et al. Variation in susceptibility of African Plasmodium falciparum malaria parasites to TEP1 mediated killing in Anopheles gambiae mosquitoes. Sci Rep. (2016) 6:20440. 10.1038/srep20440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Obbard DJ, Callister DM, Jiggins FM, Soares DC, Yan G, Little TJ. The evolution of TEP1, an exceptionally polymorphic immunity gene in Anopheles gambiae. BMC Evol Biol. (2008) 8:274. 10.1186/1471-2148-8-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Baxter RH, Chang CI, Chelliah Y, Blandin S, Levashina EA, Deisenhofer J. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci USA. (2007) 104:11615. 10.1073/pnas.0704967104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. (2001) 104:709–18. 10.1016/S0092-8674(01)00267-7 [DOI] [PubMed] [Google Scholar]
- 200.Hassan Y, Layla K, Osta MA. The mosquito melanization response is implicated in defense against the entomopathogenic FungusBeauveria bassiana. PLoS Pathog. (2012) 8:e1003029 10.1371/journal.ppat.1003029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wu C, Noonin C, Jiravanichpaisal P, Söderhäll I, Söderhäll K. An insect TEP in a crustacean is specific for cuticular tissues and involved in intestinal defense. Insect Biochem Mol Biol. (2012) 42:71–80. 10.1016/j.ibmb.2011.10.006 [DOI] [PubMed] [Google Scholar]
- 202.Junkunlo K, Prachumwat A, Tangprasittipap A, Senapin S, Borwornpinyo S, Flegel TW, et al. A novel lectin domain-containing protein (LvCTLD) associated with response of the whiteleg shrimp Penaeus (Litopenaeus) vannamei to yellow head virus (YHV). Dev Comp Immunol. (2012) 37:334–41. 10.1016/j.dci.2011.12.010 [DOI] [PubMed] [Google Scholar]
- 203.Wang XW, Xu WT, Zhang XW, Zhao XF, Yu XQ, Wang JX. A C-type lectin is involved in the innate immune response of Chinese white shrimp. Fish Shellfish Immunol. (2009) 27:556–62. 10.1016/j.fsi.2009.07.011 [DOI] [PubMed] [Google Scholar]
- 204.Liu H, Jiravanichpaisal P, Soderhall I, Cerenius L, Soderhall K. Antilipopolysaccharide factor interferes with white spot syndrome virus replication in vitro and in vivo in the crayfish Pacifastacus leniusculus. J Virol. (2006) 80:10365–71. 10.1128/JVI.01101-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lin FY, Gao Y, Wang H, Zhang QX, Zeng CL, Liu HP. Identification of an anti-lipopolysacchride factor possessing both antiviral and antibacterial activity from the red claw crayfish Cherax quadricarinatus. Fish Shellfish Immunol. (2016) 57:213–21. 10.1016/j.fsi.2016.08.037 [DOI] [PubMed] [Google Scholar]
- 206.Suraprasit S, Methatham T, Jaree P, Phiwsaiya K, Senapin S, Hirono I, et al. Anti-lipopolysaccharide factor isoform 3 from Penaeus monodon (ALFPm3) exhibits antiviral activity by interacting with WSSV structural proteins. Antiviral Res. (2014) 110:142–50. 10.1016/j.antiviral.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 207.Wang P-H, He J-G. Nucleic acid sensing in invertebrate antiviral immunity. Int Rev Cell Mol Biol. (2019) 345:287–360. 10.1016/bs.ircmb.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 208.Chen YH, Jia XT, Huang XD, Zhang S, Li M, Xie JF, et al. Two Litopenaeus vannamei HMGB proteins interact with transcription factors LvSTAT and LvDorsal to activate the promoter of white spot syndrome virus immediate-early gene ie1. Mol Immunol. (2011) 48:793–9. 10.1016/j.molimm.2010.11.020 [DOI] [PubMed] [Google Scholar]
- 209.Li H, Wang S, Chen Y, Lu K, Yin B, Li S, et al. Identification of two p53 isoforms from Litopenaeus vannamei and their interaction with NF-κB to induce distinct immune response. Sci Rep. (2017) 7:45821. 10.1038/srep45821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Huang XD, Zhao L, Zhang HQ, Xu XP, Jia XT, Chen YH, et al. Shrimp NF-κB binds to the immediate-early gene ie1 promoter of white spot syndrome virus and upregulates its activity. Virology. (2010) 406:176–80. 10.1016/j.virol.2010.06.046 [DOI] [PubMed] [Google Scholar]
- 211.Li C, Weng S, He J. WSSV–host interaction: Host response and immune evasion. Fish Shellfish Immunol. (2019) 84:558–71. 10.1016/j.fsi.2018.10.043 [DOI] [PubMed] [Google Scholar]
- 212.Liu WJ, Chang YS, Wang AH, Kou GH, Lo CF. White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J Virol. (2007) 81:1461–71. 10.1128/JVI.01880-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Huang PH, Huang TY, Cai PS, Chang LK. Role of Litopenaeus vannamei yin yang 1 in the regulation of the white spot syndrome virus immediate early gene ie1. J Virol. (2017) 91:e02314-16. 10.1128/JVI.02314-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang X, Yuan J, Sun Y, Li S, Gao Y, Yu Y, et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat Commun. (2019) 10:356. 10.1038/s41467-018-08197-4 [DOI] [PMC free article] [PubMed] [Google Scholar]