The endoplasmic reticulum (ER) is a major regulator of cellular proteostasis. However, only little is known about signaling molecules resident to this organelle. Centonze et al. identify LTK as the first ER-resident receptor tyrosine kinase and show that it stimulates secretory trafficking out of the ER.
Abstract
The endoplasmic reticulum (ER) is a key regulator of cellular proteostasis because it controls folding, sorting, and degradation of secretory proteins. Much has been learned about how environmentally triggered signaling pathways regulate ER function, but only little is known about local signaling at the ER. The identification of ER-resident signaling molecules will help gain a deeper understanding of the regulation of ER function and thus of proteostasis. Here, we show that leukocyte tyrosine kinase (LTK) is an ER-resident receptor tyrosine kinase. Depletion of LTK as well as its pharmacologic inhibition reduces the number of ER exit sites and slows ER-to-Golgi transport. Furthermore, we show that LTK interacts with and phosphorylates Sec12. Expression of a phosphoablating mutant of Sec12 reduces the efficiency of ER export. Thus, LTK-to-Sec12 signaling represents the first example of an ER-resident signaling module with the potential to regulate proteostasis.
Introduction
The secretory pathway handles a third of the proteome (Sharpe et al., 2010), and it is becoming increasingly clear that its functional organization is regulated by a wide range of signaling pathways (Pulvirenti et al., 2008; Farhan et al., 2010; Farhan and Rabouille, 2011; Zacharogianni et al., 2011; Giannotta et al., 2012; Cancino and Luini, 2013; Scharaw et al., 2016). Much has already been learned about how the secretory pathway responds to external stimuli. However, our understanding of its autoregulation, i.e., about its homeostasis-maintaining responses to stimuli from within the endomembrane system, is less developed. This is mainly due to our ignorance of signaling cascades operating locally on the secretory pathway. The probably best-understood example for autoregulation of the secretory pathway is the unfolded protein response (UPR). The UPR is induced by an accumulation of unfolded proteins in the ER, which results in increasing the expression of chaperones as well as the machinery for protein degradation, vesicle budding, tethering, and fusion (Gardner et al., 2013). A major characteristic of the UPR is that its signaling mediators localize permanently to the ER. However, this is not the case with other signaling molecules identified so far. Very recently, Gα12 was shown to be active at the ER (Subramanian et al., 2019), but only a minor fraction of Gα12 localizes to this organelle. The small GTPase Rac1 was also shown to be activated at the nuclear envelope, which is part of the ER (Woroniuk et al., 2018). Again, the vast majority of Rac1 is either in endosomes or the plasma membrane. Mutant variants of the kinase FLT3 were shown to be permanently ER localized, but these are confined to cancer driving mutants and thus not useful to decipher physiological ER-based signaling (Choudhary et al., 2009; Schmidt-Arras et al., 2009). Thus, signaling at the ER remains poorly understood, which emphasizes the importance of the quest for ER-localized or -resident signaling molecules.
COPII vesicles form at ER exit sites (ERESs) and are responsible for ferrying secretory cargo out of the ER. The COPII coat is composed of the small GTPase Sar1, the Sec23-Sec24 heterodimer, and the Sec13-Sec31 heterotetramer (Zanetti et al., 2011). Activation of Sar1 is mediated by its exchange factor, Sec12, a type II transmembrane protein, which localizes to the general ER as well as to ERESs (Montegna et al., 2012; Saito et al., 2014). ERESs were discovered as COPII decorated sites that often localize in close vicinity to the ER Golgi intermediate compartment (ERGIC; Orci et al., 1991; Appenzeller-Herzog and Hauri, 2006).
Previous siRNA screens uncovered a collection of kinases that regulate ERESs (Farhan et al., 2010; Simpson et al., 2012). Among the hits shared between the two RNAi screens, we focused on leukocyte tyrosine kinase (LTK), because it was previously reported to partially localize to the ER (Bauskin et al., 1991). Our current work identifies LTK as the first ER-resident receptor tyrosine kinase that regulates COPII-dependent trafficking and thus represents a potential druggable proteostasis regulator.
Results and discussion
LTK localizes to the ER
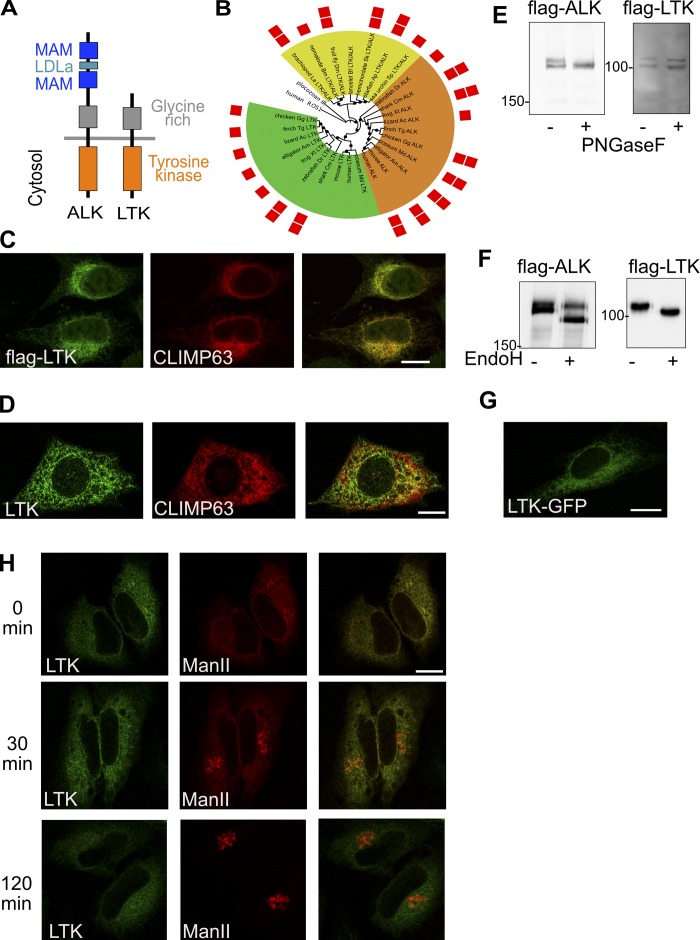
LTK is a receptor tyrosine kinase that is highly homologous to the anaplastic lymphoma kinase (ALK; Fig. 1 A). While their cytoplasmic kinase domain is 79% identical, the extracellular domain of ALK is much larger than that of mammalian LTK as it contains two MAM domains (acronym derived from meprin, A-5 protein, and receptor protein-tyrosine phosphatase mu). Analysis of LTK and ALK evolution shows that deletions of the largest part of the extracellular domain of LTK occurred only in mammals (Fig. 1 B). Non-mammalian LTK rather resembles ALK than human LTK. According to The Human Protein Atlas, LTK mRNA is found in most tissues except muscle.
Figure 1.
Subcellular localization of LTK. (A) Schematic illustrating the domains of LTK and ALK. LDLa, low-density lipoprotein (LDL) receptor class A repeat. (B) Phylogenetic tree of ALK and LTK kinase domains. Color ranges highlight invertebrate LTK/ALK-like proteins (yellow), vertebrate ALK proteins (orange), and vertebrate LTK proteins (green). Red squares indicate the presence of one or two MAM domains. Black circles mark branches with bootstrap support above 50%. Human ROS1 and placozoan insulin receptor–like kinase domains are used as outgroup. The list of abbreviations used in the figure can be found in the Materials and methods section. (C) Immunostaining of flag-tagged human LTK and endogenous CLIMP63 in HeLa cells. (D) immunofluorescence staining of endogenous LTK and CLIMP63 in HepG2 cells. (E) HeLa cells expressing flag-tagged LTK or ALK were treated with PNGase F followed by lysis and immunoblotting against flag to detect ALK or LTK. (F) HeLa cells expressing flag-tagged LTK or ALK were lysed and the lysate treated with EndoH followed by immunoblotting against flag to detect ALK or LTK. (G) HeLa cells expressing GFP-tagged LTK were imaged using live microscopy. (H) HeLa cells expressing GFP-tagged LTK and mCherry-tagged Man-II in the RUSH system were treated for 0 or 2 h with biotin followed by fixation. Scale bars are 10 mm.
LTK was reported to localize to the ER (Bauskin et al., 1991), but this was questioned by recent findings showing LTK activation by extracellular ligands (Zhang et al., 2014; Reshetnyak et al., 2015). Overexpressed flag-tagged LTK, but not ALK, colocalized with the ER marker CLIMP63 (Fig. 1 C and Fig. S1 A). Endogenous LTK also localized to the ER (Fig. 1 D). The specificity of the antibody was tested by showing that the fluorescence signal is weaker in LTK-depleted cells (Fig. S1 B). We also noticed in 10% of cells a weak colocalization of LTK with the ERES marker Sec31 (Fig. S1 C). Immunofluorescence of endogenous LTK was performed in HepG2 cells because they express high levels of LTK but are essentially ALK negative (Fig. S2 A), limiting the possibility of antibody cross-reactivity.
To corroborate the immunofluorescence results, we subjected intact cells expressing flag-tagged ALK or LTK to PNGase F treatment. PNGase F is an enzyme that cleaves glycans and is therefore expected to cause a shift in electrophoretic mobility of proteins exposed to extracellular milieu. Consistent with its absence at the cell surface, we found that LTK was insensitive to treatment of cells with PNGase F (Fig. 1 E). On the contrary, ALK, which is expressed at the cell surface, was sensitive to digestion with PNGase F (Fig. 1 E). We next treated cell lysates expressing flag-tagged LTK or ALK with endoglycosidase H (EndoH), which only digests core-glycosylated proteins that have not entered the Golgi apparatus. LTK was completely sensitive to EndoH treatment, indicating that it resides in a preGolgi compartment (Fig. 1 F). On the other hand, only 60% of the ALK pool was sensitive to EndoH (Fig. 1 F). Available antibodies do not detect endogenous LTK by immunoblotting, preventing us from performing the same analysis with endogenous LTK.
To rule out that the absence of staining of LTK at the cell surface is due to fixation artifacts, we tagged LTK with GFP and performed live imaging. LTK localization was similar as in fixed cells, and was reminiscent of the ER (Fig. 1 G). Finally, we wanted to directly test whether LTK leaves the ER using the retention using selective hooks (RUSH) assay (Boncompain et al., 2012). The RUSH assay monitors the trafficking of a fluorescently labeled reporter protein out of the ER. This reporter is retained in the ER through a streptavidin-based interaction with an ER-resident hook. Treatment with biotin relieves retention and allows the reporter to exit toward post-ER compartments. We engineered GFP-tagged LTK into the RUSH system and expressed it together with a well-described secretory RUSH reporter, Mannosidase-II (Man-II), tagged with mCherry. Initially, LTK and Man-II colocalized in the ER (Fig. 1 H). We fixed and imaged cells 30 min after biotin addition, a time point at which Man-II was entirely in the Golgi. However, LTK was still ER-localized (Fig. 1 H). Even after 2 h, LTK showed no signs of leaving the ER (Fig. 1 H), making it highly unlikely that it ever leaves the ER. Altogether, our results show that LTK is an ER-resident receptor tyrosine kinase, making it a promising candidate to regulate secretion by local ER-based signaling.
LTK regulates ER export
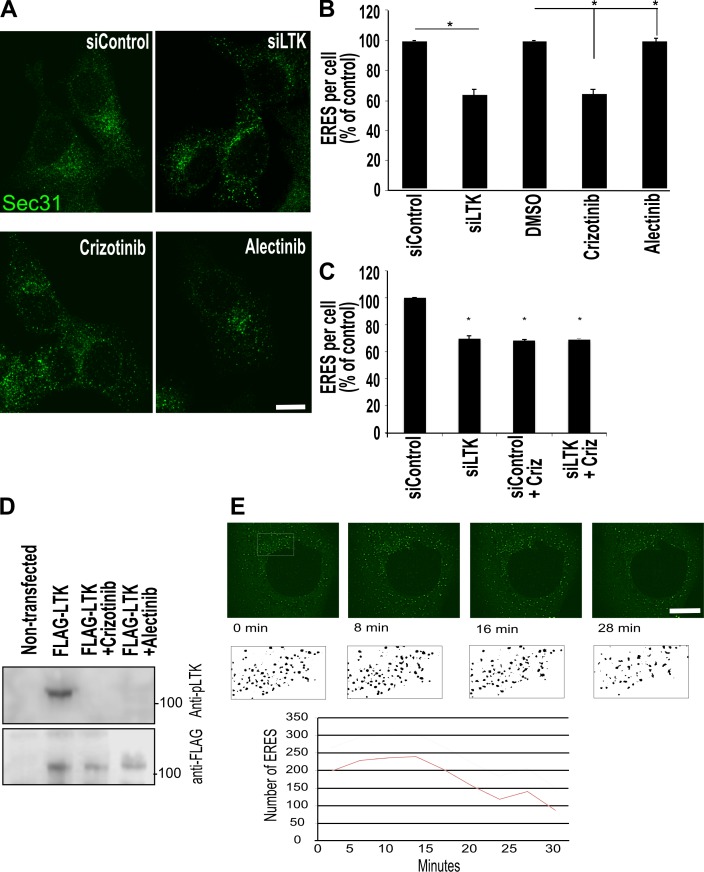
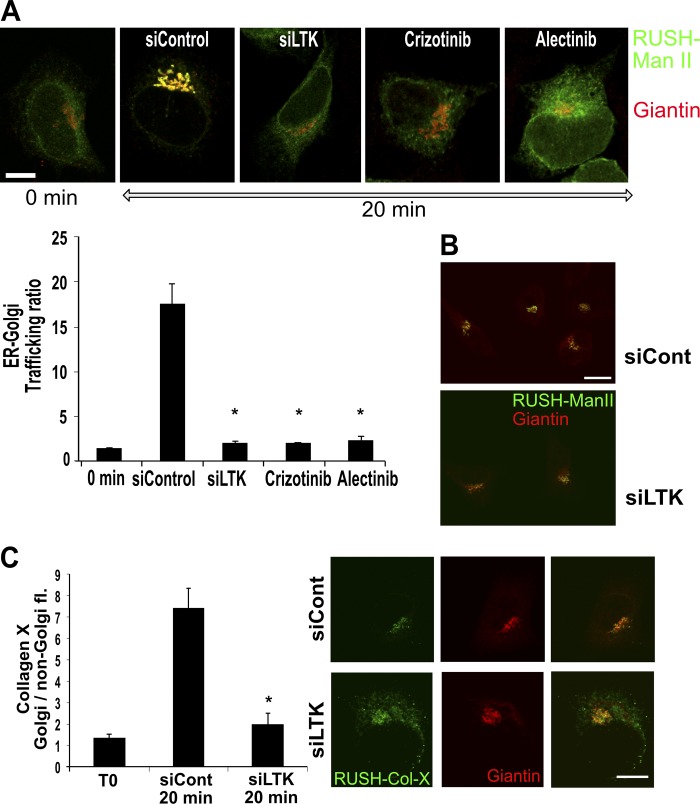
We next asked whether LTK regulates ER-to-Golgi trafficking. We chose to test this in HeLa and HepG2 cells, which are LTK positive but negative for its close relative ALK (Fig. S2 A). Knockdown of LTK (Fig. S2 B; siRNA #3 was used for all further experiments) resulted in a reduction of the number of ERESs by 30–40% in HepG2 (Fig. 2, A and B) and HeLa cells (Fig. S2 C). To support the results of the knockdown experiments, we treated HepG2 cells with two LTK inhibitors, alectinib and crizotinib. Because HepG2 cells are ALK negative (Fig. S2 A), any effect of these drugs is due to LTK inhibition. Treatment with both drugs for 30 min resulted in a reduction in the number of ERES comparable to LTK knockdown (Fig. 2 B). Notably, crizotinib had no effect on ERESs in LTK-depleted cells, supporting the notion that crizotinib affects ERESs by inhibiting LTK (Fig. 2 C). We confirmed that crizotinib and alectinib inhibited LTK autophosphorylation in our experimental system (Fig. 2 D). We also tested the effect on crizotinib in live imaging and found that the onset of ERES reduction is after ∼10 min of treatment (Fig. 2 E). To determine the effect on ER-to-Golgi trafficking, we used the RUSH assay with Man-II as a RUSH cargo (RUSH–Man-II; Boncompain et al., 2012). Silencing LTK expression or its pharmacologic inhibition resulted in a clear retardation of trafficking to the Golgi (Fig. 3 A, Video 1, and Video 2). This effect was a retardation of traffic rather than a total inhibition, because when we allowed the RUSH cargo to traffic for two hours, there was no difference between control and LTK-inhibited cells (Fig. 3 B). The effect of LTK knockdown was not limited to Man-II, but was also observed with another RUSH cargo, namely collagen X (Fig. 3 C), indicating that the effect of LTK is not limited to one type of cargo. Our results so far indicate that LTK is an ER-resident receptor tyrosine kinase that regulates ER export.
Figure 2.
LTK regulates ER export and ERESs. (A) HepG2 cells were either subjected to LTK silencing with siRNA followed by fixation and staining after 72 h, or treated with crizotinib and alectinib (1 µM) for 30 min before fixation and immunostaining against Sec31 to label ERESs. (B) Quantification of ERES number per cell displayed as percentage of control (all values are set as percentage of siControl). Data are from three independent experiments with at least 35 cells per experiment per condition. Asterisk indicates statistically significant differences at P < 0.05. (C) HepG2 cells were transfected with control or LTK siRNA. After 72 h, cells were treated with solvent or with crizotinib (1 µM) for 30 min before staining for Sec31 to determine ERES number. (D) HeLa cells expressing flag-tagged LTK were treated with solvent or with crizotinib or alectinib for 30 min before lysis and immunoblotting as indicated. P-LTK indicates immunoblotting against an antibody that detects phosphorylation on Y672. Flag immunoblotting was performed to determine equal loading. (E) HeLa cells expressing GFP-Sec16A treated with 1 µM crizotinib followed by confocal live imaging. Stills of the indicated time points are depicted. The ERESs in the boxed area are depicted in black and white to enhance visibility. The number of ERESs was counted and is displayed in the lower graph. siLTK, LTK silenced. Scale bars are 10 mm.
Figure 3.
LTK regulates ER export. (A) Representative images of HeLa cells stably expressing the GFP-RUSH-Man-II construct (Str-KDEL-Man-II-EGFP) under different conditions: 0 min, cells not treated with biotin; 20 min, cells fixed 20 min after biotin treatment; Cont, control siRNA transfected; siLTK, LTK silenced; crizotinib and alectinib indicate cells treated with 1 µM 30 min before biotin addition. Bar graph shows quantification from three independent experiments. Asterisk indicates statistically significant differences at P < 0.001. Scale bars in this figure are 15 µm. (B) Representative images of HeLa cells stably expressing the GFP-RUSH-Man-II construct (Str-KDEL-Man-II-EGFP) imaged 2 h after release of the reporter from the ER. Two conditions are depicted, control and LTK knockdown cells. (C) HeLa cells expressing the RUSH-Collagen-X construct were transfected with control or LTK siRNA. After 72 h, cells were treated with biotin and fixed immediately (T0) or after 20 min. Cells were immunostained against Giantin to label the Golgi. The increase in green fluorescence in the Golgi region relative to outside the Golgi region was measured using ImageJ. The bar graph represents the mean of four independent experiments.
LTK interacts with and phosphorylates Sec12
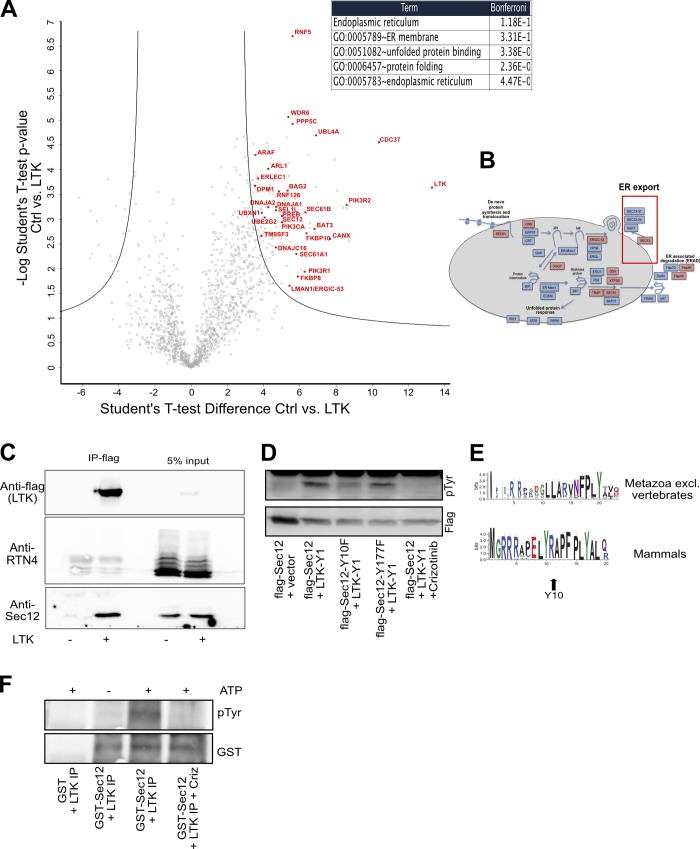
We next sought to mechanistically uncover how LTK regulates ER export. To this end, we mapped the interactome of flag-tagged LTK expressed in HEK293 cells, because they do not express endogenous LTK and can be transfected easily. The most notable enrichment within the LTK interactome is proteins of the early secretory pathway (Fig. 4, A and B; and Table S1). This is consistent with the localization of LTK to the ER. The top associated gene ontology (GO) term among the LTK interactome was “Endoplasmic reticulum” (Fig. 4 A). Among the potential LTK interaction partners identified, we focused on Sec12 (also known as PREB), due to its well-established role in ER export and the biogenesis of ERESs (Barlowe and Schekman, 1993; Montegna et al., 2012; Saito et al., 2014). Sec12 is a type II transmembrane protein that acts as a guanine nucleotide exchange factor for Sar1. Using coimmunoprecipitation, we confirmed that LTK interacts with Sec12, but not with an unrelated transmembrane protein of the ER that was not recovered in the interactome (Fig. 4 C and Fig. S3 A). Co-expression with LTK resulted in an increase in tyrosine phosphorylation of Sec12, which was crizotinib sensitive (Fig. 4 D). Two tyrosine residues in Sec12 (Y177 and Y10) were predicted by databases to be phosphorylated (PhosphoSitePlus and NetPhos3.1). Therefore, we mutated both tyrosine residues to phenylalanine, creating Sec12-Y10F and Sec12-Y177F. Sec12-Y10F was markedly less tyrosine-phosphorylated than wild-type Sec12, indicating the Y10 residue is a strong candidate site for phosphorylation by LTK (Fig. 4 D). Mutation of tyrosine 177 had no effect. We also noted that the Y10 residue in Sec12 is conserved in mammals but not in nonvertebrates (Fig. 4 E). We next purified the cytosolic domain of Sec12 and incubated it with an immunoprecipitate containing flag-LTK. Addition of ATP to the mix resulted in a crizotinib-sensitive increase of Sec12 phosphorylation (Fig. 4 F), supporting the notion that Sec12 phosphorylation is LTK-dependent. Inhibition of Src family kinases had only a marginally effect on Sec12 tyrosine phosphorylation (Fig. S3 B).
Figure 4.
LTK interacts with and phosphorylates Sec12. (A) Volcano plot of the interactome of HA-tagged LTK revealed by immunoprecipitation MS from HEK293 cells. The red labeled candidate interacting proteins are related to receptor tyrosine kinase signaling or ER-associated processes. The table indicates the top-scoring biological processes enriched among the LTK interaction partners. (B) Schematic representation of key components of the ER folding, quality control, and export machinery with red-highlighted interaction partners. Red box highlights ER export. (C) Flag-tagged LTK was immunoprecipitated followed by immunoblotting against Sec12, which was identified in the interactome and against RTN4, a transmembrane protein that we did not find in the LTK interactome. (D) HeLa cells were transfected with vectors encoding flag-tagged Sec12 or its mutants together with an empty vector or with YFP1-tagged LTK (LTK-Y1). In the last lane are lysates from cells pretreated with 1 µM crizotinib for 30 min. (E) Sequence logo to demonstrate the conservation of amino acids in Sec12 in mammals or in metazoan excluding vertebrates. (F) Purified GST-tagged cytosolic domain of Sec12 or GST were incubated a flag-LTK immunoprecipitate from HEK293 cells (LTK immunoprecipitation) in the presence or absence of ATP or crizotinib.
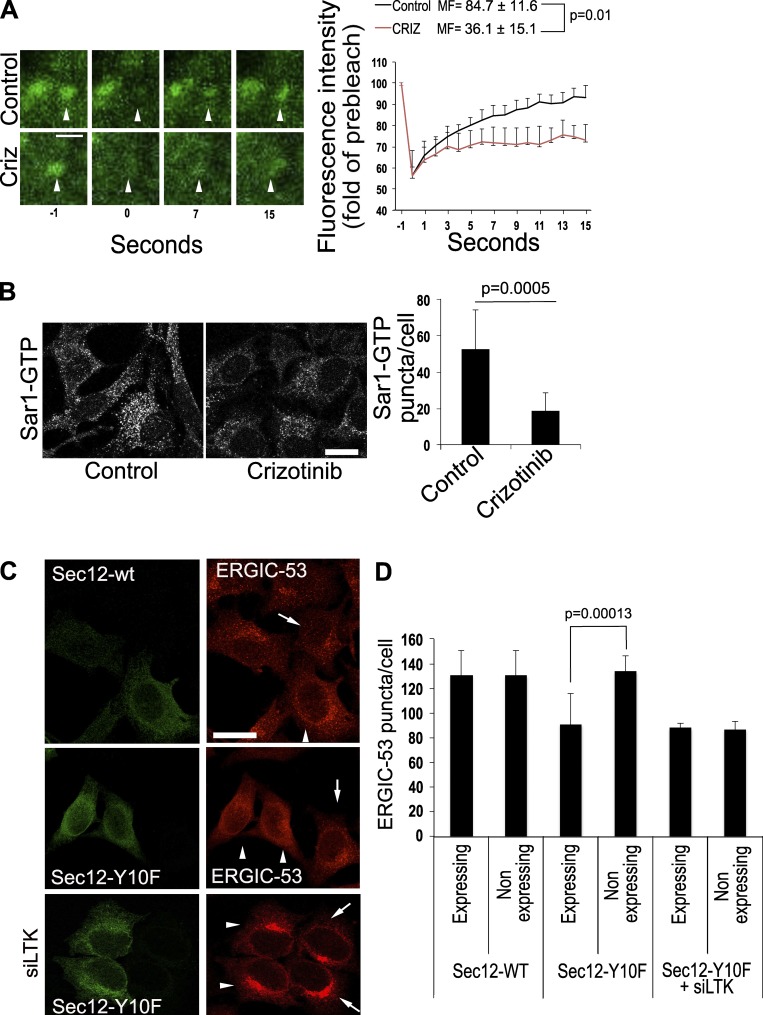
Because Sec12 is the exchange factor for Sar1, we next tested the effect of LTK inhibition on the dynamics of YFP-tagged Sar1A using FRAP microscopy. Cells were pretreated with solvent or with crizotinib for 20 min before FRAP microscopy. Inhibition of LTK reduced the mobile fraction of Sar1, indicative of a reduced exchange activity on single ERES (Fig. 5 A). No effect of crizotinib on general ER structure was detected (Fig. S3 C). We tried using the tryptophan fluorescence assay to monitor GTP exchange in Sar1 and its modulation by LTK. However, this assay cannot be used because the inclusion of ATP in the reaction (to promote Sec12 phosphorylation) distorted the assay (data not shown). Thus, we used a different approach to support the results of the FRAP assay, namely by immunostaining for Sar1-GTP–positive ERESs. This approach has been used by others previously (Venditti et al., 2012). Treatment of cells with crizotinib resulted in cells with fewer and fainter Sar1-GTP–positive puncta (Fig. 5 B), indicating that inhibition of LTK negatively affects the levels of active Sar1 on ERESs.
Figure 5.
LTK regulates Sec12 function. (A) FRAP assay of HepG2 cells expressing YFP-tagged Sar1A. Images on the left side show magnified single ERES at different time points before (−1) and directly after (0) bleaching as well as at the indicated time points after bleaching. Graph shows an evaluation of nine FRAP curves for each condition from three experiments. MF, mobile fraction. Scale bar, 1 µm. Criz indicates a condition where cells were treated with 1 µM of crizotinib for 20 min before the FRAP assay. (B) HeLa cells were treated with solvent (Control) or 1 µM crizotinib for 30 min before fixation and immunostaining against Sar1-GTP. The number of Sar1-GTP puncta was counted using ImageJ and is displayed in the bar graph on the right side of the panel. Results represent the average number of puncta per cell obtained from 100–150 cells. Statistical significance was tested using unpaired, two-tailed t test. (C) Wild-type Sec12 or its mutant Sec12-Y10F were expressed in HeLa cells immunostained for ERGIC-53 and flag. siLTK, expression of Sec12 in LTK-depleted cells. Arrows indicate nontransfected cells. Arrowheads indicate cells expressing Sec12. Scale bars, 30 µm. (D) Bar graph showing the quantification of the number of ERGIC-53 puncta per cell from three independent experiments. The graph compares cells expressing the Sec12 construct to directly adjacent nontransfected cells.
To obtain further support for a role of Sec12 phosphorylation in ERES function, we determined the number of peripheral ERGIC-53 structures in cells expressing the phosphoablating mutant of Sec12 (Sec12-Y10F). Peripheral ERGIC structures are good indicators of ERES function (Ben-Tekaya et al., 2005; Farhan et al., 2010). Expression of Sec12-Y10F resulted in a decrease in the number of ERGIC-53 puncta (Fig. 5 C). No effect of Sec12-Y10F expression was detected in LTK knockdown cells (Fig. 5 C). Altogether, we propose that Sec12 is phosphorylated in a manner dependent on LTK and that this phosphorylation affects ERES function.
Over the past decade, mounting evidence has indicated that endomembranes house a wide variety of signaling molecules such as GTPases, kinases, and phosphatases (Farhan and Rabouille, 2011; Cancino and Luini, 2013; Baschieri et al., 2014). An emerging concept of endomembrane signaling is autoregulation, which is defined as a response of a biological system that helps reestablish homeostasis. The UPR is the best understood and characterized autoregulatory response of the secretory pathway (Ron and Walter, 2007; Gardner et al., 2013), making it a useful template to compare other autoregulatory circuits with. The UPR is induced by misfolded or unfolded proteins, and its main purpose is to globally up-regulate the capacity of the endomembrane system to promote folding or degradation of these misfolded proteins. Such a broad response is expected because the purpose of the UPR is to maintain or reestablish global homeostasis of the ER. Another feature of the UPR is that its main sensors and mediators such as IRE1, ATF6, and PERK are resident to the ER. Contrary to the response of the ER to misfolded proteins, we know very little about whether and how local signaling at the ER controls the capacity of the ER to unload of folded proteins, i.e., of ER export. Very recently, a signaling cascade including Gα12 was shown to operate at ERESs (Subramanian et al., 2019). However, this signaling circuit controls the export of a small subset of proteins, and interfering with it had no effect on general protein secretion or on global ERES number. Thus, this novel pathway represents a tailored response of the ER, which is unlike the more global response of the UPR. Another difference to the UPR is that the main signaling mediator of this pathway Gα12 is not resident at the ER or ERES.
As far as LTK is concerned, our results indicate that it might be more similar to the UPR. First, LTK is resident in the ER, and second, the effect of LTK is a global regulation of trafficking and ERES number. This is in line with the observation that LTK phosphorylates Sec12, a general regulator of ERES biogenesis. Thus, LTK is a strong candidate to be a general autoregulator of ER export. Future work will need to address the question concerning what stimuli activate LTK. Because the LTK interactome contained several cargo receptors such as ERGIC-53, VIP36, ERGL, ERGIC1, and SURF4, we speculate that these cargo receptors might represent stimuli that induce LTK activity to positively regulate ER export. Previous work has suggested that secreted ligands called FAM150A and FAM150B might act as ligands for LTK and ALK (Zhang et al., 2014; Guan et al., 2015; Reshetnyak et al., 2015). However, this is not compatible with our observation that LTK is resident to the ER. A potential reason for this discrepancy is that FAM150A and FAM150B are ligands for ALK and that none of the aforementioned papers tested the effects in an ALK-free background, or they have been used with fish LTK that rather resembles ALK than mammalian LTK (Fadeev et al., 2018).
Another important question for future investigations is how LTK is deactivated. In principle, receptor tyrosine kinases can be deactivated either by dephosphorylation or by degradation. A number of ER-resident phosphatases have been described, and our LTK interactome also contains few phosphatases. Because LTK is an ER export regulator, identifying a phosphatase that regulates LTK will further expand our understanding of the regulation of ER export by signaling molecules.
The role of LTK in secretion might also be relevant for human diseases. Gain of function mutations in LTK have been observed in patients and mice with systemic lupus erythematosus (Li et al., 2004). We speculate that this gain of function mutation confers a selective advantage to autoimmune plasma cells as it allows them to cope with a higher secretory load. LTK might also represent a suitable drug target in cancer therapy, especially since cancer cells are considered to be addicted to secretion due to a high proteostatic challenge (Dejeans et al., 2015; Urra et al., 2016). This notion is supported by our observation that LTK inhibition increases the ER stress response (as measured by increased XBP1s levels) in cells treated with thapsigargin (Fig. S3 D). The investigation of the potential of LTK as a drug target will be an interesting area of future investigation.
Materials and methods
Mass spectrometry (MS)
Immunoprecipitated proteins were eluted from beads using repeated incubations with 8 M Guanidinhydrochloride at pH 8.0 and subjected to reductive alkylation (using 15 mM iodoacetamide and 5 mM DTT) and methanol/chloroform extraction followed by digestion with sequencing-grade trypsin (Promega) overnight at 37°C. Tryptic peptides were desalted and analyzed by liquid chromatography tandem MS using a NanoLC 1200 coupled via a nano-electrospray ionization source to a Q Exactive HF mass spectrometer. Peptide separation was performed according to their hydrophobicity on an in-house packed 18-cm column with 3-mm C18 beads (Dr. Maisch) using a binary buffer system consisting of solutions A (0.1% formic acid) and B (80% acetonitrile, 0.1% formic acid). Linear gradients from 7 to 38% B in 35 min were applied with a following increase to 95% B within 5 min and a reequilibration to 5% B. MS spectra were acquired using 3e6 as an AGC target, a maximal injection time of 20 ms, and a 60,000 resolution at 200 m/z. The mass spectrometer operated in a data-dependent Top15 mode with subsequent acquisition of higher energy collisional dissociation fragmentation MS/MS spectra of the top 15 most intense peaks. Resolution for MS/MS spectra was set to 30,000 at 200 m/z, AGC target to 1e5, maximum injection time to 64 ms, and the isolation window to 1.6 Th. Raw data files were processed with MaxQuant (1.6.0.1) as described previously (Cox and Mann, 2008; Cox et al., 2011) using human (UP000005640) UniProt databases, tryptic specifications, and default settings for mass tolerances for MS and MS/MS spectra. Carbamidomethylation at cysteine residues was set as a fixed modification, while oxidations at methionine and acetylation at the N terminus were defined as variable modifications. The minimal peptide length was set to seven amino acids, and the false discovery rate for proteins and peptide-spectrum matches to 1%. Perseus (1.5.8.5) was used for further analysis (Pearson’s correlation, two-sample t test) and data visualization. Functional annotation enrichment analysis was performed using the DAVID database (Huang et al., 2007) coupled to significance determination using Fisher’s exact test and correction for multiple hypothesis testing by the Benjamini and Hochberg false discovery rate.
Immunofluorescence
Cells were fixed in 3% paraformaldehyde for 20 min at room temperature. Afterward, cells were washed in PBS with 20 mM glycine followed by incubation in permeabilization buffer (PBS with 0.2% Triton X-100) for 5 min at room temperature. Subsequently, cells were incubated for 1 h with the primary antibody and after washing for another 1 h in secondary antibody diluted in 3% BSA in PBS. Cells were mounted in polyvinyl alcohol with DABCO antifade and imaged.
For Sar1-GTP staining, cells were fixed and permeabilized in ice-cold 50% methanol–50% acetone for 10 min at −20°C. Subsequently, cells were incubated for 1 h with the blocking buffer (PBS with 10% goat serum) at room temperature followed by incubation with primary antibody for 2 h and another 1 h with secondary antibody, both diluted in 3% BSA in PBS.
Cell culture and transfection
HeLa, HEK293T, and HepG2 cells were cultured in DMEM (GIBCO) supplemented with 10% FCS and 1% penicillin/streptomycin (GIBCO).
For overexpression of plasmids, cells were transfected with either Fugene 6 or with TransIT-LT1 (Mirus). For knockdown experiments, cells were reverse-transfected with 10 nM siRNA (final concentration) using HiPerfect (Qiagen) according to the manufacturer’s instructions.
Cell lysis, immunoblotting, and immunoprecipitation
Cells were washed twice with PBS and collected in lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 100 mM NaCl, 0.1% SDS, and 1% NP-40) supplemented with proteinase and phosphatase inhibitor (Pierce Protease and Phosphatase Inhibitor Mini Tablets, EDTA free). Lysates were incubated on ice for 10 min followed by clearing centrifugation at 20,000 xg at 4°C for 10 min. Supernatants were transferred into a fresh tube, and reducing loading buffer was added. Lysates were subjected to SDS-PAGE and transferred on a nitrocellulose membrane using semidry transfer. The membrane was blocked (in ROTI buffer [Roth] or 5% milk in PBS with 0.1% Tween) and probed with the appropriate primary antibodies. Subsequently, membranes were incubated with HRP-conjugated secondary antibody. Immunoblots were developed using a chemiluminescence reagent (ECL Clarity; BioRad) and imaged using ChemiDoc (BioRad).
For immunoprecipitation experiments, cells were lysed in immunoprecipitation buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 0.5% NP-40, and n-dodecyl-B-D-maltoside).
PGNase F digestion and EndoH
For PGNase F digestion, 3.2 × 105 HeLa cells were seeded into 6-well plates and the next day transfected with 1 µg plasmid DNA. 24 h later, cells were washed with PBS, and the cells were incubated in 1 ml serum-free medium and 250 U/ml PNGase F (P0704S; NEB) for 6 h. Subsequently, cells were lysed as described above.
For EndoH digestion, 2 × 106 cells were plated into in a 10-cm dish. After 24 h, cells were transfected with 3 μg plasmid DNA. The next day, cells were lysed in immunoprecipitation buffer (50 mM Tris-HCl, pH 7.4, 10% glycerol, 150 mM NaCl, 2 mM EDTA, and 0.5% Triton X-100) supplemented with proteinase and phosphatase inhibitor (Pierce Protease and Phosphatase Inhibitor Mini Tablets, EDTA free). Immunoprecipitation against flag was performed using EZview Red ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) overnight at 4°C followed by washing in immunoprecipitation buffer. Beads were incubated with 1,000 U EndoH (P0702S; NEB) according to the manufacturer’s instructions for 90 min at 37°C. Subsequently, the reaction was stopped by adding reducing sample buffer.
Microscopes and image acquisition
Imaging was performed on laser scanning confocal microscopes: LeicaSP5 and Zeiss LSM700. All images were acquired using a 63× oil immersion objective (NA 1.4).
FRAP was performed on a LeicaSP5 confocal microscope using a 63×/1.4 NA oil-immersion objective at threefold digital magnification. All experiments were performed at 37°C, and cells were maintained in complete medium supplemented with 25 mM Hepes, pH 7.4. After acquisition of a prebleach image, the ERES was bleached at 100% laser intensity for 750 ms. After bleaching, images were acquired at one image per second. Images were analyzed using ImageJ. The fluorescence intensity of the ERES before bleaching was set to 100%, and all subsequent values were normalized to it. The mobile fraction was calculated as MF = (F∞−F0)/(Fi−F0), where F∞ is fluorescence in the bleached region after recovery, Fi is the fluorescence in the bleached region before bleaching, and F0 is the fluorescence in the bleached region directly after bleaching.
ERESs were quantified as described previously (Tillmann et al., 2015).
Sequence analysis
The sequences of ALK and LTK kinase domains were aligned using the Muscle algorithm and Jalview environment (Edgar, 2004; Waterhouse et al., 2009). Phylogenetic trees were built using the PhyML program (Guindon et al., 2009) on the phylogeny.fr server (Dereeper et al., 2008) and visualized with the help of the iToL server (Letunic and Bork, 2016). Protein domains were detected using a conserved domain search (Marchler-Bauer et al., 2017). The following abbreviations were used in Fig. 1 B: alligator Am, Alligator mississippiensis; brachiopod La, Lingula anatine; chicken Gg, Gallus gallus; finch Tg, Taeniopygia guttata; frog Xt, Xenopus tropicalis; fruit fly Dm, Drosophila melanogaster; hemichordate Sk, Saccoglossus kowalevskii; lancelet Bf, Branchiostoma floridae; lizard Ac, Anolis carolinensis; mouse Mm, Mus musculus; nematode Bm, Brugia malayi; plocozoan Ta, Trichoplax adhaerens; possum Md, Monodelphis domestica; sea urchin Sp, Strongylocentrotus purpuratus; shark Cm, Callorhinchus milii; starfish Ap, Acanthaster planci; and zebrafish Dr, Danio rerio.
Purification of GST-tagged Sec12 cytosolic domain
Escherichia coli BL21 (DE3) was transformed with GST-tagged Sec12 construct. Bacteria were cultured in HSG growth medium, and induction was performed with 0.4 mM IPTG. The bacterial pellet was dissolved in lysis buffer (50 mM Tris, pH 8, 150 NaCl, 10% glycerol, 0.1% Triton X-100, and 100 μg/ml lysozyme supplemented with proteinase and phosphatase inhibitor), sonified and centrifuged at 32,000 xg for 30 min at 8°C. Supernatant was incubated with Glutathione Sepharose 4 Fast Flow (GE Healthcare) for 2 h at 4°C and washed with PBS, and beads were resuspended in buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 5% glycerol).
Kinase assay
HeLa cells expressing flag-tagged LTK were lysed, and LTK was immunoprecipitated using anti-flag M2 beads. The immunoprecipitate was resuspended in buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, and 10% glycerol). Typically, 5 × 106 cells were used. The immunoprecipitate was incubated with 1.5 µg of GST or GST-tagged Sec12 cytosolic domain for 30 min at 30°C. To induce kinase activity, 400 µM ATP was included. The reaction was stopped by adding sample buffer.
Quantitative PCR (qPCR)
The levels of LTK and ALK were determined by qRT-PCR. Total RNA was extracted using a Direct-Zol RNA kit (Zymo Research), and cDNA was reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The expression of LTK and ALK was determined using the LightCycler 480 SYBR Green I Master Mix (Roche Life Science) and normalized to GAPDH using commercially available primers (Qiagen; for LTK, QT00219877; ALK, QT00028847; and GAPDH, QT00079247).
Reagents
Antibodies
A list of all used antibodies is provided in Table S2.
Primers
A list of all used primers is provided in Table S3.
Online supplemental material
Fig. S1 (related to Fig. 1) shows the plasma membrane localization of ALK as well as a reduction of endogenous LTK labeling in LTK-depleted cells to test for antibody specificity. Finally, this figure shows costaining of endogenous LTK and Sec31 in HepG2 cells. Fig. S2 (related to Fig. 1 and Fig. 2) shows qPCR data of LTK and ALK expression in HepG2 and HeLa cells. In addition, it shows a test of knockdown efficiency of three different LTK siRNAs and their effects on ERES numbers. Fig. S3 (related to Fig. 4) shows a coimmunoprecipitation between LTK and Sec12 as well as a test of LTK phosphorylation in Src-inhibited cells. In addition, it shows a FRAP of general ER in crizotinib-treated cells as well as XBP1s levels in crizotinib-treated cells. Table S1 shows the results of the MS experiment of the LTK interactome (related to Fig. 4). Table S2 shows a list of all antibodies used in this work. A list of all primers used is shown in Table S3. Video 1 and Video 2 show live imaging of a RUSH experiment in control and crizotinib-treated cells.
Supplementary Material
Acknowledgments
Work in the Farhan laboratory was supported by grants from the Norwegian Research Council (NFR; grant number 262717), the Norwegian Cancer Society (Kreftforeningen; grant number 182815), the Anders Jahre Foundation, the Rakel-Otto-Bruun Legat, the Swiss Science Foundation, and the German Science Foundation (DFG; grant number 271101596). F.G. Centonze is supported by a PhD scholarship from the Institute of Basic Medical Sciences, University of Oslo. K. Pawlowski was supported by a Polish National Science Centre grant (2014/15/B/NZ1/03359). C. Behrends was supported by the DFG within the framework of the Munich Cluster for Systems Neurology (EXC2145 SyNergy) and the Collaborative Research Center (CRC1177) as well as by the Boehringer Ingelheim Foundation.
The authors declare no competing financial interests.
Author contributions: F.G. Centonze and V. Reiterer performed and analyzed most experiments. K. Nalbach and C. Behrends performed and analyzed the MS experiments. K. Saito provided reagents and help conceive the Sec12 experiments. K. Pawlowski performed the evolutionary LTK analysis and produced the sequence logo. H. Farhan conceived the project, acquired the funding, planned and analyzed experiments, and wrote the manuscript with input from all authors.
References
- Appenzeller-Herzog C., and Hauri H.P.. 2006. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 119:2173–2183. 10.1242/jcs.03019 [DOI] [PubMed] [Google Scholar]
- Barlowe C., and Schekman R.. 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 365:347–349. 10.1038/365347a0 [DOI] [PubMed] [Google Scholar]
- Baschieri F., Confalonieri S., Bertalot G., Di Fiore P.P., Dietmaier W., Leist M., Crespo P., Macara I.G., and Farhan H.. 2014. Spatial control of Cdc42 signalling by a GM130-RasGRF complex regulates polarity and tumorigenesis. Nat. Commun. 5:4839 10.1038/ncomms5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauskin A.R., Alkalay I., and Ben-Neriah Y.. 1991. Redox regulation of a protein tyrosine kinase in the endoplasmic reticulum. Cell. 66:685–696. 10.1016/0092-8674(91)90114-E [DOI] [PubMed] [Google Scholar]
- Ben-Tekaya H., Miura K., Pepperkok R., and Hauri H.P.. 2005. Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 118:357–367. 10.1242/jcs.01615 [DOI] [PubMed] [Google Scholar]
- Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., and Perez F.. 2012. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 9:493–498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Cancino J., and Luini A.. 2013. Signaling circuits on the Golgi complex. Traffic. 14:121–134. 10.1111/tra.12022 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Olsen J.V., Brandts C., Cox J., Reddy P.N., Böhmer F.D., Gerke V., Schmidt-Arras D.E., Berdel W.E., Müller-Tidow C., et al. . 2009. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell. 36:326–339. 10.1016/j.molcel.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Dejeans N., Barroso K., Fernandez-Zapico M.E., Samali A., and Chevet E.. 2015. Novel roles of the unfolded protein response in the control of tumor development and aggressiveness. Semin. Cancer Biol. 33:67–73. 10.1016/j.semcancer.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., et al. . 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465-9 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeev A., Mendoza-Garcia P., Irion U., Guan J., Pfeifer K., Wiessner S., Serluca F., Singh A.P., Nüsslein-Volhard C., and Palmer R.H.. 2018. ALKALs are in vivo ligands for ALK family receptor tyrosine kinases in the neural crest and derived cells. Proc. Natl. Acad. Sci. USA. 115:E630–E638. 10.1073/pnas.1719137115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H., and Rabouille C.. 2011. Signalling to and from the secretory pathway. J. Cell Sci. 124:171–180. 10.1242/jcs.076455 [DOI] [PubMed] [Google Scholar]
- Farhan H., Wendeler M.W., Mitrovic S., Fava E., Silberberg Y., Sharan R., Zerial M., and Hauri H.-P.. 2010. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J. Cell Biol. 189:997–1011. 10.1083/jcb.200912082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B.M., Pincus D., Gotthardt K., Gallagher C.M., and Walter P.. 2013. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5:a013169 10.1101/cshperspect.a013169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannotta M., Ruggiero C., Grossi M., Cancino J., Capitani M., Pulvirenti T., Consoli G.M., Geraci C., Fanelli F., Luini A., and Sallese M.. 2012. The KDEL receptor couples to Gαq/11 to activate Src kinases and regulate transport through the Golgi. EMBO J. 31:2869–2881. 10.1038/emboj.2012.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J., Umapathy G., Yamazaki Y., Wolfstetter G., Mendoza P., Pfeifer K., Mohammed A., Hugosson F., Zhang H., Hsu A.W., et al. . 2015. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. eLife. 4:e09811 10.7554/eLife.09811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Delsuc F., Dufayard J.F., and Gascuel O.. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537:113–137. 10.1007/978-1-59745-251-9_6 [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., and Lempicki R.A.. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35(suppl_2):W169-75 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., and Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242-5 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Nakamura K., Jiang Y., Tsurui H., Matsuoka S., Abe M., Ohtsuji M., Nishimura H., Kato K., Kawai T., et al. . 2004. Gain-of-function polymorphism in mouse and human Ltk: implications for the pathogenesis of systemic lupus erythematosus. Hum. Mol. Genet. 13:171–179. 10.1093/hmg/ddh020 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., et al. . 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45(D1):D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montegna E.A., Bhave M., Liu Y., Bhattacharyya D., and Glick B.S.. 2012. Sec12 binds to Sec16 at transitional ER sites. PLoS One. 7:e31156 10.1371/journal.pone.0031156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Ravazzola M., Meda P., Holcomb C., Moore H.P., Hicke L., and Schekman R.. 1991. Mammalian Sec23p homologue is restricted to the endoplasmic reticulum transitional cytoplasm. Proc. Natl. Acad. Sci. USA. 88:8611–8615. 10.1073/pnas.88.19.8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvirenti T., Giannotta M., Capestrano M., Capitani M., Pisanu A., Polishchuk R.S., San Pietro E., Beznoussenko G.V., Mironov A.A., Turacchio G., et al. . 2008. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat. Cell Biol. 10:912–922. 10.1038/ncb1751 [DOI] [PubMed] [Google Scholar]
- Reshetnyak A.V., Murray P.B., Shi X., Mo E.S., Mohanty J., Tome F., Bai H., Gunel M., Lax I., and Schlessinger J.. 2015. Augmentor α and β (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc. Natl. Acad. Sci. USA. 112:15862–15867. 10.1073/pnas.1520099112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., and Walter P.. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529. 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Saito K., Yamashiro K., Shimazu N., Tanabe T., Kontani K., and Katada T.. 2014. Concentration of Sec12 at ER exit sites via interaction with cTAGE5 is required for collagen export. J. Cell Biol. 206:751–762. 10.1083/jcb.201312062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharaw S., Iskar M., Ori A., Boncompain G., Laketa V., Poser I., Lundberg E., Perez F., Beck M., Bork P., and Pepperkok R.. 2016. The endosomal transcriptional regulator RNF11 integrates degradation and transport of EGFR. J. Cell Biol. 215:543–558. 10.1083/jcb.201601090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Arras D., Böhmer S.A., Koch S., Müller J.P., Blei L., Cornils H., Bauer R., Korasikha S., Thiede C., and Böhmer F.D.. 2009. Anchoring of FLT3 in the endoplasmic reticulum alters signaling quality. Blood. 113:3568–3576. 10.1182/blood-2007-10-121426 [DOI] [PubMed] [Google Scholar]
- Sharpe H.J., Stevens T.J., and Munro S.. 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 142:158–169. 10.1016/j.cell.2010.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.C., Joggerst B., Laketa V., Verissimo F., Cetin C., Erfle H., Bexiga M.G., Singan V.R., Hériché J.K., Neumann B., et al. . 2012. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat. Cell Biol. 14:764–774. 10.1038/ncb2510 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Capalbo A., Iyengar N.R., Rizzo R., di Campli A., Di Martino R., Lo Monte M., Beccari A.R., Yerudkar A., Del Vecchio C., et al. . 2019. Auto-regulation of Secretory Flux by Sensing and Responding to the Folded Cargo Protein Load in the Endoplasmic Reticulum. Cell. 176:1461–1476.e23. 10.1016/j.cell.2019.01.035 [DOI] [PubMed] [Google Scholar]
- Tillmann K.D., Reiterer V., Baschieri F., Hoffmann J., Millarte V., Hauser M.A., Mazza A., Atias N., Legler D.F., Sharan R., et al. . 2015. Regulation of Sec16 levels and dynamics links proliferation and secretion. J. Cell Sci. 128:670–682. 10.1242/jcs.157115 [DOI] [PubMed] [Google Scholar]
- Urra H., Dufey E., Avril T., Chevet E., and Hetz C.. 2016. Endoplasmic Reticulum Stress and the Hallmarks of Cancer. Trends Cancer. 2:252–262. 10.1016/j.trecan.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Venditti R., Scanu T., Santoro M., Di Tullio G., Spaar A., Gaibisso R., Beznoussenko G.V., Mironov A.A., Mironov A. Jr., Zelante L., et al. . 2012. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science. 337:1668–1672. 10.1126/science.1224947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., and Barton G.J.. 2009. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woroniuk A., Porter A., White G., Newman D.T., Diamantopoulou Z., Waring T., Rooney C., Strathdee D., Marston D.J., Hahn K.M., et al. . 2018. STEF/TIAM2-mediated Rac1 activity at the nuclear envelope regulates the perinuclear actin cap. Nat. Commun. 9:2124 10.1038/s41467-018-04404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharogianni M., Kondylis V., Tang Y., Farhan H., Xanthakis D., Fuchs F., Boutros M., and Rabouille C.. 2011. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 30:3684–3700. 10.1038/emboj.2011.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G., Pahuja K.B., Studer S., Shim S., and Schekman R.. 2011. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14:20–28. 10.1038/ncb2390 [DOI] [PubMed] [Google Scholar]
- Zhang H., Pao L.I., Zhou A., Brace A.D., Halenbeck R., Hsu A.W., Bray T.L., Hestir K., Bosch E., Lee E., et al. . 2014. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc. Natl. Acad. Sci. USA. 111:15741–15745. 10.1073/pnas.1412009111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.